Abstract
Background
2,3-Butanediol (2,3-BD) can be used as a liquid fuel additive to replace petroleum oil, and as an important platform chemical in the pharmaceutical and plastic industries. Microbial production of 2,3-BD by Bacillus licheniformis presents potential advantages due to its GRAS status, but previous attempts to use this microorganism as a chassis strain resulted in the production of a mix of D-2,3-BD and meso-2,3-BD isomers.
Results
The aim of this work was to develop an engineered strain of B. licheniformis suited to produce the high titers of the pure meso-2,3-BD isomer. Glycerol dehydrogenase (Gdh) was identified as the catalyst for D-2,3-BD biosynthesis from its precursor acetoin in B. licheniformis. The gdh gene was, therefore, deleted from the wild-type strain WX-02 to inhibit the flux of acetoin to D-2,3-BD biosynthesis. The acoR gene involved in acetoin degradation through AoDH ES was also deleted to provide adequate flux from acetoin towards meso-2,3-BD. By re-directing the carbon flux distribution, the double-deletion mutant WX-02ΔgdhΔacoR produced 28.2 g/L of meso-2,3-BD isomer with >99 % purity. The titer was 50 % higher than that of the wide type. A bench-scale fermentation by the double-deletion mutant was developed to further improve meso-2,3-BD production. In a fed-batch fermentation, meso-2,3-BD titer reached 98.0 g/L with a purity of >99.0 % and a productivity of 0.94 g/L–h.
Conclusions
This work demonstrates the potential of producing meso-2,3-BD with high titer and purity through metabolic engineering of B. licheniformis.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-016-0522-1) contains supplementary material, which is available to authorized users.
Keywords: Meso-2, 3-Butanediol, Bacillus licheniformis, gdh Gene, Metabolic engineering
Background
A significant research has been conducted for microbial production of 2,3-butanediol (2,3-BD), as a potential liquid fuel additive for petroleum replacement. As an important platform chemical, 2,3-BD has wide applications in the pharmaceutical and plastic industries [1, 2]. In nature, 2,3-BD exists in three isomeric forms, such as D-2,3-BD, L-2,3-BD, and meso-2,3-BD, with each isomer having its own unique applications. For example, optically active D-2,3-BD and L-2,3-BD can be used as building blocks in the synthesis of chiral compounds [3], while meso-2,3-BD can be used for producing renewable polyesters [4] and enantiomerically pure halohydrins [5], as well as microbial production of 1,3-butadiene [6], 2-butanol, and butanone [7, 8]. Natural organisms usually produce 2,3-BD in the form of a mix of two of three isomers, with the ratio of products dependent on the strains and fermentation conditions used [9]. To reduce the cost of purifying a specific 2,3-BD isomer from fermentation broth, it is desirable for the microbial strain to produce the specific target isomer without the synthesis of the other forms of 2,3-BD.
Genetic engineering has been used to produce specific 2,3-BD isomers with high purity. For example, D-2,3-BD was produced with a purity >97.5 % through an engineered Enterobacter cloacae strain to titers of 152.0 g/L [10]. Engineered Escherichia coli has also been used to produce L-2,3-BD from diacetyl with a purity >99 % [3]. A high purity (~98 %) of meso-2,3-BD was also reported through a recombinant E. coli strain [11]. However, the titer (17.7 g/L) and yield (0.18 g/g glucose) of meso-2,3-BD produced from this system were very low [11], making economic industrial application difficult.
Many microorganisms are capable of producing 2,3-BD, such as Enterobacter, Klebsiella, Serratia, and Bacillus. Among them, Bacillus licheniformis is a promising chassis strain for 2,3-BD production. It is generally regarded as safe (GRAS) and used as an expression platform for producing enzymes, amino acids, antibiotics, and other secondary metabolites [12]. Natural B. licheniformis strains usually produce a mixture of D-2,3-BD and meso-2,3-BD isomers. Figure 1 shows the proposed 2,3-BD biosynthesis pathway in natural B. licheniformis. The precursor of 2,3-BD isomers is D-(−) acetoin (D-AC), which is reduced to D-2,3-BD and meso-2,3-BD through two 2,3-butanediol dehydrogenases (BDHs), D-BDH and meso-BDH, respectively [9]. The conversion between 2,3-BD and D-AC is reversible and coupled with NAD+/NADH conversion. The enzyme meso-BDH was confirmed in our earlier study [13], while D-BDH is a putative enzyme and had not been confirmed to catalyze the conversion of D-AC to D-2,3-BD. Oxygen supply is a critical factor for AC and 2,3-BD inter-conversion. High dissolved oxygen (DO) levels lead to AC synthesis, while low DO levels result in 2,3-BD synthesis [14–16]. In addition to serving as a precursor for 2,3-BD, D-AC also serves as a carbon source by B. licheniformis when glucose is depleted from the medium [17]. This metabolism of AC is mediated by the acetoin dehydrogenase enzyme system (AoDH ES), which is encoded by the acoABCL operon. Transcription of aco operon is activated by AcoR and SigL (sigma factor), and inhibited by CcpA-based catabolite repression. It had been reported that blocking AC catabolism resulted in an accumulation of this compound, which contributed to 2,3-BD synthesis [18].
Fig. 1.
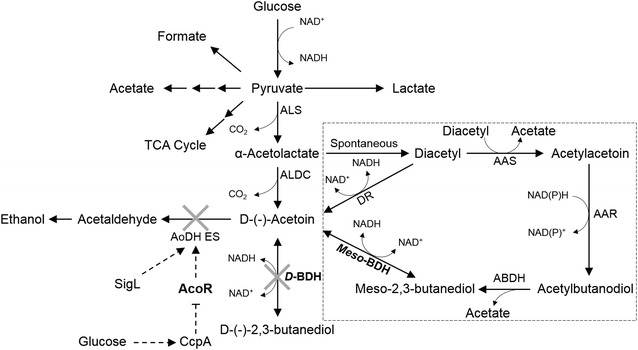
Metabolic pathway of 2,3-butanediol synthesis in B. licheniformis WX-02 (modified based on [9, 24, 41, 44]). ALS α-acetolactate synthase, ALDC α-acetolactate decarboxylase, BDH 2,3-butanediol dehydrogenase, AAS acetylacetoin synthase, AAR acetylacetoin reductase, DR diacetyl reductase, AoDH ES acetoin dehydrogenase enzyme system. The solid arrows indicate the reactions; the dashed arrows indicate the regulatory events; the dashed square marks the hypothetical 2,3-butandiol cycle; the crosses indicate the disruption of the target pathways in this work
The metabolic pathway in Fig. 1 provides guidance for engineering B. licheniformis for producing the targeted 2,3-BD isomer with high purity. Qi et al. [13] have engineered a strain to produce the pure D-2,3-BD isomer to titers of 30.8 g/L and a purity >95.4 %. However, meso-2,3-BD production by this strain had not been attempted. The aim of this work was to develop a metabolic engineering approach to produce pure meso-2,3-BD isomer by altering the carbon flux by blocking the D-2,3-BD biosynthesis and acetoin degradation pathways. In addition, the enzyme that catalyzes the conversion of D-AC to D-2,3-BD in B. licheniformis was also elucidated in this study.
