Abstract
Sequence-specific binding to DNA is crucial for targeting transcription factor-DNA complexes to modulate gene expression. The heterocyclic diamidine, DB2277, specifically recognizes a single G•C base pair in the minor groove of mixed base pair sequences of the type AAAGTTT. NMR spectroscopy reveals the presence of major and minor species of the bound compound. To understand the principles that determine the binding affinity and orientation in mixed sequences of DNA, over thirty DNA hairpin substrates were examined by NMR and thermal melting. The NMR exchange dynamics between major and minor species shows that the exchange is much faster than compound dissociation determined from biosensor–surface plasmon resonance. Extensive modifications of DNA sequences resulted in a unique DNA sequence with binding site AAGATA that binds DB2277 in a single orientation. A molecular docking result agrees with the model representing rapid flipping of DB2277 between major and minor species. Imino spectral analysis of a 15N-labeled central G clearly shows the crucial role of the exocyclic amino group of G in sequence-specific recognition. Our results suggest that this approach can be expanded to additional modules for recognition of more sequence-specific DNA complexes. This approach provides substantial information about the sequence-specific, highly efficient, dynamic nature of minor groove binding agents.
INTRODUCTION
Targeting transcription factor (TF)-DNA complexes for regulation of gene expression is very attractive for the treatment of a number of diseases (1–3). This could be done by targeting the TF or the DNA binding site (4). It has proven difficult to target TFs directly and they are frequently considered ‘undruggable’ (5). The alternative of targeting the DNA binding site of the TF with designed small molecules is, thus, gaining increasing attention (6–8). A barrier to this approach for control of gene expression in cells is that only a limited variety of small molecules bind strongly and sequence-specifically to DNA (9,10). The modification of heterocycles in classical type minor groove binders is an approach to expand the applications of minor groove binders (11,12).
The greatest variety of A•T specific minor groove binding compounds such as netropsin, DAPI, pentamidine, furamidine and Hoechst 33258 and its analogs are a complement to the highly sequence-specific hairpin polyamides (13). Many of the A•T specific compounds are taken up by cells and have a unique variety of therapeutic as well as biotechnology applications (14,15). Non-covalent binding minor groove agents have been used in therapeutic targeting of DNA in various types of cells with successful application all the way to clinical trials (16,17). As described above, many minor groove binding compounds are highly A•T specific (18) but it is essential to be able to better target mixed, A•T and G•C base pair (bp) sequences (19,20). Such novel compounds would be useful for a new class of potential drug molecules that can target promoter DNA sequences to modulate TF binding (21–23). In order to expand past the pure A•T specific compounds, new types of compounds must be designed (24–26). These modified compounds still require the correct curvature and charge to match the minor groove but they will also need functional groups that have H-bond acceptors for favorable interactions with the G-NH2 group that projects into the minor groove and presents a steric block to strong binding by A•T specific minor groove binders (27,28).
Our understanding of G•C bp recognition by small molecule minor groove binders is very limited, and for use of the DNA minor groove for new drug design and development we need a more detailed understanding of compound-DNA complexes (29–31). Compounds with aza-benzimidazole H-bond acceptors have initially been designed for G•C recognition (32). DB2277 (Figure 1) is a novel example with strong binding to an AAAGTTT binding site (32,33). To fully appreciate the binding of DB2277 to its target DNA sequence, structural information is required. NMR, in conjunction with molecular modeling, is uniquely suited to study the molecular interactions and dynamic behavior of DNA-ligand complexes (34–38). An initial NMR experiment for a DB2277 complex with an AAAGTTT sequence, however, revealed the surprising result that two species are present, even in a 1:1 complex. Interestingly, these two species were not observed in other high resolution biophysical experiments (32,33). To understand these multiple binding species and their importance for compound design, we have investigated the DB2277 complex with numerous hairpin DNA duplexes.
Figure 1.
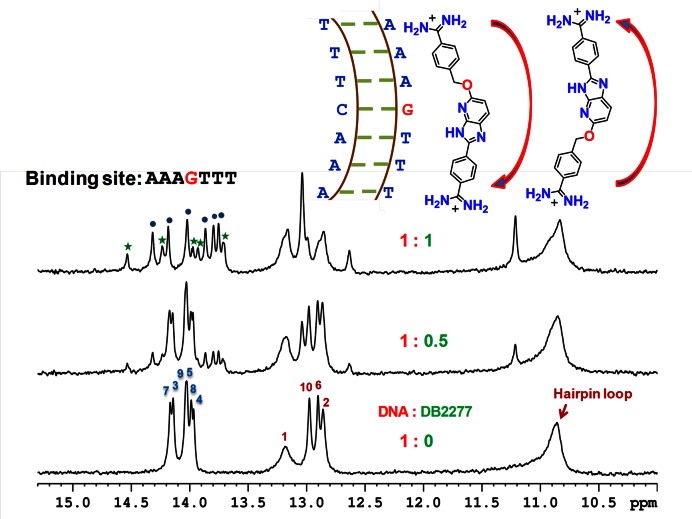
Shifts in the imino spectral peaks at different G-hp1:DB2277 ratios at 285 K. The major and minor binding species of DNA-DB2277 complex are represented by ‘ ’ and ‘
’ and ‘ ’ labels, respectively. A schematic model showing two possible orientations of the DB2277 complex with G-hp1 is at the top. The NMR sample consisted of 100 μM DNA at pH 6.9.
’ labels, respectively. A schematic model showing two possible orientations of the DB2277 complex with G-hp1 is at the top. The NMR sample consisted of 100 μM DNA at pH 6.9.
METHODS
Sample preparation and NMR experiments
A Bruker Avance 600 MHz NMR equipped with a QXI probe was used to collect NMR data. The NMR samples consisted of 100 μM DNA, 10 mM cacodylate buffer (cacodylic acid/NaOH at pH 7.0), EDTA (ethylene diamine tetra-acetic acid) and DSS (4, 4-dimethyl-4-silapentane-1-sulfonic acid) as an internal standard. In experiments involving exchangeable protons 90% H2O–10% D2O was used (39). The pH of the solution was adjusted in the range 6.6–6.8. The DNA concentrations determined using UV absorbance were around 0.5 mM in 1 ml ddH2O. Details about sample preparation and NMR methods are provided in Supplementary Information text.
