Abstract
Cisplatin is a widely prescribed anticancer drug, which triggers cell death by covalent binding to a broad range of biological molecules. Among cisplatin targets, cellular RNAs remain the most poorly characterized molecules. Although cisplatin was shown to inactivate essential RNAs, including ribosomal, spliceosomal and telomeric RNAs, cisplatin binding sites in most RNA molecules are unknown, and therefore it remains challenging to study how modifications of RNA by cisplatin contributes to its toxicity. Here we report a 2.6Å-resolution X-ray structure of cisplatin-modified 70S ribosome, which describes cisplatin binding to the ribosome and provides the first nearly atomic model of cisplatin–RNA complex. We observe nine cisplatin molecules bound to the ribosome and reveal consensus structural features of the cisplatin-binding sites. Two of the cisplatin molecules modify conserved functional centers of the ribosome—the mRNA-channel and the GTPase center. In the mRNA-channel, cisplatin intercalates between the ribosome and the messenger RNA, suggesting that the observed inhibition of protein synthesis by cisplatin is caused by impaired mRNA-translocation. Our structure provides an insight into RNA targeting and inhibition by cisplatin, which can help predict cisplatin-binding sites in other cellular RNAs and design studies to elucidate a link between RNA modifications by cisplatin and cisplatin toxicity.
INTRODUCTION
Cisplatin was discovered in 1965 as a toxic byproduct of electrolysis from platinum electrodes, which suppressed growth of Escherichia coli cells (1). Soon thereafter cisplatin was found to exhibit antitumor activity (2), and in 1978 it was approved by the Food and Drug Administration as an anticancer drug (2,3). Since then, cisplatin has become one of the most widely prescribed chemotherapeutic agents used to treat diverse types of cancer such as ovary, lung, head-and-neck, germ-cell and bladder cancer (4–6).
In eukaryotic cells, cisplatin causes cell-cycle arrest and induces apoptosis (7–10). For decades, the underlying mechanism has been primarily attributed to cisplatin cross-linking to purine nucleotides in chromosomal DNA, which was shown to prevent the cellular replication machinery to traverse through cisplatin–DNA adducts leading to apoptotic cell death (7). This model, however, has been challenged by numerous studies demonstrating that cisplatin toxicity can originate rather from multiple sources, including modifications of proteins, small peptides, lipids and RNA, than solely from modifications of chromosomal DNA (7–11). In particular, cisplatin was shown to inactivate a number of essential RNA molecules, including RNA components of translation, splicing machineries and telomeric RNA, indicating that RNA molecules may represent an underexplored class of pharmacologically relevant cisplatin targets (12–17).
Despite the growing interest, experimental studies focused at elucidation of a link between RNA modifications by cisplatin and its toxicity remain challenging. To this end, RNA modifications by cisplatin have been studied using ribosomal, transfer, messenger, spliceosomal, telomeric and other RNAs primarily by biochemical techniques, which were aimed to identify cisplatin binding sites and describe the effect of cisplatin binding on RNA activity (12–17). The only available crystal structures of RNA modified by cisplatin-derived Pt species are for tRNAPhe and were determined at a relatively low resolution (18,19). These studies roughly located cisplatin-binding sites in several RNA molecules, but, none of them provided an accurate model of cisplatin–RNA interactions. Therefore, the underlying mechanism of cisplatin recognition and inactivation of RNA molecules has remained elusive.
Here we report 2.6Å-resolution crystal structure of Thermus thermophilus 70S ribosome modified by cisplatin-derived Pt species. This is the first nearly atomic resolution structure of Pt-RNA adducts following cisplatin treatment of the ribosome. We observe nine cisplatin-derived molecules bound to the ribosome and show that the cisplatin-modified sites share common structural features, which appear to determine specific cisplatin binding to a very limited number of sites in the rRNA molecules. Consistent with previous biochemical studies, cisplatin targets two universally conserved centers of the ribosome—the mRNA channel and the GTPase activating center. Strikingly, in the mRNA channel, cisplatin forms simultaneous contacts with the ribosome and the mRNA—a feature that has been recently observed for the ribosome-targeting antibiotic amicoumacin A, which inhibits protein synthesis by tethering mRNA to the ribosome (20). In summary, our study identifies cisplatin-binding RNA motifs, provides a rationale for the cisplatin-induced inhibition of protein synthesis and reveals an unexpected similarity between the anticancer drug cisplatin and the ribosome-targeting antibiotics.
MATERIALS AND METHODS
Purification of Thermus thermophilus ribosomes and preparation of tRNAs
70S ribosomes from T. thermophilus and deacylated tRNAiMet and tRNAPhe from E. coli were purified as previously described (21). Synthetic mRNA with the sequence 5′-GGC AAG GAG GUA AAA AUG UUC UAA-3′ was obtained from Integrated DNA Technologies (Coralville, IA, USA).
Crystallization and data collection
The ribosome complex with mRNA and tRNAs was formed by programming 5 μM 70S ribosomes with 10 μM mRNA and 20 μM P- and A-site tRNA substrates, in the buffer containing 5 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM NH4Cl, and 10 mM Mg(CH3COO)2, and 6 mM β-mercaptoethanol. Next, the cisplatin (dissolved in dimethylformamide) was added to a final concentration of 500 μM and the resulting mixture was left at room temperature for additional 10 min. Next, crystallization was initiated by mixing 3 μl of the ribosome complex solution with 4 μl of the buffer containing 100 mM Tris–HCl (pH 7.6), 2.9% (w/v) PEG-20K, 7–12% (v/v) MPD, 100–200 mM arginine and 0.5 mM β-mercaptoethanol. Crystals were grown in 10 days by the vapor diffusion method in sitting drops at 19°C and were stabilized as described previously (21). In the soaking experiment, cisplatin was not included during complex formation and crystallization. Instead, it was included into the final crystal-stabilization solution at 100 μM concentration. This solution, containing ribosome crystals and cisplatin, was left at room temperature for 12 h and then the crystals were frozen and used for data collection. Diffraction data were collected using beamline 24ID-C at the Advanced Photon Source (Argonne, IL, USA). A complete dataset for each ribosome complex was collected using 0.979Å wavelength at 100K from multiple regions of the same crystal using 0.3° oscillations.
