Abstract
Mitotic chromosomes are one of the most commonly recognized sub-cellular structures in eukaryotic cells. Yet basic information necessary to understand their structure and assembly, such as their composition, is still lacking. Recent proteomic studies have begun to fill this void, identifying hundreds of RNA-binding proteins bound to mitotic chromosomes. However, by contrast, there are only two RNA species (U3 snRNA and rRNA) that are known to be associated with the mitotic chromosome, suggesting that there are many mitotic chromosome-associated RNAs (mCARs) not yet identified. Here, using a targeted protocol based on 5′-tag sequencing to profile the mammalian mCAR population, we report the identification of 1279 mCARs, the majority of which are ncRNAs, including lncRNAs that exhibit greater conservation across 60 vertebrate species than the entire population of lncRNAs. There is also a significant enrichment of snoRNAs and specific SINE RNAs. Finally, ∼40% of the mCARs are presently unannotated, many of which are as abundant as the annotated mCARs, suggesting that there are also many novel ncRNAs in the mCARs. Overall, the mCARs identified here, together with the previous proteomic and genomic data, constitute the first comprehensive catalogue of the molecular composition of the eukaryotic mitotic chromosomes.
INTRODUCTION
The structure of the eukaryotic mitotic chromosome remains one of the oldest unresolved problems in biology (1–3). While there is growing knowledge of the larger-scale changes in size and shape of the chromosomes from interphase to metaphase (4,5), our understanding of the molecular details underlying these changes is still quite rudimentary (2,3). In fact, one of the basic characteristics of any molecular-level description of a large biological complex—its composition—has only just recently begun to be fully addressed with regards to the mitotic chromosome. In particular, several proteome investigations have catalogued thousands of proteins that appear to be integral components of the metaphase chromosome (as opposed to more loosely-bound ‘hitch-hikers’ from the cytoplasm) (6–8). Although further studies are needed to validate all of these candidates, such work has identified hundreds of RNA-binding proteins associated with the mitotic chromosome (8). This high abundance of RNA-binding proteins suggests that there are likewise many non-coding RNAs (ncRNAs) associated with the mitotic chromosome.
However, to date, there is no comprehensive annotation of the ncRNA composition in the mitotic chromosome. In fact, inspection of the present literature identifies only two different species that have been confirmed to be associated with mitotic chromosomes: U3 snRNA and rRNA (9–20,21). This should be compared with the ∼400 ncRNA species, including snoRNA, that have recently been discovered to be associated with interphase chromosomes (22–27). With these, one of the critical functions of these interphase chromosome-associated RNAs (iCARs) is the maintenance of the chromatin in a more open, de-condensed state (23–25). Consistent with this notion, at least some of the more prominent iCARs, such as LINE RNA, dissociate from the chromosome during condensation prior to metaphase (25). Nonetheless, we reasoned that the striking difference in numbers between the candidate RNA-binding proteins associated with the mitotic chromosome and the identified metaphase chromosome-associated RNAs (mCARs) suggests that there are many more mCARs than are presently known, whose function is likely to be as critical as, if different from, that of the iCARs.
More generally, the recent widespread interest in detailed characterizations of ncRNAs is a result of the unexpected finding of the high extent of expression of genomes (30), with up to 98.5% of the genomic sequence being transcribed in some cases (31). Subsequent extensive annotation of these transcripts showed that only a minor fraction encodes for polypeptides: the majority is non-coding. In mice, for example, there are now more than 41 000 annotated ncRNA transcripts that have been identified (see Materials and Methods), yet there are only ∼25 000 known protein-coding genes (32). While the functions of some of these ncRNAs have been determined (33,34), including roles in regulating chromatin structure (22,24,25,33,34), the functions of most are presently not understood. For those cases in which a function has been identified, a highly effective first step was the identification of their sub-cellular localization (22,24,25,28,29,35,37–41). Indeed, identification of the chromatin association of the aforementioned iCARs was a critical early step in eventually identifying the functional consequences of this association (22,24,25).
In this study, using an optimized method to isolate metaphase chromosomes and 5′-tag sequencing, we characterize the mammalian mCAR population. In particular, we identified 1279 mCARs, nearly 3-fold more than the presently known iCARs and several-fold more than the number of candidate RNA-binding proteins on the mitotic chromosome. This population includes many lncRNAs and lincRNAs that are highly conserved, as well as a pronounced enrichment of a few, specific SINE RNAs and, somewhat unexpectedly, many snoRNAs, including some that are homologues of the iCAR snoRNAs. Yet nearly half of these mCARs are presently not annotated, suggesting that there are also many novel ncRNA in the mCAR population that have eluded prior detection. The presence of so many novel ncRNAs, together with the predominance of mCAR-specific lncRNAs and lincRNAs, points to a dramatic redistribution of the chromatin-associated RNA species from interphase to metaphase. Interestingly, fluorescence in situ hybridization (FISH) of select mCARs revealed two strikingly different classes with distinct spatial patterns: one localizes to the chromosome periphery, similar to that of U3 snRNA (9–20), while the other localizes within the DNA-containing interior, either uniformly or in a punctate pattern. Overall, the identity of these mCARs, along with the previous characterizations of the protein and genome content of the mitotic chromosomes (6–8,36), effectively constitute the first extensive catalogue of the major components of the mitotic chromosome, an essential resource for future investigations into the structure and assembly of the mitotic chromosome.
MATERIALS AND METHODS
Cell culture
Mouse 3T3 cells were cultured in Dulbecco's Minimal Essential Medium (GIBCO, Life Technologies, Carlsbad, CA, USA) supplemented with 10% heat inactivated fetal calf serum (GIBCO, Life Technologies, Carlsbad, CA, USA) and penicillin-streptomycin (GIBCO, Life Technologies, Carlsbad, CA, USA). When the cells achieved 70–80% confluence, they were treated with colcemid (100 ng/ml) for 12 h to arrest the cells at the metaphase (6,7).
