Abstract
Cell differentiation is a central process in development and in cancer growth and dissemination. OCT4 (POU5F1) and NANOG are essential for cell stemness and pluripotency; yet, the mechanisms that regulate their expression remain largely unknown. Repetitive elements account for almost half of the Human Genome; still, their role in gene regulation is poorly understood. Here, we show that the dioxin receptor (AHR) leads to differentiation of human carcinoma cells through the transcriptional upregulation of Alu retrotransposons, whose RNA transcripts can repress pluripotency genes. Despite the genome-wide presence of Alu elements, we provide evidences that those located at the NANOG and OCT4 promoters bind AHR, are transcribed by RNA polymerase-III and repress NANOG and OCT4 in differentiated cells. OCT4 and NANOG repression likely involves processing of Alu-derived transcripts through the miRNA machinery involving the Microprocessor and RISC. Consistently, stable AHR knockdown led to basal undifferentiation, impaired Alus transcription and blockade of OCT4 and NANOG repression. We suggest that transcripts produced from AHR-regulated Alu retrotransposons may control the expression of stemness genes OCT4 and NANOG during differentiation of carcinoma cells. The control of discrete Alu elements by specific transcription factors may have a dynamic role in genome regulation under physiological and diseased conditions.
INTRODUCTION
Recent evidences suggest that active transposable elements (TEs) have an important role in defining Human Genome structure and function and, consequently, in controlling development and disease (1,2). Short interspersed nuclear Alu elements (SINE) are a class of TEs highly abundant in the Human Genome that account for nearly 10% of its size (3). Alu retrotransposons derive from the 7SL RNA and are highly abundant in non-coding genomic regions including upstream promoters and gene introns (4,5). Previous studies have shown that global transposon activity varies under diverse cellular conditions; yet, very little is known regarding the mechanisms through which TEs regulate the expression of specific genes (6). In this context, a recent study revealed that an Alu element inserted in human chromosome 9p21 within the long non-coding RNA (lncRNA) ANRIL was needed to trans-regulate genes presumably involved in atherosclerosis (7). In the mouse, Anril lncRNA regulated cell proliferation and differentiation through the Cdkn2A/B gene (8).
Notably, TEs are potential carriers of binding sites for transcription factors. Genome-wide analyses have found an enrichment of binding sites for ESR1, TP53, OCT4 (POU5F1), SOX2 and CTCF in human TEs (9–11). In fact, TEs provide up to 25% of the binding sites for the pluripotency regulators OCT4 (POU5F1) and NANOG and for the chromatin remodeler CTCF in both human and mouse embryonic stem (ES) cells (10). Consequently, it appears plausible that TEs assume an important role in the control of transcriptional programs that regulate cell turnover and plasticity (10). Furthermore, certain classes of TEs were upregulated whereas others were downmodulated during the reprogramming of differentiated cells into induced pluripotent stem (iPSc) cells, thus producing an expression profile reminiscent of that of ES cells (12,13). Overall, these former studies suggest that TEs could modulate specific transcriptional programs that drive pluripotency and cell reprogramming (12). Previous work from our laboratory identified a novel B1-SINE retrotransposon (B1-X35S) widely represented in upstream regulatory regions of the mouse genome that acts as a genomic insulator blocking target gene expression (14,15). B1-X35S-dependent insulation required the interaction of transcription factors dioxin receptor (AhR) and Slug (Snai2) with their consensus sequences present in B1-X35S and the transcriptional activity of RNA polymerases III and II (15,16).
It is becoming increasingly clear that some repetitive elements are relevant for cell functioning. Recent efforts have identified repetitive sequences with the potential to regulate gene expression and to participate in the control of specific cell processes under normal and pathological conditions (15,17–19). In this work, we have investigated the functional relevance of Alu retrotransposons regulated by the dioxin receptor AHR in the differentiation of human embryonic carcinoma cells. We have focused on individual Alu elements located in the upstream regulatory regions of pluripotency genes NANOG and OCT4. AHR exerted a pro-differentiation role that was responsible, at least in part, for the repression of OCT4 and NANOG. Interestingly, such repressive mechanism appears to be mediated by non-coding RNA transcripts produced by RNA polIII from the Alu elements following AHR binding. In fact, the NANOG Alu was able to repress the expression of both OCT4 and NANOG in the absence of a differentiating stimulus. Among the mechanisms that could repress NANOG and OCT4 in differentiated carcinoma cells, processing and loading of NANOG Alu-derived transcripts into the miRNA machinery appears as a plausible possibility. In sum, we suggest that individual Alu retrotransposons could have a causal role in the control of complex cellular functions such as differentiation and pluripotency. The regulatory mechanism proposed here could also contribute to establish gene expression programs required for cellular reprogramming and for the maintenance of an undifferentiated state.
MATERIALS AND METHODS
Antibodies
The following antibodies were used: βIII-tubulin (Santa Cruz Biotechnology sc-58888, clone TUJ-1), GAP43 (Millipore AB-5220), Tau (generous gift of Dr Lorenzo-Benayas, University of Extremadura), GAPDH (Cell Signaling 2118, clone 14C10), OCT4 (Santa Cruz Biotechnology sc-5279, clone C-10), NANOG (AbCam Ab-21624), AGO2 (Millipore 03–110), AHR (ENZO Life Sciences BML-SA210 and Immunostep 130605–1) and β-Actin (Sigma A2066).
Cells lines and reagents
Human embryonic teratocarcinoma NTERA-wt and NTERA-sh cells were cultured in DMEM containing 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine at 37°C and 5% CO2 atmosphere. NTERA-wt and NTERA-sh cell lines were authenticated by DNA profiling using 8 different and highly polymorphic short tandem repeat (STR) loci (German Biological Resource Centre DSMZ). H9 human ES cells were cultured in matrigel-coated culture plates in high glucose Dulbecco's modified Eagle's medium (DMEM) knockout medium containing knockout serum replacement, L-glutamine and non-essential aminoacids at 37°C in a 5% CO2 atmosphere. Cells were tested to be mycoplasma free using the LookOut Mycoplasma detection kit (Sigma). Protein A/G-plus agarose was from Santa Cruz Biotechnology. The iScript™ Reverse Transcription Supermix was from Bio-Rad and the SYBR® Select Master Mix was from Life Technologies. The AhR agonist 6-formylindolo[3,2-b]carbazole (FICZ) was from Enzo.
Retroviral knockdown of human AHR
AHR was knocked-down in NTERA cells by retroviral transduction essentially as described (20,21) using the sh-RNA sequence 5′ TGCTGTTGACAGTGAGCGAGCA
ATGAATTTCCAAGGGAAGTAGTGAAGCCACAGATGTACTTCCCTTGGAAATTCATTGCCTGCCTACTGCCTCGGA 3′. For rescue experiments, an shRNA was synthesized targeting the 3′UTR region of the human AHR: 5′ TGCTGTTGACAGTG
AGCGAACTCTTTACCTTTATTGATATTAGTGAAGCCACAGATGTAATATCAATAAAGGTAAAGAGTGTGCCTACTGCCTCGGA 3′. shRNAs sequences were designed using the algorithm available at: http://www.stanford.edu/group/nolan/retroviral_systems/retsys.html.
Bioinformatic analysis of Alu elements containing AHR binding sites
The Human Genome was analyzed for the presence of conserved elements containing an XRE site and an E-box using the algorithm previously described (14). Two of the most abundant groups of elements (e.g. X36S and X45S) were analyzed for their presence in the 5′promoter region of pluripotency and stemness genes within a 200-bp interval from the transcription start site. The background expression of NANOG and OCT4 Alus was estimated by referring the RNA Seq data under basal cell conditions (untreated NTERA-wt cells) to the total number of AluSq and AluSx elements identified in the Human Genome.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) and double-sequential ChIP for AHR and OCT4 binding to the NANOG and OCT4 Alus were performed in NTERA-wt cells essentially as described (15,22). NTERA-sh cells were used as negative controls for ChIP experiments.
RNA immunoprecipitation (RIP)
RNA immunoprecipitation (RIP) for NANOG Alu-T1 and Alu-T2 loading into AGO2 was done essentially as described (23). Briefly, NTERA-wt cells were lysed in polysome lysis buffer containing RNase and protease inhibitors. Pre-washed protein-A/G plus agarose was incubated overnight at 4°C with 5 μg anti-AGO2 antibody or the same amount of IgG. After washing, the mRNP lysate was added and the mix briefly centrifuged to obtain the input fraction in the supernatant. Reactions were then gently rotated at room temperature for 2 h, washed and incubated for 30 min in NT2 buffer containing sodium dodecyl sulphate and proteinase K. RNA was phenol-chloroform-isoamyl alcohol extracted and ethanol precipitated. RIP was analyzed by qPCR using the oligonucleotides indicated in Supplementary Table S1. Polymerase chain reaction (PCR) reactions were also analyzed in agarose gels to determine the presence of the expected product.
RNA sequencing (RNAseq)
RNA sequencing (RNAseq) was performed in NTERA-wt and NTERA-sh cells in presence or absence of 1 μM RA. Libraries were made using RNA molecules of 75–250 bases in length isolated from agarose gels. Libraries were sequenced in an Illumina MiSeq equipment, which produced 8.32, 5.92, 7.70 and 8.79 million lectures for NTERA-wt (UT), NTERA-wt (RA), NTERA-sh (UT) and NTERA-sh (RA), respectively. FASTQ raw reads were transformed into FASTA format using a custom Perl script available on request. These 75-bp FASTA reads were integrated into a database, and NCBI Blast was applied using OCT4 and NANOG Alu full-length sequences as query. Search parameters were adjusted for the detection of sequences with the highest similarity, and normalized against the values obtained for standard Alu elements. When the analysis of identical sequences was required, ad hoc Perl scripts comparing specific sequences against the full Human Genome sequence were used.
Synthesis and cloning of NANOG and OCT4 Alus and 3′UTR sequences
Wild-type NANOG and OCT4 Alu elements and NANOG Alus mutated in the XRE site and/or A + B boxes were synthesized by GenScript and cloned in the promoter-less pUC57 vector. NANOG mRNA 3′UTR was amplified using the primers indicated in Supplementary Table S1 and cloned in the pGL2-Basic-Luc vector. NANOG 3′UTR mutants lacking the homology sequences for Alu-T1 and/or Alu-T2 were constructed by PCR from the wild-type sequence and cloned in the pGL2-Basic-Luc vector. The pUC57 vector does not contain any promoter and it is therefore indicated to assay the endogenous transcription of NANOG and OCT4 Alus by RNA polIII.
Mouse teratoma induction
Swiss male immunodeficient mice at 8–10 weeks of age (Charles Rives) were used for teratoma induction. Mice experimentation was approved and followed the regulation established by the Bioethics and Biosecurity Commission of the University of Granada. Teratoma assays were performed as described previously (24). Briefly, aliquots of 106 exponentially growing NTERA-wt and NTERA-sh cells were injected subcutaneously in the flank of the neck of each mouse. Two experiments of two mice were done for each cell line. After 7–8 weeks, the mice were killed and the tumors extracted, fixed in buffered formalin, embedded in paraffin and sectioned. Sections were stained with antibodies for the three germinal layers: endoderm (pan-cytokeratins and α-fetoprotein), mesoderm (α-smooth muscle actin) and ectoderm (acidic fibrilar acidic protein). Immunohistochemistry sections were analyzed blinded by eight different investigators and the intensity of staining estimated on a 0–10 scale.
