Abstract
Defects in DNA replication and repair are known to promote genomic instability, a hallmark of cancer cells. Thus, eukaryotic cells have developed complex mechanisms to ensure accurate duplication of their genomes. While DNA damage response has been extensively studied in tumour cells, the pathways implicated in the response to replication stress are less well understood especially in non-transformed cells. Here we show that in non-transformed cells, APC/CCdh1 is activated upon severe replication stress. Activation of APC/CCdh1 prevents new origin firing and induces permanent arrest in S-phase. Moreover, Rad51-mediated homologous recombination is also impaired under these conditions. APC/CCdh1 activation in S-phase occurs after replication forks have been processed into double strand breaks. Remarkably, this activation, which correlates with decreased Emi1 levels, is not prevented by ATR/ATM inhibition, but it is abrogated in cells depleted of p53 or p21. Importantly, we found that the lack of APC/CCdh1 activity correlated with an increase in genomic instability. Taken together, our results define a new APC/CCdh1 function that prevents cell cycle resumption after prolonged replication stress by inhibiting origin firing, which may act as an additional mechanism in safeguarding genome integrity.
INTRODUCTION
Faithful DNA replication is essential to prevent DNA damage and chromosomal instability, a hallmark of cancer (1). Replication errors induced by natural replication fork barriers such as secondary DNA structures, non-histone protein/DNA interactions and replication-transcription clashes, as well as replication stress induced by nucleotide deficiency (2) and DNA damage underlie many genome alterations that can compromise genome integrity (3–7). Interestingly, during recent years compelling evidences have arisen indicating that oncogene overexpression in non-transformed cells causes replication stress, inducing DNA damage and a permanent withdrawal from the cell cycle (8,9). This process, known as oncogene-induced senescence (OIS), is considered a tumourigenic barrier. Thus, an accurate knowledge of the DNA replication stress response in non-transformed cells is important to understand the alterations that allow OIS bypass in tumour cells, as well as to develop new cancer therapies to act specifically against transformed cells. In this regard, taking advantage of the fact that tumour cells have increased DNA replication stress, it has been proposed that novel therapeutic approaches could be developed that capitalize on the presence of DNA replication stress in cancer but not normal cells (10).
Arrested replication forks and DNA double strand breaks (DSBs) in S-phase are signalled by distinct pathways known as the DNA replication checkpoint and the DNA damage checkpoint respectively. Once activated, these intra-S-phase checkpoints promote replication fork stabilization and DNA repair, regulate cell cycle progression and, eventually, control the resumption of DNA replication, ensuring correct genome duplication (3). In mammalian cells the central players of the DNA replication checkpoint pathway are ATR and Chk1 kinases. Notably, ATR and Chk1 are also essential for correct DNA replication during normal cell cycle progression by controlling both replication fork stability and origin firing (11–15). Upon stalling of replication forks, Replication Protein A (RPA)-coated regions of single-stranded DNA are generated, which mediate the recruitment of ATR and a subset of proteins essential for its activation (16). Once activated, ATR, in complex with Claspin, phosphorylates and activates Chk1 (17). Chk1 arrests cell cycle progression and mitotic entry by down-regulation of Cdk2/Cyclin A and Cdk1/Cyclin B activities through inhibition of several isoforms of Cdc25 phosphatases (18–21) and activation of the tyrosine kinase Wee1 (22), these being positive and negative regulators of the Cdk/cyclin complexes respectively. In addition, ATR/Chk1 inhibits late origin firing after DNA replication stress while allowing activation of nearby dormant origins (23), which is important for correct global replication restart under these conditions (24). Moreover, Chk1 promotes Treslin phosphorylation, thus preventing loading of replication initiation protein Cdc45 to the origins (13). Another critical role for ATR and Chk1 in response to replication stress is the stabilization of replication forks, which prevents generation of additional DNA damage and allows faithful replication restart (25). Specifically, Chk1 prevents Mus81/Eme1 endonuclease-dependent DSB formation at the replication forks (14). However, stalled forks can eventually collapse and be processed into DSBs after prolonged replication arrest (26). In this regard, Helledays’ group showed that after a short (2 h) hydroxyurea (HU) treatment, U2OS (osteosarcoma) cells were able to restart DNA synthesis by reactivating stalled forks, while after a long period of HU treatment (24 h), forks were converted into DSBs and replication was reinitiated mainly by new origin activation. It should be noted that even though DNA synthesis could be completed by new origin firing, DSBs originated at collapsed forks would need to be repaired. Nevertheless, the reactivation of forks that have been processed into DSBs can also be achieved by a sub-pathway of homologous recombination (HR) called break-induced replication (BIR) (27–30). In response to DSBs, Mre11-Rad50-Nbs1 (MRN) complex binds to DNA and together with other helicases and nucleases such as BLM, CtIP, Exo1 and Dna2 creates a 3′ single-stranded DNA overhang that it is ultimately coated by Rad51, which promotes homology search and strand invasion, both essential steps in HR repair (31). While replication resumption can be achieved by error-free mechanisms before DSBs are present, once damaged, replication fork restart can disrupt genome integrity as BIR-mediated restart of damaged forks is highly mutagenic. Indeed, BIR-based mechanisms can explain the complexity of the chromosomal changes that occur in cancer cells (4,28,32–34). Moreover, HR in S-phase can result in epigenetic alterations (35). Additionally, altering the timing of origin activation also induces changes in epigenetic marks (36,37). Thus, restarting DNA replication after DSB formation could induce genomic instability and alterations in epigenetic determinants, favouring tumour formation and progression. Data showing that an extra allele of Chk1 limits oncogene-induced replication stress and promotes transformation (38) reflects the relevance of directing non-transformed cells to senescence instead of promoting potential error-prone repair of replication stress-induced DNA damage in order to avoid transformation.
Several studies have focused on understanding the mechanisms controlling senescence after DNA damage. Interestingly, it has recently been proposed that DNA damage-activated senescence is mainly induced in G2-phase (39). In G2, DSBs induce permanent cell cycle exit mediated by p53 and p21 (40). This cell cycle exit is rapidly determined, with p21-mediated nuclear Cyclin B1 translocation being the first detectable event of this decision, which is followed by Cdh1-bound anaphase-promoting complex/cyclosome (APC/CCdh1)-mediated Cyclin B1 degradation (41–43). Several studies have also reported unscheduled APC/CCdh1 activation in G2 upon DNA damage (44–46). APC/C is an E3 ubiquitin ligase that is involved in cell cycle regulation and becomes activated upon sequential binding of Cdc20 and Cdh1 coactivators (47,48). Cdc20 is associated with APC/C during early mitosis and mainly regulates mitotic progression, whereas Cdh1 interacts with APC/C from late mitosis onwards until the following G1/S transition (47,49). During S- and G2-phases, APC/C is kept inactive through different mechanisms: Cdk2/Cyclin A- and Cdk1/Cyclin A-mediated phosphorylation of Cdh1, which hinder association of Cdh1 with APC/C (49–52), Emi1-mediated disruption of APC/CCdh1 and APC/CCdc20 interaction with their substrates (50,53) and degradation of both Cdh1 and Ubc10 (a ubiquitin-conjugating enzyme that works with APC/C) (54). In response to DNA damage, the unscheduled activation of this ubiquitin ligase has been proposed to depend on either Emi1 down-regulation or Cdc14B phosphatase activation (45,46,55,56).
As mentioned above, the mechanisms leading to cell cycle withdrawal after DNA damage in G1 and G2 have been broadly studied. However, although irreversible arrest upon prolonged replication inhibition has been described in different cell lines (57), the circumstances and pathways leading to permanent cell cycle arrest in response to DNA replication stress in non-transformed human cells remain to be elucidated. In the current study, we have analysed the response of non-transformed human cells to HU or to DNA damage induced during S-phase. Our data shows that upon severe replication stress, the resumption of DNA replication and mitotic entry are both compromised in non-transformed cells, and that cells undergo senescence even with unreplicated DNA. This loss of ability to resume replication is due to APC/CCdh1 activation in S-phase, which causes new origin firing inhibition. Importantly, activation of this ubiquitin ligase is observed after stalled replication forks have collapsed and been processed into DSBs. Thus, our results show for the first time that APC/CCdh1 is activated after severe replication stress to inhibit new origin firing, consequently preventing cell cycle progression of non-transformed cells with damaged DNA.
MATERIALS AND METHODS
Cell culture, synchronization and drugs
hTERT-immortalized retinal pigment epithelial (hTERT-RPE) cells, human mammary epithelial MCF10A cells, colorectal cancer HCT116 cells and colorectal cancer DLD1 cells were grown in DMEM: Ham's F12 (1:1) medium (Biological Industries) supplemented with 6% fetal bovine serum (FBS, Biological Industries), except from MCF10A cells that were supplemented with 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor (Peprotech), 0.5 μg/ml hydrocortisone (Sigma-Aldrich) and 10 μg/ml insulin (Sigma-Aldrich). hTERT-immortalized foreskin BJ-5ta fibroblasts were cultured in DMEM: M199 (4:1) medium (from Biological Industries and Sigma-Aldrich respectively) supplemented with 10% FBS. Human foreskin 1306 fibroblasts and human embryonic foreskin WS1 fibroblasts were cultured in MEM (Biological Industries) medium supplemented with 10% FBS. Human cervical epithelial cancer HeLa cells, lung primary fibroblasts, squamous cancer A431 cells, pancreatic cancer HPAFII cells, colorectal cancer HT29 cells, breast cancer MCF7 cells, pancreatic cancer SW1990 cells and osteosarcoma U2OS cells were cultured in DMEM medium supplemented with 10% FBS. Peripheral blood lymphocytes were cultured in RPMI (Biological Industries) medium 10% FBS. All culture media were supplemented with 1% non-essential amino acids (Biological Industries), 2 mM L-glutamine (Sigma-Aldrich), 1 mM pyruvic acid (Sigma-Aldrich), 50 U/ml penicillin and 50 μg/ml streptomycin (both from Biological Industries).
For hTERT-RPE cells synchronization in S-phase by serum starvation, cells were first grown in FBS-containing medium until they reached full confluence. Cells were then plated diluted (1/4) in culture medium without serum during 48 h. After serum starvation cells were cultured again in FBS-containing medium during 16 h. In order to synchronize the different cell lines in S-phase by single thymidine (Sigma-Aldrich) block, cells were plated during 12 h (siRNA transfected cells) or 24 h in FBS-containing media (if not specified otherwise). Thymidine was added (1.5 mM to hTERT-RPE cells and 2.5 mM to the rest of cell lines) during 22 h after which cells were extensively washed with phosphate buffered saline (PBS) and released into fresh medium without thymidine during 2 h.
Drugs and their working concentrations were used as follows: 10 μM BrdU (Sigma-Aldrich), 50 μM EdU (Invitrogen), 25 μM CldU (Sigma-Aldrich) if not specified otherwise, 250 μM IdU (Sigma-Aldrich), 300 nM UCN-01 (Calbiochem, Merck KGaA), 10 μM SB203580 (Calbiochem, Merck KGaA), 20 μM MG132 (Selleckchem), 10 μM VE 821 (Axon Medchem), 100 nM CHIR-124 (Axon Medchem), 20 μM KU-55933 (Selleckchem), 50 μM proTAME (BostonBiochem), 10 mM HU (Sigma-Aldrich) if not specified otherwise, nocodazole (Sigma-Aldrich) (50 ng/ml for BJ-5ta and 250 ng/ml for the rest of the cell lines). Camptothecin (Sigma-Aldrich) and etoposide (Sigma-Aldrich) concentrations are indicated in the corresponding figure legend.
