Abstract
Expression of the viral virulence protein PB1-F2 during infection has been linked to NLRP3 inflammasome complex activation in macrophages and induction of early inflammatory events enhancing immunopathology during influenza disease. We sought to determine whether PB1-F2-specific NLRP3 inflammasome activation influenced the magnitude and/or robustness of the CD8+ T-cell responses specific for conserved viral antigens and subsequent virus elimination. Using murine heterosubtypic viral infection models, we showed that mice infected with virus unable to produce PB1-F2 protein showed no deficit in the overall magnitude and functional memory responses of CD8+ T cells established during the effector phase compared with those infected with wild-type PB1-F2-expressing virus and were equally capable of mounting robust recall responses. These data indicate that while expression of PB1-F2 protein can induce inflammatory events, the capacity to generate memory CD8+ T cells specific for immunodominant viral epitopes remains uncompromised.
The strength of the innate immune response to influenza A virus (IAV) infection is a key determinant in clinical outcome. IAV infection by highly pathogenic viruses can lead to excessive inflammation, enhancing disease and mortality.1, 2 Activation of innate immune components that trigger inflammation is necessary to drive adaptive immune responses essential for viral clearance. The NLRP3 inflammasome is now recognized as a major mechanism by which the innate immune system recognizes and responds to IAV infection.3, 4 The NLRP3 inflammasome is an oligomeric intracellular signaling complex that recognizes many pathogen-, host- and environment-derived factors.5 Inflammasome-induced cytokine release requires two signals: (1) activation of the prototypic inflammatory transcription factor nuclear factor-κB, typically through Toll-like receptor engagement, resulting in upregulation of components of the NLRP3 inflammasome and synthesis of pro-interleukin-1β (pro-IL-1β) and pro-IL-18; and (2) engagement of NLRP3, which induces inflammasome assembly and activation and in turn results in caspase-1 cleavage-dependent maturation and secretion of IL-1β and IL-18. IAV single-stranded RNA and proton flux via the influenza virus-encoded M2 ion channel have been shown to provide these two signals to activate the NLRP3 inflammasome.6, 7 We have recently demonstrated that the PB1-F2 protein of IAV is also a potent inducer of signal 2, resulting in IL-1β production and pulmonary inflammation early during the infection period.8
IL-1β and IL-18 not only activate monocytes, macrophages and neutrophils but have also been shown to drive the development of CD4+ T-cell adaptive responses in both mice and humans, specifically by the regulation of Th17 and Th1 responses.9, 10, 11, 12 During IAV infection of mice, inflammasome-dependent release of IL-1β and IL-18 has an important role in the recruitment of leukocytes into the lung to control infection. When NLRP3-deficient mice are infected with high doses of virulent IAV, they exhibit greater mortality compared with wild-type mice13 yet infection with the same virus at a dose that is sublethal in wild-type mice does not increase mortality. At high virus doses, the NLRP3 inflammasome is thought to mediate its protective effects through promotion of early healing and repair of the damaged lung tissue7 rather than any enhancement of humoral or cellular immunity that may contribute to clearance in the effector phase.7, 14
Despite lack of any demonstrable impact on primary CD8+ T-cell induction following IAV infection of mice deficient in the NLRP3 inflammasome, the memory and recall responses have yet to be investigated. Unlike the primary CD8+ response to IAV, establishment of memory responses are CD4+ T-cell dependent15, 16 and thus may be influenced by inflammasome-dependent production of IL-1β and IL-18. CD8+ T cells constitute a vital component of adaptive immunity for clearance of IAV infection through cytotoxic killing and secretion of antiviral cytokines. Importantly, recall of memory CD8+ T cells in the absence of specific antibody has been associated with recovery from severe influenza disease in humans infected with novel influenza subtypes, including the highly pathogenic H7N9 IAV.17 Effector CD8+ T cells release antiviral cytokines including interferon γ (IFNγ), tumor necrosis factor α (TNFα) and IL-2 and cause recruitment of other immune cells through triggering chemokine release.18, 19, 20 These cells also have the capacity to secrete anti-inflammatory IL-10 to reduce the effector response and prevent further immunopathology.21
In studies of IAV infection in B6 (H-2b) mice, effector CD8+ T-cell responses are directed against two main immunodominant epitopes, nucleoprotein (DbNP366-374) and acid polymerase (DbPA224–233).22 During primary infection, NP366-specific and PA224-specific CD8+ T-cell responses are largely co-dominant but in a secondary infection, NP366-specific responses become immunodominant over PA224-specific CD8+ T cells.23, 24 Infection with IAV in which the NP and PA-specific epitopes have been disrupted revealed that CD8+ T cells specific for the otherwise subdominant PB1-F262 can compensate for these primary immunodominant responses, but further deletion of the PB1-F262–70 epitope effectively switches the response against IAV infection to an antibody-dominated response.25 The targeting of PB1-F2 by CD8+ T cells supports the idea that this protein may have a role in influencing host immunity to IAV infection. In this study, we hypothesized that the initial inflammatory events occurring during IAV infection, triggered by PB1-F2-induced NLRP3 inflammasome activation, may influence the development of memory CD8+ T-cell responses in terms of their quality and magnitude and in turn may impact host response upon subsequent viral challenge with a serologically distinct subtype of IAV.
