Abstract
The Fanconi anemia protein SLX4 assembles a genome and telomere maintenance toolkit, consisting of the nucleases SLX1, MUS81 and XPF. Although it is known that SLX4 acts as a scaffold for building this complex, the molecular basis underlying this function of SLX4 remains unclear. Here, we report that functioning of SLX4 is dependent on its dimerization via an oligomerization motif called the BTB domain. We solved the crystal structure of the SLX4BTB dimer, identifying key contacts (F681 and F708) that mediate dimerization. Disruption of BTB dimerization abrogates nuclear foci formation and telomeric localization of not only SLX4 but also of its associated nucleases. Furthermore, dimerization-deficient SLX4 mutants cause defective cellular response to DNA interstrand crosslinking agent and telomere maintenance, underscoring the contribution of BTB domain-mediated dimerization of SLX4 in genome and telomere maintenance.
INTRODUCTION
Mutations in human SLX4 (also known as FANCP) have been linked to the genetic disease Fanconi Anemia (FA) (1,2), characterized by congenital abnormalities, increased susceptibility to cancer, and sensitivity to DNA interstrand crosslinking agents. SLX4 assembles and coordinates a nuclease toolkit to function in diverse pathways of genome maintenance, including DNA interstrand cross link (ICL) repair, DNA replication, nucleolytic processing of homologous recombination (HR) intermediates such as Holliday Junctions (HJs), management of replication stress at specific difficult-to-replicate genomic loci such as common fragile sites (CFS), and telomere maintenance (3). The multi-domain architecture of SLX4 enables it to not only bind to a wide range of DNA repair proteins, but also orchestrate their delivery and activities at the target site, each function mediated by one or more specific domain(s) of SLX4 (Figure 1A). For example, direct interaction of SLX4 with structure-specific endonucleases (SSEs) SLX1, MUS81 and XPF is mediated by the SLX4SBD (SLX1-binding domain), SLX4SAP (SAP motif, MUS81-binding region) and SLX4XBR (XPF-binding region) domains, respectively (4–7). SLX4 also coordinates dispatch and activity of its associated protein partners (8–10), and domains implicated in this include SLX4ZF (ubiquitin-binding zinc finger domain), SLX4SIMs (SUMO-interacting motifs) (11–13) and SLX4BTB (Bric-a-brac, Tramtrack and Broad complex domain) (14,15). While the ZF and XBR domains of SLX4 are critical for its function in ICL repair, the BTB domain has been shown to play a modest role in conferring cellular resistance to ICL-inducing agent mitomycin C (MMC) (12,16). Yet the mechanism behind BTB regulating this cellular function is unclear. The SIMs, which promote SUMOylation of XPF and SLX4 itself, are important for managing replication stress at genomic CFS (11,12). The SUMOylation function of SLX4 depends not only on the SIMs, but also on the BTB domain of SLX4 (12).
Figure 1.
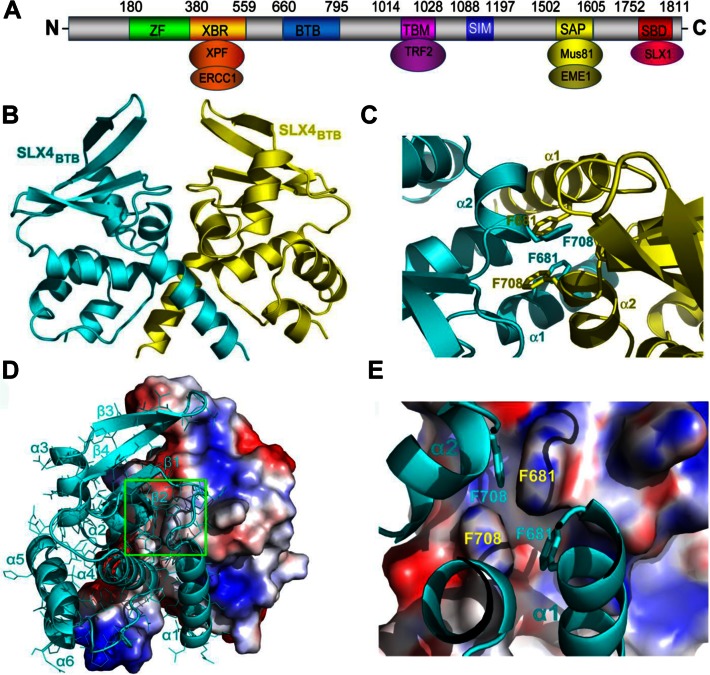
Hydrophobic contacts located in the BTB domain mediate dimerization of SLX4. (A) Schematic showing domain mapping of human SLX4 protein. ZF, ubiquitin-binding zinc finger domain; XBR, XPF-binding region; BTB, Bric-a-brac, Tramtrack and Broad complex domain; TBM, TRF2-binding motif; SIM: SUMO-Interacting Motif; SAP, SAP motif, MUS81-binding region; SBD, SLX1-binding domain. (B) Overall structure of dimeric SLX4BTB. The monomers are colored in cyan and yellow. (C) An enlarged view of the SLX4BTB dimeric interface. Residues F681 and F708 essential to preserve the dimer interface are highlighted as stick models. (D) Surface view of the hydrophobic dimerization interface. Hydrophobic, basic and acidic residues are shown in white, blue and red, respectively. (E) Close-up view of the critical hydrophobic interaction region of panel (D). Main binding surface is located in α1 and α2 helices.
SLX4 is also a critical player in telomere maintenance (7,17). Mammalian genomes are protected by nucleoprotein structures called telomeres at chromosome ends, and proper telomere maintenance is imperative for genome maintenance (18). Telomeres, which are hot spots for formation of alternate and secondary DNA structures, present an inherently challenging landscape for DNA metabolism (19). Conceivably, the longer the telomeres are, the greater is the severity of such challenges (20–22). Previously, we and others have shown that SLX4TBM (TRF2-binding motif) mediates a direct interaction between SLX4 and the TRFH domain of the telomeric protein TRF2 (TRF2TRFH) (4,7,17). The SLX4-TRF2 platform acts as a double-layered scaffold that recruits the SLX4-associated nucleases to long telomeres, where the SLX4-nuclease complex likely engages in resolving alternate DNA structural intermediates during telomere maintenance processes of replication, recombination and length homeostasis (7,17,23).
