Abstract
Genomic DNA replication is a complex process that involves multiple proteins. Cellular DNA replication systems are broadly classified into only two types, bacterial and archaeo-eukaryotic. In contrast, double-stranded (ds) DNA viruses feature a much broader diversity of DNA replication machineries. Viruses differ greatly in both completeness and composition of their sets of DNA replication proteins. In this study, we explored whether there are common patterns underlying this extreme diversity. We identified and analyzed all major functional groups of DNA replication proteins in all available proteomes of dsDNA viruses. Our results show that some proteins are common to viruses infecting all domains of life and likely represent components of the ancestral core set. These include B-family polymerases, SF3 helicases, archaeo-eukaryotic primases, clamps and clamp loaders of the archaeo-eukaryotic type, RNase H and ATP-dependent DNA ligases. We also discovered a clear correlation between genome size and self-sufficiency of viral DNA replication, the unanticipated dominance of replicative helicases and pervasive functional associations among certain groups of DNA replication proteins. Altogether, our results provide a comprehensive view on the diversity and evolution of replication systems in the DNA virome and uncover fundamental principles underlying the orchestration of viral DNA replication.
INTRODUCTION
Viruses are the most abundant biological entities on Earth, greatly outnumbering their cellular hosts (1–3). The virosphere is not only vast but also extremely diverse. Viruses infect organisms from all three domains of life and span a wide spectrum of morphological, genomic and functional complexity. Viral genome sizes range from tiny (<2 kb) to huge (>2 Mb), and their genetic information may be stored within different types of nucleic acids—DNA or RNA, single- or double-stranded (ds), circular or linear, monopartite or segmented. The more complex viruses which represent a large fraction of known viruses, just like cellular organisms, carry dsDNA genomes. Strikingly, despite mechanistic uniformity of replication of the DNA double helix, the proteins involved are not universally conserved. In cellular organisms there are two distinct types of DNA replication machineries, one in bacteria and another one in eukaryotes and archaea (4). Replicative DNA polymerases, key replication enzymes, provide an illustrative example of these differences. In bacteria they belong to the C-family (PolC), whereas eukaryotes and archaea use unrelated B-family DNA polymerases (PolB), in addition, archaea employ D-family DNA polymerases.
DNA replication systems in dsDNA viruses are even more diverse than those of their cellular hosts. This diversity manifests itself in two different ways. First, dsDNA viruses differ according to the sets of DNA replication proteins encoded in their genomes. Some viruses encode most or all of the proteins necessary for replication of their genomic DNA, while others depend entirely on the host DNA replication machinery. Second, compared to cellular organisms, dsDNA viruses employ a larger number of evolutionary solutions for at least some of the DNA replication steps. For example, in addition to DNA polymerases of the B and C families, viruses employ A-family and protein-primed B-family DNA polymerases (pPolB) for genome replication. Meanwhile, in eukaryotes A-family polymerases are limited to replication of mitochondrial DNA and in bacteria perform mostly repair-related functions (5). Replicative pPolB polymerases that utilize a protein-supplied hydroxyl group as a primer appear to be unique to viruses and other selfish genetic elements (6).
The heterogeneity in nature and assortment of viral DNA replication proteins have become apparent early on from detailed studies on a handful of model viruses, such as bacteriophages φ29, T7 and T4 as well as eukaryotic adeno-, polyoma-, papilloma- and herpesviruses (7). However, these well-studied viruses and their close relatives represent only a small fraction of currently known viruses. Furthermore, with the advent of new genome sequencing technologies and revived interest in viral ecology and diversity, the number of new virus isolates with complete genome sequences increases at an unprecedented rate. For most of these viruses no experimental data on DNA replication are (or ever will be) available. Nevertheless, the wealth of available genomic data enables us to ask a series of questions. Are there novel assortments of DNA replication proteins in new viruses identified by genomics and metagenomics studies? Are there viruses that use yet unseen replication strategies? Can we make inferences extending across dsDNA viruses in the three domains of life? Here, we set out to address these questions by performing a global computational analysis of DNA replication proteins encoded by all sequenced bacterial, archaeal and eukaryotic dsDNA viruses. Using sensitive state-of-the-art computational tools we investigated the diversity and distribution of proteins associated with major molecular functions in DNA replication, including replicative DNA helicases, primases, replicative DNA polymerases and their accessory proteins, single-stranded DNA binding (SSB) proteins, nucleases for RNA primer removal, DNA ligases and topoisomerases. Our results show that despite overwhelming diversity, there appear to be clearly detectable common patterns of assortment of viral DNA replication proteins transcending the boundaries of individual domains of life. For example, our analysis suggests the existence of a common dominating viral strategy for recruitment of the host DNA replication proteins. Our results also reveal strong links between DNA replication proteins in several functional categories.
MATERIALS AND METHODS
Virus databases
Genomes and proteomes of double-stranded DNA viruses were obtained from NCBI: ‘http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10239&sort=taxonomy’. All the genomes of dsDNA viruses were subjected to six-frame translation and all previously unassigned open reading frames (ORFs) longer than 60 residues were retained for further analysis. Family Polydnaviridae was excluded from the analysis because these viruses have a distinct genome organization (split in small segments), and their genome acts only as a vector for transmission of parasitic wasp genes (8).
Genome filtering
To obtain a more representative genome set, highly similar genomes were removed. All genomes were grouped according to their pairwise nucleotide and protein sequence similarities using LAST (9) and CLANS (10), respectively. Genomes with statistically significant local nucleotide sequence identity >70% or those whose proteomes had more than three-quarters of highly similar proteins (CLANS P-value < 1 × 10−10) were filtered out.
Identification of replication proteins
Replication proteins of dsDNA viruses were identified based on their similarity to characterized DNA replication proteins. All viral proteins were subjected to sensitive homology search using HHsearch (11). Sequence profiles of viral proteins were generated by running two iterations of either HHblits (12) or jackhmmer (part of the HMMER3 software package (13)) against the nr70 database (the non-redundant database with no more than 70% identity between any sequences) using the E-value = 1 × 10−3 inclusion threshold. Profiles generated by HHblits were then used by HHsearch to search the PDB (http://www.pdb.org/), SCOP (http://scop.mrc-lmb.cam.ac.uk/scop/) and Pfam (http://pfam.xfam.org/) databases. HHsearch results with probability >20% were extracted and clustered according to their pairwise similarity using CLANS. For each sequence, CLANS was configured to run two iterations of PSI-BLAST (14) using the E-value = 1 × 10−3 inclusion threshold against the reference database (nr70) to generate a sequence profile. The last PSI-BLAST iteration with the obtained profile was run against the database of sequences to be clustered. Next, clusters obtained with CLANS were analyzed and viral proteins that clustered with known replication proteins were assigned to that group. If there appeared to be more than one candidate for the same group of replication proteins (e.g. for replicative helicase) the analysis included additional steps. Proteins were tested for the presence of additional domains typical of DNA replication proteins (e.g. primase domain fused to a helicase). Also, gene context in the vicinity (ten genes downstream and ten upstream) was analyzed for the presence of other DNA replication proteins.
Detection of remote homology
If standard searches with PSI-BLAST, jackhmmer and HHsearch failed to produce confident homology assignments, a number of strategies were tried in order to increase sensitivity. They included splitting query sequence into putative domains, using additional databases and metagenomic sequence data as well as modifying search parameters. Putative domains were assigned based on the analysis of alignments and scores of initial HHsearch searches. Sequence databases of higher redundancy (nr filtered to >70% identity) and metagenomic data were used to enrich ‘thin’ query profiles (derived from less than five sequences). In addition, HHsearch profile databases were supplemented with curated in-house profiles for DNA replication proteins. Reciprocal searches (if A detects B, B should detect A) were used to substantiate weak matches. For increased sensitivity, HMMER3 searches were performed with the acceleration heuristic turned off (option ‘–max’).
Multiple sequence alignments
Multiple sequence alignments of potential replication proteins were examined for the presence of active site (where applicable) and other characteristic conserved regions. Multiple sequence alignments were constructed with MAFFT (15) optimized for accuracy (option ‘L-INS-i’).
Phylogenetic analysis
Sequence alignments for phylogenetic analysis were constructed from representative members of viral replication proteins and well-characterized cellular proteins found in the nr70 database. To obtain more accurate alignments, highly divergent viral proteins were supplemented with homologs from nr70 and metagenomics databases. Sequences were aligned using MAFFT (15). Alignments were manually adjusted based on secondary structure prediction, structure comparison and HHsearch profile-profile alignments. Non-conserved N and C terminal regions of DNA polymerases and primases were removed prior to analysis. In addition, trimAl (16) was used to remove 50 and 5% of columns containing the largest fraction of gaps from alignments of DNA polymerases and primases, respectively. Protein evolution model LG+G+I was applied for both groups of proteins as suggested by ProtTest (17). For every alignment we generated 500 trees and 1000 bootstrap replicates using RAxML (18). RAxML convergence test was performed to verify whether the selected number of bootstrap replicates was sufficient for the analysis.
