Abstract
The transcriptional shift from repression to activation of target genes is crucial for the fidelity of Notch responses through incompletely understood mechanisms that likely involve chromatin-based control. To activate silenced genes, repressive chromatin marks are removed and active marks must be acquired. Histone H3 lysine-4 (H3K4) demethylases are key chromatin modifiers that establish the repressive chromatin state at Notch target genes. However, the counteracting histone methyltransferase required for the active chromatin state remained elusive. Here, we show that the RBP-J interacting factor SHARP is not only able to interact with the NCoR corepressor complex, but also with the H3K4 methyltransferase KMT2D coactivator complex. KMT2D and NCoR compete for the C-terminal SPOC-domain of SHARP. We reveal that the SPOC-domain exclusively binds to phosphorylated NCoR. The balance between NCoR and KMT2D binding is shifted upon mutating the phosphorylation sites of NCoR or upon inhibition of the NCoR kinase CK2β. Furthermore, we show that the homologs of SHARP and KMT2D in Drosophila also physically interact and control Notch-mediated functions in vivo. Together, our findings reveal how signaling can fine-tune a committed chromatin state by phosphorylation of a pivotal chromatin-modifier.
INTRODUCTION
A common feature of major cell-cell signaling pathways, such as Wnt, Hedgehog and Notch, is a shift from repression to activation of target gene expression that is mediated by the same transcription factor. Gene repression and activation are normally accompanied by specific chromatin modifications that involve dynamic changes in histone tail modifications, including acetylation and methylation (1–3). However, the interplay between chromatin modifiers and signal-dependent transcription factors is poorly understood.
Notch signaling is one of a few signaling pathways that is used iteratively during development and adult tissue homeostasis (4–8). Notch-mediated transcriptional responses control key cellular decisions, such as proliferation, migration, survival and fate specification. Many progenitor cells go through several rounds of Notch activation, as exemplified during somitogenesis (9), and proper temporal and spatial control is crucial for the progressive cell fate specification and maturation of several tissues. Recently, in a Drosophila melanogaster model system, several chromatin modifiers have been shown to facilitate Notch-induced tumorigenesis (10). In humans, Notch gain-of-function mutations, e.g. in the case of acute lymphoblastic leukemia (11), chronic lymphocytic leukemia (12) or mantle cell lymphoma (13), prolong the Notch response and either contribute to or account for the pathogenesis of these leukemia/lymphomas. The involvement of Notch in these diseases emphasizes the clinical relevance of controlling Notch transcriptional responses for therapeutic purposes. However, the exact molecular mechanisms regulating the transcriptional switch from repression to activation in the Notch pathway remain far from understood.
The DNA binding transcription factor RBP-J (also known as CSL) actively represses Notch target genes in the absence of a Notch signal. The paradigmatic view of Notch signaling is that when the Notch receptor binds transmembrane ligands expressed on neighbouring cells, Notch undergoes several proteolytic cleavage events that liberate its intracellular domain (NICD) from the cell membrane. Subsequently, NICD enters the nucleus and associates with the transcription factor RBP-J and the coactivator Mastermind, which converts RBP-J from a repressor to an activator by displacing the interacting corepressors. Although static binding of RBP-J has been challenged in Drosophila (14) and recently in a genome-wide study using a murine myoblast cell line (15), there is ample genetic and functional evidence for RBP-J acting as a transcriptional repressor (16,17).
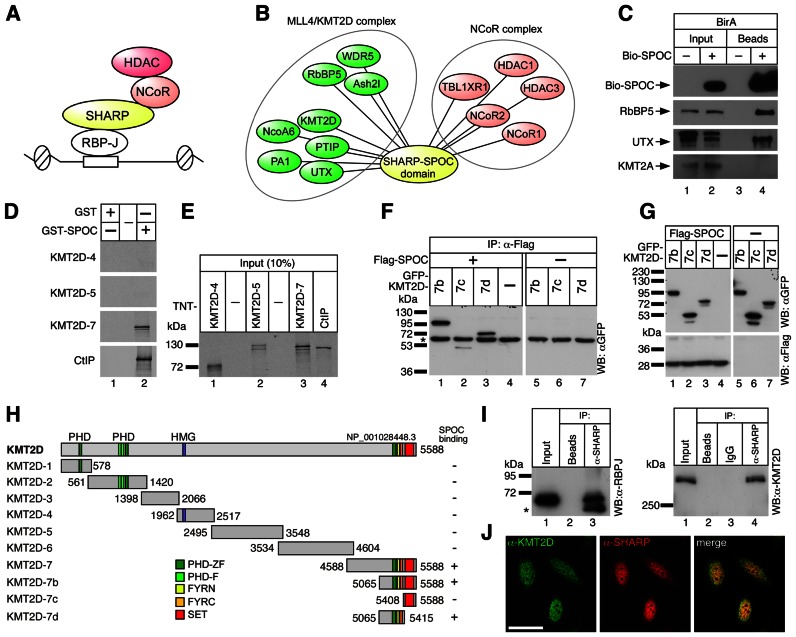
We have previously identified SHARP (SMRT/HDAC1 associated repressor protein) as a corepressor for RBP-J-mediated transcriptional regulation at Notch target genes [(17–20) and summarized in Figure 1A]. RBP-J interacts with SHARP or NICD in a mutually exclusive fashion (18,21). SHARP was originally identified in a yeast-2-hybrid screen for SMRT/NCoR binding partners (22). SMRT/NCoR recruits histone deacetylases like HDAC3 thereby mediating transcriptional repression. Interestingly, both HDAC3 and SMRT/NCoR have been implicated in Notch signaling (23,24). SHARP (in mouse also known as MINT, Msx2-interacting nuclear target protein) is a multidomain protein, containing N-terminal RNA recognition motifs, an RBP-J interaction motif and a conserved C-terminal SPOC domain. SHARP directly binds to RBP-J with high affinity (21). The SPOC domain is essential for SHARP to function as a transcriptional repressor interacting with corepressors such as SMRT/NCoR. Thus SHARP forms the bridge between the transcription factor RBP-J and the NCoR corepressor complex (Figure 1A).
Figure 1.
The KMT2D complex interacts with the SPOC-domain of SHARP. (A) Schematic representation of the RBP-J/SHARP repressor complex. SHARP interacts with RBP-J via its RBP interaction domain and with NCoR/HDAC complexes due to its C-terminal SPOC-domain. (B) SPOCome. A biotinylation-tagging approach followed by mass spectrometry was performed in pre-T cells and allowed to identify the KMT2D and NCoR complexes as interactors of the SPOC domain of SHARP. A complete list of interactors is provided in Supplementary Table S1. (C) Bio-SPOC interacts with KMT2D complex specific component UTX but not with KMT2A in pre-T cells. Streptavidin magnetic beads were used to pull-down the SPOC interactors. Precipitated proteins were analyzed by Western blot using streptavidin-HRP and specific antibodies directed against RbBP5, UTX and KMT2A. (D and E) The SPOC domain of SHARP interacts with the C-terminus of KMT2D in vitro. (D) Only the KMT2D-7 construct interacts with GST-SPOC (lane 2). CtIP, a known binding partner of SPOC, served as a positive control. (E) Input of the cell free synthesized 35S-labeled KMT2D fragments [(KMT2D-4, lane 1), (KMT2D-5, lane 2), (KMT2D-7, lane 3) and CtIP (lane 4, positive control) used in the GST pull-down experiments shown in Figure 1D. (F and G) Mapping of the SPOC-KMT2D interaction in cellular extracts. HEK293 cells were transfected with the indicated expression constructs for Flag-tagged SPOC domain of SHARP and GFP-tagged KMT2D fragments. (F) GFP-KMT2D fragments 7b (lane 1) and 7d (lane 3) coimmunoprecipitate with the SPOC domain of SHARP. A weak interaction was detected with the KMT2D-7c fragment (lane 2), which represents the SET domain of KMT2D (see also Figure 1H). HEK293 cells were transfected with the indicated expression constructs for Flag-tagged SPOC domain of SHARP and GFP-tagged KMT2D fragments. The asterisk denotes the heavy chain of the antibody used for immunoprecipitation. (G) Input control of the proteins used in (F). (H) Schematic representation of the KMT2D constructs used in (D)–(G) and their binding capacity to the SPOC-domain. Amino acid numbering is according to accession NP_001028448.3. KMT2D domains: PHD-zf, PHD-zinc finger (CCD: 197604); PHD-f, PHD-finger (CCD: 201356); HMG, HMG-box (CCD: 28965); FYRN, F/Y-rich N-terminus (CCD: 191411); FYRC, F/Y rich C-terminus (CCD: 197781); SET, SET domain (CCD: 197640). (I) Endogenous RBP-J (left panel, lane 3) and KMT2D (right panel, lane 4) interact with SHARP in HeLa cells. The asterisk denotes the heavy chain of the antibody used for immunoprecipitation. (J) KMT2D (left) and SHARP (middle) are located in the nuclei of HeLa cells and show regions of colocalization (right) as determined by confocal microscopy. Scale bar, 20 μm.
Dynamic changes in histone acetylation via histone acetyltransferases and histone deacetylases critically regulate the timing of Notch responses (17,24–26). Trimethylation of lysine 4 of histone H3 (H3K4me3) is another histone modification that has been intimately linked with transcriptional activation and that correlates significantly with histone acetylation sites (27). Notably, H3K4 trimethylation is dynamically controlled at Notch target genes by histone demethylases LSD1 (28) and KDM5A/RBP2/JARID1A (29) and a hitherto unknown methyltransferase. The H3K4 methylation mark is written by enzymes of the KMT2 (lysine methyltransferase 2) family, which are parts of different large multisubunit complexes (30). All of them share the core subunits WDR5, RbBP5 and Ash2l but whereas Menin1 is a specific subunit of the KMT2A/B complexes, UTX, PA1, PTIP and NcoA6 are specific subunits of the KMT2C/D complexes (31–33).
Here, we show that the RBP-J associated cofactor SHARP interacts with either KMT2D coactivator or NCoR corepressor complex regulating the chromatin environment at Notch target genes. KMT2D and NCoR competitively bind to the same region of the cofactor SHARP, its SPOC domain. NCoR/KMT2D balance depends on the phosphorylation status of two conserved serine residues at the C-terminus of NCoR. Our data suggest that the conversion of RBP-J from a repressor to an activator is not a simple single-step process, as previously thought, but involves a chromatin intermediate directed by cofactor SHARP. The novel dual functionality of the RBP-J/SHARP complex may facilitate a robust, yet flexible, Notch-dependent transcriptional response.
MATERIALS AND METHODS
Cell culture and preparation of cell extracts
Murine pre-T lymphoma cell line (Beko) and murine hybridoma mature T-cell line (MT) were grown in Iscove's Modified Dulbecco Medium (Gibco) supplemented with 2% FCS, 0.3 mg/l peptone, 5 mg/l insulin, nonessential aminoacids and penicillin/streptomycin. Cells were grown at 37°C with 5% CO2. pre-T cells were treated with 10 μg/ml DAPT/GSI (Alexis ALX-270–416-M025) or DMSO as control. Mature T-cells were treated with 25 μM TBB (4,5,6,7-tetrabromobenzotriazole, Sigma-Aldrich T0826), 25 μM TBCA [(E)-3-(2,3,4,5-tetrabromophenyl)acrylic acid, Millipore 218710] or DMSO as control. Cell lines HEK293 (ATCC CRL 1573), 293 T and HeLa (ATCC CCL 2) were cultivated in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal calf serum (FCS), penicillin and streptomycin.
For western blotting, EMSA and immunoprecipitation experiments whole-cell lysates were prepared as previously described (20). Protein concentrations were determined using the Bradford assay method (BioRad).
