Abstract
Salinity tolerance levels and physiological changes were evaluated for twelve rice cultivars, including four white rice and eight black glutinous rice cultivars, during their seedling stage in response to salinity stress at 100 mM NaCl. All the rice cultivars evaluated showed an apparent decrease in growth characteristics and chlorophyll accumulation under salinity stress. By contrast an increase in proline, hydrogen peroxide, peroxidase (POX) activity and anthocyanins were observed for all cultivars. The K+/Na+ ratios evaluated for all rice cultivars were noted to be highly correlated with the salinity scores thus indicating that the K+/Na+ ratio serves as a reliable indicator of salt stress tolerance in rice. Principal component analysis (PCA) based on physiological salt tolerance indexes could clearly distinguish rice cultivars into 4 salt tolerance clusters. Noteworthy, in comparison to the salt-sensitive ones, rice cultivars that possessed higher degrees of salt tolerance displayed more enhanced activity of catalase (CAT), a smaller increase in anthocyanin, hydrogen peroxide and proline content but a smaller drop in the K+/Na+ ratio and chlorophyll accumulation.
Keywords: Antioxidant enzymes, Anthocyanins, Rice, Salt stress
1. Introduction
Salinity is a common abiotic stress that severely limits crop growth and development, productivity and causes the continuous loss of arable land, which results in desertification in arid and semi-arid regions of the world (Pons et al., 2011). It is estimated that more than 800 million hectares of land throughout the world are adversely affected by high salinity (Munns and Tester, 2008). Saline soils are characterized by excess of sodium ions with dominant anions of chloride and sulfate resulting in higher electrical conductivity (>4 dS m−1) (Ali et al., 2013). In general, salinity stress induces an initial osmotic stress and subsequent toxicity as a consequence of the accumulation of ions. However, damage can also ensue as a result of excessive reactive oxygen species (ROS) such as superoxide radicals (O2−), hydrogen peroxide (H2O2) and hydroxyl radicals (•OH) produced at a high rate commonly accumulated in plant tissues due to ion imbalance and hyperosmotic stresses. ROS accumulation leads to lipid oxidation and has a negative effect on cellular metabolism and physiology, thus adversely ruining the membrane integrity (Munns et al., 2006). Salinity tolerance in glycophytic crops including rice is predominantly associated with the maintenance of ion homeostasis, particularly low Na+/K+ or high K+/Na+ ratios, through exclusion, compartmentation, and partitioning of Na+ (Blumwald, 2000). In addition to ion homeostasis strategies, many plants have evolved mechanisms to regulate the synthesis and accumulation of compatible solutes such as proline and glycine betaine, which function as osmoprotectants and have a crucial role in plant adaptation to osmotic stress through stabilization of the tertiary structure of proteins (Zhu, 2002, Munns and Tester, 2008).
Rice is the most important global food crop that feeds over half of the world population and more than 400 million people in rice producing areas of Asia, Africa and South America are still getting a major proportion of their energy requirement from rice and its derived products with the demand for food expected to increase by another 38% within 30 years (Surridge, 2004, Joseph et al., 2010). However, rice productivity in many areas is affected by salinity stress, which originates from the accumulation of underground salt and is exacerbated by salt mining, deforestation and irrigation (Akbar, 1986). Rice is generally characterized as a salt sensitive crop but the extent of its sensitivity varies during different growth and developmental stages. It is tolerant to salinity stress during germination and active tillering, whereas it displays more sensitivity during early vegetative and reproductive stages (Lutts et al., 1995, Zhu et al., 2001). A large body of literature is available for common white rice varieties, whereas data on the salt tolerance ability and physiological traits of highly nutritive pigmented rice are relatively scarce. Recently, black glutinous rice or pigmented rice has received increasing attention due to high amounts of bioactive properties and numerous health benefits. Evaluation and comparison of growth and physiological traits of rice genotypes subjected to salinity stress using principle component analysis (PCA) to explore relationship between physiological and biochemical parameters, are considered as one useful way for breeders to better understand physiological changes during development of rice varieties grown on salt-affected soils, which are essential for the development of rice with higher salt tolerant trait. The present study was therefore, conducted to explore growth and physiological changes in seedlings subjected to salinity stress in rice varieties differing in their level of salt tolerance along with classification of rice varieties with diverse growth and physiological parameters employing PCA. The study was conducted on rice seedlings of twelve genotypes, composed of four white and eight black glutinous rice cultivars. The information gained may be used to assist in the evaluation of relative field performance of different rice genotypes and characterization of contributing physiological traits that may be employed as reliable indicators for breeding and selection for salt tolerance.
2. Materials and methods
2.1. Plant materials, plant culture and salinity stress
Twelve rice (Oryza sativa L.) cultivars, including eight local black glutinous rice cultivars kindly provided by the Faculty of Agriculture of Khon Kaen University and four white rice cultivars obtained from Biotech, Thailand, were screened for their salt tolerance levels at the seedling stage. Pokkali, the salt-tolerant landrace from India, and the salt-susceptible IR29 were used as a standard check in salt screening.
For an establishment of seedlings rice seeds were surface sterilized, soaked in distilled water for 24 h and then germinated on wet filter papers embedded in Petri dishes. Subsequently three-day-old seedlings of each rice cultivar raised in Petri dishes were transplanted into pots (15 cm height × 30 cm diameter) each containing 7 kg of homogeneous mixture (11:6:3) of paddy husk, soil and farmyard manure. Fifteen seedlings were planted in each pot. After 21 days of transplanting, seedlings of each rice cultivar were subjected to salinity stress at 100 mM NaCl for 14 days. Sampling was performed at the end of the experiments and physiological changes were evaluated. Scoring of visual salt stress injury and growth reduction of rice seedlings treated with 100 mM NaCl were performed using the Standard Evaluation System of rice (SES, Table 1) (Gregorio et al., 1997). Seedling shoots and roots were separated into aerial and below ground part for fresh weight (FW) and dry weight (DW) determinations. The determination of DW was performed after drying the seedling parts in a hot-air oven at 60 °C for 3 days. Evaluations of ion concentration, proline content, total chlorophyll (TC) content, total anthocyanin (TA) content, H2O2 content and antioxidant enzyme activities (POX and CAT) were included in this study.
Table 1.
Modified standard evaluation score (SES) of visual salt injury at seedling stage (Gregorio et al., 1997).
| Score | Observation | Tolerance |
|---|---|---|
| 1 | Normal growth on leaf symptoms | Highly tolerant |
| 3 | Nearly normal growth, but leaf tips or few leaves whitish and rolled | Tolerant |
| 5 | Growth severely retarded; most leaves rolled; only a few are elongating | Moderately tolerant |
| 7 | Complete cessation of growth; most leaves dry; some plants dying | Sensitive |
| 9 | Almost all plants dead or dying | Highly sensitive |
2.2. Measurement of ion concentration
Following 3 days of oven-drying of seedling shoots at 60 °C, about 0.5 g of each dried powdered sample was digested with 10 mL of nitric acid at 300 °C, 5 mL of perchloric acid at 200 °C and 20 mL of 6 M hydrochloric acid. The concentrations of Na+ and K+ were analyzed using atomic absorption spectroscopy.