Thermal melting
Hairpin DNA oligomers binding with DB2277 were monitored by thermal melting of the DNAs and complexes (1:2 ratio of DNA:DB2277) in Tris-HCl buffer (50 mM Tris, 100 mM NaCl, 1 mM EDTA, pH 7.4) at 260 nm wavelength as previously described (32).
Surface plasmon resonance
Biosensor–surface plasmon resonance (SPR) measurements for affinity and kinetics were performed with a four-channel Biacore T200 optical biosensor system. 5′-biotin-labeled DNA sequences (G-hp2 and G-hp5 shown in Table 1) were immobilized onto streptavidin-coated sensor chips as previously described (40,41). Standard free energy (ΔG°) is calculated using the equilibrium dissociation binding constant from SPR at room temperature (ΔG° = RT ln KD). Further information about SPR measurements is provided in Supplementary Information text.
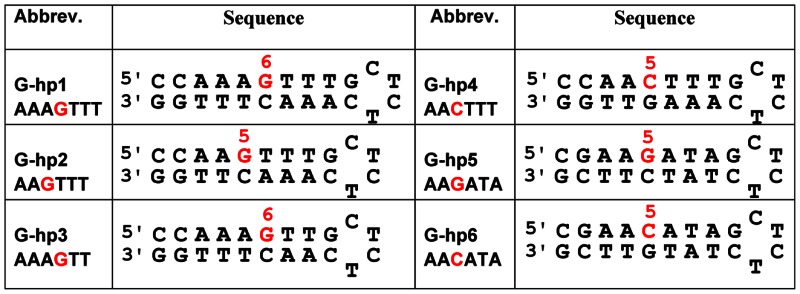
Table 1. Selected mixed-sequence hairpin DNAs.
|
Molecular modeling and docking
DB2277 was optimized at the B3LYP/6-31*G level of theory using Spartan 10 software (Wavefunction, Inc.) (42). All minimized ligands were assigned Gasteiger−Huckel charges by using Autodock vina 4.02 (43). G-hp1 duplex DNA was generated from the biopolymer-build DNA double helix from the Tripos SYBYL-X1.2 software package (44). G-hp1 was docked with minimized structure of DB2277 using AutoDock vina 4.02 (44). Further description is provided in detail in Supplementary Information text.
RESULTS AND DISCUSSION
Proton NMR spectra
The chemical shifts of DNA protons of the A•T and G•C bp have been used to monitor the binding of DB2277 with DNA. Numerous changes in the chemical shifts for DNA imino protons have been observed upon complexation with DB2277, particularly in the central region of the hairpin DNAs. We have investigated over thirty DNA sequences and results for six G-hp DNAs that illustrate different modes of sequence-specific binding of DB2277 to DNA are shown here (Table 1).
G-hp1 (AAAGTTT binding site)
The G-hp1 DNA sequence gives well-resolved spectra with six A•T and four G•C bp signals and a broad upfield band for the unpaired T loop bases (Figure 1). Assignment of the imino proton signals of free DNA shown in the Figure 1 was done using 1D-NOE experiments. Titration of G-hp1 with DB2277 revealed both upfield and downfield chemical shift changes. The addition of DB2277 to G-hp1 at the ratio of 1:0.5 (DNA:DB2277) shows both free DNA and complex resonance peaks in the spectra indicating slow exchange on the NMR timescale. No significant chemical shift changes were observed for DNA imino peaks of the terminal bp of the duplex indicating their minimal contribution to binding with DB2277. The appearance of an unexpected additional peak at 11.3 ppm was observed with the addition of DB2277. No significant changes were observed after reaching a 1:1 ratio of DNA:DB2277. At ratios above 1:1, slightly broadened peaks are probably due to some non-specific, electrostatic interaction of the excess DB2277 cation with the anionic DNA backbone. No unbound G-hp1 peaks are observed at the binding ratio of 1:1 (DNA:DB2277) suggesting a tight binding of DB2277 with the G-hp1, in agreement with SPR results (33).
For DB2277 binding with G-hp1 as a single species, six imino proton resonance peaks are expected for A•T bp at a 1:1 binding ratio. The imino proton spectra (Figure 1), however, reveal that all complex signals are doubled, even though the stoichiometry of binding is 1:1. The two sets of signals clearly indicate two binding modes for DB2277 with G-hp1, a major and a minor binding species based on signal intensity (45,39). Interestingly, other biophysical methods such as SPR, mass spectrometry, circular dichroism and isothermal titration calorimetry also indicated a 1:1 complex between DB2277 and G-hp1 (33) but these methods were not able to show any evidence of two binding orientations of DB2277. The local symmetry (similar A•T bp sequences on both sides of the central G•C bp) of G-hp1 may result in the two orientations of DB2277 with similar energy. The two modes suggest complexes with reverse orientations in the AAAGTTT sequence (Figure 1) as previously observed for the polyamide minor groove binder, netropsin, in an A•T sequence (39).
In order to understand the origin of the two bound species and to facilitate study of structural details of the DB2277-DNA complex, a more detailed analysis of DB2277 binding to different DNA sequences was undertaken. It is important to understand the origin of the different compound orientations and how they can be modulated by DNA sequences. It seemed possible that formation of a unique complex could be achieved with DNA sequences that are less symmetric than the AAAGTTT motif in G-hp1.
G-hp2 (AAGTTT binding site)
A 5′ A•T bp in G-hp1 (G-hp2) was removed to give a less symmetric binding site. The question to be addressed is whether changes of this type will give a single DB2277-DNA species with good binding affinity? Imino proton spectra of G-hp2 show one less A•T bp resonance peak (Supplementary Figure S1). At a 1:1 binding ratio of DNA:DB2277, the fraction of the minor species was reduced, but not eliminated relative to the G-hp1-DB2277 complex. The largest differences in chemical shifts between the major and minor species are observed for A4 and T6 imino protons which are adjacent to the central G. The chemical shift of T6 signal for the major species is downfield of A4 signal. It is very interesting that the minor species T6 signal is the most upfield while the minor species A4 is very downfield. In other words, the protons shift position is in an opposite way for these major and minor species, in agreement with an opposite orientation of the DB2277 for two species evident in the flipping exchange model (Figure 1). In addition, the chemical shifts for the major binding species are similar to the major species in G-hp1 complex but with one less A•T bp signal. The less symmetric binding site reduces the amount of minor species. This result indicates that the less symmetric binding site in G-Hp2 relative to G-Hp1 is an advantage for obtaining a single species. While this may seem obvious for asymmetric sites, most of the asymmetric sequences shown and not shown in this report of over thirty sequences we investigated, do not yield a single binding species.