Isolation of mouse mitotic chromosomes
To isolate highly purified mitotic chromosomes, we optimized a previously described protocol (6). In particular, the arrested mitotic cells were washed twice by carefully adding phosphate-buffered saline (PBS) and then the cells were suspended in PBS by mechanically agitating the culture dish. Following centrifugation at 120 g for 5 min, the cells were re-suspended in a hypotonic solution (75 mM KCl) at room temperature for 30 min. The cells were then collected by centrifugation at 600 g for 5 min at room temperature, and suspended in PA (polyamine) buffer (15 mM Tris–HCl, 0.2 mM spermine, 0.5 mM spermidine, 0.5 mM EGTA, 2 mM EDTA, 80 mM KCl, 20 mM NaCl, 0.1 mM PMSF) with 1 μg/ml digitonin. The cell suspension was kept on ice for 5 min, and then homogenized with a dounce homogenizer. The remaining procedures involving centrifugation were performed at 4°C. The sample was centrifuged at 190 g for 3 min, the supernatant (S1), which contains most of the chromosomes, was recovered, and then, in order to recover additional chromosomes from the remaining intact metaphase cells in the pellet, the pellet was re-suspended, homogenized twice, and centrifuged at 190 g for 3 min. The supernatant (S2) was recovered. Both supernatants S1 and S2 were combined and centrifuged at 420 g for 5 min to remove any residual cell debris. The supernatant was recovered and then centrifuged at 1750 g for 10 min. The precipitated chromosomes were then resuspended in the PA buffer (the low salt sample). For the high salt sample, the precipitated chromosomes were treated with the PA buffer with 0.2 M NaCl for 25 min on ice, centrifuged at 1750 g for 6 min and then resuspended in the PA buffer. The chromosome suspension was loaded on the sucrose gradient (2 ml each of 5%, 15%, 25%, 35%, 60% (w/v) sucrose) in the PA buffer, and centrifuged at 1100 rpm for 90 min in a SW41 Ti rotor (Beckman Coulter, Indianapolis, IN, USA). The presence of chromosomes in the different fractions was first determined by phase contrast microscopy, and then the morphology of the chromosomes was examined with confocal laser scanning microscopy (A1Si, Nikon, Tokyo, Japan) using DAPI to ensure that the gross structure of the chromosome was not significantly altered by the isolation procedure. To measure for the presence of β-actin mRNA, the entire 2 ml of the 5%, 15%, and 25% sucrose fractions were collected separately and placed in three separate tubes. However, for both the 35% and 60% sucrose fractions, we collected each in two 1 ml aliquots and placed them in four separate tubes to avoid potential contamination of β-actin mRNA (and other mRNA) in the lower 35% fraction. With each sample, an equal volume of Trizol (Invitrogen, Carlsbad, CA, USA) was added to extract the RNA, followed by incubation with 0.04 units/μl DNase I for 40 min (New England Biolabs, Ipswich, MA, USA) and then extraction with phenol/chloroform and precipitation with ethanol. RNA was then converted into cDNA using Super Script III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) with oligo dT primers. The presence of β-actin mRNA in these samples was determined using RT-PCR.
Preparation of the RNA sequencing library and sequencing using the Illumina Hiseq 2000
After finding that chromosomes free from β-actin mRNA were located within the 15–35% fractions, we pooled the chromosomes from these fractions and then extracted RNA from each sample following the same method described in the previous section. We then treated the sample with a RiboMinus™ Eukaryote Kit (Invitrogen, Carlsbad, CA, USA) to remove rRNA. The sample was next incubated with 0.5 units/μl E. coli Poly(A) Polymerase (New England Biolabs, Ipswich, MA, USA) at 37°C for 40 min to add a poly-A tail to the RNA. Finally the poly-A tailed RNA was converted into cDNA using Super Script III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) with oligo dT primers. The oligo dT was removed using 1.5 units/μl Exonuclease I (New England Biolabs, Ipswich, MA, USA) at 37°C for 40 min and the RNA was eliminated by incubating in 0.1M NaOH for 15 min at 65°C, followed by neutralization of the buffer with 0.1 M HCl. Finally, the sample was treated with 0.5 units/μl RNase ONE™ Ribonuclease (Promega, Madison, WI, USA) at 37°C for 1.5 h, extracted with phenol/chloroform and precipitated with ethanol.
An N6 adaptor 1 (N6-up: ACAGGTTCAGAGTTCTACAGTCCGACGATCTAGCAGCAGN6/N6-down: CTGCTGCTAGATCGTCGGACTGTAGAACTCTGAACCTGT-Pi) was ligated to the 3′ end of the cDNA using a DNA ligation kit (Takara, Tokyo, Japan). The ligated templates were first denatured at 94°C for 3 min, followed by incubation at 42°C for 5 min to anneal the 5′ biotinylated primer (ACAGGTTCAGAGTTCTACAGTCCGAC) to the 3′ end of the single-stranded cDNA. This biotinylated primer allowed for easy isolation with streptavidin-coated magnetic beads (M-280, Invitrogen, Carlsbad, CA, USA) at a later step. Second-stranded cDNA synthesis was performed using 5 units/μl LA Taq: 68°C for 20 min, 62°C for 2 min; then 4°C. EcoP15I (New England Biolabs, Ipswich, MA, USA) digestion (42) was performed to create a specific 5′ tag (27 bp) for each DNA template (66 bp in length). Then, the 3′ adaptor (3′ up: NNTCGTATGCCGTCTTCTGCTTG/3′ down: CAAGCAGAAGACGGCATACGA) was ligated to the recovered 66 bp fragments. We then isolated the sample with streptavidin-coated magnetic beads.
PCR (primer-1: 5′-CAAGCAGAAGACGGCATACGA-3′; primer-2: 5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA-3′) was performed to prepare a DNA library suitable for sequencing with the Illumina platform. The final PCR product was 106 bp in length. After gel extraction (QIAGEN, Frankfurt, Germany), the quality and quantity of the library was evaluated using the Agilent Technologies 2100 Bioanalyzer. Sequencing was performed using Illumina HiSeq 2000, according to the manufacturer's protocols.
Data analysis
The Illumina sequencing reads were mapped using Bowtie (version 1.0.0) (43) onto the repeat-unmasked or repeat-masked mouse reference genome mm10, allowing a maximum of two mismatches in the reads. Only uniquely mapped reads were considered for further analysis. Tag clusters were produced by grouping overlapping tags with at least two reads in the cluster that start at exactly the same base on the same strand (Supplementary Table S1). Only those clusters that were enriched in the HS sample over the LS sample were analyzed further as mCARs. BEDTools (v2.17.0) (44) was used to assign all clusters to exonic and intronic regions of the genes and to intergenic regions. We evaluated clusters for overlap with ESTs available in the UCSC Genome Browser (http://genome.ucsc.edu/). Visualizations were performed using Circos (45). Enriched clusters were similarly compared to appropriate ncRNA data sets: lncRNA database: NONCODE (V3.0); snoRNA, snRNA database: fRNAdb (V3.4); uRNA (46). These databases were also used to tabulate the number of annotated ncRNA in mice.
A track of conserved genomic regions based on a phastCons analysis of an alignment of 60 vertebrate species was downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/). We then examined the extent of the overlap of the 95 mCAR lncRNA regions with these conserved phastCons elements, using R, and compared this with the degree of overlap of the entire population of 39 132 lncRNAs in the mouse genome with the conserved regions. A similar analysis was also performed to compare the conservation of the 139 mCAR snoRNAs to that of the entire population of 1602 snoRNAs in the mouse genome.
MUSCLE was used for multiple sequence alignments of the ID4 sequences (47). Visualization of the multiple sequence alignment was performed using Unipro UGENE (48). Visualization of the phylogenetic tree was performed using MEGA (49).
A two-stage method was used to identify putative snoRNAs in the novel mCAR population: First, SnoReport (50) was used to search for a potential snoRNA sequence within −20 to +350 b of the 5′ base of a novel tag cluster, and then secondly, for putative CD snoRNAs, we examined within −5 to +25 b of the 5′ base in a tag cluster for a C box. These criteria were chosen based on manual inspection of our tag clusters and known snoRNAs. In particular, this procedure was first validated with a control set that contained 77 known CD snoRNAs. Using these criteria, we correctly identified 94% of the CD snoRNA. We next examined 1000 random mouse sequences (982 intronic and 18 intergenic) based on these criteria and predicted 10 CD. Thus, based on this, for 404 random transcripts, we would identify ∼4 CD snoRNAs. As described in the text, in our population of 404 novel ncRNA mCARs, we instead identified 21 CD snoRNAs. We examined for putative target sequences in these potential snoRNAs at the known locations using PLEXY (51). We attempted to identify putative H/ACA snoRNA in a similar way but did not find criteria that enabled the identification of significantly more putative H/ACA snoRNA in our novel ncRNA population than in the randomly chosen mouse sequences.