RESULTS
A global proteome encoded by 1574 dsDNA viral genomes (over 150 000 proteins) was analyzed for the presence of DNA replication proteins using sensitive homology detection and sequence classification techniques. Sequences were assigned to specific groups of replication proteins using several lines of evidence, including the similarity to experimentally characterized proteins, domain organization and genomic neighborhood. To reduce the bias in database coverage of viral genomes, dsDNA viruses with highly similar genomes were filtered out, retaining a single representative member per virus group (see Materials and Methods). As a result, 308 representatives belonging to 33 different viral families and including 221 bacteriophages (72%), 61 (20%) eukaryotic and 26 (8%) archaeal viruses were retained for detailed analysis (Supplementary file 1).
RNA- and protein-primed DNA replication depend on distinct sets of proteins
Protein-primed DNA replication systems appear in viruses infecting hosts from all three domains of life. However, they are confined to viruses with small (<50 kbp) genomes, consistent with previous observations (7,19). Interestingly, the complement of accessory proteins appears to be distinctively different in DNA replication systems primed by RNA and protein primers (Table 1). This divide appears to be dictated by the intrinsic properties of pPolB. Unlike other members of the B family, pPolB has a distinct subdomain (TPR1) which is responsible for the binding to a protein primer, which is covalently attached to both ends of the viral genome (20). The second pPolB-specific subdomain (TPR2) ensures intrinsic processivity and strand displacement capability (21). Therefore, in general, there is no need for the DNA helicase in protein-primed DNA replication. Viruses with protein-primed DNA polymerases often encode SSB proteins. However, these SSBs are unique with no sequence or structural relationship to the OB-fold proteins typical of RNA-primed DNA replication systems (6,22). Overall, pPolB-coding viruses have few replication proteins and make only a small fraction of a representative genome set (7%). Therefore, in further analysis we consider only RNA-primed replication systems.
Table 1. Comparison of protein- and RNA-primed DNA replication in dsDNA viruses.
| Protein-primed DNA replication | RNA-primed DNA replication | |
|---|---|---|
| Genome size | ∼10–50 (kbp) | ∼5–2500 (kbp) |
| Viruses in the representative set | 7% | 93% |
| Replication proteins | pPolB | rPolB/PolA/PolC |
| SSB | SSB (usually OB-fold proteins) | |
| Terminal protein | Helicase | |
| Primase | ||
| Primer removal protein | ||
| Ligase | ||
| Processivity factor and clamp loader | ||
| Topoisomerase |
Viral proteins within functional groups show uneven diversity and distribution
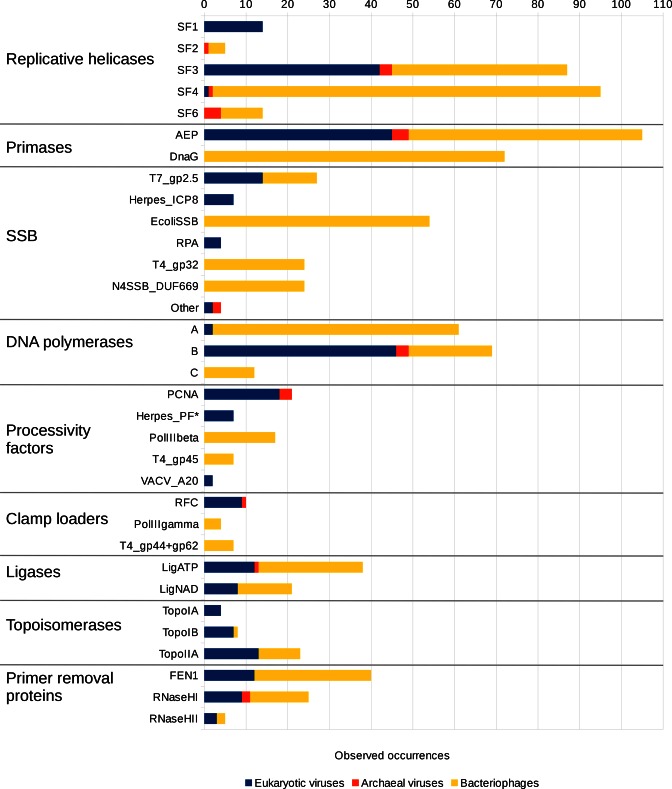
Our results show that the number of protein families associated with individual functional categories of viral DNA replication is quite variable (Figure 1). In most functional groups the diversity of viral proteins exceeds that of cellular organisms. On the other hand, the representation of individual protein families varies significantly both within a particular domain of life and across all three domains. Below we summarize our findings for every major functional group of viral DNA replication proteins, focusing on their diversity and distribution.
Figure 1.
Quantitative distribution of DNA replication protein families and the taxonomy of virus host. The length of the bar is proportional to the number of protein family members found in the representative set of dsDNA viruses. Asterisk (*) denotes herpes processivity factors that include UL42, UL44 and BMRF1 homologs.
Replicative helicases
Helicases and nucleic acid translocases are classified into six superfamilies, SF1-6 (23). SF1 and SF2 helicases have two RecA-like domains and function as monomers or dimers. The remaining four superfamilies comprise hexameric helicases. SF3 and SF6 are based on the AAA+ fold, whereas SF4 and SF5 are built around the RecA fold. Replicative helicases in cellular organisms are confined to only two superfamilies of hexameric helicases, namely, SF4 (DnaB-type) in bacteria and SF6 (MCM-type) in archaea and eukaryotes. Replicative helicases in dsDNA viruses show a significantly higher diversity. We assigned both known and putative replicative DNA helicases identified in our analysis to five out of six superfamilies (Figure 1, SF1-4 and SF6). The confident identification of viral replicative helicases is not always straightforward due to ubiquity of genes for various NTPases in the viral genomes. For example, in addition to the replicative helicase (gp41), phage T4 has at least two other helicases (Dda and UvsW). To discriminate between genuine replicative helicases and NTPases involved in other processes, we used several criteria. First, we identified proteins that grouped together with known replicative helicases (i.e. cellular DnaB and MCM, T4 phage gp41, T7 phage gp4, herpes UL5, Vaccinia D5 and polyoma TAg). Second, we investigated the domain organization of putative helicases, focusing on helicase-primase fusions, because viral replicative helicases are known to be closely associated with DNA primases (24). Third, we considered the genomic neighborhood of helicases (whether or not primases and other replication proteins are coded nearby). As a result of this stringent selection procedure, assignments in only 6 out of 215 cases are less confident (Supplementary Figure S1). These include Tetraselmis viridis virus S1 (TvV-S1), phage Ma-LMM01 and archaeal viruses HVTV-1, HGTV-1, HF1 and Acidianus filamentous virus 6 (AFV6).
We found that SF4 members are the most abundant (44%) among viral replicative helicases and that they are almost exclusively encoded by bacteriophages (Figure 1). Most of these proteins are similar to E. coli DnaB or RepA replicative helicases (25). The largest fraction of DnaB homologs (Figure 2) has a stand-alone ATPase domain. Another recurrent variant contains a helicase domain fused with the DnaG primase domain as seen in gp4 of T7 phage. Replicative helicases similar to RepA are often fused with the archaeo-eukaryotic primase (AEP) domain as exemplified by the BFK20 phage protein gp43 (26). In general, there was little ambiguity in assigning SF4 members as putative replicative helicases, except in the case of Haloarcula virus HVTV-1 of the order Caudovirales. HVTV-1 encodes a number of replication proteins (DNA polymerase, DNA clamp and its loader, AEP primase and RNase HI) characteristic of archaeal hosts. However, replicative minichromosome maintenance (MCM) helicase of the SF6 superfamily, typical of archaea, could not be identified. Instead, HVTV-1 encodes a divergent SF4 helicase (Supplementary Figure S2, gi: 443404669), the only apparent candidate for the replicative helicase.
Figure 2.
Diversity of replicative helicases domain organizations in dsDNA viruses. The number of representative sequences having specific domain organization is indicated on the left. The catalytic domain of the helicase is shown in green; primase, orange; polymerase, red; other domains, gray. αHD, α-helical domain; DUF, domain of unknown function; WH, winged-helix-turn-helix domain; ZnBD, Zn-binding domain; DBD, DNA-binding domain; Pol, DNA polymerase.
