Abstract
Active Hobo/Activator/Tam3 (hAT) transposable elements are rarely found in vertebrates. Previously, goldfish Tgf2 was found to be an autonomously active vertebrate transposon that is efficient at gene-transfer in teleost fish. However, little is known about Tgf2 functional domains required for transposition. To explore this, we first predicted in silico a zinc finger domain in the N-terminus of full length Tgf2 transposase (L-Tgf2TPase). Two truncated recombinant Tgf2 transposases with deletions in the N-terminal zinc finger domain, S1- and S2-Tgf2TPase, were expressed in bacteria from goldfish cDNAs. Both truncated Tgf2TPases lost their DNA-binding ability in vitro, specifically at the ends of Tgf2 transposon than native L-Tgf2TPase. Consequently, S1- and S2-Tgf2TPases mediated gene transfer in the zebrafish genome in vivo at a significantly (p < 0.01) lower efficiency (21%–25%), in comparison with L-Tgf2TPase (56% efficiency). Compared to L-Tgf2TPase, truncated Tgf2TPases catalyzed imprecise excisions with partial deletion of TE ends and/or plasmid backbone insertion/deletion. The gene integration into the zebrafish genome mediated by truncated Tgf2TPases was imperfect, creating incomplete 8-bp target site duplications at the insertion sites. These results indicate that the zinc finger domain in Tgf2 transposase is involved in binding to Tgf2 terminal sequences, and loss of those domains has effects on TE transposition.
Transposable elements (TEs) are discrete DNA segments that are able to move from one locus to another within genomes of host cells using a cut-and-paste mechanism1,2. Their wide distribution among all major branches of life, their diversity, and their intrinsic biological features have made TEs a considerable source of genetic innovations during species evolution3,4. Moreover, transposons may be valuable genomic tools for transgenesis, insertional mutagenesis and DNA delivery vehicles in gene therapy5,6,7,8,9. In eukaryotic genomes, DNA transposons have been classified into approximately 20 superfamilies based on amino acid sequence similarities of their encoded transposases10,11.
The hAT superfamily of transposons, named after the Drosophila element hobo, McClintock’s maize Activator and snapdragon Tam3, is widespread in plants and animals12. All hAT elements share several defining features, including terminal inverted repeats (TIRs) and subterminal repeats (STRs) at each end of the TE, and a gene encoding ~600–800 amino acid transposase that catalyzes DNA cleavage and target integration, with 8 bp target site duplications (TSDs) at both ends of the integration site during transposition13,14,15. In vertebrates, most hAT transposons are inactive, as host cells have developed the mechanism of vertical inactivation to silence and avoid the deleterious effects of active transposons on genome stability16. Thus, only a few active vertebrate elements have been discovered. Tol2, the first autonomous vertebrate hAT transposon, was identified in medaka (Oryzias latipes) and has proven active in a variety of vertebrate cell types17,18. The goldfish (Carassius auratus) Tgf2 transposon is another autonomously active vertebrate hAT transposon19,20. The Tgf2 element is 4,720 bp long21, and the full length Tgf2 transposase is 686 aa long; variant isoforms naturally occur in the goldfish due to the different starting positions of the coding frame19. Although it is capable of mediating gene transfer effectively in different teleost fish19,20,21,22, the functional domains of the Tgf2 transposase are poorly understood at the mechanistic level. The exploration of its role in the transposition process is crucial to understanding its mechanisms for catalyzing excision and transposition.
In this study, the domain architecture of Tgf2 transposase was predicted based on in silico analysis of its amino acid sequence. Two cDNAs were cloned from goldfish embryos, which encoded two truncated Tgf2 transposases with deletions of the N-terminal zinc finger domain. The biological functions of prokaryotically expressed truncated Tgf2TPases were assessed by in vitro DNA binding assay and in vivo transposition activity in zebrafish model. Our results show that the zinc finger domain in Tgf2 transposase is involved in binding to Tgf2 terminal sequences, and mutations to this domain have effects on the transgenic efficiency and integration patterns during the transposition process. Our work may facilitate the development of improved genomic tools, and provide insight into aspects of the transposition process of Tgf2 element.
Materials and Methods
Experimental animals
Ryukin goldfish (Carassius auratus) embryos were provided by the Jujin ornamental fish farm, Shanghai, China. The wild-type Tübingen strain of zebrafish (Danio rerio) maintained in our laboratory was used for mating, spawning, microinjection, and transposition activity analysis in this study. All animal experiments were performed in accordance with the Shanghai Ocean University Committee on the Use and Care of Animals and were approved by the Committee on the Use and Care of Animals at Shanghai Ocean University.
Sequence analysis and modeling of full-length Tgf2 transposase
Functional domains of the goldfish full-length Tgf2 transposase (L-Tgf2TPase, 686 amino acids) were predicted using Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/)23. A three-dimensional model of the L-Tgf2TPase monomer was generated using Phyre2 and protein structures were visualized using PyMol (www.pymol.org), based on a homology model of Hermes transposase protein24. Nuclear localization of L-Tgf2TPase was predicted on cNLS mapper (http://nls-mapper.iab.keio.ac.jp)25. Alignment of conserved amino acid sequences of DDE-based catalytic domains of hAT transposases from different species was performed with the Clustal X 1.81 program26.
Plasmid construction, prokaryotic expression and purification
Three cDNAs (2061bp, 1734 bp and 1692 bp) encoding goldfish wild type Tgf2 transposases of L- (1–686 aa.), S1- (110–686 aa.) and S2-Tgf2TPase (124–686 aa.) were previously isolated from Ryukin goldfish embryos19. In addition to the 5′-truncated region, cDNAs of both S1- and S2-Tgf2TPases were identical to those in L-Tgf2TPase. The recombinant vector pET-28a(+)-L-Tgf2TPase was constructed for L-Tgf2TPase, and was prokaryotically expressed and purified in our previous publication27. The control plasmid pET-28a(+)-L-Tgf2TPaseD228N, E648Q, mutated in the catalytic domain of Tgf2 transposon, was constructed on the basis of the plasmid pET-28a(+)-L-Tgf2TPase, using the QuikChange Lightning multi site-directed mutagenesis kit from Stratagene. The primer set for aspartic acid (D)-to-asparagine (N) mutation at position 228 was 5′-GCAACCACAACGAATTGTTGGACTGCACGTAGAAAGTCATTC-3′ and 5′-CCAACAATTCGTTGTGGTTGCAATCCATTCAACTTCACTCAT-3′. The primer set for glutamic acid (E)-to-glutamine (Q) mutation at position 648 was 5′- GGCTGCCTGTCAGAGGCTTTTCAGCACTGCAGGATTGCTTTT -3′ and 5′- AAAAGCCTCTGACAGGCAGCCGATGCGGGAAGAGGTGTATTA -3′. All reactions were performed according to the manufacturers specifications, and positive clones were examined by PCR and direct sequencing. Using S1-Tgf2TPase cDNA as template, the coding region for S1-Tgf2TPase was amplified by PCR using Pfu DNA polymerase (Stratagene, La Jolla, CA, USA) using the primer set 5′-CGCGGATCCATGAAACGTAAAATTGGCA-3′ and 5′-CCGCTCGAGTTATTCAAAGTTATAAAAACG-3′. Using S2-Tgf2TPase cDNA as template, the coding region for S2-Tgf2TPase was amplified by the primer set 5′-CGCGGATCCATGCATCCGAACTATCT-3′ and 5′-CCGCTCGAGTTATTCAAAGTTATAAAAACG-3′. These coding regions were both cloned into pMD-19T vector (Takara, Dalian, China). For expression of the truncated recombinant Tgf2TPases in E. coli, the coding regions for S1- and S2-Tgf2TPase were subcloned into the pET-28a(+) vector (Merck, Shanghai, China).