Flow cytometry
Cells were harvested and fixed in 70% ethanol for at least 2 h at −20°C before immunostaining and flow cytometry analysis. Combined analysis of DNA content (1% propidium Iodide, PI, Sigma-Aldrich) and Phospho-H3 (anti-P-H3 S10, Millipore, #06–570; 1/400) or DNA content, BrdU (anti-BrdU, Abcam, ab6326; 1/250) and MPM2 (anti-MPM2, Millipore, #05–368; 1/250) was performed as previously described (58).
Colony formation and proliferation assays
For colony formation assays, hTERT-RPE and HCT116 cells were cultured in 12-well plates, synchronized by single thymidine block in S-phase and then treated during 14 h with HU or left untreated. Cells were then extensively washed with PBS and then released into fresh medium without HU. After 12 h of release cells were plated diluted (250 cells in each well) in 6-well plates. Eight days later cells were fixed and stained with 1% crystal violet in 70% ethanol during 10 min at room temperature (RT) and the number of colonies was counted.
For proliferation assays, hTERT-RPE cells were plated in 12-well plates, synchronized in S-phase by single thymidine block and then treated during 14 h with HU or left untreated. Cells were then extensively washed with PBS and released into fresh medium without HU. Plates were harvested before the treatment, just after the treatment (0 h) and at the indicated time after release. Finally, cells were fixed in 4% paraformaldehyde- (PFA) PBS for 15 min at RT and stained with 0.25% crystal violet during 5 min. Cristal violet absorbance (λ595) was used to determine cell confluence in each condition.
Senescence-associated β-Galactosidase (SA-β-Gal) assay
SA-β-Gal staining was performed as described previously (59). Briefly, cells were fixed in 2% PFA-/0.2% glutaraldehyde-PBS for 3 min at RT and then incubated with X-gal (B4252, Sigma) containing solution for 15 h at 37°C in darkness. Images were obtained with Leica DMRB microscope.
RNA interference
Transient siRNA experiments were performed using HiPerfect Transfection Reagent (Qiagen). The following siRNA oligos were transfected at 50 nM final concentration using manufacturer's guidelines: (i) Human ON-TARGETplus SMARTpool siRNA oligos that specifically target Cdh1 (Dharmacon, L-015377-00-0005). The siRNA oligo set contains four sequences as follows: Cdh1 #6: 5′-CCACAGGAUUAACGAGAAU-3′, Cdh1 #7: 5′-GGAACACGCUGACAGGACA-3′, Cdh1 #8: 5′-GCAACGAUGUGUCUCCCU A-3′ and Cdh1 #9: 5′-GAAGAAGGGUCUGUUCACG-3′. (ii) Human ON-TARGETplus SMARTpool siRNA oligos that specifically target p21 (Dharmacon, L-003471-00-0005). The siRNA oligo set contains four sequences as follows: 5′-CGACUGUGAUGCGCUAAUG-3′, 5′-CCUAAUCCGCCCACAGGAA-3′, 5′-CGUCAGAACCCAUGCGGCA-3′ and 5′-AGACCAGCAUGACAGAUUU-3′. (iii) Human ON-TARGETplus SMARTpool siRNA oligos that specifically target p53 (Dharmacon, L-003329-00-0005). The siRNA oligo set contains four sequences as follows: 5′-GAAAUUUGCGUGUGGAGUA-3′, 5′-GUGCAGCUGUGGGUUGAUU-3′, 5′-GCAGUCAGAUCCUAGCGUC-3′ and 5′-GGAGAAUAUUUCACCCUUC-3′. (iv) Human ON-TARGETplus SMARTpool siRNA oligos that specifically target Emi1 (Dharmacon L-012434-00-0005), The siRNA oligo set contains four sequences as follows: 5′-CAACAGACACUUAAUAGUA-3′, 5′-CGAAGUGUCUCUGUAAUUA-3′, 5′-UGUAUUGGGUCACCGAUUG-3′ and 5′-GAAUUUCGGUGACAGUCUA-3′. (v) Human ON-TARGETplus non-targeting pool (Dharmacon, D-001810-10-20) was used as a negative control. The siRNA oligo set contains four sequences as follows: 5′-UGGUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAUGUUGUGUGA-3′, 5′-UGGUUUACAUGUUUUCUGA-3′ and 5′-UGGUUUACAUGUUUUCCUA-3′.
Immunoblotting
Cells were collected in 2% SDS containing 67 mM Tris-HCl (pH 6.8) buffer, run in Laemmli SDS-polyacrylamide gels and transferred to nitrocellulose membranes as previously described (58). Incubation with primary antibodies was conducted overnight at 4°C. Antibodies against the indicated proteins were used as follows: Cyclin B1 (GNS1, sc-245; 1/200), Cyclin A2 (H-432, sc-751; 1/500), Plk1 (F-8, sc-17783; 1/50), Aurora A (Cell signalling, #3092; 1/500), pRb (IF-8, sc-102; 1/500), p21 (Ab-1, OP64; 1/1000), Cdh1 (homemade against amino acids 1–225 of human Cdh1; 1/1000), p53 (Ab-5, MS-186; 1/1000), P-Chk1 S296 (Cell signalling, #2349; 1/1000), P-Chk1 S345 (Cell signalling, #2341; 1/1000), CtIP (A300–488A; 1/1000), P-Chk2 T68 (NB100-92502; 1/1000), Emi1 (37–6600; 1/100), Rad51 (H-92, sc-8349; 1/200), GAP120 (sc-63; 1/100), Cdk4 (H-303, sc-749; 1/500), Lamin B (M-20; sc-6217; 1/200), actin (Cell signalling, #69100; 1/5000) and H3 (Abcam, ab1791; 1/2000). Proteins were visualized using ECL detection system (Biological Industries).
Chromatin enriched fraction isolation
For chromatin enriched fraction isolation, we followed a protocol adapted from (60). Shortly, whole cell pellets were incubated with 0.1% triton X-100 containing buffer A (10 mM HEPES (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol) supplemented with protease and phosphatase inhibitors for 20 min on ice. After buffer removal by centrifugation (600 x g 4 min 4°C) pellets were rinsed with buffer A without triton and then incubated with buffer B (3 mM EDTA, 0.2 mM EGTA) once again supplemented with inhibitors for 10 min on ice. After buffer removal (1700 x g 5 min 4°C) pellets were washed with buffer B (600 x g 5 min 4°C) and then resuspended in 2% SDS containing 67 mM Tris-HCl (pH 6.8) buffer. Chromatin enriched fractions were finally resuspended in Laemmli buffer and analysed by immunoblotting as previously described.
Isolation of proteins on nascent DNA (iPOND)
Isolation of proteins on nascent DNA was performed as described previously (61) with a few modifications. Briefly, EdU-labelled and treated cells were fixed in 1% PFA for 10 min at RT and quenched with 0.125 mM glycine (pH 7) for 5 min at RT. Cells were harvested, pelleted by centrifugation and lysed in lysis buffer (ChIP Express kit, Active Motif) for 30 min at 4°C. Lysates were passed 10× through a 21-gauge needle, and the nuclei were pelleted by centrifugation and rinsed with PBS supplemented with protease inhibitor cocktail (PIC, Roche) before subject them to click reaction for 30 min at RT with 0.2 mM Biotin-azide (Invitrogen). After click reaction, nuclei were pelleted by centrifugation, rinsed with PBS + PIC, resuspended in shearing buffer (ChIP Express kit, Active Motif) and sonicated (Bioruptor, Diagenode) for 15 min at high intensity (30-s/30-s on/off pulses). Streptavidin-conjugated Dynabeads M-280 (Invitrogen) were washed three times with 1x blocking buffer (1% Triton X-100, 2 mM EDTA (pH 8), 150 mM NaCl, 20 mM Tris-HCl (pH 8), 20 mM beta-glycerol phosphate, 2 mM sodium orthovanadate, PIC) and then blocked for 1 h at RT with blocking buffer containing 10 mg/ml salmon-sperm DNA (Sigma-Aldrich). Lysates were then incubated with previously blocked Dynabeads (1:10) for 30 min at RT. Finally, beads were washed twice with low-salt buffer (containing 150 mM NaCl) and twice with high-salt buffer (containing 500 mM NaCl), and then resuspended in Laemmli buffer to analyse them by immunoblotting as previously described.
Immunofluorescence microscopy
For immunofluorescence microscopy cells were grown on glass coverslips and then treated as indicated. After treatment, cells were rinsed with PBS and fixed in 2% PFA-PBS for 20 min at RT. After extensive washing, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min at RT and washed with PBS during 5 min. Cells were then blocked with 3% FBS containing 0.1% Triton X-100-PBS for 1 h at RT and then incubated with the indicated antibodies diluted in blocking solution during 45 min at 37°C. The following primary antibodies were used: anti- γ-H2AX (Millipore, #05-636; 1/3000), anti-53BP1 (Abcam, ab36823; 1/500), anti-Cyclin D1 (DCS-6, sc-20044; 1/100). After extensive washing in blocking solution cells were stained with Alexa488-, Alexa555- or Alexa647-conjugated secondary antibodies (Invitrogen, 1/500 diluted in blocking solution) for 20 min at 37°C. Finally, cells were counterstained either with 1% PI-containing 0.1 mg/ml RNAse A (Fermentas) -PBS solution or with DAPI (Sigma-Aldrich). Images were obtained using Leica TCS-SL confocal microscopy and then analysed using Fiji software.
For EdU staining, previously labelled and treated cells were fixed in 4% PFA-PBS for 30 min at RT and then click reaction was performed (as previously described) for 30 min at RT with 1 μM Alexa488-azide (Invitrogen) before immunostaining.
For CldU and IdU immunofluorescence analysis, previously labelled and treated cells were fixed for 10 min in 70% ethanol at RT. Cells were then rinsed with PBS and incubated with 0.2% triton X-100 containing 2 M HCl solution for 30 min at RT. HCl was neutralized by washing three times with borate buffer. Cells were then washed twice with PBS and blocked with 1% bovine serum albumin- (BSA, Sigma-Aldrich) PBS. Finally, cells were incubated with primary anti-BrdU (Abcam, ab6326; 1/250 for CldU labelling and Becton Dickinson, 347580; 1/50 for IdU labelling) and secondary, anti-rat (Alexa488 conjugated; 1/500) and anti-mouse (Alexa647 conjugated; 1/500) antibodies diluted in blocking buffer. DNA was counterstained with DAPI.
Pulse field gel electrophoresis (PFGE)
For DNA break analysis cells were washed with PBS, trypsinized and resuspended at 8.33 × 106 cells/ml cell density in incubation buffer (0.25 mM EDTA, 20 mM NaCl, 10 mM Tris, pH 7.5). This suspension (120 μl) was mixed 1:1 with 1% low-melting point agarose (Sigma) to obtain two agarose inserts each of them containing 0.5 × 106 cells. Afterwards, cells in plugs were lysed (0.25 mM EDTA, 20 mM NaCl, 10 mM Tris, pH 7.5, 1% N-laurylsarcosyl, 1 mg/ml proteinase K) for 48 h at 50°C, washed three times with TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5) and run in 1% agarose gel (chromosomal grade; Bio-Rad) in a CHEF DR III PFGE apparatus (Bio-Rad; 120 angle; 60–240 s switch time; 4 V/cm) at 14°C for 20 h. Finally, gels were stained with SYBR® Safe and analysed using LAS-4000 system (Fujifilm).
