Abstract
Abiotic and biotic stresses cause significant yield losses in all crops. Acquisition of stress tolerance in plants requires rapid reprogramming of gene expression. SR1/CAMTA3, a member of signal responsive transcription factors (TFs), functions both as a positive and a negative regulator of biotic stress responses and as a positive regulator of cold stress-induced gene expression. Using high throughput RNA-seq, we identified ~3000 SR1-regulated genes. Promoters of about 60% of the differentially expressed genes have a known DNA binding site for SR1, suggesting that they are likely direct targets. Gene ontology analysis of SR1-regulated genes confirmed previously known functions of SR1 and uncovered a potential role for this TF in salt stress. Our results showed that SR1 mutant is more tolerant to salt stress than the wild type and complemented line. Improved tolerance of sr1 seedlings to salt is accompanied with the induction of salt-responsive genes. Furthermore, ChIP-PCR results showed that SR1 binds to promoters of several salt-responsive genes. These results suggest that SR1 acts as a negative regulator of salt tolerance by directly repressing the expression of salt-responsive genes. Overall, this study identified SR1-regulated genes globally and uncovered a previously uncharacterized role for SR1 in salt stress response.
Abiotic stresses, such as drought, cold, heat and salinity, and biotic stresses caused by pathogenic bacteria, viruses and fungi, limit plant growth and development resulting in significant yield losses in crop plants1,2,3. Acquisition of tolerance to these stresses and other adverse environmental conditions requires coordinated regulation of a multitude of biochemical and physiological changes, and a vast majority of these changes rely on stress-dependent reprogramming of gene expression4,5,6,7,8,9. The alterations in gene expression patterns are largely responsible for plants’ ability to cope with the adverse environmental factors. Previous studies have shown that Ca2+ is one of the key messengers in mediating stress responses7,10,11. Stress-induced changes in cellular Ca2+ are perceived by Ca2+ sensors such as calmodulin (CAM), which in turn regulate diverse processes including gene expression7. Signal responsive (SR) proteins, which are also referred to as CAMTAs (CAM-binding Transcriptional Activators), are a class of highly conserved Ca2+/CAM-binding transcription factors (TFs) in plants and animals12,13,14,15,16,17. In Arabidopsis there are six SR family TFs (SR1 to SR6) and expression of these genes is regulated by diverse biotic and abiotic stresses, as well as hormones12,13,18,19,20. All members of SR/CAMTA family TFs have a DNA binding domain called CG-1 at the N-terminus, which binds to CGCG or CGTG core motifs21,22,23,24, a TIG (an immunoglobulin–like fold) domain that is involved in non-specific DNA binding, several ankyrin repeats that are responsible for protein-protein interactions, followed by five tandem repeats of Ca2+-independent CAM binding domains (IQ motifs), and a Ca2+-dependent CAM binding domain7,11. SR1 (also known as CAMTA3) is one of the well-studied members of the SR family TFs. The core DNA binding motif of SR1 is part of a rapid stress response element (RSRE - VCGCGB) found in the promoters of many genes that are rapidly activated in response to stress25,26. It has been shown that SR1 can activate reporter genes driven by RSRE in a Ca2+-dependent manner26, further suggesting the role of SR1 in stress-induced gene expression through Ca2+. Recent genetic screens also confirmed that SR1 is an important component in RSRE-driven gene expression27.
Several studies with SR1 have shown that it functions as a negative regulator of plant immunity in Arabidopsis28,29,30, a positive regulator of insect resistance31,32 and cold-induced gene expression24,33. A rice CAMTA (OsCBT) also functions as a negative regulator of disease resistance against Xanthomonas oryzae and Magnaporthe grisea34. Although SR1 has been shown to play important regulatory roles in plant immunity, herbivory and cold-induced gene expression, the full set of SR1-regulated genes is largely unknown. A previous microarray study performed with wild type and SR1 mutant reported only about 100 SR1-regualted genes29. However, in that study a complemented line was not included. Here we sequenced the transcriptomes of wild type, SR1 mutant and a complemented line using RNA-seq and identified about 3000 SR1-regulated genes. By analyzing the promoters of all SR1-regulated genes for the presence of known SR1 binding sites, we identified potential direct targets of SR1. Comprehensive analysis of SR1-regulated genes confirmed its known roles and uncovered a previously uncharacterized role for SR1 in salt stress. Furthermore, our results established that SR1 is a negative regulator of salt stress.
Results
Loss of SR1 resulted in misregulation of about 3000 genes
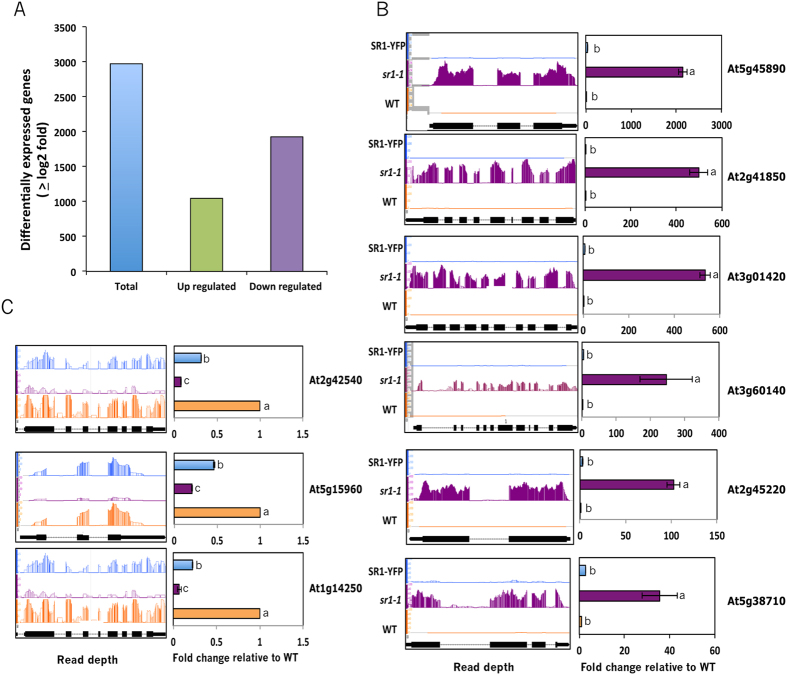
Although SR1 TF is known to regulate multiple stress responses in plants, an in-depth study of SR1-regualted genes (direct or indirect) in the genome using deep sequencing of transcriptomes has not been performed. Here we performed RNA-seq analysis of gene expression with RNA from wild type, SR1 loss-of-function mutant and a complemented line in which the mutant phenotypes are rescued28,31. Prior to RNA-seq, genotypes of all three lines were verified by genomic PCR and RT-qPCR (Supplementary Fig. S1). In the complemented line, the expression of SR1 at the protein level was also confirmed (Supplementary Fig. S1). For each line, two biological replicates were sequenced using Illumina platform. About 37 to 45 million high quality reads (FastQC quality score is >36) were obtained for each replicate (Supplementary Table S1). About 80 million reads for each line were used for gene expression analysis. Around 94% of reads from each sample were mapped to the Arabidopsis genome (TAIR10) (Supplementary Table S1). Of these, ~90 to 92% of the reads were uniquely mapped to a single location. The expression of each transcript in each sample was measured by the number of reads per kilobase per million reads (RPKM). A very high linear correlation was observed in the expression of genes among the replicates indicating that there are no significant differences in gene expression among the biological replicates (Supplementary Fig. S2). The R2 values were between 0.87 and 0.9 for the replicates of all three lines (Supplementary Fig. S2). However, there was a substantial effect of SR1 loss on gene expression as evident from linear regression values when compared to WT (Supplementary Fig. S2B). Furthermore, expression of SR1 in sr1 mutant background significantly restored gene expression changes observed in the mutant (Supplementary Fig. S2). Using the Cufflinks package we identified differentially expressed (DE) genes by comparing the transcriptomes of the mutant and wild type. A total of 2973 genes (Adj. P < 0.05 and fold change >2) were misregulated in sr1 as compared to the WT (Additional File 1, Sheet 1). Expression of about ~85% of DE genes was partially or fully restored to wild type level (Supplementary Fig. S3 and Additional File 1, Sheet 2). These results suggest that the DE genes in the mutant are either direct or indirect targets of SR1 and that the loss of this TF has substantial effect on expression of large number of genes (Fig. 1A). Among the DE genes, 1046 were up-regulated whereas 1927 were down-regulated (Fig. 1A). Using RT-qPCR we validated the expression of 9 randomly selected DE genes. The RT-qPCR results corroborated RNA-seq data and the observed changes in the mutant were fully or partially restored in the complemented line (Fig. 1B,C). In addition, expression of several other DE genes involved in salt stress was also verified by RT-qPCR (see below).
Figure 1. SR1-regulated genes in Arabidopsis.
(A) Total DE, up- or down-regulated genes. (B) RT-qPCR validation of randomly selected up-regulated genes. (C) RT-qPCR of randomly selected down-regulated genes. Left panels in (B,C) show relative sequence read abundance (Integrated Genome Browser view) as histograms in WT, sr1-1 and SR1-YFP lines. The Y-axis indicates read depth with the same scale for all three lines. The gene structure is shown below the read depth profile. The lines represent introns and the boxes represent exons. The thinner boxes represent 5′ and 3′ UTRs. Right panels in (B,C) show fold change in expression level relative to WT. WT values were considered as 1. Student t-test was performed and significant differences (P < 0.05) among samples are labeled with different letters. The error bars represent SD.