Superfamily 3 (41% of all replicative helicases) is nearly as abundant among viruses as SF4, but more diverse with respect to both sequence similarity (Supplementary Figure S3) and domain composition (Figure 2). SF3 is represented by well-studied polyoma- and papillomavirus replicative helicases, large T antigen (Tag) and E1, respectively. Other known SF3 members include the poxvirus primase-helicase D5 (27) and its homologs in large eukaryotic viruses, in particular members of the order Megavirales. Poxvirus D5 is a fusion of the N-terminal AEP primase domain and the C-terminal helicase region. The D5 group also includes phage replicative helicases typified by the phage P4 gpα protein, also representing a primase-helicase fusion (28). However, unlike in poxvirus D5, the N-terminal primase domain in gpα is of the bacterial DnaG-type. Quite often (about two-thirds of all cases) SF3 replicative helicases are fused to origin-binding domains (OBDs). For example, both Tag and E1 helicases have an OBD, which has evolved from the inactivated endonuclease domain (29). In gpα protein of phage P4 and other D5 family members, origin binding is accomplished via the C-terminal winged helix-turn-helix (WH) DNA-binding domain (28). In addition to known instances, we have newly identified D5 homologs in White spot syndrome virus (Nimaviridae; gi: 17158445) and hytrosaviruses (Hytrosaviridae; gis: 187903120 and 168804177) (Supplementary Figure S3). Other groups of putative replicative SF3 helicases do not have characterized representatives. One of these groups is similar to the VirE family (PF05272) and mainly includes phage sequences. Another group corresponds to the DUF927 family (Supplementary Figure S3 and Figure 2). In both groups, the putative helicase domains are often fused with DnaG- or AEP-like primase domains, strongly suggesting their involvement in phage DNA replication. An interesting case is represented by the putative primase-helicase proteins from the Phaeocystis globosa virus virophage and Sputnik virophage (gis: 509141013 and 195982544, respectively). These proteins have the C-terminal D5-like helicase module and an N-terminal domain (Figure 2 and Supplementary Figure S3) identified as a novel A-family polymerase implicated in both polymerase and primase activities (30).
Superfamily 6 is exemplified by extensively studied archaeal and eukaryotic replicative MCM helicases as well as RuvB proteins (23). MCM-like helicases were rather frequently found in proviruses integrated in genomes of diverse archaeal species (31), but in viral genomes these proteins so far were detected only in Halorubrum virus BJ1 and spindle-shaped Thermococcus prieurii virus 1 (32,33). Using sensitive profile-profile comparisons, we identified divergent SF6 members in several bacteriophages and archaeal haloviruses, but, curiously, not in eukaryotic viruses. The bacteriophage helicases belong to the DUF3987 family (Supplementary Figure S4), distantly related to MCM. For example, we could link the Mycobacterium phage Corndog SF6 protein (gi: 29566306) to the MCM family (PF00493) with 98% HHsearch probability. Some of these SF6 helicases are fused with AEP primase domains. Interestingly, the N4 phage protein dns (gi: 119952220), which is essential for replication (34) features a non-typical fusion. Unlike in most of primase-helicase fusions, the AEP domain is C-terminal to the helicase domain (Figure 2). In general, proteins from the DUF3987 family are often associated with AEP or DnaG primases either as fusions or as separate proteins encoded immediately upstream of the helicase. Furthermore, similarly to known replicative SF6 helicases, DUF3987 proteins possess the C-terminal winged helix-turn-helix domain (Figure 2). These observations support the notion that DUF3987 proteins act as replicative helicases in phage replication.
Although SF1 helicases are abundant in viruses of the proposed order Megavirales (35), our data show that dsDNA viruses rarely (in 7% of the cases) use helicases of this type for genome replication (Figure 1). All SF1 helicases known to be essential for replication are represented by herpesviral UL5 proteins. The UL5 helicase is related to the eukaryotic Pif1 helicase and is present in all three herpesvirus families (36). We identified TvV-S1 as the only other virus with a putative replicative SF1 helicase (Supplementary file 2, gi: 472343114).
Superfamily 2 has the fewest members assigned as replicative helicases (Figure 1), despite being one of the most abundant protein groups in all prokaryotic viruses (37,38). SF2 members from phages N15 and PY54 (gi: 9630494 and 33770544, respectively) have the DnaG primase domain fused to their N-termini (Figure 2) and their function as replicative helicases has been experimentally confirmed (39,40). Another likely candidate for the role of a replicative helicase is phage PAU protein gp68 (gi: 435844571), which has an AEP primase domain (Figure 2). The assignment of viral SF2 members as replicative helicases in the case of archaeal virus AFV6 and phage Ma-LMM01 is less certain. Both viruses have more than one helicase (Supplementary file 1) and none of them have been studied experimentally.
Primases
There are two types of primases in cellular organisms. Bacteria have the TOPRIM domain-containing primase (i.e. E. coli DnaG), which is distinct from the archaeo-eukaryotic primase (AEP) with the RNA recognition motif (RRM) fold (41). In dsDNA viruses we detected both types of primases and they were frequently fused with a replicative helicase or encoded next to it, as detailed above. We observed that viruses have either AEP or DnaG homolog. The only exception to this rule is Rhodothermus phage RM378, which encodes primases of both types (Supplementary file 1). DnaG homologs were always found in genomes that also contained replicative helicases, whereas in few cases AEP proteins (gis: 197261587, 41057246 and 472342211) were detected in genomes devoid of both replicative helicases and polymerases.
Replicases
Cellular replicases are composed of a DNA polymerase, a processivity factor (DNA sliding clamp) and its loader. Our present results on viral replicases are in close agreement with the previous observations (19). DNA polymerases of structurally related A and B families are dominant, while homologs of the unrelated C family of bacterial DNA polymerases are rare and are present only in bacteriophages (Figure 1). We did not find viral homologs of D-family DNA polymerases, specific to Archaea. DNA polymerase accessory proteins (clamps and clamp loaders) were usually detected in larger genomes. Viral processivity factors, with the exception of poxviral A20, could be assigned to one of the three groups based on their similarity to the archaeal/eukaryotic PCNA, the bacterial DNA polymerase III β-subunit (PolIIIβ) or the phage T4 gp45 protein (Figure 1). The structure of poxviral A20 is unknown, but differently from typical clamp proteins it functions as a heterodimer with the uracil-DNA glycosylase D4 (42). Viral clamp loaders according to their homology relationships mirror the three groups of viral clamp proteins (Figure 1). Our new findings include distant homologs of DNA sliding clamps in one of the largest archaeal viruses (HVTV-1) and giant Pandoraviruses as well as a complete set of DNA polymerase accessory proteins (T4-like DNA clamp and clamp loader) in Sphingomonas phage PAU (see Supplementary file 1). Initially, in phage PAU we only identified homologs of T4 clamp loader (gp44/gp62) (gi: 435844579 and 435844589, respectively), but not the clamp. However, the presence of a clamp loader strongly suggested that the DNA sliding clamp is also encoded in the genome of phage PAU. Indeed, nonstandard (acceleration heuristics turned off) HMMER searches with the T4 clamp (gp45) HMM profile against the phage PAU proteome detected a single protein (gp109) with a significant E-value (E-value<0.001). Consistent with the putative DNA clamp function, gp109 is encoded next to the DNA polymerase, has chain length (271 amino acids) and isoelectric point (pI = 4.9) both typical of a gp45-like clamp that functions with the clamp loader (19). Direct searches with gp109 identified no homologs in protein databases, but found a homologous incomplete sequence in the metagenomics database. With the addition of this sequence HHsearch could now detect T4 gp45 with a highly significant value (HHsearch probability > 95%). Altogether, these data indicate that gp109 is a bona fide homolog of the T4 DNA sliding clamp.
Single-stranded DNA binding (SSB) proteins
Typically, the SSB function at a replication fork is performed by proteins that have one or more domains of the oligonucleotide-binding (OB) fold (43). We found that in dsDNA viruses SSBs based on the OB-fold are also most common, and the majority of them can be unambiguously linked to one of the groups represented by crystal structures of E. coli SSB (44), phage T7 protein gp2.5 (45), phage T4 protein gp32 (46), herpesvirus ICP8 (47) or archaeo-eukaryotic Replication Protein A (RPA) (48). Most often dsDNA viruses have E. coli SSB homologs that appear to be confined to bacteriophages (Figure 1). Another abundant group includes homologs of T7 phage SSB (gp2.5). In line with our previous findings (49), gp2.5-like SSBs are present not only in phages, but also in eukaryotic viruses of the recently proposed order Megavirales. The ICP8 protein family distantly related to T7 SSB (49) is found only in herpesviruses. T4 gp32-like proteins form a sizeable group, which is present only in phages. Close homologs of the archaeo-eukaryotic RPA were exclusively found in Emiliania huxleyi virus 86 (EhV-86). Most likely they were acquired horizontally from the algal host (49).