Transient transfection
NANOG and OCT4 Alu transposons, DROSHA and DGCR8 dominant negative mutants (25) and NANOG 3′UTR plasmids were purified and transiently transfected in NTERA-wt or NTERA-sh cells using FuGene 6 (Promega). Aliquots of 1 μg DNA were used for the transfections essentially as described (14). Transfection continued for 48 h in experiments analyzing the expression of NANOG and OCT4 Alus or for 36 h in luciferase activity assays. H9 cells were transiently transfected with the AHR sh-RNA sequence indicated above using the Human Stem Cell Nucleofector kit in an Amaxa equipment (A-23 program) following the recommendations of the manufacturer.
Luciferase activity assays
NANOG 3′UTR constructs were amplified by RT-PCR from cellular RNA using the oligonucleotides indicated in Supplementary Table S1. The amplified 3′UTR was cloned in the XhoI-KpnI sites of the pGL2-Basic vector containing the firefly luciferase gene. This 3′UTR sequence was further modified by PCR to delete the homology region for NANOG Alu-T1, Alu-T2 or both. 3′UTR constructs were co-transfected with the pRLTK (renilla luciferase) vector to normalize luciferase activity. Firefly and renilla luciferase activities were measured after 36 h using the Dual Glow kit (Promega) in a Varioskan Flash luminometer (Thermo).
Nuclear and cytosolic cell extracts and immunoblotting
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis and immunoblotting were performed using NTERA-wt and NTERA-sh cell extracts as indicated (21). Nuclear and cytosolic cellular extracts were prepared from both cell lines as described (26).
Immunofluorescence
Immunofluorescence was done in an Olympus FV1000 confocal microscopy on cultures fixed for 20 min at room temperature in 4% paraformaldehyde (Polysciences Inc.). Fluorochromes used were Alexa 488 (NANOG, OCT4, βIII-tubulin) and Alexa 633 (AHR, Tau and GAP43). DAPI was used to stain cell nuclei. Fluorescence distribution was analyzed using the FV10 software (Olympus).
Reverse transcription and real-time RT-qPCR
Total RNA was isolated from NTERA-wt and NTERA-sh cells using the RNeasy kit (Qiagen). Reverse transcription was performed with random hexamers and the iScript™ Reverse Transcription Supermix (Bio-Rad). qPCR was done using SYBR® Select Master Mix (Life Technologies) in a Step One equipment (Life Technologies). The oligonucleotides used for RT-qPCR are indicated in Supplementary Table S1. Conventional (no quantitative PCR) was done using Biotaq DNA polymerase in a Bio-Rad T100 equipment.
Clone formation assay
Clonogenic assays in 2D were done by platting 103 NTERA-wt and NTERAsh cells in 35 mm plain tissue culture dishes basically as described (20). After 7 days, the medium was removed and clones washed in PBS, photographed and counted using the ImageJ software (version 1.45S).
Flow cytometry
The analysis and separation of cancer stem-like cells in NTERA-wt and NTERA-sh cultures was done in a MoFlo-XDP equipment (Beckman-Coulter) essentially as described (20). Both cell lines were stained for 10 min at 10°C with antibodies specific for CD133+ (CD133-PE), CD44+ (CD44-PerCP) and CD29+ (CD29-FITC). To-Pro (0.1 μM) was used to discriminate dead cells from the analyses. Cell cycle distribution was determined essentially as described (27).
Sphere formation assay
NTERA-wt and NTERA-sh cells were plated in Nunc ultra-low adherence 24-well plates (Thermo Fisher) at 3 × 104 cells/well and spheres allowed to form for 24 h. Images were acquired with an Olympus Cell-R microscope and sphere number and size quantified using the ImageJ software (version 1.45S).
Transposition assays
For transposition assays, 2 × 104 cells/35 mm2 plate were co-transfected with the driver vector pCEPL1SM-NN (containing a LINE-1) and the pBSKS-NeoIII vector harboring the NANOG or the OCT4 Alu elements. Transfectants were selected by culturing with 400 μg/ml G418 for 12–13 days.
DNA sequencing
NANOG Alu-T1 and Alu-T2 ncRNA transcripts were sequenced using the oligonucleotides indicated in Supplementary Table S1 in an Applied Biosystems 3130 automatic capillary sequencer.
Statistical analyses
Comparison between experimental conditions was done using GraphPad Prism 6.0 software (GraphPad). Unpaired two-sided Student's t-test was used to analyze differences between two experimental groups. Differences between three or more experimental conditions were analyzed using ANOVA. Data are shown as mean ± SD. Differences were considered significant at *P < 0.05; **P < 0.01; ***P < 0.001. Data analyses are indicated in the figure legends.
RESULTS
AHR knockdown leads to an undifferentiated phenotype and blocks RA-induced differentiation of human carcinoma cells
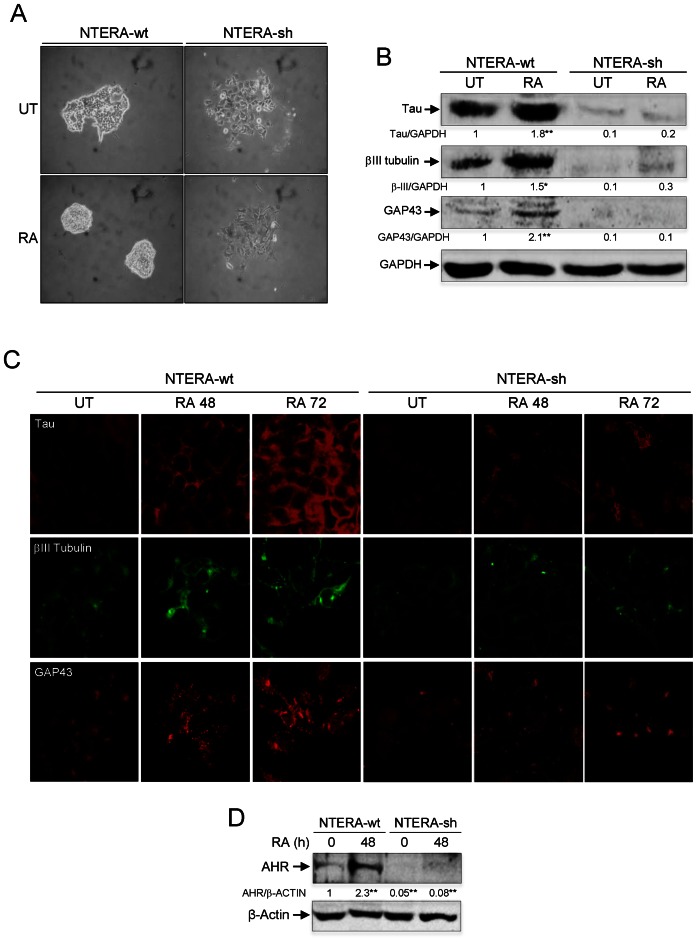
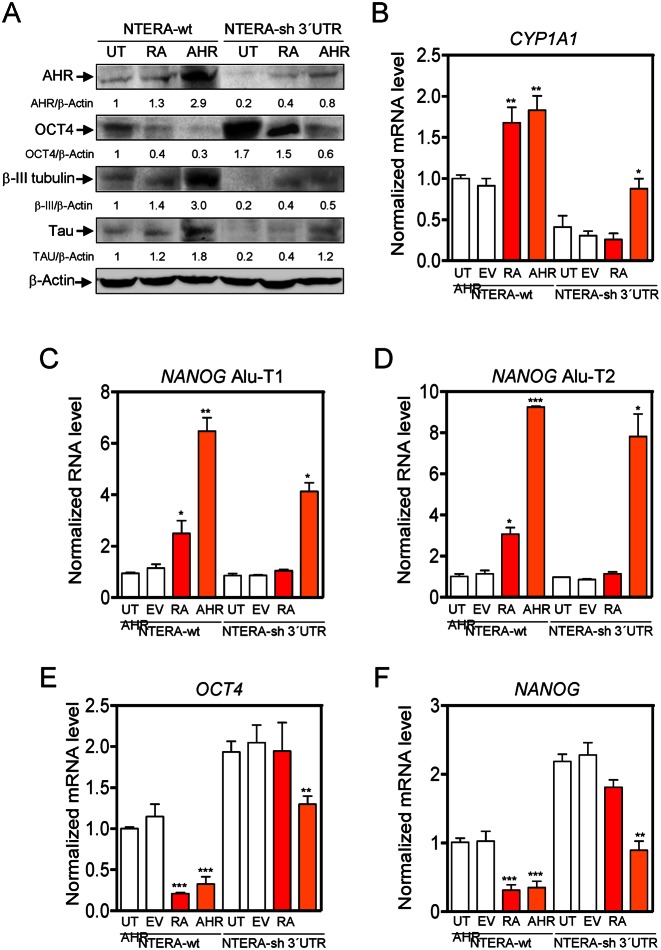
To investigate the implication of AHR in cell differentiation, we have used human pluripotent embryonic teratocarcinoma NTERA2 cells (NTERA) because they can be differentiated in culture by the addition of the endogenous molecule retinoic acid (RA) and because they can be genetically engineered to modulate target gene expression (28–30). Consistent with their pluripotent phenotype, treatment of NTERA cells growing at low cell density with RA produced highly compact clones (Figure 1A) having increased expression of the neuronal markers Tau, βIII-tubulin and GAP43 (Figure 1B and C). This differentiation phenotype of NTERA cells is consistent with that reported in previous studies (29). Notably, total cellular levels of AHR significantly increased during RA-induced differentiation of NTERA cells (Figure 1D), suggesting a role for this receptor in the process. To test this hypothesis, we used retroviral transduction to stably knockdown AHR expression in NTERA cells by means of short hairpin RNAs (NTERA-sh). This experimental approach was very efficient in reducing AHR protein to very low levels in basal and RA-treated cells (Figure 1D). To avoid caveats derived from the use of retrovirally-transduced cells, we decided to work with a pool of NTERA cells after puromycin selection rather than using individual interfered clones. AHR depletion produced spread clones (Figure 1A) with a significant reduction in the expression of neuronal markers Tau, βIII-tubulin and GAP43 (Figure 1B and C). Moreover, AHR depleted cells had an almost negligible response to RA-induced differentiation (Figure 1A–C). The undifferentiated phenotype caused by AhR downmodulation in basal and RA-treated NTERA cells was not associated with significant changes in cell cycle distribution as determined by FACS analysis (Supplementary Figure S1A). Previous reports have shown that AHR depletion could be relevant for mouse ES cell maintenance (31) and for mouse melanoma cells to develop an undifferentiated and metastatic phenotype (20,32). In agreement, AHR-depleted NTERA-sh cells had increased levels of stem-like markers CD44+/CD133+/CD29+, more potential to form spheres under low attachment growing conditions and reduced levels of differentiation-related dehydrogenase activity than wild-type NTERA cells (Supplementary Figure S1B–G). Altogether, these data suggest that AHR is involved in NTERA differentiation.
Figure 1.
AHR knockdown impairs RA-induced differentiation. (A) NTERA-wt and AHR knockdown NTERA-sh cells were grown in 2D cultures and clone formation analyzed 48 h later in absence (UT) or presence of 1 μM RA. (B) Both cell lines were left untreated (UT) or treated with 1 μM RA for 48 h. Total cell extracts were analyzed for the expression of neuronal markers Tau, βIII-tubulin and GAP43 by immunoblotting. GAPDH was used to normalize protein expression. (C) Tau, βIII-tubulin and GAP43 were also analyzed by immunofluorescence in untreated (UT) or RA-treated NTERA-wt and NTERA-sh cells using a Fluoview FV1000 confocal microscope. (D) NTERA-wt and NTERA-sh cells were left untreated or treated with 1 μM RA for 72 h and total AHR protein levels determined by immunoblotting. β-Actin was used as normalization control. Panels A–D: n = 4 biological replicates. *P < 0.05 and **P < 0.01. Data are shown as mean ± SD.
