Abstract
16S rRNA amplicon analysis and shotgun metagenome sequencing are two main culture-independent strategies to explore the genetic landscape of various microbial communities. Recently, numerous studies have employed these two approaches together, but downstream data analyses were performed separately, which always generated incongruent or conflict signals on both taxonomic and functional classifications. Here we propose a novel approach, RiboFR-Seq (Ribosomal RNA gene flanking region sequencing), for capturing both ribosomal RNA variable regions and their flanking protein-coding genes simultaneously. Through extensive testing on clonal bacterial strain, salivary microbiome and bacterial epibionts of marine kelp, we demonstrated that RiboFR-Seq could detect the vast majority of bacteria not only in well-studied microbiomes but also in novel communities with limited reference genomes. Combined with classical amplicon sequencing and shotgun metagenome sequencing, RiboFR-Seq can link the annotations of 16S rRNA and metagenomic contigs to make a consensus classification. By recognizing almost all 16S rRNA copies, the RiboFR-seq approach can effectively reduce the taxonomic abundance bias resulted from 16S rRNA copy number variation. We believe that RiboFR-Seq, which provides an integrated view of 16S rRNA profiles and metagenomes, will help us better understand diverse microbial communities.
INTRODUCTION
Microbiota are everywhere in the world, and play important roles in various ecosystems. A suite of culture-free methods have enabled us to detect the genetic landscape of microbiota (1–3). The rapid progress in high-throughput DNA sequencing provides optimism for a bright spectrum of community diversity, phylogeny and functional capacity. Up to now, there are two main sequencing strategies to explore microbial communities, namely, 16S rRNA amplicon sequencing (4,5) and shotgun metagenome sequencing (6,7). A variety of worldwide microbial projects employed these two strategies, for instance, human microbiome project (HMP) (8) and earth microbiome project (9).
Bacterial 16S rRNA has a complex and highly conserved secondary structure, including nine variable (V) regions (V1–V9) (10). Direct sequencing of these variable regions is normally served as a standard approach for assessing composition and variation of complex microbial communities. Thus, variable region selection becomes a key step to answer the question of what is present in a given specific environment. Bioinformatic analysis tools, such as Mothur (11) and QIIME (12), and curated ribosome-related database likes SILVA (13), Greengenes (14) and ribosomal database project (RDP-II) (15), have revolutionized this culture-independent investigation of microbial diversity more easily and quickly. Multiple copies of 16S rRNA within one bacterial genome ranging from 1 to 15 (16), can affect the accurate assessment of bacterial abundance. Several computational tools attempt to adjust gene copy number for 16S rRNA analysis. For example, Kembel et al. (17) presented a method that employed sequences and genomic copy number of 16S rRNA genes combined with phylogenetic placement and ancestral state estimation to adjust organismal abundances. However, they are not applicable to novel species or strains with distinct 16S rRNA copy numbers. In addition, another drawback of 16S rRNA amplicon analysis is that it cannot provide direct experimental evidence on metagenomic function, although PICRUSt (18) predicted metagenome functional content from 16S rRNA sequences based on a priorly trained model using sequenced genomes.
Whole genome shotgun (WGS) sequencing is another important method for microbiome studies, which provides a comprehensive understanding of community structure, genetic population heterogeneity and potential metabolism pathway with relatively low-cost, less time and high throughput data than before. Due to the rapid advance of sequencing technologies, WGS-based metagenomic studies are growing sharply (19). However, exponential growth in sequencing data size generally needs more computational resources and efficient tools for further analyses. Recently, a number of bioinformatic tools such as FOCUS (20), MetaPhlAn2 (21), Kraken (22), CLARK (23) and SUPER-FOCUS (24), have been developed to facilitate taxonomic profile and metabolic function analyses based on metagenomic sequences. However, shortage of reference genomes and chimeric assembly are bottlenecks of shotgun metagenome sequencing (25,26), which influence the accuracy and reliability of genomic assembly and annotation, and thus it cannot provide a consensus microbial composition compared with 16S profiles.
More recently, many metagenomic surveys have employed both 16S- and WGS-based methods together, but the downstream analyses were performed separately. As a result, taxonomy and phylogeny inferred by 16S rRNA amplicons are often inconsistent with those retrieved from shotgun metagenome sequencing (27,28). It should be noted that in metagenome assembly, highly conserved ribosomal RNA sequences tend to be misassembled together or treated as repetitive sequences and thus filtered out. Therefore, it is necessary to establish a direct connection between 16S rRNA and metagenomes. In this study, we propose a novel method RiboFR-Seq (Ribosomal RNA gene Flanking Region Sequencing) (Figure 1) for capturing both ribosomal RNA variable regions and their flanking protein-coding genes simultaneously. This approach goes beyond traditional metagenomic analysis by taking into account not only phylogenetic features of 16S rRNA typing but also metagenome-scale genes derived from the same sample. With the rapid development of read length and quality generated from high-throughput sequencing, RiboFR-Seq would be a more reliable approach to explore the microbial community.
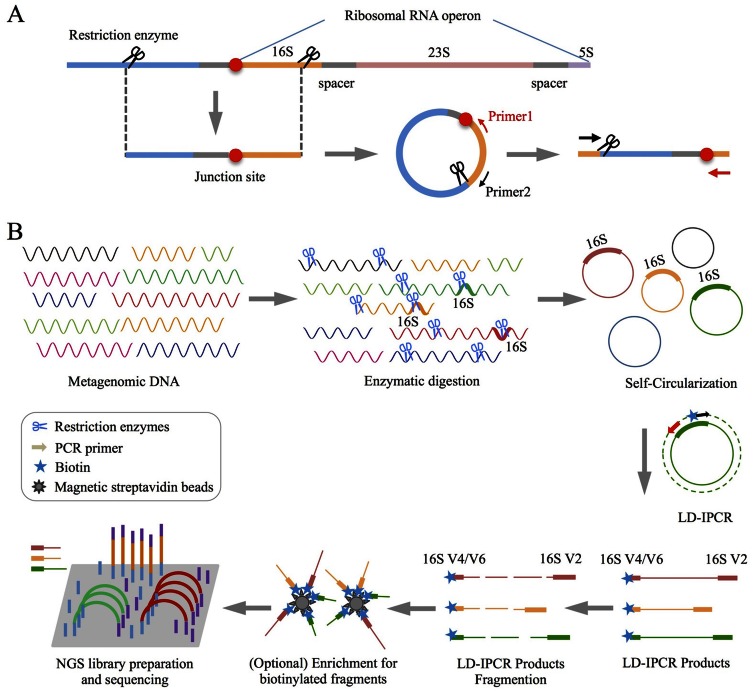
Figure 1.
An overview of RiboFR-Seq method. (A) A schematic diagram shows the process for capturing both ribosomal RNA variable regions and their flanking sequences simultaneously. (B) Experimental flowchart of RiboFR-Seq. Briefly, restriction enzymes with a 6-bp recognition site that can cleave bacterial 16S rRNA sequences were used to digest metagenomic DNA. Subsequently, the enzyme-digested DNA fragments with sticky end were self-circularized by direct intra-molecule ligation, which were then used as templates for LD-IPCR with specific inverse primers (labeled with biotin). LD-IPCR products were fragmented, of which contained biotin could be collected by magnetic streptavidin beads, then constructed NGS library for high- throughput sequencing.
MATERIALS AND METHODS
Sample collection
A healthy human volunteer was recruited, who was free of systemic diseases and other oral diseases, had no prosthetic dental appliances, had never received periodontal therapy and had not taken any antibiotics within three months prior to the sampling. About 10 ml saliva was collected in sterile plastic tube from this volunteer, and frozen at −80°C for further processing.
