Abstract
Gene structure and expression in diplonemid mitochondria are unparalleled. Genes are fragmented in pieces (modules) that are separately transcribed, followed by the joining of module transcripts to contiguous RNAs. Some instances of unique uridine insertion RNA editing at module boundaries were noted, but the extent and potential occurrence of other editing types remained unknown. Comparative analysis of deep transcriptome and genome data from Diplonema papillatum mitochondria reveals ∼220 post-transcriptional insertions of uridines, but no insertions of other nucleotides nor deletions. In addition, we detect in total 114 substitutions of cytosine by uridine and adenosine by inosine, amassed into unusually compact clusters. Inosines in transcripts were confirmed experimentally. This is the first report of adenosine-to-inosine editing of mRNAs and ribosomal RNAs in mitochondria. In mRNAs, editing causes mostly amino-acid additions and non-synonymous substitutions; in ribosomal RNAs, it permits formation of canonical secondary structures. Two extensively edited transcripts were compared across four diplonemids. The pattern of uridine-insertion editing is strictly conserved, whereas substitution editing has diverged dramatically, but still rendering diplonemid proteins more similar to other eukaryotic orthologs. We posit that RNA editing not only compensates but also sustains, or even accelerates, ultra-rapid evolution of genome structure and sequence in diplonemid mitochondria.
INTRODUCTION
DNA sequence alone does not always indicate what a genome encodes. One reason is RNA editing, the programmed alteration of a transcript, with the result that the RNA sequence differs from that of its genomic template. All kinds of transcripts can be affected by editing: mRNAs, intron RNAs, structural RNAs and regulatory RNAs. RNA editing plays an important role across the Tree of Life, and unsurprisingly, alterations in RNA editing can lead to human disease (1). In the following, we will use the term ‘RNA editing’ for processes that change the sequence of a transcript, not including chemical modifications such as pseudouridylation, 2'-O methylation, etc. (2).
RNA editing is a post-transcriptional process that changes the sequence of the precursor transcript. RNA editing can act either on full-length transcripts or on nascent RNAs prior to 3′ end formation. This latter case has been referred to as ‘cotranscriptional RNA editing’ (3), although nucleotides are changed post-transcriptionally. Traditionally, cotranscriptional RNA editing describes a scenario discovered in myxomycete (slime mold) mitochondria where changes are intimately linked to RNA synthesis, and pre-edited (nascent) transcripts seem not to exist (4,5). Therefore, in a strict sense, the term ‘RNA editing’ does not apply to myxomycetes, because not the RNA sequence is changed but rather the DNA template is ‘incorrectly’ transcribed. In fact, the term RNA editing is often employed to generically describe differences in gene versus transcript sequences, although in many cases the origin of these changes remain unknown as in dinoflagellates (6,7).
Post-transcriptional RNA editing is classified in three distinct types. The first type results in insertions or deletions (indels), by addition of new, or removal of existing, nucleotides in transcripts. The second type involves nucleotide substitutions, which are generated in situ by either deamination or (trans) amination, most commonly pyrimidine exchange (i.e. cytidine (C) to uridine (U; C-to-U) and U-to-C) and adenosine-to-inosine (A-to-I) deamination. Reverse transcriptases read Is in RNA as Gs, and similarly, the translation machinery is thought to interpret Is in mRNA as Gs (8). In structural RNAs, A-to-I replacement has consequences as well. It influences RNA folding stability, and tRNAs extend codon recognition when the altered nucleotide is part of the anticodon. Note that for tRNAs, A-to-I deamination has been traditionally classified as nucleotide modification, but is now widely considered as RNA editing (9). The third type of RNA editing causes nucleotide substitution as well, but acts exclusively on the 5′ and 3′ ends of the acceptor stem of mitochondrial tRNAs. In this case, mis-paired nucleotides are removed from one side of the helix and replaced by ones matching the complementary portion of the helix (reviewed in 10). New types of post-transcriptional indel and substitution RNA editing are the topic of this work.
RNA editing has been discovered first in mitochondria (11). It is quite common and highly diverse in this organelle. Post-transcriptional substitution of Cs by Us is most frequent, with land plant mitochondria featuring up to 2000 distinct events of this kind (12,13). Mitochondrial C-to-U editing is sporadically observed in other taxa, such as heteroloboseans (14,15) and metazoans (16,17). Also, certain plastids perform C-to-U RNA editing (7). Elsewhere, only a few such instances have been reported: notably one in an archaean tRNA (18) and a few dozen in metazoan nuclear mRNAs, nearly all within 3′ untranslated regions (19). The prototype of mammalian C-to-U editing acts on apolipoprotein B (apoB) mRNA, and remains the only case of this type that impacts a coding region (20). The inverse reaction, U-to-C substitution, occurs much more rarely than C-to-U, with the majority of sites in plant mitochondria (21). Unheard of is A-to-I editing of organellar mRNAs or rRNAs, whereas this kind of substitution is pervasive in the metazoan nucleus (22, see also 23).
Mitochondria also perform post-transcriptional insertion and deletion RNA editing, which is extremely rare in other systems. The flagship organisms are kinetoplastids (Euglenozoa), where solely Us are inserted in, or deleted from, mitochondrial pre-mRNAs, up to nearly 600 in a single gene. Kinetoplastid indel editing involves site-specific cleavage of pre-mRNAs, U-insertion or deletion and re-ligation. All steps are directed by small guide RNAs (24).
The sister clade of kinetoplastids is a group of ocean-thriving unicellular flagellates, the diplonemids. With only two genera recognized, Diplonema and Rhynchopus, diplonemids are seemingly an insignificant protist taxon. However, recent environmental explorations revealed that these organisms are among the most abundant and genetically most diverse eukaryotes in the oceans (25–27). Diplonemids are notorious for their eccentric genome architecture and gene structure in mitochondria (28). Specifically, mitochondrial genes of the type species Diplonema papillatum are systematically split in up to 11 pieces (modules) that are ∼40–550 nt long. Each such piece is encoded on one of the ∼80 distinct circular chromosomes of 6 kbp (class A) or 7 kbp (class B) length.
Chromosomes have a surprisingly regular structure (Supplementary Figure S1A; (29)). Coding regions are flanked by on average 50-nt unique sequence and together, they make up a distinctive cassette that is unique to a given chromosome. The rest of the circle (∼90%) mostly consists of repeats. Specifically, adjacent to each cassette are two ‘class-specific constant regions’ of 1–3 kbp whose sequence is conserved across all chromosomes of a given class. In addition, opposite to the cassette resides a ∼2.5-kbp ‘shared constant region’, which is common to A- and B-class chromosomes (29).
Gene modules in Diplonema mitochondria are transcribed separately as RNA precursors, then end-processed, and subsequently joined into contiguous RNAs (30). The molecular mechanism of this unique trans-splicing process is yet to be unraveled. Collectively, modules specify a relatively ‘standard’ set of 12 recognized genes, including two ribosomal RNAs (mt-rRNAs) as well as protein components of the respiratory chain, oxidative phosphorylation and the mito-ribosome (Table 1, column 1); as in kinetoplastids, tRNAs appear to be imported from the cytosol.