Streptavidin-immunoprecipitation of bio-SPOC domain of SHARP
The SPOC-domain of SHARP containing an N-terminal biotinylation tag (34) was introduced into pre-T cells (29) expressing already the biotin ligase BirA. 3 × 109 cells were centrifuged, washed in PBS and resuspended in 50 ml low salt buffer A (20 mM HEPES pH 7.9, 1 mM MgCl2, 10 mM KCl, 0.1% NP-40; add fresh 0.2 mM PMSF, 0.5 mM DTT, 1 × protease inhibitor). After 20 min incubation on ice, the samples were vortexed 10 s and centrifuged (20 000 rpm, 4°C, 30 min) to break the cell membrane. The pellet was then resuspended in 7 ml buffer C (0,2% NP-40, 20 mM HEPES, 100 mM KCl, 1 mM MgCl2 1 mM DTT, 1× Complete protease inhibitor and 0.2 mM PMSF). A 2.5 M KCl solution was dropwise added to reach a final concentration close to 400 mM KCl to allow an effective extraction of the nuclear proteins. After 20 min agitation at 4°C, the solution was transferred into ultracentrifuge tubes (Beckman) and ultracentrifuged for 60 min, 40 000 rpm at 4°C (Optima LE-80000, Beckman). 5 mg of nuclear extract were incubated with 150 μl streptavidin magnetic beads (Dynabeads M-280, Invitrogen 112.06) for 2 h at 4°C. The Dynabeads were pre-blocked at RT for 1 h with chicken egg albumin (200 μg for 150 μl beads mixed with 1 ml 1× TBS). During the blocking the NE was diluted to adjust it to optimal bead binding conditions (100 mM KCl and 10% glycerol). After binding, the beads were washed six times with streptavidin washing buffer (TBS 1×, 0.2% NP-40) using different ionic strengths (200, 250 or 300 mM NaCl) as indicated in Supplementary Table S1. Beads were resuspended in 2x SDS loading buffer, boiled for 5 min and loaded on a NuPAGE 4–12% Bis-Tris Gel (Invitrogen). After Coomassie staining (NOVEX colloidal blue staining kit, Invitrogen) individual bands were cut out and subjected to MS analysis (LC–MS).
For Western blot purposes 500 μg of nuclear extract were incubated with streptavidin magnetic beads (Dynabeads M-280, Invitrogen 112.06) in a binding buffer containing 120 mM NaCl and 0.1% NP-40. After binding, beads were washed five times with streptavidin washing buffer containing 300 mM NaCl and two times with PBS.
RbBP5 (Bethyl, A300–109A, lot. #A300-109A-2), KMT2A (Bethyl, A300-374A, lot. #A300-374A-2) and UTX (Bethyl,A302-374A, lot. #A302-374A-1) antibodies were used in western blot as follows. Briefly, membranes were blocked 1h at room temperature in 5% milk, 1× TBS, 0.1% Tween-20 and then incubated over night at 4°C with the proper antibody diluted (RbBP5 1:5000; KMT2A 1:5000 and UTX 1:2000) in 5% milk, 1× TBS, 0.1% Tween-20. After washing three times 10 min each in 1× TBS, 0.1% Tween-20, membranes were incubated with the secondary antibody (Cell Signaling, #7074S) diluted 1:5000 in 5% milk, 1× TBS, 0.1% Tween-20 for 1 h at room temperature. Finally, membranes were washed three times 10 min each in 1× TBS, 0.1% Tween-20.
Streptavidin blot was performed as follows. Briefly, membranes were blocked 1 h at room temperature in 5% BSA, 1× TBS. Membranes were incubated 1h at room temperature in streptavidin horseradish (PerkinElmer, NEL750001EA) diluted 1:20 000 in 5% BSA, 1× TBS and washed three times 15 min each in 1× TBS, 0.5 M NaCl, 0.5% Triton X-100.
On-bead digestion and liquid chromatography–mass spectrometric analysis
Direct endoproteinase digestion of SHARP protein complexes immobilized on magnetic bead supports was performed as follows: streptavidin magnetic beads from biotin–streptavidin purifications were finally washed with 1× TBS (lacking NP-40) and resuspended in 50 μl denaturation buffer (20 mM Tris–HCl, pH 8.0, 8 M urea). The urea concentration was lowered to 4 M by adding a 50 μl aliquot of 50 mM ammonium bicarbonate (ABC). The resulting bead suspension was adjusted to 1 mM DTT and protease digestion was started by the addition of 500 ng Lys-C (Wako). After one-hour incubation at 25°C beads were separated (magnetic bank, Invitrogen) and washed with 200 μl 50 mM ABC. The supernatant (containing the pre-digested peptides) and the wash were pooled (combined volume = 300 μl) and alkylated with iodoacetamide (1.8 mM final concentration) at 25°C for 2 h. Digestion was completed by addition of 500 ng trypsin (Promega) and over night incubation at 25°C. Proteolysis was stopped by adding TFA to a final concentration of 3%. The tubes were centrifuged at 20 000 g in order to pellet insoluble material. Finally, the digest was subjected to C18 ‘Stage Tip’ purification as described (35).
Desalted samples were subsequently analyzed using nanoflow (Agilent 1200 nanoLC, Germany) LC–MS/MS on a linear ion trap (LIT)-Orbitrap (LTQ-Orbitrap XL+ETD) mass spectrometer (Thermo Fisher, Germany). The MS system was calibrated with a caffeine, M-R-F-A-acetate peptide and ultramark 1621 polymer containing calibration mix (ProteoMass LTQ/FT-Hybrid ESI Pos. Mode Cal MIX, Supelco) according to the manufacturer's protocol. All steps were done in a dust-free environment, to prevent contamination with human keratin.
Peptides were eluted stepwise with a linear gradient of 10−60% MS sample buffer B at a flow rate of 250 nl/min over 120 or 140 min depending on the experiment. Data were acquired using a data-dependent ‘top 10’ method, dynamically choosing the ten most abundant precursor ions from the survey scan (mass range 350−1800 Th) in order to isolate and fragment them in the LTQ. Survey scans were acquired in the profile whereas MS/MS scans were acquired in the centroid mode and dynamic exclusion was defined by a list size of 500 features and exclusion duration of 90 s with a MMD of 10 ppm. The isolation window for the precursor ion selection was set to 2.0 Th. Precursor ion charge state screening was enabled, and all unassigned charge states as well as singly charged ions were rejected. For the survey scan a target value of 1 000 000 (1000 ms maximal injection time) and a resolution of 60 000 at m/z 400 were set (with lock mass option enabled for the 445.120024 ion), whereas the target value for the fragment ion spectra was limited to 5000 ions (150 ms maximal injection time). The general mass spectrometric conditions were: spray voltage, 2.3 kV; no sheath and auxiliary gas flow; ion transfer tube temperature, 160°C; collision gas pressure, 1.3 mTorr; normalized collision energy using wide-band activation mode; 35% for MS2. Ion selection thresholds were 500 or 1000 counts for MS2 depending on the experiment. An activation q = 0.25 and activation time of 30 ms was applied.
Raw files were processed into peak lists by DTASuperCharge 2.0b1 [part of the MSQuant 2.0b7 software environment (36)] and searched with Mascot 2.2 against the human International Protein Index protein database (IPI, version 3.65) combined with frequently observed contaminants and concatenated with the reversed versions of all sequences. Enzyme specificity was set to trypsin (with a maximum of two missed cleavages) allowing cleavage N-terminal to proline and C-terminal to aspartate. The MMD for monoisotopic precursor ions and MS/MS peaks were restricted to 5 ppm and 0.6 Da, respectively. For all searches carbamidomethylated cysteines were set as a fixed modification, whereas oxidation of methionine, deamidation of asparagine or glutamine and N-terminal protein acetylation were treated as variable modifications. Protein and peptide identifications were further analyzed and manually verified by inspection of chromatograms and spectra.
Oligonucleotides
All oligonucleotides used for plasmid construction, EMSA, qPCR and ChIP experiments are shown in Supplementary Table S4.
Constructs
The following vectors were commercially obtained: pcDNA3 (Invitrogen), pGEX6P1 (GE-Healthcare), pFN10A (Promega pOT2-TTR-AY113651 (Drosophila Genomics Resource Center SD13650; #13205), pBlue-TTR-AY069273 (Drosophila Genomics Resource Center GM10003; #4251), pCMV-sport6-hRbBP5 (Open Biosystems, #5576406), pCR4-TOPO-hKDM6A/UTX (Open Biosystems, #9051779), pOTB7-hWDR5 (Open Biosystems #3538255).
The following plasmids were described previously: pGEX6P1-mNotch-1-IC (25), pcDNA3-Flag-1, pcDNA3-mNotch-1-ΔE, pcDNA3-RBP2N, pGa981/6 and pCMV-mNotch-1-IC (18), pGex6P1-SHARP(3477–3664), pcDNA3-Flag-SHARP(3477–3664) and pSP6-CtIP (19), pcDNA3-Flag3-RBP2N-mRuby and pcDNA3-Flag3 (20), pcDNA3-GFPoStp, pGex6P1-hsRITA (37). Constructs that were newly generated by PCR-Amplification are listed in Supplementary Table S5. The PCR products were cut by restriction enzymes and ligated in the listed vectors. After annealing of the 5-prime phosphorylated oligonucleotides Bio_F and Bio R (see Supplementary Table S4) the Flag-Tag of pcDNA3-Flag-SHARP(3477–3664) was exchanged with the Bio-Tag by Acc65I and EcoRI digestion to generate pcDNA3-Bio-SPOC. The Bio-SPOC cDNA was amplified with the primers Bio-SPOC_F and BIO-SPOC_R and ligated into the retroviral vector pLXSP after BamHI/SalI digestion to generate pLXSP-Bio-SPOC. PCR was also used to generate the point mutations pcDNA3-Flag1-Sharp(3477–3664) K3516A, pcDNA3-Flag1-Sharp(3477–3664) R3552A/R3554A and pcDNA3-Flag1-Sharp(3477–3664) Y3602A via side-directed-mutagenesis. The upstream primer SPOC_F and the downstream primer SPOC_Xho_R were used for all three mutagenesis. Additionally, for inserting K3516A the primers K3516A_F and K3516A_R were designed. The double substitution of Arginine 3552 and 3554 to Alanine was performed with the primers R3552A/R3554A_F and R3552A/R3554A_R. To create the Y3602A mutation the primers Y3602A_F and Y3602A_R were used. All PCR-products were sequenced and cut EcoRI/XhoI to ligate into pcDNA3-Flag1. To obtain the three pcDNA3-Flag3-RBP-SPOC mutants the corresponding pcDNA3-Flag1-SPOC constructs were cut with PflMI and XbaI and ligated into pcDNA3-Flag3-RBP-SPOC resulting in pcDNA3-Flag3-RBP-SPOC (K3516A), pcDNA3-Flag3-RBP-SPOC (R3552A/R3554A) and pcDNA3-Flag3-RBP-SPOC (Y3602A). For SP6 driven in vitro transcription, the plasmids pCS2-Flag3-RBP-SPOC (wt) and pCS2-Flag3-RBP-SPOC (R3552A/R3554A) were made. The corresponding cDNA inserts from pcDNA3-Flag3-RBP-SPOC were subcloned into the pCS2 vector. Details about all other constructs used in this study are provided in Supplementary Tables S4 and S5.
Gene knockdown
In order to perform the UTX and NCoR1 knockdown in mature T-cells, the pLKO.1 TRC1 shRNA library (SIGMA-ALDRICH) was used. Briefly, 293 T cells were transfected with 3.33 μg of the desired shRNA construct and the packaging vectors psPAX2 (2.5 μg) and pMD26 (1 μg) using 14 μg of linear PEI (Polysciences # 23966). After 48 h of incubation, the supernatant from the transfected 293 T cells was filtered and used for infections of mature T-cells. In total, four spin infections were performed over 2 days. Approximately 36 h after the last infection, the selection of the positively infected cells was performed by adding 1 μg/ml of puromycine (Serva). The target sequences of the hairpins used in this study are listed in Supplementary Table S4.
RNA extraction, RT-PCR and qPCR from cell lines
Total RNA was purified using Trizol reagent (Ambion, 15596018) accordingly to manufacturer's instructions. 1 μg of RNA was retro-transcribed in cDNA using random hexamers and M-MuLV reverse transcriptase (NEB). qPCRs were assembled with Absolute QPCR ROX Mix (Thermo Scientific, AB-1139), gene-specific oligonucleotides and double-dye probes (see Supplementary Table S4) and analyzed using the 7300 ABI PRISM sequence detector system (Applied Biosystem). Data were normalized to the housekeeping gene glucuronidase β (GusB).
In order to measure the relative abundance of KMT2 family members′ transcripts, serial dilutions of the cDNA were used for generating a calibration curve for the housekeeping gene TBP. The calibration curve was used for calculating the expression of the members of the KMT2 family.