Escherichia coli DH5α (competent cells) was incubated aerobically at 37°C in LB (Luria broth) medium. Cells at logarithmic phase (106–107/ml) were collected and frozen at −80°C for storage.
Female gametophytes of brown algae (Saccharina japonica, SJ) were cultured at 10 ± 1°C and 5 μmol photons m−2 s−1 with a 12:12 h light/dark photoperiod. Microorganisms colonized on the surface of SJ were harvested using shaking equipment (Thermo Fisher Scientific, Waltham, MA, USA), and stored at −80°C for further processing.
Total DNA extraction
Cells of specimens were washed three times with sterile 1× phosphate buffered saline (Sangon Biotech Co., Ltd Shanghai, China). Bacterial genomic DNA was extracted with a TIANamp Bacteria DNA Kit (Tiangen Biotech Co., Ltd Beijing, China). The quantity of isolated DNA was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and the quality was checked by agarose gel electrophoresis. DNA solution was stored at −20°C for further processing.
Restriction enzyme selection and digestion
A total of 2 879 170 bacterial 16S rRNA sequences were downloaded from RDP database (29). A total of 8667 nearly full-length 16S rRNA sequences (≥1400 nt) of type strains were picked and aligned against a 16S alignment database from Greengenes (14) using Mothur v.1.32.1 (11). All aligned 16S rRNA sequences were in silico digested using 67 commercially available restriction endonucleases (Supplementary Table S1) with 6-bp recognition site by custom Python scripts. Restriction enzyme selection for further experimental applications was based on the following three criteria: (i) more than half of 8667 nearly full-length 16S rRNA sequences of type strains could be digested; (ii) only one recognition site was in most of full-length 16S rRNA sequences and the recognition site was adjacent to any of the nine ‘hypervariable regions’ (V1-V9) of 16S rRNA; and (iii) sticky ends of 16S rRNA sequences were cleaved by restriction enzymes with a 6-bp recognition site.
High molecular weight genomic DNA was digested by the selected restriction enzymes following the reaction mixture and procedures from the bundled protocols. After digestion, heat inactivation was performed according to the usage protocols of these enzymes. Enzyme-digested products were checked by agarose gel electrophoresis and stored at −20°C for further processing.
Self-circularization and linear-DNA degradation
Enzyme-digested genomic DNA fragments with sticky ends were self-circularized by direct intra-molecule ligation using T4 DNA ligase incubating at 16°C for 16 h. Then, T4 DNA ligase was heat inactivated at 65°C for 10 min. The remaining linear genomic DNA fragments were degraded by exonuclease I (ExoI, New England BioLabs (NEB), Hitchin, UK) and Plasmid-safe™ adenosine triphosphate (ATP)-dependent DNase (PSAD, Epicentre Biotech, Madison, WI, USA) by incubating at 37°C for 30 min and denaturing the enzymes at 75°C for 20 min. The circular genomic DNA solution was frozen at −20°C for further processing.
PCR primer sets and long distance-inverse PCR amplification
Nearly full-length 16S rRNA sequence of E. coli DH5α was targeted with universal primer pair 16S-8F/16S-1541R (30) (Table 1). The V4 and V6 region of 16S rRNA were individually amplified from metagenomic samples using universal primer sets 16S-V4-515F/16S-V4-806R (5) and 16S-V6-784F/16S-V6-1061R (31) (Table 1). Long-range genomic DNA fragments were obtained from circular DNA of enzyme-digested metagenome by long distance-inverse polymerase chain reaction (LD-IPCR) with specific inverse primer pairs 16S-EcoRI-515F/16S-EcoRI-338R and 16S-AatII-784F/16S-AatII-338R (Table 1, Figure 1A). Biotin-labeled primers 16S-EcoRI-515F and 16S-AatII-784F were optional based on the purpose of enrichment. The PCR programs for amplifying these target sequences were shown in Table 1. All PCR products were checked by agarose gel electrophoresis and stored at −20°C for further processing.
Table 1. PCR primer sets used in this study.
| Primer pair | sequence (5′ to 3′) | PCR program |
|---|---|---|
| 16S-8F | AGA GTT TGA TCC TGG CTC AG | 95°C 5 min, 30 × (94°C 30 s, 58°C 1 min, 72°C 1 min), 72°C 6 min |
| 16S-1541R | AAG GAG GTG ATC CAN CCR CA | |
| 16S-V4-515F | GTG CCA GCM GCC GCG GTA A | 95°C 5 min, 26 × (94°C 1 min, 50°C 1 min, 72°C 1.5 min), 72°C 10 min |
| 16S-V4-806R | GGA CTA CVS GGG TAT CTA AT | |
| 16S-V6-784F | AGG ATT AGA TAC CCT GGT A | 95°C 5 min, 26 × (94°C 40 s, 60°C 1 min, 72°C 1 min), 72°C 7 min |
| 16S-V6-1061R | CRR CAC GAG CTG ACG AC | |
| 16S-EcoRI-515F | CGT GCC AGC MGC CGC GGT AAT ACG | 94°C 1 min, 30 × (98°C 10 s, 68°C 10 min), 72°C 10 min |
| 16S-EcoRI-338R | CAC TGC TGC CTC CCG TAG GAG TNT GG | |
| 16S-AatII-784F | AGG ATT AGA TAC CCT GGT AGT CCA | |
| 16S-AatII-338R | CAC TGC TGC CTC CCG TAG GAG TNT GG |
Primer numbering relates to Escherichia coli position complementary to the 5′ end of the primer.
Last letter denotes direction: forward and reverse.
Next-generation sequencing (NGS) library preparation
The PCR products of 16S V4 (∼300 bp) and V6 (∼290 bp) region amplified from bacterial communities were purified by QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). Purified PCR products were quantified by Qubit® dsDNA HS Assay Kit with the Qubit® 2.0 Fluorometer system (Invitrogen, Life Technologies, Grand Island, NY, USA). For each metagenomic sample, 16S V4 and V6 amplicons were directly used to construct paired-end (PE) libraries using NEBNext® Ultra™ DNA Library Prep Kit.
LD-IPCR products from circular AatII-digested genomic DNA and circular EcoRI-digested genomic DNA were sheared into fragments of ∼900 and ∼500 bp in size by sonication using Covaris s220 (Covaris, Woburn, MA, USA), respectively. PE libraries were constructed according to a standard protocol with NEBNext® Ultra™ DNA Library Prep Kit. Biotin-labeled LD-IPCR products were adsorbed onto magnetic streptavidin beads and then bound to the magnet for enrichment. Subsequently, NGS libraries of these biotinylated PCR fragment were performed the same as above.
Purified metagenomic DNA sequences were fragmented into ∼180 bp by sonication using Covaris s220 (Covaris). PE libraries were constructed according to a standard protocol provided by Illumina, Inc. (San Diego, CA, USA).
Next-generation sequencing and data preprocessing
NGS libraries were quantified using a Stratagene Mx3000P Real-time PCR Cycler (Agilent, Santa Clara, CA, USA) before cluster generation in a c-Bot automated sequencing system (Illumina, Inc.). Quantified libraries with different barcodes were pooled together and sequenced on an Illumina HiSeq 2000 high-throughput sequencing instrument with 2 × 100 bp PE sequencing.
Raw image data and base-calling were performed using the standard Illumina pipeline with default parameters. Low quality reads and adaptor contaminations were filtered by Trimmomatic (32).
