Abstract
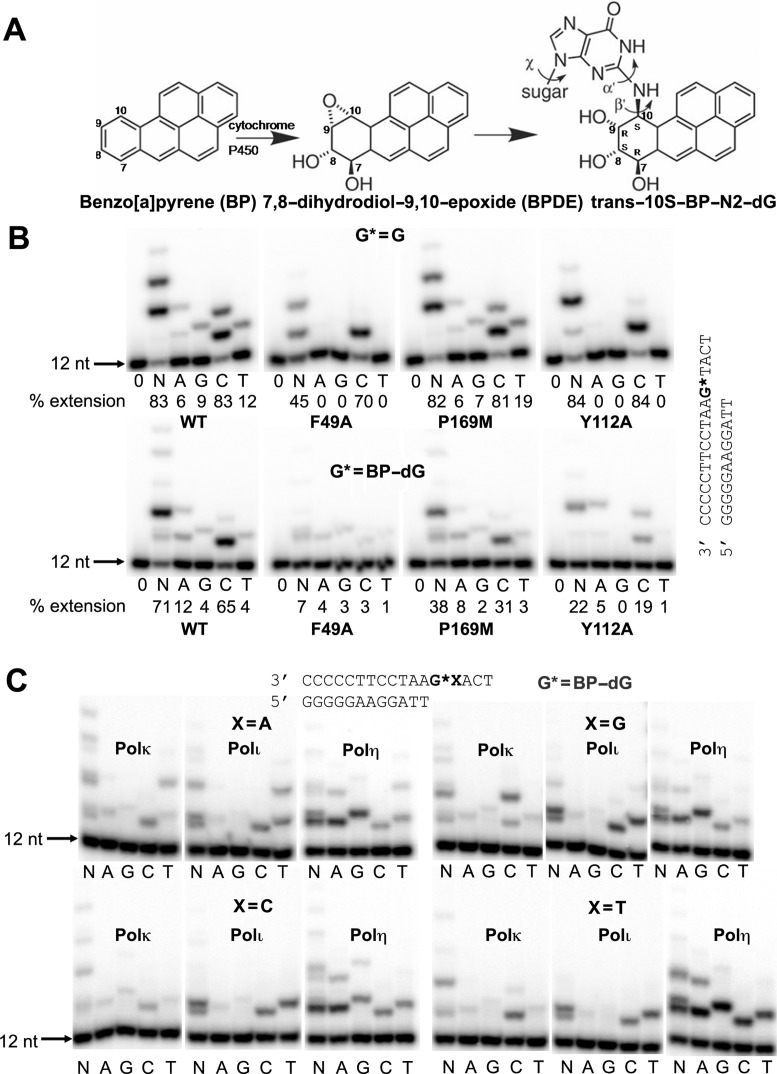
Benzo[a]pyrene (BP) is a well-known and frequently encountered carcinogen which generates a bulky DNA adduct (+)-trans-10S-BP-N2-dG (BP-dG) in cells. DNA polymerase kappa (polκ) is the only known Y-family polymerase that bypasses BP-dG accurately and thus protects cells from genotoxic BP. Here, we report the structures of human polκ in complex with DNA containing either a normal guanine (G) base or a BP-dG adduct at the active site and a correct deoxycytidine. The structures and supporting biochemical data reveal a unique mechanism for accurate replication by translesion synthesis past the major bulky adduct. The active site of polκ opens at the minor groove side of the DNA substrate to accommodate the bulky BP-dG that is attached there. More importantly, polκ stabilizes the lesion DNA substrate in the same active conformation as for regular B-form DNA substrates and the bulky BPDE ring in a 5′ end pointing conformation. The BP-dG adducted DNA substrate maintains a Watson–Crick (BP-dG:dC) base pair within the active site, governing correct nucleotide insertion opposite the bulky adduct. In addition, polκ's unique N-clasp domain supports the open conformation of the enzyme and the extended conformation of the single-stranded template to allow bypass of the bulky lesion. This work illustrates the first molecular mechanism for how a bulky major adduct is replicated accurately without strand misalignment and mis-insertion.
INTRODUCTION
The genome of cells is continuously challenged by external and internal toxic agents that damage the DNA (1). High-fidelity replicative polymerases are usually blocked by these DNA lesions, leading to replication fork stalling and threatening cell survival as well. Cells employ specialized DNA polymerases (mainly Y-family polymerases) to bypass DNA lesions and to reduce the genotoxic stress (2,3). Humans have four Y-family polymerases: polη, polι, polκ and Rev1. The first three members exhibit a unique DNA-lesion bypass and fidelity profile (4). Polη and polι replicate, in an error-free manner, the thymine-thymine (T–T) cyclobutane pyrimidine dimer and oxidative lesion 8-oxy-guanine, respectively (5–8).
Polκ accurately bypasses bulky N2-adducted deoxyguanosines, such as DNA lesions containing aromatic alkyl groups from benzo[a]pyrene (BP), 2-acetylaminofluorene (AAF) and 9-mythylanthracene (ANTH) (9–11).
Polycyclic aromatic hydrocarbons (PAHs) are widespread environmental pollutants. BP is the best characterized PAH (12,13) and is associated with human cancers such as lung, skin and colon cancers (14,15). BP is highly carcinogenic due to its property of metabolizing into highly reactive products, benzo[a]pyrene diol epoxides (BPDEs) (16,17). BP is now listed as a Group 1 carcinogen by the International Agency for Research on Cancer (IARC). In cells, anti-BPDEs are the most active metabolites (18), which react with DNA predominantly at the N2 position of guanine and generate minor groove attached bulky adducts (16,19). The dominant BPDE adduct in cells is (+)-trans-10S-BP-N2-dG (BP-dG) (Figure 1A). The bulky BP-dG strongly blocks DNA synthesis by high fidelity DNA polymerases and causes mutations in cells (20,21). Replication of the adduct occurs via translesion synthesis and is frequently associated with mutagenesis. However, polκ has been shown to bypass BPDE-induced DNA lesions efficiently in an error-free manner, preferentially incorporating the correct nucleotide dCTP in vitro (10,11,22–25), thus contributing to bulky carcinogen tolerance in cells (10,26,27). In vivo experiments of mouse embryonic fibroblasts support the important role of polκ in efficient and accurate bypass of BP-dG adducts (26–29).