Results
PB1-F2 expression by IAV enhances early inflammatory responses to infection
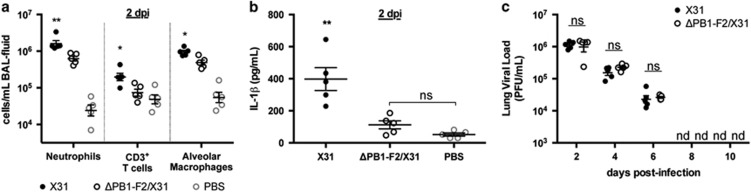
For this study, we used the well-characterized X31 IAV,26 which contains the H3 and N2 surface glycoproteins and all other viral proteins matching those of the H1N1 IAV A/Puerto Rico/8/34 (PR8), including the highly inflammatory PB1-F2 that is expressed from the PR8 PB1 gene. Previously, we utilized X31 and an otherwise isogenic virus that fails to express PB1-F2 (ΔPB1-F2/X31) to demonstrate the involvement of the PB1-F2 protein in IL-1β secretion and increased cellularity in the lungs of mice 24–72 h post-infection (h.p.i.).27 We additionally demonstrated that PB1-F2 has a direct role in activation of signal 2 of the inflammasome complex when it is produced within infected macrophages or when a peptide from PB1-F2 is phagocytosed and the phagolysosomal compartment becomes acidified. Here we used the X31 and ΔPB1-F2/X31 IAVs to determine whether the enhanced inflammation induced by PB1-F2-dependent activation of the inflammasome created an environment that was either beneficial or detrimental to the induction of subsequent CD8+ T-cell responses. Supporting our previously published findings,27, 28, 29 we showed that mice infected with ΔPB1-F2/X31 virus had reduced recruitment of neutrophils, CD3+ T cells and alveolar macrophages into the bronchoalveolar lavage (BAL) fluid at 48 h.p.i. compared with mice infected with X31 virus (Figure 1a). These mice also had diminished levels of IL-1β within the BAL fluid at 48 h.p.i. (Figure 1b) that were not significantly different from those in uninfected phosphate-buffered saline (PBS)-inoculated control mice, consistent with our earlier finding that PB1-F2 was driving the IL-1β production early after infection. Importantly, the differences in cellular infiltration and IL-1β concentrations in the BAL fluid were not due to viral load variances as the viral load and rate of clearance from the lungs did not differ between X31 and the ΔPB1-F2/X31-infected mice (Figure 1c). Weight loss over the duration of the experiment was also equivalent between groups (data not shown). In addition, although our previous findings showed that PB1-F2 protein production during infection increased cellular infiltrate in the lungs up to 72 h.p.i.,28 we showed here that the numbers of neutrophils, T cells and alveolar macrophages in the BAL had returned to those comparable to infection in the absence of PB1-F2 by 4 days post infection (d.p.i.; Supplementary Figure 1), and remained comparable until 10 d.p.i., after the virus had been cleared (Figure 1c).
Figure 1.
Effect of early IL-1β induced inflammation on viral clearance. C57BL/6 mice (n=5, female, 8 weeks old) were infected intranasally with 100 PFU of either X31 or ΔPB1-F2/X31 viruses in 50 μl, or inoculated intranasally with 50 μl PBS. On day 2 post-infection, mice were killed, BAL fluid extracted and assayed for (a) neutrophil, CD3+ T-cell and alveolar macrophage content by flow cytometry and (b) secreted IL-1β via ELISA. *P<0.05, **P<0.01 compared with all other groups, ns, not significantly different, two-way ANOVA. (c) Lungs from infected mice were also harvested at different d.p.i., homogenized and assayed for infectious virus by plaque formation in MDCK cells. Virus titers are expressed as plaque forming units per ml (PFU ml–1). nd, not detected; ns, not significantly different between infection groups, Student's unpaired T-test. Data are representative of three independent experiments.
Functional IAV-specific CD8+ T cells are induced similarly in the presence or absence of PB1-F2-dependent early inflammation
To investigate whether the early PB1-F2-dependent inflammatory response influences the induction of CD8+ T cells, mice were infected with X31 or ΔPB1-F2/X31 viruses and the total CD8+ T cells enumerated in the BAL between 2 and 10 d.p.i. An increase in the numbers of CD8+ T cells was observed from day 6 but this was not different in X31 virus- or ΔPB1-F2/X31 virus-infected mice (Figure 2a). The total numbers of splenic CD8+ T cells was also equivalent between the X31- and ΔPB1-F2/X31-infected groups (data not shown).
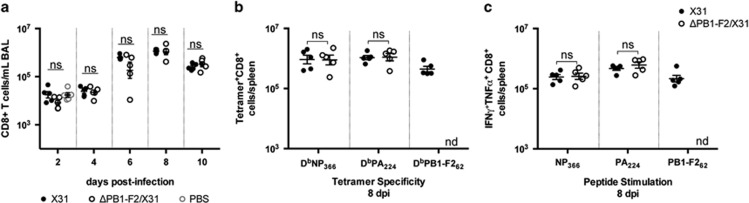
Figure 2.
Primary CD8+ T-cell induction following infection with viruses differing in PB1-F2 expression. C57BL/6 mice (n=5, female, 8 weeks old) were infected intranasally with 100 PFU of either X31 or ΔPB1-F2/X31 viruses in 50 μl, or inoculated intranasally with 50 μl PBS. (a) At different d.p.i., the numbers of CD8+ T cells in BAL fluid were determined by flow cytometry (ns, not significantly different, one-way ANOVA for three group comparison at 2 d.p.i.; Student's unpaired T-test for two group comparisons at 4–10 d.p.i.). (b) At 8 d.p.i., the numbers of splenic CD8+ T cells specific for viral epitopes NP366, PA224 and PB1-F262 were determined by tetramer staining. (c) Cytokine production by splenic CD8+ T cells was determined by intracellular cytokine staining after stimulation with NP366, PA224 and PB1-F262 peptides. Shown are the numbers of IFNγ and TNFα co-producing cells. ns, data not significant, Student's unpaired T-test; nd, no significant numbers detected. Data are representative of three independent experiments.