Among groups that have experimentally characterized SSBs without known structures, the most abundant is the group typified by the SSB protein (gi: 119952251) of E. coli phage N4 (50). The N4 SSB does not show significant sequence similarity to OB-fold proteins or any other structures, but it can be linked to a large Pfam family of unknown function (DUF669) with high confidence (98% HHsearch probability) (Supplementary Figure S5). In contrast, members of the DUF669 family often recover OB-fold proteins among the top matches albeit not with high confidence. DUF669 sequences have acidic C-terminal tails, typical of many phage and bacterial SSBs. Furthermore, viruses that have DUF669 proteins do not encode known SSBs. Taken together, these observations suggest that DUF669 viral members represent SSBs involved in DNA replication and that both DUF669 and N4-like SSBs feature highly evolved OB-fold domains. Another structurally uncharacterized group is represented by the baculoviral SSB protein (LEF3), essential for virus replication (51). Although searches with the full-length LEF3 sequences did not produce significant matches to known structures, they suggested the presence of at least three domains. When analyzed individually, the last two putative LEF3 domains showed weak similarity to RPA. Although these results are by no means definitive, they hint that LEF3 may have at least two highly diverged OB-fold domains.
Other identified viral SSB proteins are diverse and often taxon-specific, for example, homologs of poxvirus I3 (49) and archaeal virus SIRV2 protein gp17 (52).
Primer removal proteins
In cellular organisms primer removal is performed by members of distinct FEN and RNase H families. In eukaryotes, long primers are also processed by Dna2, a member of the PD-(D/E)XK nuclease family (53). In general, PD-(D/E)XK proteins perform a variety of other functions related to nucleic acid metabolism (53). Since we did not find close Dna2 homologs in dsDNA viruses, we did not assign viral proteins of PD-(D/E)XK family to the list of putative primer removal proteins. Identification of FEN and RNase H homologs revealed that the FEN family members are present only in phages and eukaryotic viruses, whereas RNase H proteins were found in viruses associated with all three cellular domains of life (Figure 1).
Ligases
DNA ligases are classified into two groups depending on whether they use ATP or NAD+ as a cofactor (54). We found that DNA ligases in dsDNA viruses are infrequent and that they are usually encoded in large genomes. Remarkably, Pandoraviruses, despite having the largest genomes among presently known dsDNA viruses, lack their own DNA ligase. The distribution analysis shows that viruses from the same taxonomic group can have DNA ligases of different types (ATP/NAD+). For example, members of families Poxviridae and Mimiviridae have either ATP or NAD+ ligase (Supplementary File 1), consistent with previous observation (55). Notably, we did not find NAD+-dependent DNA ligases in archaeal viruses. However, this might be due to the under-representation of genomes of archaeal viruses in the current databases.
Topoisomerases
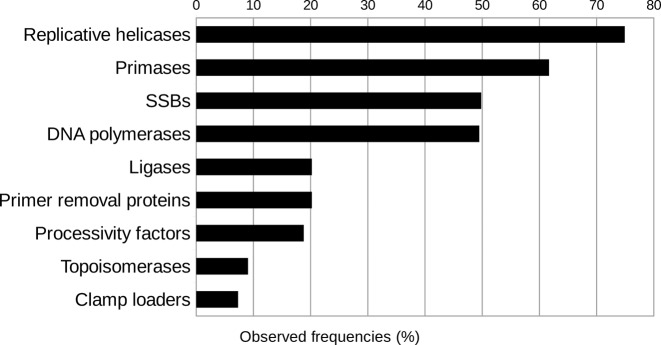
Topoisomerases are broadly grouped into two types depending on whether the enzyme introduces a single-stranded (Type I) or double-stranded (Type II) break in dsDNA. Based on sequence and structural similarities topoisomerases are further classified into families (Topo IA, IB, IC and Topo IIA and IIB) (56). DNA topoisomerases turned out to be among the least common DNA replication proteins in dsDNA viruses (Figure 3). We found DNA topoisomerases only in eukaryotic viruses and bacteriophages with large genomes (over 130 kbp). None were found in archaeal viruses (Figure 1). Most abundant among detected viral DNA topoisomerases were members of IB and IIA families. It is known that all DNA topoisomerases with the exception of IA family participate in relaxation of positively supercoiled DNA created during DNA replication (57). IA family members accounted for only 11% of all cases and, notably, were found only in genomes that already had a TopoIIA topoisomerase and, occasionally, also TopoIB.
Figure 3.
Replication proteins arranged by their overall observed frequencies (%) in genomes of dsDNA viruses.
Viral DNA replication proteins are unevenly spread across the three domains of life
Our analysis revealed that some families/superfamilies of viral DNA replication proteins are spread across all three cellular domains of life, while others are confined to particular domains (Figure 1). The ‘universal’ proteins include SF3 helicases, AEP primases, B-family DNA polymerases, RNase H and ATP-dependent DNA ligases. Although SF4 helicases, typified by bacterial replicative helicases (i.e. E. coli DnaB), seemingly also belong to this group, their ‘universal’ nature is doubtful. Nearly all of SF4 helicases are found in phages, with the exception of Haloarcula virus HVTV-1 and algal Tetraselmis viridis virus S20 (TvV-S20). In the case of HVTV-1 the assignment of a replicative helicase is ambiguous and should await experimental verification. By contrast, the uncharacterized TvV-S20 encodes a typical DnaB-like helicase; however, the virus itself might represent a case of ‘mistaken identity’. TvV-S20 encodes a number of proteins such as large and small subunits of the terminase, major capsid protein, portal protein, maturation protease, which are closely similar to those of bacteriophages but not eukaryotic viruses. This suggests that TvV-S20 might be a genuine bacteriophage associated with bacteria co-culturing with algae.
Other protein families are spread across two cellular domains at most. However, as archaeal viruses presently comprise a relatively small fraction, it might be expected that once more of their genomes become available the number of ‘universal’ protein families will also increase. Among protein families spanning two domains of life perhaps the most interesting are those that are incongruent with the corresponding proteins of their hosts. In this regard, SF6 helicases and T7-like SSBs represent intriguing cases. SF6 members are replicative helicases in archaea and eukaryotes; however, we identified helicases of this superfamily in archaeal viruses and bacteriophages, but not in eukaryotic viruses. T7-like SSBs were found in bacteriophages and eukaryotic viruses, and these SSBs seem to be specific to viruses having no close homologs in cellular organisms.
Many bacterial DNA replication protein families, including C-family DNA polymerases, homologs of bacterial DNA sliding clamp (PolIIIβ) and the subunit of bacterial clamp loader (PolIIIγ), DnaG primases and E. coli SSBs are exclusive to bacteriophages (Figure 1). However, the occurrence of bacterial-like components in bacteriophages is generally only slightly more pronounced than that of archaeo-eukaryotic homologs (Supplementary Table S1). Although some functional groups are dominated by typical bacterial homologs, as in the case of DnaB-like helicases (43% of DnaB-like helicases versus 24% of SF3 and SF6 helicases), for other groups the archaeo-eukaryotic replication proteins were detected nearly as frequently or even more frequently than the bacterial counterparts. For example, AEP primases and DnaG-like primases were detected in 26 and 32% of the genomes, respectively, whereas family B DNA polymerases and ATP-dependent ligases are two times more abundant in bacteriophages than the bacterial equivalents (PolC and NAD+-dependent ligases, respectively; Supplementary Table S1). Furthermore, bacterial and archaeo-eukaryotic components are typically intermixed within individual bacteriophage genomes. Thus, only eight bacteriophages encode purely bacterial-like assortment of major replisome components, including DnaB helicase, DnaG primase and PolC. Similarly, the three components of the archaeo-eukaryotic variety (AAA+ helicase, AEP and PolB) were identified in seven bacteriophage genomes.
Recent horizontal gene transfer does not account for the presence of archaeo-eukaryotic replication proteins in bacteriophage genomes
The abundance of archaeo-eukaryotic replication proteins in bacterial viruses as well as their provenance is puzzling. Although in none of the known bacterial lineages archaeo-eukaryotic equivalents have replaced the canonical bacterial components of the replisome, such proteins are occasionally encoded in bacterial genomes. For instance, there are two groups of PolB homologs in bacterial genomes. One group, which includes E. coli DNA polymerase II, is conserved in proteobacteria and appears to assist PolC in chromosomal replication, particularly for the lagging strand synthesis (58). The other group displays more sporadic distribution in bacteria. In principle, the two groups could represent the source of archaeo-eukaryotic replication genes in bacteriophages. To formally explore this possibility, we performed phylogenetic and genomic context analyses.