Results
Identification of the enzyme catalyzing D-AC to D-2,3-BD in B. licheniformis
To produce meso-2,3-BD with high optical purity in B. licheniformis, we proposed to block the synthesis of D-2,3-BD by knocking out the gene encoding the enzyme catalyzing the conversion of D-AC to D-2,3-BD, so that the carbon flux from D-AC would be diverted into meso-2,3-BD (Fig. 1). It was hypothesized that the reversible conversion of D-AC to D-2,3-BD was catalyzed by a putative D-2,3-butanediol dehydrogenase (D-BDH) enzyme (Fig. 1). Although gene bdhA encoding the D-BDH enzyme had been identified in B. subtilis [12]), the analysis of the B. licheniformis genome did not reveal the existence of any homologs to bdhA. Alternatively, researchers have reported that the glycerol dehydrogenase (GDH) in B. licheniformis is capable of catalyzing the in vitro conversion of D-AC to D-2,3-BD [19]. Similar activities for GDHs (DhaD and GldA) were also observed in K. pneumonia [20]. Based on these results, the gdh gene in B. licheniformis was studied for its role in D-2,3-BD biosynthesis.
A 1104-bp putative GDH encoding gene (gdh) was identified in B. licheniformis WX-02 [21]. The deduced amino acid sequence of the protein (WX-02 GDH) was compared with GDHs reported in other strains via sequence alignments through ClustalW. Comparative analysis revealed that WX-02 GDH had nearly all the conserved residues of the iron-dependent alcohol dehydrogenase (Fe-ADH), including three glycine residues that are predicted to interact with the NAD(H) cofactor and three histidine residues that coordinate an iron cofactor. This suggests that the WX-02 GDH is a member of the Fe-ADH superfamily. In addition, a strong similarity (>48 %) was found between WX-02 GDH and GDHs, exhibiting D-BDH activity from other organisms, such as Serratia marcescens [22], Klebsiella pneumoniae (DhaD, KJ206474.1), Citrobacter freundii (DhaD, P45511.1), Klebsiella oxytoca (DhaD, YP_005016612), and B. licheniformis 10-1-A (GldA, AHW84748.1).
As shown in Fig. 2a, the His-tagged recombinant GDH protein was efficiently expressed in soluble form after IPTG induction. The purified GDH protein was observed as a single band on SDS-PAGE with an approximate molecular weight consistent with that predicted from its amino acid sequence (39.5 kDa). The activity of purified GDH from WX-02 was further investigated for its catalytic activities on various substrates with coenzymes. Figure 2b showed that WX-02 GDH accepted D-AC, D-2,3-BD, meso-2,3-BD, and glycerol as substrates. However, this enzyme exhibited the highest activity towards D-2,3-BD among various substrates. Its catalytic activity on glycerol was only around 5 % of that for D-2,3-BD. The preference on D-2,3-BD as the substrate by GDH was also reported in Hansenula polymorpha [23]. These results indicated that the GDH from B. licheniformis WX-02 possessed a substrate specific catalytic activity towards D-2,3-BD. Based on these observations, we concluded that the GDH enzyme was the best candidate enzyme for catalyzing D-2,3-BD synthesis in B. licheniformis WX-02. Consequently, we decided to delete the gdh gene, so the conversion of D-AC to 2,3-BD isomers could be diverted solely towards meso-2,3-BD production.
Fig. 2.
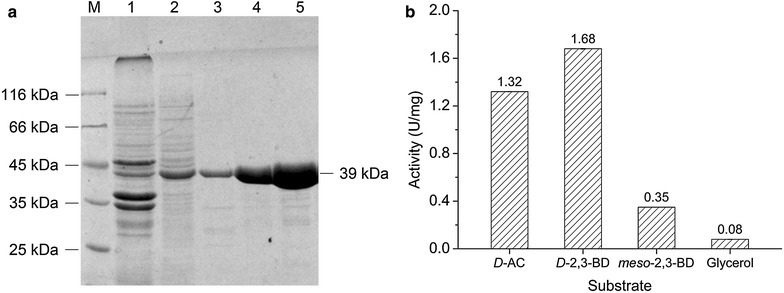
Expression and catalytic activity of GDH from B. licheniformis WX-02 (WX-02 GDH). a SDS-PAGE of the expression and purification of WX-02 GDH. The recombinant E. coli BL21(DE3)/pET-gdh was grown to the appropriate density and induced with IPTG for the production of the GDH protein. The total-cell extracts from the induced cells were separated into soluble and insoluble fractions. Proteins in the soluble fractions were purified by the Ni–NTA purification kits, and the GDH protein was purified. Lane 1 sediment of cell extracts; lane 2 supernatant of cell extracts; lane 3 purified GDH protein in 50-fold dilution; lane 4 purified GDH protein in tenfold dilution; lane 5 purified GDH protein. b Activities of purified WX-02 GDH on different substrates with corresponding coenzymes
Deletion of gdh gene for production of meso-2,3-BD with high purity
To investigate the role of GDH in the biosynthesis of 2,3-BD isomers, we constructed WX-02Δgdh, a gdh-deficient strain of B. licheniformis WX-02. As shown in Fig. 3, WX-02 produced both meso-2,3-BD and D-2,3-BD, while no D-2,3-BD was produced by WX-02Δgdh. Complementation of gdh in WX-02Δgdh (strain WX-02Δgdh/pHY-gdh) restored its ability to produce D-2,3-BD. Figure 3 also showed dramatic differences in the production of 2,3-BD and D-AC between the wild-type and the engineered strain. Compared to WX-02, production of meso-2,3-BD and D-AC increased by 66.3 and 37.8 %, respectively, in WX-02Δgdh. The results indicate that the deletion of gdh gene from the wild-type strain eliminates the synthesis of D-2,3-BD, leading to an accumulation of its precursor D-AC, which in turn promoted the conversion of D-AC to meso-2,3-BD. Meso-2,3-BD was the only 2,3-BD isomer detected in the medium, resulting in a product of high purity. It was also found that the mutant WX-02Δgdh/pHY-gdh not only restored the production of D-2,3-BD but produced an even higher titer of total 2,3-BD isomers (D-2,3-BD and meso-2,3-BD) compared to that of wild-type. Correspondingly, the D-AC produced by the complementation strain was lower than the wild-type WX-02. The distribution profile of these metabolites indicates that the high expression of gdh gene is likely a result of the strong promoter P43 and multiple copies of pHY300PLK vector.