The epitope specificity of the responding CD8+ T cells, as well as their polyfunctionality were then evaluated using splenocytes isolated from groups of infected mice at 8 and 10 d.p.i. The CD8+ T cells were analyzed for their specificity for the major epitopes NP366-374, PA224 and PB1-F262 by tetramer staining. The numbers of CD8+ T cells specific for NP366 and for PA224 induced by the two viruses were equivalent, whereas the ΔPB1-F2/X31 virus-infected group did not induce CD8+ T cells specific for the PB1-F262 epitope as expected (Figure 2b and Supplementary Figure 2a for day 8 and day 10 data, respectively). To assess any differences in CD8+ T-cell function, we measured the capacity of these cells to produce multiple cytokines following a 5 h in vitro stimulation with cognate peptide.30 Intracellular cytokine staining of NP366- and PA224-stimulated splenocytes showed that CD8+ T cells capable of producing both TNFα and IFNγ were present in mice infected with X31 or with ΔPB1-F2/X31 viruses at 8 d.p.i. (Figure 2b) and 10 d.p.i. (Supplementary Figure 2b). The PB1-F262-specific CD8+ T cells also contributed significantly to the secretion of these antiviral cytokines in X31-infected mice. As with the total CD8+ T-cell numbers in BAL (Figure 2a), there was no increase in the numbers of polyfunctional TNFα+IFNγ+ splenic CD8+ T cells specific for NP366 or PA224 in mice infected with the ΔPB1-F2/X31 virus (Figure 2b and Supplementary Figure 2b) to compensate for the absence of the PB1-F2-specific subset. CD8+ T cells capable of producing IFNγ alone or IFNγ, TNFα and IL-2 in X31 and ΔPB1-F2/X31 virus infection groups were also equivalent when stimulated by NP366 or PA224 peptides (data not shown). This may indicate that the functional capacity of the NP366- and PA224-specific T cells is sufficient for IAV clearance, which was equivalent between the infection groups (Figure 1c).
Memory CD8+ T-cell responses are not altered as a result of early PB1-F2-dependent enhancement of inflammation
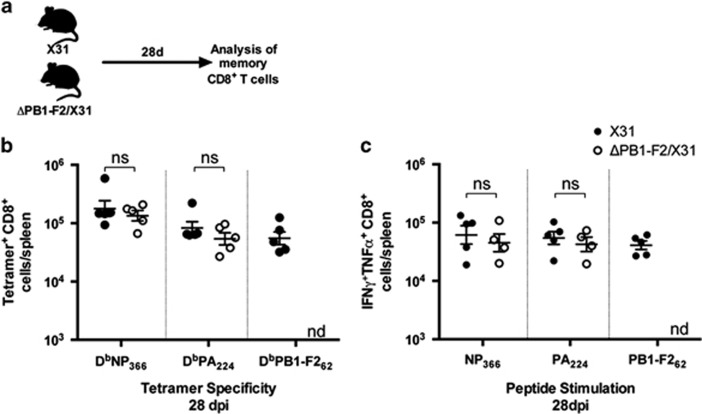
Although the primary CD8+ T-cell response appeared not to differ functionally when induced in the presence or absence of PB1-F2-dependent NLRP3 inflammasome activation, it is possible that any difference, particularly due to lack of the PB1-F262-specific CD8+ T-cell compartment in mice infected with ΔPB1-F2/X31 virus, may become apparent when the response contracts into the memory phase. We therefore sought to determine any differences the initial virus exposure has on memory CD8+ T-cell development and responsiveness toward viral antigen. Groups of mice were infected with either X31 or ΔPB1-F2/X31 virus then allowed to clear the infection and recover for 28 days (Figure 3a). Splenocytes were then assayed for the proportion of epitope-specific CD8+ T cells by tetramer staining, as well as their cytokine secretion profile after NP366, PA224 or PB1-F262 peptide stimulation. Figure 3b shows that the proportion of NP366- and PA224-specific CD8+ T cells entering the early memory phase were not different in mice initially infected with X31 or ΔPB1-F2/X31 virus. Similarly, the numbers of polyfunctional TNFα and IFNγ-secreting CD8+ T cells specific for NP366 or PA224 were no different in these memory populations (Figure 3c). As expected, polyfunctional CD8+ T cells specific for PB1-F2 antigen were present in the memory pool of mice originally infected with X31 virus but not with ΔPB1-F2/X31 virus (Figure 3c).
Figure 3.
Memory CD8+ T-cell induction following infection with viruses differing in PB1-F2 expression. (a) C57BL/6 mice (n=5, female, 8 weeks old) were intranasally infected with 100 PFU in 50 μl of either X31 or ΔPB1-F2/X31 virus. At 28 d.p.i., CD8+ T cells in the spleens were assayed for (b) epitope specificity by tetramer staining and (c) cytokine production by CD8+ T cells as in Figure 2. ns, data not significant, Student's unpaired T-test; nd, no significant numbers detected. Data are representative of two independent experiments.
