Abstract
Gene duplications play a key role in the emergence of novel traits and in adaptation. But despite their centrality to evolutionary processes, it is still largely unknown how new gene duplicates are initially fixed within populations and later maintained in genomes. Long-standing debates on the evolution of gene duplications could be settled by determining the relative importance of genetic drift vs. positive selection in the fixation of new gene duplicates. Using the Drosophila Global Diversity Lines (GDL), we have combined genome-wide SNP polymorphism data with a novel set of copy number variant calls and gene expression profiles to characterize the polymorphic phase of new genes. We found that approximately half of the roughly 500 new complete gene duplications segregating in the GDL lead to significant increases in the expression levels of the duplicated genes and that these duplications are more likely to be found at lower frequencies, suggesting a negative impact on fitness. However, we also found that six of the nine gene duplications that are fixed or close to fixation in at least one of the five populations in our study show signs of being under positive selection, and that these duplications are likely beneficial because of dosage effects, with a possible role for additional mutations in two duplications. Our work suggests that in Drosophila, theoretical models that posit that gene duplications are immediately beneficial and fixed by positive selection are most relevant to explain the long-term evolution of gene duplications in this species.
Even closely related species can differ markedly in gene content (e.g., Drosophila 12 Genomes Consortium 2007; Dumas et al. 2007). Novel genes emerge continuously over time, and many play key roles in the evolution of species-specific phenotypes (Khalturin et al. 2009; Kaessmann 2010). The importance of new genes for the evolution of novel traits cannot be overstated: They have been shown to contribute to the emergence of developmental innovations (e.g., Ragsdale et al. 2013; Florio et al. 2015), to the formation of new morphological structures (e.g., Vlad et al. 2014), and they underlie ecological adaptations (e.g., Zhang 2006; Perry et al. 2007) and domestication traits (e.g., Andersson 2013).
Although new genes can be created through a variety of structural mutations (e.g., by fusing two genes together through a deletion, inversion or translocation), the vast majority originate by duplication from previously existing genes. The study of gene duplications has a long history that spans over a century (Taylor and Raes 2004), and it has generated a substantial body of theoretical and empirical work that seeks to understand the processes by which gene duplicates evolve (Conant and Wolfe 2008; Hahn 2009; Innan and Kondrashov 2010). While these efforts have encountered many limitations, the ability to use empirical data to differentiate among competing theoretical models of gene duplication has been particularly challenging. How new gene duplicates are initially fixed within populations, and the evolutionary processes determining their subsequent maintenance in genomes, remain largely unresolved.
Most models assume that gene duplications are initially selectively neutral and fixed by genetic drift. The ultimate fate of each duplicate (i.e., whether they are maintained or lost from genomes) is determined by mutations that arise subsequent to each duplicate's fixation. These fate-determining mutations can be gain-of-function mutations that cause adaptive functional divergence between the paralogous gene copies (as in neofunctionalization models and some subfunctionalization models) (Ohno 1970; Hughes 1994; Des Marais and Rausher 2008) or loss-of-function mutations that partition the ancestral gene's functions among the duplicate copies (as the classical subfunctionalization model) (e.g., Force et al. 1999).
An alternative set of models proposes that gene duplications are immediately favored by selection and that their fixation is driven by positive selection. Immediate benefits include adaptive changes in gene expression (Kondrashov et al. 2002; Kondrashov 2012), the immediate emergence of a new function (e.g., Arguello et al. 2006), the maintenance of permanent heterozygosity (Spofford 1969), the masking of deleterious mutations (Otto and Yong 2002), or the resolution of tradeoffs between distinct ancestral gene functions (Proulx and Phillips 2006; Connallon and Clark 2011). For this second group of models, the fate of gene duplications is determined during the fixation phase (as opposed to after fixation, as assumed by the first group of models).
In principle, it should be straightforward to distinguish between these two classes of models. Because they differ in their prediction of the evolutionary force responsible for the fixation of gene duplicates (genetic drift vs. positive selection), one could distinguish between these models by studying gene duplicates while still in the polymorphic stage. Evidence for positive selection would support the second group of models, while its absence would support the first. In the case of positive selection acting on gene duplicates, because of their recent origin, it should also be possible to gather evidence on the fitness attributes of gene duplications (e.g., beneficial increases in gene expression).
Here we offer a portrait of the polymorphic phase of the evolution of new genes in Drosophila melanogaster. Utilizing five distinct globally dispersed populations, we combine whole-genome sequencing data with gene expression profiles to (1) describe the diversity and abundance of new genes segregating in these five populations, (2) understand the transcriptional consequences of gene duplications, and (3) identify the model(s) that best describe the early evolution of gene duplications. In order to address this latter point, we have asked to what extent there is evidence for positive selection driving gene duplicates to fixation and attempt to identify the likely target(s) of selection.
Results
Abundance and diversity of new genes in five continental populations of Drosophila
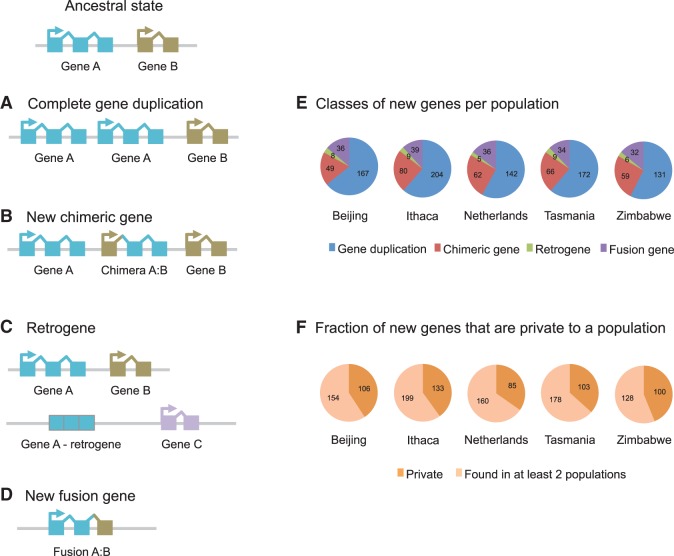
With the exception of the small number of new genes that are created de novo or are due to inversions/translocations (Ding et al. 2012; Andersson et al. 2015), new genes first appear as copy number mutations (polymorphic duplications and deletions). DNA duplications can create new genes by generating new copies of pre-existing genes (complete or partial, Fig. 1A) or by creating new genes that combine the sequences of two previously independent genes (i.e., chimeric genes) (Fig. 1B; Supplemental Fig. 1; Cardoso-Moreira and Long 2012). Duplications can also occur through an RNA intermediate (retroposition), in which case the new genes (retrogenes) have the exonic sequence of the duplicated gene but lack its introns and regulatory elements (Fig. 1C; Kaessmann et al. 2009). Finally, DNA deletions can create new genes by fusing together two previously independent genes (Fig. 1D; Long et al. 2003; Cardoso-Moreira and Long 2012).
