Abstract
Adenosine triphosphate-dependent chromatin remodeling machines play a central role in gene regulation by manipulating chromatin structure. Most genes have a nucleosome-depleted region at the promoter and an array of regularly spaced nucleosomes phased relative to the transcription start site. In vitro, the three known yeast nucleosome spacing enzymes (CHD1, ISW1 and ISW2) form arrays with different spacing. We used genome-wide nucleosome sequencing to determine whether these enzymes space nucleosomes differently in vivo. We find that CHD1 and ISW1 compete to set the spacing on most genes, such that CHD1 dominates genes with shorter spacing and ISW1 dominates genes with longer spacing. In contrast, ISW2 plays a minor role, limited to transcriptionally inactive genes. Heavily transcribed genes show weak phasing and extreme spacing, either very short or very long, and are depleted of linker histone (H1). Genes with longer spacing are enriched in H1, which directs chromatin folding. We propose that CHD1 directs short spacing, resulting in eviction of H1 and chromatin unfolding, whereas ISW1 directs longer spacing, allowing H1 to bind and condense the chromatin. Thus, competition between the two remodelers to set the spacing on each gene may result in a highly dynamic chromatin structure.
INTRODUCTION
One of the most challenging puzzles in eukaryotic cell biology is to understand how DNA is packaged into chromatin, satisfying the topological constraints imposed by the nucleus, and at the same time remaining accessible for regulatory proteins to locate their target sequences. The basic structural unit of chromatin is the nucleosome, which is composed of a histone octamer containing two molecules each of the core histones H2A, H2B, H3 and H4, around which 147 bp of DNA is wrapped in ∼1.7 turns (1). The nucleosome constitutes a barrier to transcription, replication, recombination and repair, which can be circumvented by chromatin remodeling complexes (2). They use the free energy obtained from adenosine triphosphate (ATP) hydrolysis to assemble, eject, slide or re-structure nucleosomes (3–5). Remodelers are well-conserved from yeast to human. Mutations in subunits of human remodeling complexes occur at high frequency in many cancers (4) and are associated with developmental abnormalities (6).
Genome-wide nucleosome maps for budding yeast reveal that most genes have a nucleosome-depleted region (NDR) at their promoters (7–10) and that nucleosomes are regularly spaced and phased relative to the transcription start site (TSS), usually located just inside the first (+1) nucleosome. Various chromatin remodeling complexes cooperate to organize nucleosomes on genes. RSC, a SWI/SNF-related complex, is involved in setting the size of the NDR (11–13). In vitro, RSC and SWI/SNF can eject, mobilize or remodel nucleosomes, but they do not have spacing activity (14–16). Nucleosome spacing and phasing relative to the TSS require the ISW1 and CHD1 chromatin remodeling enzymes; a third spacing enzyme, ISW2, contributes little to global phasing (17). Phasing is weaker in the absence of CHD1 or ISW1 but dramatically worse in the double mutant (17), suggesting that they might have redundant functions. However, CHD1, ISW1 and ISW2 space nucleosomes differently in vitro, forming nucleosomal arrays with spacings of ∼160, ∼175 and ∼200 bp, respectively (18–25).
Here we propose that the observed global nucleosome spacing in yeast of ∼165 bp actually represents an average of nucleosomal arrays of different spacing, specified primarily by CHD1 and ISW1. We hypothesize that genes remodeled by CHD1 have short spacing, those remodeled by ISW1 have longer spacing and genes remodeled by ISW2 have the longest spacing (Figure 1A). It follows that genes affected by more than one spacing enzyme may display intermediate spacing, resulting from competition between the enzymes. We provide evidence in support of this hypothesis by sequencing nucleosomal DNA from isw1Δ, chd1Δ and isw2Δ mutants (MNase-seq). We show that CHD1 and ISW1 compete to set the spacing on most genes. We confirm that transcription is associated with short spacing (26), but we also find that some heavily transcribed genes have extremely long spacing. In higher eukaryotes, longer nucleosome spacing is associated with the binding of linker histone (H1) and transcriptional repression through chromatin compaction (27–30). We show that H1 binding increases with nucleosome spacing, suggesting that genes with shorter spacing may be less condensed than genes with longer spacing.
Figure 1.
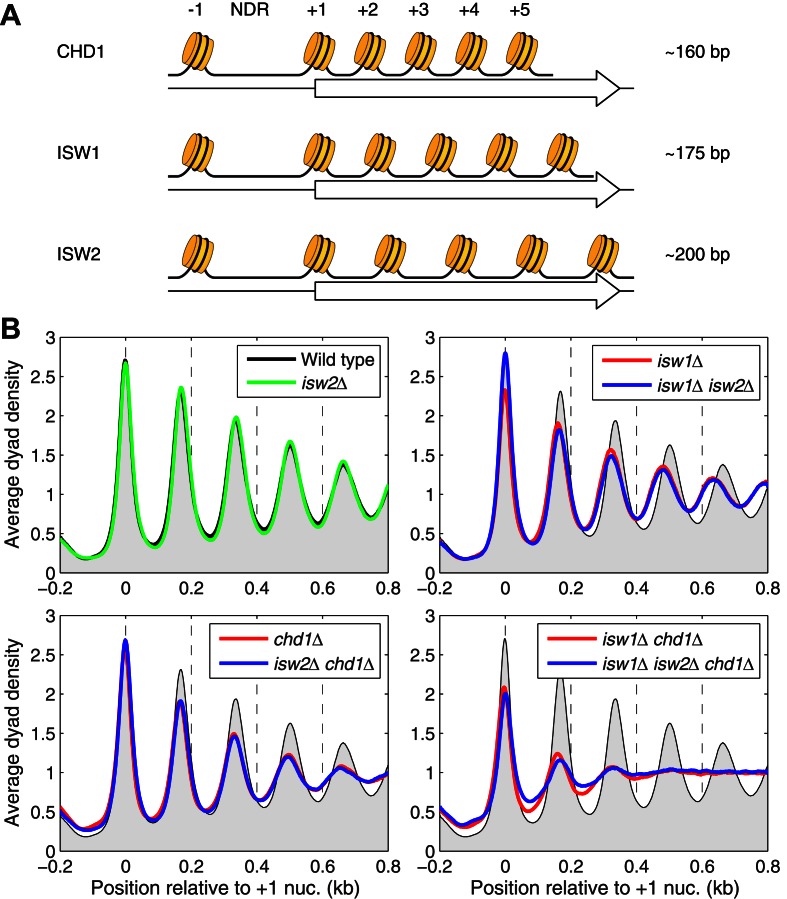
ISW1 forms arrays with longer spacing than CHD1 in vivo. (A) CHD1, ISW1 and ISW2 form nucleosome arrays with different average spacing in vitro (18,20). We propose that different genes will have different nucleosome spacing, depending on which of the three spacing enzymes act on the gene. If more than one spacing enzyme acts on a particular gene, an intermediate spacing is predicted. NDR: nucleosome-depleted region (promoter). (B) Nucleosome phasing profiles for all genes aligned on the dyad of the +1 nucleosome in wild-type cells. Phasing plots for isogenic strains corresponding to all combinations of the chd1Δ, isw1Δ and isw2Δ mutations. The sequencing depths for all datasets were adjusted to 1 read per bp. Wild-type is shown as a gray background in all panels.