Memory CD8+ T cells raised in response to IAV-lacking PB1-F2 show no deficiency in clearance of a heterosubtypic viral challenge despite the lack of a DbPB1-F262-specific population
On secondary infection with an IAV virus of a different HA and NA subtype, clonal expansion and activation of antigen-specific memory CD8+ T cells directed to common epitopes enables efficient clearance of the heterologous virus in the absence of pre-existing antibody.31 To investigate whether early enhancement of inflammation influences the robustness of immunological responses on secondary infection with IAV, groups of mice were initially infected with either X31, ΔPB1-F2/X31 or inoculated with PBS, and 28 days later infected with either the heterosubtypic PR8 virus or the corresponding ΔPB1-F2/PR8 virus engineered to negate expression of PB1-F2 protein. BAL fluid, lungs and spleens were extracted at 5 or 8 days post-secondary challenge infection for analysis (Figure 4a). The cellular content of the BAL taken at 5 days post-challenge showed no difference in the amount of leukocytes, neutrophils and alveolar macrophages in mice initially infected with either X31 or ΔPB1-F2/X31 viruses, regardless of the virus used for secondary challenge infection (Supplementary Figure 3). As the PR8 virus used for the challenge infection is highly virulent in mice not previously exposed to influenza, those in the PBS group had heightened cellular infiltrate in the BAL (Supplementary Figure 3) and enhanced viral load in the lungs (Figure 4c) compared with pre-infected mice and these mice reached the humane end point and were killed by 7 d.p.i. The groups of previously infected mice reached the same viral titer at 5 days post-secondary infection, irrespective of whether the primary infecting virus was X31 or ΔPB1-F2/X31, and no virus was detectable in the lung homogenate at 8 days post-secondary infection indicating a similar rate of clearance of the challenge virus (Figure 4c). In addition, titers of PR8 or ΔPB1-F2/PR8 in the lungs at 5 d.p.i. and the ability to clear virus by 8 d.p.i. were no different (P>0.05, Student's unpaired T-test) with respect to whether the priming virus expressed PB1-F2 or not.
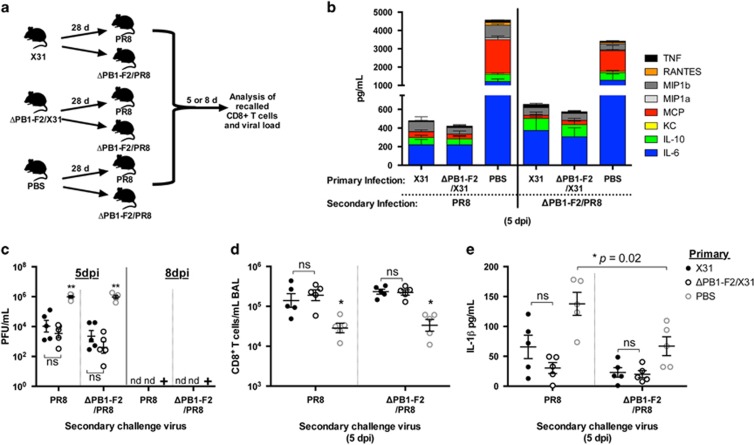
Figure 4.
Inflammatory cytokine production and viral clearance following secondary infection with viruses differing in PB1-F2 expression. (a) C57BL/6 mice (n=5, female, 8 weeks old) were infected intranasally with 100 PFU of either X31 or ΔPB1-F2/X31 viruses in 50 μl, or inoculated intranasally with 50 μl PBS. At 28 d.p.i., mice were challenged with 100 PFU of PR8 or ΔPB1-F2/PR8 virus. BAL fluid and lungs were collected at 5 or 8 days post-secondary infection and assayed for (b) chemokine and cytokine content in BAL supernatant via cytometric bead array analysis (data shown for day 5), (c) infectious viral titers of the lung homogenate (**P<0.01 one-way ANOVA, nd, no plaques detected; +, all mice in the group had reached the humane end point by 7 d.p.i. and were killed), (d) presence of CD8+ T cells in the BAL fluid via flow cytometry (*P<0.05 compared with all other groups, two-way ANOVA) and (e) IL-1β content within the BAL supernatant via ELISA (ns, not significantly different between infection groups, *P=0.02 as indicated, Student's unpaired T-test). Data are representative of three independent experiments.
Samples of BAL taken at 5 days post-secondary challenge were examined for cytokine, chemokine and cellular content. The levels of IL-6, IL-10, macrophage chemotactic protein (MCP), macrophage inflammatory protein-1β (MIP1β) and TNFα, as well as the amount of neutrophils, CD3+ T cells and alveolar macrophages were not significantly different between all infection groups, regardless of the virus used for the primary and secondary infections (Figure 4b). These levels were significantly reduced compared with mice that initially received PBS, which were undergoing a primary infection. This was especially true for the inflammatory mediators keratinocyte chemoattractant (KC), MIP1α, RANTES (regulated on activation, normal T cell expressed and secreted) and TNF in the BAL fluid, which were undetectable in previously infected mice (P<0.05 two-way analysis of variance (ANOVA)). However, mice initially infected with either X31 or ΔPB1-F2/X31 virus had elevated CD8+ T cells present in the BAL compared with mice originally exposed only to PBS, indicating an active recall of memory CD8+ T cells to the site of infection (Figure 4d). Of note, mice infected initially with X31, followed by PR8, both of which express PB1-F2, had a trend toward elevated amounts of IL-1β in their BAL fluid, however, the level was not significantly above the other infection groups (Figure 4e). Whether differences in these levels of secreted IL-1β reached statistical significance at an earlier time post-secondary infection remains to be examined. At 5 d.p.i., PBS control mice challenged with PR8 had significantly elevated IL-1β levels compared with those challenged with ΔPB1-F2/PR8 virus (Figure 4e), consistent with the data at an earlier time point for primary infection shown in Figure 1b. Only negligible levels of all cytokines and chemokines remained at 8 days post-secondary infection after the virus had been cleared (data not shown).