Quantitative reverse transcriptase PCR (qRT-PCR) of Drosophila and Xenopus genes
RNAi knock down efficiency for spen and trr, mRNA were assessed by qRT-PCR upon RNAi induction. RNAi expression was induced by hsp70-Gal4 (hsp-70>) when lethal by tub> (spen-IR, KK100153 and GD17128; trr-IR, GD4501). For heat shock induced RNAi expression, third instar larvae (L3) were shifted for 1 hour at 37°C (heat shock) followed by 1 hour at 25°C. This method understimate the depletion of the gene by RNAi but it provides a means to correlate the effectiveness of different RNAi against a given gene (as in spen-IR lines KK100153 and GD17128). For every genetic condition total RNA was isolated from 10 to 15 wandering third instar larvae (L3). All tissue samples from Drosophila melanogaster and Xenopus laevis embryos were stored in RNAlaterTissueProtect Tubes (Qiagen) until used and mRNA levels were assessed by qRT-PCR. To determine mRNA levels, was used SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen) and SYBR Green PCR Master kit (Applied Biosystems) were used according to the manufacturer's instructions. The cDNAs were amplified using specific primers, designed by using the ProbeFinder software by Roche Applied Science. Rp49 (Drosophila) and hist1h4a (Xenopus) were used as house-keeping genes for normalization. Primer sequences are shown in Supplementary Table S4. In all cases, samples were tested in triplicate and qPCR reactions were run on a 7500 Real-Time PCR System (Applied Biosystems) following the manufacture's protocol. The qPCR data were analyzed by a two-tailed unpaired t-test.
DNA transfection
HEK293 and HeLa cells were transfected using the Nanofectin transfection reagent (PAA) according to the manufacturer's instructions.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed following the Upstate Biotechnology protocol with few modifications. Briefly, cells were cross-linked in 1% formaldehyde for 10 min at room temperature and the reaction was blocked with 1/8 volume of 1 M glycine pH 8.0. Only in the case of the KMT2D, RbBP5 and UTX ChIP a pre-fixation with 10 mM dimethyladipimate (DMA) in PBS was performed 1 h at room temperature. Cells were washed twice with PBS and resuspended in SDS Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1). The cell suspension was sonicated using the Covaris System S2AFA and after dilution with ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.1, 167 mM NaCl), the chromatin was pre-cleared with 10 μl/ml pre-saturated protein-A-Sepharose beads for 30 minutes at 4°C. The chromatin was incubated over night with the proper amount of the desired antibody [anti-RbBP5 (Bethyl, A300–109A), anti-H3K4me3 (Diagenode, pAb-003–050), anti-H3K27ac (Diagenode, pAb-174-050), anti-H3 (Abcam, ab1791), anti-H3K4me1 (Abcam, ab8895), anti-(N-20) RNAPII (Santa Cruz, sc-899)] or with IgG (Santa Cruz, sc-2027) as control. Antibodies were immobilized with 40 μl pre-saturated protein-A-Sepharose beads 1 h at 4°C with shaking. Depending on the antibody, different washing conditions were used. Chromatin was eluted from beads with Elution Buffer (1% SDS, 0.1 M NaHCO3) and cross-links were reverted at 65°C over night in presence of 180 mM NaCl. After incubation with Proteinase K for 1 h at 45°C, the DNA was purified by phenol/chloroform extraction and precipitated over night at –20°C in presence of 10 μg of yeast tRNA, 10 μg glycogen and 500 μl 2-propanol. After washing with 70% EtOH and drying, the DNA was dissolved in TE pH 8.0 and analyzed by qPCR.
In the case of the KMT2D, UTX and HDAC1 ChIP, after sonication chromatin was diluted in ChIP Dilution Buffer and pre-cleared using 40 μl/ml pre-saturated protein-A-Sepharose beads (30 μl/ml in the case of the HDAC1 ChIP) for 30 min at 4°C. 50 μg of pre-cleared chromatin were incubated with 10 μg of KMT2D (Santa-Cruz, sc-292359 X, lot. #H0911), 5 μg of UTX (Bethyl, A302-374A, lot. #A302-374A-1), 2 μg of HDAC1 (Diagenode, pAb-053-050, lot. #A21–001P) antibody or rabbit IgG (Diagenode, C15410206, lot. #161013JJ or # D005) as control. Antibodies were immobilized with 40 μl pre-saturated protein-A-Sepharose beads and beads were washed once in Low Salt Washing Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 150 mM NaCl), twice in High Salt Washing Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 500 mM NaCl) and three times (twice in the case of KMT2D) in TE washing buffer (10 mM Tris–HCl, 1 mM EDTA pH 8.0). In the case of the HDAC1 ChIP, beads were washed once in Low Salt Washing Buffer, once in High Salt Washing Buffer, once in LiCl Washing Buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1 mM EDTA, 10 mM Tris–HCl pH 8.1) and three times in TE Washing Buffer. DNA was purified as previously described with the only exception that DNA was precipitated over night at −20°C in presence of 10 μg of yeast tRNA, 40 μg glycogen and 500 μl 2-propanol.
Data were normalized to positive controls (unless specified) and in the case of histone marks also to histone H3. Gene desert (chr14:97450841–97450907) was used as negative control.
Coimmunoprecipitation experiments
The coimmunoprecipitation experiments were carried out essentially as described previously (37). Briefly, 24 h after transfection cells were lysed with 600 μl CHAPS lysis buffer. Where indicated cells were incubated with 25 μM or 50 μM TBCA (CalBiochem, 50mM stock solution in DMSO) or DMSO as control for three hours prior to cell lysis. The extracts were incubated with 40 μl agarose-conjugated anti-Flag antibody (M2, Sigma) at 4°C overnight. For coimmunoprecipitation of endogenous proteins, HeLa cell extracts were incubated with 20 μl of anti-SHARP antibody [anti-SHARP.2 (18)] at 4°C overnight, followed by a 2 h incubation with 50 μl (50% slurry)sepharose conjugated protein G beads (GE healthcare). Precipitates were washed six to eight times with CHAPS lysis buffer and finally resuspended in SDS-polyacrylamide gel loading buffer. For Western blotting the proteins were resolved in SDS-polyacrylamide gels and transferred electrophoretically at room temperature to PVDF membranes (Millipore) for 1 h at 50 mA using a Tris-glycine buffer system. After blotting, the membranes were pre-blocked for 1 h in a solution of 3% milk powder in PBS-T (0.1% Tween 20 in PBS) before adding antibodies. The following antibodies were used: anti-GFP (7.1/13.1, mouse monoclonal IgG, secondary antibody peroxidase conjugated sheep anti-mouse IgG, NA931V, GE healthcare), anti-Flag (M5, Sigma; secondary antibody, NA931V, GE healthcare), anti-RBP-J (rat monoclonal IgG2a, T6709, Institute of Immunology Co., Ltd.; secondary antibody peroxidase-conjugated goat anti-rat IgG, Dianova), anti-NCoR (rabbit polyclonal IgG, A301-146A, Bethyl, secondary antibody peroxidase-conjugated donkey anti-rabbit IgG, GE healthcare), anti-AKT1 (pan) (C67E7, Cell Signaling), anti-AKT1 (p-S129) (WA-AP7141f, Biomol), anti-KMT2D (rabbit polyclonal IgG, H300, sc292359, Santa Cruz; secondary antibody peroxidase conjugated donkey anti-rabbit IgG, NA934V, GE healthcare)
ITC binding experiments of SPOC-NCoR complexes
A MicroCal VP-ITC calorimeter was used for all ITC binding studies. Experiments were carried out at 25°C in a buffer consisting of 20 mM HEPES pH 7.5 and 100 mM NaCl. The SPOC domain from murine SHARP, residues 3474–3643 (3495–3664 for human SPOC), was overexpressed as a glutathione S-transferase fusion protein in bacteria, cleaved and purified to homogeneity, using a combination of glutathione–sepharose affinity and size exclusion chromatography. The only difference in primary sequence between mouse and human SPOC resides at residue 3537, which is a serine in mouse and a threonine in human. Murine SPOC mutant R3531A/R3533A correspond to R3552A/R3554A in human SPOC. Peptides (phosphorylated and nonphosphorylated) that correspond to the conserved C-terminus of NCoR (-REPAPLLSAQYETLSDSDD) were chemically synthesized by a commercial vendor (Peptide 2.0) and purified to >95% by reverse phase HPLC. SPOC and NCoR proteins were degassed and buffer matched using size exclusion chromatography and/or dialysis. A typical experiment consisted of 15 μM of SPOC in the cell and 150 μM NCoR in the syringe. Protein concentrations were determined by both UV absorbance at 280 nm and BCA assay (Pierce). The binding data reported are the average of at least three independent experiments (n = 3). The value of c ( = Ka[M]N) for all experiments was between 1 and 245. The data were analyzed using the ORIGIN software and fit to a one-site binding model.
Xenopus laevis
Xenopus embryos were obtained by in vitro fertilization, cultured and staged according to (38). All experiments using Xenopus were performed in agreement with the German law.
RNA microinjections in Xenopus laevis
mRNA was obtained using the mMESSAGEmMACHINE SP6 Kit (Ambion) according to the manufacturers’ protocol after linearization of the vector. Embryos have been injected bilaterally at two-cell stage with 100 pg of each RNA (200–400 pg total amount).
Whole mount in situ hybridization (WMISH) in Xenopus leavis
Injected embryos were fixed at stage 16 with MEMFA (0.1 M MOPS (pH 7.4), 2 mM EGTA, 1 mM MgSO4, 4% fomaldehyde) over night at 4°C. WMISH was performed following standard protocols (39) with DIG-labeled antisense RNA probe of N-tubulin (tubb2b). The colour reaction was performed using BM purple (Roche Applied Science). After staining, the embryos were refixed in MEMFA and bleached in 30% H2O2.
Statistical analysis
If not separately specified, data are shown as mean ± SD of three independent experiments. Statistical analysis were performed using unpaired Student's t-test and P < 0.05 was considered to be statistically significant.
ChIP-Seq analysis
Notch-1 and RBP-Jκ ChIP-Seq data (# GSE29600) were analyzed using MACS14 via the Galaxy Platform. Reads were mapped to the mouse genome mm9 and default settings were used.
Luciferase assay
HeLa cells were seeded in 24-well plates at a density of 5 × 104 cells. Transfection was performed with Nanofectin reagent (see above) using 1 μg of reporter plasmid alone or together with various amounts of expression plasmid (given in the corresponding figure legends). Luciferase activity was determined from at least four independent experiments with 20 μl of cleared lysate in an LB 9501 luminometer (Berthold) by using the luciferase assay system from Promega.
In vitro protein translation and GST pull down
The in vitro protein translation and the GST pull down assays were performed exactly as previously described in (20).
Electro mobility shift assay (EMSA)
Whole cell extracts (see above) were used for electromobility gel shift assays in a binding buffer consisting of 10 mM Tris–HCl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 0.5 mM DTT, and 4% glycerol. For binding reaction, 2 μg poly(dI-dC) (Amersham) and approximately 0.5 ng of 32P-labeled oligonucleotides were added. The sequence of the doublestranded oligonucleotide FO-233 (Supplementary Table S4) corresponds to the two RBP-J-binding sites within the EBVTP-1 promoter. Super shifting of complexes was achieved by adding 1 μg of anti-RBP-J (rat monoclonal IgG2a, T6709, Institute of Immunology Co., Ltd.) antibody. The reaction products were separated using 5% polyacrylamide gels with 1× Tris–glycine–EDTA at room temperature. Gels were dried and exposed to X-ray films (Kodak).