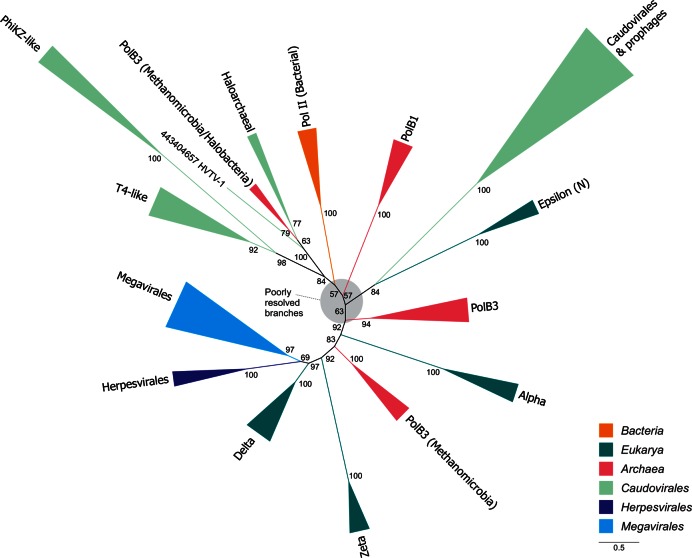
In the reconstructed maximum likelihood phylogeny of PolB proteins, all cellular homologs form well-supported clades (Figure 4, Supplementary file 3). Consistent with previous reports (59,60), all of these clades, with the exception of archaeal PolB3 (see below), are monophyletic. PolB proteins from eukaryotic viruses of the orders Megavirales and Herpesvirales form a sister clade to eukaryotic PolB of the subclass Delta. This topology has been interpreted as being suggestive of the possibility that the PolB-Delta subclass has evolved from viral proteins (59,61). However, the inverse scenario appears as likely. PolB homologs encoded by Caudovirales form several distinct clades, none of which is closely related to bacterial DNA polymerase II homologs. PolBs from T4-like and PhiKZ-like phages form well-supported sister clades, whereas PolBs from haloarchaeal Caudovirales form a clade with PolB3 proteins encoded by halophilic and methanogenic archaea (Figure 4). Importantly, in this clade, viral PolB homologs are at the base of the cellular PolB3 homologs (bootstrap support of 100), suggesting that this particular clade of PolB3 has evolved from viral proteins. This conclusion helps to rationalize the previous observation that archaeal PolB3 homologs are polyphyletic (59,60). Finally, the largest Caudovirales clade forms a sister group to eukaryotic PolB of the subclass Epsilon and includes proteins encoded both in viral and cellular genomes. Given that members of Caudovirales often reside within bacterial and archaeal genomes as prophages (62), we explored the possibility that the seemingly bacterial PolB homologs in this subgroup are encoded within prophages. This indeed turned out to be the case (Supplementary Table S2). Thus, phylogenetic analysis strongly suggests that there has been no recent horizontal transfer of cellular PolB genes into Caudovirales genomes.
Figure 4.
Maximum likelihood phylogenetic analysis of family B DNA polymerases from archaea, bacteria, eukaryotes and their respective viruses. Numbers at the branch points represent non-parametric bootstrap support (1000 replicates). Branches are color-coded and the color key is provided at the bottom right corner of the figure. Alpha, Delta, Epsilon and Zeta represent subclasses of eukaryotic PolBs. ‘N’ indicates that only the catalytic N-terminal domain of the PolB-Epsilon was considered. The scale bar represents the number of substitutions per site.
Unfortunately, due to high divergence of other groups of analyzed viral and cellular replication proteins, phylogenetic analyses were less informative and generally resulted in trees with poorly supported basal branches. Nevertheless, they are sufficient to exclude the possibility of recent horizontal gene transfer events. As a case in point, although phylogenetic analysis of viral and cellular AEP proteins has resulted in an unresolved tree (Supplementary Figure S6), AEPs encoded by mobile elements form distinct clades none of which emerge from within either archaeal or eukaryotic AEPs. Similar to PolB, a considerable number of AEP homologs were found to be encoded within mobile genetic elements, including plasmids and prophages (Supplementary Table S2). Without detailed genomic context analysis such homologs of archaeo-eukaryotic replication proteins might be mistaken for genuine bacterial proteins.
Replicative helicases are the most common replication proteins in dsDNA viruses
Replication protein complements identified across all groups of dsDNA viruses, provide a comprehensive picture of the distribution and frequency of different groups of replication proteins in the available viral genomes. Our analysis shows that replicative helicases are the most frequent (75% of genomes) replication proteins (Figure 3). We consider these results to be robust, because we analyzed only a representative set of viruses and used stringent replicative helicase assignment criteria. Even if we exclude several ambiguous cases of the assignment of replicative helicases, their observed frequency would still be the highest (73%). Moreover, replicative helicases are most abundant regardless of the virus host (Supplementary Figure S7) or viral genome size (Supplementary Figure S8). Rather unexpectedly, despite their central role in replication, DNA polymerases are considerably less common (49%), following helicases, primases (62%) and SSB proteins (50%). The frequencies of other replication proteins encoded in viral genomes are significantly lower. Some of these proteins (e.g. topoisomerases) might be dispensable for the replication of small genomes while others perhaps might be more easily replaceable by corresponding proteins of the host.
We observed that viruses lacking a replicative helicase frequently have either integrases, DNA recombination proteins or various replication initiation proteins, including homologs (and functional equivalents) of bacterial replisome organizer DnaA and helicase loader DnaC or rolling-circle replication initiation endonucleases such as gene A protein (GPA) of phage P2. Interestingly, in some archaeal viruses (AFV1, AFV2, Pyrococcus abyssi virus 1) we did not find any typical DNA replication proteins. These viruses might have highly diverged DNA replication proteins or use new replication strategies. The latter possibility is supported by the recent study on virus AFV1, which appears to employ a novel strand displacement mechanism of genome replication. However, the proteins involved in this process have not yet been identified (63).
DNA replication machineries are genome size dependent
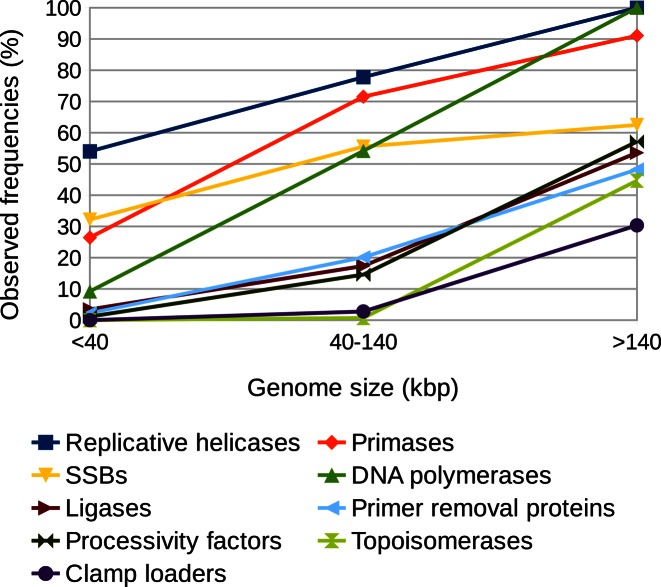
Intuitively, the completeness of the viral DNA replication machinery should correlate with the increased coding capacity of viral genome. Therefore, we next asked whether this is true and if so, how strong is this correlation. We divided dsDNA viruses into three genome size groups (<40 kbp, 40–140 kbp and >140 kbp) in line with our previous finding (19) that the composition of DNA replicases is genome size-dependent. For each group of genome sizes we counted frequencies of occurrence of replication proteins. It turned out that as the genome size increases dsDNA viruses tend to code their own replication proteins more often (Figure 5). Our data revealed the strong genome size dependency for all analyzed groups of DNA replication proteins, with the exception of SSB proteins (Figure 5). The reason why SSB proteins show only modest genome size dependency is that we did not detect SSBs in a number of large eukaryotic viruses of the Alloherpesviridae, Malacoherpesviridae, Nimaviridae, Nudiviridae, Hytrosaviridae, Asfarviridae families and phiKZ-like phages. This is rather surprising, because SSB proteins are found in all cellular and many (semi)autonomous viral DNA replication machineries (e.g. phages T7, T4, herpes, baculo and pox viruses). Since the identification of OB-fold proteins from sequence data is notoriously difficult, it is quite likely that at least some SSB proteins may have escaped detection. Another possible cause might be the non-orthologous displacement of canonical OB-fold SSBs. These considerations are supported by our previous discovery of distant homologs of T7 SSB in nucleo-cytoplasmic large DNA viruses (49) and examples of unique SSBs in poxviruses, phages and archaeal viruses (see above).
Figure 5.
Relationship between the observed frequencies of viral DNA replication proteins and the genome size of dsDNA viruses.
As expected from the determined trend, the most complete DNA replication machineries are typically present in viruses with large genomes such as the largest eukaryotic viruses (Megavirales group, except Ascoviridae family), T4-like and some other large phages and archaeal virus HVTV-1 (Supplementary Table S3).