Table 1. Genes and RNA editing sites in D. papillatum mitochondria.
| No. of editing sites | ||||||
|---|---|---|---|---|---|---|
| Genea | No. of modules | FPKMb | A-to-I | C-to-U | U-appendage (length)c | Previous (current) module designation [GenBank acc. no.] |
| atp6 | 3 | 60 413 | / | / | / | |
| cob | 6 | 199 880 | / | / | 1 (3 nt*) | / |
| cox1 | 9 | 158 604 | / | / | 1 (6 nt) | / |
| cox2 | 4 | 78 328 | / | / | 1 (3 nt*) | / |
| cox3 | 3 | 128 593 | / | / | 1 (1 nt*) | / |
| nad1 | 5 | 85 793 | / | / | 1 (16 nt*) | / |
| nad4 | 8 | 33 778 | 7 | 22 | 1 (2 nt) | / |
| nad5 | 11 | 46 198 | / | / | / | / |
| nad7 | 9 | 34 080 | 1 | / | / | / |
| nad8 | 3 | 26 772 | / | / | / | / |
| rnl | 2 | 234 706 | / | / | 1 (∼26 nt) | / |
| rns | 1 | 174 561 | 15 | 30 | 1 (8 nt*) | X3d |
| y1 | 2 | 101 526 | 4 | 7 | 1 (4 nt*) | X1-m(k-1, k)d ( = y1-m1, 2) |
| y2 | 4 | 16 226 | 1 | 2 | 2 (18 nt; 11 nt*) | / |
| y3 | 5 | 13 365 | 1 | 6 | 3 (∼28 nt; 16 nt; 1 nt*) | X2-m(k)d ( = y3-m5) [JQ314396.1] |
| y4 | 2 | 29 973 | / | / | 2 (∼29 nt; 12 nt*) | / |
| y5 | 2-3 | 45 165e | / | 18 | 1-2 (>30 nt; 1 nt*) | / |
| y6 | 2 | 21 121 | / | / | 1 (6 nt*) | / |
| Total | 82 | 44 | 70 | 17-18 (>221 nt) | ||
aGene products are: atp6, subunit 6 of ATP synthase; cob, apocytochrome b; nad1-8, subunits of NADH dehydrogenase; rnl, mt-LSU rRNA; rns, tentative mt-SSU rRNA. Gene products of y1-y6 are unknown; y1-y4 code for proteins. GenBank accession numbers of transcripts determined here are listed in Supplementary Table S6.
bFragments Per Kilobase of transcript per Million of mapped reads in library DPA2 made from poly(A) RNA, determined by Star/Cufflinks (see ‘Materials and Methods’ section).
cAsterisks indicate terminal modules.
d(30).
eFPKM average over three transcript variants, differing in their 3′-terminal region; see Figure 1.
In diplonemids, we previously noted a mode of mitochondrial RNA editing that somewhat resembles U-insertion editing in kinetoplastid mitochondria, as it involves the addition of multiple Us at 3′ ends of modules (therefore termed ‘U-appendage’ editing). For example, the module 4-transcript of the gene encoding cytochrome c oxidase subunit I (cox1) is extended by six Us that are retained in the trans-spliced mRNA, consisting of a total of nine modules (28,31). An even more spectacular U-appendage occurs during maturation of the mitochondrial large-subunit ribosomal RNA (mt-LSU rRNA). The corresponding gene (rnl) is split into two modules. At the 3′ end of the rnl module 1-transcript, ∼26 Us are added prior to trans-splicing. We showed that the U-tract-containing mt-LSU rRNA is indeed incorporated into the mito-ribosome of D. papillatum (32).
Here we examine comprehensively RNA editing in D. papillatum based on deep transcriptome sequencing data, uncovering a second type of post-transcriptional RNA editing in diplonemid mitochondria: nucleotide substitution. Remarkably, replacements include A-to-I substitutions in mRNAs and rRNAs, which has never been seen in organelles before. These nucleotide changes will be investigated experimentally. A second focus of this study is on the conservation and diversification of RNA editing pattern during the evolution of diplonemids, and possible evolutionary relationships between RNA editing in diplonemid mitochondria and those in other systems.
MATERIALS AND METHODS
Detailed descriptions of applied methods are available in Supplementary Materials and Methods.
Strains, culture, and DNA and RNA extraction
Diplonema papillatum (ATCC 50162), Diplonema ambulator (ATCC 50223), Diplonema sp. 2 (ATCC 50224) and Rhynchopus euleeides (ATCC 50226) were obtained from the American Type Culture Collection. Organisms were cultivated axenically as described earlier (31,32). To isolate mtDNA, mitochondria were enriched by differential and sucrose gradient centrifugation. Mitoribosomes were separated from whole cell lysates by kinetic glycerol-gradient ultracentrifugations (32). After extraction of RNA (33), residual DNA was removed by column purification or digestion with RNase-free DNase followed by phenol-chloroform extraction. Poly(A) RNA was enriched by a passage through oligo(dT)-cellulose.
In vitro transcription and RNase cleavage of glyoxalated RNA
The DNA templates for in vitro transcription of synthetic pre-edited or edited mt-SSU rRNA were generated either by PCR on mtDNA or by RT-PCR on purified mt-SSU rRNA. To detect inosines in RNA, we followed a protocol devised by others (34) that exploits the fact that guanosines, but not inosines, can be modified by glyoxal/borate treatment, which protects against RNase T1 cleavage (35). After glyoxalation and RNase T1-treatment, the RNA sample was deglyoxalated and then used in RT-PCR, northern blot hybridization, or primer extension assays. Oligonucleotides used as primers and hybridization probes are listed in Supplementary Table S1. For details, including deviations in electrophoretic migration behavior of certain RT-PCR products, see Supplementary Methods and https://www.protocols.io/u/matus-valach.
DNA library construction, sequencing, read processing and assembly
A genomic paired-end library was constructed from total DNA and sequenced with Illumina MiSeq. Details on libraries are compiled in Supplementary Table S2. Cutadapt version 1.2.1 (http://journal.embnet.org/index.php/embnetjournal/article/view/200) was employed for adapter clipping, quality trimming and elimination of reads shorter than 20 nt. Reads were assembled with the Celera software runCA version 8.3rc2 (http://sourceforge.net/projects/wgs-assembler/ (36)) using default parameters. Contigs originating from the mitochondrial genome were identified by sequence identity with previously determined mitochondrial chromosomes and modules (GenBank acc. nos. EU123536-8 and HQ28819-33) using BLAST at a hit-reporting threshold of 99% and then clustered with CD-HIT version 4.6 (37) employing the option -c 0.9.
RNA-Seq library construction, sequencing and read processing
We depleted D. papillatum RNA from cytosolic rRNAs and mt-LSU rRNA (32) using biotinylated oligonucleotides complementary to these rRNA species. Libraries were prepared from total cellular RNA enriched for poly(A) RNA (PA, DPA2), mitochondrial RNA (F1 from a fragmented, F2 from an un-fragmented sample) and a mito-ribosome-enriched RNA fraction (GG). Libraries from the three other diplonemids were made from total RNA depleted from cytosolic rRNAs. For details on libraries, see Supplementary Table S2.
Mapping of Illumina reads to reference sequences and calculation of FPKM
Illumina reads were mapped to reference sequences with Bowtie 2 (38). If not specified otherwise, we employed the options --local --no-unal (removing unaligned reads), and default values for the alignment and scoring parameters. Output files in sam format were subsequently transformed into ‘.bam’ files with SAMtools v1.4 (http://samtools.sourceforge.net/). Alignments were visualized with the Integrative Genomes Viewer (IGV; https://www.broadinstitute.org/igv/) (39). FPKM (Fragments Per Kilobase of transcript per Million of mapped reads) values were obtained after mapping RNA-Seq reads of the library DPA2 against the mito-transcriptome reference using TopHat v2.0.14 (https://ccb.jhu.edu/software/tophat/index.shtml) (40) or STAR version 2.4.2a (https://github.com/alexdobin/STAR) (41) with default parameters, followed by assembling mapped reads into contigs with Cufflinks (https://github.com/cole-trapnell-lab/cufflinks).
Mitochondrial reference genome sequence
From a Celera assembly of mitochondrial-genomic MiSeq Illumina reads (see above), we extracted contigs holding cassettes, i.e. those containing the distinctive left-hand and right-hand class-specific constant regions of chromosomes, but not the shared constant region (see Supplementary Figure S1A). The contig sequences were validated and polymorphisms determined simultaneously by mapping back the MiSeq reads to the contigs with Bowtie 2.
Transcript de novo assembly and function annotation
RNA-Seq reads from the PA and DPA2 libraries (Supplementary Table S2) were assembled using SOAPdenovo-Trans (42) using various kmers, and the resulting contigs were assembled again using SOAPdenovo (43) with a kmer size 127. We also used Trinity with default parameters (http://trinityrnaseq.github.io/, 44). Mitochondrial rRNA sequences were assembled from reads of library GG. Function assignment of newly detected mitochondrial transcripts was attempted with various approaches (see Supplementary Methods), but failed.
Mitochondrial transcriptome reconstruction and assignment of orphan modules
Transcripts were also reconstructed via a split-read approach developed in-house. First, RNA-Seq reads of the poly(A) library were mapped to the genome reference with Bowtie 2 (paired-end and local mode). The custom python script, findTransSplicedRNA.py, then identified read pairs whose two partners do not map to same genomic module and analyzed their sequence portions that were soft-clipped during mapping in local mode. If the soft-clipped sequence overlaps with another genomic module, then the two modules must belong to the same gene and be adjacent in the trans-spliced transcript.