Validation experiments
qPCR was performed using iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA) on the StepOnePlus™ Real-Time PCR system. Total RNA from colcemid-treated mitotic cells was isolated using Trizol whereas the cytoplasmic RNA from these cells (separated from metaphase chromosomes) was prepared following a standard protocol (8). Samples were analysed using the same reference as with the U3 snRNA enrichment measurement (β-actin). The primers used in various qPCR reactions are summarized in Supplementary Table S9.
3′ RACE was performed using Takara Taq HS (Takara, Tokyo, Japan). The amplified PCR products were cloned into the pGMT-Easy vector system (Promega, Madison, WI, USA). Several clones were sequenced and aligned to the UCSC Genome Browser to identify the transcripts associated with the 3′ ends. Gene-specific RACE primer sequences and sequencing results for clones are provided in Supplementary Tables S10 and S11.
RNA FISH analysis coupled with immunofluorescence on 3T3 cells were performed as follows. The PCR products of U3, NEAT1, MALAT1, snoRNA FR210669, snoRNA FR137451 and novel mCAR 1 with a T7 promotor sequence were used as in vitro transcription templates. Novel mCAR 1 is a novel ncRNA mCAR found in this work for which we also verified with 3′ RACE (see Supplementary Tables S10 and S11). The probe-labeling in vitro transcription was performed following a standard protocol of New England Biolabs using digoxigenin-labeled nucleotides. Cells grown on a glass coverslip were treated with colcemid (100 ng/ml) for 12 h to arrest the cells at the metaphase. The sample was then treated with a hypotonic solution (75 mM KCl) at room temperature for 8 min, followed by centrifugation using Cytospin 4 (ThermoFisher, Massachusetts, USA) at 1000 rpm for 5 min. The cells were then rinsed with PBS and fixed in 4% formaldehyde in PBS for 10 min at room temperature and washed two times with PBS, followed by permeabilization in PBS with 0.5% Triton X-100 for 5 min. The cells were then dehydrated through a series of ethanol washes and hybridized at 37°C overnight with digoxigenin-labeled probe dissolved in 50% formamide and 2X hybridization buffer (Sigma, CA, USA). Finally, the cells were washed in 50% formamide, 2X SSC at 37°C and then in 2X SSC at room temperature. Slides were blocked with 4% BSA in 2X SSC. RNA FISH signal was detected by incubating FITC-labeled anti-digoxygenin antibody (Roche, Penzberg, Germany) in 2X SSC with 4% BSA and then washed three times in 2X SSC. Slides were counterstained in ProLong® Diamond Antifade Mountant with DAPI (Invitrogen, Carlsbad, CA, USA) for 10 min and then imaged with confocal laser scanning microscopy (A1Si, Nikon, Tokyo, Japan) at a scan speed from 1/4 to 1/8 frames/s. Each RNA FISH experiment was repeated three times. Sense probes were employed as a negative control (Supplementary Table S12). The PCR primers for the RNA FISH probes are listed in Supplementary Table S12.
RESULTS
Isolation of RNA from metaphase chromosomes
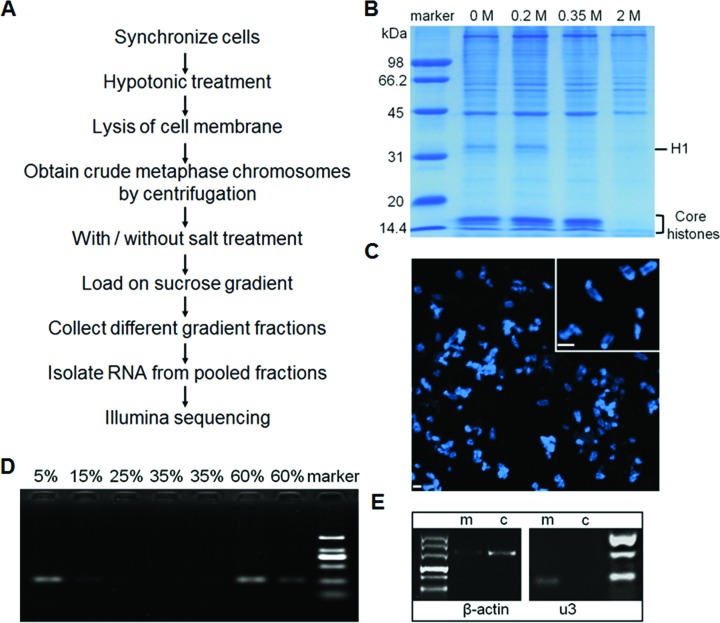
To identify the RNA composition on metaphase chromosomes, we first optimized a procedure employing sucrose gradient centrifugation to isolate mitotic chromosomes with a minimal amount of loosely bound (perhaps cytoplasmic) RNA or ribonuclear proteins (Figure 1) (21,52). Specifically, mouse 3T3 cells were first arrested at the metaphase using colcemid (6,7), and then lysed with a hypotonic solution (75 mM KCl) in which the characteristic morphology of mitotic chromosomes is retained (53). After a series of centrifugations, the metaphase chromosomes were finally isolated using sucrose gradient centrifugation (Figure 1A) (6–8). We refer to this as the low-salt (LS) sample. To enable identification of the RNAs that are most tightly bound to the metaphase chromosomes, before loading the chromosome suspension onto the sucrose gradient, we also prepared a sample that was washed with a high salt (HS) buffer (0.2 M NaCl), and, as will be described later, compared the RNAs extracted from the HS-sample with those from the LS-sample. This concentration of salt was identified as the highest concentration at which neither the protein composition associated with the metaphase chromosome (in particular the histone H1) (Figure 1B) nor the chromosome morphology (Figure 1C) are significantly altered from that in the absence of the incubation with the high salt buffer. We examined different fractions of the sucrose gradient with optical microscopy and found that only fractions from 15% to 35% (w/v) sucrose contained metaphase chromosomes (Supplementary Figure S1), and so we pooled these fractions for further examination.
Figure 1.
Purification of metaphase chromosomal-associated RNA. (A) Overview of the protocol. (B) Protein composition determined by SDS-PAGE after incubation with buffers containing the indicated increasing concentrations of salt. (C) Ultrastructure of metaphase chromosomes after incubation with the 0.2 M NaCl buffer. (D) Detection of β-actin mRNA using RT-PCR in different fractions of the sucrose gradient. (E) RT-PCR of U3 snRNA and β-actin in the mCARs (m) and cytoplasmic RNA (c) samples.
We next examined the quality of the extracted RNAs from the HS-sample. Since it is known that there is an almost complete absence of transcription during metaphase (54), it is expected that there should be very little mRNA associated with the metaphase chromosomes, other than more loosely bound ‘hitch-hikers’ from the cytoplasm. Using the highly expressed β-actin mRNA as an indicator of such ‘hitch-hiker’ RNA, we found that there is indeed nearly undetectable levels of this RNA in the HS-sample by RT-PCR, as expected (Figure 1E). To obtain a more precise understanding of the extent of enrichment of the more tightly bound RNAs, we used quantitative PCR (qPCR) to determine the degree of enrichment of U3 snRNA, which is well known to be associated with metaphase chromosomes (9–20), in the HS-sample compared to either cytoplasmic or total RNA from mitotic cells, using the β-actin mRNA as a reference. We found that U3 snRNA is enriched 252-fold and 169-fold compared to cytoplasmic RNA and total RNA (Supplementary Figure S7), respectively. Taken together, these results demonstrate that our protocol has indeed generated a highly purified population of putative mCARs, largely free from cytoplasmic RNA.