Figure 1.
Structures of BP adduct and replication of BP-dG lesion. (A) BP, its metabolite BPDE and the major BP-dG adduct in DNA. (B) Incorporation of deoxy nucleotides opposite normal G and BP-dG by wild-type (WT) polκ and mutants F49A, P169M, Y112A and Y112F at insertion stage. The reaction was carried out by adding no nucleotide (0), all four nucleotides (N) or individual nucleotides (A, G, C and T), using 5 min for undamaged G template and 15 min for BP-dG lesion template. DNA sequences used for the assays are shown on the right side of gel images. (C) Incorporation of nucleotides opposite the BP-dG adduct in four different sequence contexts and using different human Y-family polymerases: polκ, polι and polη. The BP-dG adduct is indicated as G*, and X represents either A, G, C or T. Reaction times were 10 min for polκ and polι and 5 min for polη, with 100 μM dNTPs. A general DNA sequence for the assays is shown at the top of the gel images.
Current insight into the mechanism of bypass of BP-dG lesions has been derived from the atomic resolution structures of the model Y-family polymerase Dpo4, which is the human homolog of polκ in Archaea. Dpo4 bypasses this lesion in an error-prone manner (30). Structures of Dpo4 in ternary complexes with DNA containing the BP-dG adduct have demonstrated the molecular basis for base substitutions and -1 frame shift mutations (30). In this case, the bulky adduct is flipped/looped out of the DNA helix and moved into a structural gap due to the limited space of Dpo4 on the minor groove side of the DNA substrate. Nevertheless, the Dpo4 model does not explain the mechanism of accurate replication of BP-dG adducts. Direct structural evidence is required to elucidate the molecular mechanism of error-free bypass of BP-dG in cells.
In this work, we studied the ternary structure of human polκ in complex with BP-dG adduct DNA to determine how accurately polκ replicates this bulky lesion using the dNTP containing the correct base, cytosine (C). Our structure reveals that the BP-dG adduct is well accommodated in the polκ active site without steric hindrance. Our structure-based mutagenesis study confirms that the residues surrounding the adduct and a residue in the N-clasp are important for the activity of polκ in BP-dG replication. Together, our structural and biochemical analyses provide a molecular basis for understanding how human polκ bypasses BP-dG lesions accurately and thereby helps tolerate DNA damage induced by BP in cells.
MATERIALS AND METHODS
Preparation of protein and DNA for crystallization and biochemical analysis
The human polκ construct encoding residues 1–526 was cloned into the pHIS-Parallel1 vector with hexa-histidine (6XHis) tag (31). We will refer to this N-terminal fragment as polκ for simplicity since the fragment harbors all five polymerase domains and has the same nucleotide incorporation activity and fidelity as the full-length polκ (35). The polκ mutants in this study were generated by site-directed mutagenesis using primers containing the desired mutation. Mutations were confirmed by DNA sequencing. The polκ and mutants with an N-terminal 6XHis tag were overexpressed in Escherichia coli (BL21pRARE) with 0.5 mM IPTG and purified by Ni-affinity and ion exchange chromatography. The 6XHis tag was removed by TEV protease before ion-exchange (HiTrap SP column) chromatography. A 21-mer (5′- TATGGTGATCCGCGCGGATCA -3′) and a 22-mer oligonucleotide containing a single (+)-trans-10S-BP-N2-dG (BP-dG) lesion (G*) (5′- ATGG*CTGATCCGCGCGGATCAG -3′) were synthesized by Keck Oligo Inc. (Yale University). Both modified and unmodified DNA substrates used for crystallization and biochemical assays were purified by ion-exchange chromatography (32). The mass of the modified oligonucleotide was confirmed by MALDI TOF-MS/MS analysis.
Crystallization
The ternary complexes were prepared by mixing polκ with either normal dG or BP-dG modified DNA in a 1:1.2 molar ratio in the presence of 10 mM MgCl2. The protein–DNA complexes were incubated with either 2 mM ddCTP (for normal DNA) or 2 mM dCMPNPP (designated as dCTP* for BP-dG DNA) at room temperature for 30 min before setting up crystallization trials. Crystals of polκ/BP-dG modified DNA (polκ-BPG) complex were obtained in 30% PEG400 and 0.2 M ammonium nitrate using the hanging drop vapor diffusion method at 22°C. Several rounds of streak seeding were performed to produce good quality crystals. Crystals of polκ/normal DNA (polκ-G) complex were obtained in 30% PEG400 and 0.2 M potassium acetate. For data collection, the crystals were picked from the drop and then flash frozen in liquid nitrogen. X-ray diffraction data were collected on beam line CMCF-08ID at the Canadian Light Source (CLS). Crystals belong to the space group P212121. Diffraction data were processed and scaled using MOSFLM and SCALA (33), respectively.
Structure determination and refinement
Both structures were solved by molecular replacement (MR) with PHASER (34), using the polκ ternary complex structure (PDB 2OH2) (35) as a search model. After the first round of rigid body refinement with PHENIX (36), the 2Fo–Fc electron density map demonstrates unambiguous densities for the modified guanine in the template strand, the incoming nucleotide and metal ions, which were then iteratively built into the map using the molecular graphics program COOT (37). Structure refinement was completed using PHENIX. Model inspection and manual modeling were performed using COOT. Figures were generated using PYMOL (http://www.pymol.org) (38). The data processing and refinement statistics are listed in Table 1.
Table 1. Data collection and refinement statistics.
| Polκ-G | Polκ-BPG | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 66.2, 129.3, 167.7 | 63.6, 130.2, 166.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)a | 46.2–2.60 (2.74–2.60) | 47.1–2.80 (2.95–2.80) |
| R sym or Rmerge | 6.7 (36.3) | 7.0 (67.1) |
| I/σI | 10.3 (2.7) | 12.5 (2.0) |
| Completeness (%) | 98.3 (99.8) | 98.7 (98.4) |
| Redundancy | 4.3 (4.2) | 3.3 (3.3) |
| Refinement | ||
| Resolution (Å) | 46.2–2.59 | 47.1–2.80 |
| No. reflections | 44 341 | 34 189 |
| R work/ R free | 0.200/0.239 | 0.212/0.252 |
| No. atoms | ||
| Protein | 6710 | 6747 |
| DNA | 948 | 1002 |
| Water | 103 | 113 |
| B-factors | ||
| Protein | 58.8 | 51.0 |
| DNA | 55.4 | 44.6 |
| Water | 51.1 | 41.0 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.008 | 0.006 |
| Bond angles (°) | 1.18 | 1.06 |
aValues in parentheses are for the highest resolution shell.