Differentiation promotes AHR nuclear accumulation and represses pluripotency genes OCT4 and NANOG
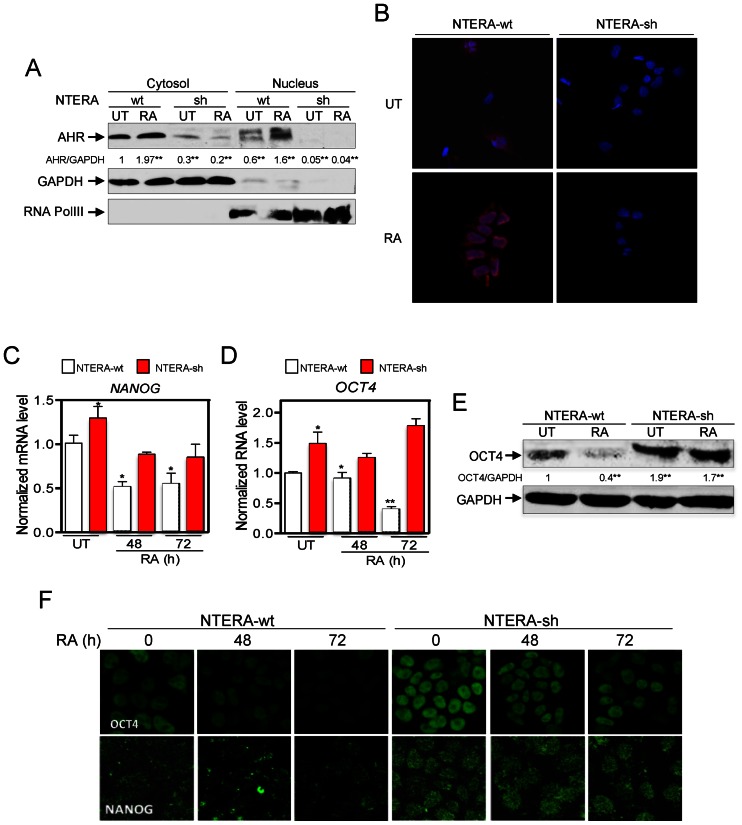
We next explored if the differentiation-dependent increase in AHR levels could lead to changes in its intracellular distribution. RA treatment promoted a robust nuclear accumulation of AHR in NTERA-wt cells (Figure 2A and B) but had a negligible effect in AHR-interfered cells, which maintained their very low basal levels of AHR (Figure 2A and B). To further analyze the implication of AHR in RA-induced differentiation, we performed long-term RA treatment (e.g. 4 weeks) of NTERA-sh cells. Under persistent differentiating conditions, AHR protein levels gradually increased and NTERA-sh cells acquired wild-type-like clone morphology and the expression of neuronal markers Tau and βIII-tubulin (Supplementary Figure S2A and B). Restoration of a differentiation response in NTERA-sh cells by AHR re-expression supports a role for AHR in preserving teratocarcinoma cell differentiation.
Figure 2.
RA-induced differentiation translocates AHR to the cell nucleus and represses OCT4 and NANOG. (A) NTERA-wt and NTERA-sh cells were left untreated or treated with 1 μM RA for 72 h. Cytosolic and nuclear extracts were prepared and analyzed for the presence of AHR in each cellular compartment by immunoblotting. GAPDH and RNA polIII were used as cytosolic and nuclear controls, respectively. (B) The intracellular distribution of AHR was determined by immunofluorescence in NTERA-wt and NTERA-sh cells left untreated or treated with 1 μM RA for 72 h in absence (UT) and presence of 1 μM RA. (C and D) NANOG and OCT4 mRNAs were quantified by RT-qPCR in both cell lines left untreated (UT) or treated with 1 μM RA for 48 or 72 h. GAPDH mRNA was used to normalize gene expression (ΔCt) and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. (E) OCT4 protein levels were analyzed by immunoblotting and normalized by GAPDH. (F) The effects of RA on OCT4 and NANOG protein expression were also determined in NTERA-wt and NTERA-sh cells by immunofluorescence. Panels A–D: n = 4 biological replicates; panels E–F: n = 6, two technical replicates in three biological replicates. *P < 0.05 and **P < 0.01. Data are shown as mean ± SD.
To address that hypothesis, we first analyzed whether the effects of AHR on cell differentiation were mediated by changes in the expression of pluripotency genes. RA treatment of NTERA-wt cells produced a large reduction in mRNA levels of the stemness-related genes NANOG and OCT4 (Figure 2 C and D, and Supplementary Figure S3A and B) without a significant effect on SOX2 and NOTCH1 (Supplementary Figure S3C and D). In agreement, protein levels of OCT4 and NANOG were also reduced by RA treatment in NTERA-wt cells as determined by fluorescence confocal microscopy and western-blot analyses (Figure 2E and F). In contrast, NTERA-sh cells had increased levels of OCT4 and NANOG mRNA and protein under basal conditions and, importantly, they were less sensitive to RA treatment (Figure 2C–F). To further support the implication of AHR in OCT4 and NANOG repression, we did treatments with the AHR ligand 6-formylindolo[3,2-b]carbazole (FICZ) in absence of RA (33). FICZ decreased OCT4 and NANOG mRNA levels in untreated NTERA-wt cells (Supplementary Figure S3E and F) but not in NTERA-sh cells (Supplementary Figure S3G and H), suggesting that AHR is involved in the mechanism that regulates OCT4 and NANOG during the differentiation of human carcinoma cells.
Pluripotency genes contain Alu retrotransposons in their upstream regulatory regions that are transcribed upon differentiation
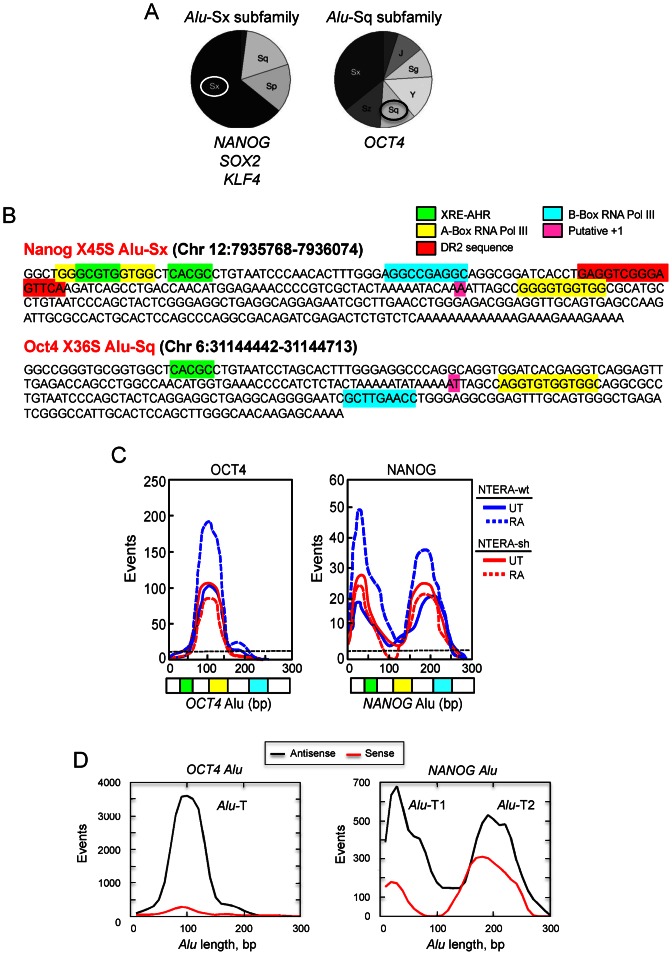
Bioinformatic analyses have identified thousands of potential AHR binding sites (XRE; Xenobiotic Responsive Element) dispersed throughout the Human Genome (14,16,34). However, their functional and biological significance remains to be determined. Using a previously developed Perl script (14), we have identified XRE sites for AHR binding that happen to be included within repetitive sequences that are located in the promoter of pluripotency genes NANOG, OCT4, SOX2 and KLF4. These repetitive elements have been assigned to the family of Alu-S retrotransposons having an XRE / E-box motif similar to that found in murine SINE-B1 repeats (14) (Figure 3A). Alu elements in the NANOG, SOX2 and KLF4 promoters belong to the Sx subfamily whereas the OCT4-Alu is included in the Sq subfamily of human retroelements (Figure 3A). Importantly, these Alus contain A and B boxes and a putative transcription start site for RNA polymerase III (RNA polIII)-dependent transcription and at least one XRE site for AHR binding (Figure 3B, NANOG and OCT4 Alus are shown). This sequence analysis suggested that these Alus could control the expression of their hosting genes through the transcription factor AHR.
Figure 3.
Human OCT4 and NANOG genes contain Alu retrotransposons in their promoters harboring AHR and RNA polIII binding sites. (A) Sequence analysis was used to determine the distribution of Alu retrotransposons among the Alu-Sx (NANOG, SOX2 and KLF4) or Alu-Sq (OCT4) subfamilies. (B) The full-length sequences of NANOG-Alu-Sx and OCT4-Alu-Sq were analyzed for the presence of XRE sites for AHR binding and A and B boxes for RNA polIII-dependent transcription. A DR2 box was also found in the NANOG-Alu. OCT4 and NANOG Alus contained the putative +1 adenosine nucleotide for RNA polIII transcription. The algorithm used for the bioinformatics analysis has been described (14). (C) Non-coding RNA libraries were prepared and sequenced as indicated in Supplementary Figure S4A. The bulk of reads obtained from each library was aligned with the sequences of OCT4 and NANOG Alus. Transcripts frequently expressed in NTERA-wt (blue) or NTERA-sh (red) in absence (UT) or presence of 1 μM RA appeared as peaks overlapping the sequence of each Alu element. The baseline expression of human AluSq and AluSx transposons is indicated by a horizontal black dotted line. (D) The sense (red line) and antisense (black line) orientation of the transcripts produced from the NANOG and OCT4 Alus was determined from the RNAseq dat. Transcripts showing complementary for either the sense or the antisense strand were plotted against the full-length Alus to produce peaks corresponding to the NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T molecules.
We considered the possibility that transcription of the Alu elements present in the promoter of these pluripotency genes could repress their expression to induce differentiation of carcinoma cells. We focus our study on NANOG and OCT4 since they were the genes more significantly affected by RA treatment in NTERA cells in an AHR-dependent manner (see Supplementary Figure S3). Transcription of Alu retrotransposons was analyzed by deep sequencing of small RNA molecules (RNAseq) isolated from NTERA-wt and NTERA-sh cells under basal and RA-differentiated conditions. Raw RNAseq data have been deposited in the Sequence Read Archive (SRA, NCBI) with submission number PRJNA301447. Previous work has shown that RNAseq is a useful tool to identify transcripts produced from individual Alu elements by RNA polIII (35). To avoid sequencing mRNAs and long non-coding RNA transcripts, libraries were made using size-selected small RNA populations (Supplementary Figure S4A, ncRNA). Bioinformatic alignment of the RNAseq data against OCT4 and NANOG Alus identified transcripts produced from the OCT4-Alu (hereafter OCT4 Alu-T) and NANOG-Alu (hereafter NANOG Alu-T1 and Alu-T2) under basal conditions in NTERA-wt cells (Figure 3C). Notably, the level of those transcripts was significantly increased by RA-induced differentiation in NTERA-wt cells but not in NTERA-sh cells (Figure 3C). Importantly, the level of NANOG and OCT4 Alu-derived transcripts was significantly higher than the predicted background level between AluSq and AluSx elements (Figure 3C, black dotted line). We have previously found that transcription of an AhR-regulated murine SINE-B1 retrotransposon takes place mainly in the antisense orientation (15). Similarly, the analysis of RNAseq data indicated that both OCT4 and NANOG Alus were preferentially transcribed in the antisense direction, despite the presence of sense transcripts produced from the NANOG Alu (Figure 3D).