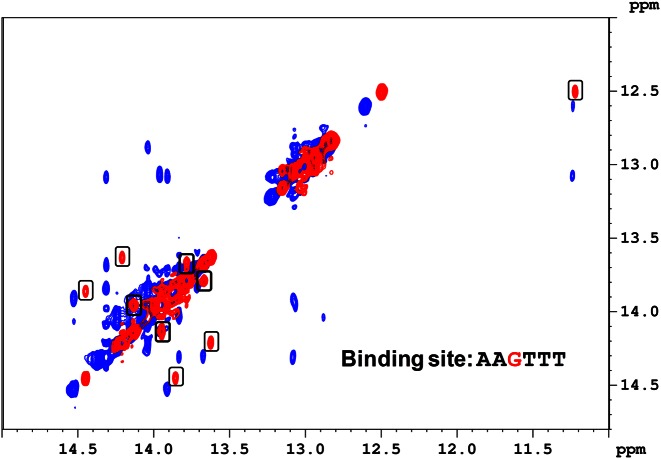
Assignment of exchangeable protons of the G-hp2-DB2277 complex was done using 1D-NOE and 2D NOESY experiments (Supplementary Figures S2–S4). The ambiguity with the imino proton assignment due to exchange peaks was resolved by performing 2D EXSY experiments (Figure 2, Supplementary Figures S5 and S12). 2D EXSY experiments are similar to very short mixing time 2D NOESY. They are useful to define which 2D cross peaks are due to exchange. Intense exchange cross peaks in the EXSY experiment showing exchange between major and minor species and NOE-build up from adjacent bp in NOESY experiment are clearly discernible in Figure 2.
Figure 2.
Overlay of imino proton region of the NOESY spectrum at 285 K (in blue) and the EXSY spectrum (in red) at 303 K of G-hp2-DB2277 complex (EXSY experiment conducted at short mixing time 50 ms whereas NOESY experiment was conducted at 150 ms mixing time). Cross peaks in boxes show exchange between major and minor species. Shifts in imino proton peaks of NOESY and EXSY are due to the temperature difference.
G-hp3 (AAAGTT binding site)
Since the removal of a 5′ A•T bp in G-hp1 improved the imino proton spectra, in the next step a 3′ terminal T•A bp was deleted from G-hp1 to remove sequence similarity in a different way. Despite similar imino proton spectra of free G-hp2 and G-hp3 DNA, their titration spectra with DB2277 were quite different (Supplementary Figures S1 and S6 respectively). Surprisingly, the change at the 3′ end of the binding site in G-hp3 resulted in sharp peaks with a significant increase in signal intensity of the minor species as compared to the G-hp2 complex. Clearly, the 3′ TTT side of the binding site is critical for the major species while the minor species is more sensitive to the 5′ AAA sequence. These observations show that the amount of the minor species is very sequence-dependent, even for two asymmetric sequences, AAGTTT and AAAGTT. Thus, the two binding orientations of DB2277 are quite sensitive to the A•T bp sequence flanking the central G•C bp.
G-hp4 (AACTTT binding site)
A different possible method to improve the results for G-hp2 is to invert the central 5′ G•C bp, again with the goal of reducing the minor species. It is surprising that this change in the central G•C bp causes the minor species to significantly increase relative to the G-hp2 complex (Supplementary Figure S7) These results indicate that the DB2277 complex is also very sensitive to the orientation of the central G•C bp. The NMR results with G-hp1, 2, 3 and 4 with DB2277 illustrate the extreme sensitivity of the complex to the DNA sequence.
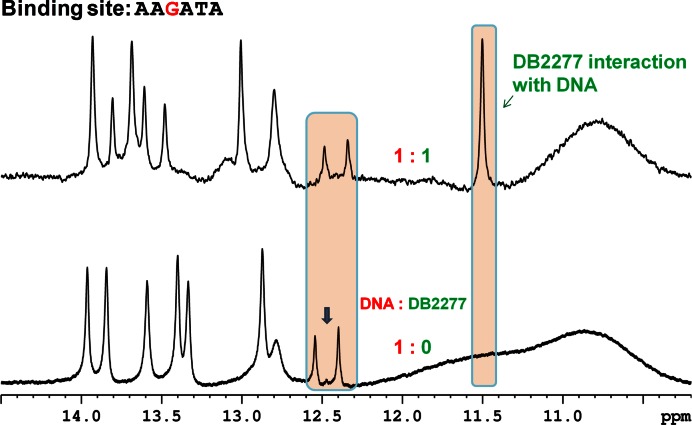
G-hp5 (AAGATA binding site)
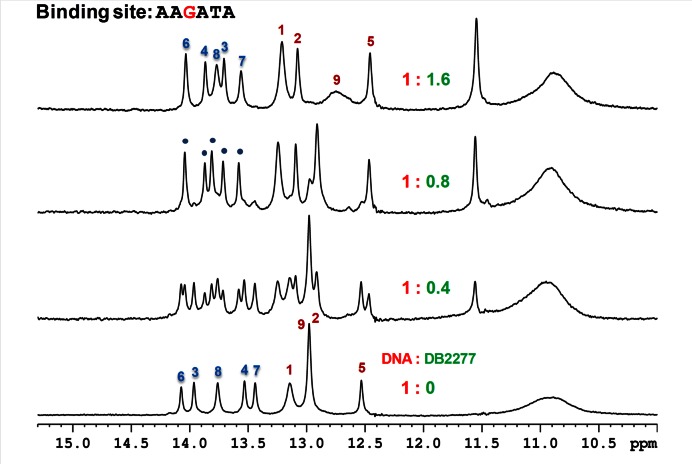
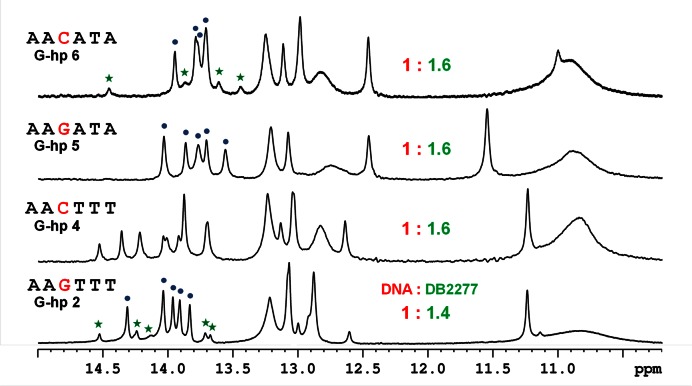
The 3′ TTT sequence clearly has a major role in directional binding with DB2277 since the removal of the T in G-hp3 causes large changes in the spectra. In G-hp5, the 3′ TTT was changed to ATA and this change yielded a single set of peaks for the complex at a 1:1 ratio. Examination of several 3′ modifications that reduce the local symmetry in the central region in different ways (data not shown) helped to pave the way towards a 3′ ATA change. At the binding ratio of 1:0.5 (DNA:DB2277), double imino resonance peaks for the five A•T bp imino protons reveal the occurrence of one binding species and the free DNA. The almost equal intensity of these binding species shows two equally populated states. Single bp resonance peaks in the imino spectral region in the G-hp5-DB2277 complex at the 1:1 binding ratio show a single binding orientation (Figure 3). The imino proton assignment of the G-hp5-DB2277 complex was done using a 2D NOESY experiment (Supplementary Figure S8). The G-hp5-DB2277 complex at the ratio of 1:1 provides single directional binding as desired. The reduction of minor species in G-hp2 and single binding species in G-hp5 is clearly seen in the spectral comparisons in Figure 4. Decrease in intensity of the peak at 14.5 ppm can be used as a probe to observe the reduction of the minor species from the G-hp1 complex to G-hp5 complex.