Primer extension assays
An 18-mer template and a 5′-32P labeled 13-mer primer were used for primer extension assays. The sequences of these oligonucleotides are shown in the respective figures. The standard replication reaction contained 50 nM DNA, 1–10 nM polκ or its mutants, 50–100 μM of either all four dNTPs or individual dNTPs in 50 mM Tris (pH 7.5), 10 mM MgCl2, 250 μg/ml BSA, 5 mM DTT and 2.5% glycerol. The reactions were carried out at 37°C for either 5 min (normal DNA) or 15 min (BP–dG DNA). The products were resolved on a 20% PAGE containing 8 M urea. Gels were visualized using PhosphorImager and the percentage of replication was calculated using ImageQuant.
RESULTS AND DISCUSSION
Polκ accurately replicates BP–dG lesion DNA
We carried out replication assays to test polκ's ability to replicate the BP–dG adduct. We used the N-terminal polymerase active portion of polκ (1-526 fragment) for the assays. Either a normal or an adducted G base was at the primer-template junction for primer extension comparison. The primer extension results show that polκ replicates the BP–dG adduct and inserts the correct C base preferentially opposite this adduct (Figure 1B). This observation fits with a recent kinetic study from Liu et al. (2014) in which the insertion efficiency of C is nearly two orders of magnitude higher than mis-insertion efficiencies (39). To compare polκ with other Y-family members and test their bypass fidelities for BP–dG in different DNA sequence contexts, we tested all three human Y-family polymerases κ, η and ι in replication assays. We used DNA substrates with different bases on the 5′ side of the BP–dG adduct. The results show polκ as the most accurate polymerase that replicates BP–dG with preferential dC insertion, independent of the DNA sequence 5′ to the BP–dG adduct (Figure 1C). Both polι and polη are highly mutagenic in BP–dG replication, regardless of the DNA sequence. Polι has a pronounced T mis-insertion, and polη prefers purine base (A and G) mis-insertions (Figure 1C), similar to previous data (10,40,41). Pronounced 5′-dependent mis-insertions opposite the BP–dG substrate, as observed with Dpo4 (30), were found to be absent when we used polι and polη. Dpo4's 5′ dependence results from BP–dG flipping/looping-out of the DNA helix in the Dpo4/BP–dG ternary complexes (30). Different replication fidelities and mutagenic patterns by the Y-family polymerases indicate that different structural mechanisms exist for BP–dG replication.
Overall ternary structures of polκ–DNA complexes
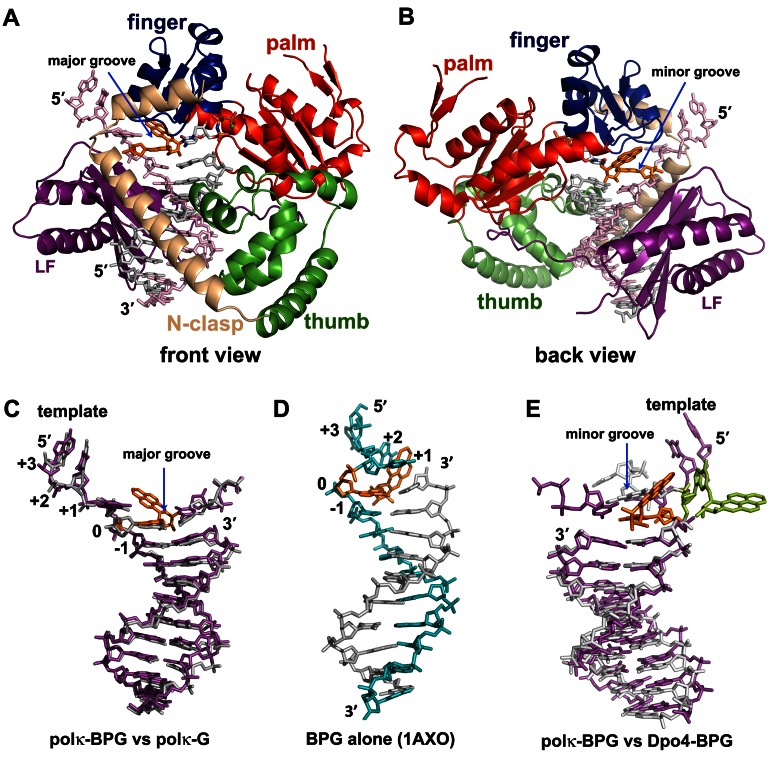
We determined the structures of polκ (residues 1–526) in complex with DNA, containing at the template position either the BP–dG adduct (polκ–BPG) or normal G (polκ–G). The structures contain a correct incoming nucleotide, ddCTP or dCMPNPP (dCTP*) at the active site. The two polκ complexes, polκ–G and polκ–BPG, were crystallized in the same crystal form that contains two ternary complexes per asymmetric unit. We refined the structures to 2.6 Å and 2.8 Å resolutions, respectively (Table 1). Polκ encircles the DNA substrate with the common Y-family polymerase domains of palm, finger, thumb and little finger (LF), as well as with a unique N-clasp domain as observed previously (Figure 2A) (35,42). The bulky BP–dG adduct resides within the active site toward the minor groove side of the DNA helix (Figure 2). No significant conformational change is caused by the bulky adduct binding to polκ. The polκ–BPG and polκ–G structures superimpose well with RMSD in a range of 0.4–0.6 Å over 423 Cα atoms in pair-wise comparisons among four independent structures. These polκ structures are also very similar to other previously reported polκ–DNA ternary complexes and superpose each other with root-mean-square deviations (RMSDs) of 0.7–0.9 Å over 423 Cα atoms. The structures superpose particularly well in the DNA binding cleft, which contacts the DNA directly (Supplementary Figure S1) (35,42), indicating that the polκ structure is stable in its DNA bound forms, regardless of the presence of BP–dG adduct and the crystal packing environments.