AHR and RNA polIII are involved in OCT4 and NANOG repression in differentiated carcinoma cells
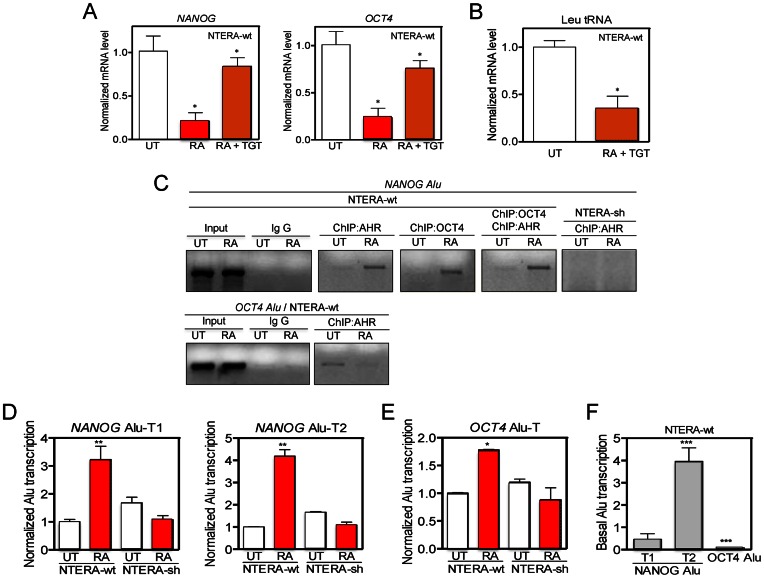
Most human Alus are transcribed by RNA polIII, which can be inhibited by the molecules tagetitoxin (TGT) (36) and WYE132 (37). TGT treatment significantly blocked the repression of NANOG and OCT4 produced by RA at the mRNA level in NTERA-wt cells (Figure 4A). Control experiments showed that TGT was a potent RNA polIII inhibitor in NTERA cells able to efficiently inhibit Leu tRNA expression (Figure 4B). In addition, NTERA-wt cells had a similar response to WYE132, which could also efficiently block the repression of NANOG and OCT4 by RA (Supplementary Figure S4B). WYE132 was an RNA polIII inhibitor in NTERA cells since it could repress Leu tRNA expression (Supplementary Figure S4C) without significantly affecting NANOG and OCT4 mRNAs in the absence of RA treatment (e.g. WYE132 did not inhibit RNA polII) (Supplementary Figure S4D). Thus, NANOG and OCT4 repression in differentiated NTERA cells appears to require AHR upregulation and the transcriptional activation of their Alu elements by RNA polIII. In agreement with such possibility, sequence analysis revealed that OCT4 and NANOG Alus contain the core motifs present in most active AluS subfamilies (Supplementary Figure S4E) (17). Furthermore, both NANOG and OCT4 Alus were able to retrotranspose in HeLa cells as determined using an established Alu engineered assay (25,38) (data not shown).
Figure 4.
OCT4 and NANOG Alus bind AHR, are transcribed by RNA polIII in differentiated cells. (A) NANOG and OCT4 mRNA levels were quantified in NTERA-wt cells left untreated (UT), treated with 1 mM RA or with 1 mM RA + 10 mM tagetitoxin (TGT) for 48 h. (B) Leu tRNA levels were measured in NTERA-wt cells untreated (UT) or treated with 1 mM RA + 10 mM TGT for 48 h. (C) Chromatin immunoprecipitation (ChIP) for AHR and OCT4 binding to the NANOG and OCT4 Alus was done in NTERA-wt cells left untreated (UT) or treated with 1 μM RA for 48 h. For specificity, one primer for the PCR reaction to amplify each Alu was located in a unique genomic sequence flanking the transposon (see Supplementary Table S1). Double ChIPs involved a first immunoprecipitation with anti-OCT4 followed by a second immunoprecipitation with anti-AHR antibody. NTERA-sh cells were used as negative controls. Input DNAs and immunoprecipitations done in absence of specific antibodies were also performed. (D and E) NANOG and OCT4 Alu-derived transcripts were quantified by RT-qPCR in absence (UT) or presence of 1 μM RA using the conditions described in Supplementary Figure S5A-C. (F) Basal levels of NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T were measured in NTERA-wt cells by RT-qPCR. GAPDH mRNA was used to normalize gene expression (ΔCt) and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. Panels A and B: n = 6, two technical replicates in three biological replicates; panel C: n = 4 biological replicates; panels D-F: n = 4, two technical replicates in two biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± SD.
As AHR is a transcription factor, we next tested if the repression of NANOG and OCT4 required AHR binding to the XRE sequences present in their upstream Alu elements (Figure 3B). ChIP experiments in NTERA-wt cells using genomic region-specific primers showed that, under basal conditions, AHR was present in the OCT4 Alu but not in the NANOG Alu (Figure 4C). Upon RA-induced differentiation, AHR was recruited to the NANOG-Alu and released from the OCT4 Alu (Figure 4C). Interestingly, sequential double AHR + OCT4 ChIP revealed that OCT4 and AHR were recruited together to the NANOG-Alu in RA-differentiated NTERA-wt cells (Figure 4C). A negligible interaction of AHR with the NANOG-Alu (Figure 4C) or the OCT4 Alu (not shown) was found in basal or RA-treated NTERA-sh cells. These results indicate that AHR binds to the NANOG Alu in differentiated cells and that AHR could be part of a transcriptional complex that also includes OCT4.
The NANOG and OCT4 Alu elements are transcribed during differentiation
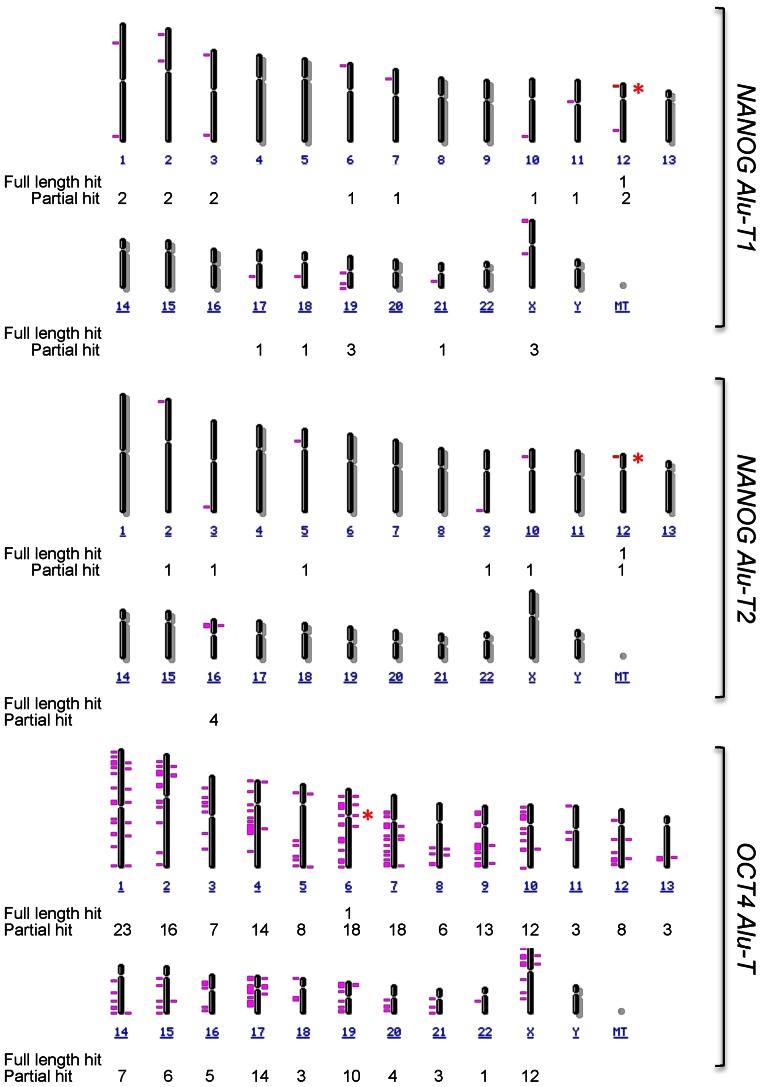
Based on the apparent prevalence of defined transcripts in differentiated NTERA cells (Figure 3C), we next used the RNAseq data to assess which amplicons may be used to detect the expression of NANOG and OCT4 Alus. Considering the repetitive nature of Alu elements, we first interrogated the Human Genome for the presence of instances identical to either NANOG or OCT4 Alus. At the current sequence resolution of the Human Genome, we did not find any other Alu elements fully homologous to those present in NANOG and OCT4 promoters. Based on this finding, we then decided to map putative sites in the Human Genome having full sequence identity with the transcripts produced from the NANOG and OCT4 Alus. This approach would give us an accurate estimation on to what extent those transcripts were produced from the NANOG and OCT4 Alus (since both elements appear at unique locations in the Human Genome) and not from other related retrotransposons. Interestingly, the amplicons predicted by RNA-seq analysis (Figure 3C) were also unique in the Human Genome for the case of NANOG Alu-T1 and Alu-T2 in chromosome 12 (Figure 5, full length hit and asterisks) and OCT4 Alu-T in chromosome 6 (Figure 5, full length hit and asterisks). Moreover, NCBI Blast analysis predicted partial sequence similarity with only 10 and 21 more Alu elements for NANOG Alu-T1 and Alu-T2 and with 214 more Alu elements in the case of OCT4, respectively (Figure 5). Based on these results, transcript-specific RT-qPCR assays were designed to quantify the expression of OCT4- and NANOG-Alu-derived transcripts (Supplementary Figure S5A and B). Amplicons were selected to cover the T1 (yellow) and T2 (red) transcripts from the NANOG Alu and the T (cyan) transcript from the OCT4 Alu (Supplementary Figure S5C). Capillary fluorescent dideoxi-sequencing was used to confirm the identity of the amplicons obtained by qPCR (Supplementary Figure S5D). The results obtained indicated that NANOG Alu-T1 and Alu-T2 (Figure 4D) and OCT4 Alu-T (Figure 4E) were significantly upregulated by RA in NTERA-wt but not in NTERA-sh cells. In addition, the OCT4 Alu-T was expressed at much lower levels than the NANOG Alu-T1 and Alu-T2 under basal cell conditions (Figure 4F). Moreover, treatment with TGT or WYE132 reduced the levels of OCT4 Alu-T, NANOG Alu-T1 and Alu-T2 transcripts in untreated NTERA-wt cells, further supporting the implication of RNA polIII in the transcription of OCT4 and NANOG Alus (Supplementary Figure S5E).
Figure 5.
Full length NANOG and OCT4 Alu-derived transcripts map to their specific genomic locations. The complete sequence of the NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T transcripts were aligned against the Human Genome to identify their fully homologous locations in each chromosome (100% sequence homology, full length hit, asterisk). Full length NANOG Alu-T1 and Alu-T2 map to chromosome 12 whereas full length OCT4 Alu map to chromosome 6. A similar analysis was done reducing the percentage of sequence homology of each Alu-derived transcript and the Human Genome (partial hit).
We next investigated the role of AHR in OCT4 and NANOG transcription. As a first approach, we treated NTERA cells with the AHR non-toxic ligand FICZ. AHR activation by FICZ increased the levels of NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T transcripts in NTERA-wt cells (Supplementary Figure S6A–C), supporting a role for this receptor in the regulation of these non-coding RNA transcripts. The effect was transient and each Alu-derived transcript returned to its basal level 24 h after FICZ treatment (Supplementary Figure S6A–C). Consistently, ChIP experiments showed that FICZ was able to induce AHR binding to the NANOG Alu in NTERA-wt cells to a level similar to that produced by RA treatment (Supplementary Figure S6D). Surprisingly, FICZ was also a potent inducer of OCT4 binding to the NANOG Alu, reaching levels significantly higher that those obtained by RA (Supplementary Figure S6D).