Crystallographic data processing and model building
The raw data were integrated and processed using the XDS software package (22). All crystals belonged to the primitive orthorhombic space group P212121 with approximate unit cell dimensions of 210Å × 450Å × 620Å and contained two copies of the 70S ribosome per asymmetric unit. The initial molecular replacement solutions were obtained by rigid body refinement, followed by positional and individual B-factor refinement. The initial search model was generated from recently published high-resolution structure of Tth 70S ribosome with bound mRNA, tRNAs and containing all post-transcriptional modifications of the rRNAs (PDB ID: 4Y4P) (23). Cisplatin modification sites in the ribosome were identified using the unbiased electron density maps (Figure 1) together with the anomalous difference Fourier maps (Supplementary Figure S1). The final model of the cisplatin-modified 70S ribosome was generated by multiple rounds of model building in COOT (24), followed by refinement in PHENIX (25). For several cisplatin modification sites, the improved electron density maps revealed conformation of individual chemical groups of the cisplatin moiety, which enabled nearly atomic description of the cisplatin–rRNA interactions. The statistics of data collection and refinement are compiled in Supplementary Table S1. All rRNA nucleotides were renumbered according to E. coli numbering system.
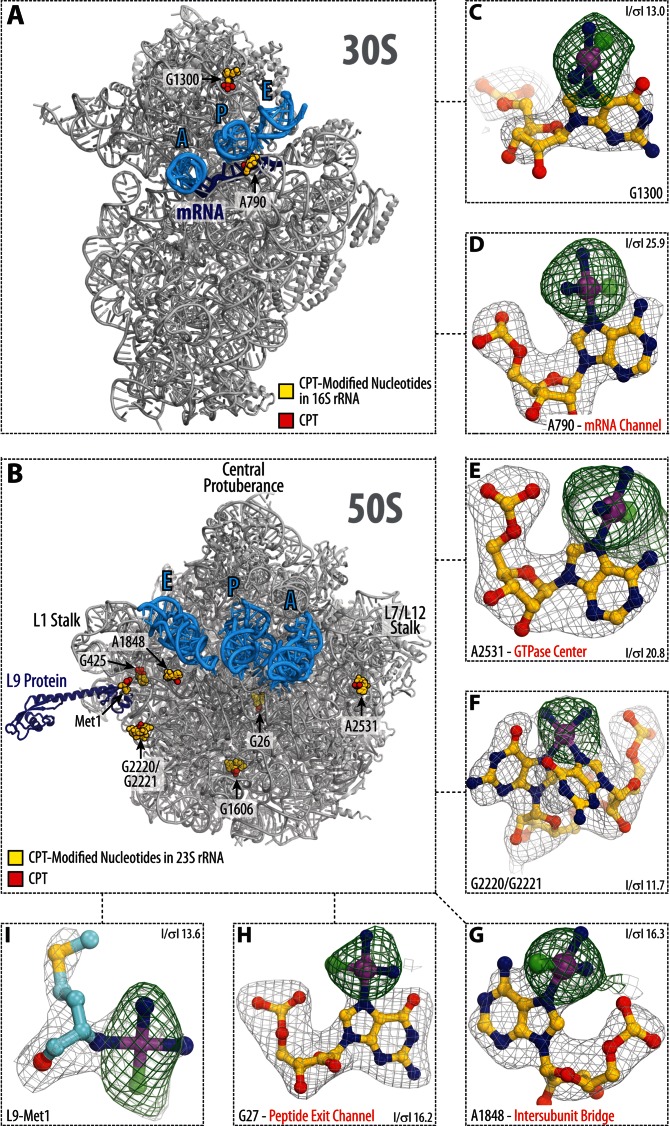
Figure 1.
Cisplatin-binding sites in the ribosome. (A and B) Overview of cisplatin-modified residues (yellow spheres) in the large (A) and small (B) subunits of the Thermus thermophilus 70S ribosome. The actual cisplatin-derived Pt species are shown as red spheres. Panels (C–I) show close-up views of the cisplatin modification sites. The refined models of cisplatin-modified nucleotides are displayed in their respective unbiased electron density maps. Gray mesh shows the Fo–Fc map after refinement with the entire cisplatin-modified nucleotides omitted (contoured at ∼3.0σ). Green mesh shows the Fo–Fc electron density map after refinement with unmodified nucleotides (contoured at ∼3.0σ). Carbon atoms are colored yellow, nitrogens—blue, oxygens—red, platinums—magenta, chlorines—green.
RESULTS
Cisplatin targets only a small fraction of rRNA bases
To gain insights into the cisplatin binding to the ribosome, we co-crystallized T. thermophilus 70S ribosomes with the mRNA, tRNAs and cisplatin, and determined a 2.6Å resolution X-ray structure of cisplatin-modified ribosome (Supplementary Table S1). The ribosome-bound cisplatin-derived Pt species appeared as prominent peaks in both the unbiased (Fo–Fc) and in the anomalous difference electron density maps (Figure 1, Supplementary Figure S1 and Table S2).
Strikingly, the maps revealed only nine cisplatin-derived Pt species in the entire ribosome, despite the fact that the ribosome harbors nearly 2200 potential modification sites (Supplementary Figure S2). Three of the observed moieties are coordinated by the N7-atoms of adenine bases: A790 of the 16S rRNA (E. coli numbering is used throughout the text; Figure 1A and D; Supplementary Figure S1), and A1848 and A2531 of the 23S rRNA (Figure 1B, E and F; Supplementary Figure S1). Five moieties are coordinated by the N7-atoms of guanine bases at positions G1300 of the 16S rRNA and G27, G425, A1606, G2220, G2221 of the 23S rRNA (Figure 1, Supplementary Figure S1). One cisplatin moiety is coordinated by the N-terminus of the ribosomal protein L9 (Figure 1I). A small total number of the cisplatin binding sites in the ribosome points to the high specificity of RNA recognition by cisplatin.
