Abstract
In opportunistic Gram-positive Staphylococcus aureus, a small protein called hibernation-promoting factor (HPFSa) is sufficient to dimerize 2.5-MDa 70S ribosomes into a translationally inactive 100S complex. Although the 100S dimer is observed in only the stationary phase in Gram-negative gammaproteobacteria, it is ubiquitous throughout all growth phases in S. aureus. The biological significance of the 100S ribosome is poorly understood. Here, we reveal an important role of HPFSa in preserving ribosome integrity and poising cells for translational restart, a process that has significant clinical implications for relapsed staphylococcal infections. We found that the hpf null strain is severely impaired in long-term viability concomitant with a dramatic loss of intact ribosomes. Genome-wide ribosome profiling shows that eliminating HPFSa drastically increased ribosome occupancy at the 5′ end of specific mRNAs under nutrient-limited conditions, suggesting that HPFSa may suppress translation initiation. The protective function of HPFSa on ribosomes resides at the N-terminal conserved basic residues and the extended C-terminal segment, which are critical for dimerization and ribosome binding, respectively. These data provide significant insight into the functional consequences of 100S ribosome loss for protein synthesis and stress adaptation.
INTRODUCTION
Protein synthesis is the most energy-consuming cellular process (1–3). Thus, suppression of translation during stress conditions is a universal adaptation mechanism used by all living organisms to mitigate wasteful energy consumption. In bacteria, the majority of non-translating ribosomes accumulate as 70S monomers and dimers of 70S (100S complex). Gammaproteobacteria possess three 70S ribosome-silencing factors that bind to the 30S subunit near the interface of the 30S and 50S subunits (4,5). In Escherchia coli, RMFEc and HPFEc (formerly YhbH) concertedly induce the formation of 100S ribosomes, whereas YfiAEc (also known as pY or RaiA) (6) stabilizes the inactive 70S ribosome analogously to PSRP1 in plant plastids (7). Crystal structures of the T. thermophilus ribosome in complex with RMFEc, HPFEc and YfiAEc have suggested that translational inactivation is due to the steric interference of these factors with the binding of tRNA, mRNA and initiation factors to the ribosomal decoding center on the 30S subunit (4). Electron microscopy studies have shown in vitro and in situ that the two 70S monomers in the 100S dimer are joined together via the 30S subunits (8–10). Of note, E. coli hibernating ribosomes are exclusively found during the stationary growth phase when nutrients are limited (11–13). Upon starvation relief, they are split into subunits and presumably are recycled for new rounds of translation by a poorly defined mechanism (11,14–16). Although an E. coli rmfEc mutant is reduced in long-term viability and stress tolerance during stationary phase growth (13,17,18), the deletion of hpfEc or yfiAEc does not appear to elicit strong cellular defects (19).
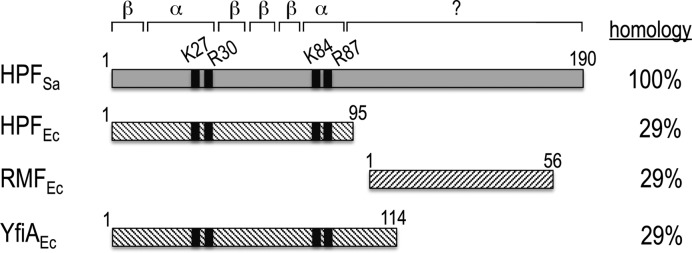
In stark contrast to E. coli, the clinically important Gram-positive bacterium Staphylococcus aureus does not contain RMF or YfiA homologs; instead, it carries only a long form of HPF (HPFSa) that is twice the size of HPFEc (Figure 1) (20–23). In fact, the long C-terminal extension, which shares some degrees of homology with RMFEc, is a hallmark of the HPF family in plant plastids and all Gram-positive bacteria (10,22,24,25). Curiously, HPFSa-mediated 100S ribosomes are present throughout all growth phases even when nutrients are abundant (23,26). The physiological relevance of these translationally incompetent ribosomes during exponential growth remains unknown. Nevertheless, the deletion of hpf in Listeria monocytogenes leads to attenuated virulence in a mouse model of infection (24) and sensitivity to prolonged antibiotic exposure (27). The Lactococcus lactis hpf mutant fails to resuscitate from nutrient-depleted growth (10).
Figure 1.
Similarity of S. aureus HPFSa to E. coli ribosome hibernating factors HPFEc, RMFEc and YfiAEc. Black patches indicate the conserved basic residues (K27, R30, K84 and R87, HPFSa numbering) on the two predicted α-helices. A question mark denotes the unknown C terminus folding.
In this study, we sought to define the role of HPFSa in a methicillin-resistant S. aureus (MRSA) strain by examining the biochemical features of HPFSa and the effect of the hpfSa null mutation on in vivo global translation. We found that the hpfSa mutant has an extremely low survival rate in long-term cultures that coincides with the rapid degradation of ribosomes. We identified HPFSa regions that are critical for ribosome dimerization and binding. Using in vivo ribosome profiling and in vitro cell-free translation, we provided experimental evidence that HPFSa attenuates translation of a subset of genes.
MATERIALS AND METHODS
Bacterial strains and reagents
Strains JE2 and NE838 are derivatives of the community-associated methicillin-resistant (CA-MRSA) S. aureus isolate USA300 (28). The hpfSa mutant strain (NE838) carries a bursa aurealis transposon insertion at 816475 that disrupts the synthesis of hpfSa. Complementing plasmid pLI50hpf was constructed by PCR to amplify the promoter and coding region of hpfSa with primers P651 (5′-CGG GAT CCA TAC AAC TGG ATT AAC AAT TCA TCG TGC AGG GTG-3′) and P627 (5′-TGA AGC TTT AAA CTT AAT TTA TTG TTC ACT AGT TTG AAT CAA GCC-3′) and the genomic DNA of JE2 as a template. The PCR fragment was then cloned into the BamHI and HindIII sites of pLI50 (29). The overexpression of recombinant His6-tagged HPF on pET28a (Novagen) in E. coli BL21(DE3) has been previously described (26). To prepare ribosomal protein S7 and L22 antibodies, the rpsG region was PCR amplified using primers P524 (5′-GGC ATA TGC CTC GTAAAG GAT CAG TAC CTA-3′) and P525 (5′-CGC TCG AGT TAC CAA CGG TAG TGA GCA AAT GCT-3′), the rplV region was PCR amplified using P522 (5′- GGC ATA TGC CTC GTA AAG GAT CAG TAC CTA-3′) and P523 (5′- CGC TCG AGT TAC CAA CGG TAG TGA GCA AAT GCT-3′) and the genomic DNA of JE2 as template. The rpsG fragment was cloned into the NdeI and XhoI sites, whereas the rplV fragment was cloned into the NheI and XhoI sites of pET28a and overexpressed in E. coli BL21(DE3). The N-terminally His6-tagged proteins were purified under native conditions as previously described (26). Polyclonal anti-S7 and anti-L22 were raised in rabbits (Josman, LLC). Polyclonal anti-HPFSa was prepared as previously described (26). S. aureus strains were routinely grown in tryptic soy broth (TSB, BD Difco), brain heart infusion (BHI, BD Difco), or chemically defined minimal medium (30) supplemented with chloramphenicol (10 μg/ml) when necessary. E. coli bearing pET28a derivatives was grown in LB supplemented with 50 μg/ml kanamycin. All strains were cultured at 37°C unless otherwise noted.