2.3. Analysis of proline
Proline content was estimated by the modified procedure of Bates et al. (1973). Approximately 0.1 g fresh weight of leaf tissues was homogenized using 5 mL of 3% aqueous sulfosalicylic acid. 2 mL of extract was used in a reaction with 2 mL of acid ninhydrin and 2 mL of glacial acetic acid and boiled at 100 °C for 1 h. The reaction was quenched by putting the tubes rapidly on ice. The resulting solutions were extracted with toluene, and the absorbance of the toluene fraction was measured at 520 nm. Proline concentration was estimated with reference to calibration curve and expressed as μg g−1 tissue FW.
2.4. Quantitative analysis of TC and TA contents
TC content was determined according to a modified method outlined by Arnon (1949). Leaf samples (0.1 g) were extracted for TC by soaking them in 10 mL of 80% acetone solution for 3 days in the dark. TC content was measured at 645 and 663 nm and was then calculated using the following equation:
where A645 and A663 represent absorbance of TC extract at 645 and 663 nm respectively, V is the total extract volume and W is the leaf fresh weight.
TA content was analyzed by the modified procedure of Abdel-Aal and Hucl (1999). Approximately 0.1 g fresh weight of leaf tissues was soaked for 72 h in 10 mL of acidified ethanol (ethanol: 1 N HCl, 85:15 v/v). The suspension was filtered through Whatman No.1 filter paper and absorbance was measured at 535 nm.
2.5. Determination of H2O2 content and antioxidant enzyme activities
For determination of H2O2 concentration, 0.1 g fresh weight of leaf tissues was extracted with 3 mL of trichloroacetic acid (0.1%, w/v) in an ice bath and centrifuged at 12,000×g for 15 min and 0.5 mL of the supernatant was added to 0.5 mL of 10 mM potassium phosphate buffer (pH 7.0) and 1 mL of 1 M potassium iodide. The absorbance of supernatant was read at 390 nm. The content of H2O2 was given on a standard curve (Velikova et al., 2000).
For extraction and assay of enzymes, 0.5 g fresh weight of leaf tissues was homogenized in 5 mL of 10 mM potassium phosphate buffer (pH 7.0) containing 4% (w/v) polyvinylpyrrolidone (PVP). The homogenate was centrifuged at 12,000×g at 4 °C for 15 min and the supernatants were immediately used for determination of enzyme activity. All steps in the preparation of the enzyme extract were carried out at 0–4 °C. An aliquot of the extract was used to determine its protein content based on the method described by Bradford (1976). A 20 μL aliquot of the supernatant was mixed with 980 μL of Bradford reagent (BioRad) and the absorbance was read at 595 nm. Protein concentration was quantified by comparison with a standard curve using bovine serum albumin.
POX activity was assayed by the method of Velikova et al. (2000). The reaction mixture contained 10 mM potassium phosphate buffer (pH 7.0), 0.2% of guaiacol and 0.04 mL of enzyme extract. To start the reaction, 3 mM of H2O2 was added to the reaction mixture and the mixture was incubated for 5 min and the absorbance was then measured at 470 nm. The activity of POX was calculated from the rate of formation of guaiacol dehydrogenation product (GDHP) using the extinction coefficient of 26.6 mM−1 cm−1 and the activity was expressed as mol GDHP min−1 mg−1 protein.
CAT activity was estimated according to the method described by Velikova et al. (2000) with minor modifications. The reaction mixture consisted of 10 mM potassium phosphate buffer (pH 7.0), 0.1 mL of enzyme extract and 0.035% of H2O2. The activity of CAT was calculated based on the rate of disappearance of H2O2, which was followed as a decline in the absorbance at 240 nm measured at 2 and 4 min after the addition of H2O2. The activity was calculated using the extinction coefficient of 40 mM−1 cm−1, and expressed as H2O2 reduced min−1 mg−1 protein.
2.6. Statistical analysis
The experiments were arranged in a randomized complete block design with five replications. The data obtained were analyzed by ANOVA and all means were separated at the p < 0.05 level using the LSD test. All calculations and data analyses were performed using the SPSS 16.0 for Windows software package. All the data obtained were converted to salt tolerance indexes before Pearson’s correlation and cluster analyses. Salt tolerance index was defined as the observed value of a target trait under a given salinity level divided by the mean value for that trait under the control (Zeng et al., 2002). PCA and Cluster analysis were performed using the FactoMineR (Factor analysis and data mining with R) package (Husson et al., 2008).
3. Results
3.1. Salinity tolerance characteristics
The salinity tolerance scores calculated for twelve rice cultivars ranged from 3.0 to 7.4. As shown in Table 2, the two white rice cultivars Pokkali and FL496 and the three black glutinous rice cultivars Niewdam Gs.no.00621, Niewdam Gs.no.21629 and KKU-LLR-065 exhibited high degrees of salinity tolerance with the salinity tolerance scores of 3.0, 3.0, 3.4, 3.4 and 3.8, respectively. Niewdam Gs.no.21427 and Niewdam Gs.no.09475 were moderately tolerant to salinity, exhibiting the salinity tolerance scores of 4.2, whereas the salinity tolerance score of KDML105 was noted at 5.0. The results also demonstrated that KKU-GL-BL-06-043, IR29 and Niewdam Gs.no.88084 were slightly susceptible to salinity with the salinity tolerance scores of 5.4, 5.4 and 5.8, respectively while the black glutinous KKU-LLR-039 was most sensitive to salt stress (salinity tolerance score = 7.4).
Table 2.
Evaluation of SES scores in rice plants following 14 days of 100 mM NaCl exposure.
| No. | Cultivars | Type of rice | Leaf color | Salinity score |
|---|---|---|---|---|
| 1 | Pokkali | White | Green | 3.0 |
| 2 | FL496 | White | Green | 3.0 |
| 3 | Niewdam Gs.no.00621 | Black glutinous | Violet | 3.4 |
| 4 | Niewdam Gs.no.21629 | Black glutinous | Green | 3.4 |
| 5 | KKU-LLR-065 | Black glutinous | Violet and green | 3.8 |
| 6 | Niewdam Gs.no.21427 | Black glutinous | Violet | 4.2 |
| 7 | Niewdam Gs.no.09475 | Black glutinous | Violet and green | 4.2 |
| 8 | KDML105 | White | Green | 5.0 |
| 9 | KKU-GL-BL-06-043 | Black glutinous | Violet and green | 5.4 |
| 10 | IR29 | White | Green | 5.4 |
| 11 | Niewdam Gs.no.88084 | Black glutinous | Green | 5.8 |
| 12 | KKU-LLR-039 | Black glutinous | Violet and green | 7.4 |
SES score: 1, Tolerant; 9, Highly susceptible.