We then decided to rescue AHR expression in AHR knockdown cells to investigate if restoring AHR could increase the content in Alu-derived transcripts while reducing the levels of NANOG and OCT4 mRNAs (Figure 6). To do that, NTERA-wt cells were retrovirally engineered to stably express an shRNA molecule targeting the 3′UTR of the human AHR (NTERA-sh 3′UTR); this approach allowed transfection of a human AHR coding sequence not targeted by the 3′UTR shRNA molecule (Supplementary Table S1). NTERA-sh 3′UTR cells had an almost complete loss of AHR expression, increased basal protein levels of OCT4 and very low expression of neuronal markers βIII-tubulin and Tau, consistent with the results presented above (Figure 6A). Similar data were obtained using two different shRNA molecules and two different pools of NTERA cells, excluding potential off-target effects in the observed phenotype. Restoring AHR expression in NTERA-sh 3′UTR cells by transient transfection mimicked the effects induced by RA with respect to the expression of OCT4 and the neuronal differentiation markers βIII-tubulin and Tau (Figure 6A). Control experiments showed that AHR produced from the transfected plasmid was transcriptionally active in NTERA-sh 3′UTR cells based on the induction of its canonical target gene CYP1A1 (Figure 6B). In agreement with data shown above, rescuing AHR expression, in absence of RA treatment, increased the levels of NANOG Alu-T1 and Alu-T2 (Figure 6C and D) and repressed the mRNA expression of OCT4 and NANOG (Figure 6E and F) in NTERA-sh 3′UTR cells. Moreover, transient transfection of AHR in wild-type NTERA cells further increased the levels of NANOG Alu-derived transcripts (Figure 6C and D) and strongly repressed OCT4 and NANOG mRNAs (Figure 6E and F). Altogether, these results provide evidences that transcripts produced from Alu elements located upstream of NANOG and OCT4 might be involved in controlling the expression of these genes during differentiation of carcinoma cells.
Figure 6.
Rescuing AHR expression in AHR knockdown cells restores the expression of NANOG Alu-derived transcripts and represses OCT4 and NANOG. (A) NTERA wild-type cells were retrovirally transduced to stably express a shRNA directed against the 3′UTR of the human AHR. The resulting NTERA-sh 3′UTR cell line was left untreated (UT), treated with 1 μM RA for 72 h (RA) or transfected with a human AHR expression vector (AHR). Total protein extracts were examined for AHR, OCT4, βIII-tubulin and Tau expression by immunoblotting. NTERA-wt cells were also analyzed in parallel experiments. β-Actin was used to normalize protein expression. (B) The expression of the AHR target gene CYP1A1 was determined by RT-qPCR. (C–F) NTERA-wt and NTERA-sh 3′UTR cells were treated as above and the levels of NANOG Alu-T1 (C), NANOG Alu-T2 (D), OCT4 mRNA (E) and NANOG mRNA (F) quantified by RT-qPCR. GAPDH mRNA was used to normalize gene expression (ΔCt) and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. Panel A: n = 4, two technical replicates in two biological replicates; panels B–F: n = 6 two technical replicates in three biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± SD.
Transcription of the NANOG Alu from its RNA polIII promoter represses OCT4 and NANOG in absence of RA
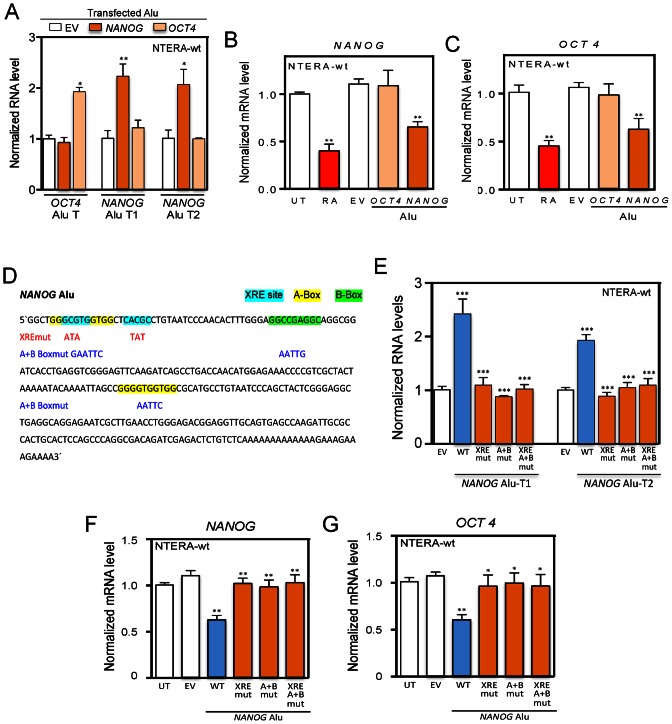
We next considered the possibility that the transcripts produced from the NANOG and OCT4 Alus could regulate the expression of their respective genes in absence of RA. To do that, each Alu element was cloned in a promoter-less plasmid (pUC57) to allow transcription from its internal RNA polIII promoter (Figure 3B, A and B boxes). We first confirmed by RT-qPCR that the OCT4 and NANOG-Alu constructs specifically expressed the corresponding Alu-T and Alu-T1 and Alu-T2 transcripts, respectively (Figure 7A). Remarkably, whereas transfection of the OCT4-Alu did not repress OCT4 nor NANOG in NTERA-wt cells, expression of the NANOG-Alu efficiently repressed the mRNA levels of both genes (Figure 7B and C), suggesting that transcripts produced from the NANOG Alu could potentially act in cis and trans to regulate NANOG and OCT4 expression. To provide additional support for the role of NANOG Alu-derived transcripts in regulating OCT4 and NANOG, we have introduced site-specific mutations in the XRE sites and in the A and B boxes for AHR and RNA polIII binding, respectively (Figure 7D). The selected mutations were highly effective in blocking NANOG Alu transcription from its internal RNA polIII promoter since the levels of NANOG Alu-T1 and Alu-T2 transcripts were largely reduced in NTERA-wt cells transfected with the promoter-less pUC57 vector harboring the XREmut, A + Bmut or XRE/A + Bmut sequences (Figure 7E). Notably, mutation of the XRE sites (XREmut), the A and B boxes (A + Bmut) or both (XREmut/A + B mut) significantly blocked the ability of the NANOG Alu to repress the mRNA expression of NANOG and OCT4 (Figure 7F and G), confirming that, in the absence of transcription from the NANOG Alu, the repressive mechanism was markedly impaired. Control experiments excluded the possibility that the effects produced by NANOG and OCT4 Alus were due to changes in AHR protein levels (Supplementary Figure S6E). In addition, repression of NANOG and OCT4 was AHR dependent since the NANOG Alu did not have a significant effect in NTERA-sh cells (Supplementary Figure S7A,B). Importantly, AHR- and RNA polIII-dependent transcription of the NANOG Alu may not have a general impact on pluripotency genes since it did not alter the mRNA levels of the reprogramming factor SOX2 (39) (Supplementary Figure S7C), the stem-cell factor and OCT4 regulator LIN28 (40) (Supplementary Figure S7D) or the pluripotency-inducing factor NR5A2 (41) (Supplementary Figure S7E). Moreover, even though the 3′UTR regions of LIN28 and NR5A2 have significant sequence homology with NANOG Alu-derived transcripts, their mRNA expression did not change by the NANOG Alu in NTERA-wt cells, further suggesting a specific effect on the NANOG mRNA.
Figure 7.
NANOG Alu represses NANOG and OCT4 expression in the absence of RA through AHR and RNA polIII. (A) NANOG and OCT4 Alus were individually transfected in NTERA-wt cells and the levels of NANOG Alu-T1, Alu-T2 and OCT4 Alu-T transcripts determined by RT-qPCR. (B and C) A promoter-less pUC57 expression vector containing the NANOG or OCT4 Alus was transfected in NTERA-wt cells and the mRNA levels of NANOG and OCT4 determined by RT-qPCR. RA was used as positive control. (D) Mutations introduced in the NANOG Alu to inactivate the XRE sites for AHR binding (cyan), the A (yellow) and B (green) boxes for RNA polIII or a combination of all of them. (E) NANOG Alu mutant constructs for AHR binding (XREmut), RNA polIII binding (A + Bmut) or both (XRE/A + Bmut) were transfected in NTERA-wt cells and the levels of NANOG Alu-T1 and Alu-T2 quantified by RT-qPCR. (F and G) Mutant constructs for the XRE sites and/or the A and B boxes in the NANOG Alu were cloned in the promoter-less pUC57 plasmid and transfected in NTERA-wt cells. Wild-type NANOG Alu (wt) and the empty plasmid (EV) were also transfected as controls. The mRNA levels of NANOG (F) and OCT4 (G) were analyzed by RT-qPCR. Wild-type NANOG Alu (wt) and the empty plasmid (EV) were used as experimental controls. GAPDH mRNA was used to normalize gene expression (ΔCt) and and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. Panels A-C; E-G: n = 6, two technical replicates in three biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± SD.
We then decided to analyze if AHR could also modulate OCT4 and NANOG expression and the levels of NANOG Alu-derived transcripts in non-transformed human ES cells. To do that, we used H9 human ES cells that were either differentiated with RA (42,43) or transiently transfected with a sh-AHR against AHR. H9 cells treated with RA significantly increased their AHR protein levels while markedly decreased their OCT4 and NANOG protein content (Supplementary Figure S8A). Downmodulation of pluripotency factors OCT4 and NANOG likely arose from a significant reduction in their mRNA levels after RA treatment (Supplementary Figure S8B). Importantly, RA treatment largely increased the expression of the Alu-T1 and Alu-T2 transcripts derived from the NANOG Alu while moderately enhanced the amount of the OCT4 Alu-T transcript (Supplementary Figure S8C). Next, H9 cells were transiently transfected with the same sh-AHR construct used to generate the stable NTERA-sh cell line analyzed above. AHR knockdown produced a significant increase in the protein levels of both OCT4 and NANOG as compared to wild-type H9 cells (Supplementary Figure S9A). Consistently, AHR downmodulation upregulated the mRNA levels of both genes in H9 cells (Supplementary Figure S9B). Furthermore, a transient reduction in AHR expression decreased the levels of NANOG Alu-T1 and Alu-T2 transcripts without having a significant effect in the OCT4 Alu-T transcript (Supplementary Figure S9C). Therefore, AHR appears to be able to regulate pluripotency factors OCT4 and NANOG and the expression of transcripts derived from the NANOG and OCT4 Alus not only in tumorigenic NTERA cells but also in untransformed H9 human ES cells.