Despite such domain-based functional analyses of SLX4, the molecular and/or structural basis underlying the functioning of the SLX4 scaffold in assembly of the nuclease toolkit remains unknown. Here we address this question by focusing on the oligomerization properties of SLX4 mediated by its BTB domain. The BTB domain is a ubiquitous eukaryotic protein motif that mediates self-oligomerization and interactions with other proteins (14,15). In proteins featuring the BTB domain, it is generally known to provide the interface for protein oligomerization and also to perform diverse functions in combination with other protein domains. However, the actual oligomeric form of SLX4, the necessity of the BTB domain for SLX4 oligomerization and the functional significance of SLX4 oligomerization all remain to be determined. In order to assess how the oligomerization feature of SLX4BTB impacts cellular functioning of the SLX4-assembled nuclease complex, here we solved the crystal structure of SLX4BTB at a resolution of 2.15 Å. We identified key hydrophobic contacts (F681 and F708) in the dimer interface, mutations in which abrogate SLX4 dimerization. Furthermore, gel filtration analysis showed that SLX4BTB exists as a dimer in solution. Using the SLX4F681R/F708R double mutant as a unique dimerization-defective tool, we showed that disruption of SLX4 dimerization adversely impacts SLX4 protein self-oligomerization and several genome maintenance functions of SLX4, including abrogation of cellular foci formation and telomere localization of SLX4 and its associated nucleases, debilitating telomere maintenance, and compromising cellular resistance to DNA ICL-inducing agent MMC. We propose that SLX4 dimerization plays a pivotal role both structurally and functionally, thereby aiding assembly and targeting of the SLX4-dependent genome maintenance nuclease toolkit.
MATERIAL AND METHODS
Protein purification and crystallization
Human SLX4BTB (residues 660–795) was expressed from a modified pET-28a vector that contained a SUMO protein fused after the N-terminal 6×His tag. Seleno-methionine labeling of the protein was achieved by expression in E. coli B834(DE3) supplemented with L-(+)-SelenoMethionine in the synthetic SelenoMethionine Expression Media (Molecular Dimensions). The protein was purified via Ni NTA affinity, followed by gel filtration chromatography on Hiload Superdex 75 column (GE Healthcare). Finally, the protein was concentrated to 35 mg/ml and stored in TN buffer (25 mM Tris-HCl pH8.0, 150 mM NaCl and 5 mM DTT).
Crystals of SLX4BTB were grown by sitting-drop vapor diffusion at 4°C under the condition of 27% PEG 400, 0.1 M HEPES pH7.5 and 300 mM CaCl2.
For detailed protein expression, purification, crystallization and structure determination, see Extended Experimental Procedures.
Analytical gel filtration chromatography
2 mg of purified wild-type or point-mutant SLX4BTB (20 mg/ml) was loaded onto a calibrated Superdex 75 column (volume 120 ml; GE Healthcare) equilibrated in 25 mM pH 8.0 Tris-HCl, 150 mM NaCl and 2 mM DTT. The column was eluted at a flow rate of 1 ml/min, and 1 ml fractions were collected, analyzed by SDS–PAGE and stained with Coomassie brilliant blue. Apparent molecular masses were calculated from the corresponding elution volumes using a calibration curve that was obtained with gel filtration standard proteins (Bio-Rad).
Yeast two-hybrid assay
Wild-type and mutant human SLX4 cDNA fragments were inserted into the modified yeast two-hybrid assay vectors pBTM116 and pACT2 (Clontech), respectively. Yeast two-hybrid constructs were co-transformed into the yeast strain L40. Cells were cultured on selective SD–Trp–Leu plates. For liquid β-galactosidase assay, yeast cells were grown in the SD–Trp–Leu liquid media, permeabilized by three cycles of freeze-and-thaw treatment, and the β-galactosidase activity was determined by measuring OD420 using ortho-Nitrophenyl-β-galactoside (ONPG) as the substrate. All readings were normalized to the density of yeast cells (OD600). All experiments were performed with three independent replicates.
Plasmid generation
Human SLX4 (wild-type and TRF2-interacting mutant), SLX1, MUS81 and XPF were cloned in various mammalian expression vectors, as described previously (7). For other plasmids info, see the Extended Experimental Procedures and Supplementary Table S1.
ShRNA-mediated SLX4 knockdown, transient transfections, immunoprecipitation and Western blot, IF-FISH, Q-FISH and CO-FISH were performed as described in (7,23). MMC sensitivity assay was performed with RA3331/E6E7/hTERT cells as described in (16).
RESULTS
SLX4 exists as a dimer, formation of which is driven by hydrophobic contacts located in the BTB domain
Primary sequence analysis of human SLX4 (data not shown) predicts the existence of a BTB oligomerization domain (14,15) in the middle region of SLX4 (Figure 1A). To examine the role of SLX4BTB in SLX4 function, we first determined the crystal structure of SLX4BTB at a resolution of 2.15 Å (Table 1). The core of SLX4BTB is made up of the characteristic BTB fold, consisting of a cluster of six α-helices capped at one end by a short four-stranded β-sheet (Figure 1B and Supplementary Figure S1A). SLX4BTB dimerizes via a hydrophobic interface that buries a total of ∼1312 Å2 solvent accessible surface area. The principle dimeric contacts between the SLX4BTB subunits are mediated by a group of hydrophobic residues from helices α1 and α2 (Figure 1C). Notably, two phenylalanine residues (F681 and F708) from each monomer pack together to form the core of the hydrophobic interface (Figure 1C–E and Supplementary Figure S1B). Multiple sequence alignment of BTB domains from different SLX4 homologs revealed that the residues that form the SLX4 dimeric interface are highly conserved across species (Supplementary Figure S2). In contrast, structure-based sequence analysis between multiple BTB domain-containing proteins showed that, although the three-dimensional structure of SLX4BTB closely resembles those of other BTB-containing proteins, the amino acid residues that mediate the dimeric contacts are highly divergent (Supplementary Figure S3). Calibrated gel-filtration chromatography showed that the elution position of SLX4BTB corresponds to a molecular weight of ∼40 kDa (Figure 2A and Supplementary Figure S4A), as expected if the crystallographic dimer interaction is preserved in solution. This result corroborates our crystallographic finding and shows that SLX4BTB exists as a dimer in solution.