Figure 2.
Polκ–DNA–dNTP ternary complexes and DNA conformations. (A and B) Overall structure of Polκ–BPG ternary complex in (A) front and (B) back views. Template is in light pink, primer in grey, BP–dG in orange and Mg++ ion in green sphere. Structure domains are in the same colors as their labels. BP–dG is in orange. (C) Superposition of DNA from polκ–BPG (magenta) and polκ–G (grey) in the front view. (D) NMR structure of BP–dG adducted DNA (no protein bound) in the same front orientation as in (C). Template is in cyan. The single stranded (ss) template strand turns back to stabilize the bulky/hydrophobic BPDE ring that is sandwiched between +1 and +2 template bases. The helix is distorted, elongated and narrowed. (E) BP–dG DNA with polκ (magenta) and with Dpo4 (light grey) in the back view. All three DNA substrates in C–E are in a B-form. The Dpo4 structure (E) shows the BP–dG adduct (light green) flipping out of the helix.
Strikingly, the BP–dG adducted DNA substrates in polκ–BPG retain a standard B-form conformation with no conformational changes, almost identical to the normal DNA substrates in polκ–G (Figure 2C and Supplementary Tables S1 and S3). Particularly, the adducted guanine maintains the anti conformation, with the BPDE ring in the minor groove and the single stranded (ss) template downstream in an extended conformation. The B-form conformation of adducted DNA observed here is different from the BP–dG adducted DNA structure, in either a protein-free form (Figure 2D) (43) or a Y-family polymerase bound form (Figure 2E) (30). The protein-free adducted DNA helix is in a distorted B-form, with a narrowed minor groove (Supplementary Table S3) and a template strand with a U-turn to protect the bulky and hydrophobic BPDE ring from aqueous solvent (Figure 2D, Supplementary Table S1) (43). For the Dpo4 bound BP–dG DNA helix, enclosure of Dpo4 on the minor groove side and the bulky/hydrophobic nature of BP–dG causes the adduct to either intercalate into the DNA helix or flip/loop out of the helix (30). In the latter case, extra helical BP–dG fits into a structural gap between the LF and finger domains to protect itself from aqueous solvent (Figure 2E) (30). Only the extra helical structures of adducted DNA in Dpo4 are in active forms for primer extension, which results in base substitution and frameshift mutagenesis. In contrast, polκ holds the adducted DNA substrate in a regular DNA conformation for accurate replication. The striking similarities in the undamaged and damaged ternary complex structures suggest that polκ is well poised to maintain a B-form DNA and insert the correct base (C) opposite G or BP–dG for accurate replication.
BP–dG resides at the active site that is open at the minor groove side of DNA
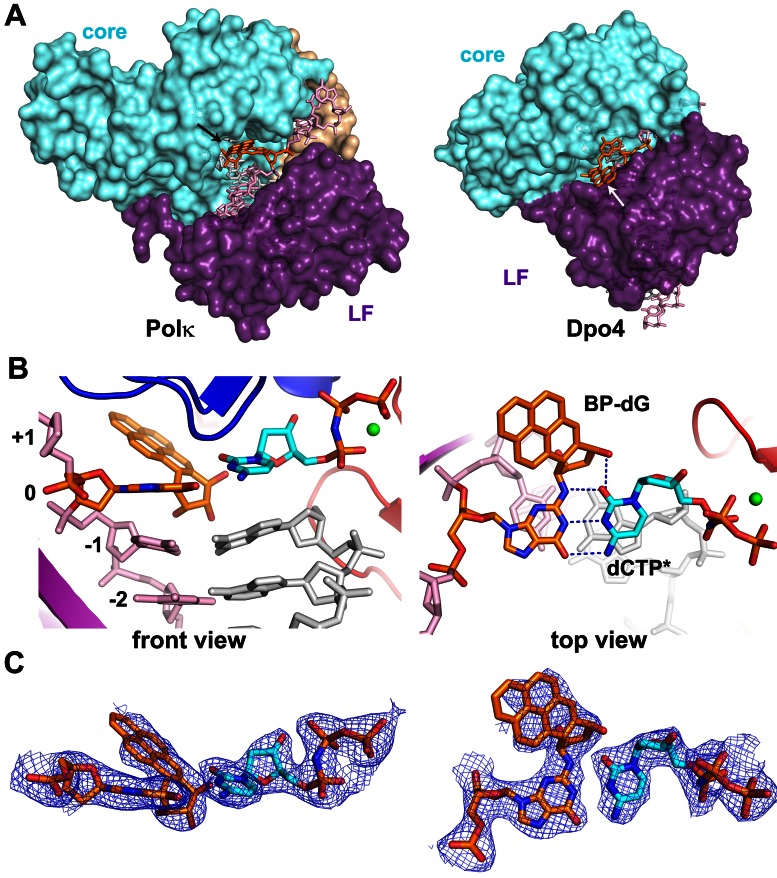
The active site of polκ is more open at the minor groove side of DNA than those in Dpo4 and other human Y-family polymerases (Figure 3A; Supplementary Figure S2). The BP–dG adduct is in a normal anti-conformation and pairs with C in a Watson–Crick base pair to allow correct replication (Figure 3B). The replicating base pair is well-defined by electron density, with an additional H-bond between the C base and O1 from BPDE (Figure 3B–C). The bulky ring is relatively mobile in the edge exposed to solvent, which is common for bulky DNA adducts which are usually not completely buried in the active sites of Y-family polymerases (30,44–46). However, the majority of the BPDE ring is covered by the electron density and well-defined for the ring orientation (Figure 3C). The 5′ orientation is well accommodated in polκ and is ready for translocation through the active site without any steric conflicts with polκ. This is in contrast to the blocking structure in the high fidelity BF polymerase (BF pol) containing the same bulky lesion (47). In the BF structure, the adducted DNA in the minor groove is severely distorted by the tight active site of BF pol, which prevents binding of incoming dNTP and disrupts the interactions between BP DNA and the polymerase for catalysis. Furthermore, in our polκ–BPG structure the incoming nucleotide is not only accommodated well in the active site, but also the 3′OH of the primer strand and Pα of the dCTP* is less than 5 Å apart. This distance is within the range of minor structural adjustments to produce a catalytically productive intermediate for catalysis. In contrast, the Dpo4 structures representing replication blockage by BP bulky lesions, the intercalated BPDE ring separates the incoming dNTPs about 10 Å from the 3′ end of the primer strands (30,44).