MATERIALS AND METHODS
Nucleosome sequencing and Pol II ChIP-seq
Yeast strains are listed in Supplementary Table S1. Wild-type and null mutants were grown to mid-log phase in synthetic complete medium containing 2% glucose. MNase-seq experiments were performed as described (31) except that Agencourt AMPure XP beads (Beckman-Coulter A63880) were used to purify adapter-ligated DNA samples and polymerase chain reaction products. Paired-end sequencing of chromatin immunoprecipitates (PESCI) was performed as described (32), but with some modifications (see Supplementary Methods).
Bioinformatic analysis
Paired-end reads (50 nt each) were aligned against the UCSC SacCer3 genome assembly using Bowtie 2 (33). After alignment of each pair of reads to the yeast genome, we obtained the length distributions for each sample. In all cases, there was a major peak at ∼150 bp, as expected (34,35). The data are summarized in Supplementary Table S3. Data analysis was performed in MATLAB using the Bioinformatics toolbox. Heat maps were smoothed with a 2D Gaussian filter (σ = 3). Raw coverage profiles were generated using BEDTools utilities (36) and viewed in IGV (37). For further analysis, nucleosome sequences in the range 120–160 bp were selected, and the locations of their dyads were inferred by calculating the midpoint coordinate. Sequencing depths were adjusted to the common value of 1 read per bp. Average profiles were smoothed using a moving average filter with a span of 21 bp. Transcript end coordinates were obtained from (38). The nucleosome spacing algorithm is described in Supplementary Methods.
RESULTS
To investigate whether the CHD1, ISW1 and ISW2 remodelers space nucleosomes differently in vivo, we constructed a set of eight isogenic yeast strains, corresponding to all possible combinations of the chd1Δ, isw1Δ and isw2Δ mutations (Supplementary Table S1). We mapped nucleosomes in all of these strains by MNase-seq, using paired-end sequencing to obtain the length of each DNA fragment, which provides more accurate nucleosome positions than single-read data (31).
ISW1 forms arrays with longer spacing than CHD1 in vivo
We examined global nucleosome spacing by analyzing nucleosome positioning relative to the midpoint of the +1 nucleosome in wild-type cells for ∼5000 yeast genes. We observed that nucleosome phasing is improved if the genes are aligned on the +1 nucleosome instead of the TSS, although our conclusions hold if the TSS is used instead (Supplementary Figure S1). As expected, wild-type cells showed a deep trough at the NDR with strong nucleosome phasing on coding regions (Figure 1B). We define nucleosome phasing as the degree of order of the nucleosomal array beginning with the +1 nucleosome, as measured by the amplitudes of the nucleosome peaks. Thus, a perfectly ordered array would have extreme peak amplitudes, whereas a completely unphased array would show no oscillations at all (a flat line). Phasing is very strong in wild-type cells, but not perfect, because the peaks have width and the troughs do not reach zero (Figure 1B). We define nucleosome spacing as the average distance between the nucleosome peaks, which we calculated by linear regression analysis using the peak values for the +1 to +5 nucleosomes (13). Consistent with previous observations (9,13,17,39), the global average spacing in wild-type cells is 166 bp with a standard deviation of 0 bp, based on the average values from two biological replicate experiments (n = 2) (Supplementary Table S2).
Both isw1Δ and chd1Δ cells show weaker nucleosome phasing than wild-type, indicated by broader peaks with reduced amplitude, and the isw1Δ chd1Δ double mutant has much worse phasing than either single mutant (Figure 1B). In contrast, isw2Δ cells do not show any obvious changes in global chromatin organization and the isw1Δ isw2Δ chd1Δ triple mutant is almost identical to the isw1Δ chd1Δ double mutant (Figure 1B). We quantified the degree of phasing using our simple mathematical description of the phasing barrier model (13). The distance between neighboring nucleosomes is fitted to a Gaussian distribution, where the mean indicates the average inter-nucleosome distance (spacing) and the standard deviation (σ) indicates the variability in the position of the next nucleosome, which is equivalent to the degree of phasing (Supplementary Figure S2A). Thus, high and narrow peaks indicate strong phasing, characterized by low values of σ. Wild-type and isw2Δ cells have the best phasing (σ = 16.0 and 14.4 for the two wild-type biological replicate experiments; σ = 15.3 and 16.1 for isw2Δ cells). Phasing is weaker in isw1Δ cells (σ = 18.9 and 18.4) and chd1Δ cells (σ = 20.2 and 19.5) and very weak in the isw1Δ chd1Δ double mutant (σ = 27.5 and 28.1) (Supplementary Figure S2A).