Stress assays
S. aureus strains were grown in tryptic soy broth (TSB) or minimal medium (MM) at 37°C until the mid-log or stationary phase, followed by exposure to various concentrations of stressors. Cell viability was measured at 1, 2 and 4 hr after stress exposure by enumerating colony-forming units (CFU). Cells treated with antimicrobials were washed twice with 1 ml of sterile 1x phosphate buffered saline (PBS) to eliminate carryover; the cells subsequently underwent serial dilution and were spotted on TSB agar plates. The final concentrations of stressors were as follows: rifampicin (5–50 μg/ml), mitomycin (0.5–2 μg/ml), kanamycin (5–50 μg/ml), streptomycin (5–50 μg/ml), tetracycline (50–200 μg/ml), 5% ethanol and hydrogen peroxide (5–20 mM). Cell survival was also evaluated under conditions of low pH (pH 4.3), osmosis (2.5 M NaCl), heat shock (52°C) and cold shock (10°C).
Purification of recombinant protein HPFSa
Recombinant HPFSa variants were purified with nickel affinity chromatography (MCLAB) under native conditions according to the manufacturer's manual with the following modifications: sodium phosphate buffer was replaced by 25 mM HEPES (pH 7.5) and NaCl was substituted with 500 mM KCl. Eluted proteins were stabilized in 25% (v/v) glycerol. His6-tag was removed by the thrombin-CleanCleave Kit (Sigma) in 1× cleavage buffer (20 mM HEPES (pH 7.5)/14 mM MgOAc) and incubated at room temperature overnight. The cleaved products were collected by centrifugation at 500 ×g for 2 min.
Preparation of salt-washed ribosomes
The hpfSa mutant was grown in 1 l of BHI at 37°C until OD600 = 0.6. The cells were poured over an equal volume of sterile ice cubes and harvested by centrifugation at 11 540 ×g (Beckman JLA 8.1 rotor) for 20 min at 4°C. The cell pellets were washed once with buffer A (20 mM HEPES (pH 7.5)/14 mM MgOAc/100 mM KOAc/6 mM 2-marcaptoethanol/0.5 M PMSF) and then resuspended in buffer B (20 mM HEPES (pH 7.5)/14 mM MgOAc/100 mM KOAc/1 mM DTT/0.5 mM PMSF) to a concentration of 0.5 g wet cells/ml. The cell pellet was flash-frozen in liquid nitrogen and pulverized on a cryomiller (Retch MM400) using eight 3 min cycles at 15 Hz in a 10-ml grinding jar with a 15-mm grinding ball. Grinding jars were re-chilled between each cycle in liquid nitrogen. Milled cells containing liquid nitrogen were placed in a −80°C freezer to allow for evaporation. The resulting milled cells were thawed in a 30°C water bath for 5–8 min and then immediately placed in an ice bath for 10 min. The lysate was centrifuged at 20 000 ×g for 10 min at 4°C. Clarified lysate was recovered and spun at 20,817 ×g at 4°C for 5 min to remove residual debris. To remove factors associated with ribosomes, equal volumes of lysate and 2× high-salt buffer (28 mM MgOAc/1 M KOAc/40 mM HEPES (pH 7.5)/1 mM EDTA) were mixed and incubated at 4°C on a rotator for 1 h. The mixture was layered on 1 M sucrose composed of 1× high-salt buffer and spun in a Beckman TLA-100 rotor at 4°C and 435 000 ×g for 30 min. The ribosome pellets were resuspended in 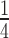 × volume of buffer B. The ribosome concentration was determined with a NanoDrop spectrophotometer (Thermo Scientific) and was calculated using 1 Abs260 = 23 pmol/ml 70S ribosome (31).
× volume of buffer B. The ribosome concentration was determined with a NanoDrop spectrophotometer (Thermo Scientific) and was calculated using 1 Abs260 = 23 pmol/ml 70S ribosome (31).
Crude lysate preparation and sucrose density gradient fractionation
TSB cultures at 37°C were harvested at 2, 28 and 96 h. Crude lysate was prepared by cryomilling as described above. Five Abs260 units of lysate were layered on 5–25% sucrose (w/v) gradient prepared with buffer C (20 mM HEPES (pH 7.5)/10 mM MgCl2/100 mM NH4Cl) and equilibrated with a BioComp Gradient Master. The gradients were spun on a Beckman Coulter L7–66 ultracentrifuge at 210 000 ×g at 4°C in a SW41 rotor for 2.5 hr. Fractionation was performed using a Brandel fractionation system equipped with a UA-6 detector. A total of 46 fractions were collected, each with a size of 200 μl. Sucrose fractions were precipitated with 10% trichloroacetic acid, and the pellets were washed once with acetone, neutralized with 50 mM Tris base containing Laemmli sample buffer and resolved using 4–20% TGX SDS-PAGE (Bio-Rad). Proteins were transferred onto a nitrocellulose membrane using a Trans-blot Turbo system (Bio-Rad). A dilution of 1/8000 of anti-HPFSa and 1/1500 of anti-S7 and anti-L22 were used for Western blots.
In vitro 100S ribosome formation
Different molar ratios of salt-washed hpf mutant ribosomes to purified HPFSa were mixed in a 100 μL reaction containing 1× binding buffer (10×: 50 mM HEPES (pH 7.5)/500 mM KOAc/100 mM NH4Cl/100 mM MgOAc). A final concentration of 0.3 μM ribosomes was routinely used in a typical experiment because no difference was observed with ribosome concentrations ranging from 0.1–0.4 μM. The mixture was incubated at 37°C for 30 min before layering on 5%-25% sucrose gradients made in buffer C. Ultracentrifugation, fractionation and immunoblotting were as described above.
Ribosome profiling
Exponentially grown S. aureus were harvested at OD600 = 0.4–0.5 after 2 min pre-treatment with 100 μg/ml of chloramphenicol. Ribosome footprints were prepared exactly as described (26), except that Ribo-Zero rRNA removal kit (Epicentre) was used in place of the MICROBExpress (Ambion). Raw FastQ sequencing data were processed using a locally installed Galaxy platform. The rRNA-less reads were aligned to the USA300_FPR3757 reference genome (GenBank CP000255.1) using Bowtie v.0.12.0. The alignment .map files were used as inputs for the modified Python scripts(32). The normalized ribosome densities, measured as reads per million reads (RPM), were visualized in MochiView (33). The gene expression levels (reads per kilobase per million mapped reads, RPKM) were calculated using Python scripts (32). Translation efficiency (TE) was calculated as the relative number of ribosome footprints to mRNA-seq reads in log2 ratios (34). Sequencing data were deposited in the NCBI GEO database with accession number GSE74197.
Hybrid in vitro translation
The hybrid in vitro translation system consists of the salt-washed S. aureus ΔhpfSa ribosome and purified translation factors from PURExpressΔribosome Kit (New England BioLabs). Ribosomes were pre-incubated with or without HPFSa in 1x binding buffer at 37°C for 30 min. HPFSa targets were translated by programming T7 promoter-containing DNA templates (final 10 ng/μl), 10 μCi Tran35S-label (MP Biomedicals) and 0.8 μM of ribosome/HPFSa or ribosome alone premix. After 1 hr incubation at 37°C, protein samples were precipitated by 4× volumes of acetone, resolved on 4–20% TGX SDS-PAGE (BioRad) and autoradiographed.