DNA fibre assay
DNA fibre assay was performed as previously described in (26) with a few modifications. Briefly, hTERT-RPE cells were pulse-labelled with CldU/IdU and treated as indicated. Labelled cells were harvested by trypsinization and resuspended in ice-cold PBS at 5 × 105 cells/ml. For DNA spreading, 2 μl of cells were spotted onto glass slides and lysed with 7 μl of spreading buffer (0.5% SDS, 200 mM Tris-HCl (pH 7.4), 50 mM EDTA). Slides were tilted (15° to horizontal), allowing a stream of DNA to run slowly down the slide, air dried and then fixed in methanol/acetic acid (3:1) solution. CldU and IdU were detected by incubating acid-treated fibre spreads with anti-BrdU monoclonal antibodies (Abcam, ab6326; 1/1000 for CldU labelling and Becton Dickinson, 347580; 1/200 for IdU labelling) diluted in blocking buffer (1% BSA in 0.1% Tween-20 -PBS) for 1.5 h at 37°C. After incubation with primary antibodies, fibre spreads were fixed with 4% PFA-PBS for 10 min at RT and finally incubated with secondary antibodies (Alexa488-conjugated anti-rat and Alexa555-conjugated anti-mouse; 1/500) for 1.5 h at 37°C. Images were obtained using Leica TCS-SL confocal microscopy with a 63× oil immersion objective and then analysed using Fiji software.
Stalled forks (only-green tracks), restarted forks (green+red tracks with a maximum gap of 2 μm between them) and new origins (only-red tracks) were quantified. The number of fibres analysed in each experiment is indicated in the corresponding figure legend.
RESULTS
Replication resumption is compromised in non-transformed human cells after severe replication stress
Given the importance of the fidelity of DNA replication for maintaining genomic stability and preventing tumour formation, we aimed to analyse the determinants of the response to replication stress in non-transformed human cells. To this end, mitotic entry was evaluated in asynchronously growing cells treated with the ribonucleotide reductase inhibitor HU (24 h) and checkpoint kinases inhibitors (added during the last 6 h). In agreement with our previous data (58,62), while inhibition of Chk1 kinase induced a high percentage of HeLa and HCT116 tumour cells to enter into mitosis with a DNA content below 4n, only a small subset of S-phase arrested cells were shown to enter into mitosis with unreplicated DNA, even after simultaneous Chk1 and p38 kinases inhibition, in all analysed non-transformed human cells (Supplementary Figure S1A and B). Similar results were obtained when HU and checkpoint kinases inhibitors were added simultaneously to S-phase synchronized non-transformed human hTERT-immortalized retinal pigment epithelial (hTERT-RPE) cells (Supplementary Figure S1C).
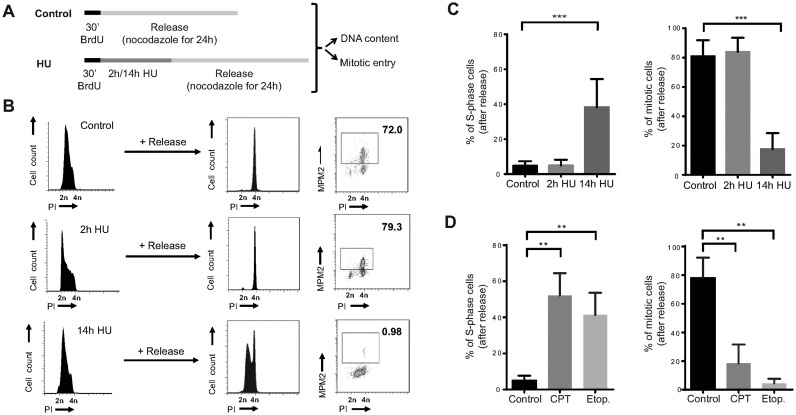
We next wondered if the observed cell cycle arrest in non-transformed human cells was reversible. To specifically study the response in S-phase, hTERT-RPE cells were pulse-labelled with bromodeoxyuridine (BrdU) before DNA synthesis was inhibited with 10 mM HU, which was shown to be the minimal dose that induced complete inhibition of DNA synthesis in our hands (Supplementary Figure S2A). Cell cycle resumption was determined by the ability of S-phase cells to restart DNA replication and to enter into mitosis after release from HU treatment in fresh media containing nocodazole (Figure 1A). Both control and briefly arrested S-phase cells were able to complete replication and enter into mitosis during the following 24 h. In contrast, prolonged treatment with HU prevented mitotic entry in 80% of the cells. Strikingly, half of the S-phase treated population was arrested with unreplicated DNA, indicating that not only mitotic entry but also replication resumption was compromised (Figure 1B and C). Similar results were obtained in another two non-transformed human cell lines, BJ-5ta (fibroblasts) and MCF10A (mammary epithelial cells) (Supplementary Figure S2B). Moreover, the same phenotype could be recapitulated after DNA damage induced by camptothecin (topoisomerase I inhibitor) and etoposide (topoisomerase II inhibitor) (Figure 1D), suggesting that severe replication stress, even the one induced by DSBs in S-phase, causes loss of replication recovery competence in non-transformed human cells.
Figure 1.
Replication resumption is compromised in non-transformed human cells after severe replication stress. (A) Schematic representation of the protocol used in (B) and (C) to analyse S-phase arrest and accumulative mitotic entry in hTERT-RPE cells after DNA replication inhibition. (B) Representative DNA profiles (PI: propidium iodide) and percentages of mitotic entry (analysed by MPM2 staining) are shown in the graphs. (C) The average percentages of BrdU positive cells that remain in S-phase (left panel) or that enter into mitosis (right panel) after release are shown in the graphs (n = 6). (D) hTERT-RPE cells were labelled during 30 min with BrdU before treating them during 14 h with camptothecin (CPT, 0.5 μM), etoposide (Etop., 50 μM) or left untreated (control). After the treatment, cells were released into fresh media with nocodazole during 24 h. DNA content and mitotic entry of BrdU positive population was analysed as in (B). The average percentages of cells that remain in S-phase (left panel) and mitosis (right panel) after release from BrdU positive population are shown in the graphs (n = 3). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (paired t-test, ** P < 0.01, *** P < 0.001).
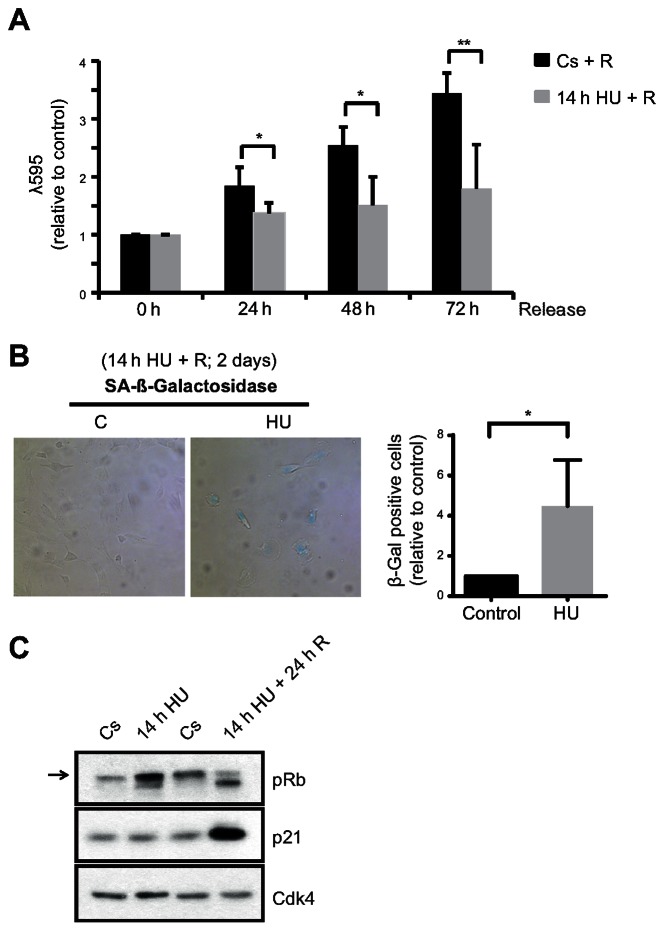
We next evaluated the long-term effect of a prolonged replication stress on cell cycle proliferation in hTERT-RPE cells. Interestingly, proliferation was impaired even 3 days after release from a 14-h HU treatment (Figure 2A). In agreement with the induction of permanent arrest in S-phase, several markers of senescence were observed in cells that had suffered prolonged replication stress, including β-galactosidase activity, p21 induction and hypophosphorylation of Retinoblastoma protein (pRb) (Figure 2B and C).
Figure 2.
hTERT-RPE cells become senescent after prolonged DNA replication inhibition. (A) S-phase synchronized (by single thymidine block) hTERT-RPE cells were treated during 14 h with HU or left untreated (Cs) and then released into fresh media. Cells were harvested at the indicated time after the treatment and stained with crystal violet (λ595) to analyse cell proliferation. The average fold increase relative to control in each time point is shown in the graphs (n = 4). (B, C) hTERT-RPE cells were synchronized in S-phase, treated during 14 h with HU or left untreated (C: control) and then harvested (for western blot) or released (R) into fresh media during 24 h (for western blot) or 48 h (for SA-β-Gal analysis). Representative images of SA-β-Gal staining (B, left panel), the average fold increase in SA-β-Gal positive cells relative to control (B, right panel) (n = 4), and whole cell lysate immunoblots (C) are shown. The arrow indicates the hyperphosphorylated band of pRb (Retinoblastoma protein). Cdk4 was used as loading control. Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (unpaired t-test, *P < 0.05, **P < 0.01).
Loss of replication recovery competence in response to prolonged DNA replication inhibition is APC/CCdh1 dependent
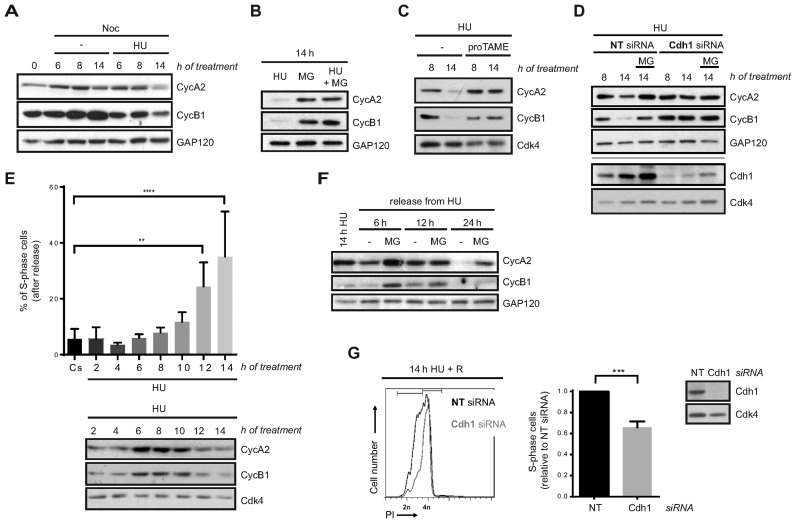
In response to DNA synthesis blockade, DNA replication and mitotic entry are prevented by inhibition of Cyclin A- and Cyclin B-associated Cdk activities, respectively (18–22). Therefore, we studied Cyclin A2 and Cyclin B1 levels in our experimental conditions. To this end, hTERT-RPE cells were synchronized in S-phase and either left untreated or treated with HU in the presence of nocodazole. As expected, unperturbed S-phase synchronized cells accumulated both cyclins as they progressed into G2 and entered into mitosis. In contrast, accumulation of Cyclin B1 was slower in HU-treated cells, as previously described (63). Surprisingly, after prolonged DNA synthesis inhibition by HU both Cyclin A2 and Cyclin B1 protein levels started to decrease although cells were still arrested in S-phase (Figure 3A and Supplementary Figure S2A). This decrease was due to proteasomal degradation as the addition of MG132 during the last 6 h of HU treatment restored their levels (Figure 3B). This was also observed when cells were synchronized by serum starvation and after damage induction in S-phase by different agents (Supplementary Figure S3A and B). Importantly, the same phenotype was observed in two additional non-transformed human cell lines, BJ-5ta and MCF10A (Supplementary Figure S3C).