Cisplatin-binding sites share common structural features
Having determined several cisplatin modification sites in the rRNAs, we attempted to elucidate what underlies specific binding of cisplatin to these particular sites. First, we assessed whether reactivity of N7-atoms in purine bases is determined by their spatial accessibility to the cisplatin molecule. For this purpose, we used the structure of a non-modified 70S ribosome from T. thermophilus and measured accessibility of all N7-atoms to a 2.2Å-radius sphere, which corresponds to the size of the cisplatin molecule (see ‘Materials and Methods’ section). We found that the 70S ribosome has more than 2200 N7-atoms with a comparable or higher solvent exposure than those of the nine actual modification sites (Supplementary Figure S2). This result indicates that selective modification of certain RNA bases by cisplatin is not defined solely by the spatial accessibility of their N7-atoms.
Previously, cisplatin was shown to target predominantly non-Watson–Crick RNA segment (26,27), and our analysis confirms this finding for modification of rRNAs (Supplementary Figure S3). Among cisplatin-modified rRNA nucleotides, only one (G425 of the 23S rRNA) forms a canonical G-C pair and is located within a regular A-form helix, whereas all others reside within bulges (A1878 in H68 of 23S rRNA), helical junctions (G1300 in h41/h42 and G1606 in H49/H51), internal loop (G27 in H2), hairpin loops (A790 in h24 and A2531 in H91) or an A-form helix distorted by a G-U pair (G2220/G2221 in H79) (Supplementary Figure S3). These and previous data suggest that irregular RNA geometry leads to structural folds that favor cisplatin binding, however identity of such folds was unclear.
To address this question, we analyzed the immediate surroundings of the cisplatin moieties within the ribosome. One common property, which we observed in all of the cisplatin-binding sites, is that cisplatin interactions with the ribosome are never limited to N7-atoms of purine bases, but involve additional contacts with adjacent rRNA nucleotides (Supplementary Table S2). We also found that these contacts are strikingly different for cisplatin moieties coordinated by guanines versus adenines.
Cisplatin modifications of guanine bases
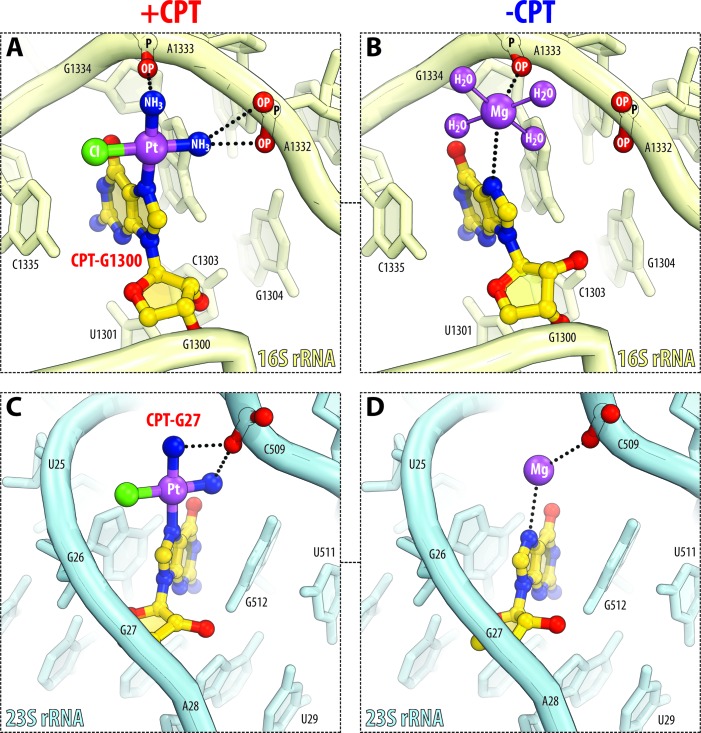
Guanines coordinate cisplatin moieties in cooperation with surrounding rRNA backbone phosphates (Figure 2A and C; Supplementary Table S2). For instance, the cisplatin moiety bound to the N7-atom of G1300 in 16S rRNA forms additional contacts with the adjacent phosphates of A1332 and A1333 via its amino groups (Figure 2A). Similar cisplatin coordination is observed for other guanine bases in the ribosome (Figure 2C and Supplementary Table S2). Importantly, the close positioning of phosphates to N7-atoms of guanine bases results from highly distorted RNA folds, rationalizing why efficient cisplatin coordination requires non-Watson–Crick geometry of RNA. We made one curious observation about modification of guanine bases by comparing the ribosome structures before and after treatment with cisplatin. If the structures of cisplatin-modified and non-modified ribosomes are superimposed, the cisplatin derivatives overlap with the hydrated Mg2+ ions (Figure 2B and D). Furthermore, these Mg2+ ions in the non-modified ribosome directly interact with the guanine N7-atoms and form similar contacts with rRNA as cisplatin moieties in the cisplatin-modified ribosome (Figure 2B and D).
Figure 2.
Cisplatin coordination by guanine bases resembles coordination of magnesium ions. (A and C) Side-by-side comparison of cisplatin-modified guanine residues in 16S rRNA (light yellow) and 23S rRNA (light blue). Panels (B and D) show the same sites in the untreated 70S ribosome (PDB ID: 4Y4P (23)). Cisplatin-modified nucleotides are highlighted in yellow. The color coding for atoms is the same as in Figure 1. Note that cisplatin binding is observed in idiosyncratic RNA folds, in which N7-atoms of purine residues normally coordinate magnesium ions (in the untreated ribosome) and are located adjacent to the RNA phosphates capable of stabilizing the cisplatin moiety by electrostatic interactions.