Recombinant vectors were then used to transform Rosetta1 (DE3) competent cells (Merck, Shanghai, China). E. coli cells containing recombinant plasmid harboring S1- or S2-Tgf2TPase were initially induced at the early log phase of culture (OD600 = 0.3–0.4) with 0.8 mM IPTG for 6 h at a low-temperature (22 °C) as previously described27. Recombinant proteins were purified with a Ni2+-affinity column in a FPLC AKTA Purifier system (GE Healthcare, Piscataway, USA). Recombinant TPases were identified using ABI 4800 Plus MALDI TOF/TOF™ (Applied Biosystems, Foster City, USA) and MS/MS ion searches using Mascot (Matrix Science Ltd, London, UK).
DNA-binding activity assay
The DNA-binding activities of truncated recombinant S1-Tgf2TPase, S2-Tgf2TPase and L-Tgf2TPaseD228N,E648Q, and previously purified full-length L-Tgf2TPase27 were evaluated by size exclusion chromatography methods used for other transposons13,24,28,29. Since transposases are proposed to bind transposon terminal regions30,31, a 50 bp DNA probe containing TIR and STR sequences was designed for a binding assay based on at the left end of Tgf2 (GenBank Accession No. HM146132, 4720bp), named as L50 (5′- CAGAGGTGTAAAAGTACTTAAGTAATTTTACTTGATTACTGTACTTAAGT-3′). Oligonucleotides were synthesized and PAGE-purified (Sangon Ltd, Shanghai China), and annealed with their complementary motifs. DNA-binding activity was determined as previously described13,24,28,29, with certain modifications. Briefly, double stranded DNAs were mixed with 20 mM Tgf2TPase at a 1:1 molar ratio of protein: DNA in a buffer containing 0.5 M NaCl. The mixture was then dialyzed into buffer (pH 7.5) containing 20 mM Tris (pH 7.5), 0.2 M NaCl and 5 mM DTT overnight. Dialyzed fluids (50 μL) were applied to a Superdex 200 column (GE Healthcare, Piscataway, USA) equilibrated with the same buffer (pH 7.5) and then eluted at 4 °C at a rate of 50 μL/min. DNA-binding activity was assessed by the formation of stable complexes under size exclusion chromatography conditions. A random 50-mer double-stranded DNA C50 (5′- CCTGTACTCACGGCATTGCCATTGGCTCGTCTACGCTAGCTCCGCGCTGA-3′) was used as a control.
Microinjection
The donor plasmid of pTgf2-EF1α-EGFP, containing 220 bp and 185 bp of the left and right ends of the goldfish Tgf2 transposon and driven by the Xenopus EF1a promoter was constructed as described previously19. A mixture of 50 pg donor plasmid (pTgf2-EF1α-EGFP) was injected alone, or with 50 pg recombinant S1-, S2- or L-Tgf2TPase into fertilized zebrafish eggs at the one- to two-cell stage (~1 nl/embryo). After injection, embryos were placed in embryo rearing medium and maintained at room temperature. EGFP fluorescence in embryos was analyzed at 12, 24 and 96 hours post fertilization (hpf) using a Nikon SMZ 1500 fluorescence microscope.
Transposition rate and insertion site analysis
Total genomic DNA was isolated from zebrafish tail fin clips (0.1 to 0.2 g,32). The primers for PCR analysis of transposition or integration rate of EGFP were: EGFP-f, 5′-ACCCTCGTGACCACCCTGAC-3′; EGFP-r, 5′-GCTTCTCGTTGGGGTCTTTGCTC-3′. PCR, cloning and sequencing were conducted as previously described19,27.
The flanking sequences of the transposon insertion sites were analyzed using the GenomeWalkerTM Universal Kit (Clontech, California, USA), as previously described19,27. For splinkerette PCR, 25 μg genomic DNA was digested for 12–16 h at 37 °C with 80 U of Stu I and EcoR V in a 100 μl reaction volume, was purified by ethanol precipitation, and then 4 μl of the digestion mix was ligated with the splinkerette adaptor overnight at 16 °C. The linker ligation was used as a template for two rounds of PCR to amplify the transposon/genome junction. The nested primers for the 5′ flanking sequences were 5′-AACAGCTCCTCGCCCTTGCTCACCAT-3′ and 5′-ACCGTCGCTGGCTTTTGTGTTACACG-3′. The nested primers for the 3′ flanking sequences were 5′- TCGGCATGGACGAGCTGTACAAGTAA-3′ and 5′-CCTCTACAAATGTGGTATGGCTGATTA-3′. The amplified fragments were cloned into the pMD19-T vector (TaKaRa, Dalian, China), transformed into DH5α E. coli cells and positive clones were examined by PCR and direct sequencing.
Statistics
Values are expressed as mean ± S.E. Differences among groups were analyzed by one-way ANOVA followed by Fisher’s Post Hoc tests or unpaired t-test. Significance was defined as p < 0.01.