Figure 3.
Shifts in the imino spectral region peaks at different G-hp5:DB2277 ratios at 285 K. A single binding orientation of DNA-DB2277 complex is represented by the labeled peaks. The NMR sample consisted of 100 μM DNA at pH 6.7.
Figure 4.
Comparison of imino proton peaks of G-hp DNA-DB2277 complexes at 285 K.
G-hp6 (AACATA binding site)
As observed with G-hp4, inversion of the central G•C in G-hp5 again results in the appearance of significant minor species signals in the spectra of DB2277-DNA complex (Supplementary Figure S9). Therefore, the AAGATA binding site (G-hp5) sequence is the only one among over thirty oligomer hairpin duplexes investigated that provided a single binding orientation with DB2277. The success with G-hp5 relative to G-hp1 is due to two sequence changes. Removal of a 5′ A base to give a less symmetric binding sequence clearly reduces the minor species. The reason for the decrease of the minor form when the 3′ TTT is changed to ATA is less clear. Subtle changes in DNA microstructure, base substituent position and fit of DB2277 to the minor groove must be responsible for an enhancement of the major species. The DB2277 complex with the AAGATA sequence is now being used for high resolution NMR analysis of the structural details of the single preferred DB2277 complex.
Duplex DNA
To test the effects of the hairpin DNA and hairpin loop, a duplex DNA (Supplementary Figure S14) with same binding site as G-hp5 (AAGATA), was prepared (strand hybridization). The binding of DB2277 with this duplex DNA is observed using imino proton NMR and the results are very similar to those with the hairpin DNA. DB2277 binds with this duplex DNA as a single species and six imino proton resonance peaks are observed as expected for A•T bp at a 1:1 binding ratio (Supplementary Figure S14). No minor species is seen in both G-hp5 and duplex DNA complex spectra. Hairpin sequences are primarily used for SPR experiments where long continuous flow can cause duplex dissociation.
Another duplex DNA (Supplementary Figure S15), twice as long as the hairpin DNA with same binding site as G-hp1 (AAAGTTT), was also prepared (strand hybridization). DB2277 binding with this duplex DNA is shown in imino proton NMR spectra in the Supplementary Figure S15 but the spectra are broadened due to the significant molecular weight increase. Close observation of spectra reveals the presence of more than seven A•T bp resonances at binding ratio of 1:1, as expected. These set of signals suggest the two binding modes of DB2277 with binding site of AAAGTTT, as also clearly observed in binding spectra of DB2277 with G-hp1 (Figure 1). Moreover, the observation of the aza-benzimidazole NH proton resonance around 11.5 ppm is also consistent with all G-hp and duplex DNA sequences.
15N-labeled central G in G-hp5
To assign the peak at 11.5 ppm, a G-hp5 sequence was synthesized having the central G fully labeled with 15N. The proton spectra of 15N-labeled G-hp5 show five A•T bp imino proton peaks, as with the G-hp5 spectra (Figure 5). Splitting of the G5 imino proton resonance at 12.5 ppm is observed in the 15N-labeled spectra due to 15N-1H coupling (88 Hz) whereas a singlet is seen for G5 in the unlabeled DNA. Splitting of the G5 proton peak reverts to a singlet upon decoupling of 15N (Supplementary Figure S10).
Figure 5.
Shift of imino proton peaks at 308 K in 1H NMR spectra of 15N labeled G-hp5. Splitting of imino proton of G-5 bp is highlighted along with the emergence of a new peak at 11.5 ppm. The NMR sample consisted of 100 μM DNA at pH 6.7.
On addition of DB2277, the additional peak emerges at 11.5 ppm as observed in all other G-hp-DNA complexes. However, this peak shows no effect of 15N-labeling, which indicates that this peak does not belong to G5. The aza-benzimidazole NH signal of DB2277 is generally downfield and exchanges with water. A strong H-bond of this proton to DNA should cause an additional downfield shift of the NH signal (46) and this signal should not be affected by 15N coupling. Moreover, in the molecular model of the G-hp1-DB2277 complex (Figure 6), the aza-benzimidazole NH of DB2277 is observed to form a strong H-bond with the carbonyl oxygen of an adjacent T base. Therefore, in this case, the signal at 11.5 ppm, not affected by 15N coupling, most likely represents the aza-benzimidazole NH proton of DB2277 that is directed to the floor of the minor groove. Support for this assignment is also provided by NMR studies done by Leupin and coworkers (46) on Hoechst 33258 with A•T sequences. In this model, the benzimidazole NH proton faces into the minor groove and forms an H-bond with the carbonyl oxygen of a T base. The chemical shifts for the labile protons of benzimidazole NH are 11.3 and 12.3 ppm in the AATT complex (46), in general agreement with an observed signal between 11 and 11.5 ppm in Figures 4 and 5.