Overall, our data are consistent with the general conclusion of Gkikopoulos et al. (17), that ISW1 and CHD1 are the primary determinants of global nucleosome spacing and phasing, whereas ISW2 is unimportant at the global level. However, we also observed that the global average nucleosome spacing is reduced by 7 bp in isw1Δ cells, to 159 +/−1 bp (n = 2) (Supplementary Table S2). The location of the +1 nucleosome is unchanged, but the peaks corresponding to the downstream nucleosomes are shifted toward the NDR (Figure 1B). This spacing change was not observed by others (17) probably because of the uncertainty in dyad positions associated with single-end sequencing data, for which assumptions must be made about nucleosomal DNA length to estimate the dyad location. The reduced spacing in isw1Δ cells accounts for the previous observation that, in isw1Δ cells, nucleosomes far from the promoter shift more than those near the promoter (40,41). This effect is due to the increasingly large shifts in position of nucleosomes farther away from the promoter: the +1 nucleosome remains in place in isw1Δ cells, but the +2 nucleosome shifts upstream by ∼7 bp and the +3 nucleosome by ∼14 bp, etc. The same reduced spacing is observed in the isw1Δ isw2Δ double mutant (159 +/−1 bp; n = 2) (Figure 1B), consistent with the absence of a global contribution from ISW2. The phasing is too weak to ascertain whether there is reduced spacing in the isw1Δ chd1Δ double mutant or the triple mutant, although the positioning of the +1 nucleosome remains strong in both mutants.
The fact that ISW2 plays no role in global spacing simplifies the predictions of our hypothesis: nucleosome spacing in isw1Δ cells is primarily due to CHD1 and, in chd1Δ cells, it is primarily due to ISW1. We predict that nucleosome spacing should decrease in the absence of ISW1 and increase in the absence of CHD1. The shorter average spacing observed in isw1Δ cells is consistent with both predictions: in the absence of ISW1, the spacing decreases to 159 bp, indicating that ISW1 is required for the longer spacing observed in wild-type cells, and the short spacing in isw1Δ cells can be attributed to CHD1, given that it is the only remaining major spacing enzyme in isw1Δ cells. However, the fact that the average spacing in chd1Δ cells is almost the same as in wild-type cells (Supplementary Table S2) is inconsistent with the simplest version of our hypothesis, because we predict that it should be longer than in wild-type cells. The weaker phasing observed in both chd1Δ and isw1Δ cells is potentially informative: it can be explained by altered spacing on subsets of genes in the mutants, because averaging the various phasing patterns would result in broader and weaker peaks. Accordingly, we addressed the possibility that subsets of genes have different average spacing.
ISW1 and CHD1 compete to set nucleosome spacing on most genes
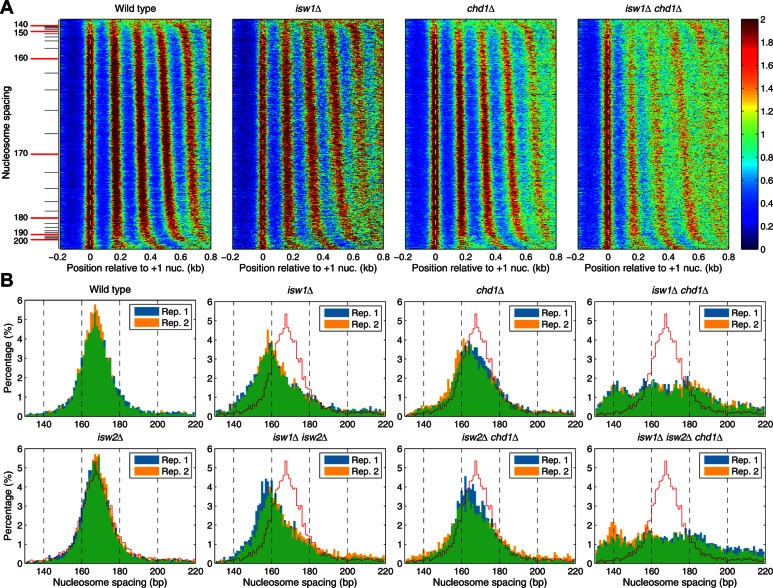
We developed an algorithm to estimate the nucleosome spacing on each gene (Supplementary Figure S3). Heat maps were constructed in which genes were sorted according to their average spacing in wild-type cells. A wide range of spacing is observed in wild-type cells, ranging from closely packed nucleosomes with no linker DNA (∼145 bp) to very long spacing (>190 bp) (Figure 2A). The small groups of genes with extreme spacing, located at the top and bottom of each heat map, have much less ordered arrays than the large majority of genes. The isw1Δ mutant is similar to wild-type but the nucleosomes are generally less well phased, especially the more distal nucleosomes (Figure 2A), as expected from the global average profile (Figure 1B). The chd1Δ heat map shows a similarly wide range in spacing, but the genes with extreme short spacing are very poorly phased relative to wild-type and isw1Δ cells, suggesting that these genes are strongly dependent on CHD1 (Figure 2A). Phasing is much more disrupted in the isw1Δ chd1Δ double mutant (Figure 2A) and in the triple mutant (Supplementary Figure S4).
Figure 2.
ISW1 and CHD1 compete to set nucleosome spacing on most genes. (A) Heat map analysis of all genes showing nucleosome dyad distributions, aligned on the +1 nucleosome dyad. Each row represents a gene. Genes are sorted in order of increasing average spacing in wild-type cells, from top to bottom. The color scale represents nucleosomal dyad density (red: high; blue: low dyad density). Equivalent heat maps for the isw2Δ strains are shown in Supplementary Figure S4. (B) Altered nucleosome spacing distributions in the mutants. Histograms of the percentage of genes having a given average spacing (1-bp bins). Data from two biological replicates are shown, indicated by blue and yellow bars; overlap is indicated by green bars.
We compared the variation in spacing in wild-type cells with that in the mutants by plotting histograms of the percentage of genes with a given average spacing (Figure 2B). Wild-type cells show a roughly symmetrical spacing distribution, with 77% of the genes having a spacing between 155 and 180 bp. The peak is at 167 bp, as expected from the global average. In isw1Δ cells, the spacing distribution is broader and the peak shifts to 158/159 bp, as expected from the global average. About 35% of genes have spacing similar to wild-type, indicating that their spacing is independent of ISW1; these genes may be affected by the residual phasing discussed above. The broader spacing range also accounts for the weaker phasing in isw1Δ cells, which is due to the averaging of the phasing patterns from genes with shortened spacing and those that are less affected. The genes with the longest spacing have the weakest phasing in isw1Δ cells (Supplementary Figure S2B). Thus, most genes in isw1Δ cells have shorter spacing than in wild-type, indicating that formation of arrays with normal spacing on these genes depends on ISW1. We infer that CHD1 is required to form the arrays with short spacing, given that it is the only other major spacing enzyme. Thus, ISW1 is associated with the formation of arrays with longer spacing than CHD1; they both act on most genes, competing to set the spacing.