Metagenome assembly and gene prediction
For shotgun metagenome sequencing, de novo assembly of short PE reads were built using the Short Oligonucleotide Analysis Package (SOAPdenovo) v2.0.1 (33) assembly method and gaps were closed by GapCloser (33) (Figure 2, right panel). MetaGeneMark v3.25 (34) was used to predict genes from assembled contigs/scaffolds (size ≥500 bp) (Figure 2, right panel). The protein sequences translated from predicted genes were used to query the non-redundant protein (NR) database (6 July 2014) by BLASTP search (E-value ≤ 1 × 10−5). For each query, top 50 best BLAST hit results were recorded. Then, the function of a query protein was categorized based on majority-rule consensus of the output results. Lowest common ancestor (LCA) method was used to determine species’ taxonomic origin (35).
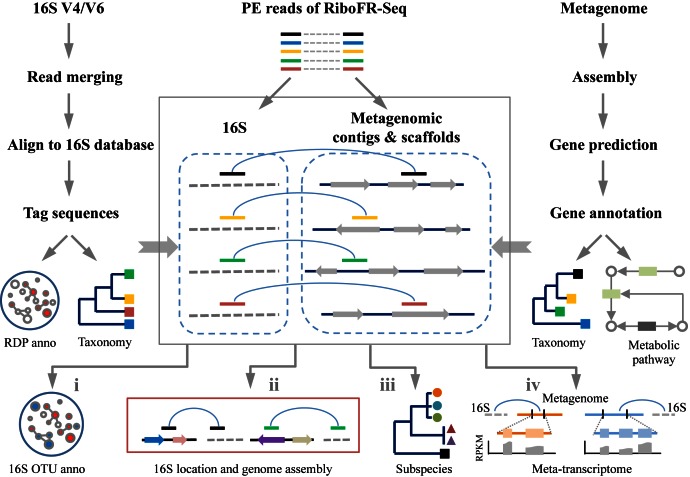
Figure 2.
Flowchart of bioinformatic analyses in RiboFR-seq. Left panel, general analysis of 16S rRNA amplicons. Right panel, bioinformatic processing of shotgun metagenome sequencing data. Median panel, RiboFR-Seq data analysis. One of PE reads from RiboFR-Seq is aligned to 16S (gray dotted line), the other is matched to metagenomic assembled contigs/scaffolds (black line with color arrows, color arrows indicate ORFs and the arrow direction indicates transcriptional orientation). Through the linkage (light blue curves) of above PE reads, 16S OTUs can be additionally annotated (i), multiple 16S rRNAs can be located in assembled genomes (ii), subspecies can be detected (iii) and expressed genes from meta-transcriptomes can be assigned (iv).
rRNA amplicons analysis
PE sequencing reads of 16S rRNA hypervariable regions (V4 or V6) were merged into long sequences by custom Perl scripts. These long sequences were aligned against a 16S alignment database from Greengenes (14) using Mothur v.1.32.1 (11) to remove chimeric sequences and convert reverse and complement sequences (Figure 2, left panel). The unique 16S V4 forward index ‘CGCGGTA’ and reverse index ‘TTAGATA’, 16S V6 forward index ‘ACCCTGG’ and reverse index ‘GTCGTCAG’ were located in filtered long sequences, and 160-bp sequences between forward index and reverse index were considered as tag sequences after removing redundancy (Figure 2, left panel). Tag sequences were clustered and binned into Operational Taxonomic Units (OTUs) based on 97% similarity (equivalent of species) using CD-HIT v4.5.4 (36), and further classified by the RDP Naïve Bayesian Classifier version 2.7 (37). Sample-based rarefaction curves were generated with OTUs in R 3.1.0 (http://www.r-project.org/). Single nucleotide variants were detected by pairwise alignment of unique tag sequences.
RiboFR-Seq data analysis
Filtered PE reads of RiboFR-Seq datasets were classified as rRNA sequences and non-rRNA sequences using SortMeRNA v2.0 (38). The PE reads that one end was rRNA sequence and the other was non-rRNA sequences were extracted from output files of SortMeRNA. For these read pairs, the rRNA read was aligned to 16S sequences and the non-rRNA read was characterized against the assembled metagenomic contigs/scaffolds by BLAT v. 36x1 (39) (E-value ≤ 10−5), respectively. Then, the taxonomic origin of this query read pair was determined based on the taxonomic classification of both rRNA-targeted read and metagenomic contig-targeted read using similar strategy like LCA (Supplementary Figure S1). Therefore, some of tag sequences unclassified by RDP classifier could be taxonomically annotated using its associated metagenomic contig assignment. Multiple copies of 16S rRNA sequences within one genome could be correctly ordered and orientated based on their coordinates in chromosome through the linkage of PE reads with one was rRNA read and the other was non-rRNA read. When these 16S rRNA copy numbers were determined, the relative abundance of certain bacteria could be recalibrated by dividing the number of 16S rRNA gene copies. Subspecies in metagenome was detected using nucleotide divergence of 16S rRNA sequences by custom Python scripts and confirmed by contigs/scaffolds associated with 16S rRNA. The computational pipeline for RiboFR-seq data analyses can be accessed at https://sourceforge.net/projects/ribofr-seq/.
RESULTS
RiboFR-Seq method
We propose a novel experimental and bioinformatic method, RiboFR-Seq, for capturing both ribosomal RNA variable regions and their flanking protein-coding genes simultaneously (Figure 1). Firstly, metagenomic DNA was digested by selected restriction enzymes and then these enzyme-digested DNA fragments were self-circularized through direct intra-molecule ligation (Figure 1A). After self-circularization, the remaining liner genomic DNA was digested using exonuclease. The long-range products were amplified from circular metagenomic DNA molecules with designed primer sets (Table 1) located in the 16S rRNA gene by long distance-inverse PCR (LD-IPCR) (Figure 1B). These amplified products were fragmented and used for library construction and PE sequencing. The filtered PE reads were aligned to the annotated reference genomic sequences. For a read pair, if one end is mapped to 16S rRNA variable region (V4 or V6) and the other end is mapped to the upstream protein-coding region, they are considered as ‘bridge read pairs’ (BRPs), which will provide a direct link between 16S rRNA taxonomic profiling and WGS metagenomic assemblies (Figure 2). This new method can be applied to make consensus annotation between 16S rRNA profiling and shotgun metagenome surveys, to accurately locate multiple 16S rRNA sequences in assembled contigs/scaffolds for aiding metagenome assembly and to detect 16S gene copy number by clustering non-ribosomal reads of BRPs along the chromosome (Figure 2).
Restriction enzyme selection and experimental verification of RiboFR-Seq on E. coli DH5α
Selection of an appropriate restriction enzyme is a key factor in RiboFR-Seq. An ideal restriction enzyme should have a 6-bp recognition site to cut sticky ends, and cleave 16S rRNA sequences in all bacterial genomes, with only one recognition site adjacent to any of nine ‘hypervariable regions’ (V1-V9) of 16S rRNA. A total of 67 commercially available restriction enzymes (Supplementary Table S1) were used to in silico digest 8667 aligned 16S rRNA sequences of type strains from RDP database. More than half of the above 16S rRNA sequences could be digested by 19 restriction enzymes (Supplementary Table S1, marked by yellow color). NcoI, SnaBI, SphI and StuI were removed from the restriction enzyme candidate sets, because the number of 16S rRNA sequences they digested was the least of these 19 restriction enzymes. About one-third of the above 16S sequences had more than two recognition sites digested by Acc65I, BspEI, EagI, KpnI, PmlI, SacII, SmaI and XmaI (Supplementary Figure S2, be similar with the recognition sites distribution of KpnI), which were thus abandoned. SspI and ZraI were also discarded since they digested sequences with blunt end, which were hard to form self-circularization especially when long-range DNA fragments were used. Combined with primer design of nine ‘hypervariable regions’ of 16S rRNA, AatII and EcoRI were finally chosen from 67 restriction enzymes to digest the metagenomic DNA, which could digest 99.7 and 77.8% of 8667 aligned 16S rRNA sequences of type strains, respectively (Supplementary Figure S2).