RESULTS
HPFSa is required for long-term viability of S. aureus
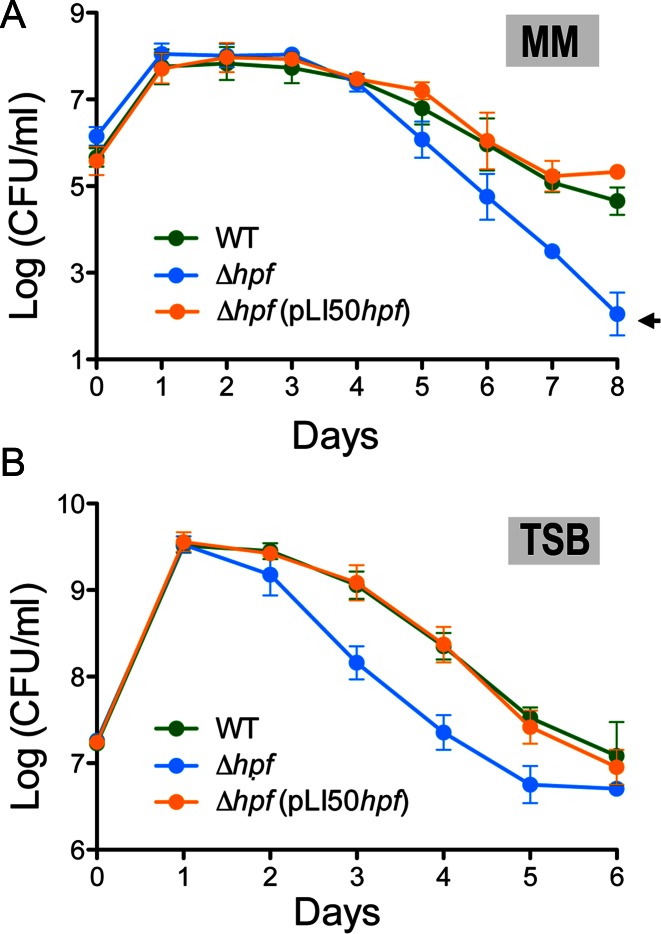
The phenotypic manifestations of hpfSa remain unexplored, and studies from other bacterial counterparts point to hibernating ribosomes playing a role in the stress response. We subjected wild-type and hpfSa mutant strains to a variety of agents that are commonly used to assess virulence. All of the tested stressors, including hydrogen peroxide, high salt, low pH, transcription inhibition (rifampicin), DNA damage induction (mitomycin) and ribosome-targeting antibiotics (kanamycin and tetracycline), sensitized the wild-type and mutant cells equally, regardless of the dosage, bacterial growth phase or medium type. However, we found that the viability of the hpfSa mutant declined rapidly in long-term minimal medium (MM) culture and eventually lost >95% of cell viability at day eight, yielding approximately 400-fold fewer CFU than the wild-type strain (Figure 2A). A similar downtrend was observed in rich tryptic soy broth (TSB) medium, although cell death was less pronounced (approximately 10-fold) than in MM (Figure 2B). In both cases, complementation of the hpfSa mutant with a plasmid-borne copy of hpfSa under control of its native promoter fully rescued the survival defect, confirming that hpfSa is essential for S. aureus long-term viability.
Figure 2.
HPFSa mediates long-term cell survival. (A) Cell viability in minimal medium (MM) over an 8-day period. (B) Cell viability in tryptic soy broth (TSB) over a 6-day period. Viability was assessed by enumerating colony-forming units (CFU) from wild-type, hpf null and complemented strain carrying pLI50hpf. Error bars indicate the standard deviation of three independent experiments. An arrow denotes the detection limit.
Loss of hpfSa sharply reduces the intact ribosome pool during the stationary phase
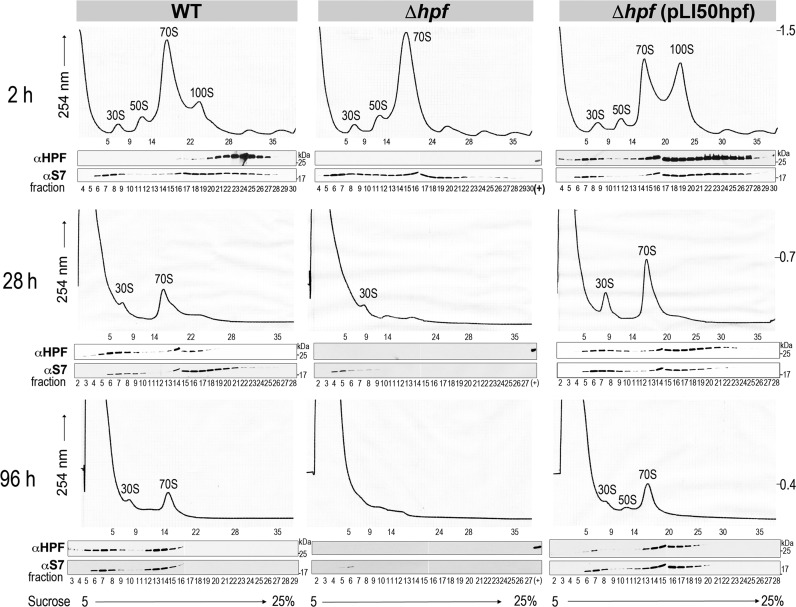
To investigate the link between 100S ribosome formation and the reduced survival rate in the hpfSa mutant, we monitored changes in ribosome species during stationary culturing by sucrose density gradient fractionation. Crude ribosomes were prepared from cells grown at different stages and extracted by cryomilling, a method known to faithfully preserve ribosome complexes and translational states (26,35,36). Consistent with previous studies (23,26), we observed an accumulation of 100S ribosomes in rich medium during early logarithmic growth (OD600∼ 0.3–0.4) (Figure 3 (top left), S1A). Western blots showed that the majority of HPFSa was associated with the 100S complex and 70S ribosome (Figure 3 (top left)). In the complemented strain in which plasmid-borne HPFSa was expressed approximately 10-fold more than wild type, the 100S ribosome peak was substantially elevated and a fraction of HPFSa also bound to the 30S subunits (Figure 3 (top right)). Curiously, the increased production of 100S ribosomes did not significantly slow bacterial growth over 24 h in the TSB culture (Supplementary Figure S1A). During the stationary phase, a substantial amount of the ribosome pool diminished in the wild-type and complemented strains (Figure 3 (middle and bottom)), presumably due to massive ribosome degradation to provide cells with energy and nutrients. The disappearance of the 100S complex and the basal 70S ribosome level in these strains suggested that 100S particles may have been redistributed to form translationally competent ribosomes. The exact sequence of 100S to 70S inter-conversion is unclear. In contrast, most of the ribosomes in the hpfSa mutant were degraded, which coincided with the onset of cell death in prolonged culture (Figure 2B). Free 30S and 50S subunits are more prone to ribonuclease attack than the 70S ribosome because of their exposed rRNA interface (37). The accumulation of 70S dimers in the wild-type cells may thus serve as a nuclease-resistant ribosome repository.
Figure 3.
Accelerated ribosome degradation in the S. aureus hpf null strain. Sucrose gradient profiles of S. aureus ribosomes harvested from TSB cultures at the logarithmic phase (top panels), the stationary phase (middle panels) and the late stationary phase (bottom panels). The y-axis corresponds to the absorbance at 254 nm of the ribosome species separated on a 5–25% sucrose gradient. Abs254 values on the right serve as a ruler to compare relative ribosome peak height. Each panel corresponds to five Abs260 units of total RNA input. The distribution of HPFSa was detected via immunoblotting. The 30S ribosomal subunit protein S7 is a marker control for fractionation. ‘(+)’ denotes loading control with the purified recombinant HPFSa.