The spacing distribution in chd1Δ cells is somewhat broader and flatter than in wild-type cells, accounting for the weaker phasing (Figure 2B), which is weakest on genes with extreme spacing (Supplementary Figure S2B). However, there is a slight shift to shorter spacing instead of the predicted shift to longer spacing. The relatively small effect of removing CHD1 suggests that it plays only a subsidiary role. If this is true, the isw1Δ chd1Δ double mutant should resemble the isw1Δ mutant, but they are very different. Spacing is heavily compromised in the double mutant, which shows a flattened distribution with no clear peaks, a much wider variation in average spacing on individual genes, more irregular spacing (Figure 2) and generally much weaker phasing (Supplementary Figure S2B). These observations can be explained if ISW1 function is partly dependent on CHD1, resulting in weakened phasing and spacing by ISW1 in chd1Δ cells. That is, ISW1 may create more regular (better phased) arrays if the nucleosomes are first spaced by CHD1.
Heavily transcribed genes have extreme nucleosome spacing
Genes with very short spacing also have poor phasing, particularly in chd1Δ cells (Figure 2A). Since heavy transcription is associated with disrupted chromatin and short spacing (26,32,34,39,42–44), we tested the relationship between nucleosome spacing and transcription. We performed ChIP-seq for the Rpb3 subunit of RNA polymerase II (Pol II) in wild-type cells, the three single mutants and the isw1Δ chd1Δ double mutant. The gene expression patterns of the isw1Δ, chd1Δ and isw2Δ single mutants are very similar to wild-type (Spearman's rank correlation, R > 0.9); very few genes showed changes >2-fold relative to wild-type, although small differences could be important to the cell (Supplementary Figure S5). Even the isw1Δ chd1Δ double mutant, which shows very poor global phasing, is not very different from wild-type (R = 0.91) (Supplementary Figure S5). The absence of major changes in gene expression patterns in these mutants is consistent with earlier work (17). We also note that Pol II density on genes encoding the subunits of all three remodeling enzymes is almost unaffected in the single mutants, indicating that loss of ISW1, CHD1 or ISW2 activity does not alter transcription of the genes encoding the other remodelers (Supplementary Figure S6).
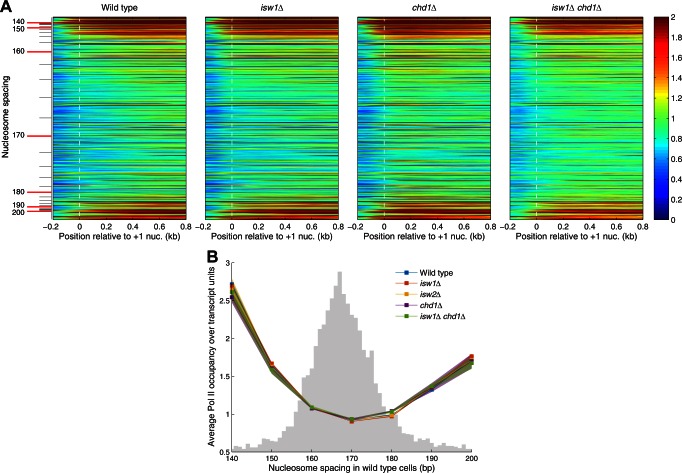
We analyzed the distribution of Pol II as a function of nucleosome spacing in wild-type cells using Pol II density heat maps in which genes were sorted from short to long spacing. A bimodal distribution of Pol II is apparent, indicating that genes with extreme spacing are the most heavily transcribed (Figure 3A). To illustrate this observation more quantitatively and to compare the mutants, we grouped the genes in 10 bp bins according to their average spacing in wild-type cells and calculated the average Pol II density for each group. This analysis confirms that heavily transcribed genes have extreme spacing in wild-type cells and shows that the same is true for all of the mutants (Figure 3B). The large majority of genes in the normal range of spacing are much less transcribed. Thus, rather surprisingly, heavy transcription is associated with very long spacing as well as very short spacing. The difference between these two classes of heavily transcribed genes may be due to CHD1, which is important for phasing on the genes with very short spacing, but not for those with very long spacing (Figure 2A).
Figure 3.
Heavily transcribed genes have extreme nucleosome spacing. Bimodal distribution of Pol II on genes as a function of nucleosome spacing. (A) Heat map analysis of all genes showing the distribution of Pol II as a function of nucleosome spacing, aligned on the +1 nucleosome dyad. Each row represents a gene. Genes are sorted in order of increasing average spacing in wild-type cells, from top to bottom. The color scale represents Pol II density (red: high; blue: low dyad density). (B) Relative Pol II occupancy on gene bodies as a function of nucleosome spacing in wild-type cells. All plots represent normalized IP/input data for the Rpb3 subunit of Pol II. Shaded areas indicate the distribution of points from two biological replicate experiments. Gray histogram indicates the wild-type spacing distribution (from Figure 2B).