It is important to note that in the structure of non-treated ribosome there are about 100 guanine N7-atoms, which coordinate hydrated magnesium ions in their inner spheres, but which are not modified by platinum in the structure of cisplatin-modified ribosome (Supplementary Figure S4). This fact indicates that inner-sphere coordination of hydrated Mg2+ ions is not the only factor that determines high reactivity of guanine bases with cisplatin. However, because Mg2+-coordinating guanines are rare (∼7% of guanines in the rRNA structure) (Supplementary Figure S4A and B), and all five modified guanines do coordinate Mg2+ ions in the structure of the non-treated ribosome, the ability to coordinate Mg2+ ions appears to be an important criterion predicting guanine reactivity with cisplatin. Importantly, this criterion suggests that guanines that do not directly coordinate Mg2+ ions should not efficiently react with cisplatin.
Cisplatin modifications of adenine bases
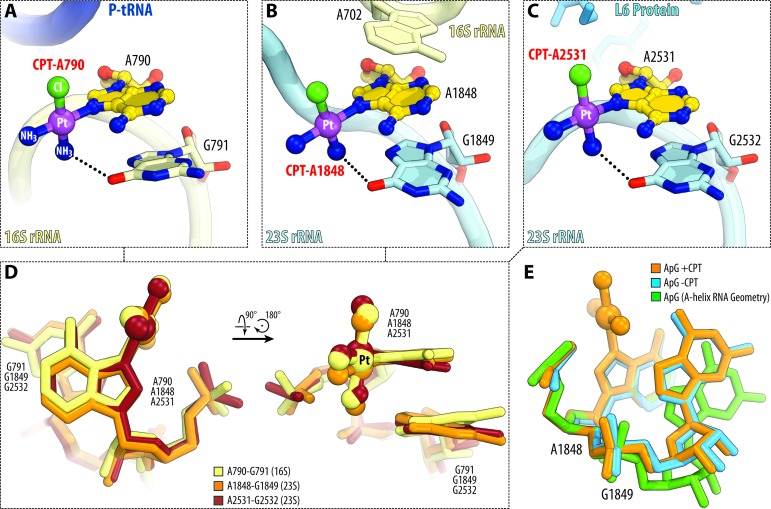
Unlike guanine bases, adenine bases coordinate cisplatin only if positioned immediately before the guanine base, in a characteristic AG-motif (Figure 3A–D). In this AG-motif, the N7-atom of the adenine base is covalently linked to platinum, whereas the 3′-adjacent guanine base provides its Hoogsteen edge (O6-atom) to interact with the cisplatin amine (dashed lines in Figure 3A–C). Notably, all cisplatin-coordinating AG-motifs in the ribosome share nearly identical geometry, which deviates from the ideal A-form RNA geometry by the 15°-turn of the G-base toward the A-base (Figure 3E, compare ‘orange’ and ‘green’ structures). At the same time, there are no apparent differences in the geometry of all AG-motifs between cisplatin-modified and non-modified ribosome (Figure 3E, compare ‘orange’ and ‘blue’ structures) indicating that their distorted geometry is likely the cause, and not the consequence, of cisplatin binding. Another notable similarity between the AG-motifs is their location at the RNA–RNA or RNA–protein interfaces (Figure 3A-C; Supplementary Table S2): nucleotide A790 of the 16S rRNA resides in the mRNA channel at the rRNA/mRNA interface, where the cisplatin-modified nucleotide contacts the mRNA molecule (Figure 3A); A1848 of the 23S rRNA resides at the 23S/16S rRNA interface, where the cisplatin moiety lies in the direct vicinity of the intersubunit bridge B7a (Figure 3B); and A2531 of the 23S rRNA lies at the interface with the ribosomal proteins L6, whose C-terminus is likely to contact the cisplatin moiety (Figure 3C).
Figure 3.
Cisplatin coordination by adenine bases occurs in a characteristic AG-motif. (A–C) Side-by-side comparison of cisplatin-coordinating adenine residues in 16S rRNA (A) and 23S rRNA (B and C). Color coding is the same as in Figure 2. Note that all three cisplatin-modified adenine residues are located immediately upstream of the guanine residues in a characteristic AG-motif. (D) Superposition of the three AG-motifs shown in panels (A–C) reveal their nearly identical geometries. (E) Superposition of A1848-G1849 (23S rRNA) before (blue) and after cisplatin modification (orange) and with the ideal A-form RNA helix (green). Superposition is based on the first adenine nucleotide in each AG-motif. Note that in the untreated 70S ribosome nucleotides A1848-G1849 deviate significantly from the ideal A-form RNA helix and do not change their relative spatial orientation as a result of modification of A1848 by cisplatin.
It is important to note, that in the ribosome structure there are a total of 343 AG-sites, among which about 12 are accessible to a cisplatin-size molecule and whose geometry is close or equivalent to those of the cisplatin-modified AG-sites (Supplementary Figure S4C and D). The fact that only a small subset of these AG-sites is modified by cisplatin indicates that the reactivity of these sites rather depends on their immediate surroundings than solely relying on the particular AG-site structure or accessibility of N7-atoms. However, because adenine modifications by cisplatin are observed exclusively within a rare type of AG-sites, the structure of this AG-site may help predict whether an adenine base is likely to react with cisplatin or not.
Probing of RNA reactivity with cisplatin in a rapid soaking experiment
We were curious to see if some of rRNA bases react with cisplatin more rapidly than the others. For this purpose, we soaked the pre-formed crystals of the T. thermophilus 70S ribosomes, bound with mRNA and A-, P- and E-site tRNAs, in a solution containing 100 μM cisplatin and determined a crystal structure at 2.8Å resolution (see ‘Materials and Methods’ section, Supplementary Table S1). Analysis of the electron density maps revealed that at much shorter incubation time (12 h compared to 10 days) and overall lower cisplatin concentration (100 μM compared to 500 μM) all three adenine nucleotides, which were modified in the co-crystallization experiment, were also modified in the soaking experiment (Supplementary Table S2). Modification of protein L9 by cisplatin was also present. However, no other sites of modification by cisplatin were observed, suggesting that adenine bases in the rRNA react faster with cisplatin than the guanine bases.