GO term enrichment of DE genes for biological processes
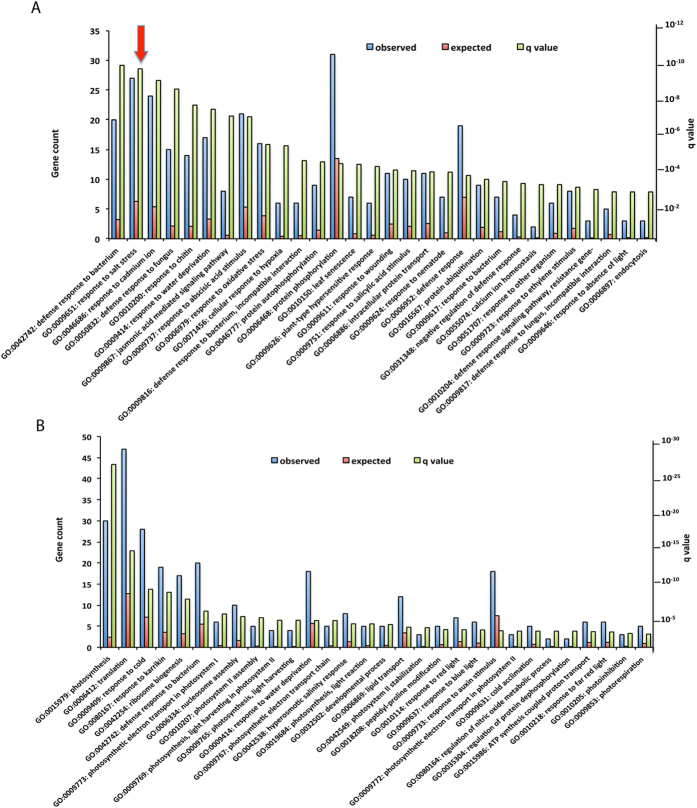
SR1 is known to function in plant immunity, herbivory and cold-regulated gene expression24,28,29,31,33. To verify if the DE genes function in these processes and to gain some insight into other functions of SR1, we performed Gene ontology (GO) enrichment analysis using the whole genome as background. Two methods, AgriGO and GeneCoDis, for singular GO term enrichment analysis yielded similar results with slight variation in the number of GO terms and the order of significance (data not shown). Results obtained with GeneCoDis are presented in Supplementary Fig. S4. A total of 81 GO terms for biological processes were enriched (Supplementary Fig. S4A and Additional File 2, Sheet 1). Consistent with the previous known functions of SR1, GO terms related to plant response to pathogens and abiotic factors were among the enriched terms. Analysis of the up- and down-regulated genes separately resulted in enrichment of 95 and 52 GO terms, respectively (Supplementary Fig. S4 and Sheets 2 and 3 in Additional File 2). Majority of the up-regulated GO terms are associated with plant defense response to biotic factors. In addition, GO terms “response to salt stress” and “response to water deprivation” are also highly enriched in the up-regulated genes. (Supplementary Fig. S4B and Additional File 2 Sheet 2). A significant enrichment of GO terms associated with abiotic factors such as “response to cold” and “response to water deprivation” was observed in down-regulated genes (Supplementary Fig. S4C and Additional File 2 Sheet3).
DE genes are enriched for SR1 binding motif
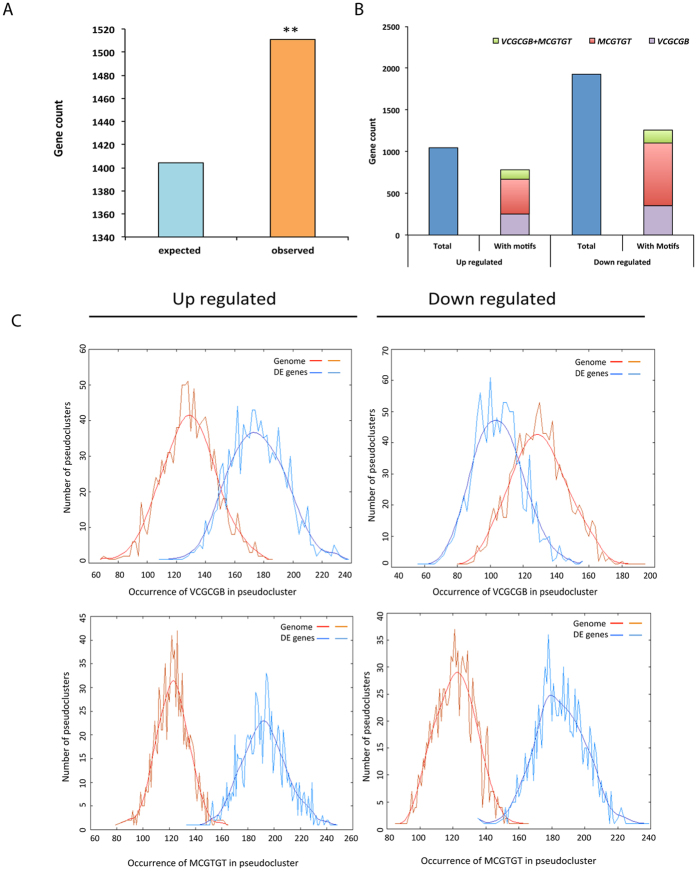
Previous studies using an oligo selection method and electrophoretic mobility shift assays showed that SR1 binds to VCGCGB (where V = A, C or G; B = C, G or T) and MCGTGT (where M = A or C) motifs in the promoter regions of SR1-regulated genes11,23,24,28,35. The rapid activation of the general stress-responsive genes is also mediated through RSRE element (VCGCGB), as promoters of many of these genes exhibit significant enrichment for this motif25,26. Here we determined whether the promoter regions of DE genes are enriched for the VCGCGB and MCGTGT motifs. As shown in Fig. 2A, both these motifs are enriched in the promoters (−1000 bp upstream of translation start site -TSS) of all DE genes (P < 0.0001). As significant enrichment for SR1 binding motifs was observed, we further checked for actual number of differentially up- or down-regulated genes that contained VCGCGB and/or MCGTGT in their promoters. Out of 1046 genes that are up-regulated, 665 (~64%) contained a minimum of one motif of either type (Fig. 2B, and Additional File 3). Of these, 37% contain VCGCGB, 39% have MCGTGT and 16% have both VCGCGB and MCGTGT (Fig. 2B). Similarly, out of 1927 down-regulated genes, 1098 (57%) have one or more of these motifs. Of these, 32% have VCGCGB, 67% have MCGTGT element and 13% have both (Fig. 2B, and Additional File 3). Together, these results indicate that a significant number (59%) of DE genes are likely direct targets for SR1. To identify if these motifs are enriched in the promoters of up- or down-regulated genes, we further analyzed the promoters using POBO analysis with upstream regions of top 500 up-regulated or down-regulated gens using the whole genome as background. This analysis revealed a significant enrichment (P < 0.0001) of both cis-elements (VCGCGB and MCGTGT) in the up-regulated genes whereas in the down-regulated genes only MCGTGT was enriched (Fig. 2C).
Figure 2. SR1-binding sites in the promoters of up- and down-regulated genes.
(A) A significant enrichment of the SR1 binding motifs (VCGCGB + MCGTGT) in the upstream (−1000 bp) of TSS of all DE genes. Asterisks on the bar represent significant overrepresentation of binding sites with a P < 0.0001. (B) Total number of up- and down-regulated genes and the number of the SR1-regulated genes that contain SR1 binding sites VCGCGB or MCGTGT or MCGCGT + VCGCGB in the −1000 bp promoter region. (C) Top panel: POBO analysis of RSRE (VCGCGB) motif in the −500 bp upstream of TSS. 1000 pseudoclusters were generated from top 500 genes from up- or down-regulated genes and genome background. The jagged lines show the motif frequencies from which the best-fit curve is derived. RSRE element is significantly overrepresented with a two-tailed P < 0.0001 in the upstream sequences of up-regulated genes but not with down-regulated genes. Bottom panel: POBO analysis of a second SR1 recognition motif (MCGTGT) using the −500 bp upstream of TSS in 1000 pseudo clusters of top 500 DE genes and genome background. The jagged lines show the motif frequencies from which the best-fit curve is derived. SR1 binding sites are significantly over represented (two-tailed P < 0.0001).
GO term enrichment of SR1 binding motif-containing genes
To understand the biological role of putative direct targets of SR1, we performed a separate GO enrichment analysis using either up- or down-regulated genes that have one or more SR1 binding motifs. In the up-regulated genes, 61 GO categories showed significant enrichment (Additional File 4). Top 30 GO categories are represented in Fig. 3A. Consistent with known function of SR1, the genes were highly enriched for the GO terms that are predominantly associated with plants response to pathogens/pests. The other highly enriched GO terms include abiotic stress and hormonal responses. One of the GO terms that is of special interest is “response to salt stress” for the following reasons: i) it is the second most enriched GO term after “response to bacterium” ii) this GO term comprises 27 genes (second most of all other categories), iii) expression of the majority of these genes is altered in opposite direction in the mutant and complemented plants (see section on salt stress below) and iv) SR1 was not previously known to be involved in salt stress.