This new designed method was firstly validated on a single laboratory strain E. coli DH5α. The nearly full length sequences of 16S rRNA were amplified from the genomic DNA of laboratory strain E. coli DH5α with primer sets 16S-8F/16S-1541R (Table 1). In consistent with in silico analysis, this 16S sequence was split into two fragments after AatII treatment, which were 1200 and 300 nt, respectively (Supplementary Figure S3). Genomic DNA of E. coli DH5α digested by AatII was smear shown in Supplementary Figure S4A. The AatII-digested genomic DNA fragments were self-circularized through sticky ends directly ligated, and linear genomic DNA fragments were decomposed by ExoI (NEB) and Plasmid-safe™ ATP-dependent DNase (Epicentre). The inverse primer pair 16S-AatII-784F/16S-AatII-338R (Table 1) was used to amplify the long-range genomic fragments from the circular DNA by LD-IPCR. Six PCR products (>2 kb) (Supplementary Figure S4B) indicated that the self-circularization and LD-IPCR were successful. LD-IPCR products were then sequenced using Sanger sequencing method to obtain the sequences of 5′ and 3′ ends, which were mapped to E. coli reference genome (K-12 substr. MG1655). One end of the sequence was aligned to V1-V2 region of 16S rRNA, the other end was split-mapped to V6 region of 16S rRNA and a functional gene (gmh B) located upstream of 16S rRNA, respectively (Supplementary Figure S4C). These results demonstrated that the newly designed method RiboFR-Seq could be manipulated successfully in molecular biological laboratory.
Application of RiboFR-Seq to microbiome from oral habitats
Because most of microbes in human saliva have been classified and sequenced, with their genomic sequences deposited to public databases, we selected salivary microbiome from a healthy human volunteer as another test dataset to evaluate the performance of this method. High-throughput sequencing of AatII-digested circularized PCR products (oral-A) from RiboFR-Seq generated 4 325 207 PE reads, and two replicates of EcoRI-digested circularized PCR products (oral-E1 and oral-E2) generated 11 327 920 and 8 559 821 PE reads, respectively. All these PE reads were aligned against 937 reference genomes from HMP, in which about 7.95% of PE reads from oral-A, 6.70 and 4.42% of PE reads from oral-E1 and oral-E2 could be classified as BRPs. These results proved our new method could be applied to metagenomic studies. The incomplete reference genome sequences in HMP may lead to the underestimation of the percentage of BRPs in all three RiboFR-Seq datasets.
We further performed 16S rRNA amplicon sequencing and shotgun metagenome sequencing of the same salivary sample from the healthy human volunteer, and used the RiboFR-Seq data to connect the 16S rRNA classification with metagenomes. After quality control, 1 409 527 and 2 042 988 clean PE reads amplified from 16S V4 and V6 region were merged based on method described in ‘Materials and Methods’ section. A total of 1 238 058 and 1 854 799 tag sequences were classified using the RDP classifier and de novo binned into 7617 and 15 620 OTUs at 97% similarity, respectively. The rarefaction curves generated with OTUs indicated sequenced PE reads of 16S variable regions were saturated (Supplementary Figure S5). Shotgun metagenome sequencing of the saliva sample resulted in 68 235 958 sequenced clean PE reads after screening out human DNA contaminants. Totally 1 476 838 contigs/scaffolds were assembled using SOAPdenovo2, and 315 723 ORFs were predicted by MetaGeneMark from contigs and scaffolds whose length longer than 500 bp, of which 90.79% were annotated (ORFs). In this metagenome assembly, only 175 contigs were found to contain partial 16S rRNA sequences with at least 200 bp. After incorporating the RiboFR-seq data, we successfully linked 16S rRNAs to 4686 contigs with at least three BRPs support.
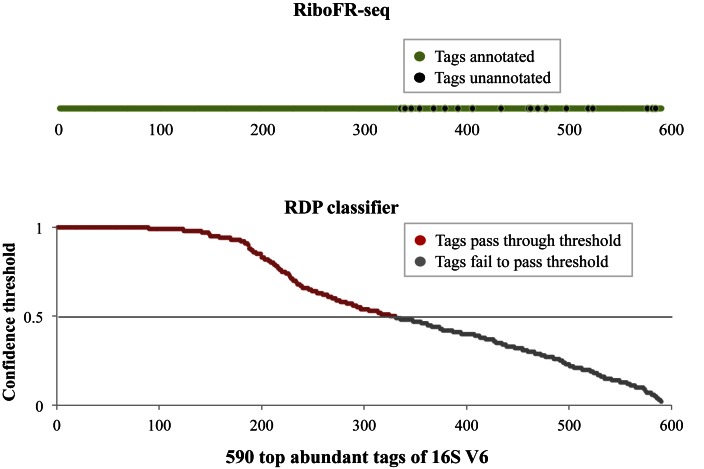
An advantage of RiboFR-Seq is that it can classify 16S sequence tags by combining classical 16S rRNA classification and the taxonomical annotation of metagenomic contigs/scaffolds connected by BRPs. About 590 top abundant tags of unique sequences acquired from the 16S rRNA V6 sequencing of oral-A were shown in Figure 3A, among these tags, 56% could be classified by RDP classifier with a confidence threshold of 50% at the genus level, whereas 89% could be classified using RiboFR-Seq by introducing additional annotation from assembled contigs (Figure 4). Similarly, the 16S V4 tags amplified from oral-E1 and oral-E2 could also be annotated at a higher percentage by RiboFR-Seq than by traditional V4 sequencing alone (Supplementary Figure S6).
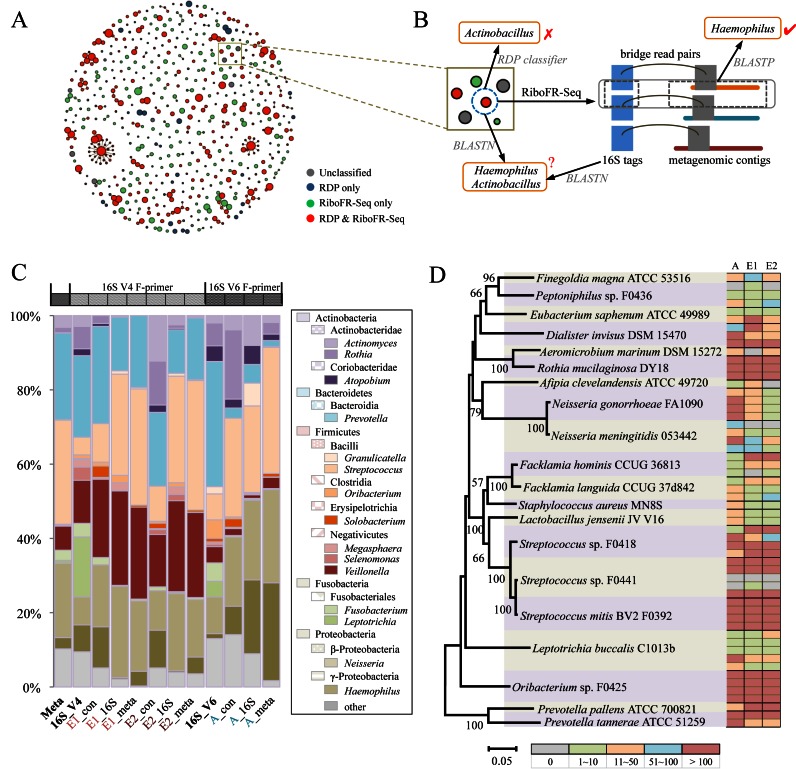
Figure 3.