Results
Molecular architecture of the goldfish full-length Tgf2 transposase
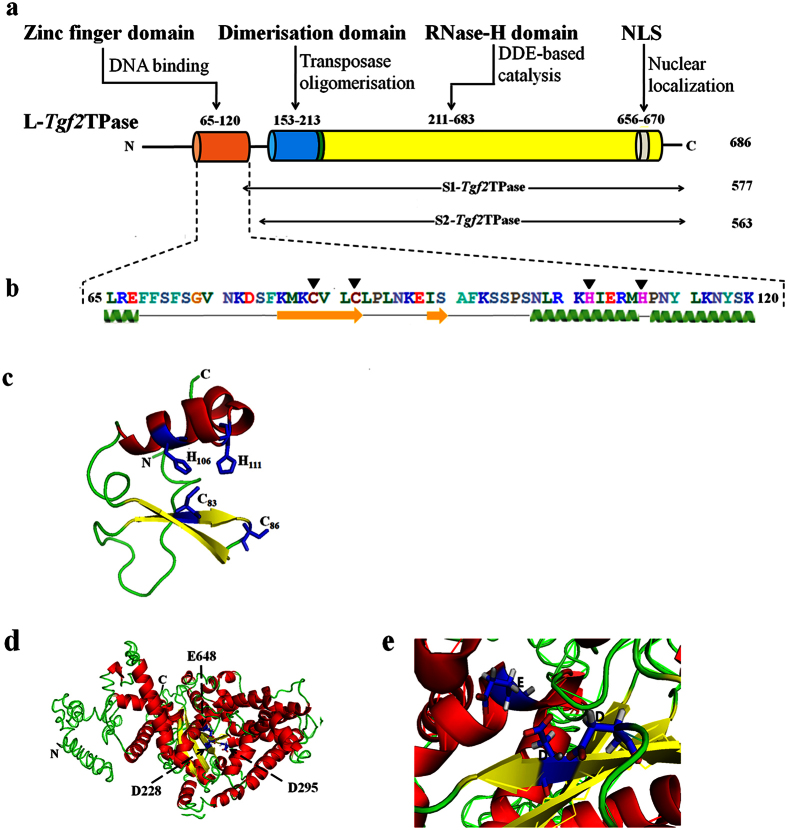
Amino acid sequence analysis on Phyre2 suggested that the full length Tgf2 transposase (L-Tgf2TPase) consisted of four functional domains (Fig. 1a). First is an N-terminal BED zinc finger domain (Cx2Cx19Hx4H, 65–120 aa) involved in DNA binding, two zinc binding residues (C83 and C86) comprise a zinc knuckle at the end of the β-hairpin, and two other residues (H106 and H111) are at the C-terminal end of the α-helix, which is composed of a β-hairpin followed by an α-helix that forms a left-handed ββα unit (Fig. 1b,c). The second presumptive domain identified is a dimerization domain defined by amino acids 153–213 (Fig. 1a), presumably involved in the formation of oligomers, as well as in DNA binding. A C-terminus RNase-H domain comprises amino acids 211 to 683 (Fig. 1a). This is presumably the core catalytic domain for DNA excision and transposition15,33,34. A 3D model of Tgf2 transposase (Fig. 1d) was constructed with 95% accuracy based on the housefly hAT Hermes transposase, the only recently available crystal structure for a hAT transposase24.
Figure 1. Molecular architecture of Tgf2 transposase.
(a) The domains of full length Tgf2 transposase (L-Tgf2TPase) (686 aa). (b) The secondary structure of the N-terminal zinc finger domain. Two conserved cysteine (C) and two conserved histidine (H) residues are indicated with dark triangles. The α-helices (green) and β-strands (yellow) are also shown. (c) The three-dimensional model of the N-terminal zinc finger domain, which consists of an α-helix (red), two antiparallel β-sheets (yellow), and a motif usually used for DNA-binding (green). Two histidine (H106, H111) and two cysteine (C83, C86) residues (blue) form a cave that can bind a zinc ion. (d) The homology model of the full length Tgf2 transposase. α-helices and β-strands are colored in red and yellow respectively. Residues of the DDE (D228, D295 and E648) active site triads are labeled. (e) Close-up view of the DDE active site.
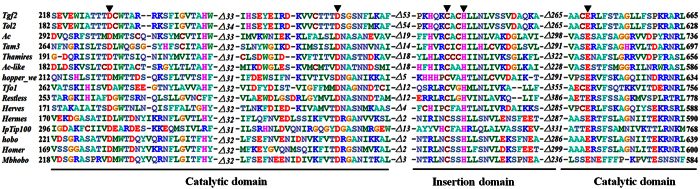
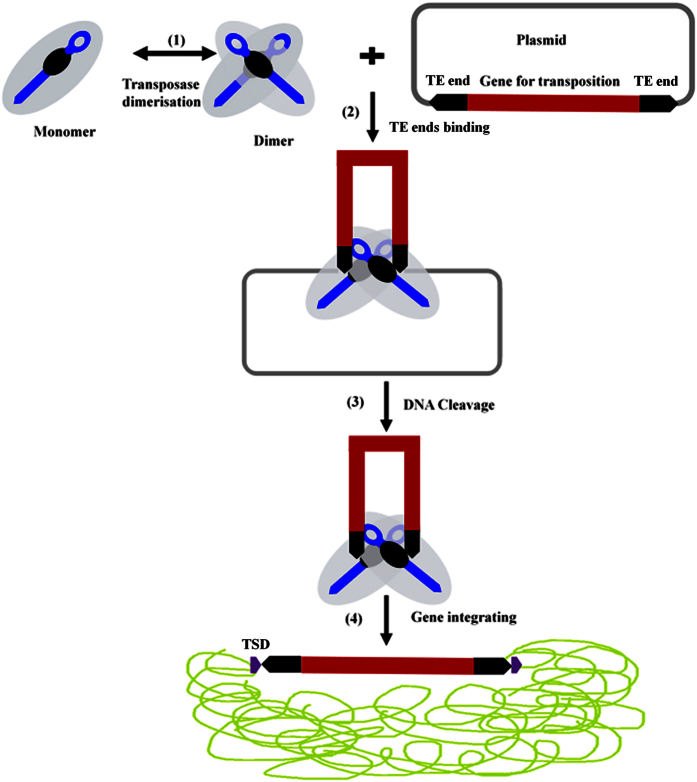
Using sequence alignment with other hAT transposases10,12,15,17,34, three conserved amino acids residues (DDE) were identified in the RNase-H catalytic domain of Tgf2 transposase (Fig. 2). Residues of the DDE (D228, D295 and E648) are extremely close in their spatial distribution (Fig. 1e). A CX2H motif within the RNase-H catalytic domain of Tgf2 transposase is also identified (Fig. 2), which functions as insertion domain for the correct positioning of the final E648 residue of the catalytic triad in the active site24,34. These conserved residues or motifs were exploited as phylogenetic characteristics to infer evolutionary relationships among hAT transposases, indicating their importance for the functioning of the enzyme15,34,35. Finally, a monopartite nuclear localization signal (NLS, 656–670 aa.) was found at the C-terminus (Fig. 1a). Based on the domain organization of the Tgf2 transposase as described above, and data on regulation from other class II transposable elements28,31,36, a Tgf2 transposition model was proposed (Fig. 3).
Figure 2. Alignment of conserved amino acid sequences of the C-terminus RNase-H domain of 15 hAT transposases.
Distances between the conserved blocks are indicated by the number of amino acid residues. Conserved DDE residues and CX2H motif are marked with dark arrows. Accession numbers: Tgf2 (Carassius auratus): AFC96942; Tol2 (Oryzias latipes): BAA87039; Ac (Zea mays): CAA29005; Tam3 (Antirrhinum majus): CAA38906; Thamires (Saintpaulia hybrid cultivar): BAJ23914; Ac-like (Amaranthus palmeri): AGH20188; hopper_we (Bactrocera dorsalis): AAL93203; Tfo1 (Fusarium oxysporum): BAA32244; Restless (Tolypocladium inflatum): CAA93759; Herves (Anopheles gambiae): AAS21248; Hermes (Musca domestica): AAC37217; IpTip100 (Ipomoea purpurea): BAA36225; hobo (Drosophila melanogaster): AAA51465; Homer (Bactrocera tryoni): AAD03082; Mbhobo (Mamestra brassicae): AAA51465.
Figure 3. Proposed model for Tgf2 transposition.