Cisplatin targets conserved functional centers of the ribosome
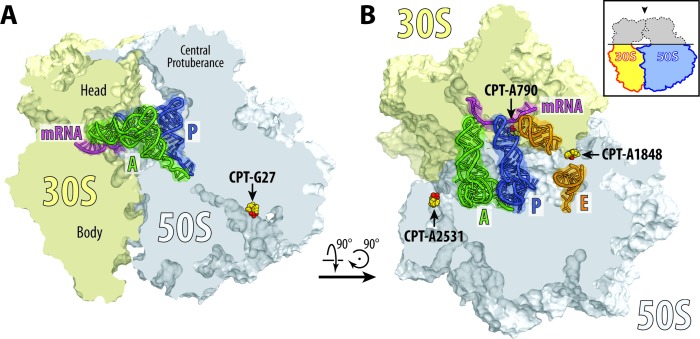
Cisplatin inactivates protein synthesis in organisms ranging from bacteria to mammals (28,29). In the current work, we found that cisplatin targets several functional centers of the ribosome including the mRNA channel, the GTPase activating center, the peptide exit tunnel, and the functionally important bridge B7a between the two ribosomal subunits (Figure 4), suggesting possible mechanisms of cisplatin-mediated inhibition of protein synthesis.
Figure 4.
Cisplatin targets functional centers of the ribosome. (A) Cisplatin modification site in the peptide exit tunnel (G26 of the 23S rRNA). (B) Cisplatin modification sites in the mRNA channel (A790 of the 16S rRNA), at the intersubunit bridge B7a (A1848 of the 23S rRNA) and in the GTPase center (A2531 of the 23S rRNA). 30S subunit is shown in light yellow, 50S subunit is in light blue. mRNA is shown in magenta and tRNAs are displayed in green for the A site, in dark blue for the P site and in orange for the E site. Middle portion of the E-site tRNA is omitted for clarity. Cisplatin-modified nucleotides are shown as yellow sphere with actual cisplatin-derived Pt species colored in red. The view in (A) is a transverse section of 70S ribosome. The view in (B) is from the top after removing the head of the 30S subunit and protuberances of the 50S subunit, as indicated by the inset.
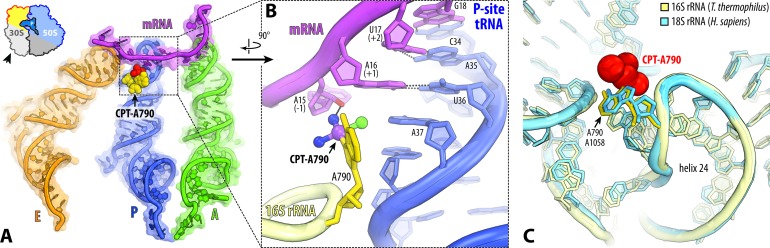
In the small ribosomal subunit cisplatin targets the universally conserved nucleotide A790 of the 16S rRNA in the mRNA channel (Figure 5A). At this location, cisplatin forms an additional contact between the ribosome and the mRNA (Figure 5B), suggesting that cisplatin might stabilize mRNA binding to the ribosomal P site and thereby inhibit protein synthesis by preventing mRNA/tRNA translocation. Notably, the A790-modification site has a universally conserved structure, suggesting that the mRNA channel could be modified by cisplatin in the same way in the ribosomes from humans and other species (Figure 5C).
Figure 5.
Cisplatin targets the mRNA channel in the ribosome. (A) Overview and (B) close-up view of the cisplatin modification site in the mRNA channel (yellow with the cisplatin moiety in red), where cisplatin modifies the universally conserved base A790 and stabilizes contacts between the ribosome and the mRNA (magenta). A-site tRNA is colored green, P-site tRNA—dark blue, E-site tRNA—orange. (C) Superposition of the 16S rRNA from Thermus thermophilus (light yellow) in the vicinity of the nucleotide A790 with the homologous region of the 18S rRNA from Homo sapiens (light blue; PDB ID: 4V6X (43)). Superposition is based on the alignment of nucleotides 787–795 from helix 24 of T. thermophilus 16S rRNA with nucleotides 1055–1063 of H. sapiens 18S rRNA. High similarity of mRNA channels suggests that cisplatin is likely to target nucleotide A1058 of the 18S rRNA (homologous to A790 in 16S rRNA) in the human ribosome.
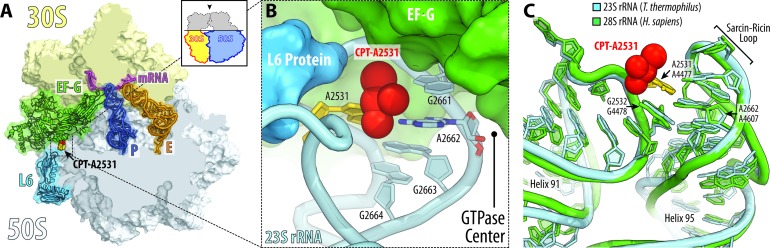
In the large ribosomal subunit, cisplatin targets residue A2531 of the 23S rRNA, which contacts the GTPase activating center (A2662 in the sarcin-ricin loop) (Figure 6A). In this position, the cisplatin moiety lies at the binding interface of elongation factor G (EF-G) and clashes with the C-terminal portion of ribosomal protein L6, suggesting that cisplatin modification of A2531 could either alter EF-G binding to the ribosome or affect GTP hydrolysis (Figure 6B). Notably, as is the case with the mRNA channel, the GTPase activating center is highly conserved across all species (Figure 6C), suggesting similar targeting of human ribosomes by cisplatin.
Figure 6.
Cisplatin modification site near the GTPase activating center of the ribosome. (A) Overview and (B) close-up view of the cisplatin position in the vicinity of the ribosome GTPase activating center. The structure of cisplatin-modified ribosome is superimposed with the structure of ribosome-bound EF-G (green) (PDB ID: 4V5F (44)). Superposition is based on the alignment of the 23S rRNA. 30S subunit is shown in light yellow, 50S subunit is in light blue. mRNA is shown in magenta and tRNAs are displayed in dark blue for the P site and in orange for the E site. tRNA for the A site is omitted for clarity. The view in (A) is from the top after removing the head of the 30S subunit and protuberances of the 50S subunit, as indicated by the inset. Note that cisplatin moiety of A2531 lies at the ribosome/EF-G interface in the immediate vicinity of the GTPase activating center (nucleotide A2662). (C) Superposition of the 23S rRNA from Thermus thermophilus (light blue) in the vicinity of the GTPase activating center with the homologous region of the 28S rRNA from Homo sapiens (green; PDB ID: 4V6X (43)). Note that due to extremely high conservation of the GTPase activating center, human ribosomes are likely to be modified by cisplatin at the same site as bacterial ribosomes.