Salivary microbiome explored by RiboFR-Seq. (A) Abundant unique sequence tags of 16S V6 amplified from salivary metagenome annotated by RDP classifier and RiboFR-Seq. Each node represents a sequence tag, and node size is proportional to tag richness. The link between nodes indicates 1 nt difference between the two linked tags. (B) An example shows that RiboFR-seq can correct a misclassified OTU by RDP classifier. (C) The abundance of 15 major genera in the salivary metagenome estimated using 12 different methods or datasets. ‘Meta’ represents taxonomic abundance based on shotgun metagenome sequencing estimated by MEGAN. ‘16S_V4’ and ‘16S_V6’ represent taxonomic abundance based on classical 16S amplicon sequencing of V4 and V6 regions, respectively. ‘E1’ and ‘E2’ represent the two replicates of RiboFR-Seq datasets with EcoRI treatment. ‘A’ represents the RiboFR-Seq dataset with AatII treatment. ‘_16S’ and ‘_meta’ represent the taxonomic abundance calculated based on 16S tags and non-rRNA reads from RiboFR-seq, respectively. ‘_con’ represents the consensus prediction of taxonomic abundance based on 16S tags and non-rRNA reads from RiboFR-seq. (D) Recovery of multiple 16S rRNA copies by RiboFR-Seq in the salivary microbiome. Left panel, phylogenetic tree of 20 most abundant species in the salivary microbiome. Right panel, the recovery of multiple 16S rRNA copies for each of the 20 genomes. Each row represents one copy of 16S rRNA gene, and the color indicates the number of BRPs that could be uniquely mapped to the corresponding 16S rRNA copy. ‘A’ represents the RiboFR-Seq dataset with AatII treatment. ‘E1’ and ‘E2’ represent the two replicates of RiboFR-Seq datasets with EcoRI treatment. For example, Oribacterium sp. F0425 has four 16S rRNA copies, and all of them could be detected by the three datasets with strong support (>100 BRPs).
Figure 4.
Genus level annotation of 590 top abundant tags from 16S V6 of oral sample by RDP classifier and RiboFR-Seq. A total of 330 tags could be annotated at genus level using RDP classifier at a confidence threshold of 50%, whereas the remaining tags were annotated with lower confidence values below the threshold. However, 89% of these tags could be classified at genus level using RiboFR-Seq by introducing additional annotation from assembled contigs.
Notably, RiboFR-Seq-based classification can correct errors in traditional 16S rRNA based taxonomic classification, in which short sequencing reads (∼100 bp) may be incorrectly assigned by RDP classifier. For example, one 16S V6 sequence tag was annotated as Actinobacillus by RDP classifier, and identified as Actinobacillus and Haemophilus with the same identities (99%) using BLASTN to search the 16S ribosomal RNA sequences database in GeneBank, but this tag was re-classified as Haemophilus by RiboFR-Seq, where the linked assembled contig was annotated to Haemophilus sp. (Figure 3B). In the other way, assembled contigs from salivary metagenome could also be specified through BRPs with annotated 16S sequence tags by RiboFR-Seq, 506 unannotated contigs/scaffolds from metagenomic assembly generated from oral-A could be classified. A total of 338 and 352 unrecognized contigs/scaffolds from oral-E1 and oral-E2 were also re-categorized, respectively.
Streptococcus, Prevotella and Neisseria were dominated in shotgun metagenome sequencing data, Prevotella, Leptotrichia and Veillonella were three major genera in 16S V4 dataset, compared with that Prevotella were absolutely dominant in 16S V6 dataset (Figure 3C). Since RiboFR-Seq based on EcoRI digestion employed an inverse PCR primer modified from the widely used forward primer in 16S rRNA V4 amplicon sequencing, sequencing datasets of oral-E1 and oral-E2 contained the V4 hypervariable region and thus were firstly compared with traditional 16S V4 data. The clean PE reads of RiboFR-Seq from oral-E1 and oral-E2 were aligned to whole genome sequences of oral bacteria deposited in public database (HMP and HOMD) (40,41), of which 83.52 and 83.66% were identified at genus level, respectively, which were slightly lower than 16S V4 (90.47%). Similar results were also found in the RiboFR-Seq data of the AatII-digested sample (87.3 versus 87.03%). Then, we estimated the abundance of the top 15 most abundant genera using 12 different methods (RDP classifier, MEGAN (35) and RiboFR-seq) or datasets (oral-E1, oral-E2 and oral-A). Among these estimations, the top 15 most abundant genera represented 83–99% of the bacteria in 12 datasets. E1_con, E2_con and A_con, which were determined based on the consensus annotation of BRPs, were more likely to reflect the real bacterial abundance (Figure 3C and Supplementary Figure S7). Moreover, two recently developed tools, CLARK and Kraken, were also used for taxonomic classification. For the salivary metagenomic data, the abundance of the top 15 most abundant genera estimated by CLARK and Kraken represented 79.87 and 83.18% of the bacteria, respectively, which were slightly lower than that of the assembled method (Supplementary Figure S8). Similar results were also found in the RiboFR-Seq data annotated with CLARK and Kraken. Although the runtime of RiboFR-Seq was longer than that of CLARK and Kraken (Supplementary Table S3–4), it required much less memory than the other two tools. In addition, RiboFR-Seq could obtain not only the taxonomic profiles like CLARK and Kraken, but also the linkage between 16S rRNA and metagenomic contigs.
rRNA copy number
Although restriction enzymes AatII and EcoRI could digest most of bacterial 16S rRNA as confirmed by both in silico (Supplementary Figure S2) and experimental approaches (Supplementary Figures S3–4), the efficiency of these two restriction enzymes on digesting different 16S rRNA copies in metagenomes was unknown. Hence, we collected 20 most abundant taxa in the sequenced salivary metagenome with 16S rRNA copy number ranging from one to five. All BRPs were aligned to genomic sequences of these 20 taxa, with one end mapped to the 16S rRNA sequence and the other end uniquely mapped to upstream flanking non-ribosomal region. The position coordinates of these two aligned reads in chromosome were recorded, and then were compared with coordinates of multiple 16S rRNA copies within one species to confirm which copy was digested. As shown in Figure 3D, 53 out of 56 16S rRNA copies from these 20 taxa were successfully digested by AatII in oral-A except for copies No. 1 of Peptoniphilus oral taxon 375 F0436, and No. 3 and No. 4 of Streptococcus F0441. Similarly, 52 and 51 copies were digested by EcoRI in oral-E1 and oral-E2, respectively (Figure 3D). One possible reason for the 16S copies No. 3 and 4 of Streptococcus F0441 could not be detected in certain samples is that these two 16S copies are truncated and are much shorter (400 and 900 bp) than the others. It should be noted that the 16S rRNA copies of the most abundant genera in salivary metagenome, such as Rothia, Streptococcus and Prevotella, were covered by more reads than those of low abundant genera. These results indicated that restriction enzymes AatII and EcoRI can digest nearly all the 16S rRNA gene copies in metagenomic DNA and thus could be used in RiboFR-Seq.