The cut-and-paste process contains four main stages: (1) pairing of transposases by dimerization or oligomerization, (2) DNA-specific binding of the transposases to the TE ends flanking the transposon, (3) cleavage of DNA strands at the ITR ends of the transposon, and (4) capture of a new locus and integration of target DNA, purple block arrows indicated target site duplications (TSDs) in the host cell.
N-terminal truncated Tgf2 transposases lose their DNA-binding activity
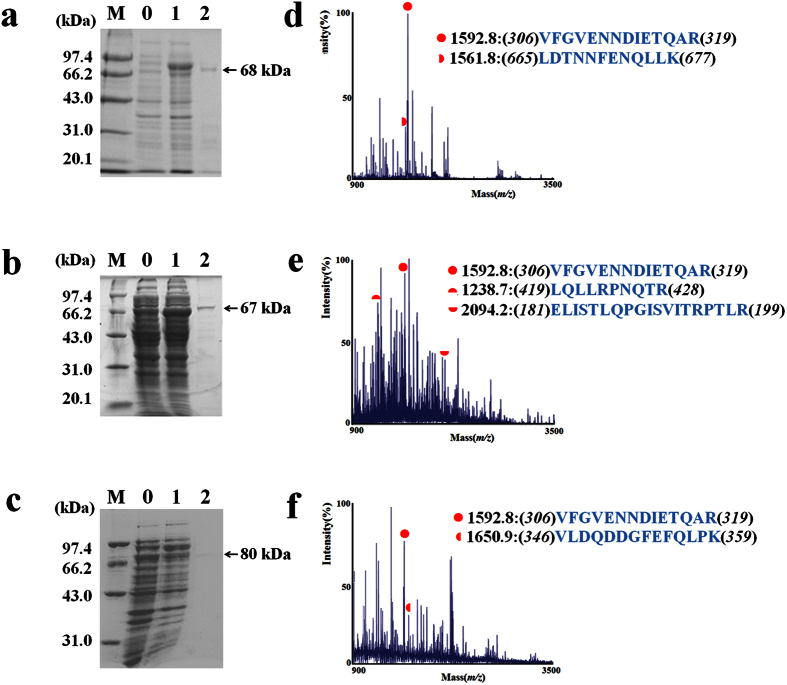
To investigate the function of the N-terminal zinc finger domain in DNA binding, two truncated Tgf2 transposase proteins, designated S1- and S2-Tgf2TPase, were prokaryotically synthesized in E. coli. The S1-Tgf2TPase was characterized by deletion of the N-terminal zinc finger domain, while S2-Tgf2TPase included an additional deletion of part of the linker region between the zinc finger domain and the dimerization domain (Fig. 1a). The double mutation L-Tgf2TPaseD228N, E648Q, with an intact N-terminal zinc finger domain, was used as control. The recombinant S1- (~68 kDa), S2- (~67 kDa) and L-Tgf2TPaseD228N, E648Q (~80 kDa) proteins were successfully expressed in a soluble form, purified with Ni2+-affinity chromatography (Fig. 4a–c), and confirmed to be Tgf2TPase components by mass spectrometry analysis following trypsin digestion (Fig. 4d–f). The recombinant L-Tgf2TPase was previously obtained27.
Figure 4. Purification and identification of recombinant Tgf2TPases.
(a–c) SDS-PAGE analysis of S1-Tgf2TPase (a) and S2-Tgf2TPase (b) with N-terminal deletions, or of mutated L-Tgf2TPaseD228N, E648Q (c) expression from E. coli strain Rosetta 1 (DE3) by vector pET-28a(+). M, standard molecular marker. 0, without IPTG. 1, with IPTG. 2, purification from eluted peak of Ni2+-affinity column. (d–f) Mass spectrometry of S1-Tgf2TPase (d), S2-Tgf2TPase (e) or L-Tgf2TPaseD228N, E648Q (f) collected from eluted peak of Ni2+-affinity column. The confirmed fragments of Tgf2TPase are indicated in red, with the corresponding peptide sequences listed.
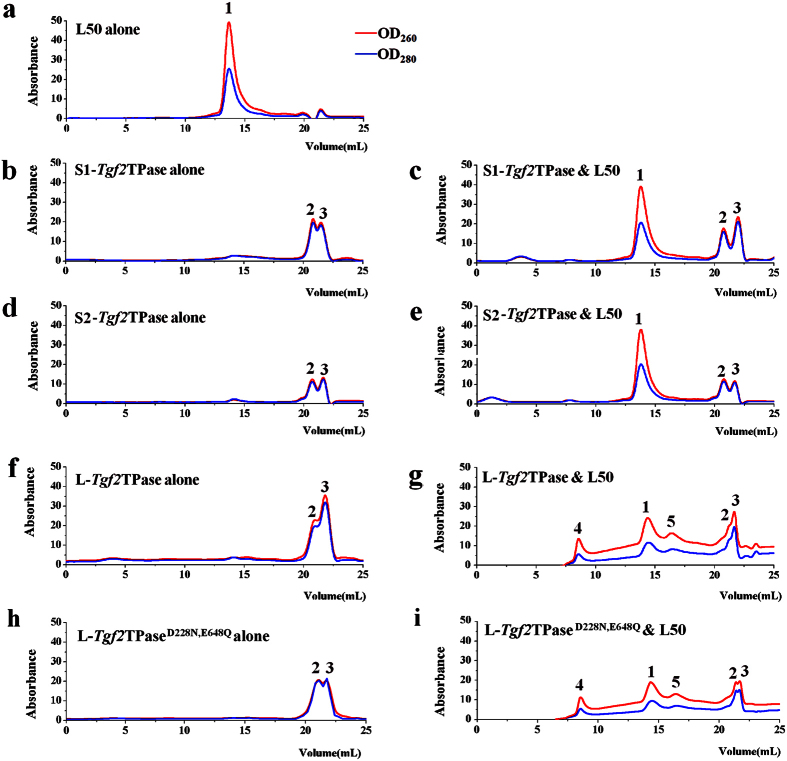
For evaluation of DNA-binding activity in vitro, the size exclusion chromatography elution profiles of Tgf2TPases were investigated. As shown in Fig. 5, S1-, S2-, L-Tgf2TPase and L-Tgf2TPaseD228N, E648Q displayed characteristic protein profiles of monomer (peak 3) and dimer (peak 2) prior to DNA binding (Fig. 5b,d,f,h), with OD260 and OD280 being approximately equal. In contrast, the L50, double-stranded DNA probe exhibited a nucleic acid profile, with OD260 higher than OD280 (peak 1 in Fig. 5a). When S1- or S2-Tgf2TPase and L50 were mixed at a 1:1 molar ratio of protein and eluted, no other complex peaks were found except for peaks 1, 2 and 3 (Fig. 5c,e), suggesting that S1- and S2-Tgf2TPase did not bind well with L50 to form DNA-Tgf2TPase complexes at the concentration ratio tested.
Figure 5. DNA-binding activity of S1-, S2- or L-Tgf2TPases analyzed by size exclusion chromatography.