Fluorescence microscopy
Cell imaging was performed by plating HeLa cells or HEK293 cells in a concentration of 105 cells cm−2 on chambered coverslips (Nunc). When required, cells were transfected with 400 ng of expression plasmids using the Nanofectin transfection reagent (see above). Cells were rinsed with PBS 24 h after transfection, fixed and permeabilized with 0.1% Triton X-100. Nonspecific immunostaining was blocked by incubating the cells in 3% BSA in PBS with 0.1% TWEEN-20. The following antibodies were used: anti-Flag, mouse monoclonal IgG (M5, F4042, Sigma), secondary antibody, Alexa-Flour-488 coupled goat anti-mouse IgG (A11011, Life Technologies), anti-SHARP.2 [rabbit polyclonal, (18)], secondary antibody, Alexa-Fluor-568 coupled goat anti-rabbit IgG, (A11011, Life Technologies), anti-MLL2/KMT2D, goat polyclonal (I18, sc68671, Santa Cruz), secondary antibody, Alexa-Fluor-488 coupled donkey anti-goat (A11055, Life Technologies). Pictures were taken using a fluorescence microscope (IX71, Olympus) equipped with a digital camera (C4742, Hamamatsu), and a 100-W mercury lamp (HBO 103W/2, Osram). The following filter set was used: Green, (Alexa-Flour-488) ex: HQ470/40, em: HQ525/50. For confocal microscopy a Leica TCS SP8-HCS microscope was used. Fluorophores were excited with the 488 nm and the 561 nm laser lines, respectively. Pictures were taken as ARY-sections in sequential scan mode.
Drosophila husbandry
The Drosophila RNAi transgenic lines used were from Vienna Drosophila RNAi Center (VDRC) and from Bloomington Stocks. Phenotype of this RNAi lines in the developing eye in the context of activated NOTCH was assessed by crossing with w1118; ey-Gal4, UAS-Dl flies as previously described (10) and the RNAi transgenic flies against Spen/SHARP and Trr/KMT2D are from NIG FLY, Fly stocks of National Institute of Genetics, (Kyoto).
Quantitative reverse transcriptase PCR (qRT-PCR) of Drosophila and Xenopus genes
RNAi knock down efficiency for spen and trr, mRNA were assessed by qRT-PCR upon RNAi induction. RNAi expression was induced by heat shock in hsp70-Gal4 (hsp-70>) transgenic larvae crossed with the RNAi of interest. In detail, third instar larvae (L3) were shifted for 1 h at 37°C (heat shock) followed by 1 h at 25°C and preserved immediately after in RNAlater (Qiagen). For every genetic condition total RNA was isolated from 10 to 15 wandering third instar larvae (L3). All tissue samples from Drosophila melanogaster and Xenopus laevis embryos were stored in RNAlaterTissueProtect Tubes (Qiagen) at −80°C until used. cDNA was prepared with SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) and qRT-PCR was performed with SYBR Green PCR Master kit (Applied Biosystems) accordingly to the manufacturer's instructions. Specific primers were designed by using the ProbeFinder software by Roche Applied Science. Rp49 (Drosophila) and hist1h4a (Xenopus) were used as house-keeping genes for normalization. Primer sequences are listed in Supplementary Table S4. Samples were analyzed in triplicate and qPCR reactions were run on a 7500 Real-Time PCR System (Applied Biosystems) following the manufacture's protocol. The qPCR data were analyzed by a two-tailed unpaired t-test.
RESULTS
SHARP directly interacts with both the NCoR corepressor and the KMT2D coactivator complexes
To dissect the molecular mechanism of chromatin-based gene regulation mediated by RBP-J/SHARP we focused our attention on the biochemical characterization of the SHARP SPOC-domain interactome (hereafter referred to as SPOCome). The highly conserved SPOC-domain of SHARP with a biotinylation-tag (bio-SPOC) was coexpressed with the biotin ligase BirA in pre-T cells. We used a streptavidin-affinity capture approach followed by mass-spectrometry (34) to characterize the SPOCome. In addition to NCoR-complex components [NCoR1 (henceforth designated as NCoR), NCoR2, TBL1XR1 and HDAC1/3], we also purified components of the KMT2D-complex (Figure 1B and Supplementary Table S1). The KMT2D-complex is known to play a role in gene activation (40). In addition to the central H3K4 methyltransferase KMT2D and the KMT2 core components Ash2L, RbBP5 and WDR5, we found specific KMT2C/D-complex components PTIP, NCoA6, histone demethylase UTX/KDM6A and PA1 (Figure 1B).
All five KMT2s (KMT2A- E) are expressed in our pre-T cell line (Supplementary Figure S1A), but only KMT2D was identified in the SPOCome. In order to validate the interaction of SHARP with the KMT2D complex, we performed coimmunoprecipitation experiments. As shown in Figure 1C, endogenous UTX, a specific component of the KMT2D complex, as well as RbBP5, copurified with the Bio-SPOC domain of SHARP. However, no copurification of KMT2A was observed reconfirming our mass spectrometric results. Since we obtained a remarkable large number of KMT2D peptides in our SPOCome, we investigated the possibility of a direct interaction between the SPOC-domain of SHARP and KMT2D (Figure 1D–H). The 530 kDa protein KMT2D was dissected into different polypeptides that were in vitro transcribed and translated (Figure 1D and E and schematic overview in Figure 1H), or transfected together with Flag-tagged SPOC domain of SHARP into HEK293 cells (Figure 1F and G and schematic overview in Figure 1H). Using purified GST-SPOC as bait, the SPOC interaction region of KMT2D was mapped to fragment KMT2D-7, which includes the C-terminal SET domain (Figure 1D and H). Of note, this C-terminal KMT2D fragment (4588–5588) interacts as strongly as the previously described SHARP-interactor CtIP (19) (Figure 1D). Furthermore, whereas GST-SPOC interacts with KMT2D-7 and 7D (Figure 1D and Supplementary Figure S1B, respectively) and with the known SPOC interactor CtIP [(19) and Figure 1D], it does not interact with the KMT2D-complex subunits RbBP5, UTX and WDR5 (Supplementary Figure S1C-E). The SHARP/KMT2D interaction was further mapped using coimmunoprecipitation experiments with constructs of Flag-SPOC and GFP-KMT2D (7b, 7c and 7d) (Figure 1F and G). GFP-KMT2D constructs 7b (Figure 1F, lane 1) and 7d (lane 3), but not 7c (lane 2), strongly interact with FLAG-SPOC, whereas no interaction is observed between Flag-SPOC and GFP-only (lane 4). In addition, when we immunoprecipitate endogenous SHARP, not only the endogenous RBP-J (Figure 1I, left panel) but also the endogenous KMT2D (Figure 1I, right panel) is copurified. Finally, by confocal microscopy we could observe colocalization between endogenous SHARP and KMT2D proteins within the cell nucleus (Figure 1J).
Taken together, these data provide evidence that the SPOC domain of the coregulator SHARP not only interacts with the NCoR corepressor complex, but also with the KMT2D coactivator complex.
Because KMT2D complex directly interacts with the SPOC domain of SHARP we investigated the occupancy of the KMT2D complex at the RBP-J-bound enhancers of Notch target genes. We first identified the enhancers of Gm266, Hes1 and Dtx1 Notch target genes by analyzing publicly available RBP-J and Notch-1 ChIP-Seq data (41) (represented in Supplementary Figure S1F). As next step we investigated by ChIP the occupancy of KMT2D complex components at the underlined enhancers in our pre-T cells. We found that KMT2D (Supplementary Figure S1G), UTX (Supplementary Figure S1H) and RbBP5 (Supplementary Figure S1I) colocalize at the enhancers of Gm266, Hes1 and Dtx1 Notch target genes.
Dynamic occupancy of the KMT2D complex and active histone marks at Notch target genes
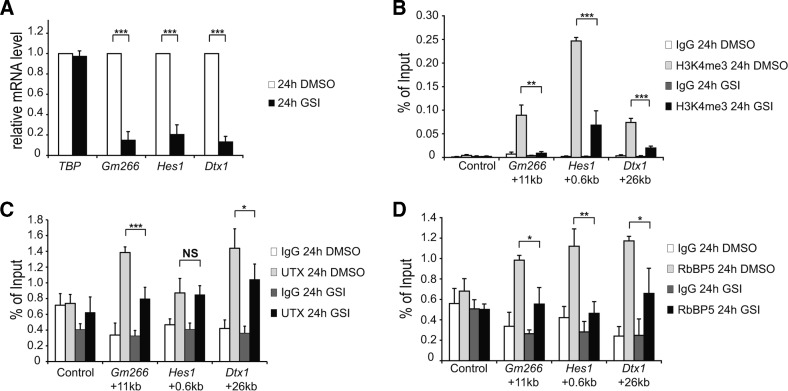
Next, we investigated the dynamics of histone marks and the KMT2D-complex components when we inhibit Notch signaling in our pre-T cells (Figure 2 and Supplementary Figure S2), which are sensitive to GSI (γ-secretase inhibition) as previously described (29). Upon GSI-treatment, transcription of the Notch target genes Gm266, Hes1 and Dtx1 was effectively inhibited (Figure 2A), which is also reflected by Notch-dependent reduction in the occupancy of RNA polymerase II (RNAPII, Supplementary Figure S2A) and H3K27 acetylation (Supplementary Figure S2B). The active mark H3K4me3 was reduced upon inhibition of Notch signaling (Figure 2B) and importantly this is not due to nucleosome depletion as measured with a pan-H3 antibody (Supplementary Figure S2D). Interestingly, the H3K4 monomethylation (H3K4me1) mark remains constant upon GSI treatment (Supplementary Figure S2C). Finally, the KMT2D-components UTX (Figure 2C) and RbBP5 (Figure 2D) are also dynamically regulated at the enhancers of Gm266, Hes1 and Dtx1 with the only exception of UTX at the enhancer of Hes1.
Figure 2.
Dynamic occupancy of KMT2D complex and H3K4me3 at the enhancer regions of Notch target genes. pre-T cells were treated for 24 h with GSI or with DMSO (control). (A) qRT-PCR showing the downregulation of the Notch target genes Gm266, Hes1 and Dtx1 in presence of GSI (black bars). Data were normalized to the housekeeping gene GusB (glucuronidase β). Shown is the mean ± SD of triplicate experiments ([***] P < 0.001, unpaired Student's t-test). (B) ChIP experiments reveal Notch-dependent changing levels of the positive histone mark H3K4me3. (C and D) Recruitment of the KMT2D complex components UTX (C) and RbBP5 (D) decline after GSI treatment. Data were normalized to the positive control (GAPDH TSS) and in the case of H3K4me3 (B), data were further normalized to histone H3 (see Supplementary Figure S2D). Shown is the mean ± SD of triplicate experiments ([NS] not significant, *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student's t-test).
RBP-SPOC, but not the NCoR-binding deficient RBP-SPOC, represses Notch-dependent transcription
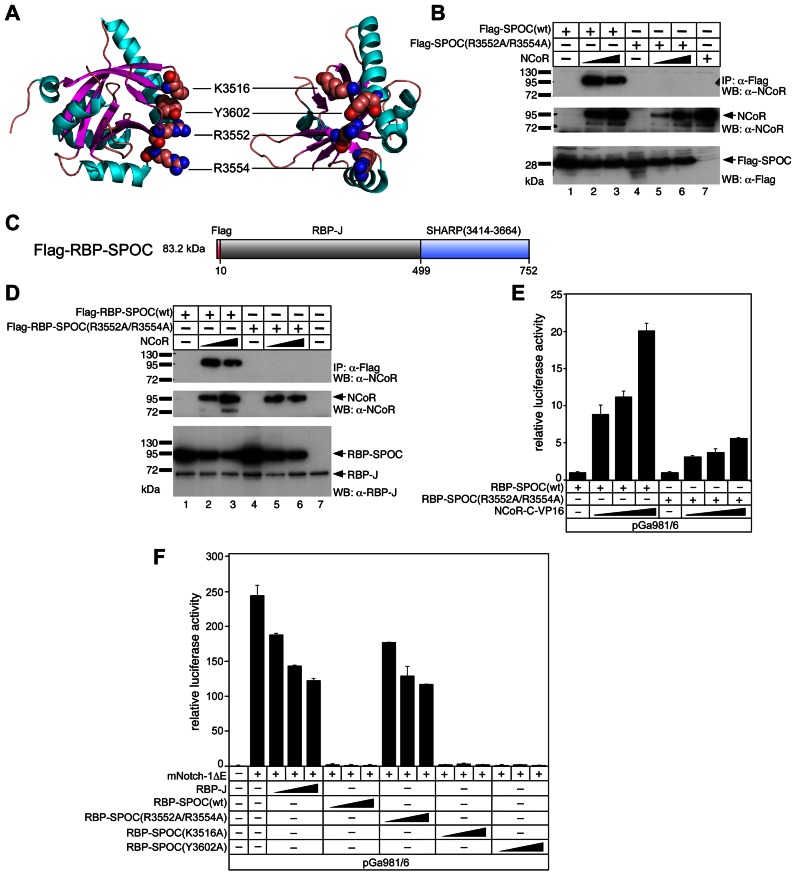
Since the C-terminal SPOC-domain of SHARP interacts with both NCoR and KMT2D we next wanted to functionally characterize SPOC. Based on the previous crystal structure of the SPOC domain (42) (Figure 3A), we generated a double mutant [SPOC(R3552A/R3554A)] that no longer interacts with NCoR (Figure 3B).