Certain functional categories of DNA replication proteins are tightly coupled
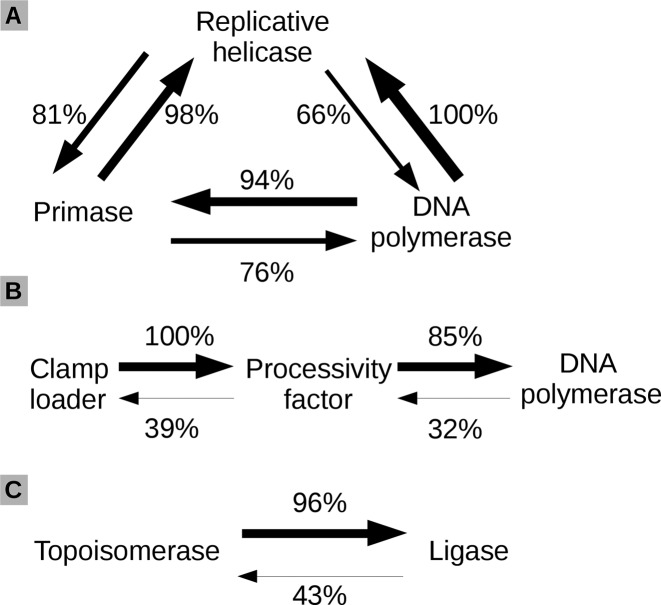
Given that complete virus-encoded replisomes are exclusive to viruses with large genomes (see above), for viruses with small and moderate genome size, the missing replication proteins have to be provided by the host cell. Thus, we asked whether virus-encoded and host-encoded replication proteins are combined at random or not. To answer this question, we investigated the correlation between the presence/absence of genes coding for DNA replication proteins in viral genomes. Certain functional categories showed strong, largely unidirectional, co-occurrence patterns indicating that the encoded complement of viral replication proteins is not random. One of the highly correlated groups involves replicative helicases, primases and DNA polymerases. Our data show that viruses which encode a DNA polymerase always have a replicative helicase (Figure 6A) and nearly always a DNA primase (98% of the cases). An ultimate testament for the coupling of the three components is presented by the two cases of the helicase, primase and polymerase fusion within the same polypeptide. One of them comprises proteins encoded by Lactococcus and Rhodococcus phages and exemplified by gp55 of Lactococcus phage 1706 (gi: 182637532), which encompasses SF3 helicase, AEP primase and PolB. Another case is exemplified by the Pas55 protein of Actinoplanes phage phiAsp2 (gi: 48697456) featuring SF3 helicase fusion with DnaG primase and PolA (Figure 2). Nevertheless, there are several notable cases of primases being absent, including phiKZ-like phages, halovirus HGTV-1 and Pseudomonas phage 119X (Supplementary file 1). In general, proteins of these viruses are highly diverged, thus it is possible that DNA primases were missed. Alternatively, these viruses might use non-canonical priming mechanisms. The presence of DNA primase is a strong indicator that a replicative helicase is also present. These two proteins are often fused together into the same polypeptide chain (41% of replicative helicases) or are encoded nearby. Almost universally (94% of cases) the primase domain appears N-terminal to the helicase module (Supplementary file 2). We observed that phages with replicative helicases of the SF4 superfamily more often code for a DNA primase as a separate gene (similarly to DnaB and DnaG in bacteria) rather than as a primase-helicase fusion (like in phage T7). Our data also show that typically DnaG primases co-occur with SF4 replicative helicases while AEPs co-occur with SF3 helicases. Alternative combinations of helicase-primase types are less common. We found SF4 replicative helicases together with AEP in only 19% of cases and SF3 with DnaG only in 14% of cases. Replicative helicases from SF6 and SF2 do not seem to show preference for a specific type of primase. SF1 replicative helicases were found only together with AEP primases. However, replicative helicases of this superfamily were only detected in herpes viruses and TvV-S1 virus. Thus, it is perhaps too early to draw firm conclusions about their preferences.
Figure 6.
Associations between DNA replication proteins based on their presence/absence in dsDNA virus genomes. An arrow pointing from the protein indicates the percent of cases when this protein is encoded together with the protein at the tip of the arrow.
Another strong, unidirectional functional link is observed between DNA polymerases, their processivity factors (DNA clamps) and clamp loaders (Figure 6B). All viruses that encode clamp loaders also have processivity factors, but less than half of the processivity factors are accompanied by clamp loaders. This apparent discrepancy can be explained by the observation that some viral processivity factors do not form closed rings (i.e. herpes UL42 and UL44 (64)) and therefore do not need to be actively loaded by clamp loaders. The presence of a processivity factor typically implies that the DNA polymerase gene is also present (85% of cases). Interestingly, viral-specific families of processivity factors (homologs of T4 gp45, herpesviral UL42, UL44, BMRF1 and poxviral A20) always co-occur with DNA polymerases. Only homologs of cellular processivity factors (PCNA and PolIIIβ) sometimes break this rule.
We were surprised to find out that the presence/absence of topoisomerases and ligases is also correlated. Although it is known that TopoIIA can function on its own (65), we found that all viruses (except Synechococcus phage S-TIM5) possessing TopoIIA also encode a DNA ligase (Figure 6C). Even the case of S-TIM5 is not a true exception: of the two subunits (GyrA and GyrB) of the bacterial-type TopoIIA, S-TIM5 has only GyrA (gi: 422936149). Another interesting coincidence is observed in Pandoraviruses, the largest known viruses. These viruses lack the canonical genes for both DNA topoisomerases and DNA ligases (66). These observations suggest that the actions of DNA ligases and topoisomerases in dsDNA viruses might be closely coordinated. Studies in vaccinia virus provide indirect support for such a coordination by showing that the viral DNA ligase interacts with cellular TopoIIA (II[α/β]) and directs it to the virus replication and assembly sites (67).
DISCUSSION
One of the outstanding questions in Biology is the origin of the two evolutionarily distinct DNA replication systems in bacteria and archaea/eukaryotes, respectively (4,68). Some of the core components in the two systems are either unrelated (DNA primases and replicative DNA polymerases) or represent homologous but apparently non-orthologous proteins (e.g. replicative DNA helicases) (4). Our results showing the relative abundance and distribution of major functional groups of DNA replication proteins among viruses infecting hosts from different domains of life might hold a clue to this riddle and shed light on the evolution of not only viral but also cellular DNA replication systems. Indeed, the potential role of viruses in the evolution of cellular DNA replication systems and possibly in the origin of the DNA itself has been recognized previously (4,68–70).
Particularly unexpected result of our analysis is that nearly all major DNA replication proteins that are generally considered to be specific to archaeal and eukaryotic systems are widespread in viruses infecting bacteria. We found that B-family DNA polymerases and AEP primases, which are specific to replication machineries of archaea and eukaryotes, are conserved in dsDNA viruses from all three domains of life. By contrast, homologs of bacterial replicative polymerase (C-family) and DnaG-like primase are confined to bacteriophages. Similarly, DNA polymerases of the A-family are largely limited to bacteriophage genomes, with a notable exception of virophages, a group of viruses that for propagation depend on large eukaryotic viruses of the family Mimiviridae.
Unwinding of the DNA duplex during the replication in bacteria and archaea/eukaryotes is performed by helicases that belong to two different superfamilies, SF4 (DnaB-like) and SF6 (MCM-like), respectively. Although conspicuously absent in eukaryotic viruses, replicative SF6 helicases are encoded in both archaeal and bacterial viruses, whereas SF4 helicases, despite constituting the most populous group of identified replicative helicases, are nearly exclusively found in bacterial viruses.
A similar trend whereby replication proteins of the archaeal/eukaryotic origin are found in bacterial viruses can be also extended to the DNA polymerase accessory proteins, DNA sliding clamps and clamp loaders. If we consider the latter proteins at the family level, it appears that dsDNA viruses are just a reflection of their hosts, i.e., PCNA and RFC are identified in archaeal and eukaryotic viruses, whereas PolIIIβ and subunits of γ/τ-complex are only observed in bacteriophages. However, if we look at distant evolutionary relationships that are only apparent at the structural level, the picture becomes different. DNA sliding clamp subunits of T4-like phages structurally are closer to PCNA rather than to PolIIIβ. More importantly, they have the same two-domain architecture as PCNA and not the three-domain architecture as PolIIIβ. In addition, the recently solved crystal structure of T4 clamp loader (gp44/62) revealed that it represents a minimal version of the archaeal/eukaryotic RFC clamp loader (71). Herpesviral clamp proteins that bind DNA directly and therefore do not need clamp loaders (72) are structurally also more similar to PCNA than to PolIIIβ. All these observations combined with our data indicate the universal spread of viral versions of archaeal/eukaryotic clamps and clamp loaders.