3.2. Growth performance
Salt stress caused a decrease in growth performance of seedlings in all rice cultivars, as shown in Table 3. A slight decrease in seedling shoot FW (SFW, 8.61%) and shoot (SDW, 13.48%) was noted for the salt tolerant Pokkali whereas the FL496 exhibited a dramatic reduction in FW and DW (56.13 and 52.21%, respectively). A decrease in RFW and RDW was observed for all rice cultivars at the seedling stage. Niewdam Gs.no.00621 demonstrated a slight drop in FW (8.97%) whereas a dramatic reduction in FW (88.62%) and DW (83.33%) was noted for Niewdam Gs.no.21427. The salt tolerance indexes of seedling FW and DW were noted to vary depending upon cultivars as presented in Table 4. The salt tolerance indexes of shoot FW and DW ranged from 0.39 to 0.91 and 0.45 to 0.87 respectively. In root, the salt tolerance indexes of FW and DW ranged from 0.11 to 0.91 and 0.17 to 0.65 respectively.
Table 3.
Comparison of growth and physiological parameters in 12 rice cultivars exposed to 100 mM NaCl.
| SFW |
SDW |
RFW |
RDW |
K+/Na+ |
TC |
TA |
Proline |
H2O2 |
POX |
CAT |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | NaCl | C | NaCl | C | NaCl | C | NaCl | C | NaCl | C | NaCl | C | NaCl | C | NaCl | C | NaCl | C | NaCl | C | NaCl | |
| Pokkali | 4.99 | 4.56 | 1.19 | 1.03 | 0.98 | 0.64 | 0.24 | 0.14 | 8.10 | 3.74 | 2.03 | 1.96 | 0.30 | 0.34 | 205.74 | 376.61 | 17.04 | 28.26 | 0.394 | 0.575 | 0.015 | 0.034 |
| FL496 | 2.77 | 1.22 | 0.63 | 0.29 | 1.07 | 0.39 | 0.17 | 0.10 | 5.42 | 2.06 | 1.70 | 1.67 | 0.29 | 0.35 | 188.08 | 482.97 | 18.85 | 33.70 | 0.673 | 1.106 | 0.018 | 0.048 |
| Niewdam Gs.no.00621 | 4.48 | 2.77 | 1.02 | 0.60 | 0.48 | 0.44 | 0.15 | 0.08 | 4.43 | 1.93 | 3.22 | 2.95 | 0.83 | 0.93 | 168.99 | 370.14 | 26.51 | 50.81 | 0.608 | 1.061 | 0.019 | 0.038 |
| Niewdam Gs.no.21629 | 4.05 | 1.57 | 1.02 | 0.57 | 0.54 | 0.13 | 0.15 | 0.04 | 4.02 | 1.83 | 1.69 | 1.54 | 0.37 | 0.42 | 277.99 | 934.79 | 21.33 | 43.29 | 0.890 | 1.716 | 0.019 | 0.031 |
| KKU-LLR-065 | 3.79 | 1.85 | 0.84 | 0.48 | 0.52 | 0.21 | 0.13 | 0.04 | 3.98 | 1.50 | 1.60 | 1.43 | 0.35 | 0.41 | 190.14 | 584.90 | 19.05 | 38.34 | 0.877 | 1.541 | 0.025 | 0.038 |
| Niewdam Gs.no.21427 | 4.98 | 2.46 | 0.98 | 0.55 | 1.79 | 0.20 | 0.24 | 0.04 | 4.02 | 0.98 | 2.13 | 2.05 | 0.58 | 0.69 | 327.25 | 1102.62 | 19.96 | 41.89 | 0.645 | 1.312 | 0.042 | 0.052 |
| Niewdam Gs.no.09475 | 6.94 | 4.47 | 1.66 | 1.11 | 1.53 | 0.58 | 0.40 | 0.19 | 7.01 | 1.84 | 2.48 | 2.38 | 0.42 | 0.49 | 120.00 | 393.02 | 25.68 | 50.01 | 0.591 | 1.133 | 0.015 | 0.023 |
| KDML105 | 3.12 | 1.58 | 0.85 | 0.38 | 0.67 | 0.30 | 0.18 | 0.07 | 3.81 | 1.26 | 1.81 | 1.17 | 0.24 | 0.30 | 176.58 | 716.44 | 15.61 | 36.90 | 0.553 | 1.356 | 0.029 | 0.044 |
| KKU-GL-BL-06-043 | 6.67 | 3.45 | 1.91 | 0.93 | 1.47 | 0.84 | 0.32 | 0.14 | 6.46 | 1.83 | 2.33 | 2.13 | 0.41 | 0.47 | 234.28 | 923.62 | 26.46 | 52.87 | 0.481 | 1.026 | 0.023 | 0.030 |
| IR29 | 4.47 | 2.01 | 1.04 | 0.49 | 1.24 | 0.53 | 0.18 | 0.06 | 7.28 | 1.35 | 2.32 | 1.91 | 0.31 | 0.40 | 243.85 | 1939.08 | 19.49 | 43.69 | 0.454 | 1.165 | 0.034 | 0.039 |
| Niewdam Gs.no.88084 | 5.48 | 3.45 | 1.20 | 0.83 | 1.11 | 0.39 | 0.22 | 0.11 | 6.62 | 1.58 | 2.41 | 2.13 | 0.40 | 0.51 | 263.49 | 935.03 | 24.96 | 57.23 | 0.588 | 1.389 | 0.026 | 0.031 |
| KKU-LLR-039 | 1.33 | 0.81 | 0.33 | 0.22 | 0.19 | 0.12 | 0.06 | 0.04 | 3.06 | 0.69 | 1.49 | 1.39 | 0.39 | 0.47 | 268.34 | 972.70 | 15.67 | 34.62 | 0.635 | 1.498 | 0.027 | 0.043 |
| Mean | 4.42 | 2.52 | 1.06 | 0.62 | 0.97 | 0.40 | 0.20 | 0.09 | 5.35 | 1.72 | 2.10 | 1.89 | 0.41 | 0.48 | 222.06 | 810.99 | 20.88 | 42.63 | 0.616 | 1.240 | 0.024 | 0.038 |
| LSD⁎ | 0.80 | 0.57 | 0.18 | 0.15 | 0.34 | 0.14 | 0.04 | 0.02 | 1.57 | 0.24 | 0.27 | 0.27 | 0.06 | 0.06 | 63.59 | 189.84 | 4.52 | 5.14 | 0.181 | 0.253 | 0.010 | 0.014 |
Note: SFW – fresh weight of shoot (g plant−1), SDW – dry weight of shoot (g plant−1), RFW – fresh weight of root (g plant−1), RDW – dry weight of root (g plant−1), TA – total anthocyanin (mg g−1 FW), TC – total chlorophyll (mg g−1 FW), proline (μg g−1 FW), H2O2 – hydrogen peroxide (μmol g−1 FW), POX – peroxidase (μmol GDHP min−1 mg−1 protein), CAT – catalase (μmol H2O2 reduced min−1 mg−1 protein).
LSD (p < 0.05).
Table 4.