ISW2 contributes to phasing and longer spacing on inactive genes
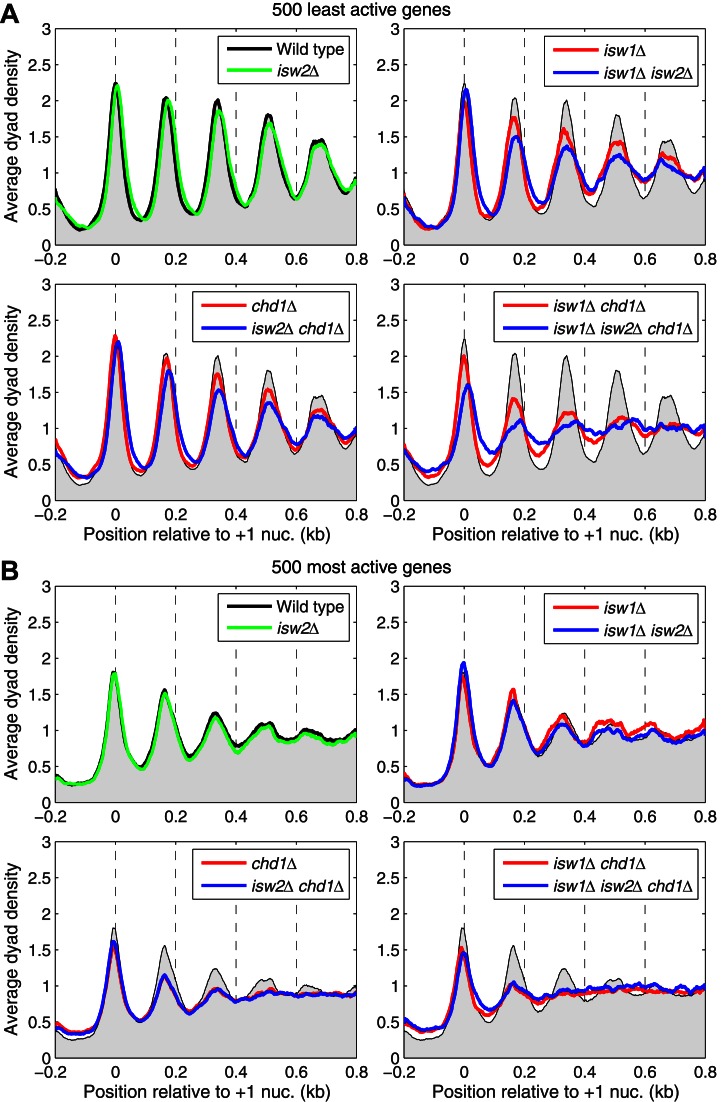
Most of the less active genes have spacings in the 160–180 bp range (Figure 3B). To determine whether there is a link between low activity and spacing, we identified the 500 least active genes in wild-type cells and subjected them to phasing analysis (Figure 4A). In wild-type cells, the least active genes show excellent phasing with similar spacing to the average (168 +/− 0 bp; n = 2) and, as for all genes, ISW2 has no effect. Phasing on the inactive genes is only slightly weaker in chd1Δ cells, suggesting that CHD1 is less important for spacing on inactive genes. In isw1Δ cells, the inactive genes show weaker phasing but they show no change in spacing, unlike the global average (compare Figure 4A with Figure 1B). The fact that the spacing does not shorten on inactive genes in isw1Δ cells is consistent with a relatively minor role for CHD1 on inactive genes.
Figure 4.
ISW2 contributes to phasing and longer spacing on inactive genes. (A) Nucleosome phasing profiles for all genes aligned on the +1 nucleosome dyad for the 500 least active genes, based on Pol II (Rpb3) occupancy on gene bodies in wild-type cells. Phasing plots for isogenic strains corresponding to all combinations of the chd1Δ, isw1Δ and isw2Δ mutations. The sequencing depths for all datasets were adjusted to 1 read per bp. Wild-type is shown as a gray background in all panels. (B) Equivalent plots for the 500 most active genes.
Although phasing in the isw1Δ chd1Δ double mutant is obviously worse than in either single mutant, it is still much better than the global average (compare with Figure 1B) and the spacing can be estimated at 176 +/− 0 bp (n = 2), which is ∼8 bp longer than for the inactive genes in wild-type cells (Figure 4A). This observation suggests that an additional spacing enzyme, which places nucleosomes farther apart, is contributing to phasing on inactive genes. A comparison of phasing on the inactive genes in the isw1Δ chd1Δ double mutant and the isw1Δ isw2Δ chd1Δ triple mutant suggests that ISW2 is involved. Phasing on the inactive genes in the triple mutant is weaker than in the double mutant (Figure 4A). This suggests that ISW2 phases nucleosomes with a spacing of 176 bp, which is longer than average, but much shorter than the spacing of ∼200 bp observed in vitro. A minor role for ISW2 is also suggested by the isw1Δ isw2Δ and isw2Δ chd1Δ double mutants, which show slightly weaker phasing than the isw1Δ and chd1Δ single mutants. Furthermore, the +1 nucleosome shows a small downstream shift of ∼5–15 bp in all isw2Δ mutants except the single mutant. Thus, ISW2 contributes to phasing on inactive genes, but its effects are masked by ISW1 and CHD1 in wild-type cells.
For comparison, we present the equivalent analysis for the 500 most active genes in wild-type cells (Figure 4B). In wild-type cells, active genes have much weaker phasing and shorter spacing than the average (162 +/− 0 bp; n = 2). However, the fact that some active genes have very long spacing implies that the weak phasing is partly due to averaging of the phasing patterns from active genes with short and long spacing; the spacing is therefore the weighted average of the two populations. The most active genes show weaker phasing in chd1Δ cells, but no change in isw1Δ or isw2Δ cells. Furthermore, the active genes have extremely weak phasing in the isw1Δ chd1Δ double mutant, indicating that both CHD1 and ISW1 play a major role in organizing the chromatin of active genes.
Linker histone binding correlates with longer nucleosome spacing and reduced transcription
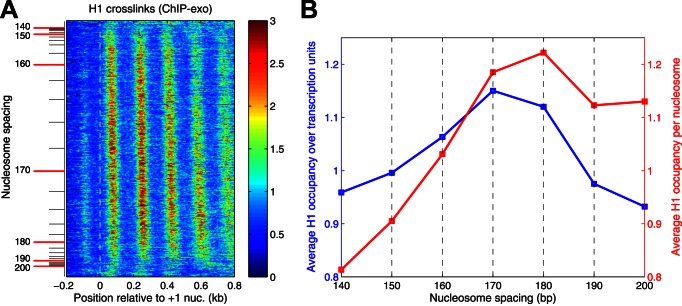
Noting the association of long spacing with H1 in higher eukaryotes, we determined how H1 distribution varies with nucleosome spacing, using ChIP-exo data reported by others (45). H1 binding is phased relative to the TSS but the H1 peaks are located over the linkers between nucleosomes rather than being coincident with them (45), suggesting that the majority of formaldehyde-induced cross-links between H1 and DNA involve the C-terminal tail domain of H1, which binds to the linker DNA, rather than the globular domain, which interacts with the nucleosome core (30). We constructed a heat map to determine how H1 distribution varies with nucleosome spacing in wild-type cells (Figure 5A). We observed the expected H1 phasing, with peaks located over the linkers on all genes except those with extremely short or extremely long spacing (top and bottom of the heat map), indicating that heavily transcribed genes have low H1 levels. Furthermore, genes with shorter spacing have weaker H1 phasing than genes with longer spacing (Figure 5A). H1 occupancy increases with nucleosome spacing up to 170 bp and then decreases (Figure 5B). The low points on the curve are the genes with extreme spacing, consistent with their high transcriptional activity. However, this calculation does not take into account that H1 binds to nucleosomes and that nucleosome density decreases with spacing. When the H1 occupancy is normalized per nucleosome, rather than per base pair, the H1 level increases with spacing until it reaches a plateau at 170–180 bp. We conclude that genes with longer spacing bind more H1.