In silico identification of polymorphic genomic sites
Variant sites in mtDNA were determined with the UnifiedGenotyper module of the Genome Analysis Toolkit (GATK) v3.3.1 (https://www.broadinstitute.org/gatk/ (45,46)) and FreeBayes (Garrison E and Marth G. (2012) In arXiv (ed.), Vol. 1207.3907v2 [q-bio.GN]). For both tools we set the ploidy to 100, the minimum number of observed variants to 2, the minimum base quality and mapping quality to 30 and the minimum allele frequency to 0.01. The output files of DNA–DNA comparison is referred to as DDd.vcf files. Only sites with at least 10% allele frequency were considered. We validated the obtained genome variants by visual inspection with IGV v2.3.40 (https://www.broadinstitute.org/igv/) (39), as well as with reads generated by Sanger and 454-FLX (Roche). For linked sites, we consolidated the allele frequencies reported by the variant caller software by calculating the mean across all linked sites.
In silico identification of RNA editing sites
We mapped RNA-Seq reads from all libraries against the genome reference, the pre-edited (virtual) and the fully edited transcriptome, using Bowtie 2 (see above) in strand-specific mode. The returned bam files were merged by the custom script mergeSAM.py. To identify DNA-RNA differences (DRds), the merged bam file was used as input for the GATK UnifiedGenotyper (45) and FreeBayes, using the same parameters as for calling genomic variants described above. To differentiate between genomic polymorphisms and RNA editing sites, DRd.vcf and DDd.vcf files were compared with the tool vcf-isec of the VCFtool kit (https://vcftools.github.io/perl_module.html#vcf-isec). The called sites were inspected and validated visually in the bam files.
Analysis of RNA processing intermediates and partially edited transcripts
Using the in-house script editpop.py (see Supplementary Methods), we analyzed the correlation between the status of RNA editing sites, and between RNA editing, module processing and trans-splicing in read pairs. U-appendage sites were analyzed by searching motifs in individual reads using GNU grep. For short internal U-insertions (1–2 nt) we requested a full match of ≥6 adjacent nucleotides in the two neighbor modules. For terminal Us, the search requested eight adjacent nucleotides in the module's 3′ end followed by ≥2 Us. To detect partially edited substitution sites, RNA-Seq reads from library DPA2 were mapped with Bowtie 2 against the pre-edited (virtual) and edited transcriptome sequences, the resulting sam alignment files were merged as described above and then parsed with the in-house script editedSitesStat.py (see Supplementary Methods) to extract the particular nucleotides present at RNA editing sites. Site positions were obtained from the vcf file Dp_mito_SNP-RNA_20160212.vcf (Supplementary File 1).
Search for cis elements and RNA trans-factors that guide RNA editing
We searched for recurrent cis-motifs near individual RNA editing sites using the in-house script editbysite.py (see Supplementary Methods). Sequence motifs in cis that flank clusters of substitution editing were searched using MEME and GLAM2 (MEME web server (47) (http://meme-suite.org/tools/meme) for ungapped and gapped motifs, respectively. Common 2D cis-motifs were searched using RNAalifold (48–50), LocARNa (51,52), RNAstructure and Multilign (48,49), with default parameters. Trans-acting guide RNAs were searched in reads of library F2 (Supplementary Table S2) employing the GNU grep utility supplied with query motifs that are reverse-complements of the edited sequence and adjacent regions. Finally, for detection of anti-sense reads with mismatches, reads of the F2 library were mapped against the sequences of pre-edited (virtual) and full-length mature transcripts, using Bowtie2 with the options --local --nofw and default mismatch settings. Resulting bam files were inspected visually.
Analysis of Nad4 protein sequences
Using MUSCLE with default parameters (40), we built a multiple alignment of Nad4 protein sequences deduced from edited and pre-edited nad4 genes from four diplonemids and 15 moderately divergent homologs from other taxa (see Supplementary Methods). From this alignment, we extracted a sub-alignment including columns 1–130, which corresponds to the N-terminal region up to the last ‘edited’ amino acid in diplonemids. Based on this alignment, protein conservation was determined with MstatsX (https://github.com/gcollet/MststX) which uses the trident statistics (41). To determine potential trans-membrane helices, the protein sequences of ‘pre-edited’ and ‘edited’ diplonemid Nad4 variants were scanned with TMHMM2.0 (53) (http://www.cbs.dtu.dk/services/TMHMM/), Phobius (54) and TMpred from the ExPASy web suite (55).
RESULTS
Assignment of modules to genes
As a first step, we compiled all cassette sequences of D. papillatum, i.e. the unique portions of mitochondrial chromosomes (Supplementary Figure S1A). Cassettes were extracted from a whole genome assembly, built with Illumina reads. These consensus sequences, 23 kbp in total, include 81 different cassettes of 166–638 nt length. In an earlier assembly (29), the cassette/module count was 75. We then determined genomic variants by mapping reads to the consensus sequences and analyzing the alignment maps by variant calling software tools. We detected nearly 100 genomic variants (with ≥10% allele frequency) spread across 37 cassettes. Variants represent 80% transitions and 20% transversions, and are mostly biallelic and linked, suggesting two versions for each chromosome (Supplementary Table S3). Finally, the consensus sequences were corrected so that they represent the majority of reads, for use as mitochondrial reference genome in the following analyses.
Mapping of RNA-Seq reads against the Diplonema mito-genome reference confirms that all predicted modules are transcribed (including allelic variants). The majority of modules are pieces of 11 previously reported, assigned mitochondrial genes. For nearly 20 modules (designated orphans), the genes they belong to were unknown at the outset of this study (e.g. modules X1 to X3 (30); Table 1).
To pinpoint the mature transcripts to which orphan gene modules belong, we assembled transcripts from RNA-Seq reads, yielding a 15-kb mitochondrial transcriptome. Orphan modules were assigned to transcripts based on sequence identity, revealing seven new genes: X3 consists of a single module, whereas the others (denoted y1, y2, etc.) are composed of two to five modules. All mitochondrial genes and their modules detected to this point are listed in Table 1 and depicted in Supplementary Figure S2.
Identification of the tentative gene for mt-SSU rRNA
Functional annotation of newly discovered transcripts was attempted initially by BLAST similarity searches in GenBank's non-redundant database, followed by searches with profile Hidden Markov Models and Covariance Models representing all known mitochondrial protein-coding genes and mitochondrial rRNAs, respectively (see ‘Materials and Methods’ section). However, no significant hit was obtained.
Gene X3 stands out because its transcript is highly abundant, with a steady-state level comparable to Diplonema's mt-LSU rRNA. In addition, the transcript is highly enriched in the library made from a mito-ribosome-enriched fraction (Supplementary Table S2), suggesting that X3 represents the elusive mitochondrial small subunit (mt-SSU) rRNA. Indeed, highly divergent structural domains (5′- and 3′-minor domains) of the SSU-RNA are recognizable (Supplementary Figure S1B), although the remainder of the secondary (2D) structure could not be modeled, and this is due to several reasons. One is that sequence similarity is very low between X3 from Diplonema and mt-SSU rRNAs from other organisms for which secondary structure models are available. Further, thermodynamics-based modeling is inconclusive, because high G + C content of the sequence generates numerous equally probable alternatives. The same issues, but to a lesser degree, were encountered when modeling mt-LSU-rRNA from D. papillatum (32).
The tentative (366 nt) mt-SSU rRNA of Diplonema is among the shortest ever reported, slightly shorter than the highly derived mitochondrial rns in certain animals (56). Yet, as of now, it cannot be ruled out that X3 represents only one of several molecules of a mitochondrial rRNA in pieces, as seen in Euglena gracilis (57), apicomplexans (58) and dinoflagellates (6), for example.
The y genes still remain unidentified. For y1 to y4, we have mass-spectrometry data demonstrating that these genes code for proteins (data not shown). Unravelling the biological role of y-genes will require detailed biochemical studies.