Salt tolerance indexesa of physiological parameters in 12 rice cultivars under salinity (100 mM NaCl).
| Cultivars | SFW | SDW | RFW | RDW | K+/Na+ | Proline | TA | TC | H2O2 | POX | CAT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pokkali | 0.91 | 0.87 | 0.65 | 0.58 | 0.46 | 1.83 | 1.12 | 0.97 | 1.66 | 1.46 | 2.24 |
| FL496 | 0.44 | 0.46 | 0.36 | 0.59 | 0.38 | 2.57 | 1.22 | 0.98 | 1.79 | 1.64 | 2.72 |
| Niewdam Gs.no.00621 | 0.62 | 0.59 | 0.91 | 0.57 | 0.44 | 2.19 | 1.13 | 0.92 | 1.92 | 1.75 | 1.97 |
| Niewdam Gs.no.21629 | 0.39 | 0.56 | 0.24 | 0.27 | 0.45 | 3.36 | 1.15 | 0.91 | 2.03 | 1.92 | 1.59 |
| KKU-LLR-065 | 0.49 | 0.57 | 0.41 | 0.29 | 0.38 | 3.08 | 1.19 | 0.89 | 2.01 | 1.76 | 1.52 |
| Niewdam Gs.no.21427 | 0.49 | 0.56 | 0.11 | 0.17 | 0.24 | 3.37 | 1.17 | 0.97 | 2.10 | 2.03 | 1.24 |
| Niewdam Gs.no.09475 | 0.64 | 0.67 | 0.38 | 0.48 | 0.26 | 3.28 | 1.17 | 0.96 | 1.95 | 1.92 | 1.59 |
| KDML105 | 0.51 | 0.45 | 0.46 | 0.37 | 0.33 | 4.06 | 1.21 | 0.64 | 2.36 | 2.45 | 1.54 |
| KKU-GL-BL-06-043 | 0.52 | 0.49 | 0.57 | 0.45 | 0.28 | 3.94 | 1.16 | 0.91 | 2.00 | 2.13 | 1.34 |
| IR29 | 0.45 | 0.47 | 0.42 | 0.33 | 0.18 | 7.95 | 1.28 | 0.82 | 2.24 | 2.56 | 1.16 |
| Niewdam Gs.no.88084 | 0.63 | 0.70 | 0.35 | 0.51 | 0.24 | 3.55 | 1.29 | 0.88 | 2.29 | 2.36 | 1.19 |
| KKU-LLR-039 | 0.61 | 0.66 | 0.64 | 0.65 | 0.23 | 3.62 | 1.20 | 0.93 | 2.21 | 2.36 | 1.63 |
Salt tolerance index was defined as the observations under salinity divided by the means of the controls.
3.3. Ion concentrations
The K+/Na+ ratios of rice seedlings were noted to decrease among all rice cultivars under salt stress (Table 3). The K+/Na+ ratio observed for IR29 dramatically decreased from 7.28 to 1.35 (5.41 folds) under salt stress whereas Pokkali demonstrated a slight reduction in the K+/Na+ ratio (2.16 folds). The salt tolerance indexes for K+/Na+ ratios noted for all rice cultivars ranged from 0.18 to 0.46 (Table 4) with Pokkali attaining the highest index.
3.4. Proline accumulation
Increment of salinity levels generally results in a reduction of seedling growth and physiological parameters. In this study, salt stress obviously induced a marked increase in proline accumulation relative to the level in the control (Table 3). A slight increment in proline accumulation was noted in the salt tolerant cultivars. The proline accumulation observed for Pokkali, FL496 and Niewdam Gs.no.00621 was 1.83, 2.57 and 2.19 folds higher than that observed for the control. Notably the salt susceptible IR29 exhibited a dramatic increase in proline accumulation (7.95 folds). Table 4 lists the salt tolerance indexes of proline accumulation among the rice cultivars examined. The results demonstrated that the salt tolerance indexes ranged from 1.83 to 7.95.
3.5. Pigment accumulations
TC and TA accumulations among twelve rice cultivars after exposure to 100 mM NaCl are presented in Table 3. It was evident that salt stress contributed to decreased TC accumulations in all rice cultivars, mostly pronounced in KDML105 (35.54%) at 100 mM NaCl. The salt tolerance indexes of TC were in the range of 0.64–0.98 (Table 4).
In general, TA accumulation was greater in black glutinous than in white rice cultivars. In the present study, TA accumulation was noted to depend on leaf color (green or violet). Subjected to 100 mM NaCl, all the rice cultivars examined accumulated more TA compared to the level in the control. TA was most accumulated in Niewdam Gs.no.88084 (28.55%) while TA accumulation slightly increased in the salt tolerant Pokkali (11.83%). The salt tolerance indexes of TA ranged from 1.12 to 1.29, as presented in Table 4.
3.6. H2O2 content and antioxidant enzyme activities
It was evident that levels of H2O2 production in Pokkali and FL496 slightly increased (1.66 and 1.79 folds, respectively) under salt stress, as presented in Table 3. By contrast, higher H2O2 production was noted in the salt susceptible cultivars (2.24, 2.29 and 2.21 folds in IR29, Niewdam Gs.no.88084 and KKU-LLR-039, respectively). The results also suggested that salinity stress caused an increase in both POX and CAT activities among all rice cultivars, but the increase in POX activity was higher in the salt sensitive IR29 (156.43%) than in the salt tolerant Pokkali (46.06%) and vice versa for CAT activity. The salt tolerance indexes of H2O2 content and antioxidant enzyme activities (POX and CAT) are presented in Table 4. The salt tolerance indexes of H2O2 concentration ranged from 1.66 to 2.36. The salt tolerance indexes of POX and CAT activities ranged from 1.46 to 2.56 and 1.16–2.72 respectively.
3.7. Pearson’s correlations
Correlation coefficients among salt tolerance indexes analyzed by Pearson’s correlation are listed in Table 5. Salt tolerance indices related to several parameters show significant correlations. The salinity tolerance scores displayed positive correlations to the POX activity (r = 0.813, p < 0.01), H2O2 (r = 0.732, p < 0.01) and TA (r = 0.514, p < 0.05), but showed a negative correlation with the K+/Na+ ratios (r = −0.782, p < 0.01) and CAT activity (r = −0.605, p < 0.05). However, no correlations were noted between the salinity tolerance score and FW and DW of shoots and roots. The K+/Na+ ratios had significantly negative correlations with the POX activity (r = −0.780, p < 0.01), proline content (r = −0.682, p < 0.01) TA (r = −0.662, p < 0.01) and H2O2 (r = −0.652, p < 0.05). TC also exhibited significantly negative correlations with H2O2 (r = −0.705, p < 0.01) and the POX activity (r = −0.640, p < 0.05). Contrarily, TA was positively correlated with both H2O2 and the POX activity. Moreover, H2O2 had a significantly positive correlation with the POX activity (r = 0.937, p < 0.01) but had a significantly negative correlation with the CAT activity (r = −0.747, p < 0.01). Likewise, the POX activity showed a negative correlation with the CAT activity.
Table 5.