Figure 3.
BP–dG and replicating base pair in the active site of polκ. (A) The open active site is shown in the back view of the polκ ternary complex (left) and compared to Dpo4 with a flipped-out DNA helix (right, PDB id: 2IA6). Proteins are in a surface representation, and DNA is shown in sticks. Core domains (finger, palm and thumb) are in cyan, N-clasp in beige and little finger (LF) in purple. The BP–dG adduct is in orange with the BPDE rings indicated by arrows. The two enzyme structures are in the same orientation, based on palm domain superposition. (B) Orthogonal views of the replicating base pair, dCTP* (cyan) represents the non-hydrolysable substrate dCMPNPP, BP-dG:dCTP*, in the active site. (C) 2Fo–Fc map contoured at 1σ level over the replicating base pair in polκ–BPG in the same orientations as in (B).
The 5′ orientated BPDE would clash into most of Y-family polymerases which have narrow or closed minor grooves (Figure 3A, Supplementary Figure S2). The molecular dynamic simulation studies of polκ containing a BP–dG lesion at the template-primer junction suggested that the BP ring needs to point toward the 3′ end to avoid steric collisions and maintain the transition states of the reaction pathway (48,49). Considering this, we tested a model with the BPDE ring pointing in 3′ orientation to exclude any possibility of multiple orientations. After refinement, there is no electron density for the 3′ oriented ring even though the B-factor of BPDE atoms increase significantly compared to the 5′ orientation (Supplementary Table S2). Furthermore, both Rwork and Rfree are higher compared to that of the 5′ oriented ring structure (Supplementary Table S2). Our structure indicates that pol κ has a unique active site that is opened at the minor groove side to accommodate the 5′ orientation BP–dG adduct in an active conformation. The 5′ orientation is consistent with the observation in the NMR structures of DNA containing the BP–dG lesion (43,50).
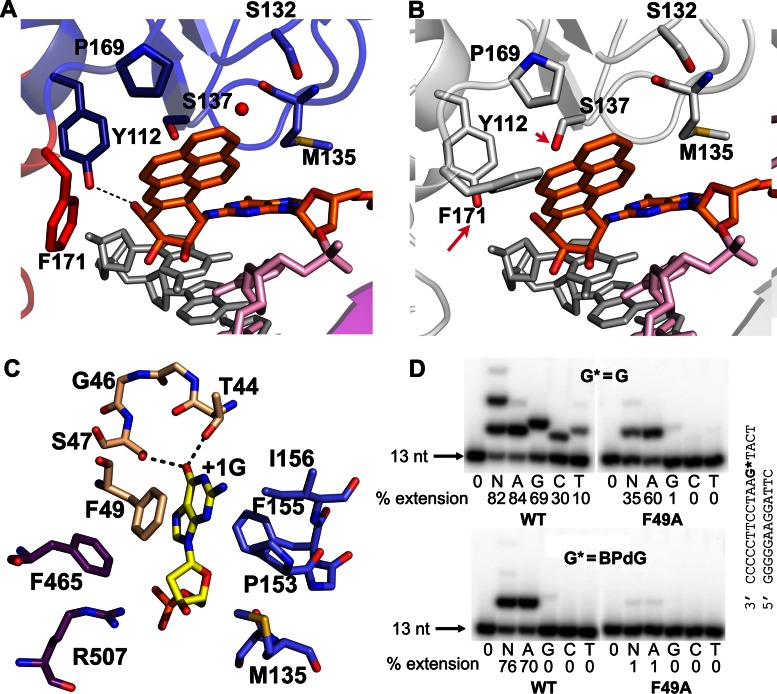
The minor groove attached BPDE ring points toward the 5′-end of the template, being packed against the finger/palm domains in one face and semi-protected by the minor groove backbone in the other face (Figures 2B, E and 3A–B). Thus, the opening of polκ to the minor groove side of DNA not only accommodates the bulky BPDE ring, but also provides solvent shielding for the hydrophobic moiety of the ring. The side chains of Tyr112, Ser137 and Pro 169 from the finger domain and the Phe171 from the palm domain form either H-bonds or van der Waals contacts with the bulky ring (Figure 4A). The bulky BPDE ring restricts Phe171 so that this amino acid adopts a specific rotamer, different from those favored in polκ–DNA complexes containing unmodified DNA (35,42). In the latter structures, Phe171 is either flexible in multiple conformations or in a conformation that is in steric conflict with the bulky BPDE ring (Figure 4B; Supplementary Figure S3). The entropic cost of Phe171 in the fixed conformation contributes to an 18-fold decrease in efficiency for dCMP incorporation opposite BP–dG by wild-type (WT) polκ relative to its F171A mutant (51). The mutational data in Liu et al.'s work also showed that the polκ mutant which contains F171W substitution has significant effects on BP–dG bypass (39), supporting the critical role of F171 in BP–dG replication.
Figure 4.
Interactions of polκ with DNA adduct. (A) The BPDE ring is stabilized by polκ residues and the backbone of the template. The red sphere is a water molecule. The template strand is in pink, the primer strand is in grey, and BP–dG is in orange. Residues are in the colors of the domains to which they belong and follow the same color scheme as in Figure 2A. (B) The BP–dG DNA from polκ–BPG superposed onto the position of the normal DNA substrates at the active site of polκ–G. F171 and S137 are different rotamers from polκ–BPG in (A). F171 would clash with the BPDE ring, and S137 would not be in contact with the ring. (C) Interactions between the +1 template G base (yellow) with polκ. All the residues are colored as in Figure 2A. (D) Incorporation of nucleotides past normal G and BP–dG by WT and mutant F49A polκ at the extension stage. Reaction conditions were the same as in Figure 1B. The substrate sequences are shown to the right of gel images.