Figure 6.
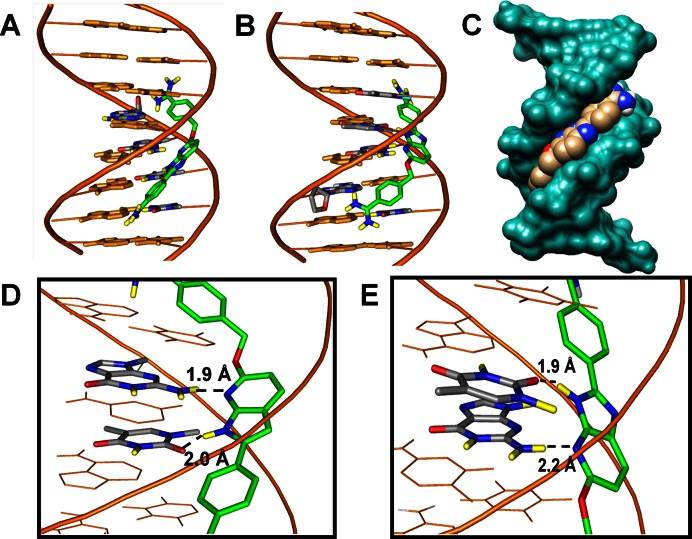
Minor groove views of two possible docked conformations of DB2277 with the AAAGTT. (A,B) The stick model in green–blue–red–yellow represents DB2277. The DNA backbone is represented as a tube form in orange, DB2277 interacting bases are in stick with gray–blue–red–yellow and other bases are in stick with orange. (C) The space filling model shows the van der Waals contacts between the DNA backbone and DB2277, shown in the minor groove with gray–blue–red atoms. (D,E) Important H-bond interactions between aza-N- with the exocyclic G6-NH2 and aza-benzimidazole NH either with N3 of A or the carbonyl group of T at bases flanking the central G.
UV-Vis thermal melting
Thermal melting experiments were performed on over thirty G-hp DNA sequences by monitoring DNA unfolding at 260 nm to screen the complexes based on their relative binding affinity with DB2277. The addition of DB2277 to the hairpin oligomers in this study resulted in Tm increases (ΔTm) of 10–14°C. The DB2277 strongly stabilizes the G-hp1-5 as clearly indicated by their ΔTm values (Supplementary Table S1) whereas, reversing the central G•C bp in G-hp6 decreases the thermal stability of the complex (ΔTm value is 8.7°C).
Surface plasmon resonance
Biosensor–SPR methods provide a quantitative estimation of the interaction of small molecules with DNA and other biomolecules (47). Very fast association and slow dissociation rates make DB2277 a very strong binder for single G•C-containing mixed sequences. SPR data (Supplementary Figure S11) reveals that DB2277 binds strongly with the single G•C bp sequence in both AAGTTT (KD = 1.3 nM) and AAGATA (KD = 4.4 nM). This represents a small difference in ΔG° for binding to these two oligomers (Δ(ΔG°) value of 0.7 kcal mol−1) that could result from a number of factors. Certainly, the increased number of species for G-hp2 contributes to the more favorable ΔG° for binding. DB2277 forms a 5-fold weaker monomeric complex with AAGATA which has DB2277 bound in one orientation.
Dynamics of bound DB2277
At a 1:1 binding ratio, only bound DNA is observed with separate peaks for the major and minor species. 2D EXSY provided clear evidence for the exchange between major and minor species of bound DB2277 and allowed evaluation of the exchange rate between both species. Strong cross peaks observed in EXSY spectra of the G-hp2-DB2277 complex shows the chemical exchange between species (Figure 2, Supplementary Figures S5 and S12). Exchange cross peaks observed in this short time experiment are not due to NOE from the adjacent bp, but due to the chemical exchange for nuclei in different environments. The exchange rate between the major and minor species is 6.8 s−1 at 303 K and 2.8 s−1 at 285 K. The exchange rate was calculated by considering the chemical exchange between major and minor species for A•T bp imino protons (A4, T6 and T7 at 303 K and A4 and T8 at 285 K) which show sufficiently resolved signals (Supplementary Table S2A and B) (48,49). The exchange rate calculated from the EXSY spectrum at 50 ms mixing time is in close agreement with the exchange rate calculated at 150 ms mixing time.
2D EXSY also provided information about the exchange of the aza-benzimidazole NH proton of DB2277. The aza-benzimidazole NH interacts with G-hp DNA in two different chemical environments as reported previously (46). A clearly visible exchange cross peak has been observed for the labile NH proton of aza-benzimidazole of DB2277 in the EXSY spectrum (Supplementary Figure S5). In addition, 1D-NOE differences in the spectra clearly show the change in signal intensity at the central G and adjacent T base on irradiating the NH proton signal of the aza-benzimidazole moiety of DB2277 resonating at 11.25 ppm (Supplementary Figure S2). The exchangeable NH proton of aza-benzimidazole in different environments characterizes the major and minor binding species and is at 11.3 ppm and at 12.7 ppm in complex spectra, respectively (Supplementary Figure S1).
An interesting comparison between macroscopic dissociation rate constants from SPR data with the total exchange rate constant from 2D EXSY for DB2277 binding with G-hp1 is shown in Table 2. The apparent half-life for the exchange between different species is around 100 ms by NMR whereas the half-life for the bound DB2277 with G-hp2 and G-hp5 is in the range of 30–60 s by SPR. Thus, the exchange between the major and minor species is much faster than the macroscopic dissociation of DB2277 from the complex. The long dissociation half-life of the complex compared to the half-life of exchange between species shows that DB2277 can flip between major and minor species while bound to the DNA. Such microscopic exchange between two orientations of bound DB2277 is clearly represented in the model shown below. In addition, the free energy change for exchange of major to minor species can be calculated using intensities of resonances originating from the different species (ΔG° = –RT ln [(IM)/(Im)]. The free energy difference between major and minor species is 0.95 ± 0.03 kcal mol−1 at 303 K and 0.97 ± 0.03 kcal mol−1 at 285 K (Supplementary Table S2B). The configurational entropy change associated with different orientations of the major and minor species is 0.4 kcal mol−1 K−1. Both dynamic amidine moieties and the –CH2O linker (as shown in the model in Figure 6) provide flexibility in the minor groove of G-hp2. This flexibility contributes to the ability of DB2277 to adopt a favorable conformation and make optimum contacts with the minor groove structure. Removal of the CH2O linker, for example, results in a loss of both selectivity and affinity (32). From the clustering histogram of 200 Autodock runs (Supplementary Figure S13), it has also been observed that the conformer in Figure 6A is the most populated and most favorable with the lowest binding energy.