Uncovering RNA editing events in mitochondrial transcripts
RNA editing sites manifest as differences between gene and transcript sequence. DNA–RNA-differences will be referred to in the following as DRds. Since mitochondrial genes in Diplonema are fragmented, we used as a reference sequence the joined gene pieces (equivalent to pre-edited full-length transcript sequences), against which we mapped RNA-Seq reads from the various libraries (Supplementary Table S2). Based on the genome/transcriptome alignment maps, we determined DRds using variant calling tools, and all these sites were visually inspected and validated. Note that ‘pre-edited’ refers to sites not yet edited, while ‘unedited’ characterizes sites that are never edited.
Two positions returned as editing sites coincide with genomic variants determined earlier. One is located in y2-module number 3 (y2-m3) (position 131 in the corresponding cassettes). It is an A/G dimorphism with a ratio of 4:6 in DNA versus 1:9 in RNA. The second site falls in rns with a C/T dimorphism of 2:8 in DNA (see Supplementary Table S3), but only U in RNA. In both cases, it cannot be distinguished whether the dimorphic sites in RNA arise from transcription of the two genomic variants or rather by transcription of the A- and C-alleles with subsequent partial RNA editing to G and T (U), respectively.
Table 1 summarizes the number and types of RNA editing sites that are frequently edited (>50%) and do not coincide with genomic variants. Positions and frequencies of sites displaying at least 5% editing are listed in Supplementary File 1.
Catalog of nucleotide insertions and substitutions in the D. papillatum mito-transcriptome
Inspection of the global landscape of RNA editing in D. papillatum mitochondria (Table 1) reveals two important features. First, post-transcriptional insertions involve only Us and no other nucleotide. Not a single event of deletion-RNA editing was detected in the 15-kb transcriptome examined. Inserted U-tracts are between 1 and ∼30 nt long and coincide with module junctions or transcript ends. Earlier we demonstrated experimentally for cox1 and mt-LSU rRNA that Us inserted at module junctions in the mature transcript are actually appended at the 3′ end of the upstream module prior trans-splicing (30,32). Our comprehensive mito-transcriptome data corroborate the 3′ appendage mechanism, since no cases of U-extensions at 5′-ends of full-length transcript were observed. Finally, 3′ U-additions are independent of the module position (5′, internal or ultimate) within the mature transcript.
The second important feature is that Diplonema mitochondria perform substitution RNA editing (Table 1), which had remained unnoticed until now. We observe more than 110 C-to-U and A-to-I substitutions. Except for one solitary substitution in nad7, sites are congregated in six clusters of 3–85 nt length, which are located in nad4, the tentative rns, and four y genes (y1, 2, 3, 5; Figure 1A). Most densely packed are the substitution editing clusters in nad4 and the tentative rns with sites often placed immediately adjacent to one another. The cluster in nad4 is 56 nt long and positioned in module 1, close to the 5′ terminus. In this transcript, a total of 29 sites undergo substitutions, notably all Cs and one half of As. The 85-nt long cluster in the tentative rns is also situated near the transcript's 5′ end and includes 45 sites; all Cs and As in this cluster are edited (Figure 1B).
Figure 1.
Substitution RNA editing of mitochondrial genes from Diplonema papillatum. (A) Module composition of edited genes. Gray and black pentagons represent modules encoded on A- and B-class chromosomes, respectively. Arrows under pentagons indicate the orientation of the module within the chromosome (see Supplementary Figure S1A). Orange stars point to clusters of substitution RNA editing. Green boxes depict post-transcriptional U-insertions with the indicated number of appended Us. ‘AAA...A’, poly(A) tail. As being part of a stop codon are shown on red background. The square bracket groups transcript isoforms of y5. (B) Sequences of substitution editing clusters. Numbers in square brackets specify the position of the depicted sequence in the corresponding cassette. Upper rows, genomic and lower rows cDNA-derived nucleotide sequence and conceptual translation (one-letter code). +, x: A-to-G and C-to-T sites, respectively. Blue and orange nucleotides, pre-edited and edited states of substitution sites, respectively.
Searches for potential cis-elements and trans-factors that guide RNA editing
We scrutinized the sequence context of mitochondrial RNA editing sites in Diplonema by searching for shared sequence motifs and 2D structure element (cis-motifs) in close vicinity of the locations where RNA editing occurs. These analyses were performed separately for sites of U-addition, A-to-I substitution, and C-to-U substitution, and for substitution clusters. However, we did not detect motifs specifically associated with either type of RNA editing site (Supplementary Figure S3A–C). The absence of discernable recurrent sequence patterns around RNA editing sites suggests that these sites are defined by specific trans factors.
We searched for potential trans-acting RNAs that guide RNA editing, postulating a population of site-specific factors with the propensity to pair with RNA editing sites. But again, convincing candidates were not detected (Supplementary Figure S3D). For both cis-elements and trans-factors, our search strategies, results and interpretations are detailed in ‘Supplementary Results and Discussion’.
Biochemically, A-to-I substitution RNA editing proceeds by deamination
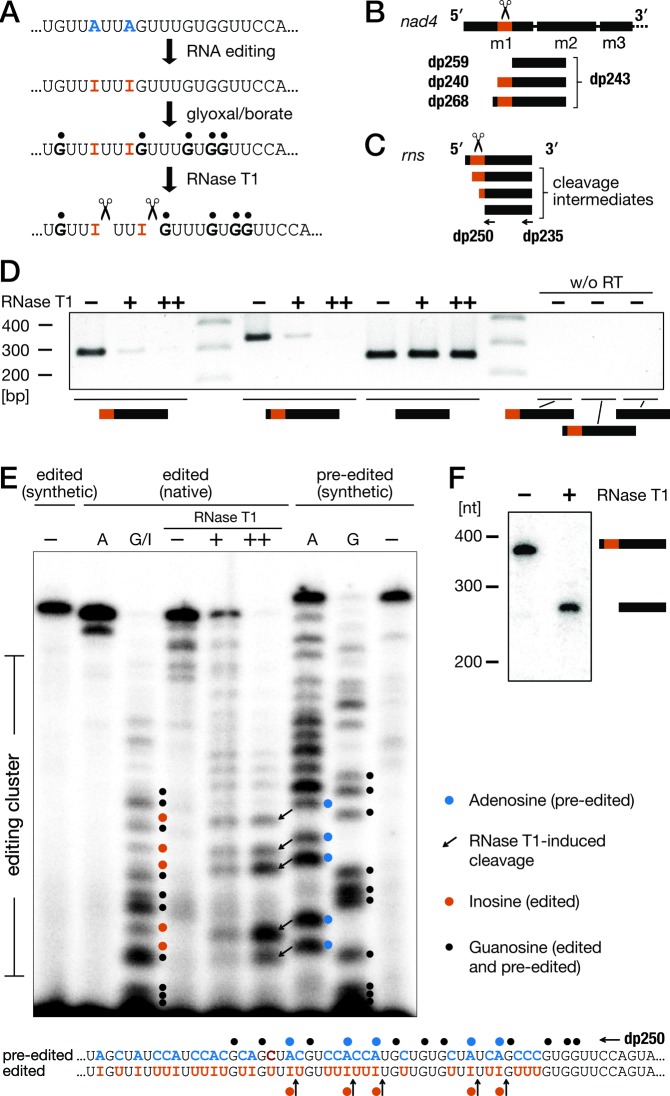
C-to-U and A-to-G differences between genomic and cDNA sequences usually originate from in situ base deamination in transcripts, but nucleotide excision and replacement is conceivable as well. In the case of deamination, a substituted G in cDNA corresponds to an inosine (I) in RNA. Therefore, we determined the presence of Is in the transcripts of two function-assigned genes that undergo substitution RNA editing in Diplonema, nad4 and the tentative rns. We treated RNA from Diplonema with glyoxal, which forms a stable adduct with G, but not I, in the presence of borate (35). RNase T1 then cleaves RNA after (unmodified) Is, while glyoxalated Gs are protected (Figure 2A–C).
Figure 2.