Fig. 3.
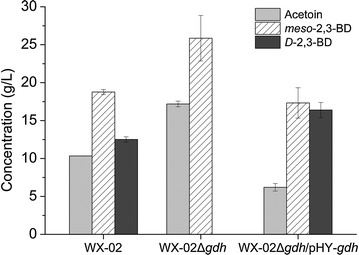
Production of acetoin and 2,3-BD isomers by B. licheniformis WX-02 and the gdh mutant strains. WX-02 wild-type strain, WX-02Δgdh the mutant strain with gdh gene knocked out from the genome of WX-02, WX-02Δgdh/PHY-gdh the mutant strain with gdh gene complemented to the knock-out strain WX-02Δgdh. The cells were grown under shake-flask culture conditions for 24 h. Data are means of three replicates, and error bars show standard deviations
Deletion of acoR gene for accumulation of acetoin
In addition to serving as the precursor for 2,3-BD synthesis, acetoin has also been reported to be used as a carbon source by B. licheniformis. This is regulated by AoDH ES (Fig. 1) when glucose was depleted [17, 24]. Acetoin dissimilation would reduce its availability for meso-2,3-BD synthesis resulting in an adverse effect on 2,3-BD production. In this work, the acoR gene encoding transcriptional activator of AoDH ES was disrupted to block the degradation of acetoin. Two separate experiments were then performed to confirm the effect of the acoR knock-out on acetoin metabolism. First, the acoR-disrupted strain WX-02ΔacoR was grown in the medium containing acetoin as the sole carbon source. The wild-type strain WX-02 was also grown in this medium as a control. As shown in Fig. 4a, the mutant WX-02ΔacoR had a very poor growth compared to that of the wild-type WX-02. The poor growth caused by the acetoin utilization deficiency was also observed in B. subtilis and B. licheniformis strains with inactivated acoABCL operons [17, 25, 26]. The results of the acetoin consumption study presented in Fig. 4a also indicated the inability of utilizing acetoin by the acoR-disrupted mutant. Second, WX-02ΔacoR and WX-02 were grown in 2,3-BD production medium containing glucose as the carbon source. As shown in Fig. 4b, the mutant WX-02ΔacoR produced 15.7 and 4.3 % more D-AC and total 2,3-BD isomers than the wild-type WX-02, respectively. The results clearly demonstrate the role of acoR in D-AC metabolism, and the deficiency of acoR contributed to D-AC accumulation, thus favoring 2,3-BD production.
Fig. 4.
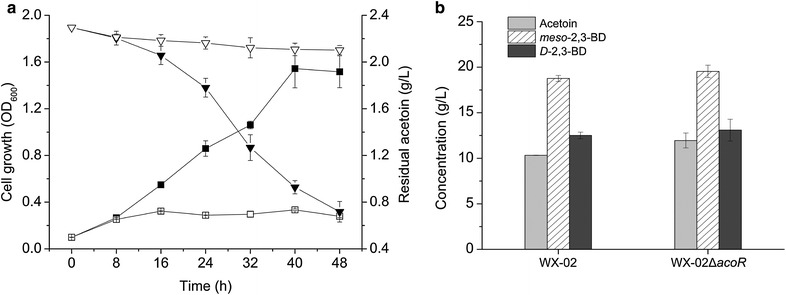
The effect of acoR knock-out on acetoin metabolism of B. licheniformis. a Time course of the growth (squares) and acetoin consumption (triangles) by the acoR-disrupted strain (WX-02ΔacoR) (open symbols) and wild-type strain (WX-02) (solid symbols) in medium with acetoin (2.3 g/L) as the sole carbon source. b The accumulation of acetoin and 2,3-BD isomers by the mutant strain (WX-02ΔacoR) and wild-type strain (WX-02) (at 24-h of culture) in medium with glucose (120 g/L) as carbon source. The cells were grown under the shake-flask culture with minerals in 2,3-BD production medium were used for each cases. Data are means of three replicates, and error bars show standard deviations
Double deletion of gdh and acoR for producing meso-2,3-BD with high purity and titer
The above results demonstrated that single deletions of gdh or acoR gene were beneficial for the production of meso-2,3-BD production in terms of both titer and purity. Compared to the wild-type strain WX-02, a gdh gene deletion resulted in the sole production of meso-2,3-BD, while deletion of the acoR gene led to an accumulation of D-AC, the precursor for 2,3-BD synthesis. To maximize meso-2,3-BD biosynthesis, we engineered a strain with both gdh and acoR genes deleted. The metabolites produced by the double-gene-deletion mutant WX-02ΔgdhΔacoR were compared to those produced by the wild-type and the single-gene deficient strains. As shown in Table 1, the mutant WX-02ΔgdhΔacoR produced 28.2 g/L of meso-2,3-BD, 50.3 % higher than that of the wild-type WX-02. The meso-2,3-BD yield and productivity of the double-deletion strain were also significantly higher with respect to the wild-type. Disruption of the gdh and acoR genes also affected the synthesis of other metabolites, such as lactic acid, acetic acid, and ethanol. Table 1 shows that the mutant strains, particularly WX-02ΔgdhΔacoR, produced lower amounts of these by-products, indicating the benefit of metabolic engineering of B. licheniformis for meso-2,3-BD production.
Table 1.
Production of various metabolites by different B. licheniformis strains
| B. licheniformis strain | Biomass (OD600) | Metabolites (g/L) | meso-2,3-BD yielda (%) | meso-2,3-BD productivityb (g/L–h) | |||||
|---|---|---|---|---|---|---|---|---|---|
| meso-2,3-BD | D-2,3-BD | Acetoin | Lactic acid | Acetic acid | Ethanol | ||||
| WX-02 | 29.60 ± 0.82 | 18.77 ± 0.33 | 12.51 ± 0.36 | 10.33 ± 0.02 | 2.21 ± 0.12 | 0.46 ± 0.06 | 0.73 ± 0.12 | 15.64 | 0.78 |
| WX-02Δgdh | 28.80 ± 0.24 | 25.86 ± 3.00 | ND | 17.19 ± 0.37 | 2.13 ± 0.02 | 0.21 ± 0.01 | 0.54 ± 0.03 | 21.55 | 1.08 |
| WX-02ΔacoR | 28.05 ± 0.88 | 19.54 ± 0.69 | 13.10 ± 1.19 | 11.96 ± 0.91 | 1.94 ± 0.07 | 0.25 ± 0.07 | 0.62 ± 0.08 | 16.29 | 0.81 |
| WX-02ΔgdhΔacoR | 30.03 ± 0.69 | 28.22 ± 0.82 | ND | 17.16 ± 0.66 | 1.63 ± 0.17 | 0.11 ± 0.04 | 0.51 ± 0.14 | 23.52 | 1.18 |
Strains were grown in 250-mL flasks containing 50-mL medium and incubated in a rotary shaker with 180 rpm at 37 °C for 24 h. The initial glucose concentration was 120 g/L, which was completely consumed by the cells at the end of culture; Data are presented as mean ± SDs of three replicates
aThe yield was calculated as the weight ratio of meso-2,3-BD produced to glucose consumed
bThe productivity was calculated by dividing maximum meso-2,3-BD concentration to the time when the cells were harvested
Optimization of meso-2,3-BD production by the mutant WX-02ΔgdhΔacoR
To this point, the previously described results demonstrated the capability of the mutant WX-02ΔgdhΔacoR to produce meso-2,3-BD with a high purity and titer. A bench-scale fermentation was further developed in both batch and fed-batch modes to explore the potential of commercial production of meso-2,3-BD by this double-deletion mutant.