In order to confirm the contribution of polκ residues that are in contact with the BPDE ring during bypass of BP–dG, we made three single mutants (P169M, Y112A, Y112F) for DNA replication analysis. P169M and Y112A mutants reduce replication activity on the BP–dG template by ∼50% and ∼70%, respectively, while Y112F does not show any effects (Figure 1B). Obviously, the Y112F mutant has a comparable side chain size as in the (WT) polκ, so replication proceeds. However, the Y112A mutant affects replication because of the absence of the aromatic side chain; in the WT polκ, this side chain contributes to BPDE binding (Figure 4A). We modeled the amino-acid substitution of P169M in the binding site of polκ and found that all three possible rotamers of Met169 would clash with the bulky ring of BP–dG in the 5′-end orientation (Supplementary Figure S4). The mutagenesis data indicates that appropriate van der Waals contacts with the BPDE ring are important for bypassing BP–dG. The mutations have little effect on the BP–dG template in the extension stage (Supplementary Figure S5) as these residues only interact with the bulky BPDE ring at the active site during insertion. Our mutagenesis data support the structural observations that the 5′ oriented bulky ring is located at the minor groove side of DNA and is stabilized by multiple residues at the insertion stage.
Noticeably, the bulky BPDE ring causes minor local disturbances in the DNA structure centered around the adduct site. The hydroxyl groups of BPDE push the incoming nucleotide and the underneath (-1) base pair off their regular positions in polκ–BPG resulting in a slightly longer base pair rise of ∼4.6 Å between the -1 base pair and the replicating base pair at the 0 position (Figure 3B and Supplementary Table S3). Furthermore, the incoming C base shows an 18° propeller twist with respect to the template BP–dG base plane, and the -1 base pair is buckled 29° (Figure 3B and Supplementary Table S3). The bulky lesion induces a local structural disturbance and reduces the stability of the substrates at the active site, which explains the blocking effects of BP–dG in our replication assays (Figure 1) and previous reports by others (10,22).
Unique domain positions create an open DNA binding cleft in polκ
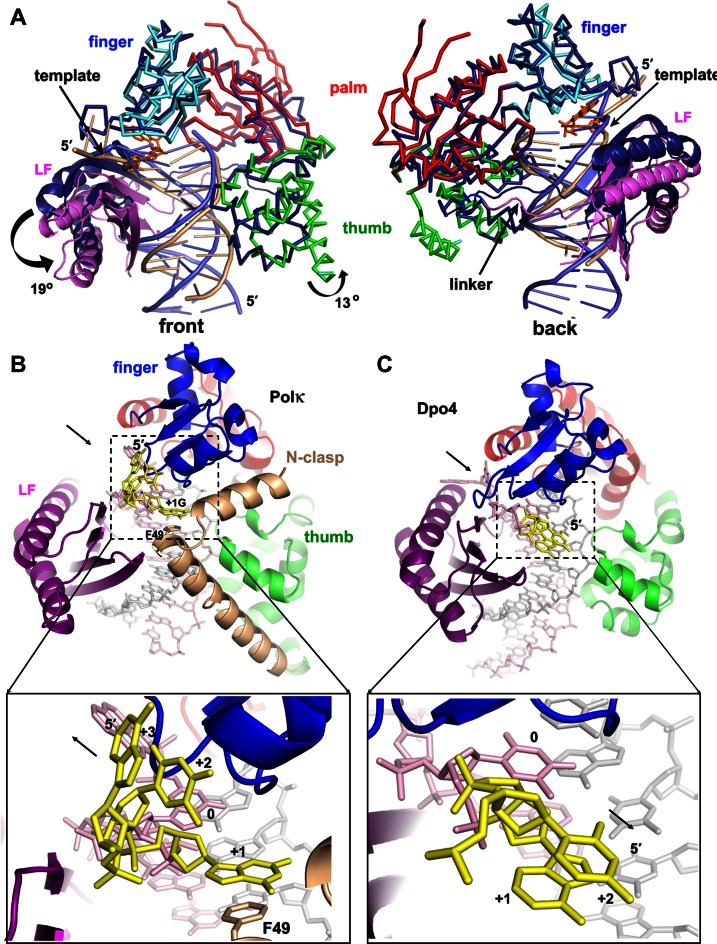
The overall open conformation of polκ is essential for bypassing of BP–dG. The spacious DNA binding cleft in polκ results from a unique domain arrangement. The finger, palm and thumb domains of Y-family polymerases form a relatively compact structural unit called the catalytic core (cyan in Figure 3A). The fourth common domain, little finger (purple in Figure 3A), is relatively flexible in its position relative to the core (52). Like all Y-family polymerases, polκ has an active site that exposes the DNA major groove to the solvent (Figure 2A). However, polκ's active site is not enclosed at the minor groove side of DNA, unlike in other Y-family polymerases (Figures 2B and 3A; Supplementary Figure S2). The opening of polκ to the minor groove side is due to the unique orientation of LF in polκ, which is different from that in the Dpo4, polη and polι ternary complexes (Figure 5A and Supplementary Figure S6). LF domains in Dpo4, polη and polι associate with the core by interacting with the finger domain (Figure 5C and Supplementary Figure S6A). In contrast, the LF domain in polκ is dissociated from its finger domain (Figure 5A) and rotates 19° away from the core domain, when compared to that of Dpo4, which allows an opening up of the minor groove side of DNA. This rotation squeezes LF into the major groove side and moves the thumb domain 13° in the same direction, which makes the DNA binding cleft fit the substrates (Figure 5A). The collective movements of the polκ LF and thumb domains shift the template DNA (beige), particularly the template strand at the active site, ∼4 Å from the usual DNA position (blue) in Y-family polymerases (Figure 5A). This DNA movement creates more room to accommodate the bulky adduct between the template strand and the core domain at the minor groove side of DNA. The linker (pink) connecting the LF and core also moves down substantially due to the 19° rotation of LF and further opens up polκ at the minor groove side (Figure 5A). Thus, unique domain positions shape the binding cleft of polκ so that it differs from other Y-family DNA polymerases, making the minor groove side to be wide enough to accommodate bulky BP–dG, but without disturbing the B-form DNA helix.
Figure 5.