Figure 3. GO term enrichment analysis.
GO term enrichment analysis for biological processes of (A) up- and (B) down-regulated genes. For each GO term, the expected and observed gene numbers along with the statistical significance (q-value) for the enrichment is presented. Observed: Number of DE genes associated with a GO term for biological processes. Expected: Number of genes expected for each GO term in the genome. “Response to salt stress” GO term is indicated with an arrow.
GO analysis with the down-regulated genes revealed enrichment for only 37 GO terms (Additional File 5). The highest enrichment for biological processes is associated with photosynthesis (Fig. 3B). Importantly, unlike the GO terms observed in up-regulated DE genes, there was a significant enrichment for GO term associated with only cold stress. Interestingly, down-regulated DE genes with SR1 binding motif also contributed towards the process of “response to bacterium” (Fig. 3B). These results indicated that genes involved in a biological process can be either up- or down-regulated by SR1 depending on the gene.
SR1 regulates the expression of other SRs
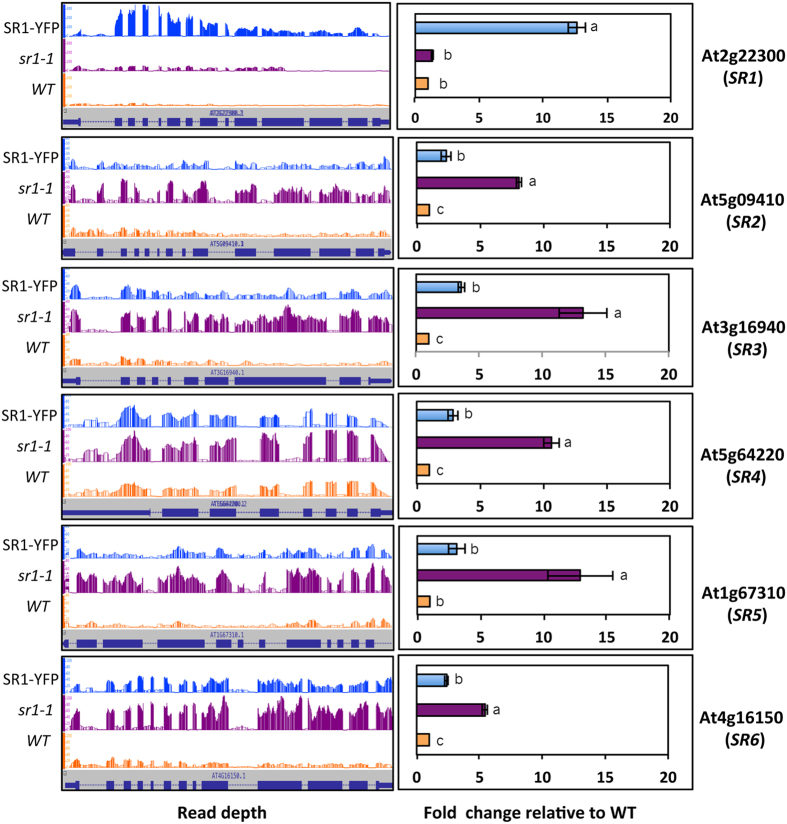
Analysis of promoters of six Arabidopsis SRs (SR1 to SR6) for the presence of SR1 binding motifs revealed that SR3, SR4, SR5 and SR6 contain one or more of these motifs (Supplementary Table S2), suggesting that their expression could be regulated by SR1. To test if any of these SRs are mis-regulated in SR1 mutant, we checked RNA-seq data for their expression. Interestingly, the expression of all five SRs (SR2 to SR6) is significantly elevated in the mutant and fully or partially suppressed in the complemented line (Fig. 4, left panel). To validate these RNA-seq results, RT-qPCR was performed and the results were in agreement with RNA-seq data (Fig. 4, right panel), indicating that SR1 suppresses the expression of other SRs.
Figure 4. SR1 represses the expression of other members of the SR family.
Expression profiles of SRs in WT, sr1-1 and SR1-YFP lines. Panels on left show relative sequence read abundance as histograms (IGB view) in WT, sr1-1 mutant and SR1-YFP. The Y-axis indicates read depth with the same scale for all three lines. The gene structure is shown below the read depth profile. The lines represent introns and the boxes represent exons. The thinner boxes represent 5′ and 3′ UTRs. Right panels show fold change in expression level relative to WT based on RT-qPCR analysis. WT values were considered as 1. Student t-test was performed and significant differences (P < 0.05) among samples are labeled with different letters. The error bars represent SD.
SR1 regulates expression of many transcription factors
The observed DE genes are likely due to direct and indirect effects of SR1; i.e., SR1 may directly bind the promoters of these genes and regulate their expression or regulate other TFs, which in turn regulate expression of down-stream genes. In Arabidopsis, there are over 1716 genes encoding TFs, which are grouped into 58 families36. Among the DE genes, we found 179 TFs belonging to 40 families (Supplementary Fig. S5 and Additional File 6). Of these families, WRKY (P < 0.0006), S1Fa like < P, 0.0007), GATA (P < 0.01), ERF (P < 0.03), EIL (P < 0.04) and ZF-HD (P < 0.04) are highly enriched (Supplementary Fig. S5A). Further examination of the TF families revealed that the genes of 33 of them contain SR1 binding sites (VCGCGB and MCGTGT) in their upstream region (−1000 bp of TSS) (Additional File 6), suggesting that they are likely direct targets of SR1. The number of TFs in each family that are affected and the direction of their expression change (up or down) in the mutant are shown in Supplementary Fig. S5B. Interestingly, expression of all TFs in certain families (e.g. WRKYs, NAC and GRAS) is up-regulated whereas all members in some other families are suppressed (e.g. ZF-HD, NF-Y3, Tri-helix and TALE) (see Supplementary Fig. S5B). The fact that expression of about 10% of all TFs is altered in the mutant suggests that many of the SR1-regulated genes in our DE list, especially those that do not contain SR1 binding motif, are likely indirect targets of SR1.
SR1 negatively regulates salt stress tolerance
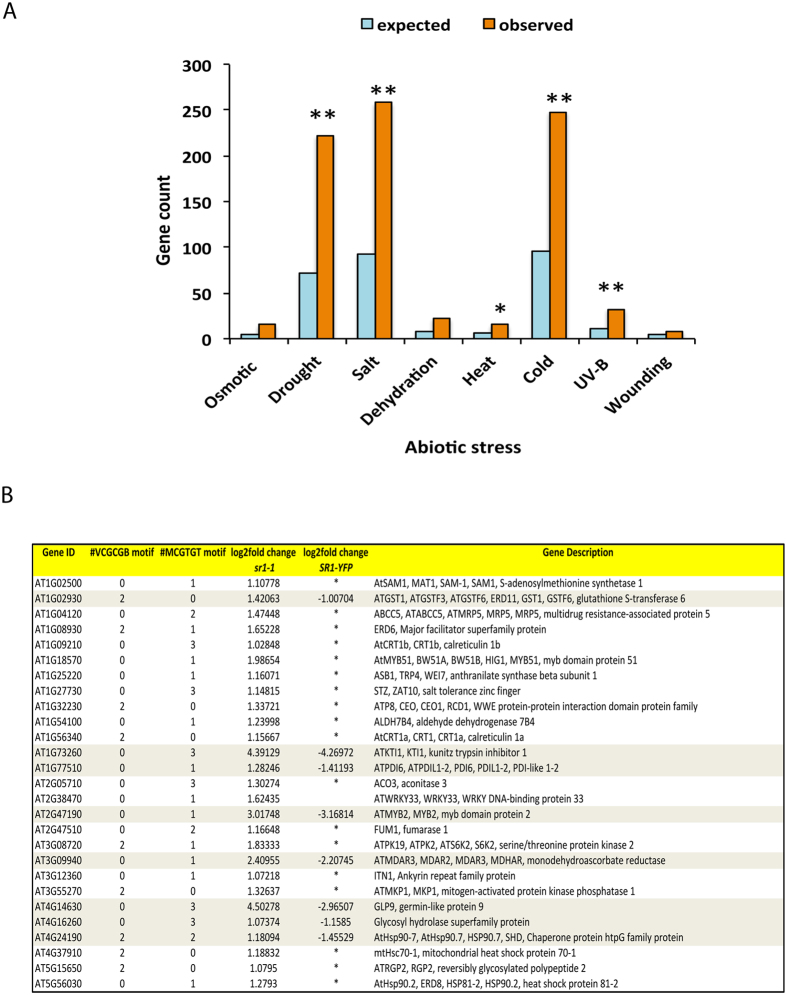
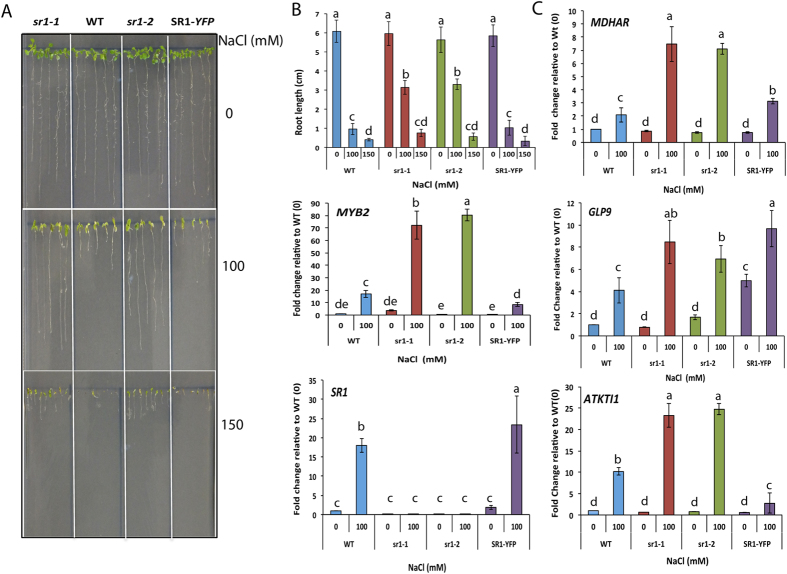
Since the promoters of a large number of DE genes contained RSRE, we performed enrichment analysis to determine if a particular stress responsive genes contributed maximally to the DE list. This analysis revealed a substantial enrichment (P < 0.001) of different abiotic stress responsive genes with a large number of them implicated in salt stress (Fig. 5A and Additional File 7). Interestingly, 27 salt-responsive genes are up-regulated in the mutant. Furthermore, in the complemented line expression of these genes was either restored to the wild type level or repressed (Fig. 5B). GO term enrichment analysis of SR1-binding motif containing up-regulated genes also showed strong enrichment of a term associated with salt stress (Fig. 3A). SR1 is known to regulate cold-induced gene expression24, but its function in salt stress is not known. We, therefore, investigated the role of SR1 in salt stress tolerance. Wild type, two homozygous loss-of-function mutants of SR1 (sr1-1 and sr1-2) and the complemented line28 were tested for salt tolerance. Root growth of all four genotypes was scored for salt tolerance by growing them on different concentrations (0, 100, 150 mM) of NaCl. Interestingly, a significant difference in the primary root length in a NaCl concentration dependent manner was observed (Fig. 6A). At 100 mM NaCl, a significant difference in root length was observed among the genotypes (Fig. 6A). A significant suppression in the primary root growth was noted in WT and SR1-YFP lines as compared to mutant lines (sr1-1 or sr1-2), indicating decreased sensitivity of mutants to salt stress (Fig. 6A, middle panel) as compared to WT and SR1-YFP. Even at 150 mM NaCl, mutants were found to be more tolerant to salt stress. These results suggest that SR1 negatively regulates salt tolerance.