Cisplatin targets two other functional sites in the large ribosomal subunit—the peptide exit tunnel (G27 of the 23S rRNA; Figure 4A) and the inter-subunit bridge B7a (A1848 of the 23S rRNA; Figure 4B). Modification of these nucleotides may hinder passage of the nascent polypeptide through the tunnel, and affect inter-subunit rotation during protein synthesis. However, both these sites are not conserved between bacteria and eukaryotes, and therefore are not likely to account for the inhibition of eukaryotic ribosomes by cisplatin.
Similarity between cisplatin and ribosome-targeting antibiotics
Protein synthesis can be inhibited by a number of small molecules, which include many ribosome-targeting antibiotics. We couldn't escape noticing that some of these antibiotics, namely amicoumacin A and edeine, bind to the ribosome in a close vicinity or even overlapping with the cisplatin-binding site in the mRNA channel. Edeine—a ribosome-targeting cationic peptide with potent anticancer activity, —binds the same 16S rRNA base as cisplatin (A790) and partially overlaps with the cisplatin position, suggesting that edeine and cisplatin binding to the ribosome are mutually exclusive (Supplementary Figure S5A) (30–33). Another ribosome-targeting drug, amicoumacin A, binds the mRNA channel a few angstroms away from the cisplatin-binding site and inhibits protein synthesis by forming simultaneous contacts with 16S rRNA and mRNA and tethering mRNA to the ribosome (Supplementary Figure S5B) (20). Since cisplatin forms similar contacts with the ribosome and the mRNA (Supplementary Figure S5B), it is possible that cisplatin and amicoumacin A exhibit the same inhibitory effects on protein synthesis.
DISCUSSION
Seeking to understand how the anticancer drug cisplatin targets RNA molecules, we determined the crystal structure of the bacterial ribosome complex with cisplatin. This structure provides the first high-resolution model of a cisplatin–RNA adduct.
In this study, we identified nine cisplatin modification sites in the ribosome and described precise positions and interactions of cisplatin with ribosome components. Consistent with previous studies of RNA modifications by cisplatin, we observe selective binding of cisplatin molecules to non-Watson–Crick segments of ribosomal RNA (reviewed in (11)). Notably, we observe a smaller number of modification sites in the ribosome when compared with the results of MALDI-MS (additional sites in helix 69 of the 23S rRNA) (34), or primer extension techniques (additional sites in helices 93, 95 and 96 of the 23S rRNA) (35). These discrepancies might be caused by several factors. First, electron density maps allow identification of cisplatin-derived Pt species only with occupancies above ∼50%. Biochemical techniques, however, are more sensitive and can detect even residual RNA modifications by cisplatin. Second, most biochemical studies explore cisplatin reactions in solution, in which ribosomes represent a mixture of different conformations, whereas our work describes cisplatin binding to a single conformation of the ribosome. Therefore, biochemical approaches can possibly detect more cisplatin modification sites due to conformational variability of ribosome in solution. Finally, the activity of cisplatin largely depends on its activation through hydrolysis, when the chloride groups of the cisplatin molecule are exchanged for water molecules (Supplementary Figure S6). It is possible that the relatively high concentration of chloride ions in our crystallization experiments (160 mM compared with physiological levels at 4–12 mM) could reduce cisplatin activation and limit cisplatin binding only to the most reactive sites in the ribosome. Apart from these discrepancies, our data agree with biochemical studies that showed cisplatin binding with conserved functional hairpins in the mRNA channel and in the vicinity of the GTPase activating center of the ribosome (35,36).
Having visualized how cisplatin modifies ribosomal RNA, we gained insights into specific recognition of RNA by cisplatin. Our structures reveal the consensus structural features of rRNA around the modification sites, in which adenine bases coordinate cisplatin that are part of the characteristic AG-motif, whereas guanine bases coordinate cisplatin in cooperation with adjacent backbone phosphates in distorted RNA folds and, in a manner, similar to inner-shell coordination of magnesium ions. The latter observation corroborates with the previous finding that cisplatin toxicity can be reduced by magnesium ions, which mask potential modification sites (37). Our study confirms this notion for guanine bases, but not for adenines, which normally do not coordinate magnesium ions, as seen in the structures of non-modified ribosomes. Collectively, comparative analysis of cisplatin-modified sites in the 70S ribosome provides the first structure-based insight into specific targeting of RNA by cisplatin. However, it is important to note that our structural study may describe only one subset of the several types of RNA modifications by cisplatin, in which Pt atom of cisplatin moiety is, most likely, retained in its dichloro- or mono-aqua form rather than the more prevalent in vivo di-aqua form. It is also important to note, that our crystal structures contain a number of AG-sites, which are structurally similar to the ones that are modified, and Mg2+-coordinating guanine bases, which are still not modified by cisplatin. This observation points to the fact that there might be other determinants of cisplatin specificity to only particular sites in RNA structure. Despite these limitations, both the structural AG-motif and Mg2+-coordination by guanine bases represent the first and only known structural signatures, whose presence in RNA correlates with cisplatin reactivity. Both of these signatures illustrate that RNA reactivity with cisplatin does not occur in RNA with ideal A-helix geometry, but instead requires highly-distorted folds, in which the cisplatin-derived Pt species forms extensive contacts with RNA molecule. This observation is curious, because highly distorted folds are the hallmarks of many active centers in RNA molecules. The fact that both the AG-motif and Mg2+-coordinating guanines are rarely occurring in RNA structures creates promise for an efficient prediction of cisplatin-reactive sites in cellular RNAs in the future.