Pearson’s correlation coefficients among physiological parameters from 12 rice cultivars exposed to 100 mM NaCl. Each square indicates the Pearson’s correlation coefficient of a pair of parameters.
| Parameters | Parameters |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Score | SFW | SDW | RFW | RDW | K+/Na+ | Proline | TC | TA | H2O2 | POX | CAT | |
| Score | 1 | |||||||||||
| SFW | −.050 | 1 | ||||||||||
| SDW | −.095 | .901⁎⁎ | 1 | |||||||||
| RFW | .088 | .602⁎ | .396 | 1 | ||||||||
| RDW | .174 | .612⁎ | .469 | .695⁎⁎ | 1 | |||||||
| K+/Na+ | −.782⁎⁎ | .213 | .275 | .284 | .104 | 1 | ||||||
| Proline | .481 | −.459 | −.517⁎ | −.232 | −.355 | −.682⁎⁎ | 1 | |||||
| TC | −.297 | .247 | .456 | −.030 | .243 | .157 | −.464 | 1 | ||||
| TA | .514⁎ | −.299 | −.363 | −.279 | .019 | −.662⁎⁎ | .649⁎ | −.385 | 1 | |||
| H2O2 | .732⁎⁎ | −.370 | −.412 | −.214 | −.276 | −.652⁎ | .611⁎ | −.705⁎⁎ | .657⁎ | 1 | ||
| POX | .813⁎⁎ | −.329 | −.423 | −.137 | −.159 | −.780⁎⁎ | .769⁎⁎ | −.640⁎ | .689⁎⁎ | .937⁎⁎ | 1 | |
| CAT | −.605⁎ | .236 | .180 | .297 | .531⁎ | .658⁎ | −.599⁎ | .369 | −.368 | −.747⁎⁎ | −.728⁎⁎ | 1 |
Note: SFW – fresh weight of shoot (g plant−1), SDW – dry weight of shoot (g plant−1), RFW – fresh weight of root (g plant−1), RDW – dry weight of root (g plant−1), TA – total anthocyanin (mg g−1 FW), TC – total chlorophyll (mg g−1 FW), proline (μg g−1 FW), H2O2 – hydrogen peroxide (μmol g−1 FW), POX – peroxidase (μmol GDHP min−1 mg−1 protein), CAT – catalase (μmol H2O2 reduced min−1 mg−1 protein).
Correlation was significant at the p < 0.05.
Correlation was significant at the p < 0.01.
3.8. Principle component analysis
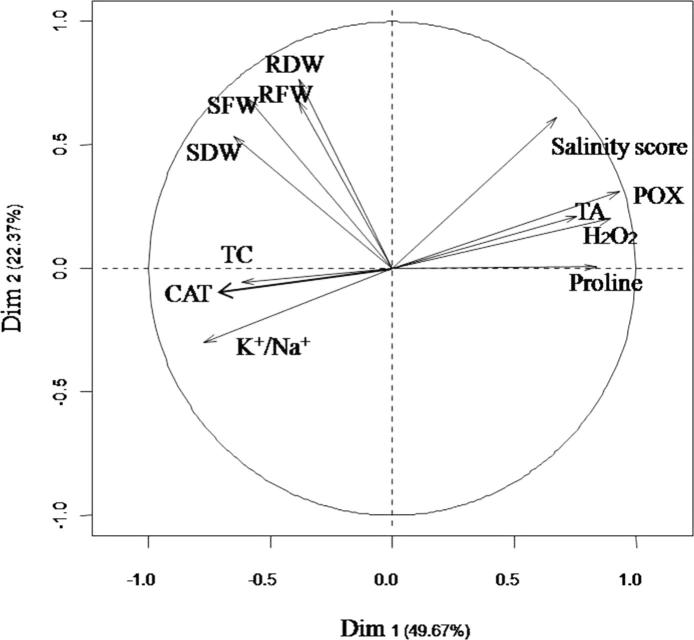
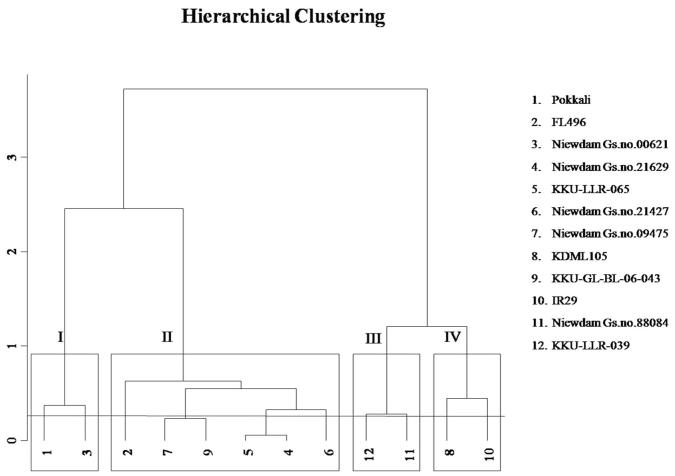
Loading plots of principle component 1 and 2 of the PCA results obtained from physiological data of twelve rice cultivars subjected to 100 mM NaCl are illustrated in Fig. 1. In this study, principle component 1 (PC1) describes 49.67% of the original information and principal component 2 (PC2) describes 22.37%. The cumulative percentage of PC1 and PC2 was 72.04%. PCA in the current study allowed for easy visualization of complex data, and the physiological parameters among twelve rice cultivars were separated by PC1 and PC2. To investigate the contributors to the principle component, the physiological loadings in PC1 and PC2 were compared. It was clear that the salinity score, POX, TA, H2O2 and proline were grouped together with positive loading on the right upper side of the biplot, suggesting that these parameters had a positive correlation among themselves. RFW, RDW, SFW and SDW were observed on the left upper side of the biplot while TC, CAT and K+/Na+ ratio were found on the left lower portion of the biplot. Hierarchical cluster analysis (HCA) was applied to search for classifiers (Fig. 2). The twelve rice cultivars were classified into four main clusters. Cluster I represented the salt-tolerant group. It is interesting to note that, other than the salt-tolerant Pokkali, the cultivar Niewdam Gs.no.00621 was the only rice cultivar that occupied this cluster. Six rice cultivars including FL496, Niewdam Gs.no.09475, KKU-GL-BL-06-043, Niewdam Gs.no.21427, Niewdam Gs.no.21629 and KKU-LLR-065 were classified into Cluster II and were considered moderately tolerant. Cluster III denoted the sensitive group, which consisted of Niewdam Gs.no.88084 and KKU-LLR-039. The salt-sensitive IR29 and KDML105 were classified into Cluster IV, which represented the highly sensitive group, and had high percentage of proline and POX activity.
Figure 1.
Loading plots of principle components 1 and 2 of the PCA results obtained from physiological data of twelve rice cultivars subjected to 100 mM NaCl.
Figure 2.
Cluster analysis of the 12 rice cultivars based on physiological parameters in salt stress condition by Hierarchical cluster analysis of the FactoMineR (Factor analysis and data mining with R) package.
4. Discussion
The modified SES of visual salt injury employed to characterize salt tolerance in twelve rice cultivars during their seedling stage development under salinity stress revealed differences in salinity tolerance both within and between groups of white rice and pigmented rice cultivars. These differences were noted to be related to the physiological parameters evaluated. Under salinity stress, root and shoot growths of all rice cultivars measured in terms of FW and DW were considerably reduced. The findings in our study that the rice roots and shoots exhibited a significant reduction in their FW and DW were consistent with Amirjani (2010) who reported that salt tolerance levels decreased as the NaCl concentration increased in rice cv. Tarom Azmoon at the seedling stage. Tatar et al. (2010) also reported salt stress significantly reduced the total dry matter of rice cultivars (Kiral and Yavuz) at the seedling stage. Likewise, Senadheera et al. (2012) observed that salt stress of 50 mM NaCl caused a significant decrease in both FW and DW of the salt-sensitive IR29 at the seedling stage.