Demonstration of inosines in substitution RNA editing clusters of Diplonema papillatum. (A) Experimental approach involving glyoxal/borate and RNase T1 treatment of transcripts. (B) Design of the RT-PCR assays for the detection of RNase T1 cleavage at unprotected inosines (Is) in the editing cluster of nad4-m1. Forward primers bind downstream (dp259), within (dp240), or upstream (dp268) of the editing cluster; the reverse primer (dp243) anneals in the downstream module. (C) Oligonucleotides designed for detection of mt-SSU rRNA; dp250 was used for the primer extension assay (see panel E), and dp235 served as a probe in northern blot hybridization (see panel F). (D) RT-PCR products after digesting glyoxalated nad4-m1 with increasing RNase T1 concentrations (0, 100, 1000 U). Yield of those RT-PCR products that overlap the edited cluster is reduced progressively, but not of those that do not overlap the cluster. w/o RT, control amplification in the absence of reverse transcriptase (RT). (E) Primer extension of mt-SSU rRNA to map RNase T1 cleavage sites at Is. As templates served the following glyoxal/borate-treated samples: in vitro transcribed substitution-edited mt-SSU rRNA (edited (synthetic)); total RNA (edited (native)); and in vitro transcribed pre-edited mt-SSU rRNA (pre-edited (synthetic)). A, sequencing lane where ddTTP was used as a chain terminator. G/I, G: ddCTP was used as dideoxy terminator. −, untreated templates. +, ++: digestion with 10 U and 50 U RNase T1, respectively, amounts which allow detection of cleavage intermediates. In the lanes labeled + and ++, bands represent reverse transcriptase-stops one nucleotide prior to I. The sequence schema at the bottom illustrates the positions of glyoxalation, predicted Is, as well as reverse-transcription stops. (For details on assay optimization and explanation of apparent size shifts, see the Supplementary Figure S4). (F) Northern blot hybridization of mt-SSU rRNA, demonstrating the expected size-reduction of the transcript by ∼110 nt after digestion with RNase T1 (1000 U) that cleaves off the 5′ portion of the rRNA.
For treated nad4 transcripts, RNase cleavage manifests as a >10-times reduction of RT-PCR amplification across the edited region compared to amplification of the adjacent unedited region (Figure 2D). In the tentative mt-rns, Is were mapped by differential RNase digestion followed by primer extension (for assay optimization, see Supplementary Figure S4). This experiment demonstrates that at least five out of 15 Gs in cDNA are indeed Is in RNA (Figure 2E). We also mapped the editing cluster with northern hybridization (Supplementary Figure S4B). After extended digestion with RNase T1, the band corresponding to the full-length transcript disappears, showing that the steady-state level of pre-edited tentative rns is extremely low (Figure 2F).
Given that A-to-I substitutions in Diplonema mitochondria occur by deamination, we presume that the same applies to C-to-U RNA editing, since deamination is the only molecular mechanism of C-to-U substitutions encountered in systems that are biochemically characterized (18,59).
RNA editing precedes trans-splicing and progresses stochastically within editing clusters
In kinetoplastid mitochondria, pre-mRNAs are transcribed full-length and subsequently edited progressively from 3′ to 5′ (24). With deep transcriptome data at hand, we examined whether the same temporal order and directionality applies to RNA editing in Diplonema mitochondria. Specifically, we examined read pairs that both span a region encompassing editing sites in a given module, as well as extend beyond the edited module. For both types, U-appendage and substitution RNA editing, we encountered two predominant transcripts forms: edited + trans-spliced and pre-edited + unprocessed, while edited + unprocessed and pre-edited + trans-spliced intermediates are extremely rare (Supplementary Table S4). An independent experiment inquiring nad4-m1 intermediates by RT-PCR confirmed this result: a pre-edited trans-spliced module was not detected among poly-adenylated transcripts (result not shown). Thus, in contrast to kinetoplastids, RNA editing in mitochondria of Diplonema takes place prior to the occurrence of a full-length transcript, and essentially in parallel with module-end processing. Further, analysis of nad4 transcript intermediates shows that edited and pre-edited substitution sites are interspersed, indicating a stochastic order in the deamination of individual sites (Figure 3). Therefore, again unlike RNA editing in kinetoplastids, there appears to be no directionality of editing progression in Diplonema.
Figure 3.
Partial substitution RNA editing in nad4-m1. RNA-Seq reads from two poly(A) libraries combined (DPA2 and PA) were analyzed for low-frequency RNA editing in the substitution editing cluster (positions 129-204 in the corresponding cassette). Editing patterns observed in RNA-Seq reads from total-RNA libraries are shown in Supplementary Figure S5. The histogram on the top indicates the editing level within the cluster. Exclusion threshold is <5% frequency, i.e. reported editing sites occur in ≥5% of the reads. Green and yellow bars represent the portion of Is and Us generated by deamination RNA editing, respectively. Arrows point to less-frequently edited sites (frequency 5–38%). Thick arrows indicate the three sites (nucleotide positions 143, 148 and 185 in cassette nad4-m1), where both editing states were confirmed to exist in nad4-mRNA (Supplementary Figure S6). Top-most nucleotide sequence, genomic sequence. Nucleotide sequence below, cDNA sequence showing the edited state of all sites observed to be edited above threshold. Lower-case letters, sites edited below 50%. +, x: predominant A-to-G and C-to-T substitution sites, edited above 50%. Chart below sequences: editing patterns observed in RNA-Seq reads. RNA and DNA sequence are identical except for red and blue squares, which indicate pre-edited As and Cs, respectively. The patterns shown are supported by at least 10 reads. The right-hand histogram represents the number of reads per editing pattern. It is the predominant form of nad4-mRNA, ranking first, that is shown in all other figures. #, the entirely pre-edited version, ranking fifth (∼6% of all reads). *, the maximally edited transcript (i.e. where all, high- and low-frequency sites are in the edited state), ranking 12th (∼1% of all reads).
Exceptionally high overall RNA editing rate, but several incompletely edited sites
In mitochondria of plants, about 15% of substitution sites (C-to-U) are partially edited (at 90% or less; 12,60–61). Our analyses show that the situation is quite different in Diplonema. Sites with partial editing are rare. For example, in 95% of RNA-Seq reads covering the nad4-m1 substitution-editing cluster, the totality of sites is either edited or pre-edited (Figure 3, Supplementary Figure S5). Among partially edited sites, there are three with nearly equal proportion of both editing states. Positions 148 (A-to-I) and 185 (C-to-U) are silent sites, while nucleotide 143 (A-to-I) occupies the first position in a codon and causes a non-synonymous amino acid substitution. An RT-PCR experiment confirms that both A-143 and I-143 exist in polyadenylated nad4 transcripts (Supplementary Figure S6). It is conceivable that both versions of nad4 mRNAs are translated, because the two alternative codons AUU and IUU specify functionally similar amino acids, Ile and Val, respectively. This finding contrasts with plant organelles, where nearly all partially edited substitution coincide with silent codon positions or fall in pseudogenes (for exceptions see e.g. (62)). It appears that incompletely edited transcripts in plant mitochondria are either not translated or, if translated, the resulting proteins are instable and readily degraded ((63); reviewed in (64)).
Crucial consequences of RNA editing for the tentative mt-SSU rRNA and nad4 protein
We examined the effect of RNA editing on the gene products of two Diplonema mitochondrial genes, tentative rns and nad4. In the tentative mt-SSU rRNA, the substitution editing cluster coincides with the 5′ domain of the 2D structure model. The seven 3′-terminal editing sites in the cluster contribute to helix h18 (Supplementary Figure S1) that contains the so-called ‘530-loop’, which is involved in A-site tRNA selection (65). This region is one of the evolutionary most conserved portions in SSU rRNAs (66). U-appendage RNA editing of rns involves addition of eight non-encoded Us at the transcript's 3′ end, and it is this U-tailed RNA that is incorporated in mito-ribosomes. Oligo-(U) tails on mt-rRNAs are also known from kinetoplastids, but the biological role of this ornament remains unclear (67 and references therein).
In nad4, ∼80% of post-transcriptionally substituted nucleotides correspond to first and second codon positions resulting in 14 non-synonymous out of 15 codon changes (Figure 4A and B). Within the editing cluster, in the stretch corresponding to amino acids 48–64 in the D. papillatum protein (referred to as Nad4), RNA editing renders the protein sequence more hydrophobic (Figure 4C and D). This outcome of deamination RNA editing has been reported repeatedly before (e.g. 61,68) but has not been recognized as an inherent consequence of nucleotide deamination. It is not the particular editing pattern, but rather the mere increase of deaminated bases (i.e. Us and Gs) in codons that leads to amino acids with a higher hydrophobicity index.
Figure 4.