(a) Elution profiles of the L50 double-stranded DNA probe alone. (b,c) Elution profiles of S1-Tgf2TPase alone (b) or a mixture of S1-Tgf2TPase and L50 (c). (d,e) Elution profiles of S2-Tgf2TPase alone (d) or a mixture of S2-Tgf2TPase and L50 (e). (f,g) Elution profiles of L-Tgf2TPase alone (f) or a mixture of L-Tgf2TPase and L50 (g). (h,i) Elution profiles of L-Tgf2TPaseD228N, E648Q alone (h) or a mixture of L-Tgf2TPaseD228N, E648Q and L50 (i). Based on elution times, peak 1 corresponds to the L50 probe and peaks 2 and 3 represent Tgf2TPase dimers and monomers, respectively. Peaks 4 and 5 are new multimeric complexes formed from presumptive binding of the L50 probe and the L-Tgf2TPase.
However, there was a marked change in the peak characteristics when the L-Tgf2TPase and L50 mixture was eluted (Fig. 5g). Two new peaks (4 and 5) were seen before the L-Tgf2TPase protein peak, accompanied by a decrease in the L50 probe peak 1 and L-Tgf2TPase peaks 2 and 3 (Fig. 5g), when compared with peak 1 in L50 alone (Fig. 5a) and peaks 2 and 3 in L-Tgf2TPase alone (Fig. 5f). Due to the formation of L-Tgf2TPase-DNA complexes, the baseline for the 260 trace was significantly raised above the 280 profile in Fig. 5g. Moreover, peak 4 eluted faster than the peak 5 and the DNA probe peak, which implied that L-Tgf2TPase likely interacted with the DNA probe in the form of oligomerization containing more than two protein molecules (Fig. 5g). Since the L-Tgf2TPaseD228N, E648Q recombinant protein has an intact N-terminal domain, like L-Tgf2TPase, peaks 4 and 5 were also found when L-Tgf2TPaseD228N, E648Q and L50 mixture was eluted (Fig. 5i). In the negative control mixture of S1-, S2-, L-Tgf2TPase or L-Tgf2TPaseD228N, E648Q with random 50-mer double-stranded DNA C50, the above mentioned changes were not seen (Fig. S1). These results suggest that the N-terminal zinc finger domain in Tgf2 transposase is involved in binding of the transposase to Tgf2 terminal sequences.
Truncated Tgf2 transposases have decreased transgenic efficiency
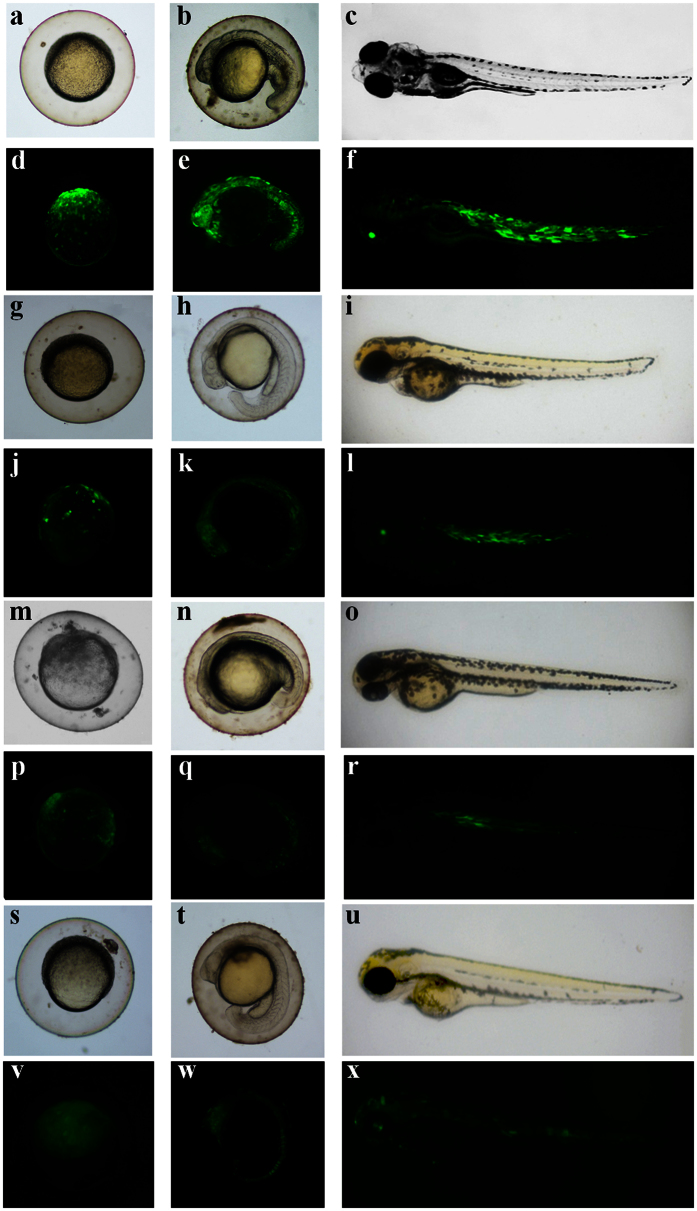
To determine if the N-terminal zinc finger domain in Tgf2 transposase has any effect on the transgenic efficiency during DNA transposition, we performed microinjection of Tgf2TPases into zebrafish embryos at the 1–2 cell stage. When 50 pg pTgf2-EF1α-EGFP was coinjected with 50 pg recombinant L-Tgf2TPase protein, an average of 68% of embryos showed strong and almost ubiquitous expression of EGFP (Table 1, Fig. 6d–f). EGFP fluorescence rates in embryos coinjected with recombinant S1- and S2-Tgf2TPase were reduced to 43% and 29% respectively (Table 1), with a weak expression of EGFP (Fig. 6j–l, p-r). In control embryos injected with donor plasmid alone or donor plasmid coinjected with recombinant L-Tgf2TPaseD228N, E648Q, 24% and 22% of embryos showed mosaic EGFP expression (Table 1); the fluorescence in most of embryos should result from a weak expression of EGFP from the donor plasmid (Fig. 6v–x). PCR analysis of the transposition rate of EGFP in 3 month old adult zebrafish was performed as previously described19,27. The integration rate of EGFP when coinjected with L-Tgf2TPase reached 56% (Table 1). In comparison, significantly decreased integration rates were detected in zebrafish coinjected with S1- (21%) and S2-Tgf2TPases (25%, p < 0.01, Table 1). The integration rate of the EGFP sequence in control embryos injected with donor plasmid alone or coinjected with recombinant L-Tgf2TPaseD228N, E648Q was 8% and 7% respectively (Table 1).