Figure 5. Abiotic stress responsive genes are over-represented in DE genes.
(A) A significant number of DE genes are associated with abiotic stress response in comparison with genome background with a P < 0.0001 (**) and P < 0.05(*). (B) SR1 regulates the expression of salt-responsive genes. List of salt-responsive genes that are enriched in the GO term “response to salt stress” is presented. Transcript levels of these genes in the mutant and complemented line and the number of SR1 binding motifs in the upstream 1000 bp of the TSS are presented. Asterisks in the table indicate that the expression level in the complemented line is restored to wild type. In case of eight other genes that are highlighted, their expression is repressed in SR1-YFP as compared to the mutant.
Figure 6. SR1 is a negative regulator of salt tolerance.
(A) Growth of seedlings of WT, sr1-1, sr1-2 and SR1-YFP on MS plates containing different concentrations of salt. Seeds were plated on ½ strength MS medium supplemented with 0, 100 and 150 mM of NaCl and were allowed to germinate and grow for two weeks. The photographs were taken after two weeks. (B) Top panel: root length was measured for each seedling for all four genotypes and plotted against the concentration of NaCl. Three biological replicates were used. Eight seedlings for each genotype per treatment for each biological replicate were included. Middle and Bottom panels: Expression levels of MYB2 and SR1 TFs under salt stress in different genotypes. Two-week-old seedlings grown on MS medium supplemented with 0 and 100 mM NaCl concentrations were used. A significant increase in the expression of these two TFs was observed. Salt-induced enhancement of MYB2 expression level was significantly higher in sr1-1 and sr1-2 lines. (C) SR1 regulates the expression of other salt-responsive genes. Expression levels of MDHAR, GLP9 and ATKTI1 in two-weeks-old seedlings exposed to 0 and 100 mM NaCl are determined by RT-qPCR. The expression levels of salt-responsive genes were normalized with ACTIN2. Fold change in expression level relative to WT controls (WT-0) is presented. WT-0 values were considered as 1. Student t-test was performed and significant differences (P < 0.05) among samples are labeled with different letters. The error bars represent SD.
SR1 suppresses the expression of salt-responsive genes
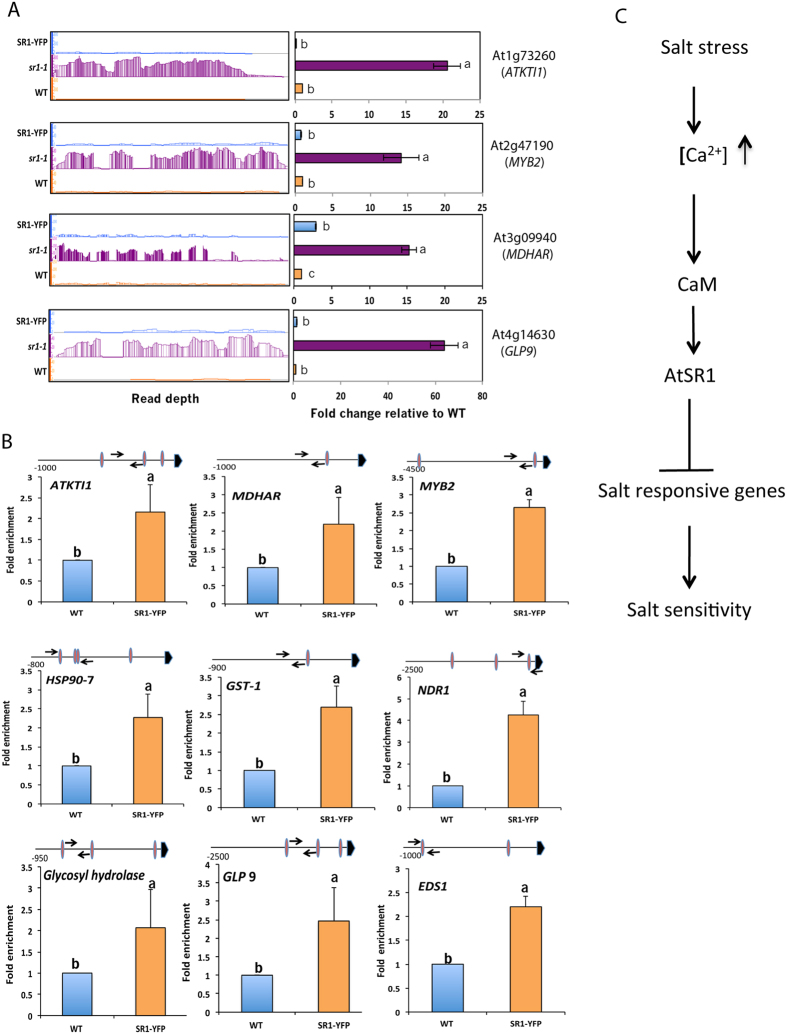
To gain further insights into the role of SR1 in salt stress, the expression level of 27 salt-responsive genes under the GO category of “response to salt stimulus” was compared in sr1-1 and SR1-YFP lines. Nineteen out of 27 salt-responsive genes were represented in both sr1-1 and SR1-YFP data sets and their expression profiles were opposite to each other (Fig. 5B). Motif analysis of upstream regions of these genes indicated that a number of them contain SR1 binding motif (VCGCGB or MCGTGT) (Supplementary Fig. S6A). Orthologs of four Arabidopsis genes (At1g73260, At2g47190, At3g09940 and At4g14630) that were previously reported to be involved in salt tolerance37,38,39,40 and contain an SR1 binding motif in their promoter were selected as representatives to analyze their expression under control and salt stress conditions. The expression pattern of these four genes was verified by RT-qPCR analysis. Expression levels of all four genes were significantly higher in both loss-of-function mutants as compared to WT or SR1-YFP in the presence of salt (Fig. 6B, middle panel and Fig. 6C), suggesting that SR1 represses the expression of these salt-responsive genes. Analysis of RNA-seq data for expression of these four genes also showed increased expression in the mutant and their expression was restored to the wild type in the complemented line (Fig. 7A, left panel). The expression pattern of these four genes was confirmed by RT-qPCR analysis (Fig. 7A, right panel).
Figure 7. SR1 regulation of salt-responsive genes.