Figure 3.
Functional and biochemical characterization of RBP-SPOC proteins. (A) Ribbon diagram of the human SPOC crystal structure [PDB1OW1] (42) with α-helices and β-strands colored cyan and magenta, respectively. Location of the four amino acids K3516, Y3602, R3552 and R3554, which were mutated to alanine in our studies, is shown. Approximately 90° views of the structure are shown. (B) Mutations in the SPOC domain disrupt the SPOC/NCoR interaction. HEK293 cells were transfected with the indicated expression constructs for Flag-tagged wildtype SPOC domain [SPOC(wt)] or mutant SPOC domain [SPOC(R3552A/R3554A)] alone or together with an NCoR expression construct. Expression was verified by Western blotting for NCoR (middle panel, lanes 2, 3, 5, 6 and 7), SPOC(wt) (lower panel, lanes 1, 2 and 3) and SPOC(R3552A/R3554A) (lower panel, lanes 4, 5 and 6). NCoR was co-immunoprecipitated with SPOC(wt) (upper panel, lanes 2 and 3) but not with SPOC(R3552A/R3554A) (upper panel, lanes 5 and 6). (C) Schematic representation of the RBP-SPOC fusion protein. The SPOC domain of SHARP (3414–3664) was fused to the C-terminus of RBP-J. (D and E) Wildtype RBP-SPOC [RBP-SPOC(wt)], but not the RBP-SPOC mutant [RBP-SPOC(R3552A/R3554A)] interacts with NCoR. (D) HEK293 cells were transfected with the indicated expression constructs for Flag-tagged RBP-SPOC(wt) or RBP-SPOC(R3552A/R3554A) mutant alone or together with an NCoR expression construct. Expression was verified by Western blotting, NCoR (middle panel, lanes 2, 3, 5 and 6), RBP-SPOC(wt) (lower panel, lanes 1, 2 and 3), and RBP-SPOC(R3552A/R3554A) (lower panel, lanes 4, 5 and 6). NCoR was coimmunoprecipitated with RBP-SPOC(wt) (upper panel, lanes 2 and 3), but not with the RBP-SPOC(R3552A/R3554A) (upper panel, lanes 5 and 6). (E) The reporter construct pGa981/6 was transfected into HeLa cells together with 100 ng of RBP-SPOC(wt) or RBP-SPOC(R3552A/R3554A) expression plasmids and increasing amounts of NCoR-VP16 (50 ng, 100 ng and 250 ng). Mean values and standard deviation (error bars) based on at least four independent experiments are shown. (−) absence or (+) presence of denoted construct. (F) RBP-SPOC(wt) but not RBP-SPOC(R3552A/R3554A), acts as a repressor in Notch mediated transcription. The reporter construct pGa981/6 (1 μg) was transfected into HeLa cells alone or together with the mNotch-1ΔE (50 ng) expression plasmid and increasing amounts (50 ng, 100 ng and 250 ng) of RBP-J, RBP-SPOC(wt) and RBP-SPOC mutants (R3552A/R3554A), (K3516A) or (Y3602A). Mean values and standard deviation (error bars) based on at least four independent experiments are shown. (−) absence or (+) presence of denoted construct.
For functional characterization at Notch target genes we took a synthetic approach and fused the SPOC-domain of SHARP directly to the transcription factor RBP-J (RBP-SPOC) (Figure 3C). RBP-J and the wild type RBP-SPOC [RBP-SPOC(wt)] fusion are expressed at similar levels (Supplementary Figure S3A), bind to the RBP-J interactors NICD and RITA (37) similarly (Supplementary Figure S3B) and are both located in the nucleus (Supplementary Figure S3C). Moreover, DNA binding of RBP-SPOC(wt) is unaffected as compared to wild type RBP-J (Supplementary Figure S3D, compare lanes 4 and 5 to lanes 2 and 3). Specific DNA binding was validated by supershifting RBP-J and RBP-SPOC(wt) complexes with anti-RBP-J antibodies (Supplementary Figure S3D lanes 3 and 6). Again, the biochemical interaction between SPOC and NCoR is lost in the RBP-SPOC double point mutant [RBP-SPOC(R3552A/R3554A)] (Figure 3D) and in line with that, only RBP-SPOC(wt), but not RBP-SPOC(R3552A/R3554A), interacts with NCoR-VP16 (Figure 3E). Consistent with the SHARP/NCoR results, the repressor function of RBP-SPOC(wt) is completely abrogated in RBP-SPOC(R3552A/R3554A), whereas two independent point mutations in the SPOC domain (K3516A or Y3602A) have no effect on RBP-SPOC repressor function (Figure 3F). At the same time, the SPOC (R35352A/R3554) mutant looses its ability to bind KMT2D (Supplementary Figure S3E), indicating that KMT2D and NCoR binds to the same residues within the SPOC domain of SHARP.
Next, we tested the SPOC double mutant in a Xenopus leavis neurogenesis assay (Supplementary Figure S4A, number of injections in Supplementary Table S2 and statistics in Supplementary Figure S4B). Control injection of Notch1-ΔE (N1ΔE), a constitutively active form of Notch, causes almost complete abrogation of tubb2b expression, a neurogenic marker (Supplementary Figure S4A, compare upper and lower left panels). RBP-SPOC(wt) enhances tubb2b expression, which suggests RBP-SPOC(wt) is antagonizing Notch signaling, and this effect can be partially reverted by coinjection of N1ΔE (Supplementary Figure S4A, compare upper and lower middle panels). Injection of the RBP-SPOC(R3552A/R3554A) does not result in the enhanced expression of tubb2b and coinjection with N1ΔE results in a Notch-like phenotype (Supplementary Figure S4A, compare upper and lower right panels). These neurogenesis phenotypes are reflected in the expression of endogenous Notch target genes, as shown by qRT-PCR experiments (Supplementary Figure S4C). Notch target genes hes5.1, hes5.2, hey1 and hes4 are upregulated by N1ΔE and repressed by RBP-SPOC(wt). Importantly, repression mediated by RBP-SPOC(wt) is lost in RBP-SPOC(R3552A/R3554A). Moreover, the RBP-SPOC(wt) repressive effect is not simply due to steric hindrance, since Notch1-ΔE can still revert the phenotype.
Together, our data reveal that the SPOC domain of SHARP supports gene repression by interacting with NCoR.
Phosphorylation-dependent interaction of the SPOC domain of SHARP with corepressor NCoR
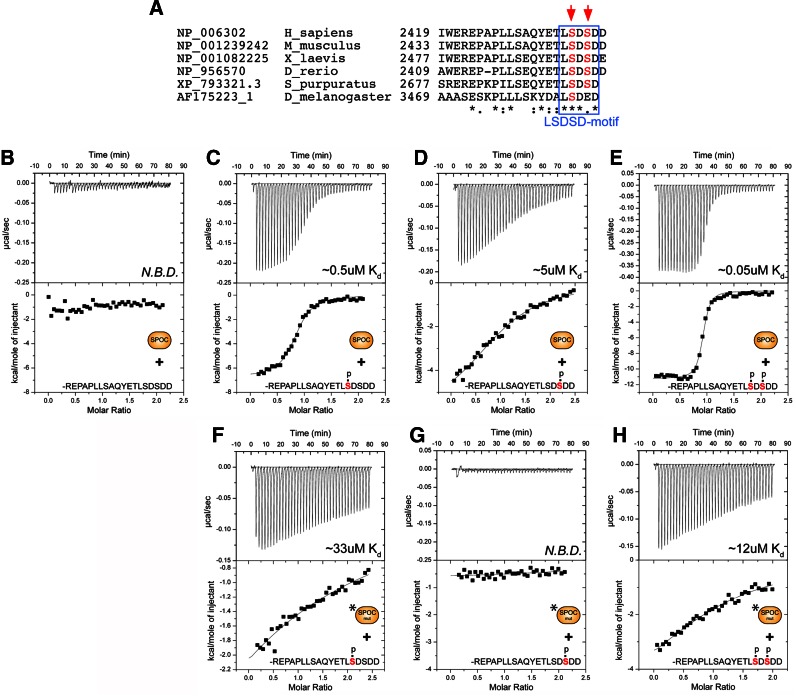
Given that both NCoR and KMT2D can interact with the SPOC domain of SHARP, we characterized the SHARP/NCoR interaction from the NCoR angle. Interestingly, the known SHARP-interacting region (42) contains a highly conserved LSDSD motif at the very C-terminus of NCoR (Figure 4A).
Figure 4.
The SPOC domain of SHARP interacts with the corepressor NCoR in a phospho-dependent manner. (A) Sequence alignment of the NCoR C-termini from Homo sapiens, Mus musculus, Xenopus laevis, Danio rerio, and Strongylocentrotus purpuratus. The conserved LSD-motif is highlighted by a blue box, phospho-serines are highlighted with red arrows. (B–H) Shown are representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for wildtype and mutant (R3552A/R3554A) SPOC proteins (represented in the figure as SPOC and SPOC mut, respectively) binding to phosphorylated and unphosphorylated NCoR peptides. The NCoR peptides used in each experiment and their affinity (Kd) for SPOC are shown. N.B.D. = no binding detected. (B) SPOC does not interact with the unphosphorylated NCoR peptide. (C and D) SPOC displays modest affinity for the two singly phosphorylated NCoR peptides. (E) SPOC binds the doubly phosphorylated NCoR peptide with high affinity. (F–H) The SPOC mutant (R3552A/R3554A) has significantly reduced affinity for the NCoR phosphopeptides. Forty titrations were performed per experiment, consisting of 7μl injections that were spaced 120 seconds apart. Complete thermodynamic binding parameters for each experiment are shown in Supplementary Table S3.
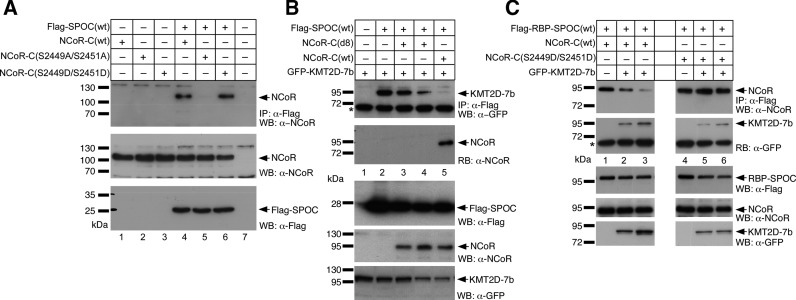
We used isothermal titration calorimetry (ITC) to systematically characterize the molecular details of SPOC–NCoR complex formation (Figure 4 and Supplementary Table S3). Previously, SPOC/NCoR binding studies have been performed in cells or cellular extracts (22,42). Surprisingly, when we performed ITC binding studies of purified recombinant SPOC with chemically synthesized peptides that correspond to the conserved C-terminus of NCoR (Figure 4A), we did not observe any binding (Figure 4B). However, using NCoR peptides that are phosphorylated on one of two conserved serine residues (…ETLSDSDDD or …ETLSDSDDD) resulted in modest affinity (Figure 4C and D); whereas, the double-phosphorylated NCoR peptide (…ETLSDSDDD) bound strongly to SPOC with a Kd of approximately 0.05 μM (Figure 4E). Importantly, two independent global phosphorylation studies demonstrated that these two serine residues in NCoR are phosphorylated in cells (43,44). Consistent with our RBP-SPOC studies, the SPOC double mutant (R3552A/R3554A) showed significantly reduced binding towards the phosphorylated NCoR peptides (Figure 4F–H). The ITC binding studies were first validated in cells by coimmunoprecipitation experiments using the wildtype SPOC-domain and the NCoR C-terminal domain (Figure 5A). When mutating the pivotal serine residues in NCoR [NCoR-C(S2449A/S2451A)] the SPOC/NCoR interaction is abrogated (Figure 5A, lane 5). Of note, the interaction strength of the phospho-mimetic mutation of NCoR [NCoR-C(S2449D/S2451D)] remains at wildtype levels [NCoR-C(wt)] (compare Figure 5A, lanes 4 and 6).