HPFSa alone is sufficient to induce the formation of 100S ribosomes in vitro
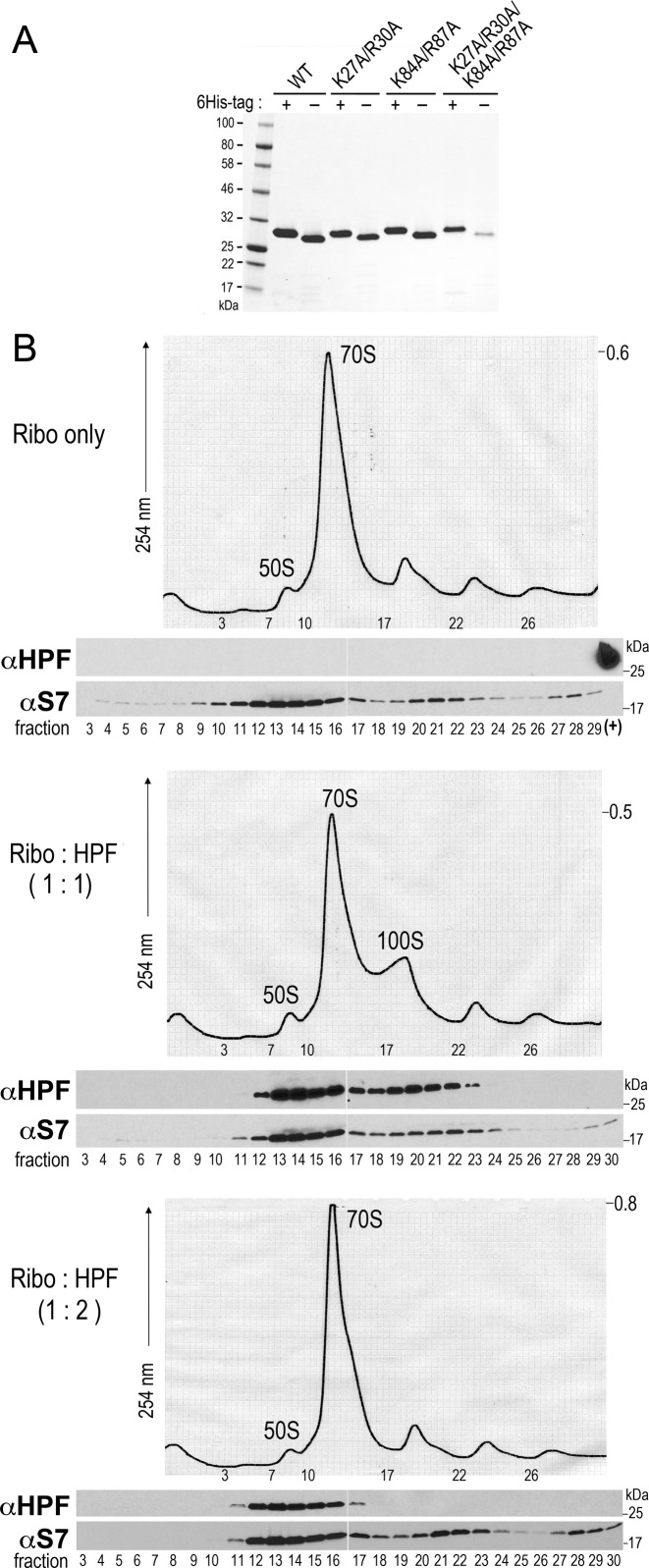
To confirm that HPFSa is the sole contributor to the formation of 100S ribosomes in vivo, we recapitulated 70S ribosome dimerization in vitro using purified HPFSa and S. aureus ribosomes isolated from the hpfSa null strain. The ribosomes were subjected to a high-salt wash to remove any associated factors. Hexahistidine-tagged HPFSa was overexpressed in E. coli and purified by nickel affinity chromatography. The affinity tag was removed by thrombin cleavage because 6His-HPFSa was prone to aggregation, possibly due to misfolding, and was nonfunctional in 100S complex formation (Figure 4A). Purified HPFSa was incubated with high-salt washed ribosomes at different molar ratios, and the ribosome profiles were obtained by sucrose density gradient fractionation. At a 1:1 ribosome to HPFSa ratio, HPFSa induced the dimerization of 70S ribosomes to a level comparable to that observed in vivo (Figure 3 (top left), 4B (middle)). Increasing the amount of HPFSa did not enhance dimerization, although immunoblotting showed that the protein effectively bound to the 70S ribosome. In fact, we found that a modest increase of the ribosome to HPFSa ratio to 1:2 completely abrogated dimerization but did not affect ribosome association (Figure 4B, bottom). The observation that a higher concentration of HPFSa is counterproductive could be attributable to HPFSa self-oligomerization or/and non-specific binding of HPFSa to the ribosome when HPFSa is in large molar excess. Nevertheless, our data demonstrate that optimal in vitro formation of the 100S ribosome occurs when HPFSa and the ribosome are present at a 1:1 molar ratio, which is in agreement with previous in vivo estimation of the E. coli 100S ribosome (23).
Figure 4.
HPFSa induces 100S dimer formation in vitro with a 1:1 ribosome to HPFSa ratio. (A) SDS-PAGE gel showing the purity of recombinant wild-type HPFSa and its mutants. ‘-’ indicates His6-tag-free proteins that have been subjected to thrombin cleavage. (B) Sucrose density gradient (5-25%) profiles of salt-washed ribosome isolated from S. aureus hpf null strain (top) and mixture of ribosomes and HPFSa in a ratio of 1:1 (middle) and 1:2 (bottom). Abs254 values on the right serve as a ruler to compare relative ribosome peak height. Association of HPFSa with ribosomes was confirmed by immunoblotting using anti-HPFSa (1:8000 dilution) and the internal control anti-S7 (1:1500 dilution). ‘(+)’ denotes a loading control with the purified recombinant HPFSa.
Ribosome dimerizing activity of HPFSa is highly sensitive to perturbation mutagenesis
Our in vitro dimerization results showed that HPFSa binding to the ribosome is not sufficient to promote 100S formation. The result prompted us to investigate the domains that are critical for dimerization. HPFEc adopts a βαβββα fold, with two basic patches each on the two α-helices (Supplementary Figure S2) (4). These positively charged residues are conserved in all HPF and YfiA homologs (Figure 1) and have been implicated in direct interactions with the 16S rRNA bases at the decoding channel (4,5). In vitro dimerization was repeated using purified HPFSa mutant proteins harboring the desired mutations in the basic patches (Figure 4A). Although the double mutation K27A/R30A on the first α-helix did not completely impair dimerization, the double mutation K84A/R87A on the second α-helix did but did not abolish ribosome binding, as a large fraction of mutant HPFSa was still associated with the 70S ribosomes (Figure 5A and B). To abrogate HPFSa binding to the ribosome, concurrent mutations (K27A/R30A/K84A/R87A) on both helices were required, as judged by the sedimentation of mutant HPFSa in the light sucrose fraction. (Figure 5B, bottom). Notably, several mutations outside the α-helices did not perturb 70S dimerization (Supplementary Figure S3). HPFSa contains a long C-terminal tail that is absent in HPFEc (Figure 1). Deletion of the last 42-amino acid segment reduced ribosome binding, whereas a larger truncation (up to 90 amino acids) exacerbated the binding defect further (Supplementary Figure S4). The data suggest that the basic residues on the second α-helix (K84/R87) are critical for dimerization, whereas both α-helices and the C-terminal tail are important for ribosome binding and/-or the stabilization of binding.
Figure 5.