NANOG and OCT4 repression by Alu-derived transcripts involves the miRNA machinery
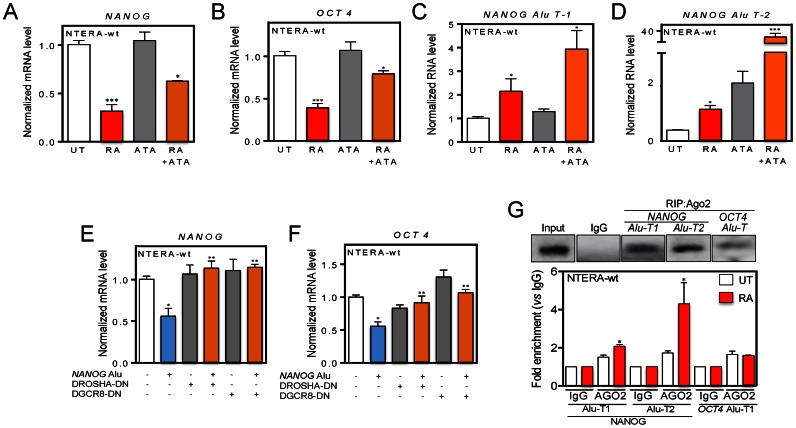
Previous studies have shown that the component of the miRNA pathway Microprocessor (DGCR8 and DROSHA) can bind transcripts produced from Alu and LINE-1 (long interspersed element-1) retrotransposons (25). It is also known that small non-coding RNAs produced by DICER/AGO3 from Alus containing the DR2 motif can modulate the proliferation of human stem cells (44). A DR2 element is defined as a 6-bp direct repeat of the retinoid acid receptor (RAR) binding site spaced by 2 nt (44). Based on these previous studies, we investigated whether the miRNA machinery was involved in the Alu-dependent regulation of NANOG and OCT4. First, we treated NTERA-wt cells with the RISC (RNA-induced silencing complex) inhibitor ATA (aurintricarboxylic acid) (45) and found that ATA significantly prevented the repression of NANOG and OCT4 caused by RA (Figure 8A and B). Consistent with the implication of NANOG Alu-derived transcripts in the mechanism, inhibition of the miRNA machinery by ATA also increased the levels of NANOG Alu-T1 and NANOG Alu-T2 in NTERA-wt cells treated with RA (Figure 8C and D). Next, we transfected plasmids expressing dominant-negative forms of DROSHA and DGCR8 in NTERA-wt cells and observed that blockade of these proteins rescued the repression of NANOG and OCT4 produced by the NANOG Alu in absence of RA (Figure 8E and F). Control experiments confirmed that dominant-negative forms of DROSHA and DGCR8 efficiently inhibited the miRNA pathway based on the accumulation of the pre-miR-24-1 (Supplementary Figure S7F). Collectively, these data suggest that transcripts produced from the NANOG Alu may act through the miRNA pathway to control NANOG and OCT4 expression, and that such process requires miRNA pathway components RISC and Microprocessor.
Figure 8.
RISC inhibition and microprocessor knockdown rescues NANOG and OCT4 expression likely through loading of NANOG Alu-derived transcripts into AGO2. (A and B) NTERA-wt cells were left untreated (UT), treated with 25 μM ATA (aurintricarboxylic acid) or with 1 μM RA + 25 μM ATA and the expression of NANOG (A) and OCT4 (B) determined as above. (C and D) NTERA-wt cells were left untreated (UT), treated with 25 μM ATA or with 1 μM RA + 25 μM ATA and the expression of NANOG Alu-T1 (C) and Alu-T2 (D) determined by RT-qPCR. The oligonucleotides used are indicated in Supplementary Table S1. (E and F) The NANOG Alu was transfected in NTERA-wt cells with or without dominant-negative forms of DROSHA or DGCR8 and their effects on NANOG (E) and OCT4 (F) expression analyzed by RT-qPCR. (G) RNA was extracted from NTERA-wt cells and RNA immunoprecipitations (RIP) for NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T transcripts performed using an anti-AGO2 antibody. RIP was quantified by qPCR using the oligonucleotides indicated in Supplementary Table S1. GAPDH mRNA was used to normalize gene expression (ΔCt) and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. Panels A and B: n = 6, two technical replicates in three biological replicates; panels C and D: n = 6, two technical replicates in three biological replicates; panels E and F: n = 4, two technical replicates in two biological replicates; panel G: n = 4, two technical replicates in two biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± SD.
As mentioned above, the DR2 element appears to be relevant for loading Alu-derived transcripts into the miRNA machinery via AGO3 (44). Since the NANOG Alu has a DR2 sequence (Figure 3B), we next tested whether AGO proteins could bind RNA transcripts produced from the NANOG and OCT4 Alus. RIP followed by RT-qPCR showed that NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T transcripts were loaded into the RISC component AGO2, and that loading increased upon differentiation of NTERA-wt cells (Figure 8G). Interestingly, cuantitative RNA analysis revealed a very low efficiency of OCT4 Alu-T loading into AGO2 even under RA-induced differentiation. Loading of NANOG Alu-T1 and Alu-T2 transcripts into AGO2 increased with time for at least 48 h after RA treatment (Supplementary Figure S7G).
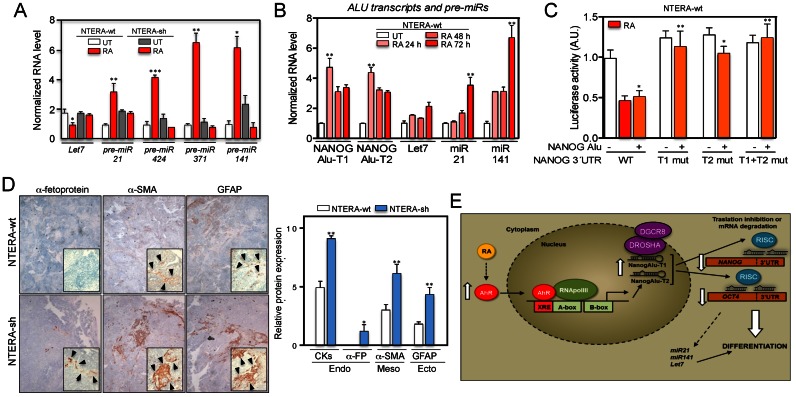
Certain TEs, including Alu retrotransposons, can regulate the generation of miRNAs (e.g. Let-7) known to be involved in cellular differentiation (46–48). The analysis of pre-miRNA expression revealed that differentiation reduced the levels of pre-Let-7 and increased pre-miR-21, pre-miR-424, pre-miR371 and pre-miR-141 in NTERA-wt but not in NTERA-sh cells (Figure 9A). Interestingly, time course experiments showed that NANOG Alu-T1 and Alu-T2 transcripts appeared early during differentiation (e.g. 24 h) whereas those pre-miR molecules reached their maximum levels only after 72 h of RA treatment (Figure 9B), suggesting that transcription of NANOG Alu-derived transcripts probably precedes the accumulation of miRNAs that could cooperate to control the differentiation of NTERA cells.
Figure 9.
NANOG Alu-derived transcripts could act through a miRNA mechanism to control NANOG and OCT4 repression during differentiation. (A) NTERA-wt and NTERA-sh cells were left untreated (UT) or treated with 1 μM RA for 48 h and the levels of pre-miRs pre-Let7, pre-miR-21, pre-miR-424, pre-miR-371, pre-miR-141 quantified by RT-qPCR using the oligonucleotides indicated in Supplementary Table S1. (B) Both cell lines were treated with 1 μM RA for the indicated times and the levels of NANOG Alu-T1 and Alu-T2, pre-Let7, pre-miR-21 and pre-miR-141 determined by RT-qPCR. (C) A NANOG 3′UTR-Luc construct and their deletion mutants lacking the homology regions for NANOG Alu-T1 (T1 mut), Alu-T2 (T2 mut) or both (T1 + T2 mut) were co-transfected with the NANOG Alu in NTERA-wt cells. Firefly luciferase activity was measured and normalized by renilla luciferase. Control experiments were done in presence of RA (dashed). (D) NTERA-wt and NTERA-sh cells were injected subcutaneously in immunodeficient Swiss nude mice (Charles River) and the tumors formed collected and analyzed by immunohistochemistry using endodermal (pan-cytokeratins and α-fetoprotein), mesodermal (α-smooth muscle actin) and ectodermal (glial fibrilar acidic protein, GFAP) markers. Immunohistochemistry sections were analyzed blinded by eight independent observers and protein expression quantified in a 0–10 scale. Arrowheads indicate positivity areas used to estimate protein expression. (E) Scheme of the mechanism proposed for the Alu-dependent repression of NANOG and OCT4 during carcinoma cell differentiation. GAPDH mRNA was used to normalize gene expression (ΔCt) and 2−ΔΔCt to calculate variations with respect to control or untreated conditions. Panels A and B: n = 6, three technical replicates in two biological replicates; panel C: n = 4, two technical replicates in two biological replicates; panel D: n = 4 mice for each cell line. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± SD.
These results lead us to consider that NANOG Alu-T1 and Alu-T2 transcripts, loaded into AGO2, could target complementary sequences in the mRNAs of NANOG and OCT4. Sequence analysis revealed that NANOG Alu-T1 and Alu-T2 transcripts were highly homologous to the 3′UTR of NANOG and significantly homologous to the 3′UTR of OCT4 (Supplementary Figure S10A and B). Conditional training analysis for the prediction of RNA secondary structures (CONTRAfold) revealed the optimal conformation of NANOG Alu-T1 and Alu-T2 transcripts in their homology regions with the 3′UTR of NANOG and OCT4 (Supplementary Figure S10C and D). Importantly, sequence analyses by RepeatMasker (http://www.repeatmasker.org) revealed that the 3′UTR of NANOG harbors an AluY element homologous to NANOG Alu-T1 and Alu-T2 transcripts. However, the 3′UTR of OCT4 did not contain Alu-derived sequences despite its partial homology with NANOG Alu-T1 and Alu-T2 (Supplementary Figure S10A,B). Other genes involved in differentiation but not regulated by Alu-derived transcripts such as SOX2 and NOTCH1 did not contain Alu-derived sequences in their 3′UTR regions, nor homology with NANOG Alu-T1 and Alu-T2. These findings suggested that the Alu present in the 3′UTR region of NANOG could interact with NANOG Alu-derived transcripts to repress NANOG expression in differentiated cells.
To analyze if NANOG Alu could regulate NANOG 3′UTR, we cloned the wild-type NANOG 3′UTR and its mutant forms lacking the homology regions for NANOG Alu-T1 and Alu-T2 transcripts in a luciferase reporter vector (Supplementary Figure S7H). Transfection of NTERA-wt cells with NANOG Alu in absence of RA repressed the activity of the wild-type NANOG 3′UTR-Luc construct (Figure 9C). However, the NANOG Alu was unable to repress the activity of NANOG 3′UTR-Luc mutants lacking the homology regions for Alu-T1, Alu-T2 or both transcripts (Figure 9C). These data led us to speculate that NANOG Alu might regulate NANOG and OCT4 at the mRNA level through the miRNA pathway. Consistent with this hypothesis, bioinformatic studies have suggested that Alu sequences located in the 3′UTR region of certain mRNAs can be recognized by miRNAs to regulate target gene expression (49).
AHR knockdown increases the formation of undifferentiated teratomas
We finally decided to validate AHR as a differentiation factor in human carcinoma cells using an in vivo teratoma assay. NTERA-wt and NTERA-sh cells were xenografted in immunocompromised Swiss mice and the teratomas formed analyzed for the presence of the three embryonic layers (39). Using this assay, we found that NTERA-wt cells generated teratomas containing endodermal (pan-cytokeratin and α-fetoprotein), mesodermal (α-smooth muscle actin) and ectodermal (glial fibrilar acidic protein, GFAP) embryonic layers (Figure 9D). Teratomas formed by NTERA-sh cells had significantly higher expression of endodermal, mesodermal and ectodermal markers than NTERA-wt cells (Figure 9D), suggesting that AHR depletion led to a more undifferentiated phenotype and an increased ability to differentiate in vivo. It is reasonable to assume that such properties of NTERA-sh cells might be due, at least in part, to their increased expression of pluripotency factors NANOG and OCT4. Notably, our hypothesis is in agreement with recent studies showing that in vivo reprogramming induces undifferentiated teratomas with increased expression of NANOG (50), and that OCT4 has a major role in reprogramming and in the generation of iPS cells (51).