Figure 5.
NCoR and KMT2D compete for binding to the SPOC domain of SHARP. (A) NCoR(wt) and NCoR-C(S2449D/S2451D) (upper panel, lane 4 and 6, respectively) but not NCoR-C(S2449A/S2451A) (upper panel lanes 5) were co-immunoprecipitated with SPOC(wt). HEK293 cells were transfected with the indicated expression constructs for Flag-tagged SPOC(wt) with NCoR-C(wt), NCoR-C(S2449A/S2451A) or NCoR-C(S2449D/S2451D) expression constructs. Expression was verified by Western blotting for NCoR proteins (middle panel, lanes 1–6) and the Flag-SPOC-domain (lower panel, lanes 4–6). (B) The C-terminal GFP- KMT2D-7b fragment coimmunoprecipitates with Flag-SPOC(wt) (upper panel, lane 2, see also Figure 1F, lane 1). The KMT2D-7b/ SPOC(wt) interaction is lost after coexpression of NCoR-C(wt) (1942–2453, upper panel, lanes 5). The SPOC(wt)/NCoR-C(wt) interaction can be verified by reblotting with an anti-NCoR antibody (RB: α-NCoR, lane 5). NCoR-C(d8) does not coimmunoprecipitate with SPOC(wt) (RB: α-NCoR, lanes 3 and 4) and does not displace KMT2D-7b from SPOC(wt) (upper panel, lanes 3 and 4). (C) NCoR(wt)/RBP-SPOC(wt) interaction is decreased after coexpression of KMT2D-7b (upper panel, lanes 2 and 3). The SPOC(wt)/KMT2D interaction can be verified by reblotting with an anti-GFP antibody (RB: α-GFP). NCoR-C(S2449D/S2451D) is not displaced from SPOC after coexpression of KMT2D-7b (upper panel, lanes 5 and 6). HEK293 cells were transfected with the indicated expression constructs for Flag- RBP-SPOC(wt), GFP-tagged KMT2D-7b, NCoR-C(wt) and NCoR-C(S2449D/S2451D). Expression was verified by Western blotting (lower panels WB: α-Flag, α-NCoR and α-GFP). The asterisk denotes the heavy chain of the antibody used for immunoprecipitation.
Together, these data identify the SPOC domain of SHARP as a novel phospho-serine interaction domain.
NCoR and KMT2D compete for the binding to the SPOC domain of SHARP
To biochemically characterize the interplay between the corepressor NCoR and the coactivator KMT2D binding to the SPOC domain of SHARP we performed coimmunoprecipitation experiments (Figure 5B and C). We compared NCoR-C(wt) to an NCoR mutant lacking the LSDSD motif [NCoR-C(d8)] for binding to SPOC and displacement of KMT2D from SPOC. As expected, NCoR-C(wt), but not the NCoR-C(d8), binds to Flag-tagged SPOC(wt) (Figure 5B, compare lanes 3 and 4 to lane 5). By co-expressing GFP-tagged KMT2D-7b (lanes 1–5), we tested whether or not NCoR and KMT2D can compete for binding to SPOC. We observed that only NCoR-C(wt) (lanes 5), but not NCoR-C(d8) (lanes 3 and 4), can displace KMT2D from SPOC. Furthermore, when overexpressing KMT2D-7b NCoR-C(wt) can be displaced from SPOC(wt) (Figure 5C, compare lane 1 with lanes 2 and 3), but not NCoR-C(S2449D/S2451D) (Figure 5C, compare lane 4 with lanes 5 and 6). This suggests a possible mechanism whereby the two opposing chromatin modifiers, KMT2D and NCoR, are competing for binding to SPOC and this competition might be regulated by phosphorylation of NCoR.
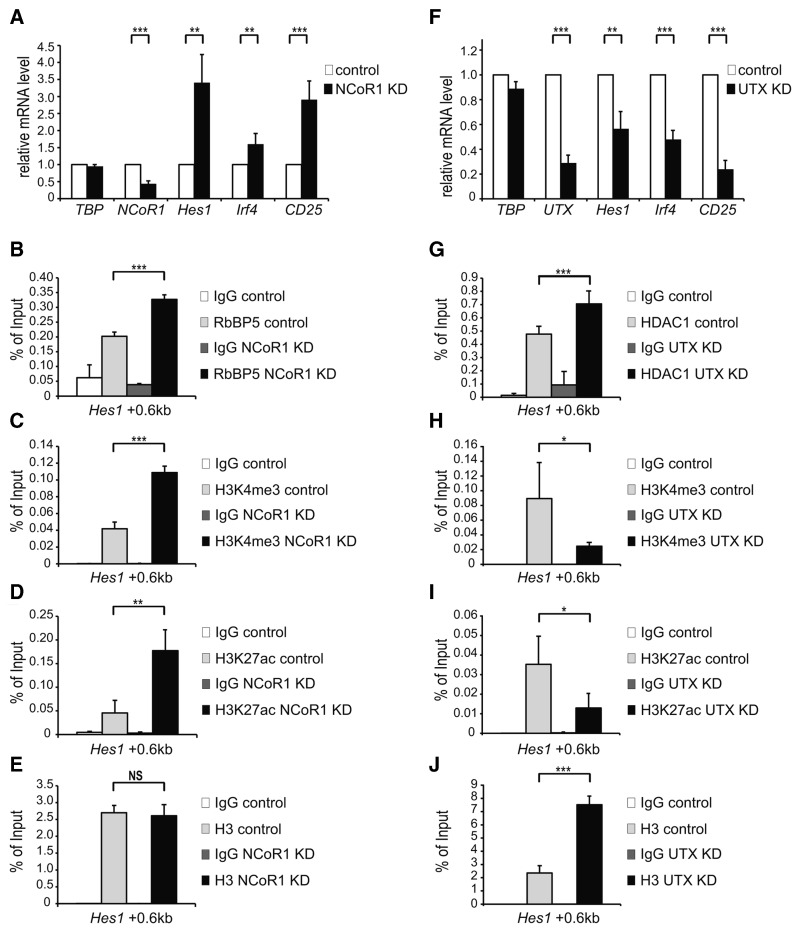
In order to analyze the balance between KMT2D and NCoR complexes at Notch target genes, we performed loss-of-function experiments of NCoR and KMT2D complexes, by knocking-down either NCoR (Figure 6A–E) or UTX (Figure 6F–J), a specific component of the KMT2D complex in a mature T-cell line which displays a low level of Notch activity. Upon knock-down of NCoR, Hes1, Irf4 and CD25 expression is increased (Figure 6A). To further characterize the NCoR knockdown, we focused on the enhancer of the well known Notch target gene Hes1 and by ChIP we observed that the occupancy of the KMT2D-component RbBP5 (Figure 6B) and corresponding H3K4me3 (Figure 6C) active mark are increased. Knocking down the HDAC-containing NCoR complex, also leads to increase in the active histone acetylation mark H3K27ac (Figure 6D), whereas overall histone H3 occupancy is unaffected (Figure 6E).
Figure 6.
Competing occupancy of the KMT2D and NCoR complexes at Notch target genes. (A) NCoR1 knockdown was performed in mature T-cells by infecting them with pLKO.1 Scramble (Control) or pLKO.1 NCoR1 Sh1. Total RNA was purified and analyzed by qRT-PCR with primers specific for Hes1, Irf4 and CD25 Notch target genes, NCoR1 or TBP (TATA box binding protein) as control. Data were normalized to the housekeeping gene GusB (glucuronidase β). Shown is the mean ± SD of duplicate experiments measured twice each (**P < 0.01, ***P <0.001, unpaired Student's t-test). (B, C and D) As measured by ChIP, knockdown of NCoR1 in mature T-cells results in increased occupancy of (B) RbBP5 and increased levels of (C) H3K4me3 and (D) H3K27ac at the Notch-dependent enhancer of Hes1 Notch target gene. (E) As revealed by ChIP using a pan H3 antibody, nucleosome occupancy is not affected at the enhancer of Hes1 Notch target gene upon knockdown of NCoR1 in mature T-cells. Data were normalized to GAPDH 0kb and further normalized to histone H3 in the case of H3K4me3 (C) and H3K27ac (D). Shown is the mean ± SD of duplicate experiments measured twice each ([NS] not significant, **P < 0.01, ***P < 0.001, unpaired Student's t-test). (F) UTX knockdown was performed in mature T-cells by infecting them with pLKO.1 Scramble (Control) or pLKO.1 UTX Sh1. Total RNA was purified and analyzed by qRT-PCR with primers specific for Hes1, Irf4 and CD25 Notch target gene, UTX or TBP (TATA box binding protein) as control. Data were normalized to the housekeeping gene GusB (glucuronidase β). Shown is the mean ± SD of three independent experiments (**P < 0.01, ***P <0.001, unpaired Student's t-test). (G, H and I) As measured by ChIP, knockdown of UTX in mature T-cells results in increased occupancy of (G) HDAC1 and decreased levels of (H) H3K4me3 and (I) H3K27ac at the Notch-dependent enhancer of Hes1 Notch target gene. (J) As revealed by ChIP using a pan H3 antibody, nucleosome occupancy is increased at the enhancer of Hes1 Notch target gene upon knockdown of UTX in mature T-cells. Data were normalized to GAPDH 0kb and further normalized to histone H3 in the case of H3K4me3 (H) and H3K27ac (I). Shown is the mean ± SD of duplicate experiments measured twice each (*P < 0.05, ***P < 0.001, unpaired Student's t-test).
Since we were unable to efficiently knockdown KMT2D, we performed knockdown of the KMT2D-specific component UTX. UTX knockdon leads to a robust downregulation of Hes1, Irf4 and CD25 (Figure 6F). Correspondingly, at the enhancer of Hes1, the NCoR-complex component HDAC1 was increased upon UTX-knockdown (Figure 6G); the active chromatin marks H3K4me3 and H3K27ac were decreased (Figure 6H and I, respectively) and the overall histone H3 levels were also increased (Figure 6J).
Taken together, these data support a model, in which the competition of NCoR-corepressor and KMT2D-coactivator complexes and the resulting NCoR/KMT2D balance affects the outcome of transcriptional responses at Notch target genes.
Functionally relevant phosphorylation of NCoR by casein kinase 2β (CK2β)
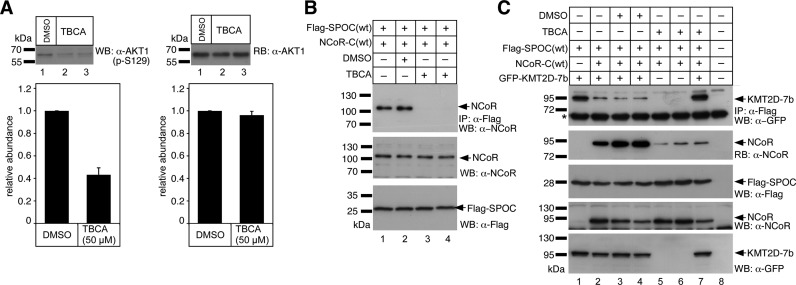
Previously, Yoo and colleagues (45) showed that the casein kinase-2β (CK2β) phosphorylates Ser-2436 but not Ser-2438 of human NCoR (accession: NP_006302, see also Figure 4A), which corresponds to Ser-2449 and Ser-2451 in the murine NCoR protein (accession: NP_001239242, see also Figure 4A). To investigate whether CK2β activity is implicated in regulating the SPOC/NCoR interaction, we performed coimmunoprecipitation experiments in the presence of the CK2β inhibitor TBCA (Figure 7). CK2β inhibition by TBCA was verified by Western blotting using an antibody against phosphorylated Ser-129 (p-S129) of AKT1 kinase, a known substrate for CK2β (46) (Figure 7A, left), and a pan-AKT1 antibody, which served as a loading control (Figure 7A right). As shown in Figure 7B (compare lanes 1 and 2 with 3 and 4), CK2β inhibition by TBCA leads to a complete loss of NCoR binding to the SPOC domain of SHARP.