(A) Expression levels of a few representative salt stress-responsive genes in WT, sr1-1 and SR1-YFP. Left Panels: relative sequence read abundance (IGB view) as histograms in wild type (WT), SR1 mutant (sr1-1) mutant and the complemented line (SR1-YFP). The Y-axis indicates read depth with the same scale for all three lines. Right panels: Expression analysis of salt-responsive genes using RT-qPCR. Panels on right show fold change in expression level relative to WT. WT values were considered as 1. Student t-test was performed and significant differences (P < 0.05) among samples are labeled with different letters. The error bars represent SD. (B) ChIP-PCR of upstream regions of salt-responsive genes containing VCGCGB or MCGTGT or MCGCGT + VCGCGB. Chromatin from 15-day-old seedlings from WT and SR1-YFP was immunoprecipitated with anti-GFP antibody and used in PCR with primers flanking the putative SR1 binding sites. The results obtained from four independent ChIP experiments were used to calculate fold enrichment. Data was normalized to DNA input levels as well as ACTIN2. The values of WT were considered as 1. Student t-test was performed and significant differences (P < 0.05) among samples are labeled with different letters. Schematic diagram over each panel shows SR1 binding sites (as oval shape) and the location of primers used in ChIP-PCR are indicated with arrows. Bold arrowhead indicates TSS. (C) Proposed model for the role of SR1 in salt stress response (see text for details).
Majority of the salt-responsive genes are known to contain cis-elements in their promoter regions to which known TFs bind. These include G box (CACGTG), N box CACG[G/A]C and NAC (CATGTG) that bind G_box bHLH, N_box_bHLH and Nac_box_NAC TFs, respectively. To understand the regulation of these salt-responsive DE genes by SR1, POBO analysis was performed for the enrichment of these cis-elements as well as RSRE (VCGCGB) element in the upstream regions of all salt-responsive genes. A significant enrichment (P < 0.0001) for VCGCGB and MCGTGT was observed in the upstream region (−1000 bp) of the salt-responsive genes that were up-regulated (Supplementary Fig. S6A). In contrast, no enrichment for MCGTGT motif was noted in the upstream regions of down-regulated genes (Supplementary Fig. S6A). Further, significant enrichment for the G box (CACGTG), N box (CACGGC) and no enrichment for NAC (CATGTG) element in the promoter regions of the up-regulated salt stress-responsive genes were observed (Supplementary Fig. S6B). Significantly, enrichment of specific sequences (ACGTGT, CCGTGT, ACGCGT, and ACGCGC) within the SR1 binding consensus motif was also observed (Supplementary Fig. S6B). In contrast to the up-regulated salt-responsive genes, a significant enrichment for only G box (CACGTG) element and the SR1 binding motif ACGTGT was found in down-regulated salt-responsive genes (Supplementary Fig. S6C). These results clearly suggest dual regulation of salt responsive genes by different TFs and preferential usage of certain cis-elements (ACGTGT, CCGTGT, ACGCGT, and ACGCGC) within the consensus motif of these transcription factors.
SR1 binds to the promoter regions of salt-responsive genes
Earlier studies have shown that SR1 binds to the promoter regions of EDS1, NDR1 and EIN3 that are involved in plant defense and ethylene signaling28,30. As shown in Supplementary Fig. S6A, there is a significant enrichment of SR1 binding sites in the DE genes that are responsive to salt stress. Consistent with this, expression levels of salt-responsive genes were significantly up-regulated in sr1-1 (Fig. 5B). Restoration of transcript levels of salt-responsive genes in the SR1-YFP line to wild type level and the presence of SR1 binding sites in their promoter regions suggest that these are potential direct targets of SR1. To further confirm that SR1 regulates the salt-responsive genes, we determined the expression levels of four of these genes in WT, sr1-1 and SR1-YFP using RT-qPCR. This analysis indicated significantly higher transcript levels of the salt-responsive genes in sr1-1 (Fig. 7A, right panel). To confirm that SR1 binds to the promoter region of these genes, we performed ChIP-PCR assays using the complemented line expressing SR1-YFP. First, we confirmed whether there is a significant enrichment for the promoters of known targets of SR1 such as EDS1 and NDR1 in the ChIP’ed DNA obtained with anti-GFP antibody. A significant enrichment for the EDS1 and NDR1 promoters was observed thus validating earlier reports (Fig. 7B). We then performed enrichment analysis for promoters of several salt-responsive genes (ATKTI1, MDAR3, HSP90-7, GST1, Glycosyl hydrolase GLP9 and MYB2) whose expression is increased in the mutant and contained one or more SR1 binding sites. Interestingly, a significant enrichment for promoters of these genes was noted in immunoprecipitated DNA (Fig. 7B), suggesting in vivo binding of SR1-YFP to these promoters and direct regulation of these genes by SR1. To address the specificity of SR1 binding to these promoters, we performed ChIP-PCR with primers corresponding to the promoter of ACTIN2, whose expression is not affected in the mutant (Supplementary Fig. S8) and also to two other genes [GRAS2 (At1g07530) and At1g15790] that are misregulated in sr1, but do not contain SR1 binding motifs. For all three genes, there was no enrichment of promoters in the ChIP’ed DNA (Supplementary Fig. S8), indicating that binding of SR1 salt-responsive genes is specific.
Discussion
SR1 regulates expression of genes involved in multiple stress responses
Recent studies using SR1 loss-of-function mutants have shown that it regulates biotic and cold stress responses24,28,29,31,33. Despite its important role in multiple stress responses, a comprehensive analysis on SR1-regulated genes is lacking. Our global transcriptome analysis using RNA-seq revealed that a large number of genes involved in diverse stress responses are regulated either directly or indirectly by SR1 (Figs 1 and 5). Previously Galon et al.29 compared the expression of genes in WT and sr1-1 using microarrays and identified only 105 DE genes (99 up-regulated and 6 down-regulated genes)29. In that study, a complemented line was not included, hence it was difficult to ascertain that these DE genes are SR1-regulated. Our study significantly differs from the former study in a number of ways. Here we used next generation sequencing that significantly increased the depth of transcriptome analysis. More importantly, the use of a complemented line in which mutant phenotypes are rescued allowed us to identify the genes that are regulated specifically by SR1 (Supplementary Fig. S1). Our study revealed thirty times more DE genes as compared to the previous study29. This huge difference in the number of DE genes is likely due to the technology used here and the depth of RNA-seq. Over half of the DE genes reported in the previous study were found in our analysis. The absence of some DE genes from a previous study in our list could be due to limitations associated with different methodologies such as probe cross hybridization in microarray or more likely due to the tissues used for DE analysis as the age of the plants used in these two studies is different. In fact, developmental regulation of expression levels of SRs has been previously reported12,13,23. Reproducibility among replicates (Supplementary Fig. S2), full or partial restoration of expression of ~85% of DE genes in our complemented line to wild type level (Supplementary Figs S2 and S3) and RT-qPCR validation of expression of a number of randomly selected DE genes indicates that the DE genes are bona fide SR1 targets. Enrichment of DE genes in multiple abiotic stress-responses indicates that SR1 plays a major role in cross-talk between multiple stress signal transduction pathways (Fig. 5 and Additional File 7). Earlier, SRs were shown to differentially respond to various stresses such as heat, cold, salinity, drought, UV and stress hormones such as ethylene and ABA18. Further, many of the SRs have been implicated for their regulatory role in abiotic stress responses19,20,26,27,33. GO analysis of the DE genes indicated high enrichment of GO terms associated with diverse cellular processes that are critical for plant responses to biotic stresses such as bacteria and fungi, and abiotic stresses including drought, cold, salt and oxidative stress. Response to hormones such as ABA and auxin was also observed (Supplementary Fig. S7 and Additional File 8). These results suggest that SR1 could function as an important integrator of variety of stress responses. Consistent with these results, SR1 is already known to play an important role in at least four different stress responses24,28,29,31,33.
SR1 binding motifs containing genes are both up- and down-regulated
Earlier studies identified CGCG and CGTG as core sequences to which SR1 binds through its CG1 DNA binding domain23. Furthermore, several studies identified VCGCGB and MCGTGT as consensus element, through which the SR1 regulates the expression of target genes24,28,33,35. Analysis of DE genes showed that >59% of SR1-regulated genes contain VCGCGB and MCGTGT elements and these motifs are significantly enriched in their promoter regions (Fig. 2). Among the genes that contain SR1 binding motif, in up-regulated genes both elements contributed towards the enrichment whereas highest representation of MCGTGT motif was observed in the down-regulated genes. Further, POBO analysis using the whole genome as a background also confirmed this observation (Fig. 2). The up-regulated genes containing SR1 binding sites, not only highly enriched for GO terms related to defense response to bacterium and fungi, but also for response to salt stress, water deprivation, and response to some hormones. In contrast, GO term enrichment of down-regulated genes that contain SR1 binding motifs exhibited significant enrichment for “response to cold” and “cold acclimation” apart from other cellular processes. This is consistent with the previous reports where SR1 was shown to function as a positive regulator of genes involved in the cold response24,33. Indeed, a preferential enrichment of either up- or down-regulated SR1 binding motif-containing genes for a biological process indicates that SR1 binds different cis-elements for regulation of different biological processes. Further, it was proposed that SR1 induced activation of the CBF2 is mediated through VCGCGB element in the promoter24.