DISCUSSION
TEs, including LINEs, SINEs and human Alus, are widespread in mammalian genomes and, although their contribution to genome evolution and dynamics is now generally accepted, very limited knowledge is available regarding their contribution to gene regulation and cell function (2,52). Alu retrotransposons derive from the 7SL RNA and are the most abundant TEs in the Human Genome. Among the different Alu lineages known, some ancient genomic AluS contain an active core sequence in terms of retrotransposition ability (17). In addition, it has been suggested that they could be active in somatic tissues as the human brain (53,54). Furthermore, there is an increasing interest in identifying Alu elements whose activation could contribute to human disease, such as that inserted into the ANRIL long non-coding RNA (7).
In this study, we have presented data suggesting that the dioxin receptor AHR regulates the transcription of Alu elements located in the upstream promoter regions of the pluripotency genes NANOG and OCT4, and that Alu-derived transcripts probably repress the expression of both genes in differentiated carcinoma cells possibly through the miRNA pathway. Our study therefore integrates the activation of specific Alu elements in the control of important cellular functions such as differentiation and pluripotency.
Here, we demonstrate that AHR acts as a differentiation factor in human embryonic teratocarcinoma cells because its stable knockdown led to a basal undifferentiated phenotype that made cells unresponsive to RA-induced neuronal differentiation. Furthermore, differentiation increased the cellular levels of AHR and promoted its nuclear translocation, thus suggesting increased transcriptional activity of this receptor. The role of AHR in differentiation is also supported by in vivo experiments, as teratomas produced by AHR-depleted NTERA cells were more undifferentiated than those from AHR-expressing NTERA cells. These results are consistent with the activation of AHR during differentiation of mouse ES cells (31) and with the inhibitory role of mouse AHR in epithelial-to-mesenchymal transition (21) and in melanoma primary tumorigenesis and metastasis (20).
AHR has been extensively characterized as a transcription factor that binds the promoter region of xenobiotic-related and xenobiotic-unrelated genes (55). It is remarkable that AHR can also interact with specific repetitive retrotransposon sequences such as the murine B1-SINE to trigger the activation of a genomic insulator eventually able to modulate gene expression (14–16). Binding to TEs appears to be a conserved property of AHR, as we show here that it can also directly binds to human AluS retrotransposons located in the promoter region of the pluripotency genes OCT4 and NANOG. However, a complex interplay between AHR binding and AluS sequences seems to occurs, since differentiation recruits AHR to the NANOG Alu while releasing it from the OCT4 Alu. One possible explanation for these results is that the increase in nuclear AHR that takes place during differentiation could shift the receptor from the OCT4 to the NANOG Alu, inducing at the same time the co-recruitment of OCT4 to the NANOG Alu. This would imply that AHR can cooperate with OCT4 in repressing NANOG expression and, thus, integrates this receptor into the proposed ‘pluripotency core regulatory circuitry’ of transcription factors that also includes SOX2 and that governs differentiation in human and mouse ES cells (56–58). In fact, AHR activation by its non-toxic ligand FICZ produced a robust binding of OCT4 to the NANOG Alu in absence of RA.
Notably, the repression of Alu-containing OCT4 and NANOG genes in differentiated NTERA cells is AHR dependent based on the lack of response shown by AHR-depleted cells, and appears to be specific since it does not take place in other examined Alu-containing pluripotency genes such as SOX2, NOTCH1, MYC and KLF4. Importantly, the repression process seemed to require the synthesis of non-coding RNA molecules since blockade of RNA polymerase III restored OCT4 and NANOG expression in an AHR-dependent manner. Notably, and as RNA polIII may transcribe 7SL RNAs and other non-coding sequences (59), we speculated that transcripts derived from the NANOG and OCT4 Alus could participate in the repression of both genes. Indeed, RNAseq and RT-qPCR experiments revealed that differentiation of NTERA cells increased the levels of transcripts derived from NANOG and OCT4 Alus, being predominant those synthesized from the NANOG Alu. In agreement with the gene expression data, transcription of these Alu elements is AHR dependent because their levels are upregulated by AHR knockdown in RA differentiated cells and increased by AHR rescue in RA-untreated cells. Such AHR-dependent mechanism regulating the expression of OCT4, NANOG and NANOG Alu-derived transcripts may be operative in non-transformed human cells since RA treatment and AHR knockdown reproduced in human H9 ES cells the same regulatory pattern to that observed in embryonic carcinoma NTERA cells. One important observation is that the Alu retrotransposons located in NANOG and OCT4 promoters have a high degree of sequence specificity, not been detectable at additional locations in the Human Genome. This property most likely determines that NANOG- and OCT4 Alu-derived transcripts have full sequence identity and full homology only to the chromosomal regions where their respective Alus are located and from which they are transcribed. Another relevant finding from this study is that expression of these Alu elements, by themselves, could regulate NANOG and OCT4 during carcinoma cell differentiation. We consider this result relevant since a deep transcriptome profiling study has shown that non-coding transcripts derived from long terminal repeats can associate to enhancers of pluripotency related genes (18,60,61). Nevertheless, the effects of NANOG and OCT4 Alus in gene repression appear to have a significant degree of specificity regarding not only to the Alu element but also their target genes. Indeed, expression of NANOG Alu from its endogenous promoter repressed both NANOG and OCT4 mRNAs in absence of a differentiation stimulus. In contrast, expression of OCT4 Alu does not significantly affect either gene under the same experimental conditions. Additionally, whereas NANOG Alu most likely regulates the NANOG gene in cis, its effects on the OCT4 gene could be done in trans. Such mechanism would resemble the one recently proposed for the long non-coding RNA ANRIL that regulates atherosclerosis genes in trans by means of an Alu element present in its sequence (7). Alternatively, the NANOG Alu could act on OCT4 by an indirect mechanism involving signaling through NANOG itself. Nonetheless, this possible functional connection between NANOG and OCT4 again supports the ‘pluripotency core regulatory circuitry’ mentioned above that would also include AHR (56–58). Finally, expression of NANOG Alu efficiently repressed NANOG and OCT4 but not other genes containing similar Alu elements such as SOX2, LIN28 and NR5A2. Although we cannot rule out at present time that other Alu loci dispersed in the Human Genome might contribute to the generation of additional ncRNAs, when combined with the ChIP data, our results strongly support that NANOG and OCT4 Alu-derived transcripts may be involved in the regulation of pluripotency.
Interestingly, recent studies in rice have shown that the plant DICER homolog DICER-like 3 can use TE RNAs to generate small interfering RNAs (siRNA) with the potential to change some agricultural traits of the plant (62). Similarly, in human cells, the Microprocessor can interact and process transcripts derived from Alus and LINE-1 to regulate retrotransposition, although it is not clear whether processed RNAs give rise to functional miRNAs that might help to regulate the expression of cellular genes (25). Notably, we found that chemical inhibition of the miRNA pathway by ATA not only rescued OCT4 and NANOG repression in differentiated cells but also increased the expression of NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T transcripts. Furthermore, dominant negative forms of Microprocessor components DROSHA and DGCR8 also blocked the repression of NANOG and OCT4 induced by the NANOG Alu in absence of RA. Intriguingly, the interaction between the miRNA pathway and transcripts derived from retrotransposons could be functionally conserved since inhibition of the Microprocessor also increased the levels of LINE1-derived transcripts and the expression of their target genes (25).
Although the mechanism by which NANOG and OCT4 Alu-derived transcripts may regulate gene expression require further experimentation, we speculate that it probably involves the interaction of Alu-derived transcripts with the 3′UTR region of the NANOG and OCT4 mRNAs. Several experiments support this model. First, NANOG Alu-T1 and Alu-T2 and OCT4 Alu-T transcripts are loaded into the RISC component AGO2 and such loading significantly increased with differentiation of NTERA cells. Second, a marked sequence similarity exists between NANOG Alu-T1 and Alu-T2 transcripts and the 3′UTR region of NANOG and OCT4 mRNAs. Third, the NANOG Alu was able to regulate the 3′UTR of NANOG but only if the homology sequences for the Alu-T1 and Alu-T2 transcripts remained intact. Finally, the levels of miRNAs involved in differentiation such as Let7 and miR141 increased with time by RA treatment in NTERA-wt but not in NTERA-sh cells and, interestingly, the expression of NANOG Alu-T1 and Alu-T2 preceded the accumulation of Let7 and miR141. It will be interesting to address the processing and identity of these Alu-derived transcripts and the mechanisms by which they probably interact with the 3′UTR regions of both genes. Our working model proposing that the interaction of Alu derived-transcripts with 3′UTRs could regulate gene expression through the miRNA pathway has been also considered in previous studies. Indeed, bioinformatic analyses have shown that certain small RNAs have the potential to target Alu-complementary sequences located in the 3′UTR of human mRNAs (49). Moreover, the FANTOM4 project has revealed that retrotransposons located in the 5′ end of mouse and human protein coding genes (as is the case for NANOG and OCT4 Alus) have the potential to be transcribed into non-coding RNAs. Importantly, this study also indicates that a large fraction of the expressed transcripts that contain retrotransposon complementary sequences in their 3′UTRs have lower expression levels than retrotransposon-free transcripts (63). Accordingly, we have found that the NANOG mRNA contains an AluY element in its 3′UTR that is highly homologous with ncRNAs generated from the AluSx located in the NANOG promoter. However, the 3′UTR of the OCT4 mRNA has only partial homology with NANOG Alu-derived transcripts, suggesting the existence of subtle differences in regulation between both genes.
In sum, we propose that AHR activates the transcription of Alu retrotransposons located in the NANOG and OCT4 upstream promoter regions in differentiated human carcinoma cells. Non-coding RNA transcripts produced from these Alus could repress NANOG and OCT4 expression probably through the miRNA pathway (Figure 9E). OCT4 and NANOG repression in differentiated cells is AHR and RNA polIII dependent and, notably, the expression of the NANOG Alu is able to repress both genes in the absence of a differentiating stimuli. This regulatory mechanism could be relevant for normal and tumor cell growth and dissemination. Moreover, the results included in this study reveal how fixed TEs can participate in Human Genome regulation by a novel mechanism involving non-coding RNAs and the miRNA pathway (Figure 9E).
ACCESSION NUMBERS
Sequence Read Archive (SRA, NCBI). Submission number PRJNA301447.
Supplementary Material
Acknowledgments
Eva Barrasa and the Servicio de Técnicas Aplicadas a las Biociencias (STAB) of the Universidad de Extremadura are greatly acknowledged for their technical support. NTERA2 cells were a generous gift of Dr Esteban Ballestar (IDIBELL, Barcelona, Spain). The anti-Tau antibody was generously provided by Dr María J. Lorenzo-Benayas (University of Extremadura, Spain). Plasmids expressing dominant negative forms of DROSHA and DGCR8 were a kind gift of Dr Narry Kim (Seoul National University, Korea).