5′-tag deep sequencing of metaphase chromosome associated RNAs
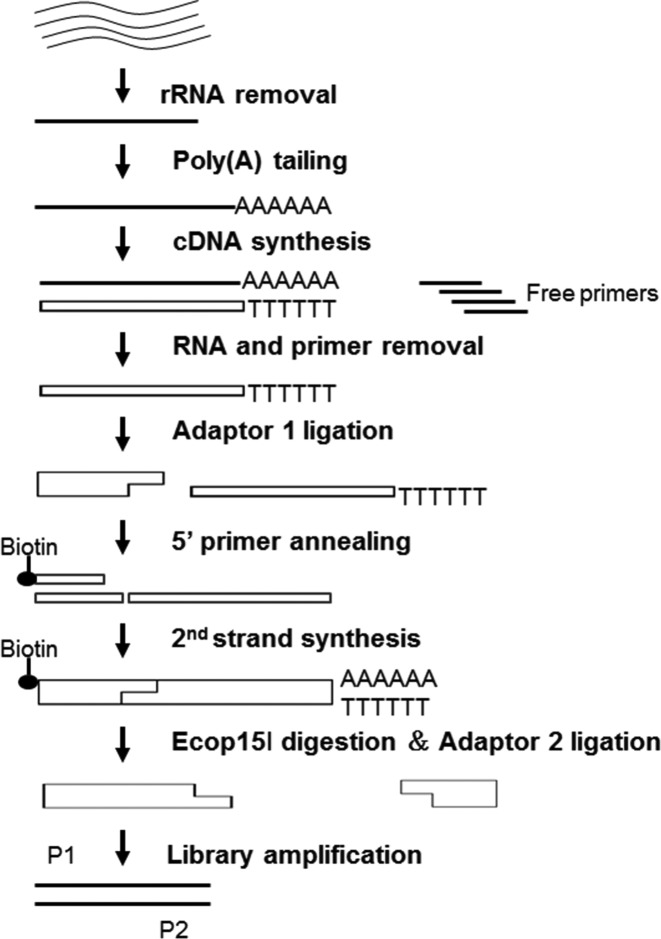
As the primary goal of this study was to identify the mitotic chromosomal RNAs from the already annotated RNAs, we reasoned that a 5′-tag sequencing strategy (Figure 2) would be sufficient to this end while providing a much higher dynamic range than other approaches (55).
Figure 2.
Strategy to prepare the 5′ tag library for Illumina sequencing.
Using the Illumina high-throughput sequencing platform, we obtained about 8 million reads that can be aligned to the repeat-unmasked genome and over 3 million reads aligned to the repeat-masked genome for each of the LS- and HS- samples (Table 1). Of these, over 2 million reads each from the LS- and HS- samples could be uniquely matched to the repeat-masked and the repeat-unmasked genomes (Table 1).
Table 1. Details of the aligned reads.
| Total aligned reads to repeat-unmasked genome-LS sample | 8004825 |
| Total aligned reads to repeat-masked genome-LS sample | 3127180 |
| Total aligned reads to repeat-unmasked genome-HS sample | 7709018 |
| Total aligned reads to repeat-masked genome-HS sample | 3075179 |
| Total unique matched reads to repeat-unmasked genome-LS sample | 2434346 |
| Total unique matched reads to repeat-masked genome-LS sample | 2154145 |
| Total unique matched reads to repeat-unmasked genome-HS sample | 3497070 |
| Total unique matched reads to repeat-masked genome-HS sample | 2339764 |
| Intronic mCAR reads | 1059675 |
| Antisense mCAR reads (antisense to intronic, exonic, UTR) | 16428 |
| Intergenic mCAR reads | 87971 |
| Exonic mCAR reads | 832 |
| UTR mCAR reads | 4024 |
| Total intronic mCARs | 306 |
| Total antisense mCARs | 126 |
| Total intergenic mCARs | 289 |
| Total exonic mCARs | 89 |
| Total UTR mCARs | 216 |
| Intronic mCARs covered by ESTs | 98% |
| Antisense mCARs covered by ESTs | 48% |
| Intergenic mCARs covered by ESTs | 23% |
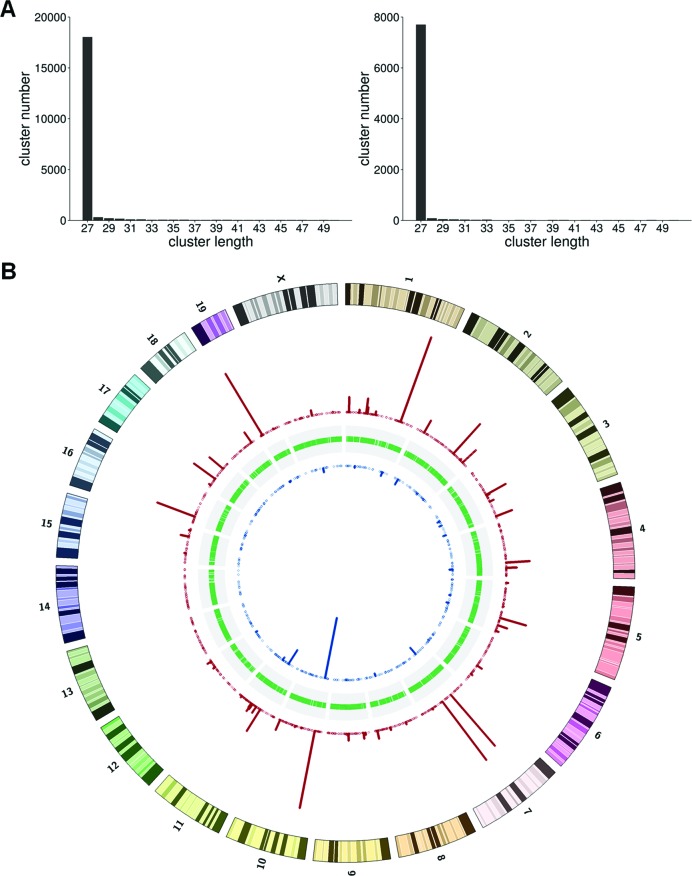
To better define individual mCARs from the sequenced tags, we clustered the tags based on their overlap and also on the requirement that at least two tags in the cluster share an identical 5′ nucleotide position (see Materials and Methods). Clusters identified in this way are predominantly 27 bp in length (Figure 3A). We considered only those clusters that are enriched in the HS-sample relative to the LS-sample (normalized by the total number of reads) as mCARs, since these are the most tightly bound transcripts. By this criterion, we finally identified 1279 mCARs, consisting of 253 in the repeat regions and 1026 in unique genomic locations (Figure 3B and Supplementary Tables S2–S5).
Figure 3.
Basic characterization of the sequenced LS and HS samples. (A) The LS (left) and HS (right) tag cluster length distributions. (B) Distribution of the chromosomal locations from which the clusters are derived. The blue (red) bars indicate cluster locations from the LS (HS) samples.