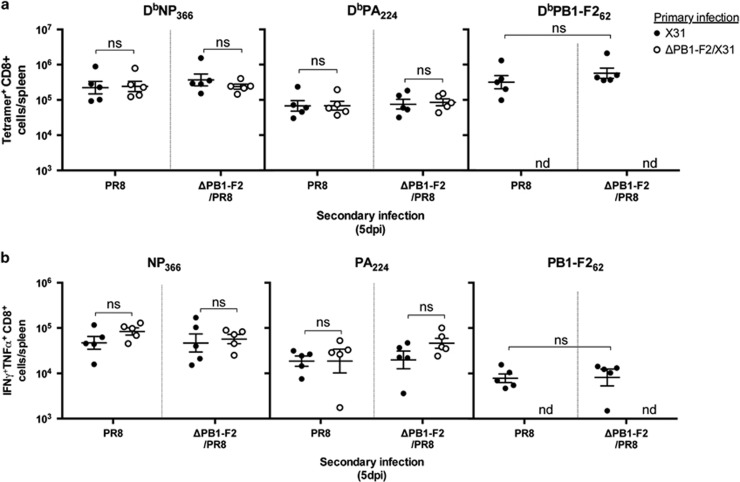
We finally evaluated whether the expression of PB1-F2 viral protein during the initial infection with influenza virus alters the robustness of the memory CD8+ T-cell responses recalled in response to the secondary challenge infection. The magnitude (Figure 5a) and polyfunctionality (Figure 5b) of the recalled CD8+ T-cell response toward NP366 and PA224 epitopes at 5 days post-challenge was no different with respect to whether the initial priming virus could express PB1-F2 or not, and this was also the case for the response at 8 days post-challenge (data not shown). Similarly, the ability of the two viruses to recall CD8+ T cells was no different. Memory CD8+ T cells specific for PB1-F2, induced in response to primary X31 infection, participated in the recall response after secondary infection with PR8 virus but notably and reproducibly, the number of these recalled cells was no different (P=0.0582 Student's unpaired T-test) to the number recalled in response to secondary infection with ΔPB1-F2/PR8 virus (Figures 5a and b), possibly reflective of antigen-nonspecific bystander activation of the memory PB1-F262-specific cells in the presence of a highly stimulatory local cytokine milieu.32 The data also reflect the fact that the magnitude (Figure 4d), dominant epitope specificity (Figure 5a) and polyfunctionality of CD8+ T-cell responses raised to either X31 or ΔPB1-F2/X31 virus are equally able (P>0.05 for each comparison, one-way ANOVA) to be recalled by the inflammatory PB1-F2-expressing challenge virus as they are by the virus lacking PB1-F2.
Figure 5.
Recall CD8+ T-cell responses following infection with viruses differing in PB1-F2 expression. C57BL/6 mice (n=5, female, 8 weeks old) were infected intranasally with 100 PFU of either X31 or ΔPB1-F2/X31 viruses in 50 μl. At 28 d.p.i., mice were challenged with 100 PFU of PR8 or ΔPB1-F2/PR8 virus. Five days later, CD8+ T cells in the spleens were assayed for (a) epitope specificity by tetramer staining and (b) cytokine production by CD8+ T cells as in Figure 2. ns, not significantly different, Student's unpaired T-test; nd, no significant numbers detected. Data are representative of three independent experiments.
Discussion
To define the possible role that PB1-F2 protein may have in induction of inflammasome activation and its influence on adaptive immunological IAV-specific CD8+ T-cell memory pools, we asked whether the magnitude and functionality of the CD8+ T cells generated in PB1-F2-producing IAV-infected mice differed from mice infected with virus unable to express this protein. We found that, despite the pro-inflammatory effects of PB1-F2 protein during the initial stages of infection, the overall magnitude of the primary effector CD8+ T-cell response was not different after infection with a virus not expressing this protein. Based on the assumption that PB1-F2 is the major contributor to early inflammation because of activation of the NLRP3 inflammasome,27 these data are consistent with the findings of Ichinohe et al.14 and Thomas et al.7 who showed no difference in primary CD8+ T-cell responses to IAV infection in wild-type and NLRP3-deficient mice. In this study, we additionally show that the memory and recall CD8+ T-cell responses were no different and were equally capable of clearing a challenge virus of a heterologous IAV subtype. This implies that if the early NLRP3-driven inflammatory response has any effect on CD4+ T-cell responses, this must be inconsequential for the role of these cells in providing help for memory CD8+ T-cell generation and expansion.15, 16
The PB1-F2 protein is encoded by a +1 alternate reading frame of the PB1 gene and was discovered while screening for influenza-specific CD8+ T-cell epitopes.33 PB1-F2 protein expression in cells has been linked to cell death, viral replication and virulence, however, these properties can differ between virus strains.34 Crucially, PB1-F2 proteins derived from twentieth century pandemic and highly pathogenic IAV strains, but not mildly virulent IAVs, trigger excessive cellular recruitment and hyper-inflammatory responses in the lungs of infected mice.28, 29 IAV is sensed in both respiratory epithelial cells and macrophages at early time points of infection, leading to NLRP3 inflammasome complex activation35 but potentially has different roles in these cell types. We previously demonstrated direct PB1-F2 activation of this complex in macrophage models in vitro and the contribution of PB1-F2 to overt inflammatory responses toward IAV in murine infection models.27 However, studies using in vitro transfection of human epithelial cell lines36 demonstrated that intracellular production of PB1-F2 protein inhibited the activation of the NLRP3 inflammasome.