Previous studies using loss- or gain-of-function alleles of SR1 have shown that it acts as a critical regulator of both basal and systemic acquired resistance28,33,41. A significant increase in the basal levels of SA in the loss-of-function mutants of SR1 has been reported24,28,33. Our gene expression analysis also indicated that 66% of the SA responsive genes have VCGCGB or MCGCG elements in their promoters indicating that they are potential direct targets of SR1. Some of these include TGA3, NAC0062, CBP60G, EDS5, WRKY8, and MPK1. Earlier CBP60g along with SARD1 had been described as key regulators of ICS1 induction and SA synthesis42,43. It has been suggested that SR1 may regulate the defense response through binding to the promoters of the genes or through activation of other repressor proteins28,29. In fact, Du et al.28 have shown direct binding of SR1 to the EDS1 promoter and repression of its expression, indicating repressive activity of this TF in regulating these genes.
SR1 suppresses the expression of other members of SR family
Loss-of-function of SR1 significantly relieved the suppressive effect of SR1 on other SRs expression. Furthermore, expression of other SRs is significantly reduced in the complemented line (Fig. 4), indicating that SR1 controls the expression of other SR genes. Regulation of expression of some of these SR genes is likely through direct binding of SR1 to cis elements (VCGCGB or MCGTGT) in their promoter. With the exception of SR2, promoters of the rest of the SRs (SR3, 4, 5 and 6) do contain the cis-elements variation of the CGCG box, which could be involved in regulation of these genes by SR1 (Supplementary Table S2). We analyzed the promoter sequence of SRs for non-canonical binding motifs (i.e. with core sequence being similar and nucleotide at 5′ and 3′ end of the element being different) and found them to contain motifs related to SR1 binding sites (Supplementary Table S2). Interestingly, elevated expression level of SR2 in sr1-1 and its down-regulation in the complemented line, even in the absence of SR1 binding motifs in its promoter region, indicate the existence of an alternate mechanism by which SR1 regulates SR2 expression. Previously, the VSP1 promoter, which does not contain a canonical SR1 binding motif, was shown to be regulated directly by SR132, thus indicating the existence of alternate regulatory pathways. Thus, it is possible that SR1 also regulates SRs through non-canonical cis-elements in their promoters.
Indirect regulation of SR1-regulated genes
Enrichment analysis for TF families in DE genes indicated highest enrichment for genes in the WRKY, EIL, ERF, ZF-HD and S1Fa TF families. The WRKY TFs, which were all up-regulated (Supplementary Fig. S5A), bind W-box in the defense genes that are primarily implicated in regulation of defense responses against pathogen infection. However, these TFs are also implicated in other cellular processes such as abiotic stresses44. Some members of this family (WRKY18, WRKY33, WRKY40, WRKY46, WRKY70, WRKY53, WRKY70 and WRKY75) have SR1 binding sites in their promoter (Additional File 6), indicating that they are likely direct targets of SR1. Given that GO terms enrichment for the “response to bacterium/pathogen” and “response to abiotic stresses” was observed in DE genes, it is possible that these TFs may regulate the expression of DE genes that do not contain an SR1 binding motif. In the down-regulated genes, the highest representation of ZF-HD, ERF, AP2, bHLH, and TCP TF families was observed, indicating that the members of these families are positively regulated in SR1. Together, these data indicate a complex network of regulation of expression of TFs by SR1.
RSRE is enriched only in up-regulated genes
Recent studies identified VCGCGB as the core element that is enriched in a majority of early-activated genes that are also regulated by Ca2+ under stress conditions25. As the RSRE element VCGCGB is identical to the binding site of SRs (VCGCGB), many studies implicated SRs in general and SR1 in particular in regulation of general stress responses. Our analysis of the promoter region of all the DE genes indicated that a significant percentage of the genes contain this element, thus establishing their role in general stress response (Fig. 2). Further, the fact that the majority of these genes are misregulated in sr1-1 and are implicated in various stress signaling pathways, confirmed the significant role played by SR1 in their regulation. Interestingly, POBO analysis indicated the enrichment of RSRE motif only in the promoter regions of the DE genes that were up-regulated, but not in genes that were down-regulated (Fig. 2C). This might be due to increased occurrence of abiotic stress responsive (with exception of cold responsive) genes in up-regulated genes and/or that the negative regulation by SR1 is not mediated through the RSRE element. As evident from Fig. 5A, significant enrichment of GO terms for the abiotic stresses such as “responses to salt stress” and “water deprivation” was observed only in the up-regulated DE genes. Furthermore, enrichment of VCGCGB motif was significantly higher in the up-regulated DE genes. Absence of enrichment for VCGCGB in the down-regulated DE genes and enrichment for GO terms “response to cold” and “cold acclimation” clearly suggest that SR1 positively regulates cold responsive genes through utilization of VCGTGT rather than VCGCGB (Figs 2C and 3, and Supplementary Fig. S6A). In fact, a significant enrichment of the SR1 binding sites, VCGTGT and VCGCGB, was noted in the early cold-responsive genes33.
SR1 confers salt sensitivity by repressing the expression of salt-responsive genes
GO analysis of the up-regulated genes that contain SR1 binding sites in their promoters exhibited significant enrichment of a GO term associated with “response to salt stress (Fig. 3A)”, suggesting a new role for SR1 in salt tolerance. Interestingly, both mutant lines of SR1 performed better in terms of root growth under increasing concentrations of NaCl when compared with the WT and SR1-YFP seedlings. Thus, our results suggest that SR1 acts as a negative regulator of seedling growth under salt stress. This negative regulation of salt stress by SR1 is similar to that observed under biotic stress28,29 and differs from that of the cold stress response24, where it functions as a positive regulator. Previously, Galon et al.19 and Pandey et al.20 identified SR2/CAMTA1, another member of SR family TF, to be a positive regulator of salt stress as mutants lacking this TF exhibited increased sensitivity to salt stress and drought stress, suggesting that SR1 and SR2 have opposing functions in salt stress.
In order to resolve the regulation (direct versus indirect) by SR1, salt-responsive genes were identified and subjected to POBO analysis for enrichment of VCGCGB in their upstream region. Analysis of the promoters of the salt-responsive DE genes revealed significant enrichment for RSRE (VCGCGB) in up-regulated genes (Supplementary Fig. S6A). Hence, it is possible that some of these genes could be direct targets of SR1. Similar analysis of promoters of down-regulated salt-responsive genes did not show enrichment of RSRE, suggesting that i) SR1 utilizes different motifs to regulate expression of these genes and/or ii) other proteins (including other SRs) might activate these genes, whose expression may be regulated by SR1.
As our data showed a negative regulatory role for SR1 in salt stress, we determined the effect of SR1 mutation on the expression levels of the genes associated with the biological process “response to salt stress”. Twenty-seven genes associated with this GO term were screened for the presence of SR1 binding sites in their promoters, their expression levels and ability of SR1 to complement their expression in SR1-YFP line. Although several genes fit these criteria (Fig. 5B), KTI1, MYB2, MDAR3, GLP9 were selected along with SR1 and their expression levels were determined in different genotypes in response to salt stress. Earlier reports have shown that overexpression of MYB2 and orthologs of other three genes confer salt tolerance38,39,45,46. Exposure to salt stress significantly enhanced their expression levels by two-fold in both WT and SR1-YFP seedlings. In contrast, >12 to 15 fold higher induction of these genes was observed in both mutant alleles of SR1. Interestingly, SR1 expression in WT and SR1-YFP was about 12 to 16 fold higher in salt-treated seedlings as compared to their respective controls (Fig. 6). Since the 35S promoter driving SR1-YFP is known to be non-responsive to salt stress, the observed increase in SR1-YFP transcript may be due to its increased stability in the presence of salt47.
Many members of different TF families are known to regulate expression of genes involved in salt stress by binding to the various cis-elements in the promoters of salt-responsive genes37,38. In response to salt stress, TFs such as G_box_BHLH and N_box_bHLH bind to the cis-element CACGTG and CACG[G/A]C, respectively, and regulate their expression48,49. In this study, we analyzed the enrichment for cis-elements to which various TFs bind in the upstream regions of salt-responsive DE genes. We compared if there are any differences in the enrichment pattern among the salt-responsive up- and down-regulated DE genes using POBO analysis. We found enrichment (P < 0.0001) for G Box (CACGTG), N box (CACGGC) but not NAC (CATGTG) in up-regulated salt-responsive genes (Supplementary Fig. S6B). Only G Box (CACGTG) enrichment was noted in the down-regulated genes (Supplementary Fig. S6C). Analysis for co-enrichment of SR1 binding motifs showed enrichment for ACGTGT, CCGTGT, ACGCGT and ACGCGC in the promoter regions of both up- and down-regulated salt-responsive genes. The observations that i) promoter regions of these genes have 1 to 3 SR1 binding sites, and ii) SR1 binds to promoters of some of these genes (Fig. 7B) provide evidence that SR1 directly regulates their expression.