Multiple alignment of nad4-m1 genomic (A) and cDNA sequences (B) from diplonemids. Genomic sites that undergo RNA editing and the entire editing cluster are highlighted in blue and orange, respectively. Sequence logos, with underscored codon triplets, show the impact of RNA editing on sequence identity. (C and D) Multiple alignment of proteins inferred from genomic (C) and fully edited transcript (D) sequences, with the editing cluster boxed. Background shading of residues indicates the extent of similarity. (E) Multiple alignment of the junction of nad4-m4 and m5 in cDNA sequences. Module boundaries were annotated based on the available genomic sequences. The Us appended to the module 4 are conserved across all examined diplonemids. (F) Multiple alignment of deduced protein sequences of the m4/m5 junction. Abbreviations: Dp, Diplonema papillatum; Da, Diplonema ambulator; Ds, D. sp.; Re, Rhynchopus euleeides.
The effect of RNA editing on Nad4's secondary structure is even more pronounced. Protein structure prediction indicates that only the ‘edited’ Nad4 has the potential to form the canonical trans-membrane helix in the N-terminal region (Supplementary Figure S7A). Finally, U-appendage RNA editing of nad4 results in the addition of two Us between the modules 4 and 5 of the trans-spliced transcript. This post-transcriptional event rectifies the reading frame and prevents premature chain termination in translation (Figure 4E and F).
Thus, for both tentative mt-SSU rRNA and Nad4 of D. papillatum, RNA-editing appears to be crucial for mitochondrial function and survival of the cell.
Comparison of RNA editing in nad4 across diplonemids
To investigate the conservation of RNA editing across diplonemids, we used nad4 as a test case. Figure 4 shows the multiple alignments of pre-edited and edited nad4 sequences and derived proteins from four diplonemids, D. papillatum, D. ambulator, D. sp. 2 and R. euleeides (69). In all taxa a cluster of substitution editing sites occurs in nad4-m1, but there are several variations to the theme. The cluster is located at different positions and it is longer in D. papillatum (55 nt) compared to that of the other diplonemids (44–53 nt). Furthermore, while every C in these clusters is edited, the proportion of A-to-I sites ranges from 7/11 in D. papillatum to 0/8 in D. ambulator and D. sp. 2; not a single substitution editing site is conserved throughout these species (Figure 4A and B). Together, non-synonymous changes of codons amount to more than 90% in D. papillatum, but to ∼50% in the other diplonemids. Remarkably, this inter-taxon diversity of substitution RNA editing results in a three times higher sequence identity between the diplonemid transcripts compared to the genes (Figure 4C and D).
In contrast to the inter-species variations in substitution sites, U-appendage RNA editing of the nad4 transcript is strictly conserved throughout the four species, with exactly two Us added post-transcriptionally between modules 4 and 5 (Figure 4E). In sum, RNA editing renders the Nad4 proteins of the examined diplonemids more similar to each other, as well as more similar to orthologs from other eukaryotes (Supplementary Table S5; Supplementary Figure S8).
DISCUSSION
RNA editing types and sites in D. papillatum mitochondria
Comprehensive comparative analysis of the mitochondrial genome and transcriptome from Diplonema uncovered two types of post-transcriptional RNA editing: (i) insertions of Us and (ii) substitutions of Cs by Us and As by Is. Together, nucleotide insertions and substitutions in Diplonema affect ∼80% of all mitochondrial transcripts and account for ∼130 RNA editing sites and ∼350 nucleotides are generated or altered by editing (Table 1).
Mitochondrial U-insertion RNA editing is extremely rare with only two other, distinct instances outside diplonemids, notably in kinetoplastids (70) (also featuring U deletions) and certain sponges (71). Substitutions of C-to-U occur more broadly in mitochondria, as well as in plastids. However, the here reported A-to-I RNA editing (of non-tRNA transcripts) is a first in organelles and the interspersed co-occurrence of A-to-I and C-to-U substitutions is unparalleled across all systems.
The distribution of RNA editing sites in Diplonema mitochondrial transcripts is conspicuously uneven. U-insertions occur either at module junctions or at the 3′end of transcripts, immediately upstream of the poly (A) tail. This is due to the particular mechanism of U-based RNA editing in this protist and consists in 3′-terminal nucleotide addition prior to trans-splicing or polyadenylation. A-to-I and C-to-U substitutions, on the other hand, are remarkably clustered with up to 45 sites per cluster and up to six sites directly adjacent to one another (Figure 1B). For comparison, in land plant organelles, congregated substitution sites (C-to-U and U-to-C) are rather exceptional (63). In metazoan nuclear transcripts, clustering is a hallmark feature of substitution editing sites (22), although intervals between sites are much larger compared to Diplonema mitochondria. Nowhere else are substitution editing sites as tightly packed as in the system investigated here.
Ribosomal RNAs are rarely ever edited, but in Diplonema mitochondria these transcripts undergo massive U-appendage and both C-to-U and A-to-I substitutions. In fact, rRNAs including Is have never been observed before (72); only one instance of an inosine derivative, O2′-methylinosine is known to occur in cytosolic rRNA of Crithidia (73). Since I has a greater repertoire of potential base pairs than either of the classical nucleotides (it pairs with A, C and U) (74), Is in rRNA probably destabilize the secondary structure of the molecule due to the larger number of alternative pairing possibilities. We speculate that the effect of 15 Is in Diplonema mt-SSU rRNA is compensated by proteins in the mito-ribosome.
How does the cellular machinery recognize RNA editing sites in Diplonema mitochondria?
In certain organisms, targets of RNA editing are recognized by a common sequence or structure element in cis. For example, Apobec-1-dependent C-to-U RNA conversion sites in the mammalian nucleus are characterized by a particular primary sequence context, such as an A + U-rich region along with a downstream 11-nt long motif (‘mooring’ sequence) (59,75). Similarly, ADAR-dependent A-to-I editing substrates in the metazoan nucleus and in viruses share a particular RNA secondary/tertiary structure (22,76).
In contrast, our analyses of the sequence context around RNA editing sites in Diplonema mitochondria did not identify common primary or secondary structure motifs in cis. Therefore, the yet elusive RNA editing machinery is probably directed by an array of distinct site-specific recognition factors acting in trans.
Our search for site-recognition factors resembling guide RNAs in trypanosome mitochondria (77) was inconclusive, suggesting that proteins might assume this task such as the pentatricopeptide repeat (PPR) trans-factors known from plant mitochondria (21). However, irrespective of the biochemical nature of the postulated recognition factors, there is a dilemma with respect to the crammed substitution RNA editing sites in clusters. How may trans-factors recognize an RNA editing site when its neighboring nucleotides are variable, since they too are subject to RNA editing? In our view, uncovering the nature of the postulated editing guides in Diplonema will require an unbiased experimental approach, notably isolation and dissection of mitochondrial complexes having in vitro RNA editing activity.
The biological role of organellar RNA editing
Editing of pre-mRNAs in organelles, including mitochondria of diplonemids, is function-critical since the large majority of events generates start codons, abolishes in-frame stop codons or changes codons to specify conserved amino acid positions (7). Indel editing of mitochondrial mRNAs is particularly essential, since it corrects frameshifts as observed in mitochondria of diplonemids and trypanosomes. Similarly function-critical is editing of tRNAs, not only for proper folding of the molecule, but often for end-processing of their precursor transcripts as well (9).
The situation is different for nuclear metazoan mRNAs where editing is typically partial. Both edited and ‘pre’-edited transcripts are translated, and the corresponding proteins play different biological roles. For example, multiple combinatorial codon changes of an mRNA may lead to a large spectrum of protein isoforms that are all encoded by a single, genomic locus. Nuclear RNA editing also acts on intronic regions or UTRs, controlling alternative splicing, efficiency of translation, transcript stability and localization (78). To summarize, RNA editing compensates disadvantageous mutations in organelles, while it is a means for diversification and regulation in the nucleus.