Figure 3.
Loss of DNA replication recovery competence in response to prolonged replication inhibition is APC/CCdh1- dependent. (A, B, C) hTERT-RPE cells were synchronized in S-phase by single thymidine block and then treated during the indicated time with HU or left untreated (-). Whole cell extracts were analysed by western blot with the indicated antibodies. (D) Non-target (NT) or Cdh1 siRNA were transfected to hTERT-RPE cells before S-phase synchronization by single thymidine block. Cells were then treated with HU during the indicated time. Whole cell extracts were prepared and analysed by western blot with the indicated antibodies. (E) S-phase synchronized hTERT-RPE cells were treated during the indicated time with HU or left untreated (Cs) and then harvested for western blot or released into fresh media with nocodazole during 24 h. DNA content was used to determine the number of cells that remain in S-phase after release (upper panel) (n = 4). Whole cell extracts were prepared and analysed by western blot with the indicated antibodies (bottom panel). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (paired t-test, **P < 0.01, ****P < 0.0001). (F) S-phase synchronized hTERT-RPE cells were treated during 14 h with HU and then harvested or released into fresh media during the indicated time. Whole cell extracts were prepared and analysed by western blot with the indicated antibodies. (G) hTERT-RPE cells were transfected with the indicated siRNA and then synchronized in S-phase. Whole cell lysates of S-phase synchronized untreated cells were harvested for western blot analysis with the indicated antibodies (right panel). S-phase synchronized cells were treated during 14 h with HU and then released (R) into fresh media with nocodazole during 24 h. DNA content (PI: propidium iodide) was analysed by flow cytometry to quantify the number of cells that remain arrested in S-phase after release. Representative DNA profiles (left panel) and the average fold increase of S-phase arrested cells in Cdh1-depleted relative to non-target siRNA transfected cells are shown in the graph (middle panel) (n = 5). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (unpaired t-test, ***P < 0.001). Where indicated, nocodazole (noc), proTAME and MG132 (MG, added during the last 6 h of treatment) were added. Cyc: cyclin. GAP120 and Cdk4 were used as loading control.
Cyclin A2 and Cyclin B1 are substrates of the APC/C ubiquitin ligase (47), which, in complex with its regulatory subunit Cdh1, has been implicated in the DNA damage checkpoint response in G2 (44–46). We then hypothesized that this E3-Ubiquitin ligase could be activated in S-phase after severe replication stress. Addition of the APC/C inhibitor proTAME (64) during HU treatment restored Cyclin A2 and Cyclin B1 levels, proving the role of APC/C in this process (Figure 3C). Moreover, Cdh1 depletion prevented Cyclin A2 and Cyclin B1 degradation after a sustained HU treatment (Figure 3D and Supplementary Figure S3D). Notice that in this case cells were synchronized in S-phase before Cdh1 depletion was evident, thus preventing DNA damage due to Cdh1 knockdown by itself (65–67). Interestingly, APC/CCdh1 activity was not specifically directed to the two cyclins, as more of its substrates, such as Plk1 and Aurora A kinases, were also degraded after prolonged HU treatment (Supplementary Figure S3E). Moreover, Cyclin A2 and Cyclin B1 levels were considerably restored after Cdh1 depletion also in camptothecin- or etoposide-treated cells (Supplementary Figure S3F) altogether indicating that APC/CCdh1 is activated in S-phase upon severe replication stress.
We noticed that APC/CCdh1 activation, measured by Cyclin A2 and Cyclin B1 degradation, started ∼12 h after HU addition, and that this timing correlated with the loss of replication recovery competence (Figure 3E). It should be taken into account that thymidine synchronization had no effect on replication restart (Figures 1 and 3E). In addition, Cyclin A2 and Cyclin B1 were still degraded following release from HU treatment, suggesting that APC/CCdh1 activity was maintained even when replication stress was removed and could be responsible for impaired resumption of the cell cycle (Figure 3F). We then predicted that inhibition of its activity could overcome the loss of replication recovery after prolonged HU treatment. In agreement with this, Cdh1 depletion significantly rescued DNA synthesis after sustained DNA replication inhibition (Figure 3G and Supplementary Figure S3G). Thus, APC/CCdh1 activity prevents DNA replication recovery after severe replication stress.
APC/CCdh1 activation in S-phase upon prolonged HU treatment is independent of ATR/ATM activity, but abrogated in the absence of p53 and p21
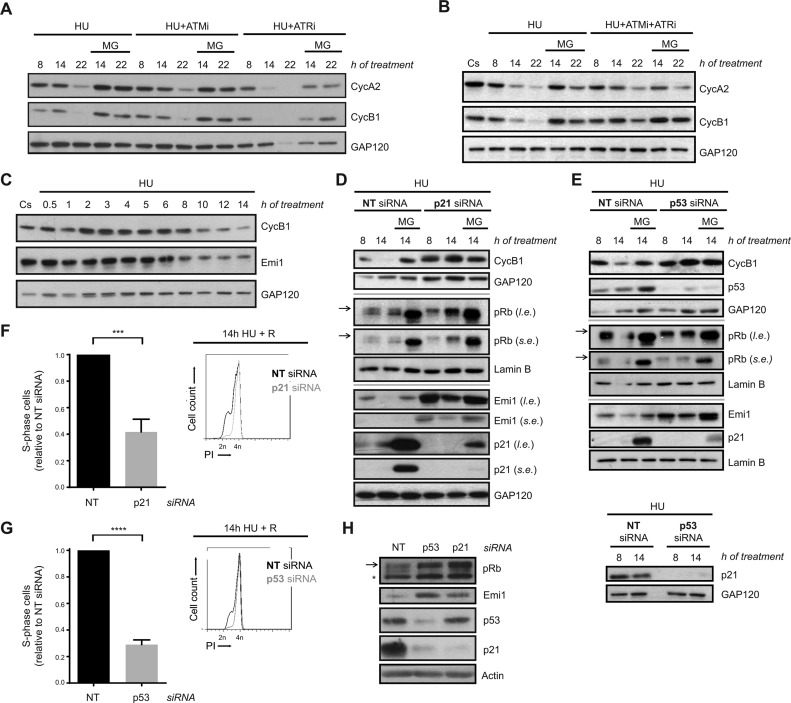
To understand the mechanism of APC/CCdh1 activation in S-phase after severe replication stress, we first analysed the contribution of ATR and ATM, the main regulators of the intra-S-phase checkpoint response, to this process. Pharmacological inhibition of ATR and ATM, alone or in combination (with VE 821 and KU-55933, respectively), did not prevent APC/CCdh1 activation in response to prolonged replication stress, as proteasome inhibition rescued Cyclin A2 and Cyclin B1 levels in all conditions (Figure 4A, B and Supplementary Figure S4A). However, while ATR inhibition alone resulted in advanced cyclins degradation, ATM inhibition did not modify Cyclin A2 and Cyclin B1 degradation dynamics. In agreement with that, Chk1 inhibition (with CHIR-124) also caused early APC/CCdh1 activation in response to HU (Supplementary Figure S4B). In this sense, APC/CCdh1 activation in response to prolonged HU treatment correlated with a decrease in Chk1 activity (as measured by its autophosphorylation at S296 (68)) (Supplementary Figure S4C). Collectively, these data indicate that ATR and ATM are not essential for APC/CCdh1 activation after prolonged replication stress, although they may modify its activation kinetics. In this regard, and consistent with APC/CCdh1 activity being essential for the loss of replication recovery competence, inhibition of these kinases could not restore DNA replication upon release from HU treatment (Supplementary Figure S4D).
Figure 4.
APC/CCdh1 activation in S-phase after prolonged HU treatment is independent of ATR/ATM activity, but abrogated in the absence of p53 and p21. (A, B) hTERT-RPE cells were synchronized in S-phase by single thymidine block and treated with HU +/- ATR (VE 821 (ATRi)) and ATM (KU-55933 (ATMi)) inhibitors alone (A) or in combination (B) during the indicated time. Whole cell lysates were analysed by western blot with the indicated antibodies. (C) hTERT-RPE cells were synchronized in S-phase and then treated with HU during the indicated time or left untreated. Whole cell lysates were analysed by western blot with the indicated antibodies. (D, E) hTERT-RPE cells were transfected with the indicated siRNA (NT: non-target) and synchronized in S-phase by single thymidine block. Cells were then treated with HU for the indicated time. Whole cell lysates were analysed by western blot with the indicated antibodies. (F, G) hTERT-RPE cells were transfected and synchronized as in (D, E) and then treated during 14 h with HU. Cells were then released (R) into fresh media with nocodazole during 24 h. DNA content (PI: propidium iodide) was analysed by flow cytometry to quantify the percentage of cells that remain arrested in S-phase after release. Representative DNA profiles (right panel) and the average fold increase of S-phase arrested cells in p21-/p53-depleted cells relative to non-target siRNA transfected cells (left panel) are shown in the graphs (n = 4). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (unpaired t-test, ***P < 0.001, ****P < 0.0001). (H) Whole cell lysates of transfected S-phase synchronized untreated cells were harvested for western blot analysis with the indicated antibodies. Arrows indicate the hyperphosphorylated band of pRb (Retinoblastoma protein). The asterisk indicates a non-specific band. Where indicated, proteasome inhibitor MG132 (MG) was added during the last 6 h of treatment. Cs: S-phase synchronized untreated cells. Cyc: cyclin. GAP120, Lamin B and actin were used as loading control. l.e.: long exposure. s.e.; short exposure.
It has been reported that the presence of Emi1 is important for keeping APC/CCdh1 inhibited during S- and G2-phases (69). Accordingly, its transcription, which is driven by E2F, starts at the G1/S transition and Emi1 levels accumulate until it is degraded at the beginning of mitosis (70,71). In this sense, we wondered whether Emi1 was downregulated after severe replication stress. Indeed, Emi1 levels decreased upon HU treatment, correlating with the timing of APC/CCdh1 activation (Figure 4C).