Mutations that affect HPFSa-mediated 100S dimer formation and binding to the ribosome. (A) Sucrose density gradient (5-25%) profiles of salt-washed ribosomes reacted with an equimolar ratio of purified HPFSa and its mutant variants. (B) Western blots showing the association of HPFSa in each ribosomal fraction. The 30S ribosomal subunit protein S7 serves as a control. A K27A/R30A double mutant has reduced 100S dimer formation. A K84A/R87A double mutant impairs dimerization but not binding. The quadruple mutant abolishes binding.
Loss of hpfSa causes unusual ribosome occupancy around start codons under nutrient-limited conditions
The paradoxical observation that 100S ribosomes are prevalent in S. aureus growing logarithmically in rich culture medium with a growth rate that is unaffected by HPFSa loss (Supplementary Figure S1A, 3 (top)) is inconsistent with the notion that HPF acts as a global ribosome inhibitor (4,20,22). It is conceivable that ribosomes are in excess in rich medium, and the subpopulation of 100S ribosomes is unable to titrate the bulk of ribosomes from normal translation, resulting in a lack of growth defect. It may be that the 100S ribosome is simply a quiescent product to conserve ribosome and has no direct role in regulating translation. Alternatively, the formation of 100S ribosomes may downregulate the translation of only a fraction of mRNAs that are not essential for exponential growth when nutrients are plentiful. This prediction is supported by the observation that when nutrients were limited, both wild-type and complemented strains were delayed in entering the stationary phase compared with the hpfSa mutant (Supplementary Figure S1B). It seems plausible that HPFsa directly participates in translational repression and leads to a slower growth rate in minimal medium. To test this model, we performed a deep sequencing-based ribosome profiling analysis to compare the translatomes of exponentially growing wild-type and hpfSa mutant strains in both minimal and rich media. Ribosome profiling captures a global snapshot of ribosome positioning and density on template mRNAs with single-nucleotide resolution (34,36). A high number of ribosome-protected footprints (RPFs) mapping to the transcriptome (mRNA-seq) at a unique position or being distributed across the transcript is indicative of ribosome stalling or active translation, respectively.
Our initial attempt to construct RPF libraries from stationary phase cells was unsuccessful. We opted for mid-log cells, as the 100S ribosomes are already prevalent at this stage. Exponentially grown S. aureus were collected at OD600 0.4–0.5. Unlike other cell lysate preparations described in this study, the cells subject to ribosome profiling were pretreated with 100 μg/ml chloramphenicol for 2 min to stabilize the polysomes (translating ribosomes) before harvest (Supplementary Figure S5). Consistent with previous observations (26), the addition of chloramphenicol appears to trap HPFSa in all ribosomal fractions (Supplementary Figure S5). Cryogenic cell disruption, construction of cDNA libraries and data analyses have been previously described in detail (26,32,36). A total of 16 RPFs and total mRNA-seq libraries were generated from the wild-type and hpfSa mutant strains, representing two independent biological replicates from rich and minimal media (Supplementary Table S1).
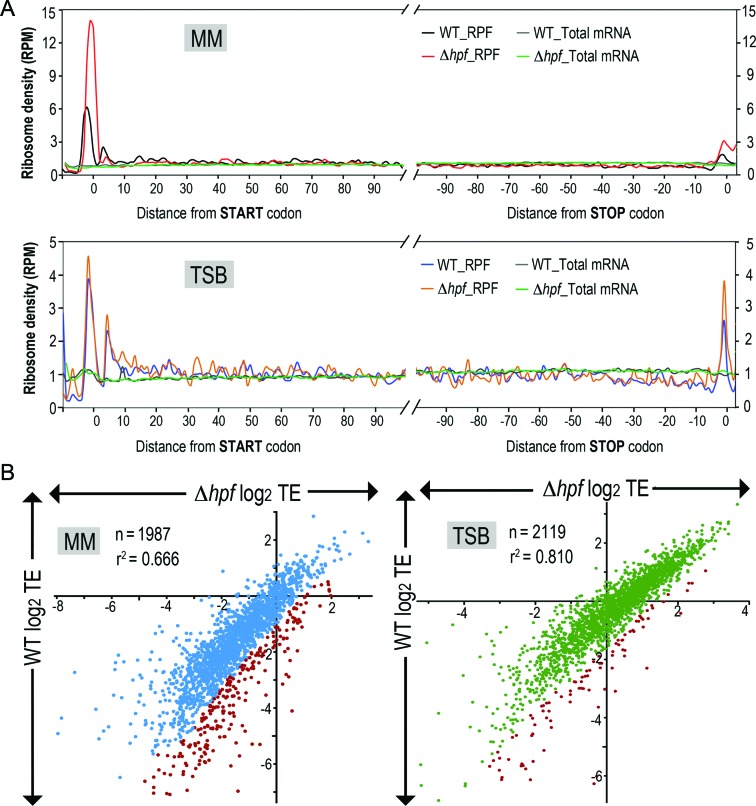
To obtain an overview of global differences in ribosome density at the beginning and end of open reading frames (ORFs), metagene analysis was performed to analyze the distribution of ribosomes along an average transcript of the most abundantly expressed genes (≥50 reads/transcript). Elevated ribosome density around the start and stop codons corresponds to slower kinetics of initiation and termination than those of the elongation rate (36). Based on the prediction that HPFSa inhibits translation, we expected that loss of HPFSa would lead either to clustering of ribosomes at the very N-terminus of ORFs, reflecting derepression of translation initiation, or to disappearance of occupancy at unique positions within ORFs, suggesting a relief of elongation arrest, or to an increase in ribosome occupancy along the transcripts, indicating elevated translation efficiency. As predicted, the loss of HPFSa disproportionately increased ribosome occupancy around start codons transcriptome-wide in cells grown in minimal medium relative to the wild-type (Figure 6A, Supplementary Figure S6). Unexpectedly, despite a feature that suggests higher translation initiation, downstream translation in the hpfSa mutant did not increase to a level that positively correlates with the elevated ribosome density at the initiation site (Figure 6A, top). The unusual accumulation of ribosomes at the 5′-end could be an artifact derived from the chloramphenicol treatment as a recent study showed that the addition of translation inhibitors could distort ribosome coverage profiles in eukaryotes (38). This is highly unlikely in our case because the same elevated ribosome density was not observed in rich medium samples (Figure 6A, bottom) and MM-grown WT samples (Figure 6A, top). In rich TSB medium, a modest increase of ribosome density along the entire transcript was observed in the hpfSa mutant, suggesting moderate inhibition of translation by HPFSa (Supplementary Figure S6).
Figure 6.