Figure 5.
Linker histone binding correlates with longer nucleosome spacing. (A) Heat map analysis of all genes showing the distribution of H1 as a function of nucleosome spacing, aligned on the +1 nucleosome. Each row represents a gene. Genes are sorted in order of increasing average spacing in wild-type cells, from top to bottom. The color scale represents H1 density (red: high; blue: low dyad density). H1 ChIP-exo data are derived from (45). (B) H1 occupancy on gene bodies relative to the genome-wide average as a function of nucleosome spacing in wild-type cells: ChIP-exo data for H1 normalized to the average occupancy per bp (blue line) or per nucleosome (red line).
DISCUSSION
Competition between ISW1 and CHD1 determines nucleosome spacing on most genes
We proposed that the three spacing enzymes identified in yeast are not functionally redundant but instead direct the formation of nucleosome arrays with different spacing, as observed with the purified remodelers in vitro (18,20–22). Thus, the spacing on a particular gene depends on which of the three enzymes act on it. We found that CHD1 and ISW1 determine the spacing on most genes, whereas ISW2 affects only the most transcriptionally inactive genes. Although in vitro studies indicated that CHD1, ISW1 and ISW2 should give spacings of ∼160, ∼175 and ∼200 bp, only CHD1 gave the expected spacing in vivo (159 bp). Nevertheless, the spacings observed in vivo are in the expected order (CHD1 < ISW1 < ISW2).
The flat spacing distribution in the isw1Δ chd1Δ double mutant confirms that ISW1 and CHD1 are the major spacing enzymes at the global level (17). The residual phasing in the double and triple mutants might indicate a contribution from another, relatively minor, spacing enzyme, perhaps INO80 (46) or it might represent positioning signals in the DNA sequence, or close-packing of nucleosomes (47). Competition between CHD1 and ISW1 should result in short spacing specified by CHD1 in isw1Δ cells and longer spacing specified by ISW1 in chd1Δ cells. The decreased average spacing in isw1Δ cells indicates that ISW1 promotes the formation of arrays with wild-type spacing and that CHD1 directs the formation of arrays with short spacing. The large shift in the spacing distribution indicates that most genes are targets of both ISW1 and CHD1. These observations indicate that a competition occurs between ISW1 and CHD1 to set the spacing on the majority of yeast genes. Although the spacing in chd1Δ cells is longer than in isw1Δ cells, it is not longer than in wild-type cells and the phasing is weaker. To explain this observation, we suggest that ISW1 activity is partially dependent on CHD1, such that CHD1 must space the nucleosomes before ISW1 can create the fully phased arrays present in wild-type cells.
In summary, we propose that CHD1 builds short-spaced nucleosomal arrays and that ISW1 converts these arrays to longer spacing (Figure 6). Consequently, removal of ISW1 reveals the short-spaced arrays due to CHD1, whereas removal of CHD1 reveals relatively poorly phased arrays with approximately wild-type spacing due to impaired ISW1 activity. In wild-type cells, ISW1 dominates the competition with CHD1 on most genes. We are probably observing a cell population average in which each gene has short spacing specified by CHD1 in some cells and longer spacing specified by ISW1 in the other cells. Nucleosome spacing on each gene may be dynamic as it is targeted by one remodeler and then the other, such that the +1 nucleosome remains in position while the downstream nucleosomes are shunted back and forth.
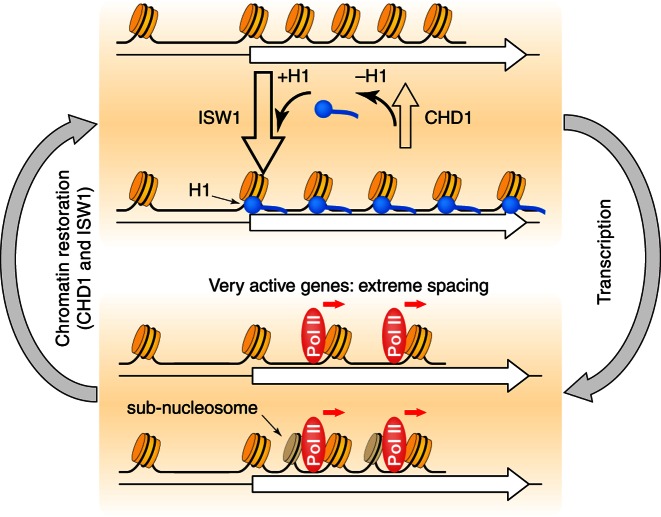
Figure 6.
Roles of the CHD1, ISW1 and ISW2 nucleosome spacing enzymes in chromatin organization. We propose that CHD1 and ISW1 compete to determine the spacing on most genes. Genes dominated by ISW1 have longer spacing resulting in linkers long enough to bind H1 (blue circles) with high affinity, resulting in more condensed chromatin. Genes dominated by CHD1 have short spacing, with linkers too short for H1 binding. Competition between CHD1 and ISW1 occurs on most genes, resulting in intermediate spacing, possibly creating a highly dynamic chromatin structure. The most inactive genes are also affected by ISW2, resulting in slightly longer spacing. Heavily transcribed genes have very disrupted chromatin, including gaps created by loss of some nucleosomes and sub-nucleosomes (darker cylinders) resulting from displacement of H2A-H2B dimers, resulting in extreme spacing.