After DNA damage, p21 reduces Emi1 expression through the regulation of pRb phosphorylation, promoting APC/CCdh1 activation in G2 (55). Additionally, p53, the upstream regulator of p21, has also been implicated in this function (46). Thus, we analysed the contribution of p53 and p21 to APC/CCdh1 activation in S-phase after replication stress. Prolonged HU treatment induced an increase in p53 levels similar to what is observed after camptothecin or etoposide treatment, whereas no induction of p21 was observed under the same conditions (Supplementary Figure S4E). However, silencing of p53 or p21 prevented Cyclin B1 degradation upon severe replication stress (Figure 4D and E) and, in agreement with our previous results, promoted replication recovery after HU removal (Figure 4F and G). Consistent with Emi1 being the determining factor for APC/CCdh1 activity in our conditions, p53 or p21 depletion induced an increase in pRb hyperphosphorylation both during a normal and a perturbed S-phase that, as expected, correlated with an increase in Emi1 levels (Figure 4D, E and H). Nevertheless, even in the absence of p53 or p21, Emi1 was degraded in response to a prolonged HU treatment as MG132 treatment restored its levels (Figure 4D and E).
Together, these results suggest that Emi1 degradation is the critical event for APC/CCdh1 activation. In addition, p53- or p21-deficient cells may become more resistant to the activation of this ubiquitin ligase upon prolonged DNA replication stress due to the presence of higher Emi1 levels, although these proteins are unlikely the molecular determinants of this process.
Lack of APC/CCdh1 activation after prolonged replication inhibition increases genomic instability
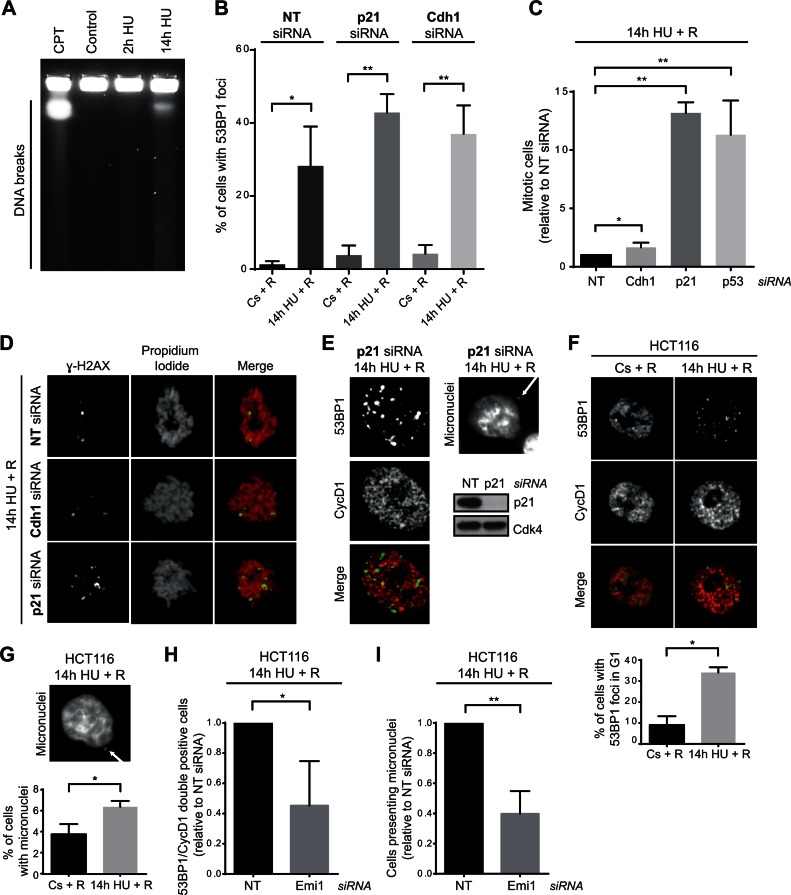
Our results showing that APC/CCdh1 was activated in S-phase preventing the resumption of replication after severe replication stress, prompted us to analyse the consequences on genomic instability of cell cycle resumption under this condition. Interestingly, DNA breakage analysis by pulse field gel electrophoresis (PFGE) and γ-H2AX/53BP1 foci presence indicated that replication forks were being processed into DSBs in response to severe replication stress (Figure 5A and Supplementary Figure S5A). Moreover, 53BP1 foci were still detected 12 h after HU removal (Figure 5B). Abrogation of APC/CCdh1 activation by p21 or Cdh1 depletion did not significantly affect the amount of cells presenting 53BP1 foci under these conditions. However, our previous results showed that while non-target siRNA transfected cells were arrested in S-phase, Cdh1 depletion allowed complete DNA duplication (Figure 3G and Supplementary Figure S3G), indicating that these cells progress to G2 with damaged DNA. In this sense, although silencing of Cdh1 slightly increased mitotic entry, the majority of cells remained arrested in G2-phase (Figure 5C and Supplementary Figure S5B). This suggests that damage caused by prolonged replication stress can be sensed by the G2 checkpoint, and that APC/CCdh1 inactivation alone cannot overcome this response. Accordingly, silencing of p53 or p21 substantially increased mitotic entry after HU removal (Figure 5C). Additionally, p53 or p21 depletion resulted in strong abrogation of senescence, while the fate of Cdh1-depleted cells did not change (Supplementary Figure S5C). Remarkably, in all cases, those cells that escaped G2/M arrest and entered into mitosis were positive for the DNA damage marker γ-H2AX (Figure 5D), highlighting the importance of inhibition of cell cycle resumption after severe replication stress. Moreover, in the absence of p21, where mitotic entry was strongly restored (Figure 5C), G1 cells (Cyclin D1 positive) with 53BP1 foci and cells with micronuclei were observed after release from HU (Figure 5E). Thus, inhibition of replication resumption by APC/CCdh1 activation in S-phase might be important to prevent genomic instability by adding a barrier prior to the G2/M response.
Figure 5.
Lack of APC/CCdh1 activity after severe replication stress correlates with an increase in genomic instability. (A) Single thymidine block synchronized S-phase hTERT-RPE cells were treated during the indicated time with HU or left untreated (control), and then harvested for DNA breakage analysis by PFGE. Cells treated during 14 h with camptothecin (CPT, 0.5 μM) were used as positive control. A representative image from three independent experiments is shown. (B) siRNA transfected and S-phase synchronized (by single thymidine block) hTERT-RPE cells were treated with HU during 14 h or left untreated (Cs), and then released (R) into fresh media during 12 h. Cells were fixed and immunostained with 53BP1 antibody. The average percentage of cells with 53BP1 foci (>6 foci) from total population are shown (n = 4). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (paired t-test, *P < 0.05, **P < 0.01). (C) siRNA transfected and S-phase synchronized (as in Figures 3G, 4F and G) hTERT-RPE cells were treated with HU during 14 h and released into fresh media with nocodazole during 24 h. The number of mitosis (MPM2 positive cells) was quantified by flow cytometry. The average fold increase of mitotic cells in Cdh1-/p21-/p53-depleted cells relative to non-target (NT) siRNA transfected cells is shown in the graph (n = 4). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (unpaired t-test, *P < 0.05, **P < 0.01). (D) hTERT-RPE cells were transfected and synchronized as in (B). S-phase synchronized cells were treated with HU during 14 h and then released into fresh media with nocodazole during 24 h before fixing and immunostaining them with χ-H2AX antibody. DNA was counterstained with propidium iodide. Representative images are shown. (E) siRNA transfected and S-phase synchronized hTERT-RPE cells were treated during 14 h with HU, and then released into fresh media during 12 h. Finally, cells were fixed and immunostained with 53BP1 and Cyclin D1 (CycD1) antibodies (left panel). Cells were counterstained with DAPI to analyse the presence of micronuclei (upper-right panel). Untreated S-phase synchronized cells were harvested and whole cell lysates were prepared to analyse them by western blot with the indicated antibodies (bottom-right panel). Cdk4 was used as loading control. (F, G) HCT116 cells were synchronized in S-phase by single thymidine block before treating them with HU during 14 h or left untreated. Cells were then released into fresh media during 12 h after which cells were fixed and immunostained with 53BP1 and Cyclin D1 antibodies. Representative images (F, upper panel) and the average percentage of cells with 53BP1 foci from G1 (Cyclin D1 positive) population (F, bottom panel) are shown (n = 3). DNA was counterstained with DAPI to analyse the presence of micronuclei. A representative image (G, upper panel) and the average percentage of cells with micronuclei (G, bottom panel) are shown (n = 3). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (paired t-test *P < 0.05). (H, I) HCT116 cells were transfected with the indicated siRNA and 4 h later thymidine was added to synchronize cells in S-phase. After synchronization, cells were treated with HU and immunostained for 53BP1 and Cyclin D1 analysis as in (F, G). The average percentage of cells with 53BP1 foci in G1 (H) and the average percentage of cells with micronuclei (I) are shown (n = 3). Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (unpaired t-test, *P < 0.05, **P < 0.01).
To further study the contribution of APC/CCdh1 activation towards safeguarding genome integrity, we aimed to analyse the effect of replication inhibition on genomic instability in cells that, in contrast to hTERT-RPE, do not activate APC/CCdh1 upon severe replication stress. As tumour cells usually present a less robust replication checkpoint response (58,62), activation of this ubiquitin ligase was first determined in a panel of nine tumour cell lines. Interestingly, from those, only one (MCF7) activated APC/CCdh1 (as measured by Cyclin A2 and Cyclin B1 degradation), indicating that tumour cell lines are predominantly deficient in APC/CCdh1 activation in S-phase in response to HU (Supplementary Figure S6A). In agreement with our previous results, this lack of APC/CCdh1 activity strongly correlated with the ability to restore DNA synthesis after severe replication stress, as six out of these eight lines were able to reinitiate DNA replication when released from prolonged HU treatment (Supplementary Figure S6B). Among them we chose HCT116 cells to study the role of APC/CCdh1 in maintaining genomic integrity upon severe replication stress. First, we validated that a high percentage of HCT116 cells were able to completely resume cell cycle once released from prolonged HU treatment, as indicated by their capacity to enter into mitosis and form colonies (Supplementary Figure S6C and D). Interestingly, a high proportion of HCT116 cells that reentered the cell cycle under these conditions acquired DNA damage and chromosome instability as measured by the increase in the percentage of G1 cells with 53BP1 foci and the percentage of cells presenting micronuclei (Figure 5F and G). To analyse if the acquisition of this instability might be abrogated by APC/CCdh1 activation in S-phase, Emi1 was depleted in synchronized HCT116 cells. As expected, Emi1 depletion during S-phase allowed the cells to activate APC/CCdh1 (Supplementary Figure S6E). In agreement with our results, resumption of DNA replication after prolonged HU treatment was compromised in Emi1-depleted cells (Supplementary Figure S6F), and consequently, the percentage of cells with 53BP1 foci in G1 decreased (Figure 5H). Likewise, a reduction in the number of cells presenting micronuclei was also observed under these conditions (Figure 5I). Collectively, these data further support the idea that APC/CCdh1 activation in S-phase in response to severe replication stress may act as an additional barrier to prevent cell cycle progression with damaged DNA, consequently contributing to safeguarding the genome integrity.