Figure 5 shows the batch fermentation profiles. The growth (Fig. 5a) and glucose consumption (Fig. 5b) of the mutant WX-02ΔgdhΔacoR were similar to the wild-type WX-02 strain. The WX-02ΔgdhΔacoR strain accumulated higher acetoin (Fig. 5c), but negligible D-2,3-BD isomer (Fig. 5d) compared to the wild-type. The meso-2,3-BD isomer produced from WX-02ΔgdhΔacoR strain was double that of the wild-type strain (Fig. 5e). The yield of meso-2,3-BD from glucose, Ymeso-2,3-BD/glucose, was 0.35 g/g for WX-02ΔgdhΔacoR, also much higher than wild-type (0.16 g/g). Finally, lactic acid was still produced as the main by-product (Fig. 5f), while acetic acid and ethanol were less than 1.0 g/L throughout the fermentation process (data not shown). It should be noted that the lactic acid produced by the mutant WX-02ΔgdhΔacoR was similar to that produced by the wide type strain under fermenter culture conditions (Fig. 5f). This trend is different from that in flask culture (Table 1), where WX-02ΔgdhΔacoR produced less lactic acid than WX-02. The reason may be due to the different operation conditions between the flask culture and the fermenter culture.
Fig. 5.
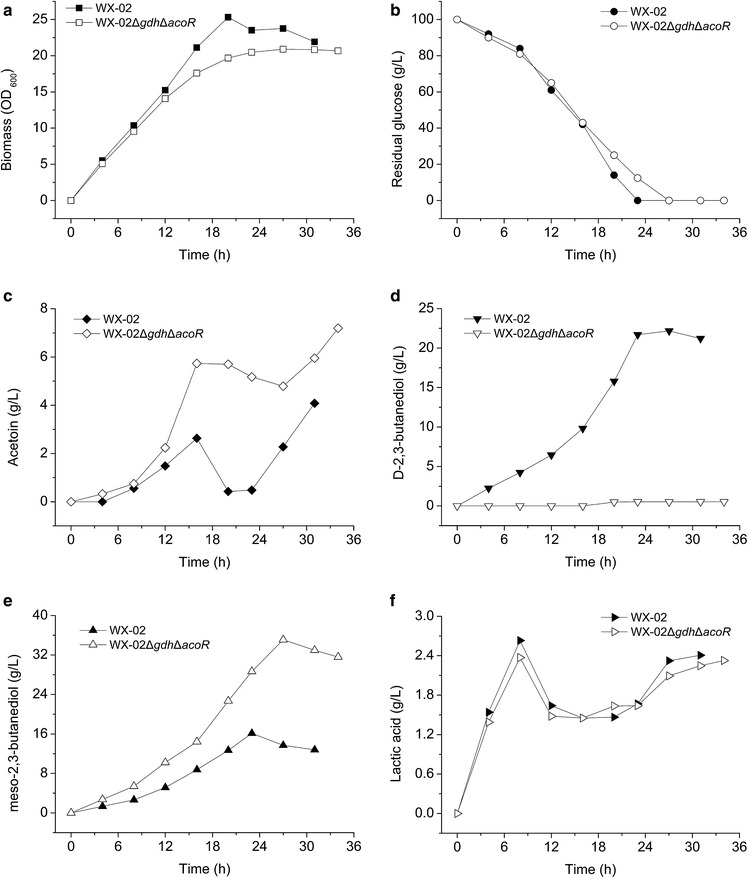
Batch fermentation profile of B. licheniformis strains WX-02 and WX-02ΔgdhΔacoR in a bench top (5-L) fermenter. a Biomass density; b residual glucose; c acetoin production; d D-2,3-BD production; e meso-2,3-BD production; f lactic acid production
Compared to the shake-flask culture results (Table 1), both the wild-type and WX-02ΔgdhΔacoR strains produced more meso-2,3-BD with less accumulation of acetoin in the fermenter culture. This was due to the use of the two-stage agitation/aeration, and thus the controlled dissolved oxygen (DO) levels, during the fermenter operation. It has been reported that DO levels play a crucial role in the reversible conversion of acetoin to 2,3-BD [14–16, 19]. In this work, the relatively high DO level generated by high aeration and agitation during the initial fermentation stage produced an increased amount of acetoin, resulting in the favorable conversion to meso-2,3-BD during the second stage when DO was intentionally reduced to a lower level.
A fed-batch fermentation using WX-02ΔgdhΔacoR was further developed to improve the meso-2,3-BD isomer titer. As shown in Fig. 6, glucose was maintained at 10-20 g/L throughout the entire culture period through periodic feedings. Meso-2,3-BD titer reached up to 98.0 g/L with a yield Ymeso-2,3-BD/glucose of 0.40 g/g and productivity of 0.94 g/L–h, which was the highest meso-2,3-BD yield reported in Bacillus species (Table 2).
Fig. 6.
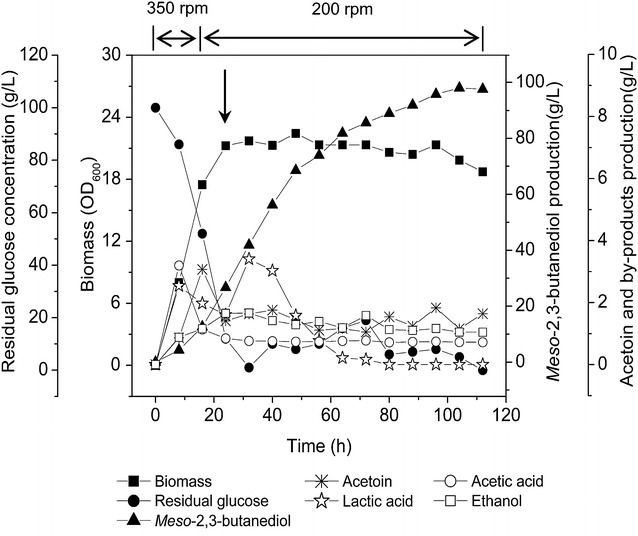
Fed-batch fermentation profiles of B. licheniformis mutant WX-02ΔgdhΔacoR in a bench top (5-L) fermenter with pH control. The tank was stirred at 350 rpm with 3-L/min aeration for first 16 h, and then decreased to 200 rpm and 1.5 mL/min for the remaining period. Glucose was fed to the medium from 24–70 h to maintain the residual glucose concentration between 10–20 g/L. Filled square Biomass; filled circle residual glucose; filled triangle meso-2,3-BD; ❊, Acetoin; ☆, Lactic acid; circle acetic acid; square ethanol. The arrow indicates the start of feeding the reactor with a concentrated glucose solution (650 g/L)
Table 2.