Our finding that viral clearance was not altered in mice infected with X31 or ΔPB1-F2/X31 viruses, or when these infected mice were challenged with an otherwise lethal dose of PR8 virus may seem at odds with the findings of Allen et al.13 and Thomas et al.,7 both of whom examined lethal PR8 infection in NLRP3 inflammasome-deficient mouse models and showed these were less likely to survive than wild-type mice. However, although the X31 virus has the internal genes from PR8 virus, the inflammation it produces is milder than that of PR8 virus, although still achieves NLRP3 inflammasome complex activation as indicated by enhanced levels of secretion of IL-1β in the BAL fluid at 2 d.p.i. In addition, the secondary infection with a lethal dose of PR8 virus was ameliorated by the pre-established CD8+ T-cell response. Our infection system is therefore more similar to the milder infection used by Ichinohe et al.14 who demonstrated that NLRP3-deficient mice were not more susceptible to death following infection with a low dose of PR8. Interestingly, mice deficient in caspase-1, ASC or IL-1R did succumb to the same low-dose PR8 infection, and this was interpreted as implicating a non-NLRP3 inflammasome in protective anti-IAV immunity. The fact that we did not observe elevated levels of IL-1β secretion in mice infected with ΔPB1-F2/X31 over those given PBS alone suggests that most, if not all IL-1β production in the BAL is induced by PB1-F2 and not by other viral triggers, at least at 2 d.p.i.
As PB1-F2 is itself a target for CD8+ T-cell responses in this mouse system, we also examined CD8+ T-cell specificity and function at the epitope level. We found that development of the immunodominant DbNP366 and DbPA224 CD8+ T cells and their functional cytokine response to peptide stimulation were the same in X31- and ΔPB1-F2/X31-infected mice during effector and memory responses at 28 d.p.i. Furthermore, despite mice initially infected with X31 virus having an additional subset of DbPB1-F262-specific CD8+ T cells this did not confer any advantage in viral clearance, suggesting either that infection with ΔPB1-F2/X31 virus led to activation of a larger proportion of CD8+ T cells recognizing other subdominant specificities, or that sufficient CD8+ T cells were produced to handle the infection regardless of the contribution from the PB1-F262-specific subset. DbPB1-F262-specific CD8+ T cells were also represented in the memory pool after infection with the PB1-F2-expressing virus and responsive upon secondary challenge with the heterologous virus. IAVs entering the human population from avian reservoirs usually express a full-length inflammatory PB1-F2 protein37 but this can be lost as the virus co-evolves with its new host. The presence of PB1-F2-specific CD8+ T-cell memory responses in humans may provide the selective pressure against production of this non-essential protein as a way by which IAVs circulating in mammalian hosts initially evolve away from the more inflammatory phenotype through mutations at amino acids implicated in PB1-F2 induced inflammation,37 then toward production of a truncated protein.
The severity of disease because of emergent influenza strains is often associated with a ‘cytokine storm' in which pulmonary involvement is associated with heightened cellular loads, chemokines, cytokines and increased epithelial damage. Our previous studies have linked PB1-F2 protein expressed by viruses containing the avian PB1 gene to enhancing these immunopathological effects through triggering NLRP3 inflammasome complex activation, recruitment of an exuberant level of neutrophils and macrophages to the site of infection and secretion of inflammatory cytokines and chemokines, and ultimately predisposing the host to secondary bacterial infections.27, 28, 29 This study demonstrates that, despite enhanced early innate inflammation as a result of PB1-F2 expression and a more diverse CD8+ T-cell repertoire toward IAV antigens including NP, PA and PB1-F2, the expression of PB1-F2 protein during infection is not critical for or detrimental to clearance of the infection nor to the establishment of protective immune responses upon secondary viral infection. Indeed these data agree with recent findings that functional memory CD8+ T cells established during oseltamivir prophylaxis for IAV infection, which reduced inflammation during the acute phase of infection, were still capable of mounting robust recall responses.38 Our study is also the first to show the converse in that, expression of the viral virulence protein PB1-F2 by a heterosubtypic challenge virus that displays heightened pathophysiology of disease, does not compromise the capacity to recall pre-existing memory CD8+ T cells essential for clearance of this serologically distinct virus. These findings highlight the importance of maintaining cross-reactive memory CD8+ T-cell pools, as these are expected to be effective at promoting viral clearance on encounter with highly inflammatory pandemic strains.
Methods
Ethics statement
All experimental procedures were approved by the University of Melbourne Animal Ethics Committee (AEC) under relevant institutional guidelines, the Prevention of Cruelty to Animals Act 1986 and associated regulations, and the Australian Code for the Care and Use of Animals for Scientific Purposes 2013.
Virus propagation and quantitation of viral titers
The X31 and ΔPB1-F2/X31 viruses were created as previously described.27 Briefly, these viruses contain the PB1, PB2, NP, PA, M and NS gene segments of A/Puerto Rico/8/34 (PR8) virus and the HA and NA gene segments of A/Aichi/2/68 (H3N2) virus. The PR8 and ΔPB1-F2/PR8 viruses were identical to the corresponding X31 and ΔPB1-F2/X31 viruses except for the HA and NA gene segments, which were derived from the serologically distinct PR8 virus (H1N1). In ΔPB1-F2 viruses, the open reading frame of PB1-F2, which is located in the second reading frame of the PB1 gene, was disrupted by altering the start codon (T120C mutation by PB1 numbering) so translation will not initiate, and inserting a stop codon after 11 and 56 residues (C153G and G291A, respectively), to ensure complete inability for production of PB1-F2 protein. In no case, did the mutations in the PB1-F2 open reading frame cause non-synonymous mutations in the PB1 open reading frame. The N40 start codon39 was intact in all viruses. All viruses were rescued by one passage in Madin Darby canine kidney (MDCK) cells, then propagated a single time in eggs. All viruses were fully sequenced to ensure no inadvertent mutations occurred during virus rescue and propagation, then characterized in tissue culture and eggs as previously described.40 Expression, or lack thereof, of PB1-F2 protein was confirmed by confocal microscopy as described previously.28, 40 The infectious viral titer was determined by the quantitation of plaques on confluent MDCK cell monolayers as previously described.41
Animal infection model
Eight- to 10-week-old female C57BL6 mice were maintained in the Specific Pathogen-Free Physical Containment Level 2 (PC2) Bioresources Facility at the Department of Microbiology and Immunology (University of Melbourne, Melbourne, VIC, Australia). All experimental inoculation procedures were conducted under general anesthesia with inhaled isofluorane at 2.5% (Baxter Healthcare Corporation NSW, Australia). Infectious agents were diluted in sterile PBS and administered intranasally to anesthetized mice (n=5) in 50 μl. Mice were monitored daily for weight loss and disease signs and killed at the pre-determined humane end point as required. The infectious dose used for all viruses in all experiments was 100 plaque-forming units (PFU). This dose of X31 virus is non-lethal for mice, whereas the same dose of PR8 virus is lethal in previously uninfected mice.
Tissue sampling of mice
BAL and lungs
BAL was extracted as previously described.29 Briefly, leukocytes (CD45+), neutrophils (Ly6G+, CD45+, F480−) and alveolar macrophages (F480+, Ly6G− MHC class IIint) were enumerated by flow cytometry using BD FACSCanto II (BD Biosciences, Franklin Lakes, NY, USA), analyzed using FlowJo X version 10.0.7r2 (Tree Star, Ashland, OR, USA) and expressed as a proportion of cellular events analyzed. The total numbers of each cell population were calculated in relation to the number of white blood cells per ml (WBC ml–1) in the original sample, determined by counting cells with the aid of a light microscope and hemocytometer using the Trypan blue exclusion method. Lungs were extracted, then homogenized using a Polytron System PT 1200 CL 230 V homogenizer (Kinematica, Lucerne, Switzerland). Viral content within the lung homogenate supernatants was then determined by the quantitation of plaques on confluent MDCK cell monolayers as previously described.41
Splenocyte collection
Spleens were collected from mice at the times indicated and pressed through a 70 μm sieve. B-cell depletion was performed by incubation of splenocytes for 45 min at 37 °C and 5% CO2 on panning plates coated with 200 μg ml–1 goat anti-mouse anti-IgM/IgG (Jackson ImmunoResearch Labs, West Grove, PA, USA).
Tetramer and phenotypic staining for CD8+ T-cell specificity
Enriched cellular suspensions of spleen and BAL were then stained for 1 h at 4 °C with tetrameric complexes of H-2Db and influenza viral peptides NP366-374 (ASNENMETM)-PE, PA224-233 (SSLENFRAYV)-APC or PB1-F262-70 (LSLRNPILV)-PE (monomers purchased from ImmunoID, University of Melbourne, Melbourne, VIC, Australia), followed by CD8α-PacificBlue (BD Pharmingen, San Diego, CA, USA). Unenriched splenic solutions were stained with anti-mouse CD8α-FITC (BioLegend, San Diego, CA, USA) for enumeration of CD8+ T cells. Based upon cell size (FSC) and granularity (SSC), approximately 500 000 cellular events were acquired on the BD LSRFortessa (BD Biosciences) and data analyzed with FlowJo X version 10.0.7r2 (Tree Star).
Peptide stimulation and intracellular cytokine staining of CD8+ T cells
Splenic cellular suspensions were examined for the production of IFNγ, TNFα and IL-2 by peptide stimulation and intracellular cytokine staining as previously described.42 Briefly, splenocytes isolated from animals were stimulated for 5 h in 5% CO2 at 37 °C with viral peptides corresponding to CD8+ T-cell epitopes NP366, PA224 and PB1-F262 in the presence of GolgiPlug (BD Biosciences) and recombinant IL-2 before surface staining with anti-mouse CD8α-PacificBlue. Cells were then fixed and permeabilized using the BD Cytofix/Cytoperm Kit (BD Biosciences) and stained intracellularly with a cocktail of anti-mouse anti-IFNγ-FITC, anti-IL-2-PE and anti-TNFα-APC (BioLegend) antibodies. Data were acquired, as described for tetramer staining (above).
Cytometric bead array and ELISA
The CBA flex set (BD Bioscience) was used as per the manufacturer's instructions to determine the cytokine concentrations of BAL supernatant. Data were acquired using FACSCanto II and analyzed with FlowJo X version 10.0.7r2 (Tree Star) with sample concentrations interpreted from standard curves using GraphPad Prism 6.0 F. For the detection of IL-1β in the BAL, 50 μl undiluted sample supernatant was used in a mouse IL-1β ELISA (BD Biosciences) according to the manufacturer's specifications.
Statistical analysis
Statistical analyses were performed using GraphPad Prism Version 6 (GraphPad Software, Inc., La Jolla, CA, USA). All graphs show mean±s.e.m.
Acknowledgments
JM is funded by the National Health and Medical Research Council of Australia (NHMRC) project grants 1026619 and 1079924; KK is a CDF2 NHMRC Fellow; KK and LB receive funding from NHMRC program grant 1071916. The authors acknowledge St Jude Children's Research Hospital (TN, USA) as the original source of the PR8 PB1 and PR8 ΔPB1-F2 plasmids.
The authors declare no conflict of interest.
Footnotes
The Supplementary Information that accompanies this paper is available on the Immunology and Cell Biology website (http://www.nature.com/icb)
Supplementary Material
References
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006; 12: 1203–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Gruta NL, Kedzierska K, Stambas J, Doherty PC. A question of self-preservation: immunopathology in influenza virus infection. Immunol Cell Biol 2007; 85: 85–92. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol 2014; 14: 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang IK, Iwasaki A. Inflammasomes as mediators of immunity against influenza virus. Trends Immunol 2011; 32: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell 2010; 140: 821–832. [DOI] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 2010; 11: 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PG et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 2009; 30: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Tate MD, MacKenzie-Kludas CJ, Pinar A, Zeng W, Stutz A et al. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog 2013; 9: e1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 2009; 27: 519–550. [DOI] [PubMed] [Google Scholar]
- Lasiglie D, Traggiai E, Federici S, Alessio M, Buoncompagni A, Accogli A et al. Role of IL-1 beta in the development of human T(H)17 cells: lesson from NLPR3 mutated patients. PLoS ONE 2011; 6: e20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA. IL-18: A TH1-inducing, proinflammatory cytokine and new member of the IL-1 family. J Allergy Clin Immunol 1999; 103: 11–24. [DOI] [PubMed] [Google Scholar]
- Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 2009; 30: 576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 2009; 30: 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 2009; 206: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J Virol 2002; 76: 12388–12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 2003; 300: 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wan Y, Qiu C, Quiñones-Parra S, Zhu Z, Loh L et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun 2015; 6: 6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Braciale T. Role of T cell immunity in recovery from influenza virus infection. Curr Opin Virol 2013; 3: 425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MQ, Stoler MH, Liu AN, Wei B, Soguero C, Hahn YS et al. Alveolar epithelial cell chemokine expression triggered by antigen-specific cytolytic CD8(+) T cell recognition. J Clin Invest 2000; 106: R49–R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Gruta N, Turner S, Doherty P. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J Immunol 2004; 172: 5553–5560. [DOI] [PubMed] [Google Scholar]
- Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nat Immunol 2011; 12: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambas J, Guillonneau C, Kedzierska K, Mintern JD, Doherty PC, La Gruta NL. Killer T cells in influenza. Pharmacol Ther 2008; 120: 186–196. [DOI] [PubMed] [Google Scholar]
- Belz GT, Xie W, Altman JD, Doherty PC. A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge. J Virol 2000; 74: 3486–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkenburg SA, Venturi V, Dang TH, Bird NL, Doherty PC, Turner SJ et al. Early priming minimizes the age-related immune compromise of CD8(+) T cell diversity and function. PLoS Pathog 2012; 8: e1002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Brown SA, Keating R, Yue W, Morris MY, So J et al. Hidden epitopes emerge in secondary influenza virus-specific CD8+ T cell responses. J Immunol 2007; 178: 3091–3098. [DOI] [PubMed] [Google Scholar]
- Kedzierska K, La Gruta NL, Turner SJ, Doherty PC. Establishment and recall of CD8+ T-cell memory in a model of localized transient infection. Immunol Rev 2006; 211: 133–145. [DOI] [PubMed] [Google Scholar]
- McAuley JL et al. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathogens 2013; 9: e1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Chipuk JE, Boyd KL, Van De Velde N, Green DR, McCullers JA. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog 2010; 6: e1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2007; 2: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Parra S, Grant E, Loh L, Nguyen TH, Campbell KA, Tong SY et al. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc Natl Acad Sci USA 2014; 111: 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty PC, Topham DJ, Tripp RA, Cardin RD, Brooks JW, Stevenson PG. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunol Rev 1997; 159: 105–117. [DOI] [PubMed] [Google Scholar]
- Tripp RA, Hou S, McMickle A, Houston J, Doherty PC. Recruitment and proliferation of CD8+ T cells in respiratory virus infections. J Immunol 1995; 154: 6013–6021. [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 2001; 7: 1306–1312. [DOI] [PubMed] [Google Scholar]
- Krumbholz A, Philipps A, Oehring H, Schwarzer K, Eitner A, Wutzler P et al. Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol 2011; 200: 69–75. [DOI] [PubMed] [Google Scholar]
- Owen DM, Gale M Jr. Fighting the flu with inflammasome signaling. Immunity 2009; 30: 476–478. [DOI] [PubMed] [Google Scholar]
- Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K et al. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun 2014; 5: 4713. [DOI] [PubMed] [Google Scholar]
- Alymova IV, Green AM, van de Velde N, McAuley JL, Boyd KL, Ghoneim HE et al. Immunopathogenic and antibacterial effects of H3N2 influenza A virus PB1-F2 map to amino acid residues 62, 75, 79, and 82. J Virol 2011; 85: 12324–12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird NL, Olson MR, Hurt AC, Oshansky CM, Oh DY, Reading PC et al. Oseltamivir prophylaxis reduces inflammation and facilitates establishment of cross-strain protective T cell memory to influenza viruses. PLoS ONE 2015; 10: e0129768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W et al. A complicated message: identification of a novel PB1-related protein translated from influenza A segment 2 mRNA. J Virol 2009; 83: 8021–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Zhang K, McCullers JA. The effects of influenza A virus PB1-F2 protein on polymerase activity are strain specific and do not impact pathogenesis. J Virol 2010; 84: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Balish AL, Katz JM. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol 2006; Chapter 15: Unit 15G 11. [DOI] [PubMed] [Google Scholar]
- Cukalac T, Valkenburg SA, La Gruta NL, Turner SJ, Doherty PC, Kedzierska K. Multiplexed combinatorial tetramer staining in a mouse model of virus infection. J Immunol Methods 2010; 360: 157–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.