Based on our work we propose a model (Fig. 7C) to explain the role of SR1 in salt stress response. Previous studies have demonstrated that exposure of plants to salt stress changes cytosolic Ca2+ levels50. In addition, Ca2+ through CAM has been shown to regulate SR1 activity28. Our work showed that AtSR1 either directly and/or indirectly suppresses the expression of salt-responsive genes that are necessary for salt tolerance there by conferring salt sensitivity. In summary, our results showed that a large number of genes that are associated with biotic and abiotic stress responses are regulated by SR1. A large fraction of these genes (~59%) contain one or more binding sites of SR1 in their promoter region, suggesting that they may be regulated directly by this TF. Our transcriptome analysis revealed a novel role for SR1 in salt stress. By analyzing growth phenotypes and salt-responsive genes we confirmed that SR1 functions in salt stress response. Further, our results showed that SR1 functions as a negative regulator of salt tolerance. These results provide novel insights into the role of SR1 in abiotic stress tolerance in general and salt stress in particular. Future studies using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) should allow identification of direct targets of SR151.
Methods
Plant Materials
Three Arabidopsis genotypes - WT (Columbia-0), two alleles of SR1 mutant (sr1-1, sr1-2) in Col-0 background, and a complemented line (SR1-YFP) – used here were developed earlier28. Surface sterilized seeds were sown in sterilized soil, allowed to germinate and grown for 40 days in a growth chamber at 21 ± 1 °C with 60% humidity, 200 μmoles/m2/sec light under day neutral condition. To test salt stress tolerance in these genotypes, surface sterilized seeds were plated on ~70 ml of ½ strength MS medium supplemented with 1% sucrose, 0.5 g/L of MES along with 0, 100 or 150 mM NaCl and 0.8% (w/v) Phytoblend in square sterile Petri dishes. The seeds were germinated and seedlings were grown vertically for two weeks to score for the seedling growth and root length. All genotypes were grown on the same plate to minimize the differences due to any changes in microenvironment. After 14 days, root length was measured and seedlings were photographed. All experiments were performed three times with a minimum of three replicates.
Western blot analysis
Leaf material was flash frozen, ground in liquid nitrogen and nuclear extracts were prepared from nuclei preparation essentially as described in Xing et al.52 with slight modifications. The pellet containing nuclei was resuspended in nuclear lysis buffer and sonicated using Covaris M220 Focused –ultrasonicator for 8 min at 7 °C with settings of peak power 75, duty factor 5 and 200 cycles/burst. The extract was clarified by centrifugation for 10 min at 16,000 g at 4 °C. Immunoprecipitation was performed essentially as described in Xing et al.52 using Chromotek GFP-TRAP_A beads. Immunoprecipitated protein was separated from beads by boiling at 95 °C for 10 min in 60 μl of 1× SDS loading buffer. Thirty μl of extract was resolved in 12% SDS gels and blotted on to a PVDF membrane. The blot was probed with anti-GFP antibody (sc-8334, Santa Cruz Biotechnology) and detected with secondary antibody conjugated with alkaline phosphatase detection system.
RNA–seq
Total RNA from leaves (collected at 4 p.m.) of 40-day-old plants of three genotypes was isolated using miRNAeasy kit (Qiagen, USA#217004). Any contaminating genomic DNA was removed using on column DNAse digestion. Ribosomal RNA was removed using a Ribozero Plant kit and the sequencing libraries were prepared from rRNA-depleted samples using TruSeq stranded RNA-seq kit (Illumina) as per manufacturer instructions and single-end sequencing of the library was done at the Genome Sequencing & Analysis Core Resource, Duke University using Illumina Hi seq 2000. All RNA-seq reads were deposited at NCBI in the GenBank sequence read archive (SRA) under the accession number SRP073518.
Mapping of the reads and identification of DE genes
The reads were aligned to the TAIR 10 version of the Arabidopsis genome using TopHat53 using default settings. The read alignments were assembled into transcriptome assembly using Cufflinks. The assemblies for each replicate were merged together using Cuffmerge utility53. Using Cuffdiff tool53 the aligned reads and merged assembly for each genotype were utilized for calculating the expression level differences of various genes. The DE genes list was computed using Cuffdiff53. Those genes that met the following criteria were considered as DE genes: i) The q-value < 0.05, ii) the fold change >2, and iii) The sum of the RPKM from the comparing genotypes >10. The common genes that are represented in one or more data sets were identified using the VENNY (http://bioinfogp.cnb.csic.es/tools/venny/) a web-based tool. Heat map of differentially expressed genes was generated using CummeRbund53. Box-and-whisker plots of DE genes were generated using the log2 transformed expression values in WT, sr1-1 and SR1-YFP with JMP Pro12 statistical software. For scatterplot analysis, the RPKM values were log2 transformed and genes with ≥1 value were used.
Bioinformatics analysis to identify SR1 binding motif-containing genes
To identify the number of DE genes with SR1 binding motifs VCGCGB and MCGTGT in their promoter, “Patmatch” (Version 1.1) utility tool (www. arabidopsis.org) was used. This tool identifies the motif on both the strands from the dataset of “TAIR10 Loci Upstream sequences-1000 bp”. 1000 bp sequence preceding the TSS was used for this analysis. Up- and down-regulated genes were included as input for scoring both type and number of SR1 binding motifs.
GO enrichment analysis
GO analysis was performed for term enrichment using GeneCodis54. Single enrichment analysis with TAIR GO annotations was performed using the hyper geometric test with Benjamin-Hochberg FDR correction with a significance of P < 0.05. The genes that are up- or down-regulated for each data set were analyzed separately.
To identify various TFs in the DE genes, a list of all TFs was obtained from Plant TF Database (version 3.0)36 and all DE genes were queried against the total TF list. TAIR 10 ID of all TF genes was used as input for identifying the genes encoding the TF and classifying them based on the similarity with Total TF family list. The TFs and the genes responsive to various abiotic stress conditions were obtained from STIFB (Stress Responsive TF Database) (http://caps.ncbs.res.in/stifdb2/). Promoters of the genes that contained cis-element for binding of the TFs that are implicated in abiotic stress response were retrieved for the analysis. DE genes were queried against the list of the genes for a specific abiotic stress. Further, on the basis of overlap of locus ID (TAIR ID) between the lists of genes, they were further categorized into different subsets.
For the promoter analysis either 500 or 1000 bp upstream of the start codon was extracted from TAIR using an online tool for bulk sequence retrieval. For the estimation of the enrichment for a particular cis-elements in the set of promoter sequences (−500 or −1000 bp) were used as input for POBO analysis (http://ekhidna.biocenter.helsinki.fi/poxo/pobo)55.
Validation of DE genes using RT-qPCR analysis
Primers for validation of DE genes using Real time qPCR (RT-qPCR) were designed using Primer Quest web tool (http://www.idtdna.com/Primerquest/Home/Index) from IDT (USA) (Supplementary Table S3). Nine DE genes were randomly selected and analyzed for their expression levels using RT-qPCR. cDNA from 40-day-old plants was prepared with SuperScript III first Strand Synthesis kit (Invitrogen), and diluted to 1:5 ratio with sterile nuclease free water, 1.5 μl of the diluted cDNA was used for each reaction. For every qPCR reaction, 5 μl of 2X LightCycler 480 SYBR Green I Master mix (Roche) was used along with 1 μl of 5 μM of each primer in a final reaction volume of 10 μl. For each genotype, cDNA from two independent biological replicates was used. Three technical replicates were used for each sample. RT-qPCR was performed in a Roche LC480 machine (Roche) using the preprogramed “SYBR green-I 96 well program”. ACTIN2 was used as a reference gene as this gene does not exhibit any difference in its expression levels among the various genotypes (Supplementary Fig. S8). Fold change in expression was calculated and plotted with respect to WT. The expression level in WT for each gene is considered as 1.
RT-qPCR analysis of salt-responsive genes
Fourteen-day-old control and salt-treated seedlings of different genotypes were collected and flash frozen in liquid nitrogen. The frozen tissues were ground to fine powder in 2 ml microfuge tubes with metal ball bearings. Total RNA was isolated using Trizol and then subjected to DNAse (Promega) treatment to remove any genomic DNA. Two μg of total RNA was used for cDNA synthesis using Superscript II reverse transcriptase (Invitrogen) as per manufacturer instructions. The cDNA was diluted 5 times and 2.5 μl/reaction was used as a template. Expression analysis was performed using RT-qPCR as described above. The data obtained was normalized with ACTIN2 and fold change in the expression level was calculated relative to WT control i.e, 0 mM NaCl. The expression level in WT control was considered as 1. A minimum of three technical replicates and three biological replicates were used for each experiment.
RNA isolated from three genotypes was used for cDNA synthesis to analyze the expression of other members of SR family (SR2-SR6). cDNA synthesis, primer design and RT-qPCR analysis were done as described above. The expression levels of the SR genes were normalized with ACTIN2 and fold change in the expression was calculated relative to WT. The values of WT were considered as 1.