A minor role for ISW2 on inactive genes
ISW2 plays only a minor role in nucleosome spacing, contributing to phasing and longer spacing only on the least active genes. The observed spacing on these genes in the isw1Δ chd1Δ double mutant is longer than average (∼176 bp), but not as long as predicted by in vitro experiments (∼200 bp) (18). Unlike CHD1 and ISW1, ISW2 has a role in setting the position of the +1 nucleosome at its target genes (48), which our data suggest are the most inactive genes. The small ISW2-dependent shift in the +1 nucleosome is in the opposite direction to that observed in RSC-depleted cells (11–13) and suggests that RSC and ISW2 might act antagonistically at inactive genes. It should be noted that we have tested only one growth condition (log phase growth in synthetic complete medium) and it is conceivable that the relative contributions of the three spacing enzymes may be quite different in other conditions.
Heavily transcribed genes and extreme spacing
Heavily transcribed genes have a wider NDR, decreased nucleosome occupancy and disrupted phasing on gene bodies (26,32,34,39,42–44). We find that active genes have extreme spacing, either very short or very long, and that only the short-spaced genes have CHD1-dependent phasing, consistent with the observation that CHD1 interacts with transcript elongation factors at transcribed genes (49). Why active genes have extreme spacing is unclear. Longer spacing would reduce the number of nucleosomes that have to be negotiated by Pol II; short spacing would be expected to aggravate the problem. The answer may lie in the reduced nucleosome occupancy observed on active genes (34), which reflects some transcription-associated loss of nucleosomes and, more often, loss of H2A-H2B dimers resulting in sub-nucleosomes (32,45,50). Thus, arrays missing the occasional nucleosome or containing some sub-nucleosomes in place of nucleosomes, resulting in a lower average nucleosome occupancy, could have a short average spacing and still be easier to transcribe (Figure 6).
Our data are consistent with a model in which ISW1 and CHD1 restore the nucleosomal array after its disruption by passage of Pol II, reducing histone exchange and cryptic initiation on gene bodies (51,52). On moderately active genes, they are able to re-organize the chromatin before a second round of transcription and therefore the chromatin remains well-organized despite the disruptive events associated with transcription. However, on highly active genes, repeated passages by Pol II may occur too rapidly for ISW1 and CHD1 to maintain the ordered chromatin structure, even though ISW1 and CHD1 densities appear to be higher on active genes (17,53,54).
Short spacing directed by CHD1 may exclude linker histone in vivo
Although H1 content and nucleosome spacing are correlated in vitro (20,27) and in vivo (28,55), loss of H1 does not have a global effect on spacing in yeast (47,56). This observation suggests that H1 does not determine spacing in yeast, although it is expressed at levels far below one molecule per nucleosome (57) and its precise function is unclear (58). In higher organisms, H1 binds to the nucleosome at the DNA entry/exit points and to the linker DNA between nucleosomes, driving chromatin condensation (59). It seems likely that yeast H1 fulfills a similar role, because H1 binding correlates with longer spacing and low transcriptional activity, perhaps facilitating chromatin condensation and repression. H1 binding may be reduced in short-spaced chromatin because the linker DNA is too short for high affinity binding (59).
We observed that genes with short spacing, determined primarily by CHD1, bind less H1 than genes with longer spacing, determined primarily by ISW1. We propose that CHD1 directs short spacing and evicts H1, resulting in partial unfolding of the chromatin fibre, whereas ISW1 directs longer spacing, allowing H1 to bind and the chromatin fibre to re-fold (Figure 6). Thus, a dynamic competition between ISW1 and CHD1 may control chromatin folding by regulating H1 binding. Supporting evidence for this model is provided by studies of Drosophila ISWI/Acf1 (both subunits of the ACF remodeling complex) and dCHD1. In vitro, short-spaced arrays made by dCHD1 exclude H1, whereas the longer-spaced arrays made by ACF accommodate H1 by increasing the spacing (20). Remodeling of nucleosomal arrays by dCHD1 is inhibited by H1 (60), potentially explaining the dominance of ISW1 in the competition with CHD1. In vivo, loss of ISWI function results in mitotic chromosome decondensation and displacement of H1 from chromatin without changing nucleosome spacing (55,61), although loss of Acf1 does reduce nucleosome spacing in embryos (62).
ACCESSION NUMBER
GEO database GSE69400.
Supplementary Material
Acknowledgments
We thank the NHLBI Core Facility (Yan Luo, Poching Liu and Jun Zhu) for paired-end sequencing. We thank Ho-Sung Rhee and Bongsoo Park for helpful discussions regarding the ChIP-exo experiments. This study utilized the high performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health.
FUNDING
Intramural Research Program of the NIH (NICHD). Funding for open access charge: Intramural Research Program of the NIH (NICHD).
Conflict of interest statement. None declared.