2,3-Butanediol (2,3-BD) production by native or engineered Bacillus strains
| Strain | Substrate | 2,3-BD isomer | Method | Concentration (g/L) | Yield (g/g) | Productivity (g/L–h) | References |
|---|---|---|---|---|---|---|---|
| Bacillus subtilis | |||||||
| AJ 1992 (WT) | Glucose | Mixture | Batch | 2.5 | 0.38 | 0.33 | [36] |
| RB03a | Glucose | Mixture | Batch | 6.1 | 0.34 | 0.41 | [37] |
| Bacillus amyloliquefaciens | |||||||
| F (WT) | Glucose | Mixture | Batch | 33.0 | 0.33 | – | [9] |
| B10-127 (WT) | Glucose | Mixture | Shake-flask | 55.8 | 0.40 | 0.80 | [38] |
| Fed-batch | 92.3 | 0.15 | 0.96 | ||||
| GARb | Crude glycerol | Mixture | Fed-batch | 102.3 | 0.44 | 1.16 | [39] |
| Bacillus licheniformis | |||||||
| Wild-type strain | Glucose | Mixture | Static flask | 8.7 | 0.47 | 0.12 | [40] |
| BL5c | Glucose | D-2,3-BD | Batch | 12.2 | 0.45 | 2.65 | [41] |
| WX-02ΔbudC | Glucose | D-2,3-BD | Shake-flask | 30.8 | 0.27 | 1.28 | [13] |
| X10 (WT) | Concentrated corn stover hydrolysate | Mixture | Fed-batch | 74.0 | 0.47 | 2.06 | [42] |
| DSM 8785 (WT) | Glucose | D-2,3-BD | Batch | 72.6 | 0.42 | 0.86 | [43] |
| Fed-batch | 144.7 | 0.40 | 1.14 | ||||
| Immobilized | 118.3 | 0.41 | 1.14 | ||||
| WX-02ΔgdhΔacoR | Glucose | Meso-2,3-BD | Shake-flask | 28.2 | 0.24 | 1.18 | This work |
| Batch | 35.1 | 0.35 | 1.30 | This work | |||
| Fed-batch | 98.0 | 0.40 | 0.94 | This work | |||
a B. subtilis 168 containing pAL10abA (PalsSD-bdhA)
b B. amyloliquefaciens B10-127 with pMA5-acr-HapII-dhaD-PbdhA-alsR
c B. licheniformis BL1Δldh
Discussion
Of the 2,3-BD isomers, meso-2,3-BD is an excellent platform chemical with numerous applications [4–8]. The ability to microbially produce meso-2,3-BD in a pure form is crucial for improving commercial viability. Examples of 2,3-BD production using native microorganisms, such as Klebsiella and Enterobacter spp., produced a mix of meso-2,3-BD and L-2,3-BD [22], whereas Bacillus spp. produced a meso-2,3-BD and D-2,3-BD mixture [19]. The resulting mixture of 2,3-BD isomers is difficult to purify with high downstream costs.
The wild-type strain B. licheniformis WX-02 produces a mix of D-2,3-BD and meso-2,3-BD [13]. It has been confirmed that different 2,3-BD isomers arise from the same precursor D-AC, and the enzyme catalyzing the reduction of D-AC to meso-2,3-BD (meso-2,3-butanediol dehydrogenases) has been identified in B.licheniformis WX-02 [13]. However, bioinformatics studies revealed that the putative gene D-2,3-BDH (D-2,3-butanediol dehydrogenases) encoding the enzyme catalyzing the reduction of D-AC to D-2,3-BD was not present in the genome of B. licheniformis WX-02. This trait is different from species, such as Paenibacillus polymyxa ZJ-9, S. cerevisiae S288c, and Clostridium beijerinckii NRRLB593, where D-2,3-BDHs were identified as the catalysts.
On the other hand, glycerol dehydrogenases (GDH) have been reported to play an important role in 2,3-BD synthesis in K. pneumoniae and S. marcescens [27, 28]. In this work, the B.licheniformis WX-02 GDH was identified. The primary structure of the enzyme is very similar to amino acid sequences of GDHs from other microbes with the D-BDH function (Additional file 1: Figure S1). This led us to explore the function of GDH as a catalyst for the conversion of D-AC to D-2,3-BD in B.licheniformis WX-02. The purified WX-02 GDH exhibited extremely high activity for D-2,3-BD (Fig. 2). Characterization and complementation of the gdh-deficient mutant B.licheniformis WX-02Δgdh further confirmed the catalytic role of WX-02 GDH in the conversion of D-AC to D-2,3-BD. A similar substrate range was observed for the GDH from other strains, such as Serratia, Klebsiella and Hansenula species [20, 23, 28].
Although phylogenetic analysis showed that the enzymes GDH and D-BDH belonged to different branches (Additional file 2: Figure S2) [29], based on homology; these two enzymes belong to the medium-chain dehydrogenase/reductase (MDR) family [20]. Other GDHs, such as O. angusta GDH, also show homology with D-BDHs (Additional file 2: Figure S2). The catalytic function of GDH in B.licheniformis WX-02 for converting D-AC to D-2,3-BD is likely due to the evolution of the enzyme [20].
The production of optically pure 2,3-BD isomers as intermediates for the pharmaceutical industry is highly desirable [20]. In this work, two metabolic engineering methods were used to produce high levels of pure meso-2,3-BD. First, the gdh gene was knocked out to block D-2,3-BD formation (Fig. 1). The engineered B.licheniformis WX-02Δgdh strain significantly increased meso-2,3-BD production and completely eliminated the production of the competing product D-2,3-BD. Second, the key transcriptional regulator AcoR in the acetoin degradation pathway was disrupted through the deletion of acoR gene. This practice proved effective in D-AC accumulation in WX-02ΔacoR; therefore, the flux from D-AC to 2,3-BD could be increased. Based on these two independent developments, a double-deletion gdh-acoR-B.licheniformis WX-02 strain was developed. The resulting strain, WX-02ΔgdhΔacoR, produced increased meso-2,3-BD titers (28.2 g/L) in shake-flask cultures, ~1.5-fold higher than the titers observed for the wild-type (Table 1). The high production of meso-2,3-BD was also accompanied by less formation of by-products, such as lactic acid, acetic acid, and ethanol (Table 1). More importantly, meso-2,3-BD was the only 2,3-BD isomer produced by the double-deletion mutant (Fig. 5; Table 1).