Effects of hpfSa null mutation on global translatome in minimal and rich culture conditions. (A) Ribosome density as a function of position. Metagene analysis of read densities in minimal medium (MM, top) and tryptic soy broth (TSB, bottom) samples. The normalized reads per million mapped reads (RPM) correspond to the average ribosome density across the most abundantly translated ORFs (>50 reads), which were aligned relative to the start and stop codons. Ribosome density was drastically elevated around start codons in the hpfSa mutant grown in MM, whereas such an increase was not observed in TSB culture. (B) Comparison between translation efficiency (TE) in the wild-type and hpfSa mutant in MM (left) and TSB (right). Each dot corresponds to a single gene. Genes with TE improvement upon hpfSa loss are shown in red. TE was moderately increased in the hpfSa mutant and more pronounced in MM cultures. TE was calculated as the log2 ratios of the RPFs to the mRNA reads that were measured in RPKM.
hpfSa null mutation increases translational efficiency of a subset of mRNAs
To evaluate how efficiently an individual transcript was translated, we calculated the TE in the wild-type and mutant strains by dividing ribosome footprints by mRNA-seq density (Figure 6B). Plotting the TEs from the hpfSa mutant against WT provides information about the changes in ribosome occupancy between strains. An improvement of hpfSa TE will be indicated by a log2 TE value of hpfSa (x-axis) that is larger than the log2 TE value of the wild-type (y-axis), whereas high R2 correlations suggest that TE is less affected by the loss of hpfSa. In general, TE in most genes remained unchanged between the wild-type and mutant. TE improvement in the hpfSa mutant was more pronounced in MM than in TSB culture (R2 = 0. 67 (MM) and 0.81 (TSB)). Consistent with the metagene analysis, we found that genes displaying high ribosome density at the 5′-end were not always correlated with the overall improvement of TE. As a result, fewer than 10% of the actively transcribed mRNAs exhibited a significant TE increase (2-fold) in the absence of HPFSa (Supplementary Tables S2 and S3). Thus, the loss of hpfSa appears to have a modest impact on overall protein output during exponential growth.
HPFSa-mediated translational inhibition in vitro
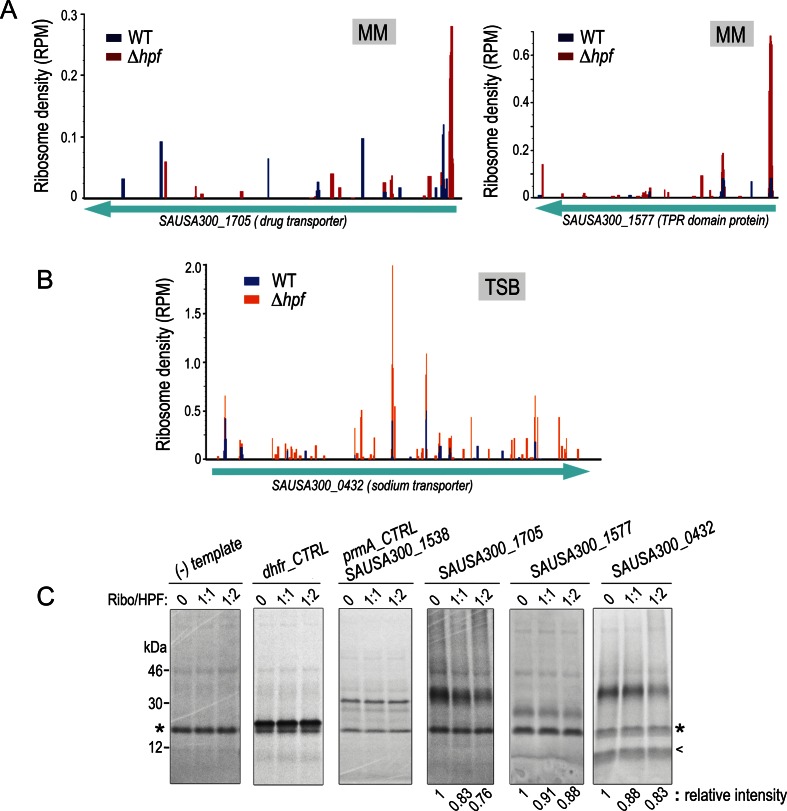
To determine the effects of HPFSa on the protein yield from transcripts that were identified from ribosome profiling, we used a cell-free hybrid translation system that combines purified E. coli translation factors and ΔhpfSa ribosomes. Because the sequence context at the 5′-end might influence the translation efficiency of individual genes, we preserved the native Shine-Dalgarno (SD) sequence and start codon in T7 promoter-driven DNA templates. Representative genes whose TEs were elevated by >2-fold were randomly selected based on known function (Figure 7A and B). Translation reactions were programmed with 35S-methionine and various molar ratios of ribosomes to HPFSa. The protein production varied because each template carried a different translation initiation signal sequence. The intensities of the anticipated protein products were quantitated by normalization to an ∼17 kDa non-specific band, whose levels remained constant in a minus template control and throughout HPFSa titration (Figure 7C). The non-specific band did not appear to be aminoacyl-tRNAs as the labile ester bond would be hydrolyzed on the alkaline SDS-PAGE. In brief, the inhibition of translation by HPFSa could be recapitulated in vitro, but the effects were less pronounced than the in vivo ribosome profiling effects, presumably due to subtle incompatibility of the purified E. coli components with the S. aureus ribosome. The inhibition was specific to HPFSa targets because the same inhibition was not seen with the non-target prmA or the heterologous E. coli dhfr.
Figure 7.
In vitro translation with S. aureus ribosomes recapitulates HPFSa-mediated translational repression. (A) Ribosome density profiles showing the strong increase in ribosome occupancy around start codons of selected genes in the hpfSa mutant grown in minimal medium (MM). (B) Ribosome density plot of genes derived from tryptic soy broth (TSB) samples that exhibit an overall increase in ribosome occupancy along the gene body. (C) In vitro translation validating the repression of translation in the genes shown in panels A and B. Translation products were resolved on an alkaline Tris-glycine-based 4–20% TGX SDS-PAGE. An asterisk depicts the non-specific band used as an internal reference for intensity quantitation. A cursor indicates a second non-specific band that was fortuitously intensified in a specific translation reaction. The predicted gene product of SAUSA300_0432 is a 50 kDa sodium transporter; it runs aberrantly faster on SDS-PAGE, which is not uncommon for membrane proteins.
DISCUSSION
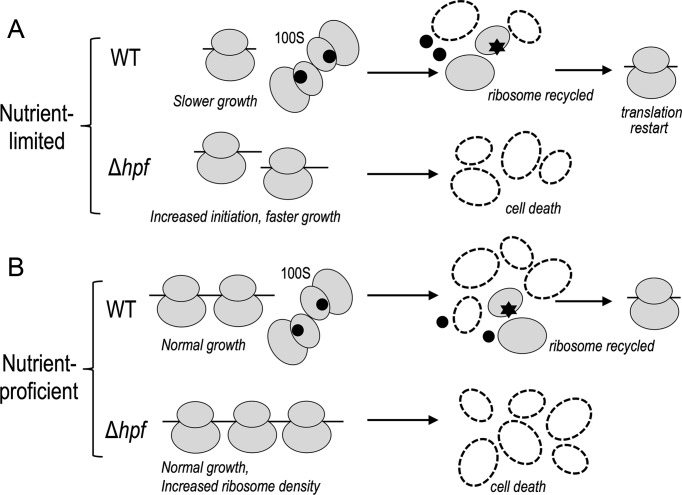
The bacterial 100S ribosome was first observed more than five decades ago (40,41). Its function has remained obscured despite its pervasiveness at different bacterial growth phases. Likewise, little is known about the biochemical properties of the factors (e.g. HPF, RMF) that promote 100S formation. Here, we integrate biochemistry and ribosome profiling to characterize the biological and biochemical features of HPFSa in the opportunistic pathogen S. aureus. HPFSa is a peculiar homolog in that it is unusually long, is constitutively expressed, and is functionally equivalent to RMFEc and HPFEc combined. Whereas 100S formation in E. coli is stationary phase-specific, it occurs in S. aureus as early as the post-lag phase. We show that the loss of hpfSa causes massive ribosome breakdown upon entering the stationary phase that correlates with the onset of cell death. It seems likely that 100S ribosomes provide a ‘readiness plan’ in advance of future growth constraints when they can be redistributed for translation reinitiation (Figure 8). Beyond their role as a ‘reservoir’, the formation of 100S ribosomes in logarithmically growing cells reduces the translation efficiency of a fraction of genes. Furthermore, structure-guided mutagenesis suggests important roles of the C-terminal segment of HPFSa in ribosome binding and of the conserved basic residues in dimerization.