Emergence and diversification of the two RNA editing types in diplonemid mitochondria
The question arises why sites of post-transcriptional U-appendage are strictly conserved, whereas sites of C-to-U and A-to-I substitutions are highly variable. We postulate the following evolutionary scenario. Both RNA editing types probably have been present already in the common ancestor of diplonemids. U-additions are likely compensating detrimental consequences of mtDNA fragmentation that has led to a multi-partite mitochondrial genome. Specifically, post-transcriptional U-appendage may fill in critical nucleotides that were lost from the ancestral genome during double-strand repair at mtDNA breakpoints. Conservation of a fragmented mtDNA throughout the descendants would therefore entail faithful conservation of U-appendage RNA editing. On the other hand, substitution RNA editing likely compensates rapid sequence evolution of diplonemid mtDNAs, explaining why nucleotide substitution pattern are species-specific. In both cases we favor the model of constructive neutral evolution (for more discussion, see Supplementary Data), which posits that prior to genome fragmentation and acceleration of sequence evolution, basic tools to add or change ribonucleotides were already in place, and needed only fine-tuning by evolutionary tinkering (79,80).
Outlook
We show that diplonemids employ unique post-transcriptional RNA editing in mitochondria involving two distinct molecular processes, U-appendage and base deamination. But what are the enzymes that perform these reactions? As a working hypothesis, we postulate that the machinery that carries out U-appendage editing in diplonemids includes components known from the editosome of trypanosome mitochondria (for a review see 81). Similarly, the enzymes for deamination RNA editing in diplonemid mitochondria might resemble either ADATs catalyzing A-to-I editing of tRNAs (9), ADARs and Apobecs responsible for A-to-I and C-to-U RNA editing of mRNAs and regulatory RNAs in the metazoan nucleus (22), or PPR-E and PPR-DYW proteins required for C/U-exchange editing in land plant organelles (21).
The latter scenario is quite plausible, because of the finding of plant-like mitochondrial RNA editing in heteroloboseans, a group that shares a common most recent ancestor with Euglenozoa (euglenids + diplonemids + kinetoplastids). Specifically, mitochondrial mRNAs of Naegleria and Acrasia undergo several C-to-U editing events and the corresponding nuclear genomes encode homologs of the PPR-DYW protein family involved in organelle RNA editing of plants (14,15).
A glance at the first draft of the nuclear genome sequence from D. papillatum identified genes that specify protein domains characteristic for TUTases, PPRs and deaminases. However, it is currently unclear whether the inferred proteins are indeed involved in mitochondrial RNA editing, or rather in basic cellular processes such as RNA turnover, RNA end processing and nucleotide metabolism.
Finally, given the unique features of RNA editing in diplonemid mitochondria, the underlying molecular mechanisms might be entirely novel and may have evolved from unexpected molecular processes. It would not be the first time that the study of protists leads to the first discovery of novel molecular mechanisms that had remained unrecognized in model systems.
ACCESSION NUMBERS
GenBank accession numbers: KU356490-570 (mtDNA cassettes, D. papillatum), KU341361-80 (mitochondrial transcripts, D. papillatum), KU341385-86 (edited + pre-edited nad4 mRNA, D. ambulator), KU341387-88 (edited + pre-edited nad4 mRNA, D. sp. 2), KU341389-90 (edited + pre-edited nad4 mRNA, R. euleeides). See listing in Supplementary Table S6.
Supplementary Material
Acknowledgments
We acknowledge G. N. Kiethega and S. Teijeiro (both Université de Montréal, Montreal, Canada) for culturing diplonemids and extracting DNA and RNA. We thank M.W. Gray (Dalhousie University, Halifax, Canada) for advice in rRNA structure modeling, Lilyanne Peras for attempting, in the context of an undergraduate summer internship, folding rRNA consensus structures, B. F. Lang and M. Sarrasin (Université de Montréal, Montreal, Canada) for critically reading the manuscript and Peter Stadler (Universität Leipzig, Leipzig, Germany) for discussions on the bioinformatics approaches.
Author contributions: G.B. conceived and orchestrated the study, and S.M. and M.V. designed and conducted the bioinformatics analyses and experimental approaches, respectively. M.A.A. assisted in DNA and RNA preparation, PCR experiments and implementation of the inosine detection assay. G.B., S.M., M.V. and C.O. performed data analysis, and the manuscript was written by G.B., together with M.V. and S.M. All authors have approved the manuscript.
FUNDING
Canadian Institute for Health Research (CIHR) operating grant [MOP-79309 to G.B.]. Funding for open access charge: CIHR [MOP-79309].
Conflict of interest statement. None declared.
REFERENCES
- 1.Maas S., Kawahara Y., Tamburro K.M., Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunin-Horkawicz S., Czerwoniec A., Gajda M.J., Feder M., Grosjean H., Bujnicki J.M. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez J., Menet J.S., Rosbash M. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol. Cell. 2012;47:27–37. doi: 10.1016/j.molcel.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gott J.M. Expanding genome capacity via RNA editing. C R Biol. 2003;326:901–908. doi: 10.1016/j.crvi.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y.W., Visomirski-Robic L.M., Gott J.M. Non-templated addition of nucleotides to the 3' end of nascent RNA during RNA editing in Physarum. EMBO J. 2001;20:1405–1414. doi: 10.1093/emboj/20.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson C.J., Norman J.E., Schnare M.N., Gray M.W., Keeling P.J., Waller R.F. Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biol. 2007;5:41. doi: 10.1186/1741-7007-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoop V. When you can't trust the DNA: RNA editing changes transcript sequences. Cell Mol. Life Sci. 2011;68:567–586. doi: 10.1007/s00018-010-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basilio C., Wahba A.J., Lengyel P., Speyer J.F., Ochoa S. Synthetic polynucleotides and the amino acid code. V. Proc. Natl. Acad. Sci. U.S.A. 1962;48:613–616. doi: 10.1073/pnas.48.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackman J.E., Alfonzo J.D. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betat H., Long Y., Jackman J.E., Morl M. From end to end: tRNA editing at 5'- and 3'-terminal positions. Int. J. Mol. Sci. 2014;15:23975–23998. doi: 10.3390/ijms151223975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benne R., Van den Burg J., Brakenhoff J.P., Sloof P., Van Boom J.H., Tromp M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 12.Grewe F., Herres S., Viehover P., Polsakiewicz M., Weisshaar B., Knoop V. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2011;39:2890–2902. doi: 10.1093/nar/gkq1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecht J., Grewe F., Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol. 2011;3:344–358. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rüdinger M., Fritz-Laylin L., Polsakiewicz M., Knoop V. Plant-type mitochondrial RNA editing in the protist Naegleria gruberi. RNA. 2011;17:2058–2062. doi: 10.1261/rna.02962911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu C.J., Sheikh S., Miao W., Andersson S.G., Baldauf S.L. Missing genes, multiple ORFs, and C-to-U type RNA editing in Acrasis kona (Heterolobosea, Excavata) mitochondrial DNA. Genome Biol. Evol. 2014;6:2240–2257. doi: 10.1093/gbe/evu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burger G., Yan Y., Javadi P., Lang B.F. Group I-intron trans-splicing and mRNA editing in the mitochondria of placozoan animals. Trends Genet. 2009;25:381–386. doi: 10.1016/j.tig.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Janke A., Paabo S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993;21:1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randau L., Stanley B.J., Kohlway A., Mechta S., Xiong Y., Soll D. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science. 2009;324:657–659. doi: 10.1126/science.1170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg B.R., Hamilton C.E., Mwangi M.M., Dewell S., Papavasiliou F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs. Nat. Struct. Mol. Biol. 2011;18:230–236. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell L.M., Wallis S.C., Pease R.J., Edwards Y.H., Knott T.J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 21.Takenaka M., Zehrmann A., Verbitskiy D., Hartel B., Brennicke A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013;47:335–352. doi: 10.1146/annurev-genet-111212-133519. [DOI] [PubMed] [Google Scholar]
- 22.Wulff B.E., Nishikura K. Substitutional A-to-I RNA editing. Wiley Interdiscip. Rev. RNA. 2010;1:90–101. doi: 10.1002/wrna.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinman C.L., Majewski J. Comment on ‘Widespread RNA and DNA sequence differences in the human transcriptome’. Science. 2012;335:1302. doi: 10.1126/science.1209658. [DOI] [PubMed] [Google Scholar]