Figure 7.
Inhibition of Casein Kinase 2β (CK2β) restores the association of KMT2D with the SPOC domain of SHARP. (A) Phosphorylation of AKT1 serine 129 (p-S129) is decreased upon CK2β inhibition by TBCA (left). HEK 293 cells were treated with DMSO (lane 1) or TBCA (lanes 2 and 3) for 3 h prior to cell lysis. p-S129 of AKT1 was tested by Western blotting. The pan-AKT1 antibody (right) served as loading control. A densitometric analysis from four independent experiments is shown in the lower panels. (B) Inhibition of CK2β activity by TBCA (lanes 3 and 4) abolishes the SPOC(wt)/NCoR(wt) interaction. (C) The C-terminal GFP- KMT2D-7b fragment coimmunoprecipitates with Flag-SPOC(wt) (upper panel, lane 1). The KMT2D-7b/SPOC(wt) interaction is reduced after coexpression of NCoR-C(wt) (1942–2453, upper panel, lanes 2, 3 and 4). The SPOC(wt)/NCoR-C(wt) interaction can be verified by reblotting (RB) with an anti-NCoR antibody (RB: α-NCoR). CK2β inhibition by TBCA reduces the SPOC(wt)/NCoR-C(wt) interaction (RB: α-NCoR, lanes 5, 6 and 7) and restores the KMT2D-7b/SPOC(wt) interaction (upper panel, compare lanes 3, and 4 with lane 7). HEK293 cells were transfected with the indicated expression constructs for Flag-SPOC(wt), GFP-KMT2D-7b, and NCoR-C(wt). Expression was verified by Western blotting (panels WB: α-Flag, α-NCoR and α-GFP). The asterisk denotes the heavy chain of the antibody used for immunoprecipitation.
CK2 inhibition by TBCA has the opposite effect on the KMT2D/SPOC interaction (Figure 7C). While NCoR-C(wt) binding to Flag-SPOC(wt) is strongly reduced in the presence of TBCA (Figure 7C, compare lanes 2 to 4 with 5 to 7), KMT2D-7b binding to Flag-SPOC(wt) is strongly increased (compare lane 2 to 4 with 7).
CK2β inhibitors were furthermore tested for their effects on chromatin marks and expression levels at the well-characterized Notch target gene Hes1 in mature T-cells. Experiments with CK2β inhibitor TBB are displayed on the left (Supplementary Figure S5A-D) and with CK2β inhibitor TBCA on the right (Supplementary Figure S5E-H). Inhibition of CK2β by TBB or TBCA leads to a 15-fold and 3-fold increase respectively, in the expression of the Notch target gene Hes1 (Supplementary Figure S5A and E). In line with the expression data, active histone marks H3K4me3 (Supplementary Figure S5B and F) as well as H3K27ac (Supplementary Figure S5C and G) are significantly increased upon CK2β inhibition by TBB or TBCA, whereas overall histone occupancy as measured by pan-H3 ChIP was slightly reduced (Supplementary Figure S5D and H). Together, these data suggest that CK2β phosphorylation of NCoR regulates the recruitment of NCoR and KMT2D to Notch target genes, which in turn affects the local histone marks and gene expression at these sites.
The Drosophila-homologs of SHARP, Spen, and KMT2D, Trr, physically and genetically interact
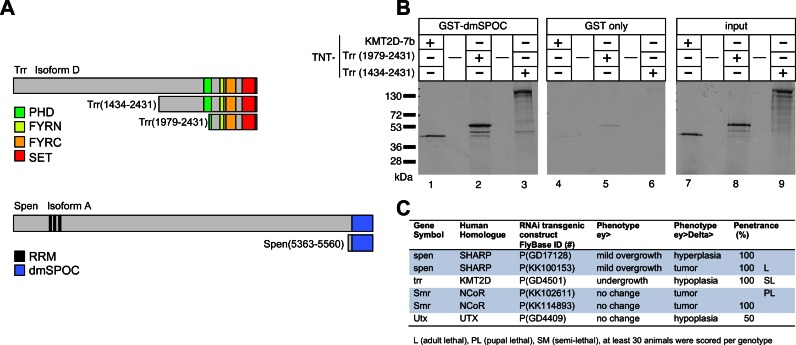
The Drosophila melanogaster homologs of SHARP (called Spen), KMT2D (called Trr) and NCoR (called SMRTER, Smr) have been linked to Notch signaling. In detail, Spen/SHARP negatively regulates Notch signaling in the developing eye (47). There is also evidence that SMRTER (48) and Trr-complex components (49) regulate the Notch signaling response. To analyze the interaction of SHARP/Spen and KMT2D/Trr (Trithorax-related) and Trr-complex components in Notch-mediated processes in vivo, we used the developing eye of Drosophila melanogaster and a well-established Notch-induced tumor model as our experimental system (10). Figure 8A represents the protein domain schematics of Trr, containing a PHD, FYRN, FYRC and a catalytic SET-domain, and of Spen, containing RNA-recognition motives (RRMs) and a SPOC-domain. We first demonstrated that Spen and Trr physically interact (Figure 8B). Using GST pulldown assays we found that the SPOC-domain of Spen/SHARP (dmSPOC) interacts with Trr (Figure 8B, left panel: Trr, 1434–2431, lane 3 and Trr 1979–2431 lane 2) and also with KMT2D-7b (Figure 8B, left panel, lane 1), but not with GST-only (Figure 8B middle panel). These data indicate that the interaction between SPOC and KMT2D is conserved in Drosophila. To investigate the functional relevance of these physical interactions in a Notch context and given the precedents that Trr and Trr-complex components regulate tissue growth (10,49), we used hairpin interfering RNA (RNAi) constructs to target these genes in vivo in the background of Notch hyperactivation (Figure 8C) and normal Notch-regulated growth and patterning (Supplementary Figure S6). Aberrant Notch activity has been found to support tumour growth and metastasis in several different animal models including Drosophila (10,49). Importantly, dysregulation of Lid, the H3K4 demethylase KDM5A/RBP2 homolog in Drosophila, enhances a Notch paradigm of tumorigenesis in vivo [29]. Alone, the overexpression of Notch-ligand Delta in the eye results in a mild eye overgrowth (10) (Supplementary Figure S6A). However, when combined with knockdown of SHARP-homolog spen, adult flies showed hyperplastic and tumoral eyes (Figure 8C and Supplementary Figure S6A and B). Tumor formation is also observed when the overexpression of Notch-ligand Delta is combined with knockdown of the NCoR-homolog Smr (Figure 8C). The eye-specific depletion of trr or the Trr-complex component, utx, by RNAi transgene expression combined with Notch-ligand Delta overexpression caused an overall eye reduction, which is the opposite phenotype as the downregulation of spen (Figure 8C and Supplementary Figure S6A and B). Altogether, our data support an evolutionary conserved role for SHARP and KMT2D as negative and positive regulators of Notch-dependent processes such as tissue growth.
Figure 8.
Physical interaction of Drosophila Spen/SHARP and Trr/KMT2D genetic interactions with Notch-induced eye growth. (A) Schematic representation of Drosophila melanogaster Trr and Spen constructs used in the GST pulldown experiments shown in panel (B). Amino acid numbering is according to accession AAN09063 (Trr, isoform D and AAF51535 (Spen, isoform A), (see also Materials and Methods for construct details). Trr and Spen domains: PHD, PHD-finger (cl02530); FYRN, F/Y-rich N-terminus (cl02650); FYRC, F/Y rich C-terminus (cl02651); SET, SET domain (cl02566); RRM, RNA recognition motif (cd00590); SPOC, Spen paralog and ortholog C-terminal domain (pfam07744). (B) The SPOC domain of the Drosophila Spen protein (dmSPOC) interacts with KMT2D and the Drosophila homolog of KMT2D (Trr) in GST-pulldown assays. Cell free synthesized and 35S labeled KMT2D-7b and Trr (1979–2431 and 1434–2431) (input, right panel) interact with GST-dmSPOC immobilized on GST sepharose beads (left panel), but not with GST alone (middle panel). (C) The eye tissue-specific depletion of spen, trr, smr, or utx using RNAi transgenic expression, alone and in the context of overexpression of Notch ligand Delta. The phenotype of mutant spen and smr can be grouped into hyperplasia whereas the mutant trr and utx display a hypoplastic phenotype. The RNAi trangenic flies are from the RNAi Collection stocks from Vienna Drosophila Resource Center (VDRC); ey> = eyeless-Gal4; ey>Delta> = eyeless-Gal4, UAS-Delta.
DISCUSSION
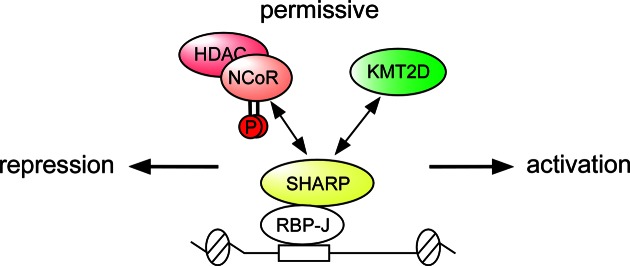
Our data suggest that the Notch core transcription complex containing RBP-J/SHARP orchestrates the dynamic methylation and acetylation of histones via a phosphorylation-dependent mechanism. So far, a two-step ‘on and off‘ model of Notch dependent transcription has been proposed (4–6,17). However, in line with our new data, we propose a pivotal intermediate step at Notch target genes as illustrated in Figure 9. RBP-J/SHARP balances histone methylation and acetylation by either interacting with the HDAC-containing NCoR complex or by interacting with the activating KMT2D-complex as revealed by changes in occupancy of NCoR and KMT2D complexes components upon depletion of UTX or NCoR, respectively (Figure 6). Since only phospho-NCoR binds to the SPOC domain of SHARP (Figures 4 and 5), signaling could directly alter the balance of chromatin marks: Positive acetylation marks can be removed by phospho-NCoR/HDAC while acquisition of active H3K4me3-methylation marks are mediated by KMT2D. Since the NICD replaces SHARP (18,21), the SHARP/NCoR or SHARP/KMT2D choice is particularly important to keep a permissive/poised state at Notch target genes before Notch activation. Once NICD is lost, the SHARP/KMT2D complex is recruited and signaling via activation of the NCoR-kinase CK2β might shift the balance more to the repressed state. As consequence, not only the active/inactive state of Notch signaling but also the state of CK2β regulates the dynamics of the active/repressive switch at Notch target genes. In line with that, a so-far unidentified NCoR-phosphatase would favour the recruitment of KMT2D. Finally, for full gene activation NICD/MAML/p300 displaces SHARP/KMT2D complex, acetylates histones and hence activates target gene expression (25,50).
Figure 9.
Model for the competing NCoR- and KMT2D-complex at Notch target genes. To keep an intermediate/permissive chromatin state, the RBP-J/SHARP complex recruits either activating KMT2D or repressing phospho-NCoR/HDAC complexes. The phospho-dependent competition between the two opposing chromatin modulators allows the incorporation of incoming signaling inputs to regulate Notch target genes responsiveness in the absence of Notch. The SHARP/NCoR interaction moves the balance versus the repressed state whereas the SHARP/KMT2D complex moves the balance versus the active state waiting for being displaced by NICD/p300.
The bivalent behavior of SHARP (also called human SPEN), interacting with both the corepressor NCoR and the coactivator H3K4 methyltransferase KMT2D, suggests that it is a ‘poising factor’ that acts either as a corepressor or coactivator. In regard to the physiological relevance of the central chromatin regulator SHARP, it is known that SHARP knockout mice are embryonic lethal (51) and that conditional SHARP knockout mice show enhanced generation of early T-cell progenitors (DN1), but also show an impaired generation of DN2 T-cells (52), which suggests an increase in the proliferation of DN1 cells (the earliest stage of T-cells) accompanied by an arrest of differentiation. In light of our new results we speculate that the aberrant T-cell development in the SHARP conditional knockout mice is due to the dysregulated chromatin context at Notch target genes. Most recently, SHARP has also been implicated in Xist-mediated X-inactivation (53–55). Thus, it will be important to evaluate whether the phospho-dependent mechanism involving NCoR and KMT2D is also implicated in X-inactivation. Our data suggest how SHARP would be involved in diseases. In fact, in recent exome-sequencing data SHARP mutations were identified in cancer (56,57) and myotonic dystrophy (58). These SHARP-mutations lead to a truncated protein lacking the C-terminal SPOC domain, which interacts with both NCoR and KMT2D. One possible scenario is that such SHARP-mutants loose the balance in chromatin regulation, thereby dysregulating a subset of Notch target genes. By site-directed mutagenesis of the SPOC-domain of SHARP we would gain further mechanistic insights into how phospho-NCoR and KMT2D recruitment is regulated. For this purpose, cocrystalisation of the SPOC domain (42) together with phosphorylated NCoR or the most recent NMR-structure of SPOC-domain in complex with phosphorylated SMRT-peptide (59) could be the basis.