Table 1. Data collection and refinement statistics for SLX4BTB X-ray crystal structure.
| SLX4BTB (Native) | SLX4BTB (SeMet-SAD) | |
|---|---|---|
| Data collection | ||
| Wavelength | 0.97861 | 0.97861 |
| Space group | P6222 | P6222 |
| Cell dimensions | ||
| a, b, c (Å) | 96.4, 96.4, 71.8 | 96.2, 96.2, 71.8 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 2.15 | 2.4 |
| R merge a | 0.089 (0.808) | 0135. (0.721) |
| I/σIa | 37.1 (1.8) | 47.3 (4.1) |
| Completeness (%)a | 99.5 (96.1) | 100.0 (100.0) |
| Redundancya | 16.8 (12.4) | 40.3 (32.4) |
| Refinement | ||
| Resolution (Å) | 41.74-2.15 | |
| No. of reflections | 11,108 | |
| R work / Rfree (%) | 20.1/24.4 | |
| No. of atoms | ||
| SLX4 | 906 | |
| Water | 32 | |
| B-factors (Å2) | ||
| SLX4 | 59.0 | |
| Water | 47.2 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (°) | 0.915 | |
| Ramanchandran plot | ||
| Favored region | 98.3% | |
| Allowed region | 100.0% | |
| Outlier region | 0.0% | |
aHighest resolution shell is shown in parenthesis.
Figure 2.
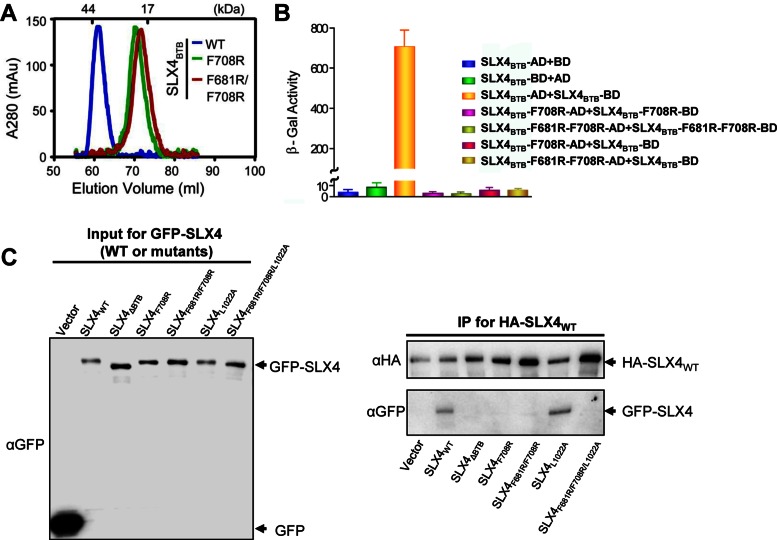
SLX4 forms a dimer. (A) Superposition of gel filtration chromatography profiles of wild-type and dimer-disrupting mutants of SLX4BTB. (B) SLX4BTB dimer formation was assessed by measuring β-galactosidase activity produced by the reporter gene in yeast two-hybrid system. The Y-axis depicts readings at OD420 with ortho-nitrophenyl-β-galactoside (ONPG) as substrate. Data are averages of three independent measurements. AD, activation domain; BD, DNA-binding domain. (C) SLX4BTB mutants impair oligomerization of SLX4 in cells. SLX4-depleted U2OS cells were transiently co-transfected with HA-SLX4WT and GFP-SLX4 (WT or mutants) plasmids. Western blot was performed on HA-SLX4 immunoprecipitates (IP) (right). Inputs for GFP-SLX4 are shown (left). GFP-SLX4WT and GFP-SLX4L1022A, but not GFP-SLX4 BTB mutants (SLX4ΔBTB, SLX4F708R, SLX4F681R/F708R and SLX4F681R/F708R/L1022A) were co-immunoprecipitated with HA-SLX4WT.
To identify key contacts driving SLX4 dimerization, we generated two arginine-substituting mutations (F681R and F708R) that locate to the hydrophobic dimer interface of SLX4BTB (Figure 1C). The mutant proteins of the isolated BTB domain were purified to homogeneity, and their oligomeric states were analyzed by gel-filtration chromatography. Both the single and double mutants of F708R and F681R/F708R completely disrupted the dimeric state of the isolated SLX4BTB, as indicated by the shift in their gel filtration profiles towards the monomer species (Figure 2A). The SLX4 dimer-disrupting effects of these mutations were also confirmed by yeast two-hybrid analyses (Figure 2B). Thus, we conclude that SLX4 dimerization in vitro is principally driven by inter-subunit hydrophobic contacts located in the BTB domain.
To ascertain the oligomerization status of SLX4 in cells, we co-transfected SLX4-depleted U2OS cells (7) with HA-SLX4 (wild type) and GFP-SLX4 (wild-type or BTB mutants). We then immunoprecipitated (IP) HA-SLX4 using anti-HA antibody, followed by probing for GFP-SLX4 via Western blot using anti-GFP antibody. GFP-SLX4WT, but not BTB domain mutants (GFP-SLX4ΔBTB, GFP-SLX4F708R and GFP-SLX4F681R/F708R) was co-immunoprecipitated with HA-SLX4WT (Figure 2C), suggesting that disruption of the dimeric interface of SLX4 leads to SLX4 protein self-oligomerization failure in cells.