REFERENCES
- 1.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Swygert S.G., Peterson C.L. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim. Biophys. Acta. 2014;1839:728–736. doi: 10.1016/j.bbagrm.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 4.Narlikar G.J., Sundaramoorthy R., Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witkowski L., Foulkes W.D. In Brief: Picturing the complex world of chromatin remodelling families. J. Pathol. 2015;237:403–406. doi: 10.1002/path.4585. [DOI] [PubMed] [Google Scholar]
- 7.Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 8.Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 9.Mavrich T.N., Ioshikhes I.P., Venters B.J., Jiang C., Tomsho L.P., Qi J., Schuster S.C., Albert I., Pugh B.F. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang C., Pugh B.F. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parnell T.J., Huff J.T., Cairns B.R. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartley P.D., Madhani H.D. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganguli D., Chereji R.V., Iben J.R., Cole H.A., Clark D.J. RSC-dependent constructive and destructive interference between opposing arrays of phased nucleosomes in yeast. Genome Res. 2014;24:1637–1649. doi: 10.1101/gr.177014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorch Y., Zhang M., Kornberg R.D. RSC unravels the nucleosome. Mol. Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 15.Saha A., Wittmeyer J., Cairns B.R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla M.S., Syed S.H., Montel F., Faivre-Moskalenko C., Bednar J., Travers A., Angelov D., Dimitrov S. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1936–1941. doi: 10.1073/pnas.0904497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gkikopoulos T., Schofield P., Singh V., Pinskaya M., Mellor J., Smolle M., Workman J.L., Barton G.J., Owen-Hughes T. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science. 2011;333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukiyama T., Palmer J., Landel C.C., Shiloach J., Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vary J.C., Fazzio T.G., Tsukiyama T. Assembly of yeast chromatin using ISWI complexes. Methods Enzymol. 2004;375:88–102. doi: 10.1016/s0076-6879(03)75006-x. [DOI] [PubMed] [Google Scholar]
- 20.Lusser A., Urwin D.L., Kadonaga J.T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 21.Stockdale C., Flaus A., Ferreira H., Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T., Bulger M., Pazin M.J., Kobayashi R., Kadonaga J.T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 23.Pointner J., Persson J., Prasad P., Norman-Axelsson U., Stralfors A., Khorosjutina O., Krietenstein N., Svensson J.P., Ekwall K., Korber P. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. EMBO J. 2012;31:4388–4403. doi: 10.1038/emboj.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torigoe S.E., Patel A., Khuong M.T., Bowman G.D., Kadonaga J.T. ATP-dependent chromatin assembly is functionally distinct from chromatin remodeling. Elife. 2013;2:e00863. doi: 10.7554/eLife.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieleg C., Ketterer P., Nuebler J., Ludwigsen J., Gerland U., Dietz H., Mueller-Planitz F., Korber P. Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density. Mol. Cell. Biol. 2015;35:1588–1605. doi: 10.1128/MCB.01070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blank T.A., Becker P.B. Electrostatic mechanism of nucleosome spacing. J. Mol. Biol. 1995;252:305–313. doi: 10.1006/jmbi.1995.0498. [DOI] [PubMed] [Google Scholar]
- 28.Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 30.Cutter A.R., Hayes J.J. A brief review of nucleosome structure. FEBS Lett. 2015;589:2914–2922. doi: 10.1016/j.febslet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole H.A., Howard B.H., Clark D.J. Genome-wide mapping of nucleosomes in yeast using paired-end sequencing. Methods Enzymol. 2012;513:145–168. doi: 10.1016/B978-0-12-391938-0.00006-9. [DOI] [PubMed] [Google Scholar]
- 32.Cole H.A., Ocampo J., Iben J.R., Chereji R.V., Clark D.J. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res. 2014;42:12512–12522. doi: 10.1093/nar/gku1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole H.A., Howard B.H., Clark D.J. Activation-induced disruption of nucleosome position clusters on the coding regions of Gcn4-dependent genes extends into neighbouring genes. Nucleic Acids Res. 2011;39:9521–9535. doi: 10.1093/nar/gkr643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole H.A., Howard B.H., Clark D.J. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12687–12692. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park D., Shivram H., Iyer V.R. Chd1 co-localizes with early transcription elongation factors independently of H3K36 methylation and releases stalled RNA polymerase II at introns. Epigenetics Chromatin. 2014;7:32. doi: 10.1186/1756-8935-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui F., Cole H.A., Clark D.J., Zhurkin V.B. Transcriptional activation of yeast genes disrupts intragenic nucleosome phasing. Nucleic Acids Res. 2012;40:10753–10764. doi: 10.1093/nar/gks870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tirosh I., Sigal N., Barkai N. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol. 2010;11:R49. doi: 10.1186/gb-2010-11-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen K., Vinayachandran V., Batta K., Koerber R.T., Pugh B.F. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field Y., Kaplan N., Fondufe-Mittendorf Y., Moore I.K., Sharon E., Lubling Y., Widom J., Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shivaswamy S., Bhinge A., Zhao Y., Jones S., Hirst M., Iyer V.R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zawadzki K.A., Morozov A.V., Broach J.R. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:3503–3513. doi: 10.1091/mbc.E09-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhee H.S., Bataille A.R., Zhang L., Pugh B.F. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell. 2014;159:1377–1388. doi: 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Udugama M., Sabri A., Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole H.A., Cui F., Ocampo J., Burke T.L., Nikitina T., Nagarajavel V., Kotomura N., Zhurkin V.B., Clark D.J. Novel nucleosomal particles containing core histones and linker DNA but no histone H1. Nucleic Acids Res. 2015;44:573–581. doi: 10.1093/nar/gkv943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitehouse I., Rando O.J., Delrow J., Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 49.Simic R., Lindstrom D.L., Tran H.G., Roinick K.L., Costa P.J., Johnson A.D., Hartzog G.A., Arndt K.M. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chereji R.V., Morozov A.V. Ubiquitous nucleosome crowding in the yeast genome. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5236–5241. doi: 10.1073/pnas.1321001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smolle M., Venkatesh S., Gogol M.M., Li H., Zhang Y., Florens L., Washburn M.P., Workman J.L. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radman-Livaja M., Quan T.K., Valenzuela L., Armstrong J.A., van Welsem T., Kim T., Lee L.J., Buratowski S., van Leeuwen F., Rando O.J., et al. A key role for Chd1 in histone H3 dynamics at the 3′ ends of long genes in yeast. PLoS Genet. 2012;8:e1002811. doi: 10.1371/journal.pgen.1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zentner G.E., Tsukiyama T., Henikoff S. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet. 2013;9:e1003317. doi: 10.1371/journal.pgen.1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chereji R.V., Morozov A.V. Functional roles of nucleosome stability and dynamics. Brief. Funct. Genomics. 2015;14:50–60. doi: 10.1093/bfgp/elu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siriaco G., Deuring R., Chioda M., Becker P.B., Tamkun J.W. Drosophila ISWI regulates the association of histone H1 with interphase chromosomes in vivo. Genetics. 2009;182:661–669. doi: 10.1534/genetics.109.102053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes A.L., Rando O.J. Comparative genomics reveals Chd1 as a determinant of nucleosome spacing in vivo. G3. 2015;5:1889–1897. doi: 10.1534/g3.115.020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freidkin I., Katcoff D.J. Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patterton H.G., Landel C.C., Landsman D., Peterson C.L., Simpson R.T. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:7268–7276. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- 59.Clark D.J., Kimura T. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 1990;211:883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- 60.Maier V.K., Chioda M., Rhodes D., Becker P.B. ACF catalyses chromatosome movements in chromatin fibres. EMBO J. 2008;27:817–826. doi: 10.1038/sj.emboj.7601902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corona D.F., Siriaco G., Armstrong J.A., Snarskaya N., McClymont S.A., Scott M.P., Tamkun J.W. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fyodorov D.V., Blower M.D., Karpen G.H., Kadonaga J.T. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.