Table 1. Transposition efficiencies of N-terminal truncated Tgf2TPases in zebrafish.
| Injection batches | No. of embryos | No. of adults | ||||
|---|---|---|---|---|---|---|
| Examined | EGFP- expressed | Expressed rate (%) | Survived | EGFP- insertion | Insertion rate (%) | |
| pTgf2-EF1α-EGFP + S1-Tgf2TPase | 88 | 32 | 36 | 30 | 7 | 23 |
| 130 | 70 | 58 | 64 | 10 | 16 | |
| 46 | 19 | 41 | 16 | 4 | 25 | |
| Average | – | – | 43 | – | – | 21b |
| pTgf2-EF1α-EGFP + S2-Tgf2TPase | 70 | 17 | 24 | 17 | 4 | 23 |
| 110 | 37 | 34 | 35 | 7 | 20 | |
| 45 | 13 | 29 | 13 | 4 | 31 | |
| Average | – | – | 29 | – | – | 25b |
| pTgf2-EF1α-EGFP + L-Tgf2TPase | 90 | 60 | 67 | 55 | 28 | 51 |
| 90 | 62 | 69 | 60 | 36 | 60 | |
| 56 | 38 | 68 | 34 | 19 | 56 | |
| Average | – | – | 68 | – | – | 56c |
| pTgf2-EF1α-EGFP + L-Tgf2TPaseD228N, E648Q | 85 | 20 | 23 | 18 | 2 | 11 |
| 93 | 21 | 23 | 19 | 1 | 5 | |
| 75 | 16 | 21 | 14 | 1 | 7 | |
| Average | – | – | 22 | – | – | 7a |
| Control | 86 | 21 | 24 | 18 | 2 | 11 |
| 95 | 25 | 26 | 22 | 1 | 4 | |
| 86 | 20 | 23 | 18 | 2 | 11 | |
| Average | – | – | 24 | – | – | 8a |
Control is injected pTgf2-EF1α-EGFP plasmid only. Different letters is significant p < 0.01.
Figure 6. Microscopy images of EGFP expression in zebrafish embryos after microinjection of S1-, S2- or L-Tgf2TPase with donor construct pTgf2-EF1α-EGFP at the 1-2-cell stage.
Light (a–c, g–i, m–o, s–u) and fluorescence (d–f, j–l, p–r, v–x) microscopy images of zebrafish embryos were shown at 12 hpf (a, d, g, j, m, p, s, and v), 24 hpf (b, c, h, k, n, q, t, and w) and 96 hpf (c, f, i, l, o, r, u, and x). Panels a–f are embryos treated with L-Tgf2TPase, panels g-l were treated with S1-Tgf2TPase, panels m-r were treated with S2-Tgf2TPase and panels s-x injected with donor plasmid pTgf2-EF1α-EGFP alone as controls.
Truncated Tgf2 transposases exhibit altered TE excision and integration
We further cloned the junctions of the integrated Tgf2 element and the surrounding genomic DNA using inverse PCR. All 83, 21, 15, 4 and 5 EGFP-transgenic zebrafish adults that survived from each transgenic group (L-, S1-, S2-Tgf2TPase, L-Tgf2TPaseD228N, E648Q and control) respectively were examined (Table 1). A total of 143 insertion sites were identified from 83 zebrafish coinjected with L-Tgf2TPase (Table 2). There were 1 to 3 genomic integration sites in the genome of the zebrafish, and the average copy number was 1.7 (143/83). Most of L-Tgf2TPase injected fish (95%, Table 2) demonstrated intact TE end integration, indicating accurate excision and insertion during transposition and creation of complete 8 bp TSD signatures adjacent to both ends of Tgf2 at the insertion sites (Fig. 7b). Among the 83 EGFP-transgenic positive zebrafish, 78 individuals (94%) had accurate insertions (Table 2). The remaining 6% (5/83) had partial deletion of transposon ends and/or plasmid backbone insertion/deletion, as well as incomplete 8 bp TSDs (Table 2; Fig. 7c), indicating imprecise excisions and insertions have occurred during transposition. In contrast, only 14% of individuals (3/21) with the S1-Tgf2TPase injections had precise transposition, while 67% of individuals (14/21) demonstrated imprecise integration, similar to the pattern in Fig. 7b,c. The remaining 19% (4/21) did not have any detectable insertion, which may be due to the absence of the primer binding region during splinkerette PCR. In all the insertion sites detected, the precise insertion rate was only 21% (4/21, Table 2). Consistently, only 13% (2/15) of individuals with the S2-Tgf2TPase injections exhibited precise genomic integration, while 67% (10/15) of individuals demonstrated imprecise insertion, similar to the pattern in Fig. 7b,c; the remaining 20% (3/15) had no detectable insertion. In all insertion sites detected with S2-Tgf2TPase injections, the precise insertion rate was only 20% (3/15, Table 2). The flanking sequences of the transposon insertion sites in 4 individuals coinjected with recombinant L-Tgf2TPaseD228N, E648Q were not detected by splinkerette PCR. Moreover, only 1 of 5 individuals from control embryos injected with donor plasmid alone had imprecise insertion (Table 2).
Table 2. Insertion accuracy and germline transmission of N-terminal truncated Tgf2TPases in EGFP-transgenic adult zebrafish at 3-month age.
| Injection batches | No. of fish examineda | No. of fish with precise insertion sites | No. of fish with imprecise insertion sites | No. of fish with undetectable insertion sites | Total precise insertion sites | Total imprecise insertion sites | Precise insertion sites/total insertion sites (%) |
|---|---|---|---|---|---|---|---|
| pTgf2-EF1α-EGFP + S1-Tgf2TPase | 21 | 3 (2) | 14 (4) | 4 (1) | 4 | 15 | 21 |
| pTgf2-EF1α-EGFP + S2-Tgf2TPase | 15 | 2 (2) | 10 (4) | 3 (0) | 3 | 12 | 20 |
| pTgf2-EF1α-EGFP + L-Tgf2TPase | 83 | 78 (71) | 5 (2) | 0 | 136 | 7 | 95** |
| pTgf2-EF1α-EGFP + L-Tgf2TPaseD228N, E648Q | 4 | 0 | 0 | 4 (1) | 0 | 0 | 0 |
| Control | 5 | 0 | 1 (1) | 4 (0) | 0 | 1 | 0 |
aThe EGFP-transgenic zebrafish from Table 1 were examined by splinkerette PCR. The number of fish demonstrating germline transmission is indicated in parentheses. Control was injected with pTgf2-EF1α-EGFP plasmid only. **p < 0.01 versus control.
Figure 7. Exogenous Tgf2 end regions and the surrounding insertion site sequences in the genome of 3 month old adult zebrafish.
(a) Sequences of Tgf2 end regions in the donor construct pTgf2-EF1α-EGFP. (b) Insertion sequences from 5 of 78 individuals listed in Table 2, with precise insertion sites after coinjection with pTgf2-EF1α-EGFP and L-Tgf2TPase. (c) Insertion sequences from all 5 samples in Table 2 with imprecise insertion sites after coinjection with pTgf2-EF1α-EGFP and L-Tgf2TPase. Precise insertion copies are shown with gray background. Lower case letters indicate the Tgf2 end sequences. The 8 bp direct repeat signature of the target DNA associated with transposition is underscored. The surrounding sequences of the insertion sites are in italic. Triangles and inverted triangles represent deletions and insertions, respectively.
EGFP-transgenic fish were then raised to maturity and crossed with wild type zebrafish, and EGFP fluorescence expression in F1 embryos was examined at 24 hpf, to determine the existence of germline transmission37. Among genomic precise integration groups of L-, S1-, and S2-Tgf2TPase, 91% (71/78), 67% (2/3), and 100% (2/2) founders were able to transmit EGFP to their F1 embryos (Table 2); EGFP fluorescence expression in F1 embryo ranged from 20% to 78% (data not shown). Although the ratio of precise transposition in S1- or S2-Tgf2TPase populations was significantly reduced, EGFP-transgenic individuals with precise integration could efficiently transmit EGFP to their offspring. Taken together, our results indicate that truncated Tgf2 transposases not only severely impair their in vivo transgenic efficiency, but also negatively impact on precision of DNA transposition.
Discussion
In this study, the domain architecture of full-length Tgf2 transposase was predicted (from N- to C-terminus) based on bioinformatics analysis. Four domains were identified: (1) an N-terminal zinc finger domain, that presumably coordinates Zn2+ through a conserved Cys2-His2 motif, and participates in binding to DNA38; (2) a dimerization domain13,24,29,39; (3) a C-terminus RNase-H domain that is a critical domain for DNA cleavage and integration15,33; and (4) a monopartite nuclear localization signal (NLS) found in the C-terminus25. These functional domains of Tgf2 transposase are consistent with the analysis of conserved amino acids from hAT transposases34. Despite very limited sequence similarity among these hAT transposases, it seems likely that they share common mechanistic and structural features24,34. Since only a few hAT transposases have been studied in detail24, our successful purification of N-terminal truncated recombinant transposases makes it possible to experimentally verify the functional domains of the Tgf2 transposase.
The BED zinc finger domain was initially described as Cx2CxnHx3–5(H/C) and have been proposed to bind DNA and to coordinate Zn2+ through a conserved CCHH or CCHC motif38. The zinc finger domain at residues 65–120 of the Tgf2 transposase has the structure Cx2Cx19Hx4H. L-Tgf2TPase and L-Tgf2TPaseD228N, E648Q with intact zinc finger domains can bind to the L50 DNA probe, which consists of the 11-bp terminal inverted repeat (TIR) and five subterminal repeats (STRs) from left end of the Tgf2 element (Fig. S2). In contrast, S1- and S2-Tgf2TPase do not bind well to probe, as suggested by size exclusion chromatography analysis. Accumulating data indicate that hAT transposases recognize their transposon tips in a bipartite manner, with weaker transposase binding to the TIRs and stronger binding by an N-terminal domain to these STRs24,34,41,43. For the Ac transposase, the zinc finger domain (Cx4Cx17Hx4H) is split into two subdomains, in which the C-terminal subdomain (Cx4C) appeared essential for binding to both sequences, while the N-terminal half (Hx4H) appears to bind to the TIRs but not to the STRs40,41. These data suggest that the zinc finger of hAT transposases is capable of binding to the TIRs and the STRs within the TE ends. Considering that S1- and S2-Tgf2TPases are distinguishable from L-Tgf2TPase only in the N-terminal region, this loss of DNA binding activity seen is likely due to the lack of the zinc finger domain.
The repeated subterminal repeats are haphazardly present within both ends of hAT transposons and are a defining feature of this superfamily24,41,42. The Tgf2 elements are found to include 17 STR copies in the L end and 18 copies in the R end (Fig. S2). The zinc finger domain interacts with the outermost subterminal repeat on each end and this is important for both cleavage and strand transfer41,43. In the present study, L-Tgf2TPase-mediated gene transfer in the adult zebrafish genome in vivo occurred at a significantly higher efficiency than that mediated by zinc finger domain truncated Tgf2TPases (p < 0.01). In agreement with our results, the N-terminal deletion IS911, Himar1 and Sleeping Beauty transposases containing the catalytic domain also showed downregulation of transposition activity44,45,46. In comparison with truncated Tgf2TPase, L-Tgf2TPase could accurately catalyze gene excision at TE ends and integration at zebrafish genome with complete TSD signatures adjacent to both ends of Tgf2 at the insertion sites. These results indicate that both transposition efficiency and accuracy of the Tgf2 system depends on zinc finger domain. The zinc finger domain interaction with both TIRs and STRs within the transposon ends could improve insertion fidelity, but the underlying mechanism is still an area of active investigation24,34.
During TE transposition, transposases tend to form transpososomes that contain multiple transposase monomers24,39. The hAT superfamily transposase Hermes can generate an unusually ring-shaped octamer in vivo, on the basis of crystal structure and negative staining electron microscopy analysis13,24. The Tc1/mariner Mos1 and Sleeping Beauty transposases can also generate oligomers containing more than two molecules5,28. Due to the presence of a dimerization domain between aa 153 and 213, our data indicate that the S1-, S2- and L-Tgf2TPase can form dimers in solution prior to DNA binding, and sequential multimerization occurs concomitant with L-Tgf2TPase-DNA complex formation. The multimeric complexes contain multiple specific DNA-binding domains. The avidity provided by multiple sites of interaction could allow a transposase to locate its transposon ends amidst a sea of chromosomal DNA24. In addition to mediating the formation of dimers, the other function of dimerization domain in hAT transposases is to perform a weak DNA-binding34. Side chains from three amino acids (R107, F109, and S110) within dimerization domain interact with the sugar phosphate backbone of the Hermes L TIR between bp 6 and 824. Moreover, the α-helices from insertion domain also bind to the Hermes transposon DNA24,34. These alternative DNA-binding motifs within the C-terminal dimerization and insertion domains are also conserved in Tgf2 transposase. Since the alternative binding is relatively weak, the binding peaks are undetectable when the truncated Tgf2TPase and L50 mixture was eluted in our in vitro DNA-binding activity assay. The additional DNA binding sequences help to explain why truncated Tgf2TPases lost the zinc finger domain still have a moderate TE integration rate, although integration occurs imprecisely.
In summary, we predicted the functional domains in the full length goldfish Tgf2 transposase by in silico analysis. The N-terminal zinc finger domain of Tgf2 transposase was found to be responsible for the DNA-binding activity towards specific Tgf2 end sequences, which had consequent effects on the gene-transfer efficiency. This DNA-binding domain is essential for mediating accurate excision and integration of the Tgf2 element during in vivo transposition. The EGFP transgenic individuals with precise integration could efficiently transmit EGFP to their offspring, indicating germline transmission have occurred during transposition. Furthermore, our results demonstrate that D228N and E648Q mutations lead to knock out of Tgf2 transposase function, which gives the experimental support that the proposed DDE motif forms the active center in the Tgf2 transposase. Our efforts in elucidating the structure of Tgf2 transposase provide insights into the transposition process and suggest application to further scientific investigations.
Additional Information
How to cite this article: Jiang, X.-Y. et al. The N-terminal zinc finger domain of Tgf2 transposase contributes to DNA binding and to transposition activity. Sci. Rep. 6, 27101; doi: 10.1038/srep27101 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation of China (31272633, 31201760, 31572220); the National High Technology Research and Development Program of China (863 Program) (2011AA100403); and the Shanghai University Knowledge Service Platform (ZF1206).
Footnotes
The authors declare no competing financial interests.
Author Contributions X.J. and S.Z. designed the study; F.H., X.S., H.X. and X.J. performed the experiments; X.D. contributed sequence analysis; X.J. and S.Z. wrote the manuscript. All authors reviewed the manuscript.
References
- Craig N. L. et al. Mobile DNA II. Washington, DC: ASM Press, pp. 12–23 (2002). [Google Scholar]
- Benjak A. et al. Genome-wide analysis of the “cut-and-paste” transposons of grapevine. PloS One 3, e3107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. & Pritham E. J. DNA transposons and the evolution of eukaryotic genomes. Ann. Rev. Genet. 41, 331–368 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. J. Eukaryotic transposable elements and genome evolution. Trends Genet. 5, 103–107 (1989). [DOI] [PubMed] [Google Scholar]
- Ivics Z. et al. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish and its transposition in human cells. Cell 91, 501–510 (1997). [DOI] [PubMed] [Google Scholar]
- Abe G. et al. Tol2-mediated transgenesis, gene trapping, enhancer trapping, and the Gal4-UAS system. Methods Cell Biol. 104, 23–49 (2011). [DOI] [PubMed] [Google Scholar]
- Aronovich E. L. et al. The Sleeping Beauty transposon system: a non-viral vector for gene therapy. Hum. Mol. Genet. 20, R14–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates L. et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753–761 (2009). [DOI] [PubMed] [Google Scholar]
- Carlson C. M. & Largaespada D. A. Insertional mutagenesis in mice: new perspectives and tools. Nat. Rev. Genet. 6, 568–580 (2005). [DOI] [PubMed] [Google Scholar]
- Arensburger P. et al. Phylogenetic and functional characterization of the hAT transposon superfamily. Genetics 188, 45–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S. et al. Repeat structure of the catfish genome: a genomic and transcriptomic assessment of Tc1-like transposon elements in channel catfish (Ictalurus punctatus). Genetica 131, 81–90 (2007). [DOI] [PubMed] [Google Scholar]
- Calvi B. R. et al. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell 66, 465–471 (1991). [DOI] [PubMed] [Google Scholar]
- Hickman A. B. et al. Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 12, 715–721 (2005). [DOI] [PubMed] [Google Scholar]
- Cui Z. et al. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J. Mol. Biol. 318, 1221–1235 (2002). [DOI] [PubMed] [Google Scholar]
- Yuan Y. W. & Wessler S. R. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA 108, 7884–7889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R. et al. Horizontal transmission, vertical inactivation, and sto-chastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12, 62–72 (1995). [DOI] [PubMed] [Google Scholar]
- Koga A. et al. Transposable element in fish. Nature 383, 30 (1996). [DOI] [PubMed] [Google Scholar]
- Kawakami K. et al. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97, 11403–11408 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. et al. Goldfish transposase Tgf2 presumably from recent horizontal transfer is active. FASEB J. 26, 2743–2752 (2012). [DOI] [PubMed] [Google Scholar]
- Cheng L. et al. The goldfish hAT-family transposon Tgf2 is capable of autonomous excision in zebrafish embryos. Gene 536, 74–78 (2014). [DOI] [PubMed] [Google Scholar]
- Zou S. et al. Cloning of goldfish hAT transposon Tgf2 and its structure. Hereditas 32, 1–6 (2010). [PubMed] [Google Scholar]
- Zhang L. et al. Characterization of four heat-shock protein genes from Nile tilapia (Oreochromis niloticus) and demonstration of the inducible transcriptional activity of Hsp70 promoter. Fish Physiol. Biochem. 40, 221–233 (2014). [DOI] [PubMed] [Google Scholar]
- Kelley L. A. & Sternberg M. J. E. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009). [DOI] [PubMed] [Google Scholar]
- Hickman A. B., et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell 158, 353–367 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S. et al. Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 106, 10171–10176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D. et al. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids. Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. Prokaryotic expression and purification of soluble goldfish Tgf2 transposase with transposition activity. Mol. Biotechnol. 57, 94–100 (2015). [DOI] [PubMed] [Google Scholar]
- Carpentier G. et al. Transposase–transposase interactions in MOS1 complexes: a biochemical approach. J. Mol. Biol. 405, 892–908 (2011). [DOI] [PubMed] [Google Scholar]
- Shibano T. et al. Recombinant Tol2 transposase with activity in Xenopus embryos. FEBS. Lett. 581, 4333–4336 (2007). [DOI] [PubMed] [Google Scholar]
- Kahlon A. S. et al. DNA binding activities of the Herves transposase from the mosquito Anopheles gambiae. Mob. DNA 2, 9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauriere L. et al. DNA binding specificity and cleavage activity of Pacmmar transposase. Biochemistry 48, 7279–7286 (2009). [DOI] [PubMed] [Google Scholar]
- Sambrook J. et al. Molecular cloning: A laboratory manual. 2nd ed. NewYork: Cold Spring Harbor, USA (1989). [Google Scholar]
- Wolkowicz U. M. et al. Structural basis of Mos1 transposase inhibition by the anti-retroviral drug raltegravir. ACS. Chem. Biol. 9, 743–751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson P. hAT transposable elements. Microbiol. Spectrum 3(4), MDNA3-0054-2014 (2015). [DOI] [PubMed] [Google Scholar]
- Michel K. et al. Does the proposed DSE motif form the active center in the Hermes transposase? Gene 298, 141–146 (2003). [DOI] [PubMed] [Google Scholar]
- Auge-Gouillou C. et al. Mariner Mos1 transposase dimerizes prior to ITR binding. J. Mol. Biol. 351, 117–130 (2005) [DOI] [PubMed] [Google Scholar]
- Guo X. et al. Tc1-like transposase Thm3 of silver carp (Hypophthalmichthys molitrix) can mediate gene transposition in the genome of blunt snout bream (Megalobrama amblycephala). G3 5, 12601–2610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposase. Trends Bicohem. Sci. 25, 421–423 (2000). [DOI] [PubMed] [Google Scholar]
- Michel K. et al. The C-terminus of the Hermes transposase contains a protein multimerization domain. Insect. Biochem. Mol. Biol. 33, 959–970 (2003). [DOI] [PubMed] [Google Scholar]
- Feldmar S. & Kunze R. The ORFa protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. EMBO J. 10, 4003–4010 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H. A. & Kunze R. Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol. Gen. Genet. 254, 219–230 (1997). [DOI] [PubMed] [Google Scholar]
- Hickman A. B. & Dyda F. Mechanisms of DNA transposition. Microbiol. Spectrum 3, MDNA3-0034-2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A. et al. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen E. et al. Truncated forms of IS911 transposase downregulate transposition. Mol. Microbiol. 62, 1102–1116 (2006). [DOI] [PubMed] [Google Scholar]
- Butler M. G. et al. The N-terminus of Himar1 mariner transposase mediates multiple activities during transposition. Genetica 127, 351–366 (2006). [DOI] [PubMed] [Google Scholar]
- Yant S. R. et al. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell. Biol. 24, 9239–9247 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.