Foci formation of SLX4 is contingent upon its dimerization
The mammalian nucleus is organized into functional foci that are factories harboring different proteins participating in the same process such as DNA replication and repair (24). Because the functioning of SLX4 in various genome maintenance pathways depends upon assembly of multiple nucleases, we investigated the importance of SLX4 dimerization in nuclear foci formation. The SLX4-complex functions in genome maintenance, but it is primarily associated with telomeres in human cells, such as U2OS that possess long telomeres which resemble difficult-to-replicate genomic CFS and face extra DNA metabolism challenges due to alternate and secondary DNA structures (7,21,22). Hence we aptly chose telomeres as a genomic testing site for the functionality of SLX4 dimerization.
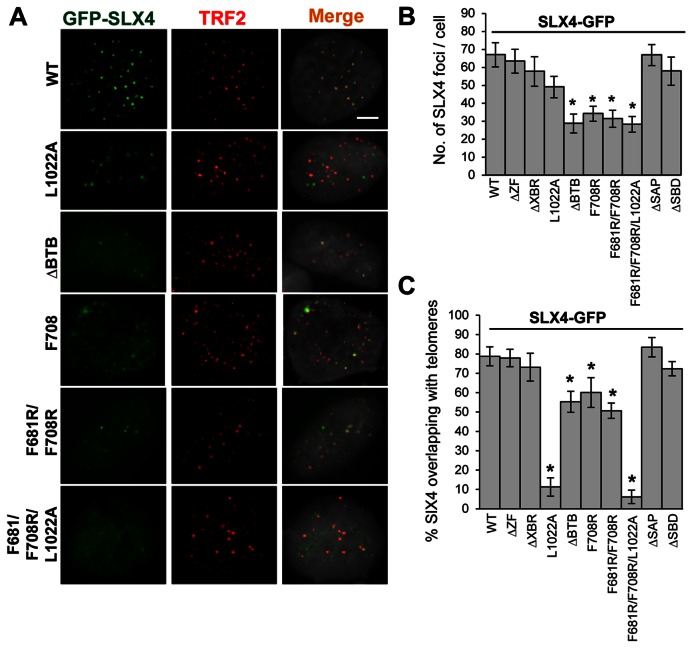
SLX4-depleted U2OS cells were transiently transfected with GFP-wild-type SLX4 or various domain truncation or point mutants of SLX4 (Figure 2C and Supplementary Figure S4B) (7). Immunofluorescence (IF) revealed that expression of dimer-disrupting mutants of SLX4 (SLX4F708R, SLX4F681R/F708R and SLX4ΔBTB) failed to rescue SLX4 foci formation (Figure 3A and B), unlike expression of other domain deletion mutants of SLX4 [SLX4ΔZF, deletion of ZF domain; SLX4ΔXBR, XPF non-interacting; SLX4ΔSAP, MUS81 non-interacting; and SLX4ΔSBD, SLX1 non-interacting (Figure 1A)], all of which restored foci formation comparable to wild type SLX4 (Figure 3B).
Figure 3.
SLX4 dimerization is necessary for foci formation of SLX4. (A) Representative IF images, showing nuclear foci formation and colocalization of moderately expressed GFP-SLX4 (wild type or mutants) with TRF2 in SLX4-depleted U2OS cells. Bar: 5 μm. (B,C) Quantification of number of SLX4 foci per cell, and percentage of SLX4 foci overlapping with telomeres. SLX4ΔBTB, SLX4F708R and SLX4F681R/F708R are SLX4BTB domain mutants that are defective in dimerization. SLX4L1022A is a SLX4TBM mutant that does not interact with TRF2. SLX4F681R/F708R/L1022A contains mutations in both BTB domain and TBM of SLX4. SLX4ΔZF, SLX4ΔXBR, SLX4ΔSAP and SLX4ΔSBD are domain deletion mutants of the ZF, XBR, SAP and SBD domains of SLX4, respectively. Approximately 30 cells/genotype were examined. Error bars: SD; P-values: Student's t-test. *P < 0.0001.
Apart from foci formation, for SLX4 to function in telomere maintenance, it must be recruited to telomeres via the direct interaction between SLX4TBM and TRF2TRFH (7). Although point mutation in the TBM of SLX4 (SLX4L1022A) abrogates telomeric localization of SLX4 (Figure 3A and C) (7), this mutant retains foci formation ability (Figure 3A and B), unlike the dimerization-defective SLX4 mutants. To delineate the necessity of SLX4BTB dimerization-dependent foci formation and SLX4TBM-dependent TRF2 interaction features of SLX4, we generated a mutant SLX4 (SLX4F681R/F708R/L1022A) that abrogates both dimer formation (mutation F681R/F708R in the BTB domain) and TRF2 interaction (mutation L1022A in the TBM). IF analyses showed that this SLX4 mutant was also impaired in foci formation, similar to the dimerization-defective SLX4 mutants (Figure 3A and B). For all the SLX4 mutants that were impaired in foci formation (SLX4F708R, SLX4F681R/F708R, SLX4ΔBTB and SLX4F681R/F708R/L1022A), very few SLX4 foci colocalized with telomeres, as compared to wild-type SLX4 (Figure 3A and C). These results collectively suggest that in vivo foci formation of SLX4 is contingent upon its dimerization, failure of which amounts to significantly reduced SLX4 presence at telomeres.
Disruption of SLX4 dimerization abrogates nuclease toolkit assembly
We have shown previously that deletion of the specific nuclease interacting regions of SLX4 (SBD, SAP or XBR) (Figure 1A) abolishes interaction with the respective endonuclease, their foci formation and telomeric localization (7). Cellular role of SLX4 in preserving genome stability is dependent upon assembly of the nuclease toolkit. Because disruption of SLX4 dimerization negated its foci formation and hence its telomeric presence, we questioned if SLX4 dimerization is necessary for building the nuclease complex and targeting the same to the required genomic site such as the telomeres.