Domain/DNA movements and the role of N-clasp: comparison of polκ with Dpo4. (A) Superposition of polκ and Dpo4 (1JX4). The proteins are mainly in Cα traces. Dpo4 is in dark blue, with DNA in light blue, and polκ is in the same domain colors as in Figure 2A, with DNA in beige. LF domains are highlighted in ribbons. The N-clasp domain was removed for clarity. The rotation direction and angles are in the front view (left panel). Arrows in the back view (right panel) indicate the shifting site of the template strand near the active site and the movement of the linker of polκ relative to Dpo4. (B) N-clasp is holding the finger/LF/thumb domains of polκ in position as a scaffold and adding additional contacts to the single-stranded template (yellow) to form a 3-way stabilization. The arrow in the upper panel indicates the dissociation of LF from the core; the lower panel's arrow indicates that the ss template strand extends from the active site for continuous replication. (C) Dpo4 structure (1JX4) in the same view. The N-clasp domain support for the ss template strand is absent in Dpo4. The black arrow in the upper panel indicates the association of LF with the core; the lower panel's arrow indicates the ss template strand turns back toward the major groove of the DNA helix.
N-clasp scaffold holds finger/LF/thumb domains in unique positions
The N-clasp plays a critical role in maintaining a unique conformation of polκ and stabilizing the ssDNA template. The N-clasp domain interacts with all three important DNA binding domains (finger/thumb/LF) and holds them together by its two α-helices arranged in an L-shape (Figure 2A). In contrast to the other Y-family polymerases, the LF domain of polκ lacks the contacts with the finger domain to associate with the polymerase core (Figures 2A and 5B). Because of the lack of direct support from the finger domain (Figure 5B), the polκ LF domain must associate with the rest of the protein through the N-clasp, which links the LF, finger and thumb domains across the major groove side of DNA (Figures 2A and 5B). The N-clasp domain holds the LF, finger and thumb domains in position for proper DNA binding. This structural observation is consistent with the indispensable role of the N-clasp observed in polκ's replication of BP–dG (39).
Furthermore, the N-clasp stabilizes the ss template (downstream of the template base), in addition to the finger and LF domains, thus forming a three-way ssDNA stabilization (Figure 5B). The 5′-ss template extends from the replicating base pair to allow continuous replication and does not fold back to protect the bulky BPDE ring at the template–primer junction, as observed in NMR structures (Figure 2D) (43). The extended conformation supported by the N-clasp domain prevents strand slippage and the ‘U-turn’ template conformation observed in Dpo4 and polι (30,45). In particular, the side chain of Phe49 from the N-clasp stacks with the +1 G base 5′ to the template BP–dG and stabilizes the ssDNA template (Figures 4C and 5B). The F49A mutation almost abolishes polκ replication activity on BP–dG DNA at both insertion (Figure 1B) as well as extension (Figure 4D) stages, but only partially reduces the polymerase activity on normal templates. The mutagenesis data indicate that the stacking from Phe49 to the +1 base is critical for template stability and is particularly essential for bypass of bulky lesion.
Interestingly, polκ (WT) shows a higher degree of fidelity when it incorporates the next nucleotide after the BP–dG lesion than a non-modified G (Figure 4D). Current structure work at the insertion stage could not provide insights into better fidelity. Further structural analysis on extension complexes will help to shed light on how polκ extends primer strand after the bulky adduct more accurately than normal DNA.
CONCLUSIONS
Human polκ is specialized to replicate the major bulky BP–dG adduct and other N2-adducted deoxyguanosines. The crystal structure of the polκ–BPG ternary complex captures DNA replication of the bulky BP adduct in the insertion stage. The unique domain arrangement of polκ opens up the active site at the minor groove side of DNA to accommodate the minor groove attached adduct. At the same time, residues along the minor groove side stabilize the hydrophobic BPDE ring and allows adducted DNA to maintain a standard B-form with Watson–Crick base pairing for accurate replication. The unique N-clasp domain supports an open conformation of polκ and stabilizes the ss template for the efficient bypass of BP–dG. This structural work reveals a new structural mechanism for bypassing bulky BP lesions and is unique when compared to that of other Y-family polymerases. This structural work answers why polκ replicates BP–dG accurately. The novel molecular model of error-free replication explains the major role of polκ in response to cytotoxicity of BP in cells (26,27,29,39).
ACCESSION NUMBERS
Coordinates and structure factors for polκ-BPG (4U7C) and polκ-G (4U6P) have been deposited in the Protein Data Bank.
Supplementary Material
Acknowledgments
We thank Dr Jane M. Sayer for providing trans-N2-dG-B[a]PDE-2 phosphoramidite for BP-dG DNA synthesis and the staff at the MX beamlines in the Canadian Light Source (CLS) for data collection.