Figure 8.
A model depicting the role of HPFSa. (A) In minimal medium, HPFSa-mediated formation of 100S dimers causes a reduction of translation capacity, and the cells grow slower than the hpfSa mutant. Upon entering the stationary phase, 100S dimers in the wild-type are converted to recyclable ribosome subunits to support a viable state, whereas all post-termination ribosomes in the hpfSa mutant are degraded, leading to cell death. (B) In rich medium, the sequestration of excess ribosomes in the form of 100S complexes does not affect cell growth, but the translation efficiency of a small set of genes is reduced. hpfSa mutation increases the active ribosome pool and increases the ribosome occupancy of specific genes. Hyperactive translation in the hpfSa mutant leads to rapid degradation of ribosomes during the stationary phase. A black circle denotes HPFSa and a star denotes an unknown factor that splits the 100S dimer.
Although the 100S ribosomes are prevalent in bacteria, translationally silent ribosomes in eukaryotes exist as 80S monomers held by the clamping protein Stm1 (43). The mammalian equivalent of 100S, a dimer of 80S monomers (110 complex), occurs infrequently except under nutrient starvation conditions (42).
The rapid degradation of ribosomes in hpf null mutants has not been reported. The degradation of S. aureus ribosomes likely involves RNase III and RNase J1/J2 (44), as implicated by the accumulation of 100S dimers in Streptomyces rnc and rnj mutants (45). Curiously, decay of the GFP-tagged E. coli ribosome is not aggravated in rmfEc and hpfEc mutants. Under our experimental conditions, the S. aureus USA300 strain enters the stationary phase much earlier than do other Gram-positive bacteria (10,22,24). The differences are likely due to the choice of medium, culture conditions and strains. The strain-specific abundance of 100S ribosomes has been observed in E. coli (47). In our hands, S. aureus RN4200 and N315 produce 30% more 100S ribosomes than the strain (USA300) we used in this study under the same conditions.
We propose a model for the role of HPFSa (Figure 8). In minimal medium, the translation of a fraction of genes is suppressed because a subpopulation of ribosomes is sequestered away from active translation, leading to a slower growth rate. The reason underlying the specific targeting of mRNA has yet to be determined. It is possible that some mRNAs are more sensitive to altered cellular concentrations of active ribosomes; e.g. the translation of genes with highly structured mRNA or weaker SD sequence may require higher concentrations of ribosome. A loss of hpfSa liberates the ribosome for translation initiation but only partially increases the overall translational output, presumably due to low cellular concentrations of charged-tRNAs and other translation factors. Both wild-type and hpfSa null strains undergo ribosome degradation, but the former retains a fraction of the functional ribosomes that are derived from the 100S pool (Figure 8A). The process by which 100S dimers are disassembled and converted to active 70S ribosome is unclear, although the participation of the initiation factors IF1 and IF3 and ribosome recycling factor RRF have been implicated in E. coli (4,11,14). In rich medium, excess unused ribosomes are preserved as silent 100S dimers. The formation of the 100S complex does not significantly affect wild-type cell growth but is important for viability during the stationary phase, when the ribosome pool is low. The absence of HPFSa increases ribosome occupancy but only modestly upregulates translation efficiency (Figure 8B). The reason is unclear, although it is possible that elevated ribosome occupancy causes ribosomal traffic jams, which in turn may perturb polysome organization (48) and limit translation.
Although an increase in ribosome density at the 5′ regions of coding sequences suggests elevated translation initiation, it is possible that secondary effects in the absence of hpfSa, such as proteotoxic stress, may lead to ribosome stalling at 5′ proximal regions (49). Why is only a subset of genes derepressed in the hpfSa null strain, particularly at the initiation phase? These genes do not belong to a specific functional category, although several important virulence factors are among the list (Supplementary Tables S2 and S3). No correlation with gene length or the mRNA's abundance was observed. Sequence context, mRNA secondary structure and codon usage bias may shape the expression levels of individual genes (50). Moreover, the first 50 codons have been shown to dictate translation efficiency (51). Our attempts to identify unique sequence signatures upstream and downstream of the start codons were unsuccessful, although we noted that the spacer between the SD sequence and start codon in HPFSa targets is highly enriched in uracil in contrast to the non-targets’ enrichment in adenine. Furthermore, mRNA secondary structures that occlude the SD sequence and initiation site appear to be overrepresented in the HPFSa-affected genes. The significance of these features remains to be determined.
The formation and stability of 100S ribosomes are sensitive to ionic strength and magnesium concentration (21). The binding of RMFEc and HPFEc induces a structural rearrangement of the 30S subunit head domain that is necessary for dimerization (4). Whereas dimerization is impaired by minor perturbation of the conserved basic patches of HPFSa, ribosome binding is abrogated when a large deletion or concurrent mutations on the two predicted α-helices are introduced. These data imply that HPFSa interacts with the ribosome at multiple sites and that a single-site defect does not abolish binding. Likewise, dimerization does not occur prior to the stabilization of ribosome binding. Previous work (10) has shown that the L. lactis HPFLl C-terminal tail (residues 126–185) is functionally equivalent to that of RMFEc. Truncation of this region does not impair ribosome binding, which is completely opposite to HPFSa. These differences suggest some idiosyncrasy of long-form HPF homologs in interacting with their cognate ribosomes.
Our data imply that HPF/100S ribosomes serve two purposes in S. aureus. The first is to suppress the translation of specific mRNA through titrating active ribosomes concomitantly with reducing energy consumption. The second is to avoid rapid ribosome turnover by dimerizing 70S monomers so that the dimers can be dissociated and reassembled into active ribosomes during stationary phase. Bacterial persistence has been linked to the suppression of macromolecule biosynthesis and translation (52–54) and S. aureus is particularly adept at establishing long-term survival inn the host, which leads to recalcitrant and relapsing infections (55). Given that the hpfSa null strain is non-viable in long-term minimal medium culture, strategies that disrupt the 100S ribosome may vastly synergize the efficacy of conventional antibiotics and may serve as a new anti-staphylococci drug target to treat recalcitrant infections.
ACCESSION NUMBER
NCBI GEO GSE74197.
Supplementary Material
Acknowledgments
We thank David Gohara and Hye-Sook Kim for providing analysis tools. S. aureus USA300 derivatives were acquired from the NIH/NIAID-funded Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources.
FUNDING
National Institutes of Health [R00GM094212 to M.N.Y.]; Edward Mallinckrodt Jr. Foundation; Saint Louis University faculty start-up fund. M.N.Y. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. Funding for open access charge: The Pew Charitable Trusts [00002920].
Conflict of interest statement. None declared.