APC/CCdh1 activation in S-phase in response to severe replication stress inhibits new origin firing
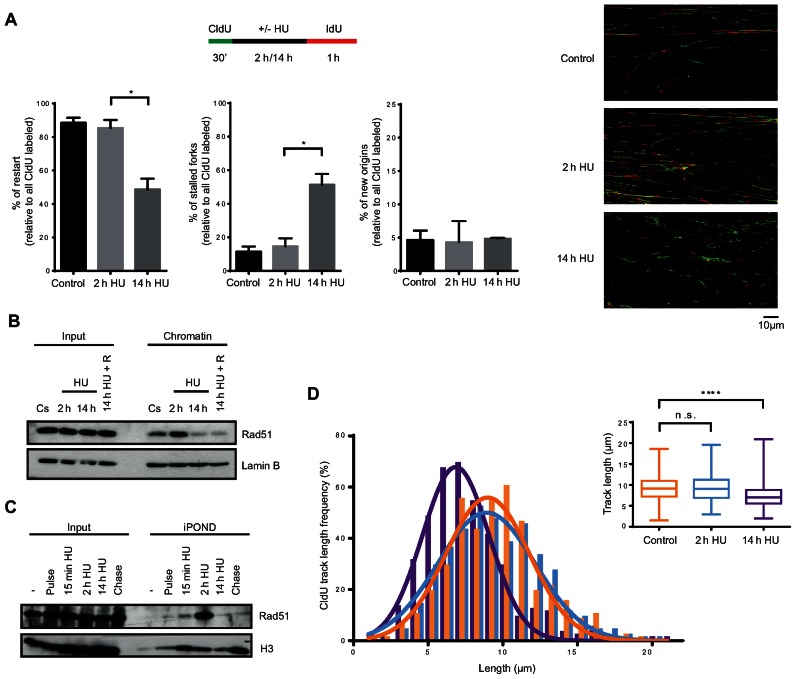
As shown above, hTERT-RPE cells are able to reinitiate S-phase after a 2-h HU treatment while this recovery is abolished after a 14-h HU treatment, when replication forks are being processed into DSBs and APC/CCdh1 is activated (Figures 1, 3E, 5A and Supplementary Figure S5A). Resumption of replication in the presence of DSBs has been reported to involve either HR mechanisms, which depend on the presence of Rad51-coated 3′ ssDNA ends, or new origin firing (26,27,72–74). To analyse whether any of these mechanisms are inhibited by APC/CCdh1 activation in S-phase, we first decided to better characterize the loss of replication recovery competence in hTERT-RPE cells. To this end, we monitored replication by single molecule DNA fibre analysis after a 2- or 14-h HU treatment. Cells were pulse-labelled with 5-chlorodeoxyuridine (CldU) for 30 min, followed by 2 h or 14 h of exposure to HU, and labelled again for a period of 60 min with 5-iododeoxyuridine (IdU). The second labelling period was extended to avoid any interference from a possible delay in restarting stalled forks in the analysis. Additionally, as a slight replication fork progression was observed during the first 15 min of HU treatment (Supplementary Figure S7A), we decided to maintain the CldU in the media during this time to avoid progression of the forks without CldU incorporation. The analysis of IdU incorporation during the rest of HU treatment confirmed that replication was completely blocked after these 15 min (Supplementary Figure S7A). As shown in Figure 6A, after 2 h HU almost all forks were able to restart DNA synthesis (the same proportion as untreated cells), whereas half of the active replication forks were stalled upon 14 h of HU treatment. Interestingly, lack of replication fork restart was not compensated by an increment in new origin firing, suggesting that this pathway was inhibited under these conditions. Moreover, the absence of gaps between green and red tracks at restarted forks confirmed that replication forks were completely blocked in the presence of HU, and allowed us to confidently quantify the number of new origins (only-red/ IdU tracks) in each condition.
Figure 6.
Replication forks of hTERT-RPE cells become inactivated after prolonged DNA replication inhibition. (A) Labelling protocol for DNA fibre analysis (upper panel). S-phase synchronized (by single thymidine block) hTERT-RPE cells were treated as indicated and then DNA fibres were prepared and labelled with anti-BrdU antibodies. The percentage of replication fork restart, stalled replication forks and new origin firing relative to total CldU labelled fibres are shown in the graphs. At least 1500 fibres from three independent experiments were counted in each condition. Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (paired t-test, *P < 0.05). Representative images of each condition are shown (right panel). (B) S-phase synchronized hTERT-RPE cells were treated during the indicated time with HU or left untreated (Cs). Cells were harvested just after the treatment or after a 30 min release into fresh media (14 h HU + R). Chromatin extracts were prepared and analysed by western blot with the indicated antibodies. Input: whole cell lysates. Lamin B was used as loading control. (C) hTERT-RPE cells were synchronized in S-phase and labelled with EdU during 15 min before treating them with HU during the indicated time. EdU was present in the media for additional 15 min in HU-treated cells. Isolated proteins on nascent DNA (iPOND) were analysed by western blot with the indicated antibodies. Input: nuclear extracts. Pulse: cells were harvested just after EdU labelling. Chase: 15 min EdU + 2 h thymidine (50 μM) chase. (-): no EdU. (D) DNA fibres from (A) were used to measure the CldU track length. Three hundred fibres were counted in each condition. CldU track length distribution and statistical analysis of CldU track distribution are shown. Box and whiskers show Min, Max, Median and first quartiles. Values marked with asterisks are significantly different (unpaired t-test, n.s.: non-statistically significant, ****P < 0.0001).
To further analyse how replication fork restart is lost after prolonged DNA replication inhibition, we monitored Rad51 association with chromatin and replication forks (by iPOND) after short and long HU treatments. Rad51 was present in chromatin and replication forks during unperturbed S-phase and its association increased after a short (2 h) HU treatment as previously described (26,75). In contrast, Rad51 disassembled from both chromatin and replication forks after prolonged inhibition of DNA synthesis, when DSBs were already present (Figures 6B, C, 5A and Supplementary Figure S5A) and was not reestablished even after release from HU (Figure 6B), suggesting that Rad51-dependent replication fork restart was impaired after prolonged, but not short, HU treatment in hTERT-RPE cells. In agreement with the already known role of Rad51 in protecting ssDNA (75–77), and with the fact that it was dissociated from replication forks after a 14-h HU treatment, CldU-labelled tracks were shorter after prolonged HU treatment, suggesting that DNA was degraded at stalled replication forks after sustained replication stress (Figure 6D).
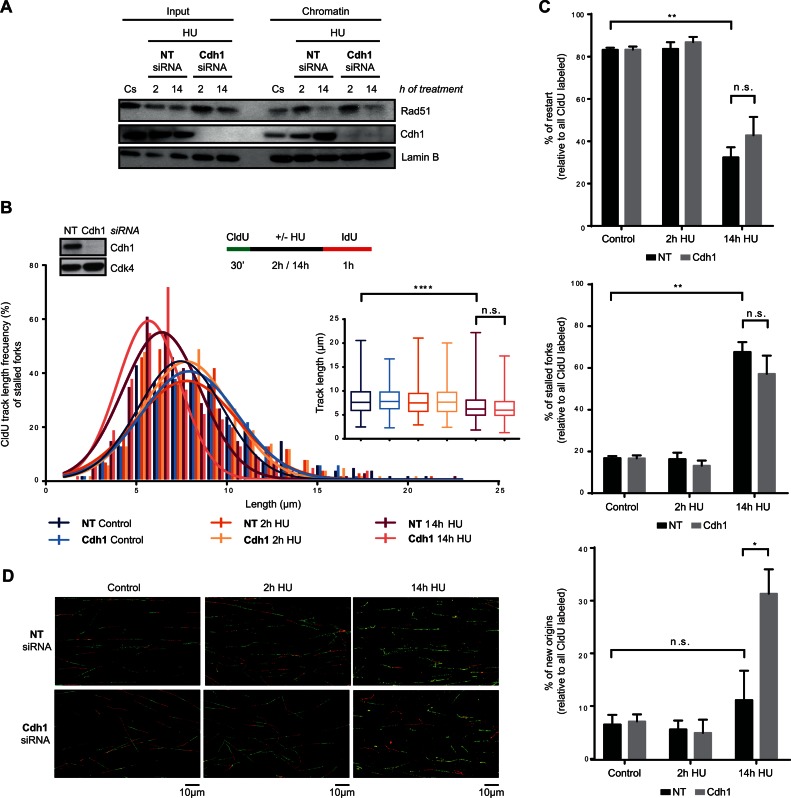
To determine how Cdh1 depletion led to the recovery of DNA replication, and consequently discern the function of APC/CCdh1 in S-phase in response to severe replication stress, we first analysed the association of Rad51 with chromatin after Cdh1 depletion. As shown in Figure 7A, Cdh1 knockdown did not rescue Rad51 levels in chromatin after a 14-h HU treatment. Consistent with this, CldU track length shortening was also observed at stalled but not restarted forks of Cdh1-depleted cells (Figure 7B and Supplementary Figure S7B). Likewise, DNA fibre analysis showed that the percentage of restarted forks did not increase after Cdh1 depletion (Figure 7C and D), altogether indicating that Rad51-dependent HR is not the mechanism used by Cdh1-depleted hTERT-RPE cells to resume S-phase after prolonged HU treatment. We then wondered whether the role of APC/CCdh1 in S-phase is to inhibit new origin firing after prolonged replication stress. As expected, the percentage of stalled forks remained unchanged while an increase in the number of new origin firing events was observed in Cdh1-depleted cells compared to non-target transfected ones (Figure 7C and D). Furthermore, the percentage of active forks (restart + new origin) after a 14-h HU treatment relative to initial ones (all CldU labelled forks) was ∼80% in Cdh1-depleted cells, indicating that the observed increase in new origin firing might be sufficient to restore DNA synthesis after prolonged inhibition of replication in these cells (Figure 7C and D). Accordingly, in the absence of Cdh1, the IdU incorporation rate (measured by the average IdU intensity per nucleus) was similar in S-phase cells treated for a short and a longer time (Supplementary Figure S7C), which supports the idea that the new origin firing observed in the absence of APC/CCdh1 activation is sufficient to reestablish the DNA replication rate. It should be considered that with the methodology used, firing of nearby dormant origins (78) cannot be distinguished from real fork restart and thus, the observed new firing events may correspond mainly to those activated in different replicons.
Figure 7.
APC/CCdh1 inhibits new origin firing in S-phase after prolonged HU treatment. (A) Non-target (NT) or Cdh1 siRNA were transfected to hTERT-RPE cells before synchronizing them in S-phase by single thymidine block. Cells were then treated with HU (or left untreated (Cs)) during the indicated time. Chromatin extracts were prepared and analysed by western blot with the indicated antibodies. Input: whole cell lysates. Lamin B was used as loading control. (B, C, D) Schematic of the labelling protocol (B, upper-right panel). hTERT-RPE cells were transfected with the indicated siRNA and synchronized in S-phase before CldU (250 μM) labelling. Whole cell lysates of S-phase synchronized untreated cells were harvested for western blot analysis with the indicated antibodies (B, upper-left panel). Cdk4 was used as loading control. CldU track length of 300 fibres from three independent experiments was measured. CldU track length distribution and statistical analysis are shown (B, bottom panel). Box and whiskers show Min, Max, Median and first quartiles. Values marked with asterisks are significantly different (unpaired t-test, n.s.: non-statistically significant, ****P < 0.001). The percentage of replication fork restart, stalled replication forks and new origin firing relative to total CldU labelled fibres is shown in the graphs (C). More than 1000 fibres from three independent experiments were counted in each condition. Means and standard deviation (bars) are shown. Values marked with asterisks are significantly different (paired t-test, n.s.: non-statistically significant, *P < 0.05, **P < 0.01). Representative images of each condition are shown (D).