Identification of ncRNA as the major component in mCARs
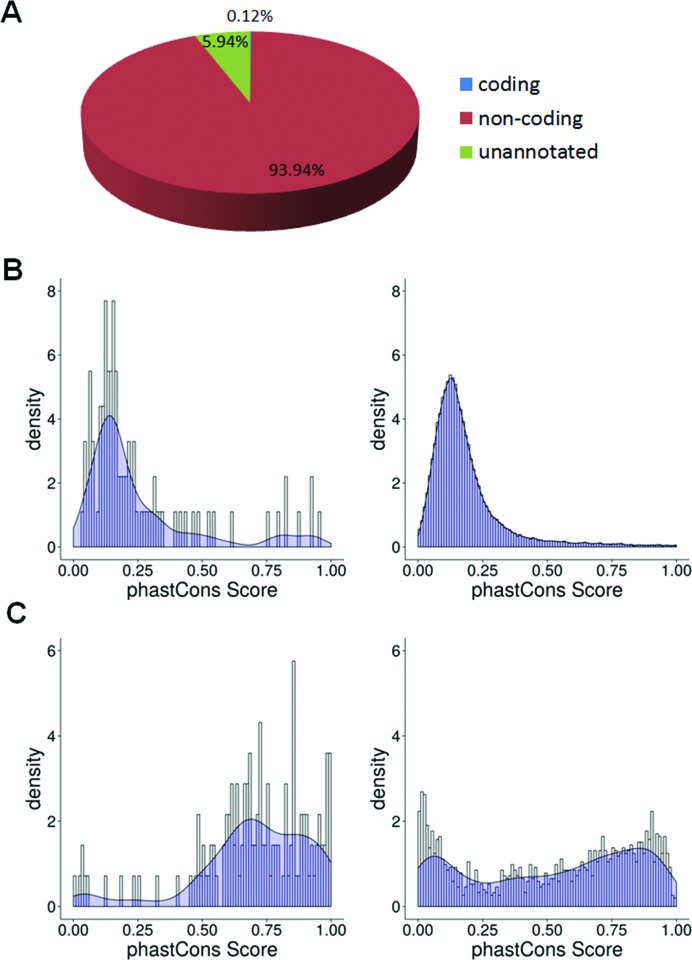
By far, most, 94.0% in terms of reads, of the mCARs originate in the annotated non-coding regions of the genome, and 5.9% of the reads are derived from unannotated regions, while only 0.1% of the reads mapped to coding regions (Figure 4A).
Figure 4.
Genomic characteristics of mCARs. (A) Pie chart showing the percentage of mCAR reads in the coding (blue), noncoding (red), or unannotated (green) region of the genome. (B) Conservation analysis of lncRNA mCAR (left) and all annotated lncRNAs (right). (C) Conservation analysis of the snoRNA mCARs (left) and all annotated snoRNAs (right).
The mCARs that map to the unique sequences of the annotated genome are dominated by two different classes (Table 2): sense or anti-sense to lnc/lincRNA (95 mCARs) and snoRNA (139 mCARs). In the former, only very few (namely MALAT-1, NEAT1, n285814, and n297068) are homologues of iCARs identified previously (22), indicating that this group of mCARs essentially represents a unique population of lnc/lincRNAs that are associated with chromatin. As for the snoRNAs, the 139 snoRNAs identified as mCARs reflect a significant proportion of all known snoRNA in the mouse genome (812) (fRNAdb (V3.4)). Interestingly, 18 of these 139 snoRNA mCARs are homologues of the human snoRNAs that are iCARs (Supplementary Table S6). We note that more than 91% of the mCARs mapped precisely to the 5′ end of known snoRNAs, attesting to the high quality of this correlation (Supplementary Table S7).
Table 2. Types of RNA in the mCARs.
| Different RNA species | Total reads | Cluster number | Reads/cluster | |
|---|---|---|---|---|
| snoRNA | 1022527 | 139 | 7356 | |
| predicted snoRNA | 4227 | 21 | 201 | |
| lncRNA | 490 | 44 | 11 | |
| lincRNA | 8803 | 38 | 232 | |
| Annotated antisense lncRNA | 183 | 13 | 14 | |
| Unannotated (42% EST coverage) | Intergenic | 86812 | 260 | 334 |
| Antisense | 16634 | 117 | 142 | |
| Intronic | 26743 | 123 | 217 | |
| Repeat ncRNA | 673697 | 253 | 2663 | |
| other ncRNA | 670 | 27 | 267 | |
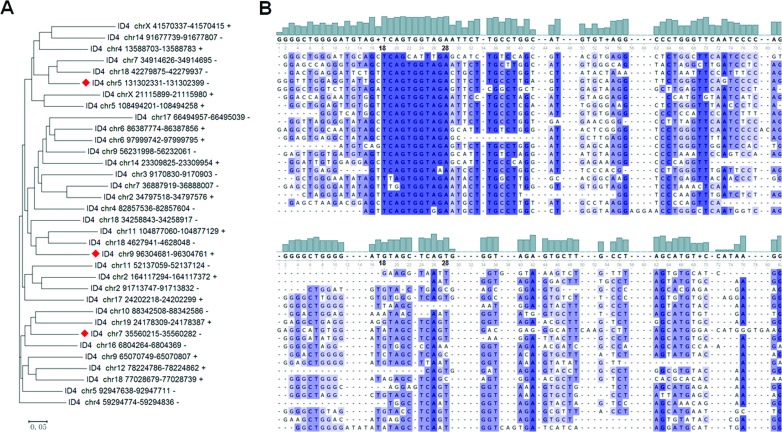
The mCARs within the repeat classes are dominated by the rRNAs (61% of total reads) and by the SINEs (37% of total reads) (Supplementary Figure S2). Although there is at least one member of each of the major types of SINEs (B1, B2, B4/RSINE, ID and MIR) in the mCARs, in terms of the numbers of species, ∼40% of the SINEs are members of the ID family, the least abundant SINE family in mice (56). Remarkably, in terms of the number of reads, only three members of the ID4 subtype account for 99.7% of all SINE mCARs, and a single ID4 member (located on chromosome 5, from 131302331 to 131302399 bp) alone accounts for 94.1% of all of the SINE mCARs reads. We evaluated whether the read count is correlated with sequence conservation in the ID4 population of mCARs, but no such correlation is apparent (Figure 5A).
Figure 5.
Conservation analysis of the ID4 mCARs. (A) Phylogenic analysis of the ID4 mCARs. The three sequences with the greatest number of reads are indicated with a red diamond. The scale bar reflects evolutionary distance. (B) Sequence comparison of 20 randomly chosen ID4 mCARs (top) and 20 randomly chosen ID4 sequences from the whole population of ID4 sequences in the genome (bottom). Similar results are obtained with the entire population of ID4 mCARs and 100 randomly chosen ID4 sequences from the whole genome population (Supplementary Figure S3).
Still, a comparison of the sequences of this specific population with those of all ID4 sequences in the mouse genome identified sequence commonalities that are not characteristic of the ID4 sequences as a whole (Figure 5B; Supplementary Figure S3), suggesting that these sequence similarities might be important for the association of these specific ID4 RNAs with metaphase chromosomes. In this regard, we identified two sequence motifs using MEME (57) in the mCAR ID4 sequences that are different from those motifs identified from all ID4 sequences (Supplementary Figure S4), although they do not match binding motifs of any presently known RNA-binding proteins (58).