Our experiments, showing how cisplatin targets active sites of the ribosome, extend our understanding of ribosome inhibition by small molecules. Together with recent studies of several ribosome-targeting antibiotics, our study changes our understanding of how small molecules act to inhibit protein synthesis. Until recently, it was thought that most of the ribosome-targeting drugs inhibit protein synthesis by altering the conformation of the ribosome or by preventing the proper binding of the ribosomal substrates. This paradigm, however, was challenged by recent structural studies of antibiotics blasticidin S (38), amicoumacin A (20), negamycin (39,40) and hygromycin A (41,42). These antibiotics were shown to tether the ligands (such as mRNA and tRNAs) to the ribosome or force their non-productive conformations. It appears from the structure of cisplatin-modified ribosome that the inhibitory activity of cisplatin might rely on a similar mechanism, in which cisplatin stabilizes interactions between the ribosome and the messenger RNA.
To date, we lack a comprehensive understanding of how cellular molecules are targeted by cisplatin. We also do not fully understand how this targeting provokes cell death and, in particular, how modification of RNA by cisplatin contributes to cisplatin-induced toxicity. Therefore, detailed knowledge of cisplatin binding sites in the ribosomal RNA may not only provide a tool for prediction of modification sites in other essential cellular RNAs, but will also help design experimental studies of the multifaceted nature of cisplatin cytotoxicity.
ACCESSION NUMBERS
The atomic coordinates and structure factors were deposited in the RCSB Protein Data Bank with the accession codes 5J4B for the T. thermophilus 70S ribosome modified by cisplatin (co-crystallization experiment) and 5J4C for the T. thermophilus 70S ribosome modified by cisplatin (soaking experiment).
Supplementary Material
Acknowledgments
We thank Melissa Moore (University of Massachusetts Medical School, USA) for bringing this project to our attention, Peter Glaser (Yale University, USA) for providing initial sample of cisplatin, Robert Grodzicki for preparation of the unmodified tRNAs and for careful reading of the manuscript. We are also thankful to the members of the Y.S.P., T.A.S. and D.S. laboratories for insightful discussions and critical feedback. We thank the staff of the Northeastern Collaborative Access Team (NE-CAT) at the Advanced Photon Source (beamline 24-ID) for technical assistance during data collection. We also thank staff at the Richards Center at Yale University for computational support.
FUNDING
Illinois State startup funds (to Y.S.P.); Howard Hughes Medical Institute and U.S. National Institutes of Health [GM022778 to T.A.S.]; U.S. National Institutes of Health [GM022854 to D.S.]; U.S. National Institute of General Medical Sciences [P41GM103403 to the Northeastern Collaborative Access Team (NE-CAT) at the Advanced Photon Source (beamline 24-ID)]; U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory [DE-AC02-06CH11357]. Funding for open access charge: Illinois State startup funds (Y.S.P.); Howard Hughes Medical Institute (T.A.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Rosenberg B., Vancamp L., Krigas T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg B., VanCamp L., Trosko J.E., Mansour V.H. Platinum compounds: a new class of potent antitumour agents. Nature. 1969;222:385–386. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg B. Anticancer activity of cis-dichlorodiammineplatinum(II) and some relevant chemistry. Cancer Treat. Rep. 1979;63:1433–1438. [PubMed] [Google Scholar]
- 4.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 5.Wheate N.J., Walker S., Craig G.E., Oun R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010;39:8113–8127. doi: 10.1039/c0dt00292e. [DOI] [PubMed] [Google Scholar]
- 6.Sava G., Bergamo A., Dyson P.J. Metal-based antitumour drugs in the post-genomic era: what comes next? Dalton Trans. 2011;40:9069–9075. doi: 10.1039/c1dt10522a. [DOI] [PubMed] [Google Scholar]
- 7.Siddik Z.H. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 8.Wang , Guo Z. The role of sulfur in platinum anticancer chemotherapy. Anticancer Agents Med. Chem. 2007;7:19–34. doi: 10.2174/187152007779314062. [DOI] [PubMed] [Google Scholar]
- 9.Basu A., Krishnamurthy S. Cellular responses to cisplatin-induced DNA damage. J. Nucleic Acids. 2010;2010:1–16. doi: 10.4061/2010/201367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D., Lippard S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug. Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 11.Chapman E.G., Hostetter A.A., Osborn M.F., Miller A.L., DeRose V.J. Binding of kinetically inert metal ions to RNA: the case of platinum(II) Met. Ions Life Sci. 2011;9:347–377. doi: 10.1039/9781849732512-00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg J.M., Sato P.H. Messenger RNA loses the ability to direct in vitro peptide synthesis following incubation with cisplatin. Mol. Pharmacol. 1988;33:611–616. [PubMed] [Google Scholar]
- 13.Hostetter A.A., Chapman E.G., DeRose V.J. Rapid cross-linking of an RNA internal loop by the anticancer drug cisplatin. J. Am. Chem. Soc. 2009;131:9250–9257. doi: 10.1021/ja809637e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman E.G., DeRose V.J. Site-specific platinum(II) cross-linking in a ribozyme active site. J. Am. Chem. Soc. 2012;134:256–262. doi: 10.1021/ja206455t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker J.P., Weiss J., Theile D. Cisplatin, oxaliplatin, and carboplatin unequally inhibit in vitro mRNA translation. Toxicol. Lett. 2014;225:43–47. doi: 10.1016/j.toxlet.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Burger A.M., Double J.A., Newell D.R. Inhibition of telomerase activity by cisplatin in human testicular cancer cells. Eur. J. Cancer. 1997;33:638–644. doi: 10.1016/s0959-8049(96)00521-7. [DOI] [PubMed] [Google Scholar]