- 24.Simpson L., Thiemann O.H., Savill N.J., Alfonzo J.D., Maslov D.A. Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6986–6993. doi: 10.1073/pnas.97.13.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukes J., Flegontova O., Horak A. Diplonemids. Curr. Biol. 2015;25:R702–R704. doi: 10.1016/j.cub.2015.04.052. [DOI] [PubMed] [Google Scholar]
- 26.de Vargas C., Audic S., Henry N., Decelle J., Mahe F., Logares R., Lara E., Berney C., Le Bescot N., Probert I., et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 27.Lara E., Moreira D., Vereshchaka A., Lopez-Garcia P. Pan-oceanic distribution of new highly diverse clades of deep-sea diplonemids. Environ. Microbiol. 2009;11:47–55. doi: 10.1111/j.1462-2920.2008.01737.x. [DOI] [PubMed] [Google Scholar]
- 28.Marande W., Burger G. Mitochondrial DNA as a genomic jigsaw puzzle. Science. 2007;318:415. doi: 10.1126/science.1148033. [DOI] [PubMed] [Google Scholar]
- 29.Vlcek C., Marande W., Teijeiro S., Lukeš J., Burger G. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 2011;39:979–988. doi: 10.1093/nar/gkq883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiethega G.N., Yan Y., Turcotte M., Burger G. RNA-level unscrambling of fragmented genes in Diplonema mitochondria. RNA Biol. 2013;10:301–313. doi: 10.4161/rna.23340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiethega G.N., Turcotte M., Burger G. Evolutionary conserved cox1 trans-splicing without cis-motifs. Mol. Biol. Evol. 2011;28:2425–2458. doi: 10.1093/molbev/msr075. [DOI] [PubMed] [Google Scholar]
- 32.Valach M., Moreira S., Kiethega G.N., Burger G. Trans-splicing and RNA editing of LSU rRNA in Diplonema mitochondria. Nucleic Acids Res. 2014;42:2660–2672. doi: 10.1093/nar/gkt1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-Ezpeleta N., Teijeiro S., Forget L., Burger G., Lang B.F. Construction of cDNA libraries: focus on protists and fungi. In: Parkinson J, editor. Methods in Molecular Biology: Expressed Sequence Tags (ESTs) Vol. 533. Totowa, NJ: Humana Press; 2009. pp. 33–47. [DOI] [PubMed] [Google Scholar]
- 34.Cattenoz P.B., Taft R.J., Westhof E., Mattick J.S. Transcriptome-wide identification of A >I RNA editing sites by inosine specific cleavage. RNA. 2013;19:257–270. doi: 10.1261/rna.036202.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morse D.P., Bass B.L. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- 36.Myers E.W., Sutton G.G., Delcher A.L., Dew I.M., Fasulo D.P., Flanigan M.J., Kravitz S.A., Mobarry C.M., Reinert K.H., Remington K.A., et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 37.Li W., Jaroszewski L., Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 38.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valdar W.S. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 42.Xie Y., Wu G., Tang J., Luo R., Patterson J., Liu S., Huang W., He G., Gu S., Li S., et al. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 2014;30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 43.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;11:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathews D.H. Using the RNAstructure Software Package to predict conserved RNA structures. Curr. Protoc. Bioinformatics. 2014;46:11–22. doi: 10.1002/0471250953.bi1204s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013;41:W471–W474. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Will S., Joshi T., Hofacker I.L., Stadler P.F., Backofen R. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA. 2012;18:900–914. doi: 10.1261/rna.029041.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith C., Heyne S., Richter A.S., Will S., Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnhammer E.L., Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- 54.Kall L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 55.Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pett W., Ryan J.F., Pang K., Mullikin J.C., Martindale M.Q., Baxevanis A.D., Lavrov D.V. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: Insight from mtDNA and the nuclear genome. Mitochondrial DNA. 2011;22:130–142. doi: 10.3109/19401736.2011.624611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spencer D.F., Gray M.W. Ribosomal RNA genes in Euglena gracilis mitochondrial DNA: fragmented genes in a seemingly fragmented genome. Mol. Genet. Genomics. 2011;285:19–31. doi: 10.1007/s00438-010-0585-9. [DOI] [PubMed] [Google Scholar]
- 58.Feagin J.E., Mericle B.L., Werner E., Morris M. Identification of additional rRNA fragments encoded by the Plasmodium falciparum 6 kb element. Nucleic Acids Res. 1997;25:438–446. doi: 10.1093/nar/25.2.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Driscoll D.M., Innerarity T.L. RNA editing by cytidine deamination in mammals. In: Bass BL, editor. RNA Editing. Vol. 34. Oxford: Oxford University Press; 2001. pp. 61–76. [Google Scholar]
- 60.Mower J.P., Palmer J.D. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol. Genet. Genomics. 2006;276:285–293. doi: 10.1007/s00438-006-0139-3. [DOI] [PubMed] [Google Scholar]
- 61.Picardi E., Horner D.S., Chiara M., Schiavon R., Valle G., Pesole G. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 2010;38:4755–4767. doi: 10.1093/nar/gkq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inada M., Sasaki T., Yukawa M., Tsudzuki T., Sugiura M. A systematic search for RNA editing sites in pea chloroplasts: an editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol. 2004;45:1615–1622. doi: 10.1093/pcp/pch191. [DOI] [PubMed] [Google Scholar]
- 63.Verbitskiy D., Takenaka M., Neuwirt J., van der Merwe J.A., Brennicke A. Partially edited RNAs are intermediates of RNA editing in plant mitochondria. Plant J. 2006;47:408–416. doi: 10.1111/j.1365-313X.2006.02794.x. [DOI] [PubMed] [Google Scholar]
- 64.Cardi T., Giegé P., Kahlau S., Scotti N. Expression profiling of organellar genes. In: Bock R, Knoop V, editors. Genomics of Chloroplasts and Mitochondria. Vol. 35. Heidelberg: Springer; 2012. pp. 323–355. [Google Scholar]
- 65.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 66.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M., et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aphasizheva I., Aphasizhev R. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol. Cell. Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mungpakdee S., Shinzato C., Takeuchi T., Kawashima T., Koyanagi R., Hisata K., Tanaka M., Goto H., Fujie M., Lin S., et al. Massive gene transfer and extensive RNA editing of a symbiotic dinoflagellate plastid genome. Genome Biol. Evol. 2014;6:1408–1422. doi: 10.1093/gbe/evu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roy J., Faktorova D., Benada O., Lukes J., Burger G. Description of Rhynchopus euleeides n. sp. (Diplonemea), a free-living marine euglenozoan. J. Eukaryot. Microbiol. 2007;54:137–145. doi: 10.1111/j.1550-7408.2007.00244.x. [DOI] [PubMed] [Google Scholar]
- 70.Simpson L. The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication, and evolution. Annu. Rev. Microbiol. 1987;41:363–382. doi: 10.1146/annurev.mi.41.100187.002051. [DOI] [PubMed] [Google Scholar]
- 71.Lavrov D.V., Adamski M., Chevaldonné P., Adamska M. Extensive mitochondrial mRNA editing and unusual mitochondrial genome organization in calcaronean sponges. Curr. Biol. 2016;26:86–92. doi: 10.1016/j.cub.2015.11.043. [DOI] [PubMed] [Google Scholar]
- 72.Alseth I., Dalhus B., Bjoras M. Inosine in DNA and RNA. Curr. Opin. Genet. Dev. 2014;26:116–123. doi: 10.1016/j.gde.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 73.Gray M.W. O2'-Methylinosine, a constituent of the ribosomal RNA of Crithidia fasciculata. Nucleic Acids Res. 1976;3:977–988. doi: 10.1093/nar/3.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murphy F.V.t., Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 75.Blanc V., Park E., Schaefer S., Miller M., Lin Y., Kennedy S., Billing A.M., Ben Hamidane H., Graumann J., Mortazavi A., et al. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 2014;15:R79. doi: 10.1186/gb-2014-15-6-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hough R.F., Bass B.L. Adenosine deaminases that act on RNA. In: Bass BL, editor. RNA Editing. Oxford: Oxford University Press; 2001. pp. 77–108. [Google Scholar]
- 77.Aphasizhev R., Aphasizheva I. Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer. Wiley Interdiscip. Rev. RNA. 2011;2:669–685. doi: 10.1002/wrna.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stoltzfus A. On the possibility of constructive neutral evolution. J. Mol. Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- 80.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 81.Göringer H.U. ‘Gestalt,’ composition and function of the Trypanosoma brucei editosome. Annu. Rev. Microbiol. 2012;66:65–82. doi: 10.1146/annurev-micro-092611-150150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.