ChIP-PCR
For chromatin immunoprecipitation (ChIP) assays, 15 day-old seedlings of WT and SR1-YFP were grown on ½ MS medium with 1% sucrose under 16/8 h day/night cycle at 21 °C. ChIP assay was performed as described by Werner Aufsatz with modifications using GFP-Trap_A beads (http://www.abcam.com/protocols/chip-using-plant-samples—arabidopsis). Briefly, nuclear extract was prepared from formaldehyde cross-linked (1%) seedlings of WT and SR1-YFP as above and diluted with ChIP dilution buffer and pre-cleared with bab-20 agarose beads. The pre-cleared nuclear extract was further incubated with GFP–Trap_A beads for 15 h at 4 °C on rotatory wheel. The beads were collected by centrifugation and washed sequentially with 1 ml of low salt wash buffer, 1 ml of high salt wash buffer, 1 ml LiCl wash buffer and 1 ml TE buffer. Each wash was carried out by resuspending the beads in wash buffer and rotating on a wheel at 4 °C for 5 min and centrifuging at 2500 g for 2 min, and the supernatant was discarded. The protein-DNA complex was eluted twice with 250 μl of elution buffer (1% SDS and 0.1 M NaHCO3) and reverse cross-linked by incubating the eluate at 65 °C for 6–8 h followed by 3 h of proteinase K treatment at 45 °C with gentle shaking. The DNA was purified using phenol:chloroform/isoamyl alcohol and precipitated using absolute ethanol followed by washing with 75% ethanol. Air-dried DNA pellet was resuspended in 70 μl of TE buffer with RNAse A (10 μg/ml). The precipitated DNA was used for qPCR with the primers specific to a region of promoter in the target genes. Data was normalized to DNA input levels as well as ACTIN2. The results obtained from four independent ChIP experiments were used to calculate fold enrichment. The values of WT were considered as 1.
Additional Information
How to cite this article: Prasad, K. V. S. K. et al. Global gene expression analysis using RNA-seq uncovered a new role for SR1/CAMTA3 transcription factor in salt stress. Sci. Rep. 6, 27021; doi: 10.1038/srep27021 (2016).
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Science Foundation (MCB #5333470) to A.S.N.R. We thank James Craven for his comments on the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions A.S.N.R. conceived and directed the project. K.V.S.K.P., A.A.E.A. and A.S.N.R. designed the experiments. K.V.S.K.P. performed RNA-seq, ChIP-PCR and all bioinformatics analysis with the DE gene list. A.A.E.A. performed all experiments pertinent to salt stress and RT-qPCR analysis of gene expression. D.X. analyzed RNA-seq data and generated DE gene list. K.V.S.K.P. and A.S.N.R. wrote the manuscript. A.A.E.A. and D.X. read and commented on the manuscript.
References
- Boyer J. S. Plant productivity and environment. Science 218, 443–448 (1982). [DOI] [PubMed] [Google Scholar]
- Dhlamini Z. et al. Status of research and applications of crop biotechnologies in developing countries: Preiminary assessment. Food and Agriculture organization of the United Nations (2005). [Google Scholar]
- Qin F., Shinozaki K. & Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52, 1569–1582 (2011). [DOI] [PubMed] [Google Scholar]
- Hirayama T. & Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant Journal 61, 1041–1052 (2010). [DOI] [PubMed] [Google Scholar]
- Hadiarto T. & Tran L. S. Progress studies of drought-responsive genes in rice. Plant Cell Rep 30, 297–310 (2011). [DOI] [PubMed] [Google Scholar]
- Winfield M. O., Lu C., Wilson I. D., Coghill J. A. & Edwards K. J. Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnol J 8, 749–771 (2010). [DOI] [PubMed] [Google Scholar]
- Reddy A. S., Ali G. S., Celesnik H. & Day I. S. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol 67, 225–257 (2005). [DOI] [PubMed] [Google Scholar]
- Lopez-Maury L., Marguerat S. & Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 9, 583–593 (2008). [DOI] [PubMed] [Google Scholar]
- Lecourieux D., Raneva R. & Pugin A. Calcium in plant defence-signalling pathways. New Phytologist 171, 249–269 (2006). [DOI] [PubMed] [Google Scholar]
- Poovaiah B. W., Du L., Wang H. & Yang T. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol 163, 531–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S., Reddy V. S. & Golovkin M. A calmodulin binding protein from Arabidopsis is induced by ethylene and contains a DNA-binding motif. Biochem. Biophys. Res. Commun. 279, 762–769 (2000). [DOI] [PubMed] [Google Scholar]
- Yang T. B. & Poovaiah B. W. An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. Journal of Biological Chemistry 275, 38467–38473 (2000). [DOI] [PubMed] [Google Scholar]
- Bouche N., Scharlat A., Snedden W., Bouchez D. & Fromm H. A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861 (2002). [DOI] [PubMed] [Google Scholar]
- Han J. et al. The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell 127, 847–858 (2006). [DOI] [PubMed] [Google Scholar]
- Song K. et al. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125, 453–466 (2006). [DOI] [PubMed] [Google Scholar]
- Finkler A., Ashery-Padan R. & Fromm H. CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett 581, 3893–3898 (2007). [DOI] [PubMed] [Google Scholar]
- Yang T. & Poovaiah B. W. A calmodulin-binding CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058 (2002). [DOI] [PubMed] [Google Scholar]
- Galon Y. et al. Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232, 165–178 (2010). [DOI] [PubMed] [Google Scholar]
- Pandey N. et al. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14, 216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa e Silva O. CG-1, a parsley light-induced DNA-binding protein. Plant Mol Biol 25, 921–924 (1994). [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Isono T. & Sato M. H. Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell Physiol. 44, 975–981 (2003). [DOI] [PubMed] [Google Scholar]
- Yang T. B. & Poovaiah B. W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. Journal of Biological Chemistry 277, 45049–45058 (2002). [DOI] [PubMed] [Google Scholar]
- Doherty C. J., Van Buskirk H. A., Myers S. J. & Thomashow M. F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J. W. et al. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLos Genet 3, 1800–1812 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn G. et al. A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J 80, 82–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson M. et al. Distinct roles for mitogen-activated protein kinase signaling and CALMODULIN-BINDING TRANSCRIPTIONAL ACTIVATOR3 in regulating the peak time and amplitude of the plant general stress response. Plant Physiol 166, 988–996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L. et al. Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158 (2009). [DOI] [PubMed] [Google Scholar]
- Galon Y. et al. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582, 943–948 (2008). [DOI] [PubMed] [Google Scholar]
- Nie H. Z. et al. SR1, a Calmodulin-Binding Transcription Factor, Modulates Plant Defense and Ethylene-Induced Senescence by Directly Regulating NDR1 and EIN3. Plant Physiol 158, 1847–1859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K. et al. The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol 53, 2008–2015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Xi J., Du L., Suttle J. C. & Poovaiah B. W. Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol Biol 79, 89–99 (2012). [DOI] [PubMed] [Google Scholar]
- Kim Y., Park S., Gilmour S. J. & Thomashow M. F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J 75, 364–376 (2013). [DOI] [PubMed] [Google Scholar]
- Koo S. C. et al. The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Molecules and cells 27, 563–570 (2009). [DOI] [PubMed] [Google Scholar]
- Choi M. S. et al. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J Biol Chem 280, 40820–40831 (2005). [DOI] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G. & Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42, D1182–1187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J. H. et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in arabidopsis. J. Biol. Chem. 280, 3697–3706 (2005). [DOI] [PubMed] [Google Scholar]
- Srinivasan T., Kumar K. R. & Kirti P. B. Constitutive expression of a trypsin protease inhibitor confers multiple stress tolerance in transgenic tobacco. Plant Cell Physiol 50, 541–553 (2009). [DOI] [PubMed] [Google Scholar]
- Eltayeb A. E. et al. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225, 1255–1264 (2007). [DOI] [PubMed] [Google Scholar]
- Wang T. et al. Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLos one 8, e61722, 10.1371/journal.pone.0061722 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing B. et al. Brush and spray: a high-throughput systemic acquired resistance assay suitable for large-scale genetic screening. Plant Physiol 157, 973–980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107, 18220–18225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W. et al. The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol 163, 1741–1751, 10.1104/pp.113.227108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. et al. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819, 120–128, 10.1016/j.bbagrm.2011.09.002 (2012). [DOI] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S. & Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana S. et al. Overexpression of monodehydroascorbate reductase from a mangrove plant (AeMDHAR) confers salt tolerance on rice. J Plant Physiol 169, 311–318 (2012). [DOI] [PubMed] [Google Scholar]
- Chung J. S., Zhu J. K., Bressan R. A., Hasegawa P. M. & Shi H. Reactive oxygen species mediate Na+ -induced SOS1 mRNA stability in Arabidopsis. Plant J 53, 554–565, 10.1111/j.1365-313X.2007.03364.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S., O’Shea C., Jensen M. K. & Skriver K. Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14, 5842–5878, 10.3390/ijms14035842 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E. & Quail P. H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Trewavas A. J. & Knight M. R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J 12, 1067–1078 (1997). [DOI] [PubMed] [Google Scholar]
- Long C. et al. Ataxia and Purkinje cell degeneration in mice lacking the CAMTA1 transcription factor. Proc Natl Acad Sci USA 111, 11521–11526, 10.1073/pnas.1411251111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Wang Y., Hamilton A., Ben-Hur A. & Reddy A. S. N. Transcriptome-wide identification of RNA targets of Arabidopsis serine/arginine protein 45 (SR45) uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell In press, /10.1105/tpc.15.00641 (2015). [DOI] [PMC free article] [PubMed]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas-Madrid D., Nogales-Cadenas R. & Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res 40, W478–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M. & Holm L. POBO, transcription factor binding site verification with bootstrapping. Nucleic Acids Res 32, W222–229 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.