- 17.Colangelo D., Ghiglia A.L., Viano I., Cavigiolio G., Osella D. Cis-[Pt(Cl)2(pyridine)(5-SO3H-isoquinoline)] complex, a selective inhibitor of telomerase enzyme. Biometals. 2003;16:553–560. doi: 10.1023/a:1023428315746. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes D., Piper P.W., Clark B.F. Location of a platinum binding site in the structure of yeast phenylalanine transfer RNA. J. Mol. Biol. 1974;89:469–475. doi: 10.1016/0022-2836(74)90476-8. [DOI] [PubMed] [Google Scholar]
- 19.Sundaralingam M., Rubin J.R., Rao S.T. X-ray studies on the interaction of the anticancer agent cis-[Pt(NH3)2cl2] to tRNAphe. A mechanism for the formation of the intrastrand cross-link to adjacent guanines in DNA. Prog. Clin. Biol. Res. 1985;172:175–184. [PubMed] [Google Scholar]
- 20.Polikanov Y.S., Osterman I.A., Szal T., Tashlitsky V.N., Serebryakova M.V., Kusochek P., Bulkley D., Malanicheva I.A., Efimenko T.A., Efremenkova O.V., et al. Amicoumacin A inhibits translation by stabilizing mRNA interaction with the ribosome. Mol. Cell. 2014;56:531–540. doi: 10.1016/j.molcel.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polikanov Y.S., Steitz T.A., Innis C.A. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014;21:787–993. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. XDS. Acta Cryst. Sect. D Biol. Cryst. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015;22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Cryst. Sect. D Biol. Cryst. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. Sect. D Biol. Cryst. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papsai P., Aldag J., Persson T., Elmroth S.K. Kinetic preference for interaction of cisplatin with the G-C-rich wobble basepair region in both tRNAAla and MhAla. Dalton Trans. 2006;2006:3515–3517. doi: 10.1039/b603833f. [DOI] [PubMed] [Google Scholar]
- 27.Hagerlof M., Papsai P., Hedman H.K., Jungwirth U., Jenei V., Elmroth S.K. Cisplatin and siRNA interference with structure and function of Wnt-5a mRNA: design and in vitro evaluation of targeting AU-rich elements in the 3′ UTR. J. Biol. Inorg. Chem. 2008;13:385–399. doi: 10.1007/s00775-007-0327-6. [DOI] [PubMed] [Google Scholar]
- 28.Heminger K.A., Hartson S.D., Rogers J., Matts R.L. Cisplatin inhibits protein synthesis in rabbit reticulocyte lysate by causing an arrest in elongation. Arch. Biochem. Biophys. 1997;344:200–207. doi: 10.1006/abbi.1997.0198. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg J.M., Sato P.H. Cisplatin inhibits in vitro translation by preventing the formation of complete initiation complex. Mol. Pharmacol. 1993;43:491–497. [PubMed] [Google Scholar]
- 30.Szer W., Kurylo-Borowska Z. Effect of edeine on aminoacyl-tRNA binding to ribosomes and its relationship to ribosomal binding sites. Biochim. Biophys. Acta. 1970;224:477–486. doi: 10.1016/0005-2787(70)90580-0. [DOI] [PubMed] [Google Scholar]
- 31.Kurylo-Borowska Z. Edeines. In: John WCaF, Hahn E., editors. Mechanism of Action of Antimicrobial and Antitumor Agents. Vol. 3. NY: Springer; 1975. pp. 129–140. [Google Scholar]
- 32.Contreras A., Carrasco L. Selective inhibition of protein synthesis in virus-infected mammalian cells. J. Virol. 1979;29:114–122. doi: 10.1128/jvi.29.1.114-122.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pioletti M., Schlunzen F., Harms J., Zarivach R., Gluhmann M., Avila H., Bashan A., Bartels H., Auerbach T., Jacobi C., et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dedduwa-Mudalige G.N., Chow C.S. Cisplatin targeting of bacterial ribosomal RNA hairpins. Int. J. Mol. Sci. 2015;16:21392–21409. doi: 10.3390/ijms160921392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborn M.F., White J.D., Haley M.M., DeRose V.J. Platinum-RNA modifications following drug treatment in S. cerevisiae identified by click chemistry and enzymatic mapping. ACS Chem. Biol. 2014;9:2404–2411. doi: 10.1021/cb500395z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hostetter A.A., Osborn M.F., DeRose V.J. RNA-Pt adducts following cisplatin treatment of Saccharomyces cerevisiae. ACS Chem. Biol. 2012;7:218–225. doi: 10.1021/cb200279p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durlach J., Bara M., Guiet-Bara A., Collery P. Relationship between magnesium, cancer and carcinogenic or anticancer metals. Anticancer Res. 1986;6:1353–1361. [PubMed] [Google Scholar]
- 38.Svidritskiy E., Ling C., Ermolenko D.N., Korostelev A.A. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12283–12288. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polikanov Y.S., Szal T., Jiang F., Gupta P., Matsuda R., Shiozuka M., Steitz T.A., Vazquez-Laslop N., Mankin A.S. Negamycin interferes with decoding and translocation by simultaneous interaction with rRNA and tRNA. Mol. Cell. 2014;56:541–550. doi: 10.1016/j.molcel.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olivier N.B., Altman R.B., Noeske J., Basarab G.S., Code E., Ferguson A.D., Gao N., Huang J., Juette M.F., Livchak S., et al. Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome. Proc. Natl. Acad. Sci. U.S.A. 2014;111:16274–16279. doi: 10.1073/pnas.1414401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polikanov Y.S., Starosta A.L., Juette M.F., Altman R.B., Terry D.S., Lu W., Burnett B.J., Dinos G., Reynolds K.A., Blanchard S.C., et al. Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol. Cell. 2015;58:832–844. doi: 10.1016/j.molcel.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaminishi T., Schedlbauer A., Fabbretti A., Brandi L., Ochoa-Lizarralde B., He C.G., Milon P., Connell S.R., Gualerzi C.O., Fucini P. Crystallographic characterization of the ribosomal binding site and molecular mechanism of action of Hygromycin A. Nucleic Acids Res. 2015;43:10015–10025. doi: 10.1093/nar/gkv975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 44.Gao Y.G., Selmer M., Dunham C.M., Weixlbaumer A., Kelley A.C., Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.