Figure 1.
Characterization of the set of new genes segregating in the GDL. (A–D) Schematic representation of the different classes of new genes investigated in this study created by duplications (A–C) and deletions (D). Details on the formation of chimeric genes (B) in Supplemental Figure 1. (E) Counts of the different classes of new genes in each population. (F) Number of new genes that are private to one population versus those found in more than one population.
Because new genes correspond to a subset of all copy number variants (CNVs), our first step was to identify the set of CNVs that are segregating in the “Global Diversity Lines” (GDL), a reference set of 84 D. melanogaster inbred lines derived from five continents (Grenier et al. 2015). These lines were fully sequenced to an average depth of ∼12.5× and consist of 13–19 lines each from the five following populations: Ithaca (United States), Netherlands, Beijing (China), Tasmania, and Zimbabwe. The set of SNPs, small indels, and inversions segregating in these lines is publicly available (Grenier et al. 2015). We identified CNVs by integrating the results of three independent CNV-detection pipelines: Pindel (split-read detection) (Ye et al. 2009), an in-house pipeline designed around BLAT (split-read detection) (Cardoso-Moreira et al. 2012), and Delly (paired-end detection) (Rausch et al. 2012). The initial set of calls were subjected to filters (Supplemental Fig. 2), and the false-positive rate was estimated by PCR to be 12% (see Methods; Supplemental Table 1). Our final CNV data set consists of 2221 duplications, 56,562 deletions, and 3850 insertions relative to the reference genome and varying in size between 25 bp and 25 kb (our chosen size limits, see Methods) (Supplemental Table 2).
In agreement with previous work, we find that purifying selection is pervasive across the CNV data set (e.g., Emerson et al. 2008; Zichner et al. 2013). By comparing the location of the CNVs in our data set with those of a control data set created by randomly shuffling the coordinates of the CNVs across the genome, we observed that CNVs are strongly depleted among coding regions and UTRs and that this depletion is significantly stronger for deletions/insertions than for duplications (P < 2.2 × 10−16) (Supplemental Table 3). Most notably, we observed that although partial gene duplications are significantly depleted in our data set (P = 3 × 10−12), there is a clear excess of complete gene duplications (Supplemental Table 3). Given the size of the duplications in our data set, we would expect that ∼5% would encompass complete genes; instead, 14% of the duplications create new complete gene duplications (P < 2.2 × 10−16) (Supplemental Table 3). Expanded details on how purifying selection and demography shape the patterns of CNV across the genome and between populations are described in the Supplemental Material.
Our next step was to identify the subset of CNVs that create new genes. We identified 795 polymorphic new genes segregating in the 84 lines. Each population carries between 228 and 334 polymorphic new genes (Fig. 1E), and 35%–44% of these genes are private (i.e., exclusive) to each population (Fig. 1F). The distribution of the different classes of new genes is similar across all five populations: The majority (57%–64%) correspond to complete gene duplications, followed by chimeric genes (19%–26%), gene fusions (12%–15%), and finally retrogenes (2%–3%) (Fig. 1E).
Complete gene duplications
There are 491 complete gene duplications in our data set that were created by 336 independent duplications (some duplications encompass more than one gene). These gene duplications are enriched for genes associated with drug metabolism, both through the cytochrome P450 pathway and through “another pathway using other enzymes” (KEGG pathway analysis, P = 5.4 × 10−8 and P = 0.004, respectively after Holm-Bonferroni correction). In total, there are 19 different duplicated genes involved in drug metabolism, half of them private to one of the populations. It is important to note that genes in these two drug metabolism pathways are also enriched among the set of coding deletions, suggesting the genes in these pathways either experience higher mutation rates (Cardoso-Moreira and Long 2010; Cardoso-Moreira et al. 2011) and/or are less deleterious when varying in copy number than other classes of genes.
The set of gene duplications includes 45 genes (9% overall) that have been independently duplicated more than once. This is not unexpected because there are duplication hotspots in the Drosophila genome (Cardoso-Moreira and Long 2010; Cardoso-Moreira et al. 2012). Gene ontology and pathway enrichment analyses did not reveal any particular functional features associated with this group of genes. Of the 45 genes, 35 were independently duplicated twice, and eight genes were independently duplicated three times. Additionally, CG10996 (involved in the metabolism of carbohydrates) and Prosbeta5R2 (involved in the cellular response to DNA damage) were duplicated five and seven times, respectively. Both genes have increased in copy number within individual lines (Supplemental Figs. 3, 4).
Chimeric and fusion genes
There are 213 duplications that create new chimeric genes by juxtaposing the sequences of two genes on the same strand (Fig. 1B). There is, however, a similar number of duplications that juxtapose genes on opposite strands (n = 204). In principle, only chimeric structures between genes on the same strand have a chance of being (or becoming) functional. Therefore, the absence of a bias favoring novel chimeric structures between genes on the same strand suggests that our data set is not enriched with potentially functional new chimeric genes. Gene ontology and pathway enrichment analyses did not reveal functional features associated with the parental genes of chimeric genes.
Gene fusions differ from chimeric genes in that the two parental genes are lost (Fig. 1D). For this reason, gene fusions are expected to be more deleterious than chimeric genes. Unlike what we observed for chimeric genes, we found a significant excess of candidate gene fusions between genes on the same strand versus on the opposite strand: 71 versus 33, respectively (P-value = 0.0003, one-sample proportions test). We also noticed that many candidate fusions (but not chimeric genes) occur between adjacent paralogous genes. This alerted us to the possibility that gene conversion between adjacent duplicated genes could be underlying the signal of a deletion, when in fact a segment of DNA from one duplicate was copied into the other (Arguello and Connallon 2011). Distinguishing between gene conversion events and true deletions is straightforward: Only in the latter case should we find an absence of reads mapping to the predicted deleted region. By applying this additional criterion (see Supplemental Material), we arrived upon a final set of 31 fusions between genes in the same strand and 13 between genes in the opposite strand, which still reflects a significant bias favoring fusions between genes on the same strand (P = 0.01). Among the 31 fusions, five occur between pairs of sensory genes (a significant enrichment, P = 0.004 after Bonferroni correction), including the previously described Or22a:Or22b fusion (Aguadé 2009).