Altogether, our results show that APC/CCdh1 is activated in S-phase after prolonged HU treatment, preventing new origin firing in hTERT-RPE cells and thus contributing to the inhibition of replication resumption after severe replication stress.
DISCUSSION
Recovery from mild replication stress can be achieved with a low risk of introducing changes into the genome, whereas after severe replication stress that induces fork collapse, reinitiation of DNA replication may promote genomic alterations and epigenetic changes (28,30,33–37,72). Thus, although recovery from severe replication stress might be beneficial for unicellular organisms, as it will allow further proliferation, it would not be advantageous for multicellular organisms. In this sense, based on studies mainly done in tumour cells, it has been proposed that under these circumstances reinitiation of DNA replication occurs simply to allow cells to arrive at G2, where DNA damage is expected to be repaired, and if this fails, cells will be driven to apoptosis or senescence (79). Data presented here show that after prolonged DNA replication inhibition that induces DSBs in S-phase, non-transformed human cells are not competent to resume DNA replication and progress into G2 due to APC/CCdh1 activation, which leads to a permanent cell cycle exit from S-phase. Activation of APC/CCdh1 during G2 as a long-term response to genotoxic agents has been described (45,46,55), but to our knowledge, this is the first work showing APC/CCdh1 activation during S-phase in response to severe replication stress.
In agreement with previous data from tumour cells, hTERT-RPE cells reinitiate DNA synthesis by fork restart following a short HU treatment, while after prolonged HU treatment, which leads to DSBs accumulation, fork restart is strongly abrogated (26). Strikingly, whereas in tumour cells new origin firing allows reactivation of replication after prolonged HU treatment, we show here that origin firing is inhibited in hTERT-RPE cells, and consequently, the resumption of DNA replication upon HU removal is strongly compromised. Interestingly, our data indicates that APC/CCdh1 activation in S-phase after severe replication arrest is the responsible for inhibition of new origin firing during recovery. The kinases Cdk2/Cyclin A (80) and Cdc7/Dbf4 (81) are essential for the induction of origin firing in S-phase, both regulatory subunits being well-known APC/CCdh1 substrates (82,83). In this sense, we have shown that Cyclin A2, together with other APC/CCdh1 substrates, is degraded after prolonged treatment with HU as well as upon its removal. Therefore, we propose that the role of APC/CCdh1 in preventing new origin firing during recovery is mediated by Cyclin A degradation, although we cannot exclude that degradation of other substrates, such as Dbf4, is also implicated.
Rad51 is essential to protect ssDNA and mediate fork restart at stalled replication forks (72,75–77). Accordingly, the association of Rad51 with replication forks increased upon short HU treatment, after which replication was reinitiated. However, after prolonged HU treatment Rad51 dissociated from replication forks. Consistent with previous reports, we found that Rad51 dissociation correlated with nascent DNA degradation at stalled but not at restarted forks. Loss of Rad51 would explain the lack of fork restart under this condition, as both Rad51-mediated fork restart and BIR-mediated restart may be hindered. After prolonged HU treatment, Cdh1 depletion did not restore Rad51 recruitment to the chromatin, and neither did it have any effect on nascent DNA degradation, although, consistent with recently published data (84), CtIP nuclease was degraded by APC/CCdh1 (unpublished observation). Thus, although we cannot rule out a role for APC/CCdh1 in controlling DNA processing and Rad51 dissociation at stalled replication forks, we propose that its main function in S-phase is to inhibit new origin firing and thereby prevent the only mechanism left to complete DNA replication.
Due to the relevance of APC/CCdh1 in cell cycle progression, evolution has led to a very tight regulation of its activity: inhibitory phosphorylation of Cdh1 by Cdk1 and Cdk2 (51,85,86) and inhibition by Emi1 binding, the transcription of which is regulated by pRb/ E2F (69). Its activation in G2 upon genotoxic stress has been shown to be ATM-, p53- and p21-dependent (46,55,56). In contrast, we show here that APC/CCdh1 activation in S-phase is not prevented by ATR/ATM inhibition. In relation to p53 and p21, although the lack of these proteins prevents APC/CCdh1 activation in S-phase, we suggest that this is probably due to the elevated Emi1 levels already present in untreated p53- and p21-depleted cells. Consistent with this, p21 protein levels were not upregulated after a 14-h HU treatment, at which point APC/CCdh1 substrates degradation was readily detected. Thus, we propose that Emi1 degradation is more likely the trigger of APC/CCdh1 activation in S-phase. Interestingly, it has recently been shown that Chk1 can phosphorylate and inhibit Cdh1 activity after replication stress (82,87). Notably, APC/CCdh1 activation by prolonged HU treatment correlated in time with a decrease in Chk1 phosphorylation. Thus, it is also conceivable that a decrease in Chk1 activity collaborates in the initial trigger of APC/CCdh1 activation. Consistent with this, ATR or Chk1 inhibition resulted in advanced APC/CCdh1 activation. Nevertheless, it should be considered that Chk1 could also have an activating effect on APC/CCdh1 through Cdc25 inhibition and hence Cdk inactivation.
Additionally, the advanced APC/CCdh1 activation observed upon inhibition of ATR/Chk1 may also be caused by an increase in the number of DSBs. In this sense, ATR/Chk1 activation during replication stress prevents DSB generation, by inhibiting origin firing and Mus81 nuclease, among others (14,88). A decrease in Chk1 activity can occur through different mechanisms, such as Chk1 and Claspin degradation (89,90). Although we did not specifically analyse the mechanism by which Chk1 is inactivated, we observed a decrease in Chk1 autophosphorylation mainly after prolonged HU treatment, correlating with the processing of replication forks into DSBs. Chk1 inactivation in the presence of replication stress might be an adaptive mechanism to allow replication to continue through late origin firing or BIR-mediated restart. Since such mechanisms may induce epigenetic changes and genomic rearrangements, and taking into account that APC/CCdh1 activation correlates in time with Chk1 inactivation, we suggest that APC/CCdh1 inhibits new origin firing once Chk1 cannot perform this role.
After prolonged inhibition of replication, Cdh1-depleted cells duplicated their DNA despite having DNA lesions, and mostly arrested in G2. These data support recent findings indicating that, although APC/CCdh1 is activated in G2 upon DNA damage, p53 and p21 are sufficient to induce permanent cell cycle exit from G2 (42,43). Importantly, the few cells that eventually escape this G2 checkpoint enter into mitosis with damaged DNA (γ-H2AX foci). Furthermore, p21- and p53-depleted cells released from severe replication stress are able, on the one hand, to reinitiate replication due to the inability to activate APC/CCdh1 in S-phase, and on the other hand, to enter into mitosis due to the lack of a functional G2 checkpoint. Consequently, APC/CCdh1 activation in S-phase may work as an earlier additional mechanism to the G2 DNA damage checkpoint to avoid proliferation of cells with damaged or inaccurately replicated DNA.
In addition to the loss of ability to complete DNA duplication, we also observed the induction of different senescence markers, such as pRb hypophosphorylation, p21 upregulation and SA-β-Gal activity after prolonged inhibition of DNA replication. However, we were unable to analyse the real contribution of APC/CCdh1 to the induction of senescence in S-phase cells in our conditions, as Cdh1-depleted cells undergo senescence in G2 in a p53 and p21-dependent manner, and their depletion prevents APC/CCdh1 activation after severe replication stress. Nevertheless, several lines of evidence indicate that APC/CCdh1 activation by itself is sufficient to induce senescence (56,91,92). Thus, we suggest that APC/CCdh1 could reinforce the permanent cell cycle exit and senescence observed in S-phase.
It is now well accepted that oncogene expression induces replication stress leading to activation of the DNA damage response and senescence, which acts as tumourigenic barrier (8,9). Interestingly, our data suggest that APC/CCdh1 is a new element of this barrier. Thus, one should expect that avoiding APC/CCdh1 activation in S-phase would benefit tumour cells. Consistently, we found that tumour cell lines are predominantly deficient in APC/CCdh1 activation in S-phase in response to HU, and that this correlates with the ability to resume DNA replication after this treatment. In addition, data from HCT116 cells show that this may allow them to resume cell cycle and proliferate whilst bearing damaged DNA, as observed by the presence of 53BP1 foci in the next G1, and in chromosomal abnormalities such as micronuclei. Our data indicate that p53 inactivation, p21 down-regulation or Emi1 overexpression are, among others, alterations that may abrogate this novel replication stress response and consequently favour genomic instability and tumourigenesis. It has been described that Emi1 inhibition itself can activate APC/CCdh1 (93,94). Consistent with this reports, Emi1 depletion in synchronized HCT116 allowed APC/CCdh1 activation in S-phase. Interestingly, proving a role of APC/CCdh1 in preventing genomic instability upon replication stress, this resulted in impaired replication resumption after HU treatment and a decrease in the number of G1 cells with DSBs and cells with micronuclei.
In conclusion, our work provides evidence of the participation of APC/CCdh1 in inducing a permanent cell cycle exit from S-phase through inhibition of new origin firing after prolonged DNA replication stress, thereby contributing to the maintenance of genome integrity. Consequently, we propose that alterations in this response would be beneficial for tumour progression. Promisingly, and although further studies are needed, these differences in the response between non-transformed and tumour cells could provide an opportunity to develop new therapeutic strategies that specifically target cancer cells.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory for discussion and technical help and to advanced optical microscopy unit of CCiT-UB for technical assistance.