Finally, archaeal/eukaryotic proteins participating in the lagging strand synthesis, most notably ATP-dependent DNA ligase, are also prevalent in bacteriophage genomes. Taken together, our results reveal that bacteriophages, in particular members of the order Caudovirales, besides the bacteria-specific DNA replication proteins encode nearly entire suite of proteins thought to be specific to archaeal and eukaryotic systems, including B-family DNA polymerases, AEP primases, SF6 replicative helicases, PCNA-like clamps and RFC-like clamp loaders, as well as ATP-dependent DNA ligases. Although the corresponding genes in bacteriophage genomes are distributed sporadically and are often intermixed with genuine bacterial-like replisome components, some of the functional categories in their frequency match (e.g., AEP) or even surpass (PolB and ATP-dependent ligases) their bacterial counterparts. What could be the origin of these proteins in bacteriophage genomes and what does this tell us about the evolution of cellular DNA replication machineries? Phylogenetic analysis of PolB and AEP proteins shows that bacteriophage-encoded homologs are deeply branched within the respective phylogenies. This strongly suggests that these and, possibly, other archaeo-eukaryotic replication protein genes have not been acquired by bacteriophages from their contemporary hosts. The alternative possibility that there has been trans-domain host switch whereby an archaeal or eukaryotic virus evolved to infect a bacterial host appears highly unlikely. First, most viruses have a rather narrow host range and interdomain virus transfers have never been reported. Second, the archaeo-eukaryotic replication proteins are widespread and abundant in bacteriophages indicating that their presence is not spurious but is rather a reflection of a long evolutionary history of the corresponding genes in phage genomes. It is thus conceivable that bacterial viruses that encode the archaeo-eukaryotic complement have acquired these genes prior to the divergence of cellular organisms into the three contemporary domains of life. The ancient origin of Caudovirales draws independent support from the studies on the structure and assembly of their virions. It has been demonstrated that bacterial members of the Caudovirales are evolutionarily related to tailed dsDNA viruses infecting archaea (31) and to eukaryotic members of the order Herpesvirales (73). The outstanding question then is what was the composition of the DNA replication apparatus in the Last Universal Cellular Ancestor (LUCA)? Several alternative scenarios have been proposed over the years (4,68–70,74). One of the possibilities is that LUCA had either proto-archaeal or proto-bacterial DNA replication system and subsequent non-orthologous gene displacement of the key components in one of the primary cellular lineages resulted in the current dichotomy (68,70). However, which of the two DNA replication systems is ancestral remains unclear. The following observation might be relevant for addressing this conundrum: bacteriophages encompass both bacterial and archaeal/eukaryotic DNA replication systems as well as specific DNA replication systems not found in cellular organisms, whereas archaeal and eukaryotic viruses are largely devoid of bacteria-specific DNA replication proteins. Such lack of bacterial-like replication proteins in archaeal and eukaryotic viruses suggests that these proteins have evolved in the context of bacterial viruses following their divergence from viruses of archaea and eukaryotes. Based on these observations we suggest that LUCA had the archaeo-eukaryotic-like replication machinery which is universally present in contemporary viruses infecting hosts from all three domains of life.
Of special interest are DNA replication proteins that are exclusive to viruses. Although many virus-specific replication proteins display very narrow distribution and are restricted to a particular group of viruses, particularly among archaeal viruses, some are nearly ubiquitous. Most notable among these are SF3 helicases and T7-like SSB. SF3 helicases are widespread in viruses infecting hosts in all three domains of life. It is intriguing that viral SF3 helicases tend to co-occur or be fused with AEP primases, another ‘universal’ viral protein. Similarly, T7-like SSB, featuring a distinct OB-fold domain, is present in both bacteriophages and eukaryotic viruses of the order Megavirales (49). Moreover, an OB-domain most similar to that of T7-like SSB is present in ICP8, a large multidomain SSB protein of herpesviruses. Given that so far no OB-domain containing SSBs have been identified in archaeal viruses, T7-like SSB appears to represent the most widely distributed viral SSB.
There is a clear correspondence between the viral genome size and the completeness of the DNA replication machinery encoded by a viral genome. Thus, only viruses with the largest genomes can provide (nearly) all components required for their genome replication. The majority of viruses, however, have to recruit certain components from the host. In this respect, our data showing that helicases are the most frequently encoded viral DNA replication proteins is somewhat unexpected. Not only are these proteins abundant but are also very diverse. While cellular organisms for genome replication utilize helicases of only two different superfamilies, viruses make use of enzymes falling into five of the six known helicase superfamilies. The ubiquity of viral helicases implies that these proteins are best suited for recruitment of the lacking replisome components from the host. This might be explained by the fact that replicative helicases are the first of the replication fork proteins loaded onto origins of replication (75). Furthermore, helicases might also be involved in recognition of replication origins (76) through the fusion with DNA-binding domains (Figure 2). Thus, it appears that a replicative helicase represents an optimal solution for efficient assembly of the whole replisome on the viral DNA template. Consistently, we found that viruses which encode their own primases and/or DNA polymerases almost always (98 and 100% of the cases) encode also a helicase. Interestingly, the inverse co-occurrence is not nearly as stringent, i.e., viruses that encode replicative helicases might lack the genes for viral primases and DNA polymerases and likely rely on the corresponding proteins of the host. This suggests that viral versions of primases and DNA polymerases are necessarily recruited to the replication fork by the viral helicase.
Strong co-evolution is also observed among DNA polymerases and their accessory proteins. Clamp loaders are never encoded without their cognate clamps suggesting that they co-evolve as a functional module. The same is true for viral-specific families of processivity factors and DNA polymerases, but not for homologs of cellular processivity factors (PCNA and PolIIIβ) which sometimes can be encoded in a viral genome in the absence of the DNA polymerase gene. Since processivity factors cannot be expected to distinguish viral from cellular DNA, this raises a question as to the role of such stand-alone PCNA and PolIIIβ homologs. These exceptions notwithstanding, the co-occurrence patterns are a direct reflection of the steps in the assembly of DNA replicase: the clamp loader loads the clamp, which in turn tethers DNA polymerase to the DNA enabling processive DNA synthesis. Less clear is the importance of the observed co-occurrence between topoisomerases and DNA ligases. Although direct experimental data on this issue is lacking, our data strongly suggest that viral topoisomerases and DNA ligases are linked functionally and perhaps physically.
These and other patterns identified in our study reveal the fundamental logic of viral DNA replication that seems to be applicable to dsDNA viruses infecting hosts in all domains of life. Moreover, the uncovered patterns may be useful for estimating DNA replication complement of new viral genomes or guiding the search for certain components. Thus, the genome size may already give a clue as to which DNA replication proteins to expect. Similarly, the presence of proteins in specific functional categories may encourage the search for the ‘missing’ components. For example, a DNA sliding clamp may be difficult to identify from the sequence data alone as it is not an enzyme and therefore does not feature a conserved active site. However, if the viral genome encodes a clamp loader, which is relatively easy to detect due to the conserved ATP-binding motifs, this is a strong indication that the clamp should also be present. The newly detected phage PAU homolog of the T4 DNA sliding clamp in the present study is an example of practical application of this idea.
Supplementary Material
FUNDING
European Social Fund under the Global Grant measure [VP1-3.1-ŠMM-07-K-03-004]. Funding for open access charge: Research Council of Lithuania.
Conflict of interest statement. None declared.