REFERENCES
- 1.Buttgereit F., Brand M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995;312:163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell J.B., Cook G.M. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szaflarski W., Nierhaus K.H. Question 7: optimized energy consumption for protein synthesis. Orig. Life Evol. Biosph. 2007;37:423–428. doi: 10.1007/s11084-007-9091-4. [DOI] [PubMed] [Google Scholar]
- 4.Polikanov Y.S., Blaha G.M., Steitz T.A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vila-Sanjurjo A., Schuwirth B.S., Hau C.W., Cate J.H. Structural basis for the control of translation initiation during stress. Nat. Struct. Mol. Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 6.Agafonov D.E., Kolb V.A., Spirin A.S. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2001;2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma M.R., Donhofer A., Barat C., Marquez V., Datta P.P., Fucini P., Wilson D.N., Agrawal R.K. PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G) J. Biol. Chem. 2010;285:4006–4014. doi: 10.1074/jbc.M109.062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato T., Yoshida H., Miyata T., Maki Y., Wada A., Namba K. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure. 2010;18:719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz J.O., Brandt F., Matias V.R., Sennels L., Rappsilber J., Scheres S.H., Eibauer M., Hartl F.U., Baumeister W. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J. Cell Biol. 2010;190:613–621. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puri P., Eckhardt T.H., Franken L.E., Fusetti F., Stuart M.C., Boekema E.J., Kuipers O.P., Kok J., Poolman B. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol. Microbiol. 2014;91:394–407. doi: 10.1111/mmi.12468. [DOI] [PubMed] [Google Scholar]
- 11.Maki Y., Yoshida H., Wada A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells. 2000;5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 12.Wada A., Igarashi K., Yoshimura S., Aimoto S., Ishihama A. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem. Biophys. Res. Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 13.Yamagishi M., Matsushima H., Wada A., Sakagami M., Fujita N., Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida H., Ueta M., Maki Y., Sakai A., Wada A. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells. 2009;14:271–280. doi: 10.1111/j.1365-2443.2008.01272.x. [DOI] [PubMed] [Google Scholar]
- 15.Aiso T., Yoshida H., Wada A., Ohki R. Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J. Bacteriol. 2005;187:1951–1958. doi: 10.1128/JB.187.6.1951-1958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wada A., Yamazaki Y., Fujita N., Ishihama A. Structure and probable genetic location of a ‘ribosome modulation factor’ associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Sharoud W.M., Niven G.W. The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology. 2007;153:247–253. doi: 10.1099/mic.0.2006/001552-0. [DOI] [PubMed] [Google Scholar]
- 18.Niven G.W. Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch. Microbiol. 2004;182:60–66. doi: 10.1007/s00203-004-0698-9. [DOI] [PubMed] [Google Scholar]
- 19.Ueta M., Yoshida H., Wada C., Baba T., Mori H., Wada A. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells. 2005;10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 20.Ueta M., Ohniwa R.L., Yoshida H., Maki Y., Wada C., Wada A. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J. Biochem. 2008;143:425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida H., Wada A. The 100S ribosome: ribosomal hibernation induced by stress. Wiley Interdiscip. Rev. RNA. 2014;5:723–732. doi: 10.1002/wrna.1242. [DOI] [PubMed] [Google Scholar]
- 22.Ueta M., Wada C., Daifuku T., Sako Y., Bessho Y., Kitamura A., Ohniwa R.L., Morikawa K., Yoshida H., Kato T., et al. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells. 2013;18:554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 23.Ueta M., Wada C., Wada A. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells. 2010;15:43–58. doi: 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 24.Kline B.C., McKay S.L., Tang W.W., Portnoy D.A. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J. Bacteriol. 2015;197:581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagami K., Nanamiya H., Kazo Y., Maehashi M., Suzuki S., Namba E., Hoshiya M., Hanai R., Tozawa Y., Morimoto T., et al. Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. MicrobiologyOpen. 2012;1:115–134. doi: 10.1002/mbo3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis A.R., Gohara D.W., Yap M.N.F. Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15379–15384. doi: 10.1073/pnas.1410356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKay S.L., Portnoy D.A. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 2015;59:6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fey P.D., Endres J.L., Yajjala V.K., Widhelm T.J., Boissy R.J., Bose J.L., Bayles K.W. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4 doi: 10.1128/mBio.00537-12. doi:10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C.Y., Buranen S.L., Ye Z.H. Construction of single-copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 30.Pattee P.A., Neveln D.S. Transformation analysis of three linkage groups in Staphylococcus aureus. J. Bacteriol. 1975;124:201–211. doi: 10.1128/jb.124.1.201-211.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G, editor. Ribosomes and protein synthesis: a practical approach. NY: Oxford University Press; 1990. pp. 1–29. [Google Scholar]
- 32.Becker A.H., Oh E., Weissman J.S., Kramer G., Bukau B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat. Protoc. 2013;8:2212–2239. doi: 10.1038/nprot.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Homann O.R., Johnson A.D. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 2010;8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaCava J., Molloy K.R., Taylor M.S., Domanski M., Chait B.T., Rout M.P. Affinity proteomics to study endogenous protein complexes: pointers, pitfalls, preferences and perspectives. Biotechniques. 2015;58:103–119. doi: 10.2144/000114262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh E., Becker A.H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R.J., Typas A., Gross C.A., Kramer G., et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zundel M.A., Basturea G.N., Deutscher M.P. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerashchenko M.V., Gladyshev V.N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy B.J. Variations in bacterial ribosomes. Biochim. Biophys. Acta. 1960;39:563–564. [Google Scholar]
- 41.Tissieres A., Watson J.D. Ribonucleoprotein particles from Escherichia coli. Nature. 1958;182:778–780. doi: 10.1038/182778b0. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 43.Van Dyke N., Baby J., Van Dyke M.W. Stm1p, a ribosome-associated protein, is important for protein synthesis in Saccharomyces cerevisiae under nutritional stress conditions. J. Mol. Biol. 2006;358:1023–1031. doi: 10.1016/j.jmb.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 44.Bonnin R.A., Bouloc P. RNA degradation in Staphylococcus aureus: diversity of ribonucleases and their impact. Int. J. Genomics. 2015;2015:395753. doi: 10.1155/2015/395753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones S.E., Leong V., Ortega J., Elliot M.A. Development, antibiotic production, and ribosome assembly in Streptomyces venezuelae are impacted by RNase J and RNase III deletion. J. Bacteriol. 2014;196:4253–4267. doi: 10.1128/JB.02205-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shcherbakova K., Nakayama H., Shimamoto N. Role of 100S ribosomes in bacterial decay period. Genes Cells. 2015;20:789–801. doi: 10.1111/gtc.12273. [DOI] [PubMed] [Google Scholar]
- 47.Wada A., Mikkola R., Kurland C.G., Ishihama A. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J. Bacteriol. 2000;182:2893–2899. doi: 10.1128/jb.182.10.2893-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandt F., Etchells S.A., Ortiz J.O., Elcock A.H., Hartl F.U., Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 49.Han Y., Gao X., Liu B., Wan J., Zhang X., Qian S.B. Ribosome profiling reveals sequence-independent post-initiation pausing as a signature of translation. Cell Res. 2014;24:842–851. doi: 10.1038/cr.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kudla G., Murray A.W., Tollervey D., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 52.Cho J., Rogers J., Kearns M., Leslie M., Hartson S.D., Wilson K.S. Escherichia coli persister cells suppress translation by selectively disassembling and degrading their ribosomes. Mol. Microbiol. 2015;95:352–364. doi: 10.1111/mmi.12884. [DOI] [PubMed] [Google Scholar]
- 53.Ehrenberg M., Bremer H., Dennis P.P. Medium-dependent control of the bacterial growth rate. Biochimie. 2013;95:643–658. doi: 10.1016/j.biochi.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 54.Scott M., Gunderson C.W., Mateescu E.M., Zhang Z., Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 55.Proctor R.A., Kriegeskorte A., Kahl B.C., Becker K., Loffler B., Peters G. Staphylococcus aureus Small Colony Variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front Cell Infect Microbiol. 2014;4:99. doi: 10.3389/fcimb.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.