16S rRNA copy number varies greatly among bacteria (16). As a result of this variation, currently available 16S rRNA-based analytic approaches tend to overestimate the relative abundance of taxa in an environmental sample based on the 16S rRNA gene sequences. However, the RiboFR-Seq method determined 16S rRNA gene copy number within one species by clustering the non-ribosomal reads of BRPs, which could be used to adjust the species richness. 16S V6 sequences of oral microbiome assigned to 10 species were extracted and analyzed by normal methods without adjustment (Supplementary Figure S9). Then, these sequences were re-analyzed using two different 16S copy number adjustments methods, RiboFR-Seq and RDP Classifier V2.11 which was trained with the 16S gene copy number data from the Ribosomal RNA Database (rrnDB) (42). As shown in Supplementary Figure S9, before and after 16S rRNA copy number adjustment, the richness of these 10 species changed greatly. For example, the abundance of Neisseria gonorrhoeae FA 1090 was declined from 55 808 per million reads to 13 952 per million reads. Moreover, the 16S copy number detected by RiboFR-Seq with BRPs was more accurate than that estimated from rrnDB based on known genomes. For rrnDB-based adjustment, if one species was not recorded in rrnDB, the 16S copy number would be assigned with mean gene copy number of its parent taxa. For instance, Prevotella pallens ATCC 700821 was not collected in rrnDB, thus the 16S copy number of this species was assigned as 4, which was the average 16S copy number of genera Prevotella calculated from rrnDB, but actually only one 16S copy is present in Prevotella pallens ATCC 700821, which is consistent with RiboFR-Seq prediction.
Analysis of microbiome from novel natural habitats by RiboFR-Seq
To extend the application of this new method to microbial communities in novel environments with limited reference genomes, the undiscovered microflora colonized on the surface of SJ has been taken as another sample. Both 16S rRNA amplicon sequencing and shotgun metagenome sequencing were performed using the genomic DNA from microbes associated with SJ female gametophytes.
Altogether 116 497 788 clean PE reads were generated from shotgun metagenome sequencing of the SJ sample after screening out host DNA contaminants. Totally 52 173 contigs/scaffolds were assembled using SOAPdenovo2 and 205 318 ORFs were predicted by MetaGeneMark from contigs/scaffolds (length ≥ 500 bp), of which 185 325 were annotated using BLASTP against the NCBI NR database. A total of 1 813 568 and 1 798 489 of 16S V4 and V6 PE reads passed through quality control, respectively. About 88.12 and 86.48% of these high-quality PE reads were merged into long sequences and taken as tags and then de novo binned into 5368 and 6870 OTUs at a distance level of 3%. The rarefaction curves illustrated that the number of 16S sequences collected in this study was adequate (Supplementary Figure S5B). Metagenome sequencing of AatII-digested circularized products from RiboFR-Seq resulted in 6 735 510 clean PE reads, and replication control 1 and 2 of EcoRI-digested circularized products generated 10 657 788 and 9 468 582 clean PE reads, respectively.
As shown in Figure 5A, only 58.08% of metagenomic reads could be assigned to the top 25 most abundant genera and 89.22% of 16S rRNA V4 and 88.62% of 16S rRNA V6 reads could be classified to these 25 genera. Compared with taxonomy based on sequencing reads, 66.74% of metagenomic reads could be assigned to the top 25 most abundant genera by Kraken, which was higher than that of the assembled method (58.08%) and 41.62% metagenomic reads could be assigned to these 25 most abundant genera by CLARK (Supplementary Figure S10). As expected, the taxonomic profiles estimated based on shotgun metagenomic reads and 16S rRNA amplicon sequencing were distinct (Supplementary Figures S10 and 11). The main reason for this discrepancy is that OTUs or assembled contigs/scaffolds derived from novel bacterial communities had much lower sequence similarity with known 16 rRNA genes or reference genomes, respectively. For example, the genus Ilumatobacter was dominated (11.25%) in the shotgun metagenome sequencing data. In contrast, this genus only ranked the second and third most abundant genera in 16S V4 and V6 data (16.19 and 13.32%), respectively. Alteromonas was the most abundant genus in the 16S V4 and V6 datasets, which occupied 41.16 and 39.57% of total reads, respectively, compared with 7.61% in shotgun metagenome sequencing data.
Figure 5.
Microflora colonized on the surface of Saccharina japonica (SJ) revealed by RiboFR-Seq (A) Normalized percentage of 25 major genera shows the microbial diversity in three datasets from SJ microflora, Meta (shotgun metagenome sequencing data), 16S_V4 (16S V4 amplicon data) and 16S_V6 (16S V6 amplicon data). (B) Consensus classification of 16S V6 OTUs and contigs/scaffolds by RiboFR-Seq. Left panel, 20 abundant OTUs of 16S V6, area of solid circle (OTU richness). Right panel, the contigs/scaffolds linked with 16S OTUs by BRPs, with area of solid circle representing average coverage of contigs/scaffolds and gradient blue color representing contig/scaffold number. The dark red solid lines indicate linkage between 16S V6 OTUs and contigs/scaffolds, with the width representing the number of BRPs. (C) Abundant unique sequence tags of 16S V6 amplified from SJ microflora annotated by RDP and RiboFR-Seq. Each node represents a sequence tag, and node size is proportional to tag richness. The link between nodes indicates one nucleotide difference between the two linked tags. (D) Multiple 16S rRNA sequences connected with contigs/scaffolds of three taxa from metagenomic assembly. Red nodes represent 16S rRNA genes, and gray nodes represent metagenomic contigs/scaffolds.
We applied RiboFR-Seq to this SJ metagenomic sample and tried to integrate above two surveys to make a consensus classification based on BRPs. As shown in Figure 5B, the OTUs clustered from 16S amplicon sequencing and contigs/scaffolds assembled from shotgun metagenome sequencing were connected by BRPs. Twenty most abundant OTUs of 16S V6 were annotated by RDP classifier, and contigs/scaffolds were queried to the NR database using BLASTP for classification (Figure 5B). The annotation of these two datasets were distinct, but the BRPs of RiboFR-Seq directly linked the two parts and re-classified the OTUs and contigs/scaffolds at a consensus taxonomic level by reducing the biases from the two annotation strategies. For this unexplored SJ microflora, about a half of BRPs could make a consensus classification using a LCA approach (Supplementary Figure S1, ‘Materials and Methods’ section). The remaining BRPs linked the 16S rRNA or contigs/scaffolds with no annotation because of lacking reference genomes. The most abundant unique sequence tags (∼600 read supports) of 16S V6 in SJ were shown in Figure 5C. Sixty-five percent of these tags could be classified by RiboFR-Seq based on AatII-digested, 42% were identified by RDP classifier, and only 28% could be annotated by both methods. Similar findings were also present in the EcoRI-digested sample (Supplementary Figure S12). Although the ratio of unclassified tags in SJ was much higher than that of the salivary metagenome, the RiboFR-Seq approach could assign many more 16S V4 and V6 tags to certain taxa using BRPs than traditional methods. Our findings demonstrated that for bacterial communities from less explored environments, where taxonomic profiles based on 16S rRNA and metagenome were rather different, RiboFR-Seq provided a useful solution for more accurate taxonomic classification.