FUNDING
Canadian Institutes of Health Research [Operating grant MOP-67128 to H.L.]; Cancer Research Society [Operating grant to H.L.]. Funding for open access charge: Institutional funding, Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E.C., Wagner R., Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 3.Goodman M.F. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 4.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 6.Johnson R.E., Prakash S., Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 7.Johnson R.E., Washington M.T., Prakash S., Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 8.Kirouac K.N., Ling H. Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase iota. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3210–3215. doi: 10.1073/pnas.1013909108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song I., Kim E.J., Kim I.H., Park E.M., Lee K.E., Shin J.H., Guengerich F.P., Choi J.Y. Biochemical characterization of eight genetic variants of human DNA polymerase kappa involved in error-free bypass across bulky N-2-Guanyl DNA adducts. Chem. Res. Toxicol. 2014;27:919–930. doi: 10.1021/tx500072m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Wu X., Guo D., Rechkoblit O., Wang Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Rep. 2002;1:559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 11.Choi J.Y., Angel K.C., Guengerich F.P. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J. Biol. Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 12.Phillips D.H. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 13.Phillips D.H. Polycyclic aromatic hydrocarbons in the diet. Mutat. Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 14.Denissenko M.F., Pao A., Tang M., Pfeifer G.P. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 15.Pfeifer G.P., Denissenko M.F., Olivier M., Tretyakova N., Hecht S.S., Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 16.Cheng S.C., Hilton B.D., Roman J.M., Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem. Res. Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 17.Peltonen K., Dipple A. Polycyclic aromatic hydrocarbons: chemistry of DNA adduct formation. J. Occup. Environ. Med. 1995;37:52–58. doi: 10.1097/00043764-199501000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Newbold R.F., Brookes P. Exceptional mutagenicity of a benzo(a)pyrene diol epoxide in cultured mammalian cells. Nature. 1976;261:52–54. doi: 10.1038/261052a0. [DOI] [PubMed] [Google Scholar]
- 19.Jerina D.M., Chadha A., Cheh A.M., Schurdak M.E., Wood A.W., Sayer J.M. Covalent bonding of bay-region diol epoxides to nucleic acids. Adv. Exp. Med. Biol. 1991;283:533–553. doi: 10.1007/978-1-4684-5877-0_70. [DOI] [PubMed] [Google Scholar]
- 20.Shibutani S., Margulis L.A., Geacintov N.E., Grollman A.P. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-BPDE (7, 8-dihydroxy-anti-9, 10-epoxy-7, 8, 9, 10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 21.Alekseyev Y.O., Romano L.J. In vitro replication of primer-templates containing benzo[a]pyrene adducts by exonuclease-deficient Escherichia coli DNA polymerase I (Klenow fragment): effect of sequence context on lesion bypass. Biochemistry. 2000;39:10431–10438. doi: 10.1021/bi000552u. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Yuan F., Wu X., Wang M., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki N., Ohashi E., Kolbanovskiy A., Geacintov N.E., Grollman A.P., Ohmori H., Shibutani S. Translesion synthesis by human DNA polymerase kappa on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-N(2)-BPDE (7, 8-dihydroxy-anti-9, 10-epoxy-7, 8, 9, 10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 24.Huang X., Kolbanovskiy A., Wu X., Zhang Y., Wang Z., Zhuang P., Amin S., Geacintov N.E. Effects of base sequence context on translesion synthesis past a bulky (+)-trans-anti-B[a]P-N2-dG lesion catalyzed by the Y-family polymerase pol kappa. Biochemistry. 2003;42:2456–2466. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- 25.Rechkoblit O., Zhang Y., Guo D., Wang Z., Amin S., Krzeminsky J., Louneva N., Geacintov N.E. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 26.Ogi T., Shinkai Y., Tanaka K., Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avkin S., Goldsmith M., Velasco-Miguel S., Geacintov N., Friedberg E.C., Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 28.Shachar S., Ziv O., Avkin S., Adar S., Wittschieben J., Reissner T., Chaney S., Friedberg E.C., Wang Z., Carell T., et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi X., Slater D.M., Ohmori H., Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 30.Bauer J., Xing G., Yagi H., Sayer J.M., Jerina D.M., Ling H. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheffield P., Garrard S., Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 32.Shanagar J. Purification of a synthetic oligonucleotide by anion exchange chromatography: method optimisation and scale-up. J. Biochem. Biophys. Methods. 2005;64:216–225. doi: 10.1016/j.jbbm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lone S., Townson S.A., Uljon S.N., Johnson R.E., Brahma A., Nair D.T., Prakash S., Prakash L., Aggarwal A.K. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.DeLano W.L. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 39.Liu Y., Yang Y., Tang T.S., Zhang H., Wang Z., Friedberg E., Yang W., Guo C. Variants of mouse DNA polymerase kappa reveal a mechanism of efficient and accurate translesion synthesis past a benzo[a]pyrene dG adduct. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1789–1794. doi: 10.1073/pnas.1324168111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank E.G., Sayer J.M., Kroth H., Ohashi E., Ohmori H., Jerina D.M., Woodgate R. Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase iota. Nucleic Acids Res. 2002;30:5284–5292. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Wu X., Guo D., Rechkoblit O., Geacintov N.E., Wang Z. Two-step error-prone bypass of the (+)- and (-)-trans-anti-BPDE-N2-dG adducts by human DNA polymerases η and κ. Mutat. Res. 2002;510:23–35. doi: 10.1016/s0027-5107(02)00249-x. [DOI] [PubMed] [Google Scholar]
- 42.Vasquez-Del Carpio R., Silverstein T.D., Lone S., Swan M.K., Choudhury J.R., Johnson R.E., Prakash S., Prakash L., Aggarwal A.K. Structure of human DNA polymerase kappa inserting dATP opposite an 8-OxoG DNA lesion. PLoS One. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng B., Gorin A., Hingerty B.E., Geacintov N.E., Broyde S., Patel D.J. Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction. Biochemistry. 1997;36:13769–13779. doi: 10.1021/bi970069s. [DOI] [PubMed] [Google Scholar]
- 44.Ling H., Sayer J.M., Plosky B.S., Yagi H., Boudsocq F., Woodgate R., Jerina D.M., Yang W. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirouac K.N., Ling H. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase iota. EMBO J. 2009;28:1644–1654. doi: 10.1038/emboj.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirouac K.N., Basu A.K., Ling H. Replication of a carcinogenic nitropyrene DNA lesion by human Y-family DNA polymerase. Nucleic Acids Res. 2013;41:2060–2071. doi: 10.1093/nar/gks1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu G.W., Huang X., Luneva N.P., Geacintov N.E., Beese L.S. Structure of a high fidelity DNA Polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 48.Jia L., Geacintov N.E., Broyde S. The N-clasp of human DNA polymerase kappa promotes blockage or error-free bypass of adenine- or guanine-benzo[a] pyrenyl lesions. Nucleic Acids Res. 2008;36:6571–6584. doi: 10.1093/nar/gkn719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lior-Hoffmann L., Wang L.H., Wang S.L., Geacintov N.E., Broyde S., Zhang Y.K. Preferred WMSA catalytic mechanism of the nucleotidyl transfer reaction in human DNA polymerase kappa elucidates error-free bypass of a bulky DNA lesion. Nucleic Acids Res. 2012;40:9193–9205. doi: 10.1093/nar/gks653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosman M., de los Santos C., Fiala R., Hingerty B.E., Singh S.B., Ibanez V., Margulis L.A., Live D., Geacintov N.E., Broyde S., et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sassa A., Niimi N., Fujimoto H., Katafuchi A., Gruz P., Yasui M., Gupta R.C., Johnson F., Ohta T., Nohmi T. Phenylalanine 171 is a molecular brake for translesion synthesis across benzo[a]pyrene-guanine adducts by human DNA polymerase kappa. Mutat. Res. 2010;718:10–17. doi: 10.1016/j.mrgentox.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Wong J.H., Fiala K.A., Suo Z., Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J. Mol. Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.