The molecular mechanism how KMT2D interacts with SPOC remains to be further elucidated. It is possible, that KMT2D also needs to be phosphorylated to interact with the SPOC domain of SHARP.
Taken together, our data propose that there are two steps required for Notch dependent gene activation: (i) Displacement of the NCoR/HDAC complex facilitated by the KMT2D complex and subsequently (ii) NICD recruitment and displacement of the SHARP/KMT2D complex resulting in full gene activation. Since the association of RBP-J/SHARP with NCoR is phospho-dependent, this is one of the first examples how integration of different environmental signals regulates chromatin directly. Changing the composition of chromatin modifying complexes by phosphorylation would be an elegant molecular mechanism to process diverse signaling inputs into alternations of gene transcription.
Supplementary Material
Acknowledgments
We want to thank Dr M. Laschak (Ulm), Dr M. Kracht and J. Soelch (Giessen) for providing us with plasmids. We thank Dr D. van Essen for providing us with the mature T-cell line. We want to thank M. Krötschel, S. Fietzeck, C. Grubisic, P. Käse, T. Schmidt-Wöll, P. Dietmann, S. Schirmer and R. Rittelmann for excellent technical assistance. We are grateful to Drs G.Suske (Marburg), M. Lachner (Freiburg), B. Baumann (Ulm) and R. Liefke (Boston) for critical reading.
FUNDING
Collaborative research [TRR81]; Heisenberg program [BO 1639/5-1] by the Deutsche Forschungsgemeinschaft (DFG) and the Max-Planck society, regarding [to T.B.]; Excellence Initiative of the German Federal and State Governments [GSC4, Spemann Graduate School Freiburg, to B.D.G.]; DFG [collaborative research grant SFB 1074/A3 to F.O.]; BMBF (research nucleus SyStAR), regarding [to F.O. and M.K.]; International Graduate School in Molecular Medicine, Ulm [GSC270 to W.C. and M.R.]; National Institutes of Health Grant [CA178974], regarding [to R.A.K]; Leukemia and Lymphoma Society [to R.A.K.]; Spanish Grants [SAF2012-35181, SEV-2013-0317, PROMETEO II/2013/001]; Botin Foundation [to M.D.]; American Heart Association [to K.J.C.]. Funding for open access charge: DFG collaborative research [TRR81].
Conflict of interest statement. None declared.
REFERENCES
- 1.Allis C.D., Jenuwein T., Reinberg D. Epigenetics. 2007. [Google Scholar]
- 2.Campos E.I., Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Guruharsha K.G., Kankel M.W., Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopan R., Ilagan M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray S.J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell. Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 7.Radtke F., Fasnacht N., Macdonald H.R. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Borggrefe T., Lauth M., Zwijsen A., Huylebroeck D., Oswald F., Giaimo B.D. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFbeta/BMP and hypoxia pathways. Biochim. Biophys. Acta. 2015;1863:303–313. doi: 10.1016/j.bbamcr.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Aulehla A., Pourquie O. Oscillating signaling pathways during embryonic development. Curr. Opin. Cell Biol. 2008;20:632–637. doi: 10.1016/j.ceb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Ferres-Marco D., Gutierrez-Garcia I., Vallejo D.M., Bolivar J., Gutierrez-Avino F.J., Dominguez M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 11.Weng A.P., Ferrando A.A., Lee W., Morris J.P.t., Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 12.Puente X.S., Pinyol M., Quesada V., Conde L., Ordonez G.R., Villamor N., Escaramis G., Jares P., Bea S., Gonzalez-Diaz M., et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kridel R., Meissner B., Rogic S., Boyle M., Telenius A., Woolcock B., Gunawardana J., Jenkins C., Cochrane C., Ben-Neriah S., et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- 14.Krejci A., Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castel D., Mourikis P., Bartels S.J., Brinkman A.B., Tajbakhsh S., Stunnenberg H.G. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanigaki K., Honjo T. Two opposing roles of RBP-J in Notch signaling. Curr. Top. Dev. Biol. 2010;92:231–252. doi: 10.1016/S0070-2153(10)92007-3. [DOI] [PubMed] [Google Scholar]
- 17.Borggrefe T., Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oswald F., Kostezka U., Astrahantseff K., Bourteele S., Dillinger K., Zechner U., Ludwig L., Wilda M., Hameister H., Knochel W., et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knochel W., Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salat D., Liefke R., Wiedenmann J., Borggrefe T., Oswald F. ETO, but not leukemogenic fusion protein AML1/ETO, augments RBP-Jkappa/SHARP-mediated repression of notch target genes. Mol. Cell. Biol. 2008;28:3502–3512. doi: 10.1128/MCB.01966-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanderWielen B.D., Yuan Z., Friedmann D.R., Kovall R.A. Transcriptional repression in the Notch pathway: thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J. Biol. Chem. 2011;286:14892–14902. doi: 10.1074/jbc.M110.181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y., Downes M., Xie W., Kao H.Y., Ordentlich P., Tsai C.C., Hon M., Evans R.M. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pajerowski A.G., Nguyen C., Aghajanian H., Shapiro M.J., Shapiro V.S. NKAP is a transcriptional repressor of notch signaling and is required for T cell development. Immunity. 2009;30:696–707. doi: 10.1016/j.immuni.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao H.Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C.R., Evans R.M., Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oswald F., Tauber B., Dobner T., Bourteele S., Kostezka U., Adler G., Liptay S., Schmid R.M. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulligan P., Yang F., Di Stefano L., Ji J.Y., Ouyang J., Nishikawa J.L., Toiber D., Kulkarni M., Wang Q., Najafi-Shoushtari S.H., et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell. 2011;42:689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borggrefe T., Liefke R. Fine-tuning of the intracellular canonical Notch signaling pathway. Cell Cycle. 2012;11:264–276. doi: 10.4161/cc.11.2.18995. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Scully K., Zhu X., Cai L., Zhang J., Prefontaine G.G., Krones A., Ohgi K.A., Zhu P., Garcia-Bassets I., et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 29.Liefke R., Oswald F., Alvarado C., Ferres-Marco D., Mittler G., Rodriguez P., Dominguez M., Borggrefe T. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohan M., Herz H.M., Shilatifard A. SnapShot: Histone lysine methylase complexes. Cell. 2012;149:498. doi: 10.1016/j.cell.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M., et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C.M., Nakamura T., Mazo A., Eisenbach L., Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Nuland R., Smits A.H., Pallaki P., Jansen P.W., Vermeulen M., Timmers H.T. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell. Biol. 2013;33:2067–2077. doi: 10.1128/MCB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 36.Mortensen P., Gouw J.W., Olsen J.V., Ong S.E., Rigbolt K.T., Bunkenborg J., Cox J., Foster L.J., Heck A.J., Blagoev B., et al. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 2010;9:393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 37.Wacker S.A., Alvarado C., von Wichert G., Knippschild U., Wiedenmann J., Clauss K., Nienhaus G.U., Hameister H., Baumann B., Borggrefe T., et al. RITA, a novel modulator of Notch signalling, acts via nuclear export of RBP-J. EMBO J. 2011;30:43–56. doi: 10.1038/emboj.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieuwkoop P.D., Faber J. Normal Table of Xenopus laevis (Daudin) Amsterdam: North Holland Publishing Company; 1956. [Google Scholar]
- 39.Hemmati-Brivanlou A., Frank D., Bolce M.E., Brown B.D., Sive H.L., Harland R.M. Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development. 1990;110:325–330. doi: 10.1242/dev.110.2.325. [DOI] [PubMed] [Google Scholar]
- 40.Lee J.E., Wang C., Xu S., Cho Y.W., Wang L., Feng X., Baldridge A., Sartorelli V., Zhuang L., Peng W., et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H., Zou J., Zhao B., Johannsen E., Ashworth T., Wong H., Pear W.S., Schug J., Blacklow S.C., Arnett K.L., et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ariyoshi M., Schwabe J.W. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003;17:1909–1920. doi: 10.1101/gad.266203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dephoure N., Zhou C., Villen J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., Gygi S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 45.Yoo J.Y., Choi H.K., Choi K.C., Park S.Y., Ota I., Yook J.I., Lee Y.H., Kim K., Yoon H.G. Nuclear hormone receptor corepressor promotes esophageal cancer cell invasion by transcriptional repression of interferon-gamma-inducible protein 10 in a casein kinase 2-dependent manner. Mol. Biol. Cell. 2012;23:2943–2954. doi: 10.1091/mbc.E11-11-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Maira G., Salvi M., Arrigoni G., Marin O., Sarno S., Brustolon F., Pinna L.A., Ruzzene M. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005;12:668–677. doi: 10.1038/sj.cdd.4401604. [DOI] [PubMed] [Google Scholar]
- 47.Doroquez D.B., Orr-Weaver T.L., Rebay I. Split ends antagonizes the Notch and potentiates the EGFR signaling pathways during Drosophila eye development. Mech. Dev. 2007;124:792–806. doi: 10.1016/j.mod.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heck B.W., Zhang B., Tong X., Pan Z., Deng W.M., Tsai C.C. The transcriptional corepressor SMRTER influences both Notch and ecdysone signaling during Drosophila development. Biol. Open. 2012;1:182–196. doi: 10.1242/bio.2012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herz H.M., Madden L.D., Chen Z., Bolduc C., Buff E., Gupta R., Davuluri R., Shilatifard A., Hariharan I.K., Bergmann A. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol. Cell. Biol. 2010;30:2485–2497. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung C., Mittler G., Oswald F., Borggrefe T. RNA helicase Ddx5 and the noncoding RNA SRA act as coactivators in the Notch signaling pathway. Biochim. Biophys. Acta. 2013;1833:1180–1189. doi: 10.1016/j.bbamcr.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda K., Han H., Tani S., Tanigaki K., Tun T., Furukawa T., Taniguchi Y., Kurooka H., Hamada Y., Toyokuni S., et al. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- 52.Tsuji M., Shinkura R., Kuroda K., Yabe D., Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1610–1615. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moindrot B., Cerase A., Coker H., Masui O., Grijzenhout A., Pintacuda G., Schermelleh L., Nesterova T.B., Brockdorff N. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 2015;12:562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monfort A., Di Minin G., Postlmayr A., Freimann R., Arieti F., Thore S., Wutz A. Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells. Cell Rep. 2015;12:554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McHugh C.A., Chen C.K., Chow A., Surka C.F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A., et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens P.J., Davies H.R., Mitani Y., Van Loo P., Shlien A., Tarpey P.S., Papaemmanuil E., Cheverton A., Bignell G.R., Butler A.P. Whole exome sequencing of adenoid cystic carcinoma. J. Clin. Invest. 2013;123:2965–2968. doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossi D., Trifonov V., Fangazio M., Bruscaggin A., Rasi S., Spina V., Monti S., Vaisitti T., Arruga F., Fama R., et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J. Exp. Med. 2012;209:1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dansithong W., Jog S.P., Paul S., Mohammadzadeh R., Tring S., Kwok Y., Fry R.C., Marjoram P., Comai L., Reddy S. RNA steady-state defects in myotonic dystrophy are linked to nuclear exclusion of SHARP. EMBO Rep. 2011;12:735–742. doi: 10.1038/embor.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mikami S., Kanaba T., Takizawa N., Kobayashi A., Maesaki R., Fujiwara T., Ito Y., Mishima M. Structural Insights into the Recruitment of SMRT by the Corepressor SHARP under Phosphorylative Regulation. Structure. 2014;22:35–46. doi: 10.1016/j.str.2013.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.