Accurate placement of ribosomal RNA operons is a significant challenge in microbial genome assembly, particularly in metagenomic studies. RiboFR-Seq could accurately locate multiple 16S rRNA sequences through the unique connection between 16S rRNA sequences and metagenomic contigs/scaffolds using BRPs. We retrieved contigs/scaffolds of three species from metagenomic assembly and positioned multiple 16S rRNA sequences by the direct linkage of BRPs, which could assist to further bacterial genome finishing (Figure 5D). The draft genome of one species from above was constructed and identified as Devosia sp., in which two copies of 16S rRNA sequences of this Devosia sp. were successfully pinpointed in the assembled genome (Supplementary Figure S13).
With the rapid advancement of high-throughput sequencing technologies and wide application of metagenomic approaches, we believe RiboFR-Seq, which provides a direct link between 16S rRNA and metagenome, can help us better understand the microbial communities.
DISCUSSION
This study presents a novel experimental and bioinformatic framework, RiboFR-Seq. The main advantage of RiboFR-Seq is that it can provide a direct link between 16S rRNA taxonomic profiles and metagenomes using ‘BRPs’, of which one end could be mapped to 16S rRNA variable region (V4 or V6) and the other end could be mapped to the upstream protein-coding regions. RiboFR-Seq can determine 16S rRNA copy numbers within one species to reduce the bias of 16S profiling, and can unbiasedly classify 16S amplicons and metagenomic contigs. Combined with classical amplicon sequencing and shotgun metagenomic sequencing, RiboFR-Seq can link the annotations of 16S rRNA and metagenomic contigs to make a consensus classification, and can accurately locate multiple 16S rRNA sequences through BRPs and thus can assist to metagenomic assembly and binning.
Taxonomic classification in metagenomic studies
To explore the community structure, particularly the microbiome from novel niches, the first and most important step is to detect ‘who's there?’. 16S rRNA amplicon analysis and shotgun metagenome sequencing are the two most common approaches. The 16S rRNA genes are widespread in prokaryotes, which is the most commonly used marker gene to explore microbial community diversity. Shotgun sequencing of metagenomes increases our knowledge of taxonomic classification as well as genetic and functional variability. Moreover, community composition characterized by shotgun metagenome sequencing and 16S rRNA amplicon sequencing were always not identical (Figure 3C), which probably arose directly from insufficient sequencing depth and incomplete assembly in shotgun metagenome sequencing, and PCR primer sets used in 16S rRNA amplicon sequencing not suitable to cover all bacteria.
Single cell sequencing technology partly resolves the above problem. Cell sorting of microbial populations by flow cytometry (43) and microfluidics (44) makes a single cell separation successfully. Whole genome amplification such as multiple displacement amplification (45) or multiple annealing and looping-based amplification cycles (46) coupling with high-throughput sequencing has completed the genome assembly (47–49). Specific assembly tools for single cell genome such as SPAdes (50) and IDBA-UD (51), boost the applications of single cell sequencing more widely. However, there are about 500–700 different bacterial species in human oral cavity (52), more than 1000 species in gut (53) and 2.5 × 106 colony-forming units per gram of soil (54), separating every bacterial cell from natural environment is a huge project for most laboratories. In addition, whole genome amplification contamination is inevitable, and plenty of reagents are also burden for researchers.
To deal with above mentioned problems, we designed a novel method RiboFR-Seq, which could link 16S rRNA and metagenomic contigs/scaffolds to characterize species composition from natural environments. As demonstrated in the human salivary microbiota and unexplored bacterial epibionts of marine kelp, RiboFR-Seq could detect the vast majority of bacteria not only in well-studied microbiomes but also in novel communities with limited reference genomes, and provided an enhanced view on microbiota profiles by combining with traditional 16S rRNA and metagenomic sequencing.
Horizontal gene transfer (HGT) is the process in which mobile genetic material is shared between organisms, which is an important factor in the evolution of bacteria, and plays important role in transmission of antibiotic resistance and virulence (55). Currently available methods for detecting HGT events are mainly based on phylogenetic conflict. The RiboFR-Seq approach may be applicable to studying HGT events in metagenomic samples. For example, BRPs between potential mobile elements and 16S rRNA gene can be obtained from the circularized enzyme-digested DNA fragments using high throughput sequencing. The phylogeny of 16S rRNA reads from BRPs can be used to determine their taxonomic origin, while the conflicting phylogenetic signal derived from the linked metagenomic genes may indicate potential HGT events.
Restriction enzyme selection
Restriction enzymes (endonuclease) are a kind of specific enzymes which can cut DNA at or near unique recognition sites. Nowadays, about 4000 biochemically or genetically characterized restriction enzymes have been discovered and over 600 of which are commercially available for diverse research communities (56). In this study, selection of an appropriate restriction enzyme is a key factor for RiboFR-Seq, implementing in the first step of this novel method to digest metagenomic DNA. The restriction enzymes AatII and EcoRI in silico covering the abundant genera or species were selected from about 200 restriction endonucleases, and their efficiencies had been verified using RiboFR-Seq data from human salivary sample and microflora colonized on SJ. These two restriction enzymes have different recognition sites, which could digest 16S rRNA sequences at different position (Supplementary Figure S2), and cover 99.7% (AatII) and 77.8% (EcoRI) of 8667 16S rRNA sequences of type strains. In further applications, researchers can choose the restriction enzymes employed in RiboFR-Seq based on purpose of studies, not just to cleave 16S rRNA sequences of most abundant genera/species, but to cut 16S rRNA from some specific genera or species. If one single restriction endonuclease with a conserved cleavage site is not satisfied for research goals, two or three restriction enzymes could be mixed to digest the total metagenomic DNA. It is noteworthy that some restriction enzymes are sensitive to DNA site-specific methylation modified at m4C, m5C and m6A (57), which are commonly found in bacterial genomes. Therefore, usage of methylation-sensitive restriction endonucleases should be very careful in RiboFR-Seq. For example, XbaI could not cleave the genomic DNA of E. coli GM2163 with deleted dam and dcm at recognition sequences TCTAGATC and GATCTAGA (58). As more and more restriction enzymes have been discovered and commercially available, RiboFR-Seq would be suitable for broad applications in further metagenomic studies.
Application of RiboFR-Seq with development of technologies
In RiboFR-Seq method, the truly useful PE reads are those one read aligned to 16S sequences and the other matched the adjacent upstream genes or spacer sequences flanking the same ribosomal operon, which can build a bridge to directly link 16S rRNA profiling to metagenomic survey. About 7.95% of clean PE reads of RiboFR-Seq data from AatII-digested genomic DNA of human salivary microbiome were the ‘bridge reads’, and the remaining PE reads were either partial sequences of 16S rRNA or neighboring flank DNA fragments of the same ribosomal operon. In order to retrieve these ‘bridge reads’ more efficiently, the forward LD-IPCR primer 16S-AatII-784F could be modified with a 5′-end biotin label. After the fragmentation of the long-range PCR products amplified with biotin-labelled primers, the BRPs could be enriched using streptavidin-coated magnetic beads. Using this enrichment step will undoubtedly increase the percentage of bridge reads and improve the application of RiboFR-Seq.
Nevertheless, there are many factors that can influence the effectiveness of RiboFR-Seq. The most important one is the length of NGS short reads which is the cornerstone of the linking bridge. One short ‘bridge read’ may align perfectly to plenty of reference sequences from different species or genus. These multiple alignments complicate the connections that are hardly to be resolved in further analysis. However, with the rapid advancement of high throughput sequencing technologies, Illumina platforms can produce up to 2 × 250 bp or even 2 × 300 bp read pairs. These longer reads can undoubtedly reduce the ratio of multiple mappings in RiboFR-Seq, although reads assembly and homology searches are still challenging. Over the past few years, the third generation sequencers based on single-molecule approach have been commercially available. PacBio RS SMRT system achieves much longer read length (∼10 kbp) than other current technologies, and has been recently applied to full-length 16S rRNA sequencing and genome finishing for bacteria in several studies (59,60). However, the major limitations of this sequencing technology are its high single-pass raw read error rate (10–15%) and high cost. With the rapid development of sequencing technologies, the third generation sequencers based on single-molecule approach will promote the application of RiboFR-Seq in metagenomic subjects. We believe that RiboFR-Seq will facilitate our understanding of various microbial communities by integrating classical rRNA amplicon and shotgun metagenomic analyses.