Identification of novel snoRNAs in the abundant unannotated ncRNAs in mCARs
A striking number of mCARs—500—from the total population of 1279 mCARs did not overlap with the annotated regions of the genome. These genomic regions exhibit an EST coverage of 44%, are on average 263 kb from the nearest known transcription start site (TSS) (Supplementary Figure S5), and exhibit a read abundance comparable to those of the lincRNA mCARs (Table 2). Thus, these mCARs likely represent previously unknown ncRNAs.
Owing to the identification of many known snoRNAs in the mCARs, we hypothesized that at least some of these novel transcripts might be previously unknown snoRNAs. We thus examined these mCARs for well-established snoRNA sequence characteristics using SnoReport and indeed identified 21 that are possibly novel C/D box snoRNA (Supplementary Table S8). Further inspection of these putatively novel snoRNA with PLEXY resulted in the identification of the potential target of these snoRNAs for 17 of the predicted CD snoRNAs (Supplementary Figure S6, Table S5). While future efforts are needed to confirm these predictions, there are thus at least 488 novel ncRNA mCARs whose function is not presently characterized.
Extensive conservation of the mCARs
Previous studies found that iCARs exhibit evolutionary conservation (22). To determine whether the mCARs were similarly well conserved, we examined the degree of conservation of the fully-annotated 95 lncRNA mCARs compared to the entire population of mouse lnc/lincRNA among 60 vertebrate species using phastCons. The overall distribution of the lnc/lincRNA mCARs clearly shows many sequences exhibiting phastCons scores that are greater than the entire population of lncRNAs (P < 0.01) (Figure 4B). Thus the lnc/lincRNA mCARs are indeed well conserved. A similar comparison of the conservation of the snoRNA mCARs to that of all snoRNAs in the mouse genome also shows a greater degree of conservation of the snoRNA mCARs (Figure 4C). Further, we also note that 84% of the 139 known snoRNA and 67% of the predicted snoRNA in the mCARs have readily identifiable homologues in humans (Supplementary Tables S7 and S8). Therefore, overall, the two most dominant species of mCARs associated with unique sequences of the annotated genome are well conserved in mammals.
Validation of mCARs
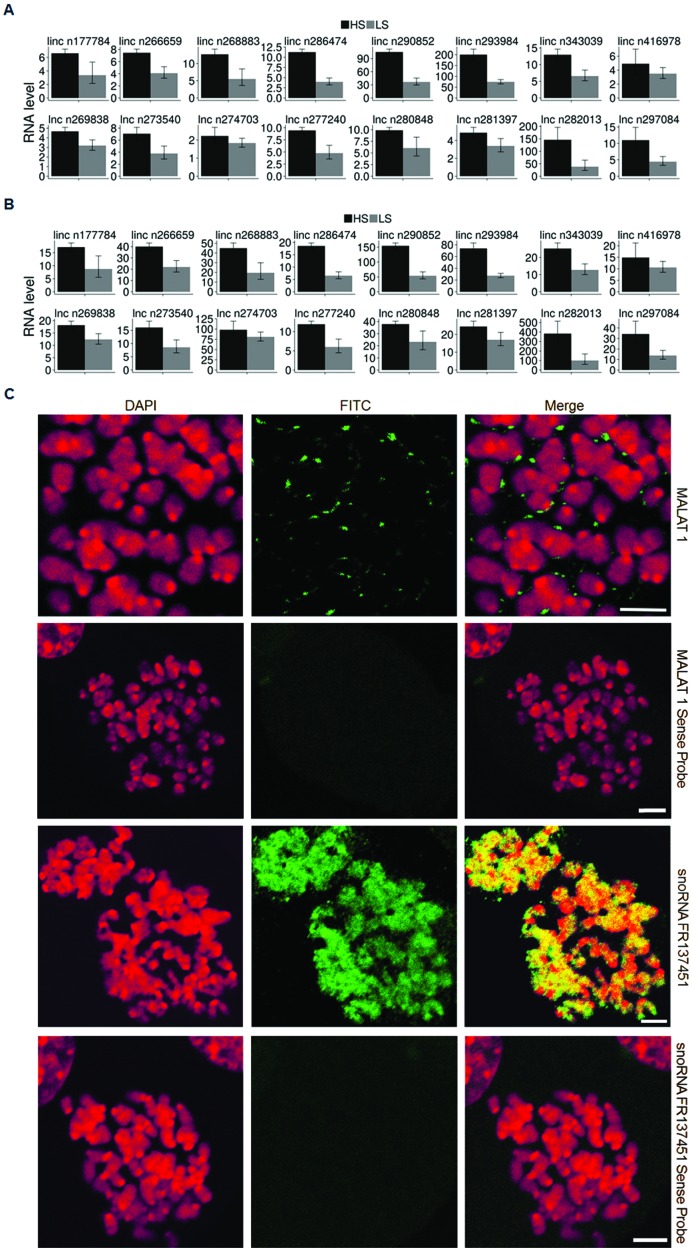
Finally, we examined 16 randomly selected lncRNAs mCARs for enrichment in the LS- and HS- samples using qPCR to confirm that the transcript was indeed found associated with the metaphase chromosomes as predicted from the 5′-tag sequencing results. As shown in (Figure 6A and B), whether compared to metaphase cell total RNA or cytoplasmic RNA, there is a marked enrichment of the lncRNAs in both the LS- and the HS-samples, with a greater enrichment in the HS-sample in almost every case. Similar observations were also found with 8 randomly chosen snoRNA or novel (snoRNA) mCARs using qPCR and by 3′RACE (Supplementary Figure S7).
Figure 6.
(A) qPCR validation for the HS- and LS- samples relative to metaphase cell total RNA. (B) qPCR validation between the HS-, LS- samples and cytoplasmic RNA. (C) RNA FISH validation of the mCAR localization of MALAT1 and snoRNA FR137451 to the mitotic chromosome. RNA FISH using sense probes was used as a negative control. Scale bar: 5 μm.
In addition, we also performed RNA FISH to verify the localization of the mCARs on the mitotic chromosomes. We investigated U3 snRNA as a positive control since it is well-known to be associated with the perichromosomal region (9–20), NEAT1 and MALAT1 since these are well-studied lncRNAs (59), two randomly chosen snoRNA mCARs since these are the most abundant species of mCARs, and a novel ncRNA identified in this work. In each case, the mCAR was found to be clearly associated with the mitotic chromosome (Figure 6C and Supplementary Figures S8, S9 and S10), in complete agreement with the sequencing results. Interestingly though, we note that these results show two different classes of mCAR localizations: those (NEAT1 and MALAT1) that, like U3 snRNA (9–20), are associated within the perichromosomal layer, and those (both snoRNAs and the novel ncRNA) that are localized within the DNA-containing interior. We note that the most intriguing of these is the novel ncRNA, which is distributed over the entire chromosome except within the centromeric region (Supplementary Figure S10), in stark contrast to a more punctate pattern of the snoRNAs.
DISCUSSION
There is perhaps no more widely known sub-cellular biological structure than the mitotic chromosome, yet this knowledge has largely long been limited to its overall shape and size: details at the molecular-scale have been limited to a few specific proteins and their general effect on the structure (60). Recent systems-wide characterizations of the protein content have revealed a much larger spectrum of candidates than previously thought, which has subsequently led to new discoveries of localization and function (6–8). Included within this population are hundreds of RNA-binding proteins, suggesting that there might likewise be a similar, large number of RNA species associated with the mitotic chromosome. The results presented here, which we believe is the first comprehensive profiling of the RNA content in the mitotic chromosome, indeed confirm this expectation.