REFERENCES
- 1.Forterre P., Prangishvili D. The major role of viruses in cellular evolution: facts and hypotheses. Curr. Opin. Virol. 2013;3:558–565. doi: 10.1016/j.coviro.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Koonin E.V., Dolja V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013;3:546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle C.A. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Leipe D.D., Aravind L., Koonin E.V. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornberg A., Baker T.A. Dna Replication. Sausalito: University ScienceBooks; 2005. [Google Scholar]
- 6.Salas M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 7.DePamphilis M., Bell S. Genome Duplication. London: Garland Science; 2010. [Google Scholar]
- 8.Federici B.A., Bigot Y. In: Evolutionary Biology - Concepts, Molecular and Morphological Evolution. Pontarotti P, editor. Berlin Heidelberg: Springer; 2010. pp. 229–248. [Google Scholar]
- 9.Frith M.C., Hamada M., Horton P. Parameters for accurate genome alignment. BMC Bioinformatics. 2010;11:80. doi: 10.1186/1471-2105-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frickey T., Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 11.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 12.Remmert M., Biegert A., Hauser A., Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 13.Eddy S.R. Accelerated profile HMM searches. PLoS. Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darriba D., Taboada G.L., Doallo R., Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazlauskas D., Venclovas Č. Computational analysis of DNA replicases in double-stranded DNA viruses: relationship with the genome size. Nucleic Acids Res. 2011;39:8291–8305. doi: 10.1093/nar/gkr564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamtekar S., Berman A.J., Wang J., Lazaro J.M., de Vega M., Blanco L., Salas M., Steitz T.A. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 21.Blanco L., Bernad A., Lazaro J.M., Martin G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 22.Kanellopoulos P.N., Tsernoglou D., van der Vliet P.C., Tucker P.A. Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein. J. Mol. Biol. 1996;257:1–8. doi: 10.1006/jmbi.1996.0141. [DOI] [PubMed] [Google Scholar]
- 23.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 24.Ilyina T.V., Gorbalenya A.E., Koonin E.V. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J. Mol. Evol. 1992;34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 25.Scherzinger E., Ziegelin G., Barcena M., Carazo J.M., Lurz R., Lanka E. The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J. Biol. Chem. 1997;272:30228–30236. doi: 10.1074/jbc.272.48.30228. [DOI] [PubMed] [Google Scholar]
- 26.Halgasova N., Mesarosova I., Bukovska G. Identification of a bifunctional primase-polymerase domain of corynephage BFK20 replication protein gp43. Virus Res. 2012;163:454–460. doi: 10.1016/j.virusres.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Moss B. Poxvirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a010199. doi: 10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeo H.J., Ziegelin G., Korolev S., Calendar R., Lanka E., Waksman G. Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins. Mol. Microbiol. 2002;43:855–867. doi: 10.1046/j.1365-2958.2002.02796.x. [DOI] [PubMed] [Google Scholar]
- 29.Koonin E.V., Dolja V.V., Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479C-480C:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer L.M., Abhiman S., Aravind L. A new family of polymerases related to superfamily A DNA polymerases and T7-like DNA-dependent RNA polymerases. Biol. Direct. 2008;3:39. doi: 10.1186/1745-6150-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krupovic M., Forterre P., Bamford D.H. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 2010;397:144–160. doi: 10.1016/j.jmb.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Gorlas A., Koonin E.V., Bienvenu N., Prieur D., Geslin C. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ. Microbiol. 2012;14:503–516. doi: 10.1111/j.1462-2920.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krupovic M., Gribaldo S., Bamford D.H., Forterre P. The evolutionary history of archaeal MCM helicases: a case study of vertical evolution combined with hitchhiking of mobile genetic elements. Mol. Biol. Evol. 2010;27:2716–2732. doi: 10.1093/molbev/msq161. [DOI] [PubMed] [Google Scholar]
- 34.Guinta D., Stambouly J., Falco S.C., Rist J.K., Rothman-Denes L.B. Host and phage-coded functions required for coliphage N4 DNA replication. Virology. 1986;150:33–44. doi: 10.1016/0042-6822(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 35.Yutin N., Wolf Y.I., Raoult D., Koonin E.V. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kazlauskas D., Venclovas Č. Herpesviral helicase-primase subunit UL8 is inactivated B-family polymerase. Bioinformatics. 2014;30:2093–2097. doi: 10.1093/bioinformatics/btu204. [DOI] [PubMed] [Google Scholar]
- 37.Kristensen D.M., Waller A.S., Yamada T., Bork P., Mushegian A.R., Koonin E.V. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J. Bacteriol. 2013;195:941–950. doi: 10.1128/JB.01801-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigel C., Seitz H. Bacteriophage replication modules. FEMS Microbiol. Rev. 2006;30:321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 39.Mardanov A.V., Strakhova T.S., Ravin N.V. RepA protein of the bacteriophage N15 exhibits activity of DNA helicase. Dokl. Biochem. Biophys. 2004;397:217–219. doi: 10.1023/b:dobi.0000039467.76197.d9. [DOI] [PubMed] [Google Scholar]
- 40.Ziegelin G., Tegtmeyer N., Lurz R., Hertwig S., Hammerl J., Appel B., Lanka E. The repA gene of the linear Yersinia enterocolitica prophage PY54 functions as a circular minimal replicon in Escherichia coli. J. Bacteriol. 2005;187:3445–3454. doi: 10.1128/JB.187.10.3445-3454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer L.M., Koonin E.V., Leipe D.D., Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanitsa E.S., Arps L., Traktman P. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 2006;281:3439–3451. doi: 10.1074/jbc.M511239200. [DOI] [PubMed] [Google Scholar]
- 43.Murzin A.G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo. J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raghunathan S., Ricard C.S., Lohman T.M., Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollis T., Stattel J.M., Walther D.S., Richardson C.C., Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shamoo Y., Friedman A.M., Parsons M.R., Konigsberg W.H., Steitz T.A. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 47.Mapelli M., Panjikar S., Tucker P.A. The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding. J. Biol. Chem. 2005;280:2990–2997. doi: 10.1074/jbc.M406780200. [DOI] [PubMed] [Google Scholar]
- 48.Bochkareva E., Korolev S., Lees-Miller S.P., Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO. J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazlauskas D., Venclovas Č. Two distinct SSB protein families in nucleo-cytoplasmic large DNA viruses. Bioinformatics. 2012;28:3186–3190. doi: 10.1093/bioinformatics/bts626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi M., Miller A., Cho N.Y., Rothman-Denes L.B. Identification, cloning, and characterization of the bacteriophage N4 gene encoding the single-stranded DNA-binding protein. A protein required for phage replication, recombination, and late transcription. J. Biol. Chem. 1995;270:22541–22547. doi: 10.1074/jbc.270.38.22541. [DOI] [PubMed] [Google Scholar]
- 51.Yu M., Carstens E.B. Identification of a domain of the baculovirus Autographa californica multiple nucleopolyhedrovirus single-strand DNA-binding protein LEF-3 essential for viral DNA replication. J. Virol. 2010;84:6153–6162. doi: 10.1128/JVI.00115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y., Kragelund B.B., White M.F., Peng X. Functional characterization of a conserved archaeal viral operon revealing single-stranded DNA binding, annealing and nuclease activities. J. Mol. Biol. 2015;427:2179–2191. doi: 10.1016/j.jmb.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shuman S. DNA ligases: progress and prospects. J. Biol. Chem. 2009;284:17365–17369. doi: 10.1074/jbc.R900017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yutin N., Koonin E.V. Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: a case of hidden complexity. Biol. Direct. 2009;4:51. doi: 10.1186/1745-6150-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forterre P., Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banach-Orlowska M., Fijalkowska I.J., Schaaper R.M., Jonczyk P. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol. Microbiol. 2005;58:61–70. doi: 10.1111/j.1365-2958.2005.04805.x. [DOI] [PubMed] [Google Scholar]
- 59.Filee J., Forterre P., Sen-Lin T., Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 60.Makarova K.S., Krupovic M., Koonin E.V. Evolution of replicative DNA polymerases in archaea and their contributions to the eukaryotic replication machinery. Front Microbiol. 2014;5:354. doi: 10.3389/fmicb.2014.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villarreal L.P., DeFilippis V.R. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Virol. 2000;74:7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krupovic M., Prangishvili D., Hendrix R.W., Bamford D.H. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 2011;75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pina M., Basta T., Quax T.E., Joubert A., Baconnais S., Cortez D., Lambert S., Le Cam E., Bell S.D., Forterre P., et al. Unique genome replication mechanism of the archaeal virus AFV1. Mol. Microbiol. 2014;92:1313–1325. doi: 10.1111/mmi.12630. [DOI] [PubMed] [Google Scholar]
- 64.Weller S.K., Coen D.M. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb. Perspect Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt B.H., Osheroff N., Berger J.M. Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nat. Struct. Mol. Biol. 2012;19:1147–1154. doi: 10.1038/nsmb.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Philippe N., Legendre M., Doutre G., Coute Y., Poirot O., Lescot M., Arslan D., Seltzer V., Bertaux L., Bruley C., et al. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 67.Lin Y.C., Li J., Irwin C.R., Jenkins H., DeLange L., Evans D.H. Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J. Virol. 2008;82:5922–5932. doi: 10.1128/JVI.02723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forterre P. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 1999;33:457–465. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]
- 69.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 70.Koonin E.V. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol. Direct. 2006;1:39. doi: 10.1186/1745-6150-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelch B.A., Makino D.L., O'Donnell M., Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuccola H.J., Filman D.J., Coen D.M., Hogle J.M. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]
- 73.Rixon F.J., Schmid M.F. Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr. Opin. Virol. 2014;5:105–110. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 74.Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bell S.P., Kaguni J.M. Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect Biol. 2013;5:a010124. doi: 10.1101/cshperspect.a010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costa A., Onesti S. The MCM complex: (just) a replicative helicase? Biochem. Soc. Trans. 2008;36:136–140. doi: 10.1042/BST0360136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.