A two-stage aeration/agitation control method had previously proven as an efficient way to enhance the 2,3-BD isomers production. This was accomplished by controlling the dissolved oxygen (DO) levels during the fermentation. The high DO levels generated by relatively high agitation speeds favored acetoin production [14, 15, 30]. Conversely, a lower DO level was generated by decreasing the agitation speed which favored the conversion of AC to 2,3-BD [16]. Here, we used a similar strategy to improve the production of meso-2,3-BD isomer by WX-02ΔgdhΔacoR in a bench-scale fermentation system. The use of fed-batch fermentation resulted in meso-2,3-BD titers of 98.0 g/L and a productivity of 0.94 g/L–h. As a comparison, other researchers have reported titers of 73.8 g/L of meso-2,3-BD with 95.5 % purity using a recombinant E. coli [2]. Therefore, the meso-2,3-BD titers obtained in this work, together with its high purity (>99.0 %), indicate that these are the highest levels of microbial meso-2,3-BD produced so far. Table 2 further summarizes the production of various 2,3-BD isomers by Bacillus strains. The titer, yield, and purity of meso-2,3-BD isomer obtained from this study are among the highest reported in the literature, indicating that the great potential of commercially producing this important 2,3-BD isomer using the engineered B. licheniformis strains reported in this work.
Conclusions
The present study reveals the role of GDH in D-2,3-BD biosynthesis in B. licheniformis. Engineered B. licheniformis with deletions of the gdh and acoR genes was able to produce the meso-2,3-BD isomer with high titers. The fed-batch fermentation developed in this work resulted in highest titer (98.0 g/L) ever reported for meso-2,3-BD. This work demonstrates the potential of using engineered B. licheniformis, as a chassis for the industrial production of pure meso-2,3-BD.
Methods
Strains and plasmids
The strains and plasmids used in this study were listed in Table 3. Escherichia coli DH5α and BL21(DE3) were used as hosts for cloning and gene expression, respectively. All B. licheniformis strains were derived from the wild-type WX-02 (CCTCC M208065) [31]. The vectors pET-28a(+) and pHY300PLK were used for protein expression in E. coli BL21(DE3) and B. licheniformis WX-02, respectively. The T2(2)-ori vector was a shuttle plasmid for gene deletions in B. licheniformis, with a temperature-sensitive replicon from B. subtilis to promote single-crossover events [32].
Table 3.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/description | Source |
|---|---|---|
| Strains | ||
| B. licheniformis | ||
| WX-02 | CCTCC M208065, wild-type strain | Laboratory stock |
| WX-02Δgdh | gdh-knock-out mutant of WX-02 | This work |
| WX-02Δgdh/pHY-gdh | recombinant strain of WX-02Δgdh, harboring pHY-gdh, Tcr | This work |
| WX-02ΔacoR | acoR knock-out mutant of WX-02 | This work |
| WX-02ΔgdhΔacoR | gdh and acoR double-deletion mutant of WX-02 | This work |
| E. coli | ||
| DH5α | F– Φ80d/lacZΔM15, Δ (lacZYA-argF) U169, recA1, endA1, hsdR17 (r –K, m +K), phoA, supE44, λ-, thi-1, gyrA96, relA1 | Laboratory stock |
| BL21(DE3) | F–, ompT, hsdS B (r −B m −B), gal, dcm (DE3) | Laboratory stock |
| BL21(DE3)/pET-gdh | E. coli BL21(DE3) harboring pET-gdh, Kanr | This work |
| Plasmids | ||
| pET-28a(+) | E. coli expressing vector, Kanr | Laboratory stock |
| pET-gdh | pET-28a(+) carrying gdh gene | This work |
| pHY300PLK | E. coli-Bacillus shuttle vector, Ampr in E. coli, Tcr in both E. coli and B. subtilis | Laboratory stock |
| pHY-gdh | pHY300PLK carrying gdh gene | This work |
| T2(2)-ori | E. coli-B. licheniformis shuttle vector, OripUC/Orits, Kanr | Laboratory stock |
| T2Δgdh | T2(2)-ori derivative containing homologous arms for gdh-knock-out | This work |
| T2ΔacoR | T2(2)-ori derivative containing homologous arms for acoR knock-out | This work |
Tc r tetracycline resistance, Kan r kanamycin resistance gene, Amp r ampicillin resistance gene, Ori ts thermosensitive replication origin. For recombinant mutants, 100-μg/mL Ampicillin, 20-μg/mL kanamycin, or 20-µg/mL tetracycline were used when necessary
Medium preparation and culture conditions
Escherichia coli and B. licheniformis seed cultures were performed in 250-mL flasks containing 50-mL LB medium at 37 °C on a rotary shaker (180 rpm). For 2,3-BD production, the seed culture with a OD600 at 4.0 was inoculated (1 %, v/v) into either 250-mL flasks containing 50-mL medium or into a 5-L stir tank fermenter (Guo Qiang Bioengineering Equipment Company, Ltd. China) with a working volume of 3 L. The medium consisted of (per liter): glucose 120 g, corn steep liquor 33 g, (NH4)2SO4 9.0 g, K2HPO4 1.0 g, MgSO4 1.5 g, NaCl 0.5 g, ZnCl2 0.12 g, FeCl3 0.001 g, and MnSO4 0.001 g, with an initial pH adjusted to 7.0 before autoclaving at 121 °C for 15 min [13]. Fermentations were carried out at 37 °C. The batch and fed-batch fermentation were operated under optimized conditions as follows: the pH was maintained at 6.0 with 6-M HCl or 7.5-M ammonia hydroxide; aeration, and agitation were, respectively, controlled at 3 L/min and 350 rpm for the first 16 h, and 1.5 L/min, and 200 rpm for the rest of fermentation. In the fed-batch operation, in addition to the above conditions used, a concentrated glucose solution (650 g/L) was added to the fermenter at a rate of 15 mL/h from 24–70 h to maintain the glucose concentration in the medium at a level of 10–20 g/L. As a result, the total glucose added to the fermenter was equivalent to 150 g/L.
Construction of the gene deletion mutants of B. licheniformis
The mutants with single-gene deletions of gdh or acoR and a mutant with both gdh and acoR deleted were developed. Table 4 lists the primers used which were designed based on sequence information available in the NCBI database [GenBank accession No. AL009126.3 (B. subtilis 168) and CP012110.1 (B. licheniformis WX-02)].
Table 4.