In fact, we find that there are ∼1300 mCARs, several-fold more than the number of candidate RNA-binding proteins associated with the mitotic chromosome and 3-fold more than the number of CARs presently known (22,24). As these mCARs are as firmly bound to the chromosome as the histone H1 (Figure 1B), we expect that these RNAs are indeed integral components of the mitotic chromosome. The striking abundance of these species is unexpected based on previous work of both lncRNA and iCARs (22,24). These previous studies showed that the ncRNA species that were either localized to the nucleus or were tightly associated with chromosomes at interphase were found not to be bound to the mitotic chromosome (25,35), suggesting that there may be less RNA bound to chromosomes at metaphase. We find however that there is a large number of mCARs, the majority of which are different from the iCARs. Together, these results indicate that there is a massive redistribution of the CARs at some stage before the onset of mitosis, with the iCARs dissociating from and the mCARs binding to the chromosomes by mitosis. Presumably, this process is then reversed after mitosis, possibly during chromosome decondensation in the daughter cells.
Future work will be needed to determine the functional consequences of the chromosomal association of the mCARs. Two of the lncRNA mCARs, MALAT-1 and NEAT1, are components of nuclear speckles and paraspeckles, respectively, but other RNA components of these nuclear compartments (including U1 and U2 snRNA) are not found in the mCARs, indicating that association with these nuclear compartments per se is not specifically related to an association with metaphase chromosomes. Similarly, while many snoRNAs localize to the nucleolus, as does the U3 snRNA, there are other RNA components of the nucleolus (namely the U8 RNA and U17 RNA) that are not mCARs, indicating that association of an RNA with this nuclear compartment also does not guarantee association with metaphase chromosomes. Interestingly, as mentioned earlier, many snoRNAs have also been found to associate with chromatin in the interphase, where they are thought to maintain the chromatin in a de-condensed structure (24). Of these, 18 are homologues of those that are mCARs (Supplementary Table S6), suggesting the possibility of a similar (though counter-intuitive) function when associated with the more condensed metaphase chromosome, perhaps functioning to accelerate decondensation following mitosis.
An intriguing finding in our work is the high abundance of the specific members of the ID4 family of SINE repeats (Supplementary Table S5). Different types of SINE repeats are not believed to be conserved among different species, although there are some that have evolved from similar genes (58). In particular, the abundant human Alu SINEs are derived from the same gene as the mouse B1/B2 SINE repeats, some of which are mCARs, although the ID family of repeats appears to be rodent-specific (61–63). Thus whether there are abundant species-specific functions of the mCARs or whether other SINEs such as Alu sequences perform similar roles in humans that these ID4 ncRNAs perform in mouse will require further investigation. We note though that there are no LINE-type repeats in the mCARs, consistent with a recent study (25).
The markedly high number of mCARs that are possibly novel ncRNAs is an unexpected observation of this work. One reason for this unusually large number is likely the fact that we focused on a sub-cellular structure – the mitotic chromosome – that had not been studied in such detail before (for its RNA content). It is thus likely that there are other novel ncRNAs that remain to be identified that are associated with hitherto less commonly studied sub-cellular structures.
The mCARs identified here, together with the proteomic and genomic data already documented, constitute the first comprehensive catalogue of the composition of the mitotic chromosome in mammalian cells. Just how these various components contribute to the self-assembly—or disassembly—of one of the most well-recognized subcellular structures in biology will be fascinating to resolve.
DATA AVAILABILITY
Datasets were deposited in the Sequence Read Archive of the National Center for Biotechnology Information under accession number SRP061962.
Supplementary Material
Acknowledgments
We thank Dr Xiaowei Li for assistance in imaging, Le Zhang for the images from the sucrose gradient, and Ke Fang, Hongmei Li, and Xuecheng Chen for technical assistance in the course of this study. The authors are also grateful for the generous support from Nikon Instruments (Shanghai) Co., Ltd.
FUNDING
School Foundation of the Tianjin Medical University [134-201301]; National Basic Research Program of China [2013CB932800, 2014YQ090709]; National Natural Science Foundation of China [91129000, 11374207, 91229108, 31370750, 21273148 and 21303104]; Science and Technology Commission of Shanghai Municipality [15142201200], Shanghai Jiao Tong University [YG2012MS58]; K.C. Wong Education Foundation (to H.K.). Funding for open access charges provided by National Natural Science Foundation of China grants.
Conflict of interest statement. None declared.
REFERENCES
- 1.Flemming W. Zellsubstanz, Kern und Zelltheilung. Leipzig: Verlag Von F.C.W. Vogel; 1882. [Google Scholar]
- 2.Kschonsak M., Haering C.H. Shaping mitotic chromosomes: from classical concepts to molecular mechanisms. BioEssays. 2015;37:755–766. doi: 10.1002/bies.201500020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vagnarelli P. Chromatin reorganization through mitosis. Adv. Protein Chem. Struct. Biol. 2013;90:179–224. doi: 10.1016/B978-0-12-410523-2.00006-7. [DOI] [PubMed] [Google Scholar]
- 4.Liang Z., Zickler D., Prentiss M., Chang F.S., Witz G., Maeshima K., Kleckner N. Chromosomes progress to metaphase in multiple discrete steps via global compaction/expansion cycles. Cell. 2015;161:1124–1137. doi: 10.1016/j.cell.2015.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mora-Bermúdez F., Gerlich D., Ellenberg J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 2007;9:822–831. doi: 10.1038/ncb1606. [DOI] [PubMed] [Google Scholar]
- 6.Uchiyama S., Kobayashi S., Takata H., Ishihara T., Hori N., Higashi T., Hayashihara K., Sone T., Higo D., Nirasawa T., et al. Proteome analysis of human metaphase chromosomes. J. Biol. Chem. 2005;280:16994–17004. doi: 10.1074/jbc.M412774200. [DOI] [PubMed] [Google Scholar]
- 7.Fukui K., Uchiyama S. Chromosome protein framework from proteome analysis of isolated human metaphase chromosomes. Chem. Rec. 2007;7:230–237. doi: 10.1002/tcr.20120. [DOI] [PubMed] [Google Scholar]
- 8.Ohta S., Bukowski-Wills J.C., Sanchez-Pulido L., Alves F.L., Wood L., Chen Z.A., Platani M., Fischer L., Hudson D.F., Ponting C.P., et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azum-Gélade M.C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gas N. Cell cycle redistribution of U3 snRNA and fibrillarin. J. Cell Sci. 1994;107:463–475. doi: 10.1242/jcs.107.2.463. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Verdun D., Gautier T. The chromosome periphery during mitosis. BioEssays. 1994;16:179–185. doi: 10.1002/bies.950160308. [DOI] [PubMed] [Google Scholar]
- 11.Yasuda Y., Maul G.G. A nucleolar auto-antigen is part of a major chromosomal surface component. Chromosoma. 1990;99:152–160. doi: 10.1007/BF01735332. [DOI] [PubMed] [Google Scholar]
- 12.Gautier T., Dauphin-Villemant C., Andre C., Masson G., Arnoult J., Hernandez-Verdun D. Identification and characterization of a new set of nucleolar ribonucleoproteins which line the chromosomes during mitosis. Exp. Cell Res. 1992;200:5–15. doi: 10.1016/s0014-4827(05)80065-5. [DOI] [PubMed] [Google Scholar]