Retrogenes
Retrogenes can be identified when apparent deletions perfectly match the limits of the introns of a given gene. We identified 17 polymorphic retrogenes segregating in the five populations, of which only eight are complete (i.e., all coding exons duplicated) (Supplemental Table 4). Surprisingly, one of the polymorphic retrogenes was formed from the sequences of two parental genes, Cf2 and Pen, that appear to have been coretroposed. We located the insertion site of one polymorphic retrogene (CG33969 retrogene) and confirmed its structure by PCR and Sanger sequencing (Supplemental Fig. 5). This polymorphic retrogene was confirmed to be only a partial duplication; it is flanked by direct repeats, does not have a poly-A tail, and was inserted within an intron of a gene on another chromosome.
In order for retrogenes to be heritable, the retroposition has to occur in the germline, which means that the probability that a given gene generates a duplicate retrogene depends on its expression level in the germline (Kaessmann et al. 2009). Consistent with this expectation, we observed that the parental genes of retrogenes are all highly expressed in the germline (expanded details are in Supplemental Material and Supplemental Table 4). Finally, we observed that the 17 polymorphic retrogenes originate from genes that have no bias in terms of chromosomal location, in agreement with previous work (Schrider et al. 2011). Thus, unlike fixed retrogenes, which show a clear bias for originating from X-linked genes (Betrán et al. 2002), polymorphic retrogenes originate with similar probabilities from genes located on all major chromosomes. This result favors the hypothesis that retrogenes derived from X-linked parental genes are more likely to be favored (and therefore retained) by selection (e.g., Vibranovski et al. 2009).
The transcriptional consequences of gene duplications
What fraction of this abundant and diverse set of new genes actually impacts fitness? Gene duplications are expected to impact fitness when they lead to changes in gene expression that affect physiology (usually increases in expression). We can therefore investigate the potential phenotypic impact of gene duplications by evaluating how often they lead to changes in gene expression. We generated gene expression profiles for whole adult male flies for 83 of the 84 GDL using microarrays (Methods). Out of the 491 genes completely duplicated in our data set, we have expression profiles for 288 (59%). The remaining genes either were not represented on the arrays or did not have detectable expression. Of the 288 genes, 121 (42%) have significantly higher expression in the lines carrying the duplication (5% FDR, Methods), and seven genes (2%) have significantly lower expression. These results suggest that approximately half of the gene duplications translate into significant changes in gene expression and that reduced expression is rare. There are two possible reasons for why approximately half of gene duplications do not lead to changes in gene expression levels. First, some gene duplications may not contain all of the regulatory elements and are therefore not truly “complete.” Support for this scenario comes from a set of nine genes that were independently duplicated twice and that have discordant expression changes. In seven of the nine, the significant expression changes are observed for the duplication that encompasses a larger fraction of the 5′ region of the ancestral gene, suggesting the other duplication shows no change in expression because not all regulatory elements were duplicated. Second, compensation or buffering effects can prevent total gene expression from directly reflecting gene copy number. These compensation/buffering effects have been previously described in Drosophila and other species (e.g., McAnally and Yamplosky 2009; Stenberg et al. 2009; Zhang et al. 2010).
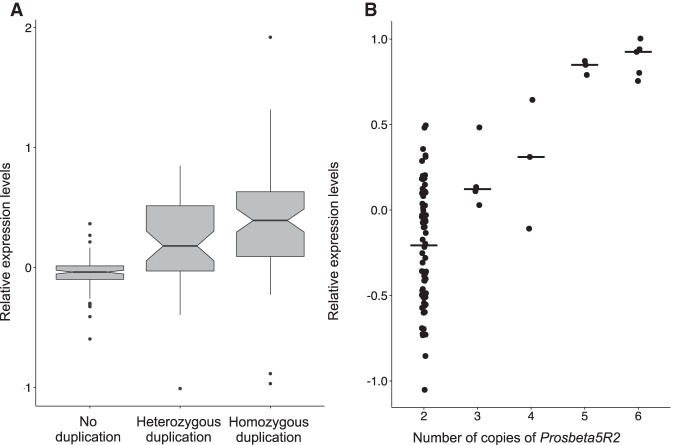
Although the GDL are mostly homozygous, the presence of inversions prevented the full inbreeding of the lines and created blocks of heterozygosity (Grenier et al. 2015). We took advantage of these “heterozygous blocks” to study gene expression changes when gene copy number increases from 2n to 3n (heterozygous duplications) and from 2n to 4n (homozygous duplications). Figure 2A focuses on the set of 132 gene duplications that are singletons (i.e., found in only one of the GDL) and contrasts the expression levels of these genes in lines without the duplications to lines carrying heterozygous and homozygous gene duplications. Two main observations stem from this analysis. First, there is a stepwise increase in gene expression with the increase in gene copy number (i.e., gene expression is highest in the lines carrying homozygous duplications, followed by lines carrying heterozygous duplications, and finally lines not carrying duplications). This step-wise increase is also observed for the several independent duplications of Prosbeta5R2 (Fig. 2B) and for three other genes with multiple independent duplications segregating in the same lines (Supplemental Fig. 6, A–C, but see also D). Second, although gene expression levels significantly increase with copy number, this increase is lower than expected if there was a direct linear relationship (slope = 1) between gene expression and gene copy number. Heterozygous duplications have median expression levels that are 13% higher (when 50% would be expected), and homozygous duplications have median expression levels 31% higher (when 100% would be expected).
Figure 2.
Relationship between gene copy number and gene expression. (A) Expression levels for genes duplicated in only one line in lines not carrying the duplication and in lines carrying heterozygous and homozygous duplications. (B) An increase in gene copy number is associated with an increase in the expression levels of Prosbeta5R2. The ancestral diploid state for this gene is two copies and its copy number can be as high as six in lines carrying three homozygous duplications.
In order to further test for the existence of buffering/compensation effects, we reanalyzed the set of singleton gene duplications by limiting the analysis to the gene duplications exhibiting significant increases in gene expression. Within this subset, we found that heterozygous duplications exhibit a median increase in expression levels of 50% (Supplemental Fig. 7), the expected result in the absence of buffering/compensation effects and similar to what was found in a previous study examining the transcriptional consequences of a similar increase in gene copy number (Stenberg et al. 2009). In contrast, we found that homozygous duplications show only a median increase of 53% (Supplemental Fig. 7), supporting the existence of buffering/compensation effects such that changes in copy number from 2n to 4n lead to less than twofold increases in gene expression. We investigated the possibility that the lack of buffering observed for heterozygous duplications was the consequence of having less power to detect changes in gene expression for heterozygous than homozygous duplications. But we found this hypothesis to be unlikely: We detect a similar percentage of homozygous and heterozygous duplications leading to significant increases in gene expression (55% and 41%, respectively, one-sided Fisher's exact test P-value = 0.095). Therefore, our data support the existence of buffering effects in Drosophila for gene duplications, but only when the increase in copy number is from 2n to 4n.
Just as we observe compensatory effects that buffer gene copy number at the transcriptional level, similar effects could operate at the translational level. This translational buffering could preclude gene duplications from significantly changing protein levels and, therefore, from impacting fitness. To investigate this, we evaluated the fitness impact of gene duplications that lead to expression changes by asking if they tend to be more deleterious than gene duplications that do not lead to expression changes. Because deleterious mutations are kept at low frequencies within populations, if gene duplications that lead to expression changes tend to be deleterious, we would expect to find a higher percentage of them segregating as singletons (i.e., present in a single line) compared with those not associated with expression changes. This is in fact what we observe. The percentage of singletons is significantly higher when gene duplications are associated with expression changes: 54% (69/128) vs. 39% (63/160) (Fisher's exact test, P = 0.017).
Evidence for positive selection driving the fixation of gene duplications
With a set of polymorphic new genes identified, we then asked if any show evidence for positive selection. We expected among the set of polymorphisms segregating in at least 10% of the lines in a population that the majority would be neutral, with a small fraction being potentially beneficial. The population dynamics of these two mutational classes differ, with beneficial mutations expected to spend significantly less time as polymorphic in a population, reaching higher frequencies (and fixation) faster than neutral mutations (Gillespie 2004). As a consequence, positively selected mutations are expected to be enriched among the set of mutations that are close to fixation or that have been recently fixed in a given population (Otto and Yong 2002). This leads to a clear prediction: If a significant fraction of gene duplications is under positive selection, they should be significantly enriched among the set of all high-frequency duplications.
For each of the five populations, we quantified the fraction of duplications that encompass noncoding regions (intronic or intergenic), partial gene duplications, chimeras (between genes on the same strand), and complete gene duplications within frequency bins (singleton, 10%–25%, 25%–50%, 50%–75%, and 75%–100%). Figure 3A shows the autosomal duplication data combined over the five populations. As predicted under a positive selection model, we found that complete gene duplications are significantly enriched among all duplications segregating in at least 75% of the lines in a given population (Fisher's exact test, P = 0.02 vs. all duplications, P = 0.03 vs. intronic/intergenic duplications). Though gene duplications correspond to 14% of all duplications, they constitute 32% of the duplications segregating at the highest frequencies (i.e., 75%–100%). We obtain a similar enrichment when considering each population separately (Fig. 3B) or when testing the X Chromosome and autosomes combined (P = 0.004 vs. all duplications, P = 0.04 vs. intronic/intergenic duplications) (Supplemental Fig. 8).
Figure 3.
Gene duplications are significantly enriched among all high-frequency duplications. (A) For bins of increasing frequency, we plot the fraction of duplications (per bin) that overlap different genomic contexts. The data shown represent the combined data from the five populations for the subset of autosomal duplications. The asterisks indicate that gene duplications are significantly enriched among all duplications segregating in at least 75% of the lines (Fisher's exact test, P = 0.02 vs. all duplications, P = 0.03 vs. intronic/intergenic duplications). (B) Subset of A showing for the five populations (in gray) and for the combined data (in red) the fraction of duplications that corresponds to gene duplications across the different frequency bins.
Complete gene duplications are the only class of new genes showing evidence for being under positive selection. In striking contrast, polymorphic chimeric genes are almost absent from the set of duplications reaching 50% of the lines (Fig. 3A). As for polymorphic retrogenes, only five of the 17 are segregating in >25% of the lines in one of the populations (Supplemental Table 4). A particularly interesting case is the eIF-4E retrogene, found only in Tasmania (the parental gene is a translation initiation factor), where it segregates in roughly half the lines (eight of 18). There is also little evidence suggesting that positive selection is acting on the set of gene fusions as all are found at low frequencies, with four exceptions: the Or22a:Or22b fusion, which has been previously proposed to be under direct or indirect selection (Aguadé 2009) (see also Supplemental Material), and three other high-frequency fusions, all fusions of tandem small nucleolar RNAs (snoRNAs). It is important to note that although we only found evidence for positive selection acting on complete gene duplications as a class, this does not preclude positive selection acting on individual new genes created by different mechanisms (e.g., the Or22a:22b fusion).
Critically, only ∼1% of all duplications are segregating in 75%–100% of the lines in at least one population (32 duplications in total, 22 autosomal). To preclude the potential for biasing our findings in the direction of favoring a role for positive selection in the fixation of gene duplications, we have excluded from our analyses (depicted in Fig. 3; Supplemental Fig. 8) the duplications associated with the Cyp6g1 gene that are already known to be under positive selection. Cyp6g1 is arguably the best example of a gene undergoing recent and strong selection in D. melanogaster. Insertions of the Accord transposable element in the 5′ region of Cyp6g1 are associated with an increased expression of this gene and with a significant increase in the resistance to the insecticide DDT (Daborn et al. 2002). In addition to the Accord insertion, this locus carries copy number mutations and additional transposable element insertions that were proposed to underlie adaptive phenotypic variation (Schmidt et al. 2010). We identified three duplications overlapping Cyp6g1 (and five small deletions) in the GDL, though only one encompasses the entire coding sequence of the gene (Supplemental Fig. 9A). Two of the duplications always cosegregate in non-African lines where they are either fixed or close to fixation (93% frequency in Beijing, 94% in Tasmania, 95% in Ithaca, and 100% in Netherlands) (Supplemental Fig. 9B). These duplications are associated with a significant increase in the expression levels of Cyp6g1 (Supplemental Fig. 9C).
Table 1 provides a description of the high-frequency gene duplications segregating in the GDL. These duplicated genes do not cluster within a specific ontology class but instead have a variety of biological functions. With the exception of the duplication of CG14810, which is only present in Zimbabwe, the remaining gene duplications are detected in all five populations, suggesting that they were created before the migration out of Africa, ∼16,000 yr ago (Stephan and Li 2007). Although some of these gene duplications show high levels of population differentiation, overall they are not enriched in the upper tail of the Fst distribution (a measure of population differentiation).
Table 1.
Characteristics of high-frequency gene duplications (and the Prosbeta5R2 duplications)

Three of the high-frequency duplications are of genes that have additional independent complete gene duplications segregating in our data set, a significant enrichment compared with the data set as a whole (only 9% of genes have been independently duplicated, Fisher's exact test P = 0.02). The additional duplications of CG34002 and CG7966 are present in lines already carrying a duplication, suggesting independent expansions of the original duplication. For CG10996, we detected five independent complete gene duplications (Supplemental Fig. 3) that, when combined, lead to an increase of the percentage of lines carrying a duplication of this gene (Supplemental Table 5). This led us to ask if there were examples of genes where the aggregated frequencies of all independent duplications would reach at least 75% of the lines in any one population. The one instance identified was of a duplicated pseudogene (CR43086) that is itself an older duplication of the gene CG34002 (one of the high-frequency gene duplications in our data set). We found an additional four genes (plus another pseudogene of CG34002) where the aggregated frequencies of all independent duplications reached at least 50% frequency in one population (Supplemental Table 5). Among these genes is Prosbeta5R2, which is remarkable for being predicted to have been independently duplicated seven times (Supplemental Fig. 4). In the Netherlands population, complete gene duplications of Prosbeta5R2 are found in 58% of the lines; however, outside this population, duplications of this gene are rare or absent (Supplemental Fig. 4). Because this gene has undergone such a dramatic expansion and in a single population, and because of the clear correlation between expression levels and copy number (Fig. 2B), we consider Prosbeta5R2 to be a strong candidate for being under positive selection.
Population genetic signals of positive selection for high-frequency gene duplications
In addition to reducing the time that variants spend as polymorphisms, the action of positive selection can also be indirectly observed through its impact on patterns of linked polymorphisms (e.g., Hohenlohe et al. 2010; Vitti et al. 2013). One of the strongest signals of positive selection for very young variants (i.e., those not yet fixed in the species such as the gene duplications in our study) is a significant increase in the extent of linkage disequilibrium (LD) between SNPs flanking the selected region. Therefore, in order to further test for signals of positive selection, we investigated the levels of LD surrounding the high-frequency duplications in our data set.
As previously described, LD varies between populations, between the X and the autosomes, and between pericentromeric and normally recombining regions of chromosomes (Grenier et al. 2015). Therefore, we investigated the distribution of population-specific LD among SNPs located 5′ and 3′ for all duplications in our data set that were within regions of normal recombination (i.e., nonpericentromeric). This allowed us to estimate the background LD distribution (calculated separately for the X and autosomes). We then compared the distributions of LD flanking all high-frequency duplications to the background distribution. If high-frequency gene duplications are under positive selection, the expectation is that they will show a significantly higher LD than the background distribution. We would also expect that only high-frequency gene duplications will show this elevated LD; i.e., high-frequency noncoding duplications should show similar levels of LD to the background distribution. Finally, we would expect that the increase in LD would be consistent with the frequency of the duplication in the population analyzed. Agreement between elevated LD and the high frequency of a duplication in a population provides additional confidence that the duplication is the direct target of selection (as opposed to being linked to a variant under selection).
For all non-African populations, the duplications displaying the highest levels of LD are the two duplications associated with the Cyp6g1 gene, a gene well known to be associated with insecticide resistance (Supplemental Figs. 10–13). As expected, in Zimbabwe, where these duplications segregate at low frequency, the levels of LD are similar to the background distribution (Supplemental Fig. 14). In addition to the Cyp6g1 locus, LD is significantly elevated around seven autosomal duplications at high frequency: three partial gene duplications and four complete gene duplications (Table 1; Fig. 4; Supplemental Figs. 10–14). None of the seven high-frequency noncoding duplications show elevated levels of LD. Critically, all complete gene duplications have higher LD in the populations where they are segregating in high frequency. For example, the only population where the independent duplications of Prosbeta5R2 reach appreciable frequencies is the Netherlands, which is also the only population in which we observe a strong increase in LD (Supplemental Figs. 10–14). For the remaining three gene duplications, a clear signal of elevated LD is only observed for a subset of the populations that carry the duplications in high frequency (Table 1). Unlike what we observe for complete gene duplications, only a single partial gene duplication shows agreement between elevated LD and high frequency (ID_39508). For the other two partial gene duplications, elevated LD is observed in populations where the duplications are absent, suggesting they are not the direct target of selection.
Figure 4.
High levels of LD flanking three duplications in the Ithaca population are suggestive of positive selection. Supplemental Figures 10–14 depict the background LD distribution and the distributions of LD surrounding all high-frequency duplications for each of the five populations.
In summary, out of 18 autosomal high-frequency duplications (four noncoding high-frequency duplications were excluded because they are pericentromeric), we detected elevated levels of LD that are suggestive of direct positive selection for: two Cyp6g1 duplications, four of six complete gene duplications, one of four partial gene duplications, and zero of the seven noncoding duplications. No evidence for elevated levels of LD in the expected population was observed for the seven X-linked high-frequency duplications (two gene duplications and five noncoding duplications).
An additional hallmark of positive selection is a decrease in nucleotide diversity flanking the variant under selection (“selective sweep”) (e.g., Hohenlohe et al. 2010; Vitti et al. 2013). For the set of 32 high-frequency duplications, plus the Prosbeta5R2 duplications, we compared the population-specific levels of nucleotide diversity flanking the duplications in the lines carrying the duplication to the lines not carrying the duplication. We found evidence for reduced levels of diversity flanking the two Cyp6g1 duplications (Supplemental Fig. 15), the duplication of CG9186 (Supplemental Fig. 16), the duplication of CG7966 (Supplemental Fig. 17), the duplications of Prosbeta5R2 (Fig. 5; Supplemental Fig. 18), a partial gene duplication (ID_19743) and an intergenic duplication (ID_53971). It is important to note that the signals for elevated LD and reduced diversity for the same duplication often come from different populations (Table 1).
Figure 5.
Reduced levels of nucleotide diversity flanking two gene duplications are suggestive of positive selection. (A) Comparison of diversity levels in lines carrying the CG7966 duplication (n = 7, solid line) and not carrying this gene duplication (n = 12, dashed line) in the Ithaca population. The gray box marks the limits of the duplication; and the dotted horizontal line, the median diversity levels for this chromosome in this population (data for the other populations in Supplemental Fig. 17). (B) Comparison of diversity levels in lines carrying the highest-frequency duplication of Prosbeta5R2 (n = 9, solid line) and not carrying this gene duplication (n = 10, dashed line) in the Netherlands population (data for the other populations in Supplemental Fig. 18).
Overall, combining data on LD and nucleotide diversity, we found evidence for positive selection acting on five of eight complete gene duplications plus the Cyp6g1 duplications. This suggests that about half of the gene duplications that are fixed or close to fixation in at least one of the populations were driven to higher frequencies by positive selection. Encouragingly, this proportion matches the observed excess of gene duplications among high-frequency variants: We see approximately double the count of gene duplications compared with what is expected from the overall proportion of gene duplications in our data set (14% in the full data set vs. 32% among high-frequency variants).
Potential targets for positive selection
Our analyses have provided evidence that positive selection has been responsible for the rise in frequency of approximately half of the high-frequency gene duplications, but what might be the target of selection? One possible benefit of gene duplications is an advantageous increase in gene expression. We have expression data for five of the six gene duplications carrying signals of positive selection, and in all five instances, we observe significantly higher expression levels within the lines carrying the duplications. Given that only 38% of nonsingleton gene duplications are associated with expression changes, this is significantly more than expected by chance (binomial test, P = 0.008). Our data therefore support the hypothesis that beneficial increases in dosage confer higher fitness and contribute to the fixation of gene duplications.
A nonmutually exclusive alternative is that one or both of the gene copies carry additional mutations that confer a new function or optimize a previously existing function. Because the regulatory elements of these genes are not fully known, we cannot evaluate whether there have been changes in the regulatory sequences that provide one or both copies with different expression profiles. We can, however, investigate the possibility that their protein sequences have changed. To this end, we identified all SNPs, indels, and CNVs that appeared after the gene duplication event and that have the potential to affect the protein sequence (amino acid changing mutations or more radical changes). For four of the six gene duplications, either there are no additional mutations capable of changing the protein sequence, or a candidate mutation is segregating at very low frequency in the lines carrying the duplication. For the other two gene duplications, we found variants with the potential to affect gene function. The duplication of CG34002 is either fixed or close to fixation in all populations analyzed (89%–100% frequency). However, outside of Africa some lines are carrying smaller deletions that inactivate one or both copies of this gene (some lines carry two overlapping deletions implying each is linked to a different copy of the gene) (Supplemental Fig. 19). In addition, there is extensive copy number variation in the region spanning CG34002 and two downstream pseudogenized older duplicates, with many of the variants having reached appreciable frequencies worldwide (Supplemental Fig. 19). The functional and fitness implications of the CG34002 duplication remain unclear, but dosage cannot be excluded (CG34002 was not present on the microarray). The other gene duplication carrying additional mutations capable of affecting function is that of CG9186, which has been shown to be crucial for the organization of lipid particles and, therefore, for fat storage (Thiel et al. 2013). A beneficial increase in dosage is plausible for this locus as the duplication leads to a significantly higher expression level. However, there are also four additional mutations segregating with the majority of the lines carrying the duplication (three amino acid substitutions and a coding indel). None of these four mutations truncate the protein or otherwise disrupt the reading frame (Supplemental Fig. 20). Interestingly, however, they do lead to amino acid changes in positions that are highly conserved throughout insects and mammals (Thiel et al. 2013). Unfortunately, the current data cannot resolve how these mutations are distributed between the two copies. CG9186 is the best candidate for a gene duplication being under positive selection for more than a beneficial increase in dosage.
In summary, beneficial increases in gene dosage could potentially explain why all high-frequency gene duplications identified in this study are being driven to fixation by positive selection. However, our data also hints at the possibility that additional mutations can occur after the duplication event that allow one or both duplicates to gain or optimize gene function.
Discussion
We have studied the polymorphic phase of gene duplication in D. melanogaster with the aim of identifying the models that best describe the evolution of gene duplications in this species. We have attempted to distinguish between the different models by identifying the evolutionary force(s) responsible for driving new gene duplications to fixation: Whereas a group of models posits that gene duplications are neutral and therefore fixed by genetic drift, another group posits that gene duplications are immediately beneficial and therefore fixed by positive selection (Conant and Wolfe 2008; Hahn 2009; Innan and Kondrashov 2010). Our work found that about half of the high-frequency gene duplications segregating in the GDL show signs of being under positive selection, suggesting that in Drosophila, approximately half of the gene duplications are fixed by positive selection with the other half fixed by genetic drift. The evolutionary trajectories of these two groups of gene duplications are expected to be different. After fixation, the gene duplications fixed by genetic drift will continue to evolve neutrally, and the most likely outcome is that one of the copies will become a pseudogene. On the other hand, those gene duplications that were fixed by positive selection will be under purifying selection, which will extend their half-life, thereby significantly increasing the probability that mutation(s) capable of creating a new function will appear and therefore that a new function will evolve (neofunctionalization). This result is in agreement with the empirical observations that even closely related Drosophila species show significant differences in gene content (e.g., Drosophila 12 Genomes Consortium 2007) and that new gene duplications can quickly evolve new functions (Chen et al. 2010; Kaessmann 2010).
Our work also suggests that advantageous increases in gene expression may provide a common explanation for the fitness benefits of gene duplications. Selection for increased dosage does not necessarily have to be for the gene's primary activity; it could also be for trace side activity that becomes beneficial after an environmental change (Bergthorsson et al. 2007). More generally, our work supports the assertion so often made but lacking strong empirical support: When gene duplications lead to changes in gene expression, they are generally deleterious (e.g., Kondrashov and Kondrashov 2006; Kondrashov 2012). These signatures can be seen on both ends of the site frequency spectrum: Gene duplications leading to expression changes are enriched among the set of low- and high-frequency duplications. Does knowing that gene dosage is a main reason why gene duplications are fixed impact our expectations regarding the long-term fate of gene duplications in Drosophila? While dosage impacts fitness, the duplicates will always face purifying selection, and loss-of-function mutations will be purged. Beneficial mutations that do not interfere with the duplicates’ function will, however, be allowed, and so the duplicates can show some limited functional divergence. If higher dosage stops being under selection and the two duplicates are functionally completely redundant, the most likely outcome is that one will be lost. However, if the duplicates have diverged and acquired new functions, then both will still be under purifying selection, and both duplicates (now not fully redundant) will be retained in the genome. At this point, each duplicate is free to evolve toward its optimum expression level, although there may still be constraints associated with shared functions. Therefore, once dosage stops being under selection, the summed transcript abundance of the two duplicates can follow different trajectories, including a significant decrease in the amount of gene expression of the duplicates as observed in some studies (e.g., Qian et al. 2010).
What can these observations made in Drosophila teach us about the evolution of gene duplications in other species? Because the number of gene duplications fixed or close to fixation in Drosophila is necessarily small, our estimate that approximately half of gene duplications are fixed by positive selection is inevitably bounded by large confidence intervals. Still, it is close to the fraction of amino acid mutations that are fixed by positive selection in Drosophila (∼40%–50%) (Sella et al. 2009). Therefore, it is likely that the same principles that apply to the fixation of amino acid mutations apply to gene duplications and that, in species with smaller effective population sizes (such as humans), a greater fraction of gene duplications will be fixed by genetic drift. If true, then the early evolutionary dynamics of gene duplications in species like Drosophila will be quite distinct from mammalian species (and other species with low effective population sizes): Drosophila should experience high rates of neofunctionalization, while mammalian species will maintain gene duplicates mostly due to the division among the duplicates of the ancestral gene functions (i.e., through subfunctionalization).
Methods
Identification of CNVs segregating in the D. melanogaster GDL
The GDL consist of 84 lines derived from five world populations: Beijing, China (15 lines); Ithaca, US (19 lines); Netherlands (19 lines); Tasmania (18 lines); and Zimbabwe (13 lines) (Greenberg et al. 2010). All 84 lines were inbred for 12 generations and are fully homozygous except in regions associated with inversions and termed “heterozygous blocks” (Grenier et al. 2015). We identified CNVs segregating in these lines by employing three independent CNV detection pipelines and accepting a CNV call when supported by at least two of the pipelines (Supplemental Material). The three pipelines used were Pindel (Ye et al. 2009), an in-house pipeline around BLAT (Cardoso-Moreira et al. 2012), and Delly (Rausch et al. 2012). The parameters used to run each of these pipelines are described in the Supplemental Material. The set of CNVs was filtered to exclude variants with breakpoints associated with transposable elements or other classes of repeats (Supplemental Material) and to only include variants between 25 bp and 25 kb (Supplemental Fig. 2). We excluded variants >25 kb (103 putative deletions and 36 putative duplications) because the read coverage within these variants did not support copy number decreases/increases (they may correspond to other structural mutations and/or gene conversion events).
We empirically evaluated the quality of our calls by PCR (and for a subset of the calls by Sanger sequencing, Supplemental Table 1). For duplications, we designed divergent primers within the predicted boundaries that only allow DNA amplification in the presence of a tandem duplication. For insertions and deletions, the primers were designed to flank the CNVs such that their presence leads to larger (for insertions) or smaller (for deletions) PCR bands than in a control line not carrying the CNVs (that additionally showed the expected size according to the reference genome). We obtained diagnostic PCR results for 27 duplications and 72 deletions (Supplemental Table 1). Of these, three of 27 duplications and nine of 72 insertions/deletions were determined to be false positives (11% and 13%, respectively).
Our CNV detection pipelines identify CNVs by comparison with the reference genome, which means that a small fraction of our calls are expected to correspond to novel variants carried by the reference genome. We polarized our set of CNV calls (i.e., determined whether the CNVs correspond to the ancestral or derived states) using the approaches described in the Supplemental Material. Our final genome annotations were done using FlyBase release 5.52 (dos Santos et al. 2015). In order to determine if there was a paucity or excess of CNVs overlapping different genomic contexts (i.e., intergenic and intronic regions, UTR, coding exons, and complete genes), we randomly shuffled 100 times the CNV coordinates within each chromosome using BEDTools's shuffleBed (Quinlan and Hall 2010). We then applied to these shuffled segments the transposable element/repeat filter also applied to the set of CNV calls. We ended up with about 95 times the number of original calls. By using these data, we determined how often the shuffled segments overlapped the different genomic contexts and contrasted these results with those observed for our set of CNV calls.
We identified gene duplications, chimeras, and fusions by identifying the new gene configurations described in Figure 1. The gene and pathway enrichment analyses were performed using FlyMine 40.0 (Lyne et al. 2007) and applying the Benjamini-Hochberg correction for multiple comparisons. The identification of retrogenes is described in detail in the Supplemental Material.
Determining the transcriptional consequences of gene duplications
We generated expression profiles for 83 of the 84 GDL (no data for ZW155) using two-channel spotted oligonucleotide arrays (69 bp oligos; Operon Array-Ready Oligo Set for the Drosophila Genome). The profiles were generated using total RNA isolated from 15 whole adult males (RNeasy 96 kit, Qiagen), and one to three replicates were created per line (only four lines had no replicate; the median and mean number of replicates is three). The reference RNA used in all array hybridizations consisted of a pool made from individual RNA samples from all lines. The final expression values are in the form of the log2 (test line/reference) and represent the median across the replicates (Supplemental Table 6). We applied two filters to these data. The first removed all genes identified as being lowly expressed or not expressed at all (Supplemental Material). The second removed all probes with potential cross-hybridization effects (Supplemental Material). When genes were represented by more than one probe in the array, we used the median value across all probes.
We used a permutation test to establish cutoffs for a significant change in gene expression between lines carrying different gene copy numbers. For the set of complete gene duplications, we shuffled their frequencies enough times to end up with about 25,000 data points (i.e., we randomly changed the identity of the lines carrying a given gene duplication while maintaining its frequency in the set of 84 lines). We removed all instances where the shuffling recapitulated the original data. Because gene expression data are only available for 59% of the genes duplicated, we ended up with about 14,000 data points (each corresponding to a gene-frequency combination). We then calculated the difference between the median expression of the lines assigned as carrying a gene duplication and those assigned as not carrying a gene duplication. As expected, the median (and mean) of this normal distribution is centered around zero with the 5% and 95% tails being −0.39 and 0.43, respectively. We then calculated, for the real set of gene duplications, the difference between the median gene expression of lines carrying and not carrying a gene duplication. We classified gene duplications as leading to significant changes in gene expression when this difference was smaller than −0.39 (significant decrease in expression) or larger than 0.43 (significant increase in expression).
Detecting signals of positive selection surrounding high-frequency duplications
We calculate the levels of LD (as measured by r2) between all SNPs located within 5 kb (for the non-African populations) or within 2.5 kb (for Zimbabwe) upstream of and downstream from all high-frequency duplications segregating in our data set using VCFtools (version 0.1.12a) (Danecek et al. 2011). We performed these calculations separately for each population and used the following parameters: –geno-r2 and –ld-window-bp 5000. We selected a smaller window for the Zimbabwe population because LD breaks significantly faster in this population (Grenier et al. 2015). We estimated diversity levels (as measured by π, the average number of nucleotide differences per site between two DNA sequences randomly chosen from one population) for 500 bp windows within 50 kb upstream of and downstream from all high-frequency duplications using VCFtools (version 0.1.12a) (Danecek et al. 2011). π was estimated separately for each population and within each population for lines carrying and not carrying a high-frequency duplication so long as there were at least five lines in each condition. We calculated Fst using the unbiased approach of Weir and Clark (1984), which allows for unequal sample sizes across populations. We defined the tail of Fst for each pairwise comparison as corresponding to the 10 most extreme Fst values, which corresponded to 16–43 duplications depending on how many duplications were given the same Fst estimate.
Identifying additional mutations segregating within high-frequency gene duplications
We identified all SNPs and small indels segregating within the set of high-frequency gene duplications using VCFtools. These variants lead to the appearance of a heterozygous call in what would otherwise be a homozygous region (because the reads from both duplicates map to the same genomic location in the reference genome). In the set of calls generated for the GDL, heterozygous variants located outside of heterozygous blocks (formed by inversions) are coded as noncalls. The information regarding the conservation of amino acid positions in CG9186 was taken from Thiel et al. (2013). The annotation of the SNPs and indels was generated using SnpEff (Cingolani et al. 2012) and obtained from Grenier et al. (2015).
All statistical analyses were done using the statistical package R (R Core Team 2014). The plots were generated using the following R packages: ggplot2 (Wickham 2009), gridExtra (https://CRAN.R-project.org/package=gridExtra), reshape2 (Wickham 2007), mapplots (https://CRAN.R-project.org/package=mapplots), mapproj (https://CRAN.R-project.org/package=mapproj), and scales (https://CRAN.R-project.org/package=scales). R and the associated packages were run using the application Rstudio (RStudio Team 2015).
Data access
The CNV calls from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/Traces/sra/) and added to accession number SRP050151. The expression data from this study have been submitted to EMBL-EBI ArrayExpress (http://www.ebi.ac.uk/arrayexpress/experiments/) under accession number E-MTAB-4580.
Acknowledgments
We thank Tim Connallon, Liang Leng, Maria Vibranovski, and Yong Zhang for comments and critical reading of the manuscript. This work was supported in part by the National Institutes of Health grant R01 DK074136 to A.G.C. and L.G.H. M.C.M. was supported by a post-doctoral fellowship from the Portuguese Foundation for Science and Technology (cofinanced by POPH/FSE) and by a Novartis post-doctoral fellowship. J.R.A. was supported by a Cornell Center for Comparative and Population Genomics Fellowship and by a Novartis post-doctoral fellowship.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.199323.115.
References
- Aguadé M. 2009. Nucleotide and copy-number polymorphism at the odorant receptor genes Or22a and Or22b in Drosophila melanogaster. Mol Biol Evol 26: 61–70. [DOI] [PubMed] [Google Scholar]
- Andersson L. 2013. Molecular consequences of animal breeding. Curr Opin Genet Dev 23: 295–301. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Jerlström-Hultqvist J, Näsvall J. 2015. Evolution of new functions de novo and from preexisting genes. Cold Spring Harb Perspect Biol 7: a017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JR, Connallon T. 2011. Gene duplication and ectopic gene conversion in Drosophila. Genes 2: 131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JR, Chen Y, Yang S, Wang W, Long M. 2006. Origination of an X-linked testes chimeric gene by illegitimate recombination in Drosophila. PLoS Genet 2: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Andersson DI, Roth JR. 2007. Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci 104: 17004–17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E, Thornton K, Long M. 2002. Retroposed new genes out of the X in Drosophila. Genome Res 12: 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira MM, Long M. 2010. Mutational bias shaping fly copy number variation: implications for genome evolution. Trends Genet 26: 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Long M. 2012. The origin and evolution of new genes. Methods Mol Biol 856: 161–186. [DOI] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Emerson JJ, Clark AG, Long M. 2011. Drosophila duplication hotspots are associated with late-replicating regions of the genome. PLoS Genet 7: e1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Arguello JR, Clark AG. 2012. Mutation spectrum of Drosophila CNVs revealed by breakpoint sequencing. Genome Biol 13: R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950. [DOI] [PubMed] [Google Scholar]
- Connallon T, Clark AG. 2011. The resolution of sexual antagonism by gene duplication. Genetics 187: 919–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, Tijet N, Perry T, Heckel D, Batterham P, et al. 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765. [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhou Q, Wang W. 2012. Origins of new genes and evolution of their novel functions. Ann Rev Ecol Syst 43: 345–363. [Google Scholar]
- dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM; FlyBase Consortium. 2015. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 43: D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, Pollack JR, Sikela JM. 2007. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res 17: 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631. [DOI] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, et al. 2015. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347: 1465–1470. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH. 2004. Population genetics: a concise guide, 2nd ed Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- Greenberg AJ, Hackett SR, Harshman LG, Clark AG. 2010. A hierarchical Bayesian model for a novel sparse partial diallel crossing design. Genetics 185: 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier JK, Arguello JR, Moreira MC, Gottipati S, Mohammed J, Hackett SR, Boughton R, Greenberg AJ, Clark AG. 2015. Global diversity lines: a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 5: 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered 100: 605–617. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Phillips PC, Cresko WA. 2010. Using population genomics to detect selection in natural populations: key concepts and methodological considerations. Int J Plant Sci 171: 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. 1994. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci 256: 119–124. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res 20: 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet 10: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. 2009. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet 25: 404–413. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279: 5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Kondrashov AS. 2006. Role of selection in fixation of gene duplications. J Theor Biol 239: 141–151. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. 2002. Selection in the evolution of gene duplications. Genome Biol 3: RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet 4: 865–875. [DOI] [PubMed] [Google Scholar]
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, Mclaren P, North P, et al. 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol 8: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnally AA, Yampolsky LY. 2009. Widespread transcriptional autosomal dosage compensation in Drosophila correlates with gene expression level. Genome Biol Evol 2: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer, New York. [Google Scholar]
- Otto SP, Yong P. 2002. The evolution of gene duplicates. Adv Genet 46: 451–483. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet 39: 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx SR, Phillips PC. 2006. Allelic divergence precedes and promotes gene duplication. Evolution 60: 881–892. [PubMed] [Google Scholar]
- Qian W, Liao BY, Chang AY, Zhang J. 2010. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet 26: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Ragsdale EJ, Müller MR, Rödelsperger C, Sommer RJ. 2013. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell 155: 922–933. [DOI] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team. 2015. RStudio: integrated development for R. RStudio, Boston, MA: http://www.rstudio.com/. [Google Scholar]
- Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, Martin J, Daborn PJ, Goddard ME, Batterham P, et al. 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Stevens K, Cardeño CM, Langley CH, Hahn MW. 2011. Genome-wide analysis of retrogene polymorphisms in Drosophila melanogaster. Genome Res 21: 2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet 5: e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford JB. 1969. Single-locus modification of position-effect variegation in Drosophila melanogaster. II. Region 3c loci. Genetics 62: 555–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P, Lundberg LE, Johansson AM, Rydén P, Svensson MJ, Larsson J. 2009. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet 5: e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W, Li H. 2007. The recent demographic and adaptive history of Drosophila melanogaster. Heredity (Edinb) 98: 65–68. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38: 615–643. [DOI] [PubMed] [Google Scholar]
- Thiel K, Heier C, Haberl V, Thul PJ, Oberer M, Lass A, Jäckle H, Beller M. 2013. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J Cell Sci 126: 2198–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M. 2009. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet 5: e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC. 2013. Detecting natural selection in genomic data. Annu Rev Genet 47: 97–120. [DOI] [PubMed] [Google Scholar]
- Vlad D, Kierzkowski D, Rast MI, Vuolo F, Dello Ioio R, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, et al. 2014. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Weir B, Clark CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2007. Reshaping data with the reshape package. J Stat Software 21: 1–20. [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York. [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2006. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat Genet 38: 819–823. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B. 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol 8: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zichner T, Garfield DA, Rausch T, Stütz AM, Cannavó E, Braun M, Furlong EE, Korbel JO. 2013. Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing. Genome Res 23: 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]