Table 2. Kinetic and equilibrium parameters calculated using SPR and NMR.
| Complex | k a (M−1s−1)a | k d (s−1)a | K D (nM)a | 1/kd (s)a | k ex (s−1)b | 1/kex (s)b |
|---|---|---|---|---|---|---|
| G-hp2-DB2277 | (1.4 ± 0.8) × 107 | 0.02 ± 0.01 | 1.3 ± 0.1 | 55.5 | 6.8 ± 1.3 | 0.15 |
| G-hp5-DB2277 | (0.6 ± 0.4) × 107 | 0.03 ± 0.02 | 4.4 ± 0.3 | 33.5 | – | – |
aDetermined from SPR.
bDetermined from NMR at short mixing time (50 ms) at 303 K (39).
The rapid, microscopic dynamics of DB2277 and netropsin (39) suggest that such dynamics may be a common feature of minor groove binding compounds, even those with quite low KD values. The flipping motion of DB2277 in different orientations is seen in the model in Figure 6 and is surprising for a compound bound to a DNA sequence.
Molecular modeling of G-hp1-DB2277 complex
DB2277 fits quite well into the minor groove of the AAAGTTT binding site with an optimum curvature to follow the groove shape. Models of the two possible orientations (Figure 1) represented in Figure 6 are in general agreement with the experimental results and predict the observed strong binding of DB2277 into this sequence. The flexible –CH2O– linker is essential for the aza-benzimidazole moiety to track along the minor groove curvature and predicts a H-bond with the exocyclic G-NH2 that projects into the minor groove of the binding site. The H-bond distance of aza-N with G-NH2 is 1.9 Å and 2.2 Å in conformers represented in Figure 6A and B, respectively. The NH of the aza-benzimidazole moiety also nicely projects towards the minor groove of the binding site and can form H-bonds with the carbonyl group of 5′ T7 on the same strand and 3′ T5 on the complementary strand as shown in Figure 6A and B respectively (T = O—H-N distances are 2.0 Å and 1.9 Å, respectively). As shown in the model in Figure 6C, DB2277 stacks well with the walls of the minor groove to form van der Waals interactions that stabilize the complex. The two phenyl-amidine groups stack parallel to both sides of the minor groove and provide an extended binding interaction with the flanking bases. Both amidine groups also dynamically form strong H-bonds with both N3 of A and O2 of T at the flanking side of mixed DNA sequences (Figure 6). Their inherent dynamic nature also allows the amidines to form H-bonds to deoxyribose O and the negatively charged phosphate backbone. The –CH2O– linker of DB2277 molecules in this model is serving primarily to add needed flexibility and appropriate distances to the groups of DB2277 to allow optimum DNA interactions.
In summary, an imino proton NMR investigation on different single G•C bp-containing DNA sequences with an aza-benzimidazole compound, DB2277, revealed that rationally designed diamidine molecules can act as very sequence-specific probes at the minor groove of mixed sequences of DNA. The strong binding efficiency and highly dynamic binding nature of DB2277-G-hp complexes allow the ligand to form two reversed oriented microstructures in the minor groove of G-hp sequences. A key finding here is that the large dissociation off-rate of G-hp2 complex allows the DB2277 to microscopically rearrange in the complex without its complete dissociation from target DNA. The study also illustrates the different orientation preferences of DB2277 in G-hp DNA due to different microstructures and base substituent positions and how well the nature of the minor groove can mesh with the functional groups of the DB2277. Another major finding of a DNA sequence with a single species orientation for DB2277 with good binding affinity is essential for detailed NMR structural studies of the DB2277-DNA complex. The examination of binding spectra of DB2277 with 15N-labeled G5 clearly shows the pivotal role of the exocyclic G-NH2 in H-bonding with the aza-benzimidazole moiety of DB2277. This research has provided exciting information about sequence specificity and exchange behavior of DNA minor groove binders which will help expand the scope of rationally designed molecules for DNA minor groove recognition.
Supplementary Material
Acknowledgments
The authors thank Sarah Nguyen, Sarah Laughlin and Carol Wilson for manuscript assistance.
Footnotes
Present address: Yun Chai, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, China.
FUNDING
National Institutes of Health [GM111749 to W.D.W and D.W.B]. Funding for open access charge: National Institutes of Health [GM111749 to W.D.W and D.W.B].
Conflict of interest statement. None declared.