FUNDING
Ministerio de Economia y Competitividad [SAF2013–42742-R to N.A., SAF2013-49149-R to R.F.]; Red Temática de investigación cooperativa en cáncer [RD 12/0036/0049 to N.A.]; FPI-fellowship from Ministerio de ciencia e innovación (to A.E. and A.L.); FI-fellowship from Generalitat de Catalunya (to S.F.) Funding for open access charge: MINECO/SAF2013-42742-R and University of Barcelona contribution.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branzei D., Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 4.Dereli-Öz A., Versini G., Halazonetis T.D. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol. Oncol. 2011;5:308–314. doi: 10.1016/j.molonc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrell R.A., McClelland S.E., Endesfelder D., Groth P., Weller M.-C., Shaikh N., Domingo E., Kanu N., Dewhurst S.M., Gronroos E., et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaillard H., García-Muse T., Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 7.Macheret M., Halazonetis T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015;10:425–428. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- 8.Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.-V.F., Kolettas E., Niforou K., Zoumpourlis V.C., et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 9.Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P.G., Bensimon A., et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 10.Murga M., Campaner S., Lopez-Contreras A.J., Toledo L.I., Soria R., Montaña M.F., D'Artista L., Schleker T., Guerra C., Garcia E., et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shechter D., Costanzo V., Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 12.Syljuåsen R.G., Sørensen C.S., Hansen L.T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo C., Kumagai A., Schlacher K., Shevchenko A., Shevchenko A., Dunphy W.G. Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication. Mol. Cell. 2015;57:492–505. doi: 10.1016/j.molcel.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forment J. V, Blasius M., Guerini I., Jackson S.P. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS One. 2011;6:e23517. doi: 10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petermann E., Woodcock M., Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nam E.A., Cortez D. ATR signalling: more than meeting at the fork. Biochem. J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chini C.C.S., Chen J. Human claspin is required for replication checkpoint control. J. Biol. Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- 18.Peng C.Y., Graves P.R., Thoma R.S., Wu Z., Shaw A.S., Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez Y., Wong C., Thoma R.S., Richman R., Wu Z., Piwnica-Worms H., Elledge S.J. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 20.Mailand N., Falck J., Lukas C., Syljuâsen R.G., Welcker M., Bartek J., Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 21.Boutros R., Dozier C., Ducommun B. The when and wheres of CDC25 phosphatases. Curr. Opin. Cell Biol. 2006;18:185–191. doi: 10.1016/j.ceb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Kumagai A., Dunphy W.G. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol. Biol. Cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge X.Q., Blow J.J. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 2010;191:1285–1297. doi: 10.1083/jcb.201007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibarra A., Schwob E., Méndez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trenz K., Smith E., Smith S., Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 28.Smith C.E., Llorente B., Symington L.S. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 29.Errico A., Costanzo V. Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns. EMBO Rep. 2010;11:270–278. doi: 10.1038/embor.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anand R.P., Lovett S.T., Haber J.E. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X., Heyer W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lydeard J.R., Jain S., Yamaguchi M., Haber J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 33.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costantino L., Sotiriou S.K., Rantala J.K., Magin S., Mladenov E., Helleday T., Haber J.E., Iliakis G., Kallioniemi O.P., Halazonetis T.D. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaratiegui M., Castel S.E., Irvine D.V., Kloc A., Ren J., Li F., de Castro E., Marín L., Chang A.-Y., Goto D., et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Méchali M., Yoshida K., Coulombe P., Pasero P. Genetic and epigenetic determinants of DNA replication origins, position and activation. Curr. Opin. Genet. Dev. 2013;23:124–131. doi: 10.1016/j.gde.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Hiratani I., Gilbert D.M. Replication timing as an epigenetic mark. Epigenetics. 2009;4:93–97. doi: 10.4161/epi.4.2.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.López-Contreras A.J., Gutierrez-Martinez P., Specks J., Rodrigo-Perez S., Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J. Exp. Med. 2012;209:455–461. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johmura Y., Shimada M., Misaki T., Naiki-Ito A., Miyoshi H., Motoyama N., Ohtani N., Hara E., Nakamura M., Morita A., et al. Necessary and sufficient role for a mitosis skip in senescence induction. Mol. Cell. 2014;55:73–84. doi: 10.1016/j.molcel.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Baus F. Permanent cell cycle exit in G2 phase after DNA damage in normal human fibroblasts. EMBO J. 2003;22:3992–4002. doi: 10.1093/emboj/cdg387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charrier-Savournin F.B., Château M.-T., Gire V., Sedivy J., Piette J., Dulic V. p21-Mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Mol. Biol. Cell. 2004;15:3965–3976. doi: 10.1091/mbc.E03-12-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krenning L., Feringa F.M., Shaltiel I.A., van den Berg J., Medema R.H. Transient Activation of p53 in G2 Phase Is Sufficient to Induce Senescence. Mol. Cell. 2014;55:59–72. doi: 10.1016/j.molcel.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Müllers E., Silva Cascales H., Jaiswal H., Saurin A.T., Lindqvist A. Nuclear translocation of Cyclin B1 marks the restriction point for terminal cell cycle exit in G2 phase. Cell Cycle. 2014;13:2733–2743. doi: 10.4161/15384101.2015.945831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudo T., Ota Y., Kotani S., Nakao M., Takami Y., Takeda S., Saya H. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. EMBO J. 2001;20:6499–6508. doi: 10.1093/emboj/20.22.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiebusch L., Hagemeier C. p53- and p21-dependent premature APC/C-Cdh1 activation in G2 is part of the long-term response to genotoxic stress. Oncogene. 2010;29:3477–3489. doi: 10.1038/onc.2010.99. [DOI] [PubMed] [Google Scholar]
- 47.Peters J.-M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 48.Pesin J.A., Orr-Weaver T.L. Regulation of APC/C activators in mitosis and meiosis. Annu. Rev. Cell Dev. Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramer E.R., Scheuringer N., Podtelejnikov A. V, Mann M., Peters J.M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller J.J., Summers M.K., Hansen D. V, Nachury M. V, Lehman N.L., Loktev A., Jackson P.K. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lukas C., Sørensen C.S., Kramer E., Santoni-Rugiu E., Lindeneg C., Peters J.M., Bartek J., Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- 52.Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J.-M. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimann J.D.R. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 2001;15:3278–3285. doi: 10.1101/gad.945701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benmaamar R., Pagano M. Involvement of the SCF complex in the control of Cdh1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- 55.Lee J., Kim J.A., Barbier V., Fotedar A., Fotedar R. DNA damage triggers p21WAF1-dependent Emi1 down-regulation that maintains G2 arrest. Mol. Biol. Cell. 2009;20:1891–1902. doi: 10.1091/mbc.E08-08-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi A., Imai Y., Yamakoshi K., Kuninaka S., Ohtani N., Yoshimoto S., Hori S., Tachibana M., Anderton E., Takeuchi T., et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol. Cell. 2012;45:123–131. doi: 10.1016/j.molcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Borel F., Lacroix F.B., Margolis R.L. Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J. Cell Sci. 2002;115:2829–2838. doi: 10.1242/jcs.115.14.2829. [DOI] [PubMed] [Google Scholar]
- 58.Llopis A., Salvador N., Ercilla A., Guaita-Esteruelas S., Barrantes Idel B., Gupta J., Gaestel M., Davis R.J., Nebreda A.R., Agell N. The stress-activated protein kinases p38α/β and JNK1/2 cooperate with Chk1 to inhibit mitotic entry upon DNA replication arrest. Cell Cycle. 2012;11:3627–3637. doi: 10.4161/cc.21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Debacq-Chainiaux F., Erusalimsky J.D., Campisi J., Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 60.Méndez J., Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aranda S., Rutishauser D., Ernfors P. Identification of a large protein network involved in epigenetic transmission in replicating DNA of embryonic stem cells. Nucleic Acids Res. 2014;42:6972–6986. doi: 10.1093/nar/gku374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodríguez-Bravo V., Guaita-Esteruelas S., Salvador N., Bachs O., Agell N. Different S/M checkpoint responses of tumor and non tumor cell lines to DNA replication inhibition. Cancer Res. 2007;67:11648–11656. doi: 10.1158/0008-5472.CAN-07-3100. [DOI] [PubMed] [Google Scholar]
- 63.Florensa R., Bachs O., Agell N. ATM/ATR-independent inhibition of cyclin B accumulation in response to hydroxyurea in nontransformed cell lines is altered in tumour cell lines. Oncogene. 2003;22:8283–8292. doi: 10.1038/sj.onc.1207159. [DOI] [PubMed] [Google Scholar]
- 64.Zeng X., Sigoillot F., Gaur S., Choi S., Pfaff K.L., Oh D.-C., Hathaway N., Dimova N., Cuny G.D., King R.W. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.García-Higuera I., Manchado E., Dubus P., Cañamero M., Méndez J., Moreno S., Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 66.Greil C., Krohs J., Schnerch D., Follo M., Felthaus J., Engelhardt M., Wäsch R. The role of APC/CCdh1 in replication stress and origin of genomic instability. Oncogene. 2015 doi: 10.1038/onc.2015.367. doi:10.1038/onc.2015.367. [DOI] [PubMed] [Google Scholar]
- 67.Sigl R., Wandke C., Rauch V., Kirk J., Hunt T., Geley S. Loss of the mammalian APC/C activator FZR1 shortens G1 and lengthens S phase but has little effect on exit from mitosis. J. Cell Sci. 2009;122:4208–4217. doi: 10.1242/jcs.054197. [DOI] [PubMed] [Google Scholar]
- 68.Kasahara K., Goto H., Enomoto M., Tomono Y., Kiyono T., Inagaki M. 14-3-3gamma mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 2010;29:2802–2812. doi: 10.1038/emboj.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu J.Y., Reimann J.D.R., Sørensen C.S., Lukas J., Jackson P.K. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat. Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- 70.Guardavaccaro D., Kudo Y., Boulaire J., Barchi M., Busino L., Donzelli M., Margottin-Goguet F., Jackson P.K., Yamasaki L., Pagano M. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev. Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 71.Margottin-Goguet F., Hsu J.Y., Loktev A., Hsieh H.M., Reimann J.D.R., Jackson P.K. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 72.Petermann E., Helleday T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 73.Anglana M., Apiou F., Bensimon A., Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 74.Blow J.J., Ge X.Q., Jackson D.A. How dormant origins promote complete genome replication. Trends Biochem. Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fragkos M., Ganier O., Coulombe P., Méchali M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015;16:360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 79.Bartek J., Lukas C., Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 80.Gillespie P.J., Blow J.J. Clusters, factories and domains: the complex structure of S-phase comes into focus. Cell Cycle. 2010;9:3218–3226. doi: 10.4161/cc.9.16.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heller R.C., Kang S., Lam W.M., Chen S., Chan C.S., Bell S.P. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamada M., Watanabe K., Mistrik M., Vesela E., Protivankova I., Mailand N., Lee M., Masai H., Lukas J., Bartek J. ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 2013;27:2459–2472. doi: 10.1101/gad.224568.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.den Elzen N., Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lafranchi L., de Boer H.R., de Vries E.G.E., Ong S.-E., Sartori A.A., van Vugt M.A.T.M. APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J. 2014;33:2860–2879. doi: 10.15252/embj.201489017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mitra J., Enders G.H., Azizkhan-Clifford J., Lengel K.L. Dual regulation of the anaphase promoting complex in human cells by cyclin A-Cdk2 and cyclin A-Cdk1 complexes. Cell Cycle. 2006;5:661–666. [PubMed] [Google Scholar]
- 86.Sørensen C.S., Lukas C., Kramer E.R., Peters J.M., Bartek J., Lukas J. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol. Cell. Biol. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blasius M., Forment J. V, Thakkar N., Wagner S.A., Choudhary C., Jackson S.P. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toledo L.I., Altmeyer M., Rask M.-B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 89.Freire R., van Vugt M.A.T.M., Mamely I., Medema R.H. Claspin: timing the cell cycle arrest when the genome is damaged. Cell Cycle. 2006;5:2831–2834. doi: 10.4161/cc.5.24.3559. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y.-W., Otterness D.M., Chiang G.G., Xie W., Liu Y.-C., Mercurio F., Abraham R.T. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 91.Shankar S.R., Bahirvani A.G., Rao V.K., Bharathy N., Ow J.R., Taneja R. G9a, a multipotent regulator of gene expression. Epigenetics. 2013;8:16–22. doi: 10.4161/epi.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fujita T., Liu W., Doihara H., Wan Y. Regulation of Skp2-p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. Am. J. Pathol. 2008;173:217–228. doi: 10.2353/ajpath.2008.070957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verschuren E.W., Ban K.H., Masek M.A., Lehman N.L., Jackson P.K. Loss of Emi1-dependent anaphase-promoting complex/cyclosome inhibition deregulates E2F target expression and elicits DNA damage-induced senescence. Mol. Cell. Biol. 2007;27:7955–7965. doi: 10.1128/MCB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Machida Y.J., Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.