- 13.Gautier T., Fomproix N., Masson C., AzumGelade M.C., Gas N., Hernandez-Verdun D. Fate of specific nucleolar perichromosomal proteins during mitosis: cellular distribution and association with U3 snoRNA. Biol. Cell. 1994;82:81–93. doi: 10.1016/s0248-4900(94)80010-3. [DOI] [PubMed] [Google Scholar]
- 14.Van Hooser A.A., Yuh P., Heald R. The perichromosomal layer. Chromosoma. 2005;114:377–388. doi: 10.1007/s00412-005-0021-9. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez-Garcia L.F., Segura-Valdez M.L., Ochs R.L., Rothblum I.L., Hannan R., Spector D.L. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westendorf J.M., Konstantinov K.N., Wormsley S., Shu M.D., Matsumoto-Taniura N., Pirollet F., Klier F.G., Gerace L., Baserga S.J. M phase phosphoprotein 10 is a human U3 small nucleolar ribonucleoprotein component. Mol. Biol. Cell. 1998;9:437–449. doi: 10.1091/mbc.9.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiMario P.J. Cell and molecular biology of nucleolar assembly and disassembly. Int. Rev. Cytol. 2004;239:99–178. doi: 10.1016/S0074-7696(04)39003-0. [DOI] [PubMed] [Google Scholar]
- 18.Strasburger E. Ueber den Theilungsvorgang der Zellkerne und das Verhältniss der Kerntheilung zur Zelltheilung. Arch. Mikroskop. Anat. 1882;21:476–588. [Google Scholar]
- 19.Gautier T., Robert-Nicoud M., Guilly M.N., Hernandez-Verdun D. Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J. Cell Sci. 1992;102:729–737. doi: 10.1242/jcs.102.4.729. [DOI] [PubMed] [Google Scholar]
- 20.Booth D.G., Takagi M., Sanchez-Pulido L., Petfalski E., Vargiu G., Samejima K., Imamoto N., Ponting C., Tollervey D., Earnshaw W.C., et al. Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. eLife. 2014;3:e01641. doi: 10.7554/eLife.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donev R.M., Djondjurov L.P. Macromolecular and ultrastructural organization of the mitotic chromosome scaffold. DNA Cell Biol. 1999;18:97–105. doi: 10.1089/104454999315484. [DOI] [PubMed] [Google Scholar]
- 22.Mondal T., Rasmussen M., Pandey G K, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caudron-Herger M., Muller-Ott K., Mallm J., Marth C., Schmidt U., Fejes-Toth K., Rippe K. Coding RNAs with a non-coding function. Nucleus. 2011;2:410–424. doi: 10.4161/nucl.2.5.17736. [DOI] [PubMed] [Google Scholar]
- 24.Schubert T., Pusch M.C., Diermeier S., Benes V., Kremmer E., Imhof A., Langst G. Df31 Protein and snoRNAs Maintain Accessible Higher-Order Structures of Chromatin. Mol. Cell. 2012;48:434–444. doi: 10.1016/j.molcel.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Hall L.L., Carone D.M., Gomez A.V., Kolpa H.J., Byron M., Mehta N., Fackelmayer F.O., Lawrence J.B. Stable C0T-1 Repeat RNA Is Abundant and Is Associated with Euchromatic Interphase Chromosomes. Cell. 2014;156:907–919. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E., et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6 doi: 10.1038/ncomms8743. doi:10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner M., Ruthenburg A. Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 2015;12:1089–1098. doi: 10.1016/j.celrep.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatt D.M., Pandya-Jones A., Tong A. J., Barozzi I., Lissner M., Natoli G., Black D.L., Smale S.T. Transcript dynamics of pro-inflammatory genes uncovered by RNA-Seq analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilgner H., Knowles D.G., Johnson R., Davis C.A., Chakrabortty S., Djebali S., Curado J., Snyder M., Gingeras T.R., Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 31.Mattick J.S. RNA regulation: a new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 32.Guenet J.L. The mouse genome. Genome Res. 2005;15:1729–1740. doi: 10.1101/gr.3728305. [DOI] [PubMed] [Google Scholar]
- 33.Wang K.C., Chang H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Gene Dev. 2013;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran N., Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 37.van Heesch S., van Iterson M., Jacobi J., Boymans S., Essers P.B., Bruijn E., Hao W., MacInnes A.W., Cuppen E., Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:1–12. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark M.B., Mattick J.S. Long noncoding RNAs in cell biology. Semin. Cell. Dev. Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Taft R.J., Simons C., Nahkuri S., Oey H., Korbie D.J., Mercer T.R., Holst J., Ritchie W., Wong J.J., Rasko J.E.J., et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 2010;17:1030–1034. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- 40.Taft R.J., Glazov E.A., Cloonan N., Simons C., Stephen S., Faulkner G.J., Lassmann T., Forrest A.R.R., Grimmond S.M., Schroder K., et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 41.Nolte E.N.M., Buermans H.P.J., Waasdorp M., Stoorvogel W., Wauben M.H.M., Hoen P.A.C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi H., Lassmann T., Murata M., Carninci P. 5′end–centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012;7:542–561. doi: 10.1038/nprot.2012.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:1–10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer T.R., Wilhelm D., Dinger M.E., Solda G., Korbie D.J., Glazov E.A., Truong V., Schwenke M., Simons C., Matthaei K.I., et al. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okonechnikov K., Golosova O., Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 49.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hertel J., Hofacker I.L., Stadler P.F. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- 51.Kehr S., Bartschat S., Stadler P.F., Tafer H. PLEXY: efficient target prediction for box C/D snoRNAs. Bioinformatics. 2010;27:279–280. doi: 10.1093/bioinformatics/btq642. [DOI] [PubMed] [Google Scholar]
- 52.Adlakha R.C., Sahasrabuddhe C.G., Wright D.A., Lindsey W.F., Rao P.N. Localization of mitotic factors on metaphase chromosomes. J. Cell Sci. 1982;54:193–206. doi: 10.1242/jcs.54.1.193. [DOI] [PubMed] [Google Scholar]
- 53.MacLeod R.A.F., Kaufmann M., Drexler H.G. Cytogenetic harvesting of commonly used tumor cell lines. Nat. Prot. 2007;2:372–382. doi: 10.1038/nprot.2007.29. [DOI] [PubMed] [Google Scholar]
- 54.Prescott D.M., Bender M.A. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- 55.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kramerov D.A., Vassatzky N.S. Origin and evolution of SINEs in eukaryotic genomes. Heredity. 2011;107:487–495. doi: 10.1038/hdy.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cook K.B., Kazan H., Zuberi K., Morris Q., Hughes T.R. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2010;39:D301–D308. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swedlow J.R., Hirano T. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell. 2003;11:557–569. doi: 10.1016/s1097-2765(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 61.Tsirigos A., Rigoutsos I. Alu and B1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS Comp. Biol. 2009;5:e1000610. doi: 10.1371/journal.pcbi.1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J., Kass D.H., Deininger P.L. Transcription and processing of the rodent ID repeat family in germline and somatic cells. Nucleic Acids Res. 1995;23:2245–2251. doi: 10.1093/nar/23.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim J., Deininger P.L. Recent amplification of Rat ID sequences. J. Mol. Biol. 1996;261:322–327. doi: 10.1006/jmbi.1996.0464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets were deposited in the Sequence Read Archive of the National Center for Biotechnology Information under accession number SRP061962.