REFERENCES
- 1.Raskatov J.A., Nickols N.G., Hargrove A.E., Marinov G.K., Wold B., Dervan P.B. Gene expression changes in a tumor xenograft by a pyrrole-imidazole polyamide. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16041–16045. doi: 10.1073/pnas.1214267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson W.D., Tanious F.A., Mathis A., Tevis D., Hall J.E., Boykin D.W. Antiparasitic compounds that target DNA. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding P., McFarland K.A., Jin S., Tong G., Duan B., Yang A., Hughes T.R., Liu J., Dove S.L., Navarre W.W., et al. A novel AT-rich DNA recognition mechanism for bacterial xenogeneic silencer MvaT. PLoS Pathog. 2015;11:1–26. doi: 10.1371/journal.ppat.1004967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez J., Mosquera J., Couceiro J.R., Vazquez M.E., Mascarenas J.L. The AT-Hook motif as a versatile minor groove anchor for promoting DNA binding of transcription factor fragments. Chem. Sci. 2015;6:4767–4771. doi: 10.1039/c5sc01415h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto Y. Chemical control of gene expression. Yakugaku Zasshi. 1994;114:357–373. doi: 10.1248/yakushi1947.114.6_357. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen B., Hamelberg D., Bailly C., Colson P., Stanek J., Brun R., Neidle S., Wilson W.D. Characterization of a novel DNA minor-groove complex. Biophys. J. 2004;86:1028–1041. doi: 10.1016/s0006-3495(04)74178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavda S., Liu Y., Babu B., Davis R., Sielaff A., Ruprich J., Westrate L., Tronrud C., Ferguson A., Franks A., et al. Hx, a novel fluorescent, minor groove and sequence specific recognition element: design, synthesis, and DNA binding properties of p-Anisylbenzimidazole-imidazole/pyrrole-containing polyamides. Biochemistry. 2011;50:3127–3136. doi: 10.1021/bi102028a. [DOI] [PubMed] [Google Scholar]
- 8.Kang J.S., Meier J.L., Dervan P.B. Design of Sequence-Specific DNA Binding Molecules for DNA Methyltransferase Inhibition. J. Am. Chem. Soc. 2014;136:3687–3694. doi: 10.1021/ja500211z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Kumar A., Boykin D.W., Wilson W.D. Sequence and length dependent thermodynamic differences in heterocyclic diamidine interactions at AT base pairs in the DNA minor groove. Biophys. Chem. 2007;131:1–14. doi: 10.1016/j.bpc.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto M., Bando T., Morinaga H., Kawamoto Y., Hashiya K., Sugiyama H. Sequence-specific DNA recognition by cyclic pyrrole-imidazole cysteine-derived polyamide dimers. Chem. Eur. J. 2014;20:752–759. doi: 10.1002/chem.201302482. [DOI] [PubMed] [Google Scholar]
- 11.Alniss H.Y., Salvia M.-V., Sadikov M., Golovchenko I., Anthony N.G., Khalaf A.I., MacKay S.P., Suckling C.J., Parkinson J.A. Recognition of the DNA minor groove by thiazotropsin analogues. Chembiochem. 2014;15:1978–1990. doi: 10.1002/cbic.201402202. [DOI] [PubMed] [Google Scholar]
- 12.Barrett M.P., Gemmell C.G., Suckling C.J. Minor groove binders as anti-infective agents. Pharmacol. Ther. 2013;139:12–23. doi: 10.1016/j.pharmthera.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Baraldi P.G., Bovero A., Fruttarolo F., Preti D., Tabrizi M.A., Pavani M.G., Romagnoli R. DNA minor groove binders as potential antitumor and antimicrobial agents. Med. Res. Rev. 2004;24:475–528. doi: 10.1002/med.20000. [DOI] [PubMed] [Google Scholar]
- 14.Xinbo Z., Zhang S.C., Dejun S., Jiang H., Wali A., Pass H., Fernandez-Madrid F., Harbut M.R., Naimei T. New insight into the molecular mechanisms of the biological effects of DNA minor groove binders. PLoS One. 2011;6:1–12. doi: 10.1371/journal.pone.0025822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matovu E., Stewart M.L., Geiser F., Brun R., Mäser P., Wallace L.J.M., Burchmore R.J., Enyaru J.C.K., Barrett M.P., Kaminsky R., et al. Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryotic. Cell. 2003;2:1003–1008. doi: 10.1128/EC.2.5.1003-1008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paine M.F., Wang M.Z., Generaux C.N., Boykin D.W., Wilson W.D., De Koning H.P., Olson C.A., Pohlig G., Burri C., Brun R., et al. Diamidines for human African trypanosomiasis. Curr. Opin. Investig. Drugs. 2010;11:876–83. [PubMed] [Google Scholar]
- 17.Sturk L.M., Brock J.L., Bagnell C.R., Hall J.E., Tidwell R.R. Distribution and quantitation of the anti-trypanosomal diamidine 2,5-bis(4-amidinophenyl)furan (DB75) and its N-methoxy prodrug DB289 in murine brain tissue. Acta Trop. 2004;91:131–143. doi: 10.1016/j.actatropica.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Munde M., Kumar A., Nhili R., Depauw S., David-Cordonnier M.-H., Ismail M.A., Stephens C.E., Farahat A.A., Batista-Parra A., Boykin D.W., et al. DNA minor groove induced dimerization of heterocyclic cations: compound structure, binding affinity, and specificity for a TTAA site. J. Mol. Biol. 2010;402:847–864. doi: 10.1016/j.jmb.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nanjunda R., Wilson W.D. Current Protocols in Nucleic Acid Chemistry. NY: Wiley; 2012. Binding to the DNA minor groove by heterocyclic dications: from AT-specific monomers to GC recognition with dimers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munde M., Kumar A., Peixoto P., Depauw S., Ismail M.A., Farahat A.A., Paul A., Say M.V., David-Cordonnier M.-H., Boykin D.W., et al. The unusual monomer recognition of guanine-containing mixed sequence DNA by a dithiophene heterocyclic diamidine. Biochemistry. 2014;53:1218–1227. doi: 10.1021/bi401582t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Chai Y., Kumar A., Tidwell R.R., Boykin D.W., Wilson W.D. Designed compounds for recognition of 10 base pairs of DNA with two at binding sites. J. Am. Chem. Soc. 2012;134:5290–5299. doi: 10.1021/ja211628j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahimian M., Kumar A., Say M., Bakunov S.A., Boykin D.W., Tidwell R.R., Wilson W.D. Minor groove binding compounds that jump a GC base pair and bind to adjacent AT base pair sites. Biochemistry. 2009;48:1573–1583. doi: 10.1021/bi801944g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munde M., Ismail M.A., Arafa R., Peixoto P., Collar C.J., Liu Y., Hu L., David-Cordonnier M.-H., Lansiaux A., Bailly C., et al. Design of DNA minor groove binding diamidines that recognize GC base pair sequences: a dimeric-hinge interaction motif. J. Am. Chem. Soc. 2007;129:13732–13743. doi: 10.1021/ja074560a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt R.A., Munde M., Kumar A., Ismail M.A., Farahat A.A., Arafa R.K., Say M., Batista-Parra A., Tevis D., Boykin D.W., et al. Induced topological changes in DNA complexes: influence of DNA sequences and small molecule structures. Nucleic Acids Res. 2011;39:4265–4274. doi: 10.1093/nar/gkq1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen B., Neidle S., Wilson W.D. A role for water molecules in DNA−ligand minor groove recognition. Acc. Chem. Res. 2009;42:11–21. doi: 10.1021/ar800016q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao Y., Lee M.P.H., Parkinson G.N., Batista-Parra A., Ismail M.A., Neidle S., Boykin D.W., Wilson W.D. Out-of-shape DNA minor groove binders: induced fit interactions of heterocyclic dications with the DNA minor groove. Biochemistry. 2005;44:14701–14708. doi: 10.1021/bi051791q. [DOI] [PubMed] [Google Scholar]