Primers used in this study
| Primer name | Sequence 5′ → 3′a |
|---|---|
| Primers for gdh cloning | |
| gdh-F | CATGCCATGGGCATGTCAAAATCAGTAAAATC |
| gdh-R | CCGCTCGAGATCGTGATAAGATTCTGC |
| Primers for gdh deletion | |
| Δgdh-A-F | GCTCTAGAGATCTGACGACGAAAACAAG |
| Δgdh-A-R | ACGGAAACGGCTTTTCGTCTAGGTAATTCCCCCTTCACTATC |
| Δgdh-B-F | GATAGTGAAGGGGGAATTACCTAGACGAAAAGCCGTTTCCGT |
| Δgdh-B-R | GCGAGCTCATTATTATGGAGTACCGTGGATC |
| Δgdh-F | TCTATCTGACCTTTGATAACGGCT |
| Δgdh-R | GAATTTAGTGGTTCTGATCTGCATG |
| Primers for acoR deletion | |
| ΔacoR-A-F | GGGGTACCTGCCTCAGGCAGAGACATG |
| ΔacoR-A-R | CGTCATGCTTTGGGCGCCGAGAGGTCCATCCTCTCCTCATGCTTTTTC |
| ΔacoR-B-F | GAAAAAGCATGAGGAGAGGATGGACCTCTCGGCGCCCAAAGCATGACG |
| ΔacoR-B-R | CCGCTCGAGGGGTAAATGTGCTTCCTCCGCCT |
| ΔacoR-F | CGGAGTCCAATATGACAGG |
| ΔacoR-R | TCTGATATGAGCCATGACG |
| Primers for gdh complementation | |
| P43-gdh-TamyL-1 | GCTCTAGAGCGGAATTTCCAATTTCATG |
| P43-gdh-TamyL-2 | GATTTTACTGATTTTGACATGTGTACATTCCTCTCTTACC |
| P43-gdh-TamyL-3 | GGTAAGAGAGGAATGTACACATGTCAAAATCAGTAAAATC |
| P43-gdh-TamyL-4 | CGTCCTCTCTGCTCTTCTATCTTTTAATCGTGATAAGATTCTG |
| P43-gdh-TamyL-5 | CAGAATCTTATCACGATTAAAAGATAGAAGAGCAGAGAGGACG |
| P43-gdh-TamyL-6 | CGCGGATCCGATCACCCGCGATACCGTC |
aRestriction sites highlight in bold. Underline stands for the overlap region for splicing by overlapping extension PCR (SOE-PCR)
To prepare the gdh-knock-out mutant, in-frame deletion of gdh gene in the genome of WX-02 was performed based on the method described previously [13]. Briefly, the 5′- and 3′-flanking sequences were amplified from the WX-02 genome using the primer pairs Δgdh-A-F/Δgdh-A-R and Δgdh-B-F/Δgdh-B-R, respectively. The two fragments were ligated by spliced overlap extension PCR (SOE-PCR) with the primers Δgdh-A-F and Δgdh-B-R. The fused fragment (1140 bp) was digested with XbaI and SacI, and inserted into T2(2)-ori, generating plasmid T2Δgdh. Transformation of B. licheniformis WX-02 with the knock-out plasmid T2Δgdh was performed via electroporation based on the previous protocols [33]. The kanamycin-resistant transformants were picked and verified by PCR with the primers T2-VF and T2-VR. A positive clone was incubated in LB medium containing 20-μg/mL kanamycin at 45 °C for 8 h, then streaked onto LB agar with kanamycin for another 8-h incubation to obtain single-crossover recombinants. The recombinants were grown in LB medium at 37 °C with serial subcultures to promote homologous recombination. The kanamycin sensitive colonies resulting from a double crossover event were confirmed by PCR with the primers Δgdh-F and Δgdh-R (Additional file 3: Figure S3). The mutant strain was designated as WX-02Δgdh. The acoR deletion mutant (WX-02ΔacoR) and gdh-acoR double-deletion mutant (WX-02ΔgdhΔacoR) were prepared using the similar procedures described above. The two mutants were confirmed through PCR with the primer pairs ΔacoR-F/R and Δgdh-F/R and ΔacoR-F/R, respectively (Additional file 3: Figure S3).
Construction of the gene complementation strain of B. licheniformis WX-02Δgdh
The P43 promoter was PCR-amplified from chromosomal DNA of B. subtilis 168 with the primers P43-gdh-TamyL-1 and 2 (Table 4). The gdh gene and terminator of the amyL gene were amplified from WX-02 genomic DNA with the primers P43-gdh-TamyL-3 to 6. The three fragments were fused by SOE-PCR with the primers P43-gdh-TamyL-1 and 6 (Table 4), and cloned into the XbaI and BamHI sites of pHY300PLK, forming the plasmid pHY-gdh. The expression vector pHY-gdh was then transformed into B. licheniformis WX-02Δgdh via electroporation based on a previously described method [34]. Positive transformants with tetracycline resistance were verified by PCR with the primers P43-gdh-TamyL-1 and 6 (Additional file 4: Figure S4), and was designated as WX-02Δgdh/pHY-gdh.
Cloning and expression of gdh gene in E. coli BL21(DE3)
The gdh gene was amplified from the genome of B. licheniformis WX-02 with the primers gdh-F and gdh-R (Table 4). The amplified product was ligated into the vector pET-28a(+) at NcoI and XhoI sites, resulting in the recombinant plasmid designated as pET-gdh. The plasmid was transformed into E. coli BL21(DE3) resulting in the recombinant strain E. coli BL21(DE3)/pET-gdh (Additional file 5: Figure S5) for protein expression. E. coli BL21(DE3)/pET-gdh was cultured in LB medium containing 20-μg/mL kanamycin at 37 °C to reach cell density (OD600) of 1.0. IPTG was then added to the culture to a final concentration of 0.2 mM. Cells were grown for another 6 h and harvested by centrifugation at 8000 g for 15 min (4 °C). The cell pellets were disrupted with an Ultra-high Pressure Continuous Flow Cell Disrupter at 1000–1500 bar and 4 °C. The cell lysate was centrifuged at 12,000 g for 30 min (4 °C) to remove residual cell debris. The enzyme in the supernatant was purified using a Ni–NTA purification kits. The purity and identity of the protein were determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Assay of activity and stereospecificity of glycerol dehydrogenase (GDH)
The activity of glycerol dehydrogenase (GDH) was assayed by measuring the change in absorbance at 340 nm corresponding to the oxidation of NADH or reduction of NAD+ at 37 °C. The assay was performed in Z buffer (pH 7.0) containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 10 mM DTT [12]. We used either 4-mM NAD+ or 0.2-mM NADH as a coenzyme, dependent on the substrate used. Based on the enzyme (BDH or GDH) to be measured, substrates were glycerol, D-AC, D-2,3-BD and meso-2,3-BD, at a concentration of 50 mM. The reactions with different substrates were as follows:
Here, one unit of activity was defined as the amount of enzyme required to consume or produce 1 μmol of NADH per minute. The specific activity of GDH was defined as the enzyme unit (U) divided by the amount of enzyme protein (mg). Protein concentrations were determined by the Bradford method [35].