FUNDING
Ministerio de Economía y Competitividad [BFU2011-22678, SAF2014-51813-R to P.M.F-S.]; Junta de Extremadura [GR10008, GR15008]; Red Temática de Investigación Cooperativa en Cáncer (RTICC); Carlos III Institute; Spanish Ministry of Economy and Competitiveness [RD12/0036/0032]; FPI Fellowship from the Junta de Extremadura (to A.M.H.); Marie Curie IRG project (FP7-PEOPLE-2007-4-3-IRG: SOMATIC LINE-1, in part to A.M.); CICE-FEDER-P09-CTS-4980, CICE-FEDER-P12-CTS-2256, Plan Nacional de I+D+I 2008–2011 and 2013–2016 (FIS-FEDER-PI11/01489, FIS-FEDER-PI14/02152), PCIN-2014-115-ERA-NET NEURON II (to J.L.G.P.); European Research Council [ERC-Consolidator ERC-STG-2012-233764]; International Early Career Scientist grant from the Howard Hughes Medical Institute [IECS-55007420]; European Union FEDER program. Funding for open access charge: Ministerio de Economía y Competitividad [BFU2011-22678, SAF2014-51813-R to P.M.F-S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Beck C.R., Garcia-Perez J.L., Badge R.M., Moran J.V. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hancks D.C., Kazazian H.H., Jr Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Kriegs J.O., Churakov G., Jurka J., Brosius J., Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Versteeg R., van Schaik B.D., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holdt L.M., Hoffmann S., Sass K., Langenberger D., Scholz M., Krohn K., Finstermeier K., Stahringer A., Wilfert W., Beutner F., et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasmant E., Sabbagh A., Vidaud M., Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 9.Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G., Chew J.L., Ruan Y., Wei C.L., Ng H.H., et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunarso G., Chia N.Y., Jeyakani J., Hwang C., Lu X., Chan Y.S., Ng H.H., Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 11.Wang T., Zeng J., Lowe C.B., Sellers R.G., Salama S.R., Yang M., Burgess S.M., Brachmann R.K., Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedli M., Turelli P., Kapopoulou A., Rauwel B., Castro-Diaz N., Rowe H.M., Ecco G., Unzu C., Planet E., Lombardo A., et al. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014;24:1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wissing S., Munoz-Lopez M., Macia A., Yang Z., Montano M., Collins W., Garcia-Perez J.L., Moran J.V., Greene W.C. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum. Mol. Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roman A.C., Benitez D.A., Carvajal-Gonzalez J.M., Fernandez-Salguero P.M. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1632–1637. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roman A.C., Gonzalez-Rico F.J., Molto E., Hernando H., Neto A., Vicente-Garcia C., Ballestar E., Gomez-Skarmeta J.L., Vavrova-Anderson J., White R.J., et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21:422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roman A.C., Gonzalez-Rico F.J., Fernandez-Salguero P.M. B1-SINE retrotransposons: establishing genomic insulatory networks. Mob. Genet. Elements. 2011;1:66–70. doi: 10.4161/mge.1.1.15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fort A., Hashimoto K., Yamada D., Salimullah M., Keya C.A., Saxena A., Bonetti A., Voineagu I., Bertin N., Kratz A., et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat. Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 19.Hung T., Pratt G., Sundararaman B., Townsend M.J., Chaivorapol C., Bhangale T., Graham R.R., Ortmann W., Criswell L.A., Yeo G. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science. 2015;350:455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contador-Troca M., Alvarez-Barrientos A., Barrasa E., Rico-Leo E.M., Catalina-Fernandez I., Menacho-Marquez M., Bustelo X.R., Garcia-Borron J.C., Gomez-Duran A., Saenz-Santamaria J., et al. The dioxin receptor has tumor suppressor activity in melanoma growth and metastasis. Carcinogenesis. 2013;34:2683–2693. doi: 10.1093/carcin/bgt248. [DOI] [PubMed] [Google Scholar]
- 21.Rico-Leo E.M., Alvarez-Barrientos A., Fernandez-Salguero P.M. Dioxin receptor expression inhibits basal and transforming growth factor beta-induced epithelial-to-mesenchymal transition. J. Biol. Chem. 2013;288:7841–7856. doi: 10.1074/jbc.M112.425009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Duran A., Ballestar E., Carvajal-Gonzalez J.M., Marlowe J.L., Puga A., Esteller M., Fernandez-Salguero P.M. Recruitment of CREB1 and histone deacetylase 2 (HDAC2) to the mouse Ltbp-1 promoter regulates its constitutive expression in a dioxin receptor-dependent manner. J. Mol. Biol. 2008;380:1–16. doi: 10.1016/j.jmb.2008.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenenbaum S.A., Carson C.C., Lager P.J., Keene J.D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez-Aranda I., Ramos-Mejia V., Bueno C., Munoz-Lopez M., Real P.J., Macia A., Sanchez L., Ligero G., Garcia-Parez J.L., Menendez P. Human induced pluripotent stem cells develop teratoma more efficiently and faster than human embryonic stem cells regardless the site of injection. Stem Cells. 2010;28:1568–1570. doi: 10.1002/stem.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heras S.R., Macias S., Plass M., Fernandez N., Cano D., Eyras E., Garcia-Perez J.L., Caceres J.F. The Microprocessor controls the activity of mammalian retrotransposons. Nat. Struct. Mol. Biol. 2013;20:1173–1181. doi: 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey-Barroso J., Alvarez-Barrientos A., Rico-Leo E., Contador-Troca M., Carvajal-Gonzalez J.M., Echarri A., Del Pozo M.A., Fernandez-Salguero P.M. The Dioxin receptor modulates Caveolin-1 mobilization during directional migration: role of cholesterol. Cell Commun. Signal. 2014;12:57. doi: 10.1186/s12964-014-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulero-Navarro S., Pozo-Guisado E., Perez-Mancera P.A., Alvarez-Barrientos A., Catalina-Fernandez I., Hernandez-Nieto E., Saenz-Santamaria J., Martinez N., Rojas J.M., Sanchez-Garcia I., et al. Immortalized mouse mammary fibroblasts lacking dioxin receptor have impaired tumorigenicity in a subcutaneous mouse xenograft model. J. Biol. Chem. 2005;280:28731–28741. doi: 10.1074/jbc.M504538200. [DOI] [PubMed] [Google Scholar]
- 28.Alfonso R.J., Gorrono-Etxebarria I., Rabano M., Vivanco M., Kypta R. Dickkopf-3 alters the morphological response to retinoic acid during neuronal differentiation of human embryonal carcinoma cells. Dev. Neurobiol. 2014;74:1243–1254. doi: 10.1002/dneu.22201. [DOI] [PubMed] [Google Scholar]
- 29.Elizalde C., Campa V.M., Caro M., Schlangen K., Aransay A.M., Vivanco M., Kypta R.M. Distinct roles for Wnt-4 and Wnt-11 during retinoic acid-induced neuronal differentiation. Stem Cells. 2011;29:141–153. doi: 10.1002/stem.562. [DOI] [PubMed] [Google Scholar]
- 30.Mancino M., Esposito C., Watanabe K., Nagaoka T., Gonzales M., Bianco C., Normanno N., Salomon D.S., Strizzi L. Neuronal guidance protein Netrin-1 induces differentiation in human embryonal carcinoma cells. Cancer Res. 2009;69:1717–1721. doi: 10.1158/0008-5472.CAN-08-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko C.I., Wang Q., Fan Y., Xia Y., Puga A. Pluripotency factors and Polycomb Group proteins repress aryl hydrocarbon receptor expression in murine embryonic stem cells. Stem Cell Res. 2013;12:296–308. doi: 10.1016/j.scr.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Contador-Troca M., Alvarez-Barrientos A., Merino J.M., Morales-Hernandez A., Rodriguez M.I., Rey-Barroso J., Barrasa E., Cerezo-Guisado M.I., Catalina-Fernandez I., Saenz-Santamaria J. Dioxin receptor regulates aldehyde dehydrogenase to block melanoma tumorigenesis and metastasis. Mol. Cancer. 2015;14:148. doi: 10.1186/s12943-015-0419-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wincent E., Amini N., Luecke S., Glatt H., Bergman J., Crescenzi C., Rannug A., Rannug U. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J. Biol. Chem. 2009;284:2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- 34.Sartor M.A., Schnekenburger M., Marlowe J.L., Reichard J.F., Wang Y., Fan Y., Ma C., Karyala S., Halbleib D., Liu X., et al. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ. Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conti A., Carnevali D., Bollati V., Fustinoni S., Pellegrini M., Dieci G. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res. 2015;43:817–835. doi: 10.1093/nar/gku1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vassylyev D.G., Svetlov V., Vassylyeva M.N., Perederina A., Igarashi N., Matsugaki N., Wakatsuki S., Artsimovitch I. Structural basis for transcription inhibition by tagetitoxin. Nat. Struct. Mol. Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shor B., Wu J., Shakey Q., Toral-Barza L., Shi C., Follettie M., Yu K. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J. Biol. Chem. 2010;285:15380–15392. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Qiu C., Ma Y., Wang J., Peng S., Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabut M., Samavarchi-Tehrani P., Wang X., Slobodeniuc V., O'Hanlon D., Sung H.K., Alvarez M., Talukder S., Pan Q., Mazzoni E.O., et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 42.Liu A., Prenger M.S., Norton D.D., Mei L., Kusiak J.W., Bai G. Nerve growth factor uses Ras/ERK and phosphatidylinositol 3-kinase cascades to up-regulate the N-methyl-D-aspartate receptor 1 promoter. J. Biol. Chem. 2001;276:45372–45379. doi: 10.1074/jbc.M105399200. [DOI] [PubMed] [Google Scholar]
- 43.Liu C., Zhong Y., Apostolou A., Fang S. Neural differentiation of human embryonic stem cells as an in vitro tool for the study of the expression patterns of the neuronal cytoskeleton during neurogenesis. Biochem. Biophys. Res. Commun. 2013;439:154–159. doi: 10.1016/j.bbrc.2013.07.130. [DOI] [PubMed] [Google Scholar]
- 44.Hu Q., Tanasa B., Trabucchi M., Li W., Zhang J., Ohgi K.A., Rose D.W., Glass C.K., Rosenfeld M.G. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat. Struct. Mol. Biol. 2012;19:1168–1175. doi: 10.1038/nsmb.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan G.S., Chiu C.H., Garchow B.G., Metzler D., Diamond S.L., Kiriakidou M. Small molecule inhibition of RISC loading. ACS Chem. Biol. 2012;7:403–410. doi: 10.1021/cb200253h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borchert G.M., Holton N.W., Williams J.D., Hernan W.L., Bishop I.P., Dembosky J.A., Elste J.E., Gregoire N.S., Kim J.A., Koehler W.W., et al. Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins. Mob. Genet. Elements. 2011;1:8–17. doi: 10.4161/mge.1.1.15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bussing I., Slack F.J., Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Gu T.J., Yi X., Zhao X.W., Zhao Y., Yin J.Q. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics. 2009;10:563. doi: 10.1186/1471-2164-10-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smalheiser N.R., Torvik V.I. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Abad M., Mosteiro L., Pantoja C., Canamero M., Rayon T., Ors I., Grana O., Megias D., Dominguez O., Martinez D., Manzanares M., et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- 51.Radzisheuskaya A., Silva J.C. Do all roads lead to Oct4? the emerging concepts of induced pluripotency. Trends Cell Biol. 2014;24:275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodier J.L., Kazazian H.H. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 53.Baillie J.K., Barnett M.W., Upton K.R., Gerhardt D.J., Richmond T.A., De Sapio F., Brennan P.M., Rizzu P., Smith S., Fell M., et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills R.E., Bennett E.A., Iskow R.C., Devine S.E. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Barouki R., Coumoul X., Fernandez-Salguero P.M. The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 2007;581:3608–3615. doi: 10.1016/j.febslet.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 56.Chambers I., Tomlinson S.R. The transcriptional foundation of pluripotency. Development. 2009;136:2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Do D.V., Ueda J., Messerschmidt D.M., Lorthongpanich C., Zhou Y., Feng B., Guo G., Lin P.J., Hossain M.Z., Zhang W., et al. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013;27:1378–1390. doi: 10.1101/gad.221176.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawkins K., Joy S., McKay T. Cell signalling pathways underlying induced pluripotent stem cell reprogramming. World J. Stem Cells. 2014;6:620–628. doi: 10.4252/wjsc.v6.i5.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paule M.R., White R.J. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X., Sachs F., Ramsay L., Jacques P.E., Goke J., Bourque G., Ng H.H. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 61.Wang J., Xie G., Singh M., Ghanbarian A.T., Rasko T., Szvetnik A., Cai H., Besser D., Prigione A., Fuchs N.V., et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 62.Wei L., Gu L., Song X., Cui X., Lu Z., Zhou M., Wang L., Hu F., Zhai J., Meyers B.C., et al. Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3877–3882. doi: 10.1073/pnas.1318131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T., et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.