Even though rice is able to thrive on salt-affected soils due to its ability to grow well in standing water that can help leach salts from topsoil, salt tolerance in rice varies considerably across genotypes that is due to different levels of ion homeostasis strategies rice genotypes have evolved to cope with excess Na+ (Blumwald, 2000). Rice plants with salt tolerant traits generally have high levels of maintenance of ion homeostasis, particularly low Na+/K+ or high K+/Na+ ratios, through exclusion, compartmentation and partitioning of Na+ in shoots or roots (Blumwald, 2000). In the present study, the highest K+/Na+ ratio was noted in the salt tolerant Pokkali whereas the lowest ratio was observed for the salt sensitive IR29 (Table 3). The findings in our study are in good agreement with one previous study in rice at the seedling stage (Tatar et al., 2010) that the highest K+/Na+ ratio was found in leaves and roots of the salt-tolerant IR4630 whereas it was the lowest in the salt-susceptible IR31785 under salt stress. Moreover, Senadheera et al. (2012) observed that the rice cultivar FL478 was able to maintain significantly higher K+/Na+ ratio in comparison to the cultivar IR29, which had lower K+/Na+ ratio in roots during their exposure to salt stress. Similar results were observed for the salt tolerant wheat cultivars at the seedling stage (Esfandiari et al., 2011) where the cultivars that were able to grow under salt stress conditions demonstrated a considerable increase in the K+/Na+ ratios. Several previous studies have suggested that the K+/Na+ ratios can be used as an important physiological selection criterion for salt tolerance in many plant species such as tomato (Juan et al., 2005), chickpea (Tejera et al., 2006) and barley (Türkyilmaz et al., 2011). Exclusion of Na+ from the shoots is often classified as the most essential feature of salt tolerance in plants (Munns and Tester, 2008). To reduce Na+ in the shoots plant can either minimize the entry from the root symplast to reduce loading, or maximize Na+ retrieval from the xylem (Davenport et al., 2007), or export Na+ from the leaf into the phloem (Berthomieu et al., 2003).
Apart from ion homeostasis, proline accumulation is another well-known mechanism that has been evolved to cope with drought or salinity stress in a number of plant species. Proline plays crucial roles in protecting the subcellular structures and mediating osmotic adjustment in stressed condition (Parvaiz and Satyawati, 2008, Rao et al., 2013). Furthermore, it is likely to display diverse adaptive roles including protection of cellular functions by scavenging ROS (Smirnoff and Cumbes, 1989), storage form of carbon to provide energy required for recovery (Hare and Cress, 1997) and acting as a signal molecule regulating reproductive development (Mattioli et al., 2008). Although the beneficial effects of overproduction of proline during water and salinity stress have been elucidated in transgenic rice plants (Su and Wu, 2004), the actual roles of proline accumulation are still unclear (Verbruggen and Hermans, 2008). Based on the findings in our study, it is interesting to note that increased level of proline accumulation in rice plants subjected to salinity stress did not correspond to the extent of improved salinity resistance (Table 3, Table 4). Lutts et al. (1999) demonstrated that over-accumulation of proline was related to a symptom of salt injury rather than an indicator of salt resistance. The findings in our study that increased proline accumulation did not contribute to improved salt tolerance in rice were inconsistent with Ghosh et al. (2011) who reported that the salt-tolerant Pokkali and Nonabokra exhibited several folds increase in proline in rice seedlings under salt stress. In a similar way, Kong-ngern et al. (2012) observed that the salt-sensitive KDML 105 accumulated the highest amount of root proline followed by the salt-sensitive Pathumthani 60 and the moderately tolerant Luang Anan and the tolerant Pokkali under salinity stress. In addition, Kanawapee et al. (2013) supported that under salt stress the highly susceptible cultivars accumulated the highest level of proline than the tolerant cultivars. However, Igarashi et al. (1997) suggested that proline accumulation was related to the degree of salt tolerance and rice plants accumulating high level of proline under salt stress were able to maintain higher green leaf area (Pongprayoon et al., 2008).
At the seedling stage a tendency in TC accumulation was somewhat similar among the twelve rice cultivars examined (Table 3, Table 4). However, in an attempt to compare a reduction in chlorophyll content in leaves of 13 rice cultivars subjected to salt stress, Cha-um et al. (2010) found that chlorophyll accumulation was dissimilar between the salt tolerant cultivars and the salt sensitive ones. On the contrary, the results in our study suggested that all the rice cultivars were induced to synthesize more anthocyanins under salinity stress. Similar results that anthocyanins were more produced in eight indica rice genotypes under salt stress were observed by Chutipaijit et al. (2011) who found that synthesis of anthocyanins was induced in all rice genotypes but with different levels, where the salt tolerant cultivars showed higher anthocyanin concentrations compared with the salt sensitive ones. Roychoudury et al. (2008) further demonstrated that the highest level of anthocyanin production was observed for the salt tolerant Nonabokra under salt stress. Anthocyanins are thought to minimize oxidative damage (Hughes et al., 2005) and act as antioxidants by neutralizing ROS directly (Kytridis and Manetas, 2006).
Salinity stress generally induces damage to plant tissues as a result of excessive ROS like H2O2 produced at a high rate owing to ion imbalance and hyperosmotic stresses. ROS accumulation leads to lipid oxidation and has a negative effect on cellular metabolism and physiology, thus detrimentally affecting the membrane integrity (Munns et al., 2006). In the present study, production of H2O2 was observed to be higher in the salt sensitive cultivars than in the salt tolerant. The findings in our study are in good agreement with Shahid et al. (2012) who found that salt stress enhanced H2O2 accumulation in the salt sensitive genotypes of pea. Antioxidant enzymes like POX and CAT play an important role in plant adaptation to stress conditions (Misra and Gupta, 2006). The results in our study revealed that the POX and CAT activities were found to increase in all rice cultivars subjected to salinity stress, but increased levels of their activities did not contribute to the extent of enhanced salt resistance, as observed for proline accumulation in our current study (Table 3, Table 4). It is interesting to note that the salt sensitive cultivars tended to have higher levels of POX, whereas the salt tolerant cultivars were likely to have higher levels of CAT. The findings in our study were consistent with previous studies in rice (Chawla et al., 2013). On the other hand, Dionisio-Sese and Tobita (1998) found that under salt stress the salt sensitive cultivars showed an increase in the POX activity. In plants, chloroplasts, mitochondria and peroxisomes contain a number of enzymes involved in the detoxification of ROS. Both CAT and APOX (or POX in many cases) are responsible for the conversion of H2O2 to water, however APOX is more effective than CAT (Anderson et al., 1995).
PCA is the most frequently used multivariate method that can be used to classify samples with diverse biological status, origin or quality (Liu et al., 2010, Low et al., 2012, Kim et al., 2013). The combination of PCA and HCA can classify twelve rice cultivars into four major groups using various physiological parameters to discriminate their level of salt tolerance (Figure 1, Figure 2). PCA analysis was able to clearly exclude the salt sensitive cultivars from other cultivars, which was supported well by the observations made by Sorkheh et al. (2012). Mazid et al. (2013) successfully clustered 41 rice genotypes differing in bacterial blight resistance into six major groups by means of PCA and cluster analysis, employing 13 morphological traits. Clusters I and II demonstrated lower salinity scores, proline content, POX and H2O2. Rice cultivars placed in Cluster I were however, more tolerant to salinity in terms of relatively low levels of those traits. Of all the rice cultivars evaluated, only Niewdam Gs.no.00621 was considered the most salt tolerant cultivar and was therefore placed in the Pokkali group. Cluster III and IV consisted of the cultivars that showed higher salinity scores, proline content, H2O2, POX and TA than those placed in Clusters I and II, thereby indicating the salt sensitive groups. These results suggest that cluster analysis and PCA analysis are useful tools for classification of rice cultivars into different salt tolerance groups based on their changes in physiological characteristics and antioxidant enzymes activity (POX and CAT) in response to salinity stress.
In conclusion, salt stress induced physiological changes in all rice cultivars during their seedling stage. As reported in previous studies, the K+/Na+ ratio is a reliable indicator of salt stress tolerance in rice. This study has provided an evidence that K+/Na+ is highly correlated with several physiological parameters including salt injury scores, proline, anthocyanin, H2O2, POX and CAT activity which could be used as supplementary or alternative indicators for salt tolerance screening. The black glutinous rice Niewdam Gs.no.00621 was identified as the most salt tolerant cultivar, as indicated by cluster analysis, which can be used as target cultivar for selection and breeding programs for salt tolerance improvement in the future study. The results also suggest that physiological parameters and antioxidant enzymes can be used as primary tools in the screening for salinity tolerance in rice.
Acknowledgements
This research was supported by a Grant from the Food and Functional Food Cluster under the National Research University Project of Thailand’s Office of the Higher Education Commission and research funding from Khon Kaen University to the corresponding author and Salt-tolerant Rice Research Group, Khon Kaen University. The authors are grateful to Dr. Poramet Buntaung of the Faculty of Agriculture of Khon Kaen University and Dr. Teerayut Toojinda of Biotech, Thailand for kindly providing the rice seed material used in this study.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Sumitahnun Chunthaburee, Email: sumitahnun@kkumail.com.
Anoma Dongsansuk, Email: danoma@kku.ac.th.
Jirawat Sanitchon, Email: jirawat@kku.ac.th.
Wattana Pattanagul, Email: wattana@pattanagul.com.
Piyada Theerakulpisut, Email: piythe@kku.ac.th.
References
- Abdel-Aal E.M., Hucl P.A. Rapid method for quantifying total anthocyanins in blue aleurone and purple pericarp wheats. Cereal Chem. 1999;76:350–354. [Google Scholar]
- Akbar M. Breeding for salinity tolerance in rice. In: IRRI, editor. Salt-affected Soils of Pakistan, India and Thailand. International Rice Research Institute; Manila, Philippines: 1986. pp. 39–63. [Google Scholar]
- Ali S., Gautam R.K., Mahajan R., Krishnamurthy S.L., Sharma S.K., Singh R.K., Ismail A.M. Stress indices and selectable traits in SALTOL QTL introgressed rice genotypes for reproductive stage tolerance to sodicity and salinity stresses. Field Crops Res. 2013;154:65–73. [Google Scholar]
- Amirjani M.R. Effect of NaCl on some physiological parameters of rice. Eur. J. Biol. Sci. 2010;3:6–16. [Google Scholar]
- Anderson M.D., Prasad T.K., Stewart C.R. Changes in isozyme profiles of catalase, peroxidase, and glutathione reductase during acclimation to chilling in mesocotyls of maize seedlings. Plant Physiol. 1995;109:1247–1257. doi: 10.1104/pp.109.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D.I. Copper enzymes in isolated chloroplasts. Polyphenyloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L.S., Waldren R.P., Tare I.D. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. [Google Scholar]
- Berthomieu P., Conejero G., Nublat A., Brackenbury W.J., Lambert C., Savio C., Uozumi N., Oiki S., Yamada K., Cellier F., Gosti F., Simonneau T., Essah P.A., Tester T., Véry A.A., Sentenac H., Casse F. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003;22:2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000;12:431–434. doi: 10.1016/s0955-0674(00)00112-5. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cha-um S., Ashraf M., Kirdmanee C. Screening upland rice (Oryza sativa L. ssp. indica) genotypes for salt-tolerance using multivariate cluster analysis. Afr. J. Biotechnol. 2010;9:4731–4740. [Google Scholar]
- Chawla S., Jain S., Jain V. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.) J. Plant Biochem. Biotechnol. 2013;22:27–34. [Google Scholar]
- Chutipaijit S., Cha-um S., Sompornpailin K. High contents of proline and anthocyanin increase protective response to salinity in Oryza sativa L. spp. Indica. Aust. J. Crop Sci. 2011;5:1191–1198. [Google Scholar]
- Davenport R.J., Munoz-Mayor A., Jha D., Essah P.A., Rus A., Tester M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007;30:497–507. doi: 10.1111/j.1365-3040.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- Dionisio-Sese M.L., Tobita S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998;135:1–9. [Google Scholar]
- Esfandiari E., Enayati V., Abbasi A. Biochemical and physiological changes in response to salinity in two durum wheat (Triticum turgidum L.) genotypes. Not. Bot. Hort. Agrobot. Cluj-Napoca. 2011;39:165–170. [Google Scholar]
- Gregorio, G.B., Dharmawansa, S., Mendoza, R.D., 1997. Screening rice for salinity tolerance. IRRI Discussion Paper Series NO.22, International Rice Research Institute, Manila, Philippines, pp. 2–23.
- Ghosh N., Adak M.K., Ghosh P.D., Gupta S., Sen Gupta D.N., Mandal C. Differential responses of two rice varieties to salt stress. Plant Biotechnol. Rep. 2011;5:89–103. [Google Scholar]
- Hare P.D., Cress W.A. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997;21:79–102. [Google Scholar]
- Hughes N.M., Burkey K.O., Neufeld H.S. Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytol. 2005;168:575–587. doi: 10.1111/j.1469-8137.2005.01546.x. [DOI] [PubMed] [Google Scholar]
- Husson F., Le S., Josse J. FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 2008;25:1–18. [Google Scholar]
- Igarashi Y., Yoshiba Y., Sanada Y., Yamaguchi-Shinozaki K., Wada K., Shinozaki K. Characterization of the gene for Δ1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and the salt tolerance in Oryza sativa L. Plant Mol. Biol. 1997;33:857–865. doi: 10.1023/a:1005702408601. [DOI] [PubMed] [Google Scholar]
- Joseph B., Jini D., Sujatha S. Biological and physiological perspectives of specificity in abiotic salt stress response from various rice plants. Asian J. Agric. Sci. 2010;2:99–105. [Google Scholar]
- Juan M., Rivero R.M., Romero L., Ruiz J.M. Evaluation of some nutritional and biochemical indicators in selecting salt-resistant tomato cultivars. Environ. Exp. Bot. 2005;54:193–201. [Google Scholar]
- Kanawapee N., Sanitchon J., Srihaban P., Theerakulpisut P. Physiological changes during development of rice (Oryza sativa L.) varieties differing in salt tolerance under saline field condition. Plant Soil. 2013;370:89–101. [Google Scholar]
- Kim J.K., Park S.-Y., Lim S.-H., Yeo Y., Cho H.S., Ha S.-H. Comparative metabolic profiling of pigmented rice (Oryza sativa L.) cultivars reveals primary metabolites are correlated with secondary metabolites. J. Cereal Sci. 2013;57:14–20. [Google Scholar]
- Kong-ngern K., Bunnag S., Theerakulpisut P. Proline, hydrogen peroxide, membrane stability and antioxidant enzyme activity as potential indicators for salt tolerance in rice (Oryza sativa L.) Int. J. Bot. 2012;8(2):54–65. [Google Scholar]
- Kytridis V.P., Manetas Y. Mesophyll versus epidermal anthocyanins as potential in vivo antioxidants: evidence linking the putative antioxidant role to the proximity of oxyradical source. J. Exp. Bot. 2006;57:2203–2210. doi: 10.1093/jxb/erj185. [DOI] [PubMed] [Google Scholar]
- Liu Z.-Y., Shi J.-J., Zhang L.-W., Huang J.-F. Discrimination of rice panicles by hyperspectral reflectance data based on principal component analysis and support vector classification. J. Zhejiang Univ. Sci. B. 2010;11:71–78. doi: 10.1631/jzus.B0900193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low M., Deckmyn G., Op de Beeck M., Blumenrother M.C., Osswald W., Alexou M., Jehnes S., Haberer K., Rennenberg H., Herbinger K., Haberle K.H., Bahnweg G., Hanke D., Wieser G., Ceulemans R., Matyssek R., Tausz M. Multivariate analysis of physiological parameters reveals a consistent O3 response pattern in leaves of adult European beech (Fagus sylvatica) New Phytol. 2012;196:162–172. doi: 10.1111/j.1469-8137.2012.04223.x. [DOI] [PubMed] [Google Scholar]
- Lutts S., Kinet J.M., Bouharmont J. Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 1995;46:1843–1852. [Google Scholar]
- Lutts S., Majerus V., KInet J.M. NaCl effects on proline metabolism in rice (Oryza sativa) seedlings. Physiol. Plant. 1999;105:450–458. [Google Scholar]
- Mattioli R., Marchese D., D’Angeli S., Altamura M.M., Costantino P., Trovato M. Modulation of intracellular proline levels affects flowering time and inflorescence architecture in Arabidopsis. Plant Mol. Biol. 2008;66:277–288. doi: 10.1007/s11103-007-9269-1. [DOI] [PubMed] [Google Scholar]
- Mazid M.S., Rafii M.Y., Hanafi M.M., Rahim H.A., Latif M.A. Genetic variation, heritability, divergence and biomass accumulation of rice genotypes resistant to bacterial blight revealed by quantitative traits and ISSR markers. Physiol. Plant. 2013;149(3):432–447. doi: 10.1111/ppl.12054. [DOI] [PubMed] [Google Scholar]
- Misra N., Gupta A.K. Effect of salinity and different nitrogen sources on the activity of antioxidant enzymes and indole alkaloid content in Catharanthus roseus seedlings. J. Plant Physiol. 2006;163:11–18. doi: 10.1016/j.jplph.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Munns R., James R.A., Lauchli A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006;57:1025–1043. doi: 10.1093/jxb/erj100. [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Parvaiz A., Satyawati S. Salt stress and phyto-biochemical responses of plants – a review. Plant Soil Environ. 2008;54:89–99. [Google Scholar]
- Pongprayoon W., Cha-um S., Pichakum A., Kirdmanee C. Proline profiles in aromatic rice cultivars photoautotrophically grown in response to salt stress. Int. J. Bot. 2008;4:276–282. [Google Scholar]
- Pons R., Cornejo M.-J., Sanz A. Differential salinity-induced variations in the activity of H+-pumps and Na+/H+ antiporters that are involved in cytoplasm ion homeostasis as a function of genotype and tolerance level in rice cell lines. Plant Physiol. Biochem. 2011;49:1399–1409. doi: 10.1016/j.plaphy.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Rao P.S., Mishra B., Gupta S.R. Effect of salinity and alkalinity on grain quality of tolerant, semi-tolerant and sensitive rice genotypes. Rice Sci. 2013;20:284–291. [Google Scholar]
- Roychoudury A., Basu S., Sarkar S.N., Sengupta D.N. Comparative physiological and molecular responses of common aromatic indica rice cultivar to high salinity with non-aromatic indica rice cultivars. Plant Cell Rep. 2008;27:1395–1410. doi: 10.1007/s00299-008-0556-3. [DOI] [PubMed] [Google Scholar]
- Shahid M.A., Balal R.M., Pervez M.A., Abbas T., Ashfaq M., Ghazanfar U., Afzal M., Rashid A., Garcia-Sanchez F., Mattson N.S. Differential response of pea (Pisum sativum L.) genotypes to salt stress in relation to the growth, physiological attributes antioxidant activity and organic solutes. Aust. J. Crop Sci. 2012;6:828–838. [Google Scholar]
- Smirnoff N., Cumbes Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry. 1989;28:1057–1060. [Google Scholar]
- Senadheera P., Tirimanne S., Maathuis F.J.M. Long term salinity stress reveals variety specific differences in root oxidative stress response. Rice Sci. 2012;19(1):36–43. [Google Scholar]
- Sorkheh K., Shiran B., Rouhi V., Khodambashi M., Sofo A. Salt stress induction of some key antioxidant enzymes and metabolites in eight Iranian wild almond species. Acta. Physiol. Plant. 2012;34:203–213. [Google Scholar]
- Su J., Wu R. Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Sci. 2004;166:941–948. [Google Scholar]
- Surridge C. Rice cultivation: feast or famine? Nature. 2004;428:360–361. doi: 10.1038/428360a. [DOI] [PubMed] [Google Scholar]
- Tatar O., Brueck H., Gevrek M.N., Asch F. Physiological responses of two Turkish rice (Oryza sativa L.) varieties to salinity. Turk. J. Agric. For. 2010;34:451–459. [Google Scholar]
- Tejera N.A., Soussi M., Lluch C. Physiological and nutritional indicators of tolerance to salinity in chickpea plants growing under symbiotic conditions. Environ. Exp. Bot. 2006;58:17–24. [Google Scholar]
- Türkyilmaz B., Aktaş L.Y., Güven A. Salinity induced differences in growth and nutrient accumulation in five barley cultivars. Turk. J. Field Crops. 2011;16:84–92. [Google Scholar]
- Velikova V., Yordanov I., Edreva A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant Sci. 2000;151:59–66. [Google Scholar]
- Verbruggen N., Hermans C. Proline accumulation in plants: a review. Amino acids. 2008;35:753–759. doi: 10.1007/s00726-008-0061-6. [DOI] [PubMed] [Google Scholar]
- Zeng L., Shannon M.C., Grieve C.M. Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica. 2002;127:235–245. [Google Scholar]
- Zhu G.Y., Kinet J.M., Lutts S. Characterization of rice (Oryza sativa L.) F3 populations selected for salt resistance. I. Physiological behaviour during vegetative growth. Euphytica. 2001;121:251–263. [Google Scholar]
- Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]