Analyses of metabolites during fermentation
Cells were harvested from the fermentation broth via centrifugation at 12,000g for 10 min. The cell free media was kept for analysis. The cell pellets were washed twice with deionized water and re-suspended with deionized water for determining cell density at 600 nm (OD600). Residual glucose concentration was measured enzymatically by a Bio-analyzer (SBA 40C, Shandong Academy of Sciences, China). Acetic acid, lactic acid, and ethanol were analyzed using an Agilent 1260 HPLC system. Analyte separation was achieved using Zorbax SB-Aq (4.6 mm ID × 25 0 mm, 5 μm) column with 99 % 20 mM Na2HPO4 and 1 % acetonitrile as the mobile phase (pH 2.0) at a flow rate of 0.5 mL/min. The column and detector temperatures were set at 35 °C; the injection volume was 10 μL and the detection wavelength was 210 nm. D-2,3-BD, meso-2,3-BD, and D-AC were analyzed by gas chromatography (Agilent Technologies 7890A) using the method described previously [13].
Authors’ contributions
YQ designed the study, performed the data analysis, and coordinated the manuscript draft and revision. JZ and LL executed the experimental work and wrote the manuscript. ZW participated in the design of the study and drafted the manuscript. CN participated in the data analysis and helped to revise the manuscript. SW and SC conceived the study, and coordinated the manuscript draft and revision. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Program on Key Basic Research Project (973 Program, No. 2015CB150505), and the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (No. 2013AA102801-52).
Availability of supporting data
Yes.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Yes.
Ethical approval and consent to participate
Yes.
Funding
Funding for this project has been provided by the National Program on Key Basic Research Project (973 Program, No. 2015CB150505), and the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (No. 2013AA102801-52). The funding body had no role in the design of the study, nor in the collection, analysis, or interpretation of data.
Abbreviations
- BD
butanediol
- AC
acetoin
- BDH
2,3-butanediol dehydrogenase
- Gdh
glycerol dehydrogenase
- AoDH ES
acetoin dehydrogenase enzyme system
- AmyL
amylase
- bp
base pairs
- OD
optical density
- SOE-PCR
splicing with overlapping extension PCR
- DO
dissolved oxygen
Additional file
10.1186/s13068-016-0522-1 Multiple sequence alignments of GDH from B. licheniformis WX-02 (WX-02 GDH) with GDHs from other strains.
10.1186/s13068-016-0522-1 Phylogenetic analysis of amino acid sequences of GDHs and D-BDHs from different strains.
10.1186/s13068-016-0522-1 Confirmation of the recombinant strains of B. licheniformis by PCR amplification.
10.1186/s13068-016-0522-1 Confirmation of the mutant B. licheniformis WX-02Δgdh/pHY-gdh strain by PCR amplification.
10.1186/s13068-016-0522-1 Analysis of the recombinant plasmid pET-gdh (A), and confirmation of the recombinant strain E. coli BL21(DE3)/ pET-gdh by PCR amplification (B).
Contributor Information
Yimin Qiu, Email: yimin_qiu@163.com.
Jinyan Zhang, Email: swjszjy@126.com.
Lu Li, Email: lilu045@163.com.
Zhiyou Wen, Email: wenz@iastate.edu.
Christopher T. Nomura, Email: ctnomura@esf.edu
Shuilin Wu, Email: shuilin.wu@hubu.edu.cn.
Shouwen Chen, Phone: +86 027-87280670, Email: MEL212@126.com.
References
- 1.Chu H, Xin B, Liu P, Wang Y, Li L, Liu X, Zhang X, Ma C, Xu P, Gao C. Metabolic engineering of Escherichia coli for production of (2S,3S)-butane-2,3-diol from glucose. Biotechnol Biofuels. 2015;8:143. doi: 10.1186/s13068-015-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Y, Chu H, Gao C, Tao F, Zhou Z, Li K, Li L, Ma C, Xu P. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab Eng. 2014;23:22–33. doi: 10.1016/j.ymben.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Wang Y, Zhang L, Ma C, Wang A, Tao F, Xu P. Biocatalytic production of (2S,3S)-2,3-butanediol from diacetyl using whole cells of engineered Escherichia coli. Bioresour Technol. 2012;115:111–116. doi: 10.1016/j.biortech.2011.08.097. [DOI] [PubMed] [Google Scholar]
- 4.Gubbels E, Jasinska-Walc L, Koning CE. Synthesis and characterization of novel renewable polyesters based on 2,5-furandicarboxylic acid and 2,3-butanediol. J Polym Sci Part A Polym Chem. 2013;51:890–898. doi: 10.1002/pola.26446. [DOI] [Google Scholar]
- 5.Liu R, Berglund P, Högberg H-E. Preparation of the four stereoisomers of 3-bromo-2-butanol or their acetates via lipase-catalysed resolutions of the racemates derived from dl- or meso-2,3-butanediol. Tetrahedron Asymmetry. 2005;16:2607–2611. doi: 10.1016/j.tetasy.2005.06.028. [DOI] [Google Scholar]
- 6.Shin HD, Yoon SH, Wu J, Rutter C, Kim SW, Chen RR. High-yield production of meso-2,3-butanediol from cellodextrin by engineered E. coli biocatalysts. Bioresour Technol. 2012;118:367–373. doi: 10.1016/j.biortech.2012.04.100. [DOI] [PubMed] [Google Scholar]
- 7.Ghiaci P, Norbeck J, Larsson C. 2-Butanol and butanone production in Saccharomyces cerevisiae through combination of a B12 dependent dehydratase and a secondary alcohol dehydrogenase using a TEV-based expression system. PLoS ONE. 2014;9:e102774. doi: 10.1371/journal.pone.0102774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghiaci P, Lameiras F, Norbeck J, Larsson C. Production of 2-butanol through meso-2,3-butanediol consumption in lactic acid bacteria. FEMS Microbiol Lett. 2014;360:70–75. doi: 10.1111/1574-6968.12590. [DOI] [PubMed] [Google Scholar]
- 9.Ji XJ, Huang H, Ouyang PK. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol Adv. 2011;29:351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Li K, Wang Y, Chen C, Xu Y, Zhang L, Han B, Gao C, Tao F, Ma C, Xu P. Metabolic engineering of Enterobacter cloacae for high-yield production of enantiopure (2R,3R)-2,3-butanediol from lignocellulose-derived sugars. Metab Eng. 2015;28:19–27. doi: 10.1016/j.ymben.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Ui S, Okajima Y, Mimura A, Kanai H, