Abstract
A three-dimensional chromatin state underpins the structural and functional basis of the genome by bringing regulatory elements and genes into close spatial proximity to ensure proper, cell-type–specific gene expression profiles. Here, we performed Hi-C chromosome conformation capture sequencing to investigate how three-dimensional chromatin organization is disrupted in the context of copy-number variation, long-range epigenetic remodeling, and atypical gene expression programs in prostate cancer. We find that cancer cells retain the ability to segment their genomes into megabase-sized topologically associated domains (TADs); however, these domains are generally smaller due to establishment of additional domain boundaries. Interestingly, a large proportion of the new cancer-specific domain boundaries occur at regions that display copy-number variation. Notably, a common deletion on 17p13.1 in prostate cancer spanning the TP53 tumor suppressor locus results in bifurcation of a single TAD into two distinct smaller TADs. Change in domain structure is also accompanied by novel cancer-specific chromatin interactions within the TADs that are enriched at regulatory elements such as enhancers, promoters, and insulators, and associated with alterations in gene expression. We also show that differential chromatin interactions across regulatory regions occur within long-range epigenetically activated or silenced regions of concordant gene activation or repression in prostate cancer. Finally, we present a novel visualization tool that enables integrated exploration of Hi-C interaction data, the transcriptome, and epigenome. This study provides new insights into the relationship between long-range epigenetic and genomic dysregulation and changes in higher-order chromatin interactions in cancer.
Genomic structural alterations, including copy-number variations (CNVs) and translocations are common in cancer, including prostate cancer (Kluth et al. 2014), leading to de-regulation of gene expression. Epigenetic alterations are also prevalent in cancer and encompass coordinated changes in DNA methylation, nucleosome positions, and histone modifications (Plass et al. 2013; Timp and Feinberg 2013); however, the relationship between the cancer genome, epigenome, and transcriptome is still in its infancy. Traditionally, there was a focus on understanding how the epigenetic machinery controls promoters, and therefore, much is known about promoter epigenetic aberrations in cancer cells, particularly at CpG islands. More recently, it was established that distal regulatory elements including enhancers (Akhtar-Zaidi et al. 2012; Taberlay et al. 2014) and insulators (Taberlay et al. 2014) are also subject to epigenetic remodeling. Enhancer-promoter connections provide an additional layer of epigenetic transcriptional control and depend on close spatial proximity of genomic elements in three-dimensional space. Interestingly, cancer cells have been shown to display differential spatial interactions across the well-studied 8q24 region containing MYC in prostate cancer (Jia et al. 2009; Pomerantz et al. 2009; Ahmadiyeh et al. 2010; Du et al. 2015) and differential enhancer usage at 9q22 in thyroid cancer (He et al. 2015). The overexpression of an oncogenic fusion protein, ERG, in normal prostate cells is also associated with global changes in chromatin organization (Elemento et al. 2012; Rickman et al. 2012). Our previous work demonstrating widespread epigenetic changes of both enhancers and insulators (Taberlay et al. 2014) across cancer genomes suggests that changes in the cancer “interactome” may therefore be important in the context of widespread, global genetic and epigenetic dysregulation and changes to the gene expression programs in carcinogenesis.
The ability to detect DNA interactions and model three-dimensional chromatin structures has been enabled by chromosome conformation techniques such as 3C (Dekker et al. 2002) and Hi-C, its global derivative (Lieberman-Aiden et al. 2009). Insight from chromosome conformation studies has revealed that the interactome contributes to the overall higher-order hierarchical structure of the genome, which is built from highly organized functional domains and territories (Dixon et al. 2012, 2015; Nora et al. 2012; Filippova et al. 2014). In particular, chromosomes favor the formation of topologically associated domains (TADs) separated by boundary regions (Dixon et al. 2012). TADs are highly interactive chromatin substructures of approximately one megabase pairs (1 Mb) in size. Within them are smaller domains called sub-TADs that often contain genes of similar expression and epigenetic profile (Yaffe and Tanay 2011) while retaining the classical hierarchical organization of the chromosome. These compartments are obvious during inter-phase (Naumova et al. 2013) and, interestingly, appear highly conserved between cell types (Dixon et al. 2012, 2015; Nora et al. 2012) despite a large degree of genomic flexibility that must exist between TAD boundaries to establish and maintain cell-type–specific gene expression programs. These findings imply that the basic substructure of a genome is retained across all phenotypes; however, it remains unclear whether this extends to include cells of tumorigenic origin and whether an atypical interactome may drive the gene expression differences distinguishing normal from cancer cells.
The concordant inactivation of adjacent genes due to long-range epigenetic silencing (LRES) has been observed in various cancer types, including colorectal, bladder, non-small cell lung cancer, breast, prostate, and Wilms’ tumor (Frigola et al. 2006; Stransky et al. 2006; Hitchins et al. 2007; Novak et al. 2008; Seng et al. 2008; Dallosso et al. 2009; Rafique et al. 2015). In prostate cancer, we have shown that LRES is primarily characterized by regional gains of repressive histone marks and loss of active histone marks encompassing tumor suppressor or cancer-associated genes (Coolen et al. 2010). More recently, we demonstrated that concordant activation of adjacent genes also occurs in prostate cancer, and this is due to long-range epigenetic activation (LREA) (Bert et al. 2013). In contrast to LRES domains, we found that LREA domains were associated with simultaneous gains in active histone marks and loss of repressive histone marks, and these domains commonly harbor oncogenes (Bert et al. 2013). While our previous work highlights that epigenetic remodeling in cancer can occur across large domains, the potential impact on higher-order chromatin interactions has not been characterized.
Results
Topological-associated domains are smaller and more numerous in the cancer cell genome
To test the hypothesis that three-dimensional chromatin organization is disrupted in cancer cells, we performed the first comparative analysis of Hi-C chromosome conformation capture sequencing data sets generated from normal prostate epithelial cells (PrEC) and two prostate cancer cell lines (PC3 and LNCaP). We generated over 4.61 billion reads of Hi-C data from three cell lines (Supplemental Table S1). Hi-C interactions from replicate experiments were mapped to the human reference genome (hg19), normalized, and corrected for bias (see Supplemental Material), then used for further analysis. These libraries were validated using a chromosome conformation capture (3C) quantitative PCR assay across the kallikrein gene locus (Supplemental Fig. S1A). This locus encompasses adjacent epigenetically silenced and activated domains that are bound by CTCF (Bert et al. 2013), and we confirmed a high degree of consistency between 3C and Hi-C derived interactions in all cell lines.
We next explored the segmentation of the normal and cancer genomes into topologically associated domains (Dixon et al. 2012, 2015; Nora et al. 2012). Using a hidden Markov model based on the directionality index (Dixon et al. 2012) at 40-kb resolution, we identified 317 TADs with a mean size of 7.86 Mb in PrEC cells, 622 TADs with a mean size of 3.92 Mb in PC3 cells, and 1111 TADs with a mean size of 2.32 Mb in LNCaP cells (Fig. 1A; Supplemental Tables S2–S4). Interestingly, the size of TADs was significantly smaller in both prostate cancer cells compared to normal PrEC (ANOVA, η2 = 0.23, P ≤ 0.0001) (Supplemental Table S5). Additionally, there was a significant difference in domain sizes between PrEC and LNCaP (d = 1.56 [95% CI: 1.42–1.7], t-test marginal P < 0.0001), between PrEC and PC3 (d = 0.83 [95% CI: 0.7–0.97], t-test marginal P < 0.0001), and between LNCaP and PC3 (d = 0.6 [95% CI: 0.5–0.7], t-test marginal P < 0.0001). By visualizing topological domains as two-dimensional interaction matrices, we observed that large domains characteristic of normal cells were often subdivided into smaller domains (“sub-domains”) in cancer cells (Fig. 1B; Supplemental Fig. S1B–D). Indeed, the number of cancer-associated domains (identified in PC3 and LNCaP cells) found to be subdomains of normal cells (PrEC) was significantly higher than expected by chance (permutation test [n = 100], P ≤ 0.01 [∼56% overlap compared to ∼49% at random for PC3 and ∼70% overlap compared to ∼58% at random for LNCaP]). Despite the differences in number and size of domains, we found that normal and cancer genomes are approximately equally partitioned into TADs, boundaries, and unorganized chromatin as previously described (Dixon et al. 2012). The distribution of partitions was not significantly different between cell types (χ2 test, P = 0.43) (Fig. 1C) with TADs covering ∼76% (range, 75.37%–77.01%) of each prostate genome, domain boundaries accounting for ∼4% (range, 1.95%–6.86%), and ∼20% of the genome designated “unorganized” chromatin (range, 16.47%–21.02%) in these three cell types. Together, our data show that the general features of the chromatin interactome are consistent between normal and cancer cells, namely the segmentation into well-defined TADs covering ∼80% of the genome (Fig. 1C). However, our results indicate that the number and size of these domains differ (Fig. 1A,B), creating subdomains in cancer cells.
Figure 1.
Topologically associated domains (TADs) are smaller but maintained across the cancer cell genome. (A) Number and average size (in Mb) of TADs identified in normal prostate epithelial cells (PrEC) and prostate cancer cells (PC3 and LNCaP). (B) Chromatin interaction heat maps from PrEC, PC3, and LNCaP cells visualized as two-dimensional interaction matrices in WashU Epigenome Browser. TADs observed in cancer cells are often “merged” in normal cells, and large domains found in normal cells are frequently occupied by more than two domains in cancer cells. The interaction data are aligned with RefSeq genes and CTCF binding sites. An example from Chromosome 1 is shown. (C) Proportion of the genome (%) organized into TADs, associated with domain boundaries, or unorganized chromatin.
Cancer cells acquire unique domain boundaries enriched for CTCF binding and H3K4me3
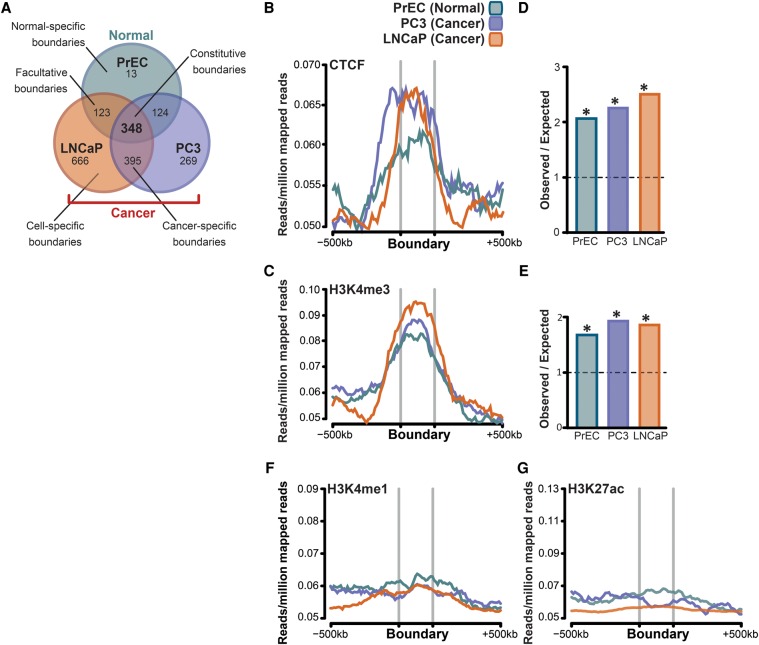
Topological domain boundaries represent the region of strict division between TADs. Stability is evident among normal cell types (Dixon et al. 2012, 2015), including our normal prostate epithelial cells (∼25% overlap compared to ∼15.2% at random, permutation test [n = 100], P ≤ 0.01) (Supplemental Table S6). Thus, we were prompted to investigate whether this extends to include cells of tumorigenic origin. Comparing the locations of boundaries between our prostate data sets, we found that the majority (97.86%) of boundaries present in normal cells were also present in at least one cancer cell type (Fig. 2A). Of these, ∼80% were “constitutive” (present in all three cell types), while ∼20% were “facultative,” as they were maintained in only one of the two cancer cell lines studied (Fig. 2A). In contrast, 41.54% of PC3 and 30.74% of LNCaP boundaries were shared with normal cells, consistent with our observation that cancer cells have a greater number of domains (Fig. 1A). A substantial fraction of new boundaries identified in cancer cells were “cancer-specific” (shared between both prostate cancer cell lines; 34.7% of PC3 and 25.78% of LNCaP boundaries), while the remainder were “cell-specific” (23.6% of PC3 and 43.4% of LNCaP boundaries). Together, these data suggest that new boundaries acquired during transformation can be distinct and not always stably maintained (Fig. 2A).
Figure 2.
Cancer cells acquire unique topological domain boundaries that retain CTCF binding and enrichment for H3K4me3. (A) Venn diagram showing that the majority of domain boundaries that are present in normal cells (PrEC) are also present in cancer cells (constitutive boundaries; PC3 and LNCaP), while ∼20% of boundaries were maintained in only one of the two cancer cells (cell-type–specific boundaries). (B,C,F,G) Genome-wide average distribution of CTCF, H3K4me3, H3K4me1, and H3K27ac binding around the domain boundaries in PrEC, PC3, and LNCaP cells. (D) Fold enrichment of CTCF binding at the domain boundaries in PrEC, PC3, and LNCaP cells compared to that expected by chance. (E) Fold enrichment of H3K4me3 binding at the domain boundaries in PrEC, PC3, and LNCaP cells compared to that expected by chance.
We then explored what factors are contributing to the formation and stability of boundaries in normal and cancer cells by examining the enrichment of a variety of histone modifications, chromatin binding proteins, and the distribution of transcriptional start sites (TSS) around the identified boundaries. We found that boundaries were highly enriched for CTCF (Fig. 2B) and H3K4me3 (Fig. 2C) in all cell lines. We calculated that enrichment of CTCF (Fig. 2D) and H3K4me3 (Fig. 2E) was significant in all cell types, approximately twofold greater than expected by chance (Benjamini–Hochberg adj. P < 0.0001). We noted that the proportion of total CTCF binding sites located within boundaries ranged from ∼6.56% in normal to ∼13.19% in cancer cells, but this was not significantly different (χ2 P = 0.27) (Supplemental Fig. S2A). The proportion of total H3K4me3 ranged from 5.12% to 12.02% between cell types but was not significantly different (χ2 P = 0.21) (Supplemental Fig. S2A). As has been reported by others (Dixon et al. 2012), we found that transcription start sites (Benjamini–Hochberg adj. P = 0.0002) and several Gene Ontology (GO) terms, most notably relating to “housekeeping functions” such as cellular proteins, RNA and protein binding, and the intra-cellular part (Supplemental Fig. S2B), were also strongly enriched around the boundaries. The GO terms were highly similar between normal-specific and cancer-specific boundary regions, despite the increased number of boundaries, and therefore overlapping genes, in cancer cells. In contrast, enhancer-associated marks (H3K4me1 and H3K27ac) were not enriched at the boundaries (Fig. 2F,G).
Copy-number variants are associated with the formation of new domain boundaries in cancer cells
The genomes of cancer cells retain the ability to form domains, with adjacent boundaries being enriched for CTCF and H3K4me3 but devoid of H3K4me1 and H3K27ac, similar to normal cells. It is well-established that the genomes of prostate cancer cells can have recurrent copy-number alterations (Williams et al. 2014; Boutros et al. 2015). Therefore, we asked whether CNVs could provide an explanation for the formation of new, smaller TADs and their associated boundaries. CNVs were identified using either a GenomeWide SNP 6.0 Array (PrEC and LNCaP cells) (Robinson et al. 2010) or Mapping 250k Sty Array (PC3 and LNCaP cells) (Rothenberg et al. 2010). We then reduced the potential bias due to large-size structural variants by including only chromosomal variants up to 10 Mb that were present in both LNCaP and PC3 cells. Sixty CNVs satisfied our criteria with an average size of 3.8 Mb (range 19 kb–9.7 Mb). Most of the overlapping CNVs were deletions (73.3%) relative to normal PrEC cells. Next, we asked whether CNVs present in cancer cells were associated with cancer-specific domain boundaries. Notably, we found that ∼70% (n = 44) of the CNVs that were shared between LNCaP and PC3 cells were located at newly formed cancer-specific domain boundaries. For example, a 400- to 600-kb cancer-specific deletion on Chromosome 17p13.1 is associated with the establishment of a new domain boundary in both prostate cancer cell lines and a resulting change in local interactions across this region (Fig. 3). Interestingly, this locus harbors the TP53 gene and is commonly deleted (14.8%) in all prostate cancers (Kluth et al. 2014). Supplemental Figure S3, A–D highlights additional examples of prostate cancer CNVs (both deletions and insertions) that are associated with the formation of new TAD boundaries and the resulting bifurcation of larger TADs into distinct smaller TADs. In order to demonstare the potential functionality of TAD-altering CNVs, we next sought to determine the potential effect on nearby genes. We identified genes that were located at cancer-specific domain boundaries overlapping CNVs common between PC3 and LNCaP (n = 44) and compared their expression between normal prostate and prostate tumor samples in a TCGA prostate adenocarcinoma (PRAD) RNA-seq data set (level 3) (normal n = 50, tumors n = 278) (The Cancer Genome Atlas Research Network 2015). Overall, out of 445 genes located in CNVs involving novel boundaries, 96 (21.53%) showed differential expression between normal and tumor samples (FC > 1.5). Representative examples of differentially expressed genes and Kaplan-Meier curves for recurrence-free survival (RFS; log-rank test) (Taylor et al. 2010) are shown in Supplemental Figure S4.
Figure 3.
Copy-number variants (CNVs) are associated with the formation of new domain boundaries in cancer cells. CNV regions visualized as relative copy-number estimates for each cell type (presented in green) are aligned with chromatin interaction heat maps (presented as normalized interaction counts visualized in WashU Epigenome Browser) and TADs, demonstrating that cancer-specific domain boundaries are located at regions of CNVs in cancer cell lines. The location of RefSeq genes in the region of copy-number variation is indicated below. An example from Chromosome 17 is shown, where a 400- to 600-kb deletion encompassing the TP53 locus that is present in both cancer cell lines is associated with establishment of a new domain boundary and a resulting change in local interactions across this region.
Acquisition of new, cancer-specific loops explains the majority of differential interactions that occur within topological domains
Using the diffHiC package (Lun and Smyth 2015), we next identified differential interactions between normal and cancer cells by comparing the normalized Hi-C interaction data from the normal cell line (PrEC) to both cancer cell lines (PC3 and LNCaP) independently. Read pairs from each Hi-C experiment were aligned to the reference genome (hg19) and processed into interaction counts for bin pairs that were filtered and normalized. These counts were then analyzed using methods in the edgeR package to test for significant differences in interaction intensity between cell lines (Lun and Smyth 2015). The differential chromosomal interactions detected at 100-kb and 1-Mb bin sizes are presented in Supplemental Tables S7–S9. We excluded inter-chromosomal interactions from further analysis to avoid bias driven by potential genomic rearrangements present in cancer cells. We then defined anchor points of chromatin interactions as the genomic locations where an interaction is present (Fig. 4A; Lun and Smyth 2015). We detected 2188 anchor points that differed between PrEC and PC3 and 2832 differential anchors between PrEC and LNCaP cell lines (FDR < 5%) (Supplemental Tables S10, S11). Notably, nearly 72% of all differential interactions (2036) identified in a comparison of PrEC with PC3 were also found in a comparison of PrEC with LNCaP (Fig. 4B), suggesting that the majority are common, cancer-specific interactions. The whole-chromosome differential anchor pattern suggests that large chromosomes may be predisposed to forming new interactions compared to small chromosomes (Supplemental Fig. S5A); however, it is possible that this may reflect the tendency of larger chromosomes to be more interactive (Supplemental Fig. S5B). Interestingly, differential interactions were enriched in cancer cells (67.7% in PC3 and 54.7% in LNCaP) (Fig. 4C). The most significant interactions identified at 100-kb resolution (N = 86) (Supplemental Table S12) are predominantly (84.88%) located within topological domains, in agreement with previous publications (Supplemental Fig. S5C; Dixon et al. 2012, 2015).
Figure 4.
New, cancer-specific interactions explain the majority of differential interactions, which remain within topological domains. (A) Anchor points of chromatin interactions were defined as the genomic locations where an interaction is present. Genomic coordinates of anchor points were exported into a BED file for further analysis. (B) Venn diagram showing anchor points of differential interactions between PrEC and PC3 and between PrEC and LNCaP cell lines (FDR < 5%) and the overlap between them. (C) Differential interactions are enriched in cancer cells. (D) Differential interactions are enriched for enhancers that were marked by CTCF (enhancer + CTCF), promoters marked by CTCF (promoter + CTCF), as well as distal CTCF sites (CTCF). ChIP-seq chromatin states were classified using the ChromHMM hidden Markov model, and data are presented as fold change between the observed enrichment and that expected by random chance. Enrichment was considered significant if the q value < 0.05. (E) Differential interactions are associated with significantly altered gene expression in cancer cells (χ2 OR = 2.482 [95% CI = 1.141–5.400], P = 0.0273). (F) Majority of genes located at the anchor points of differential interactions have increased expression in cancer cells.
Differential cancer interactions are enriched for enhancers, promoters, and CTCF-occupied genomic regions
We next sought to determine whether anchor points of differential interactions gained in cancer cells were enriched for regulatory elements. We utilized ChIP-seq data to generate signatures of key histone modifications (H3K4me1, H3K4me3, H3K27me3, H3K27ac) as well as CTCF and then applied the multivariate hidden Markov model, ChromHMM (Ernst and Kellis 2010, 2012) to annotate the epigenomes of each normal- and cancer-cell type into seven distinct chromatin states (enhancer + CTCF, promoter + CTCF, CTCF, enhancer, promoter, repressed, and transcribed). We then intersected the known locations of differential anchor points at 100-kb resolution with each of the ChromHMM states and asked whether any states were overrepresented. We found that enhancers that were marked by CTCF (enhancer + CTCF) had the greatest change, with ∼2.5-fold higher enrichment than would be expected by chance (Benjamini–Hochberg adj. P = 0.002) (Fig. 4D). A significant enrichment of promoters marked by CTCF (promoter + CTCF) (Benjamini–Hochberg adj. P = 0.02) as well as distal CTCF sites (CTCF) (Benjamini–Hochberg adj. P = 0.0007) was also observed at differential anchor points. We found 2.2-fold enrichment for promoter + CTCF and 1.8-fold enrichment for CTCF at differential anchor points, suggesting that functional elements occupied by CTCF may facilitate new interactions in cancer cells.
Finally, we addressed the question whether changes in local chromatin interactions could affect expression of nearby genes. We identified genes that were located within anchor points of differential interactions and compared their expression between normal (PrEC) and cancer (PC3) cells. Novel cancer-specific chromatin interactions (Supplemental Table S11) were associated with altered gene expression (χ2 OR = 2.482 [95% CI: 1.141–5.4], P = 0.0273) (Fig. 4E). For example, we found that the increased contact frequencies at the KCNMA1 locus in cancer cells (PC3 vs. PrEC fold change = 7.11) resulted in a more than 10-fold increase in KCNMA1 expression (log2FC = 3.38). Similarly, expression of PRDX6 that is located at a gained differential interaction (PC3 vs. PrEC fold change = 5.05) was up-regulated in cancer cells by almost 10-fold (log2FC = 3.3). Together, 80% of all differential interactions in cancer cells correlate with differential expression of genes located at anchor points (Fig. 4F) (log2FC ≥ 1.5, q < 0.05), consistent with their enrichment for enhancers and promoters. This implies that altered chromatin interactions lead to de-regulation of gene expression and may contribute to increased expression of oncogenes in cancer cells. To validate this further, we tested if expression of genes located within differential interactions was also altered in the TCGA prostate cohort (The Cancer Genome Atlas Research Network 2015) (normal n = 50, tumors n = 278). We determined that expression of 34 genes (51.5%) was significantly increased in tumor samples (log2FC ≥ 1; t-test P < 0.0001) (Supplemental Fig. 6A,B). Three representative examples show increased expression of RBM25, SH3RF2, and SCHIP1 in tumor versus normal samples (t-test P < 0.0001) and significant association with survival (Kaplan-Meier survival curves for RFS, log-rank p) (Supplemental Fig. 6C,D; Taylor et al. 2010).
Differential cancer interactions are coincident with altered cancer epigenetic patterns
Our results above highlight that differential chromatin interactions are enriched at enhancers and promoters; however, these potential new three-dimensional loops remain difficult to visualize in a linear genome browser. Chromosome conformation is in its relative infancy compared with other molecular biology tools to explore cellular attributes, and the volume and complexity of these data pose considerable challenges. Therefore, we developed Rondo, a web-based interface for visualizing Hi-C data, which minimizes visual noise by hierarchical clustering and grouping individual connections into larger groups that are progressively separated as more detailed information is provided. Rondo also displays gene tracks and epigenome data sets, such as histone modification marks, allowing biological interpretation of Hi-C data. Using Rondo, we visualized differential interactions alongside histone modifications and then intersected these with gene expression data to determine the functional output of atypical interactions in cancer cells. Figure 5 shows an example of a new cancer differential chromatin interaction that creates a novel loop between the enhancer and promoter of NRP1, resulting in cancer gene activation. In normal prostate cells NRP1 is not active, and the putative enhancer is marked by residual H3K4me1 but is devoid of H3K27ac, H3K4me3, and CTCF binding in normal cells. The promoter of NRP1 is also marked by H3K4me1 in normal cells and has low levels of H3K4me3, H3K27ac, and CTCF. This epigenetic signature is consistent with repressed (Fig. 5A) but permissive enhancer-promoter pairs (Taberlay et al. 2011). In cancer cells, a marked increase in H3K4me1, H3K4me3, and H3K27ac signal is observed (Fig. 5B), together with CTCF binding. Interestingly, this epigenetic remodeling was not restricted to the NRP1 gene boundaries and also encompassed neighboring enhancers and cancer-associated genes, such as ITGB1 (Fig. 5B). The H3K4me1 mark was lost from the genomic region located 3′ to the NRP1 enhancer (encompassing PARD3), without subsequent acquisition of other active or repressive marks (Fig. 5B). We found that the expression of ITGB1, NRP1, and PARD3 genes were all significantly increased in the cancer cells (Fig. 5C), suggesting that functional elements and long-range epigenetically regulated domains may be altered concomitant with the interactome.
Figure 5.

Differential interactions are enriched for enhancers, promoters, and CTCF-occupied genomic regions and may explain the unique epigenetic programs of normal and cancer cells. Chromatin interactions visualized in the Rondo interactive analysis tool. Anchor points of differential interactions are visualized simultaneously with ChIP-seq (H3K27ac, H3K4me1, H3K4me3), RefSeq genes, and RNA-seq data (circular tracks) inferring functionality (both active and repressive) of interactions. (A) In this example from Chr10:33,000,000–35,000,000, epigenetic remodeling (activation) of genes located at the differentially interacting region can be observed at 100-kb resolution. The promoter and a putative enhancer of the NRP1 gene are both marked by residual H3K4me1 in normal cells. Very little H3K4me3 (dark green) and H3K27ac (light green) is observed. (B) In contrast, these regulatory elements display a marked increase in H3K4me1, H3K4me3, and H3K27ac ChIP signal in PC3 prostate cancer cells, indicative of increased gene activity. This occurs concomitant with a new interaction depicted by the purple line in the cancer cells. (C) Genes at differential interaction anchor points are significantly overexpressed concomitant with a new interaction only present in the cancer cells (*q value < 0.0001).
Differential interactions occur in long-range epigenetically de-regulated domains in cancer cells
Epigenetic remodeling in prostate cancer cells can occur across large domains that involve concordant transcriptional de-regulation of regions that encompass multiple adjacent genes (Coolen et al. 2010; Bert et al. 2013). Interestingly, genes within long-range epigenetically de-regulated domains exhibit unidirectional expression patterns but can display divergent chromatin signatures (Coolen et al. 2010; Bert et al. 2013), suggesting that local changes in chromatin organization and abnormal chromatin interactions may be associated with long-range epigenetic activation (Bert et al. 2013) and repression (Coolen et al. 2010) in prostate cancer. To investigate if abnormal chromatin interactions also occur at long-range epigenetically de-regulated domains, we examined if LRES and LREA regions we previously identified in LNCaP cells (Coolen et al. 2010; Bert et al. 2013) were located at anchor points of differential interactions (at 1-Mb resolution) between normal PrEC cells and LNCaP prostate cancer cells (Supplemental Tables S8, S10). We found that more than half of LRES regions (61.7%; 29 out of 47 identified LRES regions; minimum overlap > 90 kb) were located at regions that showed differential interactions between PrEC and LNCaP cells. In addition, a large proportion of LREA regions (65.7%; 23 out of 35 identified LREA regions; minimum overlap > 200kb) were located at genomic regions that showed differential interactions between PrEC and LNCaP cells.
To characterize the relationship between long-range epigenetic activation and silencing and chromatin organization, we tested the significance of the observed overlap between LRES and LREA regions and differential interactions (Heger et al. 2013). Changes in gene expression between normal and cancer cell lines were reconfirmed with RNA-seq. Using Rondo, we visualized gene expression, histone modifications, and differential interactions overlapping known LRES (Fig. 6; Supplemental Fig. S7) and LREA (Fig. 7; Supplemental Fig. S8) domains with an observed over expected fold change > 2. An example of differential interactions and epigenetic marks between normal PrEC cells and LNCaP cancer cells within an LRES region is shown in Figure 6. In normal cells, the ∼1-Mb region (Chr1:207,900,000–209,800,000) harboring the genes PLXNA2, MIR205, and CAMK1G is actively expressed, as indicated by RNA-seq (Fig. 6A), whereas in cancer this region is silenced (Fig. 6B). Gene silencing across the LRES region is associated with a gain of the H3K27me3 mark and loss of the active H3K4me3 and H3K27ac chromatin marks in the cancer cells (Fig. 6A,B). Notably, in normal prostate cells there are strong interactions between the active PLXNA2 and MIR205 loci, in particular, interactions between putative active enhancers heavily marked by H3K27ac (Fig. 6A). Another interaction can be observed at Chr1:208,800,000–209,300,000 in the normal cells, in the inter-genic region that is bounded by a discrete H3K27me3 border signal. In cancer cells, the chromatin interactions are strikingly different; the higher-order interactions linking PLXNA2 with a putative enhancer element in normal cells are no longer evident, and instead, a new local interaction is formed, suggesting that at the locus harboring the inactive PLXNA2 and neighboring CD34 the gene chromatin is more condensed. Moreover, the epigenetically silenced MIR205 locus no longer interacts with the PLXNA2 locus in the cancer cells but instead interacts with the large inter-genic region at Chr1:208,800,000 that is enriched in H3K27me3. CD46 borders the LRES region and is highly expressed in both PrEC and LNCaP cells, carries the expected complement of active histone modifications in both cell types, and displays no change in interactions (Fig. 6A,B). In addition to displaying the chromatin interactions in two cell types independently, Rondo also has the capacity to combine the multiple layers of interactions and display the change in chromatin marks and gain and loss of chromatin interactions (Fig. 6C). Additional examples of alterations in chromatin interactions commonly associated with gene repression and the formation of new localized interactions within the LRES domains are shown in Supplemental Figure S7.
Figure 6.
Long-range epigenetically silenced (LRES) domains occur at differential interactions in cancer cells. The majority of LRES regions overlap differential interactions in cancer cells. Anchor points of differential interactions are visualized in Rondo simultaneously with ChIP-seq (H3K27ac, H3K4me3, and H3K27me3), RefSeq genes, and RNA-seq data. (A) In normal PrEC cells, three chromatin interactions (teal) are present across the LRES region within Chr1:207,900,000–209,800,000, which contains highly expressed genes (RNA-seq) and enriched levels of active histone marks (H3K4me3, dark green; H3K27ac, light green). (B) In LNCaP cancer cells, new interactions are observed in this LRES region and occur with concomitant loss of gene expression (RNA-seq) and a marked decrease in active marks (H3K4me3, dark green; H3K27ac, light green) and increase in repressive H3K27me3 (pink). (C) A combined view shows normal and cancer epigenomes, expression and interaction data simultaneously. Those interactions unique to PrEC (normal; teal) and LNCaP (cancer; orange) are evident, while the shared interaction is shown in yellow. The circular tracks depict gene expression (RNA-seq) and histone marks (H3K4me3, dark green; H3K27ac, light green; H3K27me3, pink). Teal lines in the circle depict a loss of chromatin interactions in cancer, and the orange lines depict a gain of interaction in the cancer cells.
Figure 7.
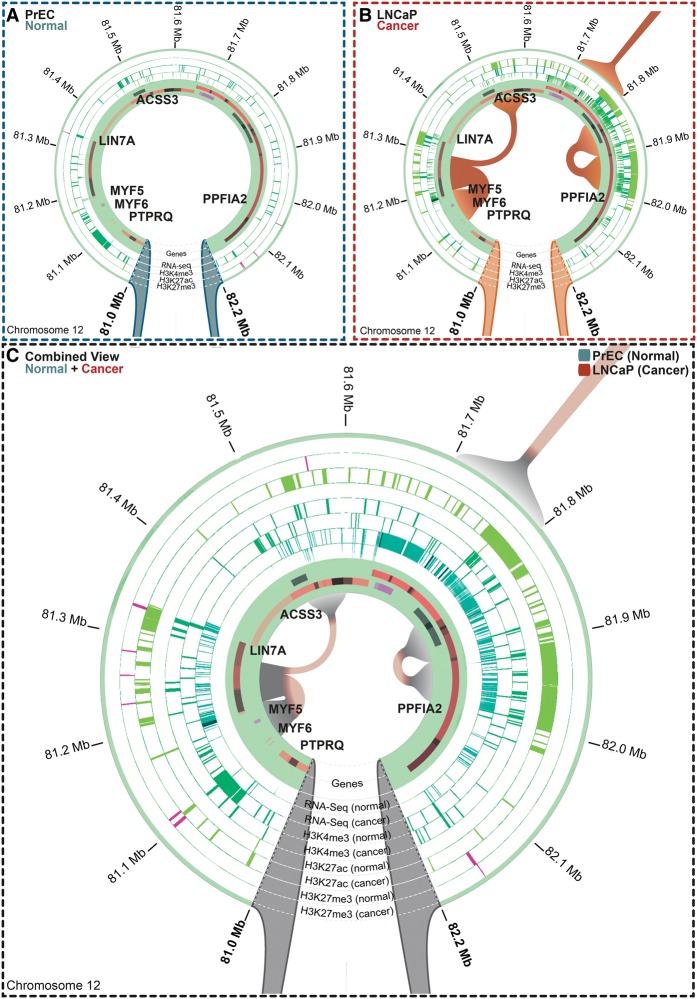
Long-range epigenetically activated (LREA) domains occur at differential interactions in cancer cells. The majority of LREA regions overlap differential interactions in cancer cells. Anchor points of differential interactions are visualized in Rondo simultaneously with ChIP-seq (H3K27ac, H3K4me3, and H3K27me3), RefSeq genes, and RNA-seq data. (A) In normal PrEC cells, there are no interactions evident across the LREA region within Chr12:81,000,000–82,200,000, which is consistent with the low levels of H3K4me3 (dark green), absence of H3K27ac (light green), and gene inactivity (RNA-seq, dark cyan). (B) In LNCaP prostate cancer cells, three new, distinct interactions (orange) are observed. Most of the genes are highly expressed (RNA-seq) and active marks (H3K4me3, dark green; H3K27ac, light green) are acquired. This region is devoid of repressive H3K27me3 marks (pink). (C) A combined view shows normal and cancer epigenomes, expression and interaction data simultaneously. Only interactions present in LNCaP (cancer; orange) are evident, with no shared interactions. The circular tracks depict gene expression (RNA-seq) and histone marks (H3K4me3, dark green; H3K27ac, light green; H3K27me3, pink).
To determine if there are also alterations in three-dimensional chromatin interactions in LREA domains associated with gene expression and epigenetic changes, we visualized the Hi-C interaction data overlapping previously identified LREA regions identified in prostate cancer (Bert et al. 2013). An example of alterations in interactions and epigenetic marks between normal PrEC cells and LNCaP cancer cells within an LREA region is shown in Figure 7. In cancer cells, the ∼1.2-Mb region (Chr12:81,000,000–82,200,000) harboring the genes PTPRQ, MYF6, MYF5, LIN7A, ACSS3, and PPFIA2 is hyperactivated, as indicated by RNA-seq (Fig. 7B), whereas in normal prostate cells this region is relatively inert (Fig. 7A). Regional gene activation is associated with an enrichment of H3K4me3 and H3K27ac chromatin marks and cancer-specific acquisition of new chromatin interactions between the MYF loci and LIN7A (Fig. 7A,B). Notably, a new interaction within the PPFIA2 gene body (Chr12:81,800,000–82,000,000) is also formed, and this spans two potentially discrete enhancer regions heavily enriched for H3K27ac (Fig. 7B). The combination view of epigenetic and chromatin interaction changes between normal and cancer cells is shown in Figure 7C, which highlights the acquisition of interactions at regions that also gain putative enhancer activation marks H3K27ac. Additional examples of alterations in chromatin interactions commonly associated with gene activation and the formation of new localized interactions within the LREA domains, in particular at H3K27ac loci of enrichment, are shown in Supplemental Figure S8. Together, these data indicate that long-range epigenetic dysregulation in cancer is also associated with an alteration in higher-order chromatin interactions commonly at regulatory loci.
Discussion
Cancer is commonly associated with widespread changes to the genome and epigenome that result in both localized and regional changes in gene expression. However, it is not known if changes to the cancer genome and epigenome are also associated with changes in the three-dimensional architecture of the cell. Here, we performed the first comparative analysis of chromatin organization between prostate cancer and normal cells using Hi-C chromosome conformation capture sequencing integrated with epigenomic and gene expression data to determine if chromatin interactions are altered in cancer. Surprisingly, we found that the overall spatial organization of chromatin in prostate cancer cells (PC3 and LNCaP) is highly similar to normal cells (PrEC), and retains the higher-order characteristics observed in embryonic and various somatic cells (Dixon et al. 2012, 2015; Nora et al. 2012). Notably, topologically associated domains are present in both normal and cancer cells, suggesting that on a large scale, chromosomes are folded similarly and that they retain their physical positions within the nucleus (Cremer and Cremer 2010) of cancer cells. It still remains unclear how TADs are established and maintained, although a role for the insulator-binding CTCF protein is likely since it is enriched at domain boundaries (Dixon et al. 2012; Zuin et al. 2014), it drives directionality of DNA interactions (Vietri Rudan et al. 2015), and its disruption is linked to gene expression changes (Guo et al. 2015). However, a proportion of TAD boundaries are devoid of CTCF binding (Dixon et al. 2012), indicating that it does not function in isolation to prevent TADs from becoming undefined. Our data suggest that the presence of H3K4me3, but not H3K4me1 or H3K27ac, may help define TADs in both normal and cancer cells. Interestingly, we noticed that both CTCF and H3K4me3 are often already present, in normal cells (PrEC), at the genomic regions that become the new boundaries in cancer cells, potentially serving as “bookmarks.”
Even though we found that the overall spatial organization of chromatin was similar, we identified a number of distinct differences in the three-dimensional organization of chromatin in the cancer cells. First, we found that the size of the TADs is smaller in cancer, with a size range of ∼2–4 Mb relative to ∼8 Mb TADs in the normal prostate cells. These smaller TADs commonly resided within the normal TAD architecture rather than forming new TADs. In fact, we found that the majority (97.86%) of domain boundaries that were present in normal cells were also present in cancer cells. Interestingly, over half of the new smaller subdomain TADs that were acquired in both prostate cancer cell lines shared similar new boundaries, suggesting a cancer-specific process in three-dimensional reorganization. That cancer cells have a higher number of smaller domains is interesting and may reflect key characteristics of transformed cells. Despite multiple studies, the mechanisms underlying the formation of topological domain boundaries remain unknown. It has been previously suggested that genomic deletions at TAD boundaries in normal cells result in a change in local chromatin organization and in a formation of new domain boundaries (Nora et al. 2012; Ibn-Salem et al. 2014). We therefore investigated the role of genomic sequence variation in establishing TAD boundaries in cancer cells. Strikingly, we found that a large proportion of cancer-specific domain boundaries occur at regions of copy-number variation that were observed in both cancer cell lines. Our results show that copy-number alterations that are common in prostate cancer are related to the establishment of new TAD boundaries. In particular, the TP53 locus is commonly deleted (14.8%) in prostate cancer (Kluth et al. 2014; Boutros et al. 2015), and here, we show that deletion of this locus is associated with alteration of the local three-dimensional architecture and the gain of a new TAD boundary. Therefore, copy-number alterations in cancer that occur within a TAD may lead to a disruption of local chromatin interactions within a domain and result in establishment of a new domain boundary at that region. As it is known that TAD boundaries are crucial for controlling the action of enhancers on genes in adjacent TADs (Nora et al. 2012; Dowen et al. 2014; Symmons et al. 2014), our findings may help explain the myriad of altered gene expression profiles in cancer cells. Indeed, all disease phenotypes caused by genomic deletions potentially involve perturbed topological domain function. However, it remains to be answered how genomic rearrangements in cancer genomes affect TAD organization and if that may result in altered gene expression. In addition, cause and directionality between formation of new TAD boundaries and CNVs remain to be determined.
Second, we demonstrated that new cancer-specific interactions occur within the smaller TADs. Notably, ∼70% of all differential interactions were shared between the two different prostate cancer cell lines, suggesting that the majority are common, cancer-specific interactions. We found that these cancer differential interactions were enriched for enhancers, promoters, and CTCF-occupied genomic regions. It is interesting to speculate that functional elements, especially ones occupied by CTCF, may facilitate new chromatin interactions in cancer cells. Third, we found that cancer-specific differential interactions may explain the unique epigenetic programs of cancer cells. Enhancers are known to engage in interactions that exist in a cell-type–specific manner, and our results support the hypothesis that enhancer dynamics play a role in regulating local interaction differences between normal and cancer cells. In particular, we found that a change in enhancer and promoter functionality as predicted by a change in H3K27ac and H3K4me3 marks was associated with a change in chromosome interactions and gene expression patterns, for example, at the NRP1 gene locus. NRP1 encodes one of the two neuropilins, which contain specific protein domains that allow them to participate in several different types of signaling pathways that control cell migration. Interestingly, overexpression of NRP1 has been reported to predict distant relapse after radical prostatectomy in clinically localized prostate cancer (Talagas et al. 2013).
Finally, we found that differential interactions also occur within long-range epigenetically de-regulated domains in cancer cells. We previously showed that epigenetic remodeling in prostate cancer cells occurs across large domains that involve concordant de-regulation of regions that encompass multiple adjacent genes (Coolen et al. 2010; Bert et al. 2013). Here, we show that these regulatory regions within the long-range epigenetically de-regulated domains exhibit local changes in chromatin organization and abnormal chromatin interactions in both LRES and LREA regions. Notably, we found new interactions at putative enhancers associated with gene activation in the cancer cells in LREA regions and loss of interactions at putative enhancer elements in cancer cells that are silenced in cancer in LRES regions. New local interactions were also commonly found in cancer cells associated with the formation of large repressed H3K27me3 domains, suggesting the formation of more condensed chromatin. Our findings indicate that long-range epigenetic dysregulation in cancer is also associated with an alteration in higher-order chromatin interactions commonly at regulatory loci that potentially results in atypical promoter-enhancer and enhancer-enhancer interactions in cancer cells.
Changes in higher-order chromatin structure have been noted at developmental loci in mammalian cells (Chambers et al. 2013), linked to gene functions that impact phenotype (Chen et al. 2015), respond to hormonal alterations (Le Dily et al. 2014), and can be associated with divergent histone marks (Lan et al. 2012; Huang et al. 2015). The implementation of a novel visualization tool has allowed us to explore the Hi-C interaction in relation to the transcription and epigenetic changes between normal and prostate cancer cells. We now show for the first time that alterations in the three-dimensional chromatin state potentially underpin the functional basis of the cancer genome by changing the spatial architecture of regulatory elements and proximity to genes resulting in aberrant cancer gene expression. This work provides a new insight into long-range genetic and epigenetic dysregulation and higher-order chromatin interactions in cancer.
Methods
Prostate cell lines
Normal human prostate epithelial cells (PrEC) were obtained from Lonza, and prostate cancer cell lines (PC3 and LNCaP) were obtained from the American Type Culture Collection (ATCC). Both were cultured under recommended conditions at 37°C and 5% CO2.
ChIP-seq, RNA-seq, and copy-number estimates
ChIP-seq experiments were performed as previously described (Taberlay et al. 2014). Additional information regarding ChIP-seq and RNA-seq experiments is provided in the Supplemental Material. Copy-number estimates for LNCaP and PrEC cell lines were taken from published data (Robinson et al. 2010). Copy-number data from Mapping 250k Sty arrays for PC3 and LNCaP cell lines were obtained from GEO (GSM827569) and processed as described previously (Bengtsson et al. 2008, 2009).
Hi-C chromosome conformation capture
Hi-C experiments were performed based on the original protocol by Lieberman-Aiden et al. (2009) with minor modifications, described in Supplemental Material.
Preparation of Hi-C libraries
Hi-C libraries were prepared using a customized protocol. Additional information is provided in the Supplemental Material. Resulting libraries were run on the HiSeq 2500 (Illumina) platform configured for 100-bp paired-end reads according to the manufacturer's instructions. All Hi-C libraries were processed through the NGSane framework v0.5.2 (Buske et al. 2014) available from GitHub using the “fastqc,” “hiccup,” and “fithicaggregate” modules as described in Supplemental Material. All Hi-C data sets reported here can be explored interactively via Rondo at www.rondo.ws.
Identification of topologically associated domains
For the identification of TADs, the corrected Hi-C contact matrices were binned at 40-kb resolution, iteratively corrected, and the domains were called according to a previously developed MATLAB (v.2.3) script called “domain-caller” that is based on the directionality index (DI) and the hidden Markov model (HMM) (Dixon et al. 2012).
Identification of differential interactions between normal and cancer cell lines
We used a diffHiC (v.1.0.1) Bioconductor R package to detect differential genomic interactions in Hi-C data between normal and cancer cell lines (Lun and Smyth 2015). Additional information is provided in the Supplemental Material.
Data access
Raw and processed Hi-C, ChIP-seq, and RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE73785.
Acknowledgments
We thank members of the Clark Laboratory for helpful discussions and careful reading of the manuscript. In particular, we thank Dr. Brigid O'Gorman for help with figure preparation and Dr. Tim Peters for help with statistics analysis. S.J.C. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow #1063559; P.C.T. is a Cancer Institute NSW Early Career Development Fellow #12/ECF/2-33; F.A.B. is a NHMRC Early-Career Research Fellow #1092170. This work was supported by Cure Cancer Australia Foundation Project Grant #1070613 and NHMRC Project Grants #1011447 and #1051757.
Author contributions: P.C.T. and J.A.-K. designed and performed experiments, interpreted data, and wrote the manuscript. A.T.L.L. performed differential data analysis. F.A.B., E.Z., and D.C.B. processed the data. K.S. designed the visualization tool Rondo. S.A.B. performed experiments; K.A.G. and C.M.G. produced and processed the RNA-seq data. G.K.S., C.S., and S.I.O. contributed to interpretation of data. S.J.C. conceived and coordinated the overall study, participated in interpretation of data, and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.201517.115.
References
- Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M, et al. 2010. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci 107: 9742–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336: 736–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson H, Irizarry R, Carvalho B, Speed TP. 2008. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics 24: 759–767. [DOI] [PubMed] [Google Scholar]
- Bengtsson H, Wirapati P, Speed TP. 2009. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics 25: 2149–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert SA, Robinson MD, Strbenac D, Statham AL, Song JZ, Hulf T, Sutherland RL, Coolen MW, Stirzaker C, Clark SJ. 2013. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell 23: 9–22. [DOI] [PubMed] [Google Scholar]
- Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. 2015. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 47: 736–745. [DOI] [PubMed] [Google Scholar]
- Buske FA, French HJ, Smith MA, Clark SJ, Bauer DC. 2014. NGSANE: a lightweight production informatics framework for high-throughput data analysis. Bioinformatics 30: 1471–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2015. The molecular taxonomy of primary prostate cancer. Cell 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers EV, Bickmore WA, Semple CA. 2013. Divergence of mammalian higher order chromatin structure is associated with developmental loci. PLoS Computat Biol 9: e1003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen J, Muir LA, Ronquist S, Meixner W, Ljungman M, Ried T, Smale S, Rajapakse I. 2015. Functional organization of the human 4D Nucleome. Proc Natl Acad Sci 112: 8002–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen MW, Stirzaker C, Song JZ, Statham AL, Kassir Z, Moreno CS, Young AN, Varma V, Speed TP, Cowley M, et al. 2010. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat Cell Biol 12: 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. 2010. Chromosome territories. Cold Spring Harb Perspect Biol 2: a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, Sarkar A, Barasch J, Vuononvirta R, Jones C, et al. 2009. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet 5: e1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159: 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Yuan T, Schilter KF, Dittmar RL, Mackinnon A, Huang X, Tschannen M, Worthey E, Jacob H, Xia S, et al. 2015. Prostate cancer risk locus at 8q24 as a regulatory hub by physical interactions with multiple genomic loci across the genome. Hum Mol Genet 24: 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Rubin MA, Rickman DS. 2012. Oncogenic transcription factors as master regulators of chromatin topology: a new role for ERG in prostate cancer. Cell Cycle 11: 3380–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova D, Patro R, Duggal G, Kingsford C. 2014. Identification of alternative topological domains in chromatin. Algorithms Mol Biol 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. 2006. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet 38: 540–549. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Li W, Liyanarachchi S, Srinivas M, Wang Y, Akagi K, Wang Y, Wu D, Wang Q, Jin V, et al. 2015. Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. Proc Natl Acad Sci 112: 6128–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Webber C, Goodson M, Ponting CP, Lunter G. 2013. GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29: 2046–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Lin VA, Buckle A, Cheong K, Halani N, Ku S, Kwok CT, Packham D, Suter CM, Meagher A, et al. 2007. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res 67: 9107–9116. [DOI] [PubMed] [Google Scholar]
- Huang J, Marco E, Pinello L, Yuan GC. 2015. Predicting chromatin organization using histone marks. Genome Biol 16: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibn-Salem J, Kohler S, Love MI, Chung HR, Huang N, Hurles ME, Haendel M, Washington NL, Smedley D, Mungall CJ, et al. 2014. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol 15: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, et al. 2009. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet 5: e1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluth M, Harasimowicz S, Burkhardt L, Grupp K, Krohn A, Prien K, Gjoni J, Hass T, Galal R, Graefen M, et al. 2014. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer 135: 1369–1380. [DOI] [PubMed] [Google Scholar]
- Lan X, Witt H, Katsumura K, Ye Z, Wang Q, Bresnick EH, Farnham PJ, Jin VX. 2012. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res 40: 7690–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dily F, Bau D, Pohl A, Vicent GP, Serra F, Soronellas D, Castellano G, Wright RH, Ballare C, Filion G, et al. 2014. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev 28: 2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, Smyth GK. 2015. diffHic: a Bioconductor package to detect differential genomic interactions in Hi-C data. BMC Bioinformatics 16: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. 2013. Organization of the mitotic chromosome. Science 342: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. 2008. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res 68: 8616–8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. 2013. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 14: 765–780. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. 2009. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nature Genet 41: 882–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafique S, Thomas JS, Sproul D, Bickmore WA. 2015. Estrogen-induced chromatin decondensation and nuclear re-organization linked to regional epigenetic regulation in breast cancer. Genome Biol 16: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman DS, Soong TD, Moss B, Mosquera JM, Dlabal J, Terry S, MacDonald TY, Tripodi J, Bunting K, Najfeld V, et al. 2012. Oncogene-mediated alterations in chromatin conformation. Proc Natl Acad Sci 109: 9083–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Stirzaker C, Statham AL, Coolen MW, Song JZ, Nair SS, Strbenac D, Speed TP, Clark SJ. 2010. Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res 20: 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg SM, Mohapatra G, Rivera MN, Winokur D, Greninger P, Nitta M, Sadow PM, Sooriyakumar G, Brannigan BW, Ulman MJ, et al. 2010. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res 70: 2158–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seng TJ, Currey N, Cooper WA, Lee CS, Chan C, Horvath L, Sutherland RL, Kennedy C, McCaughan B, Kohonen-Corish MR. 2008. DLEC1 and MLH1 promoter methylation are associated with poor prognosis in non-small cell lung carcinoma. Br J Cancer 99: 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Vallot C, Reyal F, Bernard-Pierrot I, de Medina SG, Segraves R, de Rycke Y, Elvin P, Cassidy A, Spraggon C, et al. 2006. Regional copy number–independent deregulation of transcription in cancer. Nat Genet 38: 1386–1396. [DOI] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. 2014. Functional and topological characteristics of mammalian regulatory domains. Genome Res 24: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. 2011. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell 147: 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. 2014. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res 24: 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talagas M, Uguen A, Garlantezec R, Fournier G, Doucet L, Gobin E, Marcorelles P, Volant A, DE Braekeleer M. 2013. VEGFR1 and NRP1 endothelial expressions predict distant relapse after radical prostatectomy in clinically localized prostate cancer. Anticancer Res 33: 2065–2075. [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timp W, Feinberg AP. 2013. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 13: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. 2015. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 10: 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JL, Greer PA, Squire JA. 2014. Recurrent copy number alterations in prostate cancer: an in silico meta-analysis of publicly available genomic data. Cancer Genet 207: 474–488. [DOI] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. 2011. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nature Genet 43: 1059–1065. [DOI] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, et al. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci 111: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]