- 27.Munde M., Lee M., Neidle S., Arafa R., Boykin D.W., Liu Y., Bailly C., Wilson W.D. Induced fit conformational changes of a ‘reversed amidine’ heterocycle: optimized interactions in a DNA minor groove complex. J. Am. Chem. Soc. 2007;129:5688–5698. doi: 10.1021/ja069003n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He G., Vasilieva E., Davis G., Jr, Koeller K.J., Bashkin J.K., Dupureur C.M. Binding studies of a large antiviral polyamide to a natural HPV sequence. Biochimie. 2014;102:83–89. doi: 10.1016/j.biochi.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul A., Nanjunda R., Kumar A., Laughlin S., Nhili R., Depauw S., Deuser S.S., Chai Y., Chaudhary A.S., David-Cordonnier M.H., et al. Mixed up minor groove binders: convincing A·T specific compounds to recognize a G·C base pair. Bioorg. Med. Chem. Lett. 2015;25:4927–4932. doi: 10.1016/j.bmcl.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satam V., Babu B., Patil P., Brien K.A., Olson K., Savagian M., Lee M., Mepham A., Jobe L.B., Bingham J.P., et al. AzaHx, a novel fluorescent, DNA minor groove and G•C recognition element: Synthesis and DNA binding properties of a p-anisyl-4-aza-benzimidazole-pyrrole-imidazole (azaHx-PI) polyamide. Bioorg. Med. Chem. Lett. 2015;25:3681–3685. doi: 10.1016/j.bmcl.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 31.Wei D., Wilson W.D., Neidle S. Small-molecule Binding to the DNA Minor Groove Is Mediated by a Conserved Water Cluster. J. Am. Chem. Soc. 2013;135:1369–1377. doi: 10.1021/ja308952y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chai Y., Paul A., Rettig M., Wilson W.D., Boykin D.W. Design and synthesis of heterocyclic cations for specific DNA recognition: From AT-rich to mixed-base-pair DNA sequences. J. Org. Chem. 2014;79:852–66. doi: 10.1021/jo402599s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul A., Chai Y., Boykin D.W., Wilson W.D. Understanding mixed sequence DNA recognition by novel designed compounds: the kinetic and thermodynamic behavior of azabenzimidazole diamidines. Biochemistry. 2015;54:577–587. doi: 10.1021/bi500989r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rettig M., Germann M.W., Wang S., Wilson W.D. Molecular basis for sequence-dependent induced DNA bending. Chembiochem. 2013;14:323–331. doi: 10.1002/cbic.201200706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fede A., Billeter M., Leupin W., Wuthrich K. Determination of the NMR solution structure of the Hoechst 33258-d(GTGGAATTCCAC)2 complex and comparison with the X-ray crystal structure. Structure. 1993;1:177–186. doi: 10.1016/0969-2126(93)90019-d. [DOI] [PubMed] [Google Scholar]
- 36.Trotta E., Paci M. Solution structure of DAPI selectively bound in the minor groove of a DNA T.T mismatch-containing site: NMR and molecular dynamics studies. Nucleic Acids Res. 1998;26:4706–4713. doi: 10.1093/nar/26.20.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conte M.R., Jenkins T.C., Lane A.N. Interaction of minor-groove-binding diamidine ligands with an asymmetric DNA duplex. Eur. J. Biochem. 1995;229:433–444. doi: 10.1111/j.1432-1033.1995.0433k.x. [DOI] [PubMed] [Google Scholar]
- 38.Rydzewski J.M., Leupin W., Chazin W. The width of the minor groove affects the binding of the bisquaternary heterocycle SN-6999 to duplex DNA. Nucleic Acids Res. 1996;24:1287–1293. doi: 10.1093/nar/24.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rettig M., Germann M.W., Ismail M.A., Batista-Parra A., Munde M., Boykin D.W., Wilson W.D. Microscopic rearrangement of bound minor groove binders detected by NMR. J. Phys. Chem. B. 2012;116:5620–5627. doi: 10.1021/jp301143e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen B., Tanious F.A., Wilson W.D. Biosensor-surface plasmon resonance: quantitative analysis of small molecule–nucleic acid interactions. Methods. 2007;42:150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Nanjunda R., Munde M., Liu Y., Wilson W.D. Real-time monitoring of nucleic acid interactions with biosensor-surface plasmon resonance. In: Wanunu M, Tor Y, editors. Methods for Studying DNA/Drug Interactions. Boca Raton: CRC Press-Taylor & Francis Group; 2011. [Google Scholar]
- 42.Spartan’10 Tutorial and User's Guide. Irvine: Wavefunction, Inc; 2010. [Google Scholar]
- 43.Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 44.Trott A., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis E.A., Munde M., Wang S., Rettig M., Le V., Machha V., Wilson W.D. Complexity in the binding of minor groove agents: netropsin has two thermodynamically different DNA binding modes at a single site. Nucleic Acids Res. 2011;39:9649–9658. doi: 10.1093/nar/gkr699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fede A., Labhardt A., Bannwarth W., Leupin W. Dynamics and binding mode of Hoechst 33258 to d(GTGGAATTCCAC)2 in the 1:1 solution complex as determined by two-dimensional proton NMR. Biochemistry. 1991;30:11377–11388. doi: 10.1021/bi00112a004. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y., Wilson W.D. Quantitative analysis of small molecule-nucleic acid interactions with a biosensor surface and surface plasmon resonance detection. Methods Mol. Biol. 2010;613:1–23. doi: 10.1007/978-1-60327-418-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., Germann M.W. Characterization of secondary amide peptide bond isomerization: thermodynamics and kinetics from 2D NMR spectroscopy. Biopolymers. 2011;95:755–762. doi: 10.1002/bip.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bain A.D. Chemical exchange in NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2003;43:63–103. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.