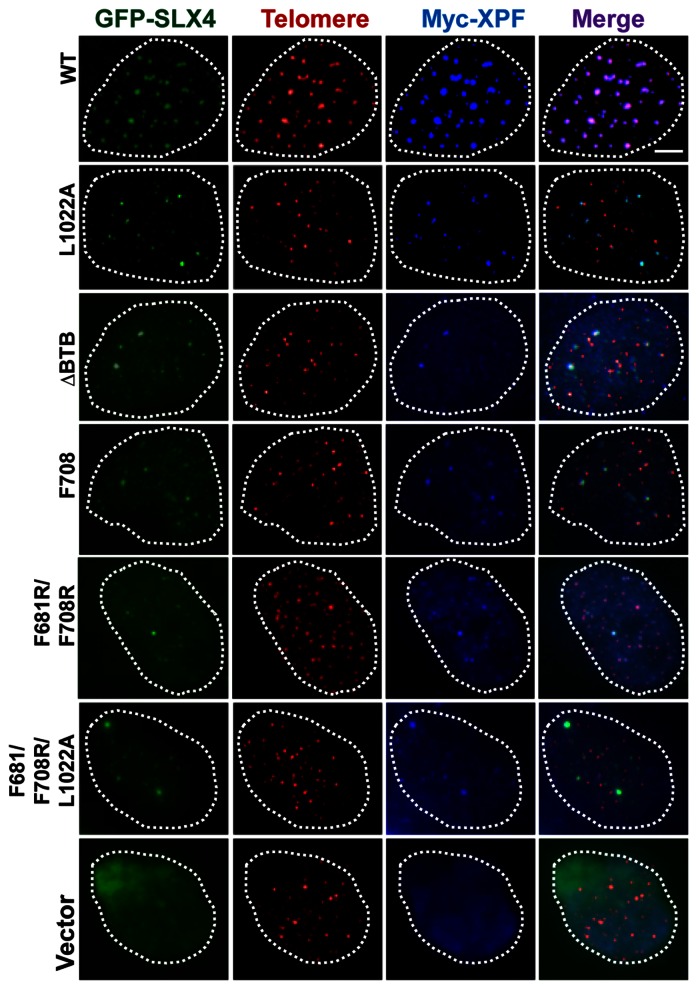
SLX4-depleted U2OS cells were transiently transfected with GFP-vector, GFP-SLX4WT, or dimer-disrupting mutants of GFP-SLX4 (SLX4F708R, SLX4F681R/F708R and SLX4ΔBTB), along with Myc-nuclease (SLX1/MUS81/XPF) (7). IF coupled to telomere FISH (IF-FISH) revealed abrogation of foci formation and telomeric presence of XPF (Figure 4), SLX1 and MUS81 (data not shown) in cells expressing SLX4F708R, SLX4F681R/F708R and SLX4ΔBTB, unlike in cells expressing wild type SLX4. This suggests that assembly/foci formation of the nucleases depends not only on their specific interaction with SLX4 (7), but also on dimerization of SLX4. Interestingly, the TRF2 non-interacting SLX4L1022A mutant retained the ability to form nuclear foci and so did the associated nucleases in presence of this mutant. These SLX4 and nuclease foci colocalized with one another, but did not colocalize with the telomeric foci (Figure 4) (7). Thus, the likely reason why SLX4 and its associated nucleases are not detected at telomeres in the dimerization-defective SLX4 mutants (as compared to wild type SLX4) is because of the inability of these mutants to form nuclear foci and assemble the nuclease complex.
Figure 4.
Disruption of SLX4 dimerization causes failure of nuclease toolkit assembly at telomeres. Representative IF-FISH images, showing that the nuclease XPF form discrete nuclear foci that colocalize with SLX4 foci and telomeres in U2OS cells expressing wild-type SLX4, but not in SLX4ΔBTB, SLX4F708R, SLX4F681R/F708R and SLX4F681R/F708R/L1022A mutants. IF-FISH was performed on SLX4-depleted U2OS cells transiently expressing GFP (vector), GFP-SLX4WT or GFP-SLX4 mutants together with Myc-XPF. Bar: 5 μm.
Telomere defects arise from failure of SLX4 to dimerize
The SLX4-nuclease complex is required at long telomeres that face greater challenges during DNA metabolism (7,17,21,22). The SLX4-nuclease complex is a regulator of telomere length maintenance, is necessary to avert telomere replication defects (manifested as fragile telomeres), and controls nucleolytic processing-dependent homologous recombination (HR) at telomeres (7,23,25). For SLX4 to regulate telomere maintenance, the SLX4-assembled nuclease toolkit needs to be recruited to telomeres via interaction of SLX4 with TRF2 (7). Since telomeric localization of SLX4 and assembly of the SLX4-nuclease complex were adversely impacted by negation of SLX4 dimerization, we next investigated the functional ramifications of abrogating SLX4 dimerization on maintenance of telomeres.
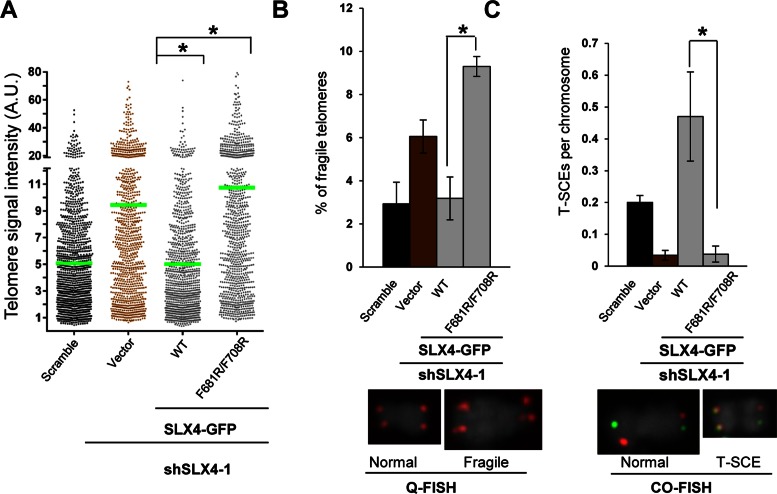
We assessed telomere length and fragile telomeres via Quantitative FISH (Q-FISH) and telomere sister chromatid exchange (T-SCE) events via chromosome orientation FISH (CO-FISH) in SLX4-depleted U2OS cells transiently expressing GFP-wild type or dimerization-defective mutant SLX4. SLX4 depletion and expression of wild type and mutant proteins in SLX4-depleted U2OS cells were shown previously (7) (Figure 2C and Supplementary Figure S4B). Unlike wild type SLX4, cells expressing the SLX4F681R/F708R mutant were defunct in restoring telomere length (Figure 5A) and exhibited elevated level of fragile telomeres (Figure 5B) and decreased T-SCE events (Figure 5C). Collectively, our results imply that telomere maintenance is contingent upon dimerization of SLX4, which mediates foci formation/assembly of a functional SLX4-nuclease complex that can be effectively targeted to telomeres.
Figure 5.
Disruption of SLX4 dimerization causes telomere defects. (A) Telomere length, (B) frequency of fragile telomeres, and (C) T-SCEs per chromosome are all adversely impacted when SLX4 dimerization is disrupted in cells. U2OS cells stably depleted of SLX4 (shSLX4) were transiently transfected with GFP (vector) or GFP-SLX4 wild type or mutant plasmids. Mean telomere length for each genotype is derived from 30 metaphases and is indicated in green in panel (A). The frequencies of individual telomeres were plotted against the telomere signal intensity using arbitrary units (A.U.). Representative fragile and T-SCE positive chromosome images are shown in panels B and C, respectively. Two groups were compared using a one-way ANOVA in panel (A) and a Student's t-test in panels (B) and (C). * P < 0.0001.
SLX4 dimerization contributes towards cellular resistance to MMC
ICLs are dangerous lesions in DNA that can block replication forks. Processing and removal of ICLs involve several different DNA metabolism pathways, including the FA pathway (26). Depletion of SLX4 sensitizes human cells to the cytotoxic effects of ICLs (4–6). SLX4 domains implicated in its ICL repair function include SLX4XBR (the domain that interacts with XPF-ERCC1, an important SSE in ICL repair), SLX4ZF and SLX4BTB (Figure 1A) (1,16). Expression of BTB domain deletion mutant of SLX4 in SLX4-null human fibroblasts RA3331/E6E7/hTERT showed partial rescue of MMC sensitivity (16). To specifically assess the contribution of SLX4 dimerization in SLX4 BTB-dependent cellular resistance to ICL-inducing agent MMC, we expressed the dimerization-defective point mutant of SLX4 (SLX4FF681R/F708R) in the SLX4-null RA3331/E6E7/hTERT cells, and monitored cell survival after subjecting the cells to MMC treatment (Supplementary Figure S5). As expected, expression of wild type SLX4 rescued cell survival in contrast to the empty vector. In comparison to wild type SLX4, the SLX4F681R/F708R mutant partially rescued MMC sensitivity, similar to what has been reported previously for the SLX4ΔBTB mutant (16). This suggests that the MMC sensitivity observed upon deletion of the BTB domain is plausibly attributable to a defect in dimerization of SLX4.
DISCUSSION
SLX4 is a highly versatile genome maintenance protein functioning in multiple and diverse pathways, including in ICL repair, nucleolytic resolution of HR intermediates, managing replication stress at CFS and telomere maintenance (3). The common requirement for all these SLX4 functions is the assembly, delivery and coordination of the nucleases SLX1, MUS81-EME1 and /or XPF-ERCC1 to the specific genome loci in the relevant context. Here we provide structural and functional insights into a molecular basis that enables SLX4 to function optimally in the cell. We show that dimerization of SLX4, mediated by hydrophobic contacts within its BTB domain, contributes toward foci formation of SLX4, assembly and regulation of the nuclease toolkit and toward SLX4-dependent genome / telomere maintenance functions.
Although BTB is generally known to provide the interface for oligomerization in many proteins, different BTB domains vary in their oligomerization state and protein–protein interaction behavior, which in turn affect the properties of the proteins containing them. For example, while the BTB domain in zinc finger proteins can homodimerize, heterodimerize and recruit transcription factors, the BTB domain in ion channel proteins primarily supports tetramerization (14). Such differential behavior may be attributed to the high sequence variability of amino acid residues within the interaction interface of BTB domains in different proteins (Supplementary Figure S3A).
Our SLX4BTB structure together with gel filtration analysis show that SLX4 exists as homodimer, as opposed to higher oligomers (Figures 1 and 2). The SLX4BTB dimer structure conforms to the overall similar arrangement of BTB-containing proteins, characterized by a unique interaction surface that contains a few conserved hydrophobic residues that are buried and exposed residues that are variable (Supplementary Figure S2). Key hydrophobic contacts (F681 and F708) in SLX4BTB act as an architectural lynchpin, driving its dimerization (Figures 1 and 2). These key residues are highly conserved in SLX4 across species (Supplementary Figure S2). Interestingly, a rare breast cancer mutation (G700R) has been reported in this highly conserved region of the interaction interface of SLX4BTB (27) in which the G700 residue is completely conserved in SLX4BTB across species (Supplementary Figure S2). It is noteworthy that unlike the SLX4-associated nuclease SLX1 in which homodimerization has been shown to inactivate SLX1 nuclease activity (28), formation of SLX4 homodimer is required for SLX4 function, as shown in this study.
The failure of the dimerization-defective SLX4 mutants (F681R/F708R, F708R and ΔBTB) to form foci and to present at telomeres (Figure 3) and also abrogate foci formation of the SLX4-associated nucleases SLX1, MUS81 and XPF (Figure 4) implicates dimerization of SLX4 to be essential in vivo in properly assembling the SLX4-nuclease toolkit at a genomic site. Importantly, for the SLX4-nuclease complex to function in telomere maintenance, two distinct requirements must be fulfilled: (i) SLX4 must be able to assemble the nuclease toolkit. This is contingent upon foci formation of SLX4 and its associated nucleases both of which are abrogated by mutations in the SLX4 BTB domain that disrupt dimerization of SLX4 (Figures 3 and 4). (ii) SLX4-assembled nuclease toolkit must be recruited to telomeres. This, as we demonstrated previously (7), is contingent upon direct interaction between the TBM of SLX4 and the TRFH domain of TRF2, and point mutation in SLX4TBM (SLX4L1022A) or TRF2TRFH (TRF2F120A) independently disrupts the SLX4-TRF2 interaction. It is noteworthy that the TRF2 non-interacting SLX4L1022A mutant retains the ability to form nuclear foci and so do the associated nucleases in presence of this mutant (Figures 3 and 4) (7). But these SLX4 or nuclease foci do not colocalize with the telomeric foci because the SLX4-TRF2 interaction is disrupted in the SLX4L1022A mutant (Figures 3 and 4) (7). In short, while SLX4BTB-dependent dimerization mediates nuclease toolbox assembly, SLX4TBM-dependent interaction with TRF2 mediates recruitment of the SLX4-nuclease toolbox to telomeres. Thus, the apparent inability of SLX4 and the nucleases to be at telomeres in presence of dimerization-defective SLX4 mutants is because of their failure to form foci and assemble the nuclease toolkit. In fact, the SLX4 mutant SLX4F681R/F708R/L1022A that abrogates both BTB-dependent foci formation and TBM-dependent TRF2 interaction is also not detected at telomeres because it is unable to form foci (Figures 3 and 4).
Telomeres, which resemble common fragile sites (29), are prone to formation of secondary structures. SLX4 has been shown to preferentially localize to telomeres in human cells harboring long telomeres, where the complex is required to avert defects in telomere length homeostasis, replication and recombination, likely via engaging in resolution of branched telomeric DNA intermediates (7,23). Hence, in this study, we chose telomeres as a model genomic testing site to appraise the consequences of abrogation of SLX4 dimerization on its cellular functions. Indeed, the dimerization-deficient SLX4F681R/F708R mutant gave rise to defects in telomere length regulation, replication and recombination (Figure 5). This is plausibly attributable to the mutant's inability to assemble the nuclease complex at telomeres (Figures 3 and 4). A recent study showed that sumoylation activity of SLX4 is required to alleviate loci-specific replication stress such as at genomic CFS, and that the SUMOylation function of SLX4 depends not only on the SIMs, but also on the BTB domain of SLX4 (12). Because telomeres resemble CFS (29), it remains plausible that the sumoylation function of the SLX4BTB domain may also contribute to telomere maintenance.
ICLs in DNA pose a direct physical block to DNA transactions, and the SLX4 complex is one of the many proteins required for repair of these lesions (30). Cells depleted of SLX4 exhibit increased sensitivity to DNA ICL-inducing agent MMC (4–6). Deletion of SLX4 BTB domain has been shown to cause mild sensitivity to MMC (16). Because the BTB domain has versatile functions, serving as the interface for self-oligomerization and interaction with other proteins, we utilized the mutant that specifically disrupts SLX4 dimerization to investigate the impact of dimerization on SLX4-mediated MMC resistance. The dimerization defective point mutant SLX4F681R/F708R recapitulated the effect of the SLX4ΔBTB mutant in exhibiting mild cellular sensitivity to MMC (Supplementary Figure S5), underscoring the contribution of the dimerization feature of SLX4BTB in imparting cellular resistance to MMC and in ICL repair. The nuclease XPF, an essential player in SLX4-dependent ICL repair, binds to SLX4 via the XBR (also known as MLR) domain of SLX4 (Figure 1) (4–7). This interaction is critical for cellular sensitivity to MMC treatment (16) and telomere homologous recombinational events, T-SCE (7). Guervilly et al.'s recent report (12) suggests that the BTB domain of SLX4 may also play a role, albeit minor, in binding XPF. Hence, it is possible that the slight contribution of the BTB domain towards XPF binding may play a minor role in the defective cellular response to MMC and in telomere defects such as T-SCE that are observed in SLX4 BTB domain mutants (Supplementary Figure S5 and Figure 5).
Thus, functional versatility of SLX4 may be accomplished by its multi-domain architecture where each function is majorly mediated by a specific domain, sometimes in conjunction with minor aid from other domains. For example, while SLX4XBR domain (XPF-binding region) is primarily responsible for cellular response to MMC sensitivity (16), the SLX4SAP and SLX4SBD domains (MUS81 and SLX1-interacting regions respectively) play no or minor role in MMC sensitivity, but they are essential for HJ resolution and common fragile site expression including telomeres (9,16,23,31). Similarly, while BTB domain-mediated SLX4 dimerization plays a partial role in cellular MMC sensitivity, it is an essential necessity for assembling the functional nuclease toolkit at common fragile sites such as telomeres.
In summary, our results indicate that in vivo, malfunction in BTB domain-mediated dimerization of SLX4 results in failure to properly assemble the nuclease complex at the genomic site such as the telomeres. This dimerization defect also jeopardizes cellular response to MMC and telomere maintenance. We propose that dimerization of SLX4 acts as a structural and functional pivot, contributing to optimal cellular functioning of the SLX4-assembled nuclease toolkit.
ACCESSION NUMBER
4ZOU (SLX4BTB).
Supplementary Material
Acknowledgments
We sincerely thank Drs. Agata Smogorzewska and J. Wade Harper for the RA3331/E6E7/hTERT cell line, pHAGE vector, and the templates of SLX4 and its interacting nucleases.
Author contributions: Y.L. and M.L. conceived and supervised the project. B.W. purified proteins and performed crystallization and yeast-two hybrid assays. J.W. and Y.C. determined the crystal structure. G.C. and K.W. generated plasmids and optimized crystallization conditions. J.Y., J.S., K.H. and P.C. performed cell-based and biochemical assays; J.Y., B.W., J.S., K.H., Y.L. and M.L. analyzed the data; Y.L., J.S. and M.L. wrote the manuscript.
FUNDING
Intramural Research Program of the National Institutes of Health, National Institute on Aging; Ministry of Science and Technology of China [2013CB910402 to M.L.]; National Natural Science Foundation of China [31330040 to M.L.]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010201 to M.L.]; U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences [DE-AC02-06CH11357]. Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Institute on Aging; Ministry of Science and Technology of China [2013CB910402 to M.L.]; National Natural Science Foundation of China [31330040 to M.L.]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010201 to M.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M.A., Steltenpool J., Oostra A.B., Eirich K., Korthof E.T., Nieuwint A.W., et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y. Nuclease delivery: versatile functions of SLX4/FANCP in genome maintenance. Mol. Cells. 2014;37:569–574. doi: 10.14348/molcells.2014.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svendsen J.M., Smogorzewska A., Sowa M.E., O'Connell B.C., Gygi S.P., Elledge S.J., Harper J.W. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fekairi S., Scaglione S., Chahwan C., Taylor E.R., Tissier A., Coulon S., Dong M.Q., Ruse C., Yates J.R., 3rd, Russell P., et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz I.M., Hain K., Declais A.C., Gardiner M., Toh G.W., Sanchez-Pulido L., Heuckmann J.M., Toth R., Macartney T., Eppink B., et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Wan B., Yin J., Horvath K., Sarkar J., Chen Y., Wu J., Wan K., Lu J., Gu P., Yu E.Y., et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep. 2013;4:861–869. doi: 10.1016/j.celrep.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castor D., Nair N., Declais A.C., Lachaud C., Toth R., Macartney T.J., Lilley D.M., Arthur J.S., Rouse J. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Mol. Cell. 2013;52:221–233. doi: 10.1016/j.molcel.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner E., Kim Y., Lach F.P., Kottemann M.C., Smogorzewska A. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 2013;5:207–215. doi: 10.1016/j.celrep.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyatt H.D., Sarbajna S., Matos J., West S.C. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang J., Garner E., Hallet A., Nguyen H.D., Rickman K.A., Gill G., Smogorzewska A., Zou L. Noncovalent Interactions with SUMO and Ubiquitin Orchestrate Distinct Functions of the SLX4 Complex in Genome Maintenance. Mol. Cell. 2015;57:108–122. doi: 10.1016/j.molcel.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guervilly J.H., Takedachi A., Naim V., Scaglione S., Chawhan C., Lovera Y., Despras E., Kuraoka I., Kannouche P., Rosselli F., et al. The SLX4 Complex Is a SUMO E3 Ligase that Impacts on Replication Stress Outcome and Genome Stability. Mol. Cell. 2015;57:123–137. doi: 10.1016/j.molcel.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Prieto R., Cuijpers S.A., Luijsterburg M.S., van Attikum H., Vertegaal A.C. SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO Rep. 2015;16:512–519. doi: 10.15252/embr.201440017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Torrado R., Yamada D., Defossez P.A. Born to bind: the BTB protein-protein interaction domain. BioEssays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 15.Stogios P.J., Downs G.S., Jauhal J.J., Nandra S.K., Prive G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y., Spitz G.S., Veturi U., Lach F.P., Auerbach A.D., Smogorzewska A. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121:54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson J.S., Tejera A.M., Castor D., Toth R., Blasco M.A., Rouse J. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell reports. 2013;4:853–860. doi: 10.1016/j.celrep.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 19.Paeschke K., McDonald K.R., Zakian V.A. Telomeres: structures in need of unwinding. FEBS Lett. 2010;584:3760–3772. doi: 10.1016/j.febslet.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taboski M.A., Sealey D.C., Dorrens J., Tayade C., Betts D.H., Harrington L. Long telomeres bypass the requirement for telomere maintenance in human tumorigenesis. Cell Rep. 2012;1:91–98. doi: 10.1016/j.celrep.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y.L., Zhang F., Sun B., Du J., Sun C., Yuan J., Wang Y., Tao L., Kota K., Liu X., et al. Telomerase enzymatic component hTERT shortens long telomeres in human cells. Cell Cycle. 2014;13:1765–1776. doi: 10.4161/cc.28705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossiello F., Herbig U., Longhese M.P., Fumagalli M. d'Adda di Fagagna. Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin. Genet. Dev. 2014;26:89–95. doi: 10.1016/j.gde.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarkar J., Wan B., Yin J., Vallabhaneni H., Horvath K., Kulikowicz T., Bohr V.A., Zhang Y., Lei M., Liu Y. SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates. Nucleic Acids Res. 2015;43:5912–5923. doi: 10.1093/nar/gkv522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spector D.L. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 25.Saint-Leger A., Koelblen M., Civitelli L., Bah A., Djerbi N., Giraud-Panis M.J., Londono-Vallejo A., Ascenzioni F., Gilson E. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle. 2014;13:2469–2474. doi: 10.4161/cc.29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niedernhofer L.J., Lalai A.S., Hoeijmakers J.H. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Landwehr R., Bogdanova N.V., Antonenkova N., Meyer A., Bremer M., Park-Simon T.W., Hillemanns P., Karstens J.H., Schindler D., Dork T. Mutation analysis of the SLX4/FANCP gene in hereditary breast cancer. Breast Cancer Res. Treat. 2011;130:1021–1028. doi: 10.1007/s10549-011-1681-1. [DOI] [PubMed] [Google Scholar]
- 28.Gaur V., Wyatt H.D., Komorowska W., Szczepanowski R.H., de Sanctis D., Gorecka K.M., West S.C., Nowotny M. Structural and mechanistic analysis of the Slx1-Slx4 endonuclease. Cell Rep. 2015;10:1467–1476. doi: 10.1016/j.celrep.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cybulski K.E., Howlett N.G. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle. 2011;10:1757–1763. doi: 10.4161/cc.10.11.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarbajna S., West S.C. Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 2014;39:409–419. doi: 10.1016/j.tibs.2014.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.