Supplementary Material
Acknowledgments
Author contributions: F.Z. designed the method. Y.Z. performed experiments. Y.Z., P.J. and F.Z. analyzed the data. Y.Z., J.W. and F.Z. wrote the manuscript. All authors discussed the results and commented on the manuscript.
FUNDING
National Natural Science Foundation of China [91531306, 31301031]; General Financial Grant from China Postdoctoral Science Foundation [2012M520019].
Conflict of interest statement. None declared.
REFERENCES
- 1.Kazor C.E., Mitchell P.M., Lee A.M., Stokes L.N., Loesche W.J., Dewhirst F.E., Paster B.J. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Temmerman R., Scheirlinck I., Huys G., Swings J. Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2003;69:220–226. doi: 10.1128/AEM.69.1.220-226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su C., Lei L., Duan Y., Zhang K.-Q., Yang J. Culture-independent methods for studying environmental microorganisms: methods, application, and perspective. Appl. Microbiol. Biotechnol. 2012;93:993–1003. doi: 10.1007/s00253-011-3800-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Qian P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh J., Byrd A.L., Deming C., Conlan S., Kong H.H., Segre J.A., Program N.C.S. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C., Knight R., Gordon J.I. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert J.A., Meyer F., Antonopoulos D., Balaji P., Brown C.T., Brown C.T., Desai N., Eisen J.A., Evers D., Field D. Meeting report: the terabase metagenomics workshop and the vision of an Earth microbiome project. Stand. Genomic Sci. 2010;3:243–248. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravorty S., Helb D., Burday M., Connell N., Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole J.R., Chai B., Farris R.J., Wang Q., Kulam S.A., McGarrell D.M., Garrity G.M., Tiedje J.M. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchet V., Huot H., Goldstein R. Molecular genetic basis of ribotyping. Clin. Microbiol. Rev. 2008;21:262–273. doi: 10.1128/CMR.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kembel S.W., Wu M., Eisen J.A., Green J.L. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput. Biol. 2012;8:e1002743. doi: 10.1371/journal.pcbi.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Thurber R.L.V., Knight R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forde B.M., O'Toole P.W. Next-generation sequencing technologies and their impact on microbial genomics. Brief. Funct. Genomics. 2013;12:440–453. doi: 10.1093/bfgp/els062. [DOI] [PubMed] [Google Scholar]
- 20.Silva G.G.Z., Cuevas D.A., Dutilh B.E., Edwards R.A. FOCUS: an alignment-free model to identify organisms in metagenomes using non-negative least squares. PeerJ. 2014;2:e425. doi: 10.7717/peerj.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., Tett A., Huttenhower C., Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 22.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ounit R., Wanamaker S., Close T.J., Lonardi S. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 2015;16:236. doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva G.G.Z., Green K.T., Dutilh B.E., Edwards R.A. SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics. 2016;32:354–361. doi: 10.1093/bioinformatics/btv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bragg L., Tyson G.W. Metagenomics using next-generation sequencing. Methods Mol. Biol. 2014;1096:183–201. doi: 10.1007/978-1-62703-712-9_15. [DOI] [PubMed] [Google Scholar]
- 26.Albertsen M., Hugenholtz P., Skarshewski A., Nielsen K.L., Tyson G.W., Nielsen P.H. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 27.Warinner C., Rodrigues J.F.M., Vyas R., Trachsel C., Shved N., Grossmann J., Radini A., Hancock Y., Tito R.Y., Fiddyment S., et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 2014;46:336–344. doi: 10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gevers D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker G.C., Smith J.J., Cowan D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Andersson A.F., Lindberg M., Jakobsson H., Bäckhed F., Nyrén P., Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2140. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopylova E., Noé L., Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 39.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T., Yu W.H., Izard J., Baranova O.V., Lakshmanan A., Dewhirst F.E. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:1–10. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoddard S.F., Smith B.J., Hein R., Roller B.R., Schmidt T.M. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller S., Nebe‐von‐Caron G. Functional single‐cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 2010;34:554–587. doi: 10.1111/j.1574-6976.2010.00214.x. [DOI] [PubMed] [Google Scholar]
- 44.Najah M., Griffiths A.D., Ryckelynck M. Teaching single-cell digital analysis using droplet-based microfluidics. Anal. Chem. 2012;84:1202–1209. doi: 10.1021/ac202645m. [DOI] [PubMed] [Google Scholar]
- 45.Lasken R. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem. Soc. Trans. 2009;37:450–453. doi: 10.1042/BST0370450. [DOI] [PubMed] [Google Scholar]
- 46.Lu S., Zong C., Fan W., Yang M., Li J., Chapman A.R., Zhu P., Hu X., Xu L., Yan L., et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pamp S.J., Harrington E.D., Quake S.R., Relman D.A., Blainey P.C. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell A.G., Campbell J.H., Schwientek P., Woyke T., Sczyrba A., Allman S., Beall C.J., Griffen A., Leys E., Podar M. Multiple single-cell genomes provide insight into functions of uncultured deltaproteobacteria in the human oral cavity. PLoS One. 2013;8:e59361. doi: 10.1371/journal.pone.0059361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rinke C., Lee J., Nath N., Goudeau D., Thompson B., Poulton N., Dmitrieff E., Malmstrom R., Stepanauskas R., Woyke T. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat. Protoc. 2014;9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- 50.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Y., Leung H., Yiu S.M., Chin F.Y.L. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 52.Bik E.M., Long C.D., Armitage G.C., Loomer P., Emerson J., Mongodin E.F., Nelson K.E., Gill S.R., Fraser-Liggett C.M., Relman D.A. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharon I., Morowitz M.J., Thomas B.C., Costello E.K., Relman D.A., Banfield J.F. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23:111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giagnoni L., Magherini F., Landi L., Taghavi S., Modesti A., Bini L., Nannipieri P., Renella G. Extraction of microbial proteome from soil: potential and limitations assessed through a model study. Eur. J. Soil Sci. 2011;62:74–81. [Google Scholar]
- 55.Soucy S.M., Huang J., Gogarten J.P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 56.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McClelland M., Nelson M., Raschke E. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1994;22:3640–3659. doi: 10.1093/nar/22.17.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson D., Walsh P.R., Bonventre J.A. Restriction endonucleases. Mol. Biol. Probl. Solver. 2001;9:225–266. [Google Scholar]
- 59.Bashir A., Klammer A.A., Robins W.P., Chin C.-S., Webster D., Paxinos E., Hsu D., Ashby M., Wang S., Peluso P. A hybrid approach for the automated finishing of bacterial genomes. Nat. Biotechnol. 2012;30:701–707. doi: 10.1038/nbt.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mosher J.J., Bowman B., Bernberg E.L., Shevchenko O., Kan J., Korlach J., Kaplan L.A. Improved performance of the PacBio SMRT technology for 16S rDNA sequencing. J. Microbiol. Methods. 2014;104:59–60. doi: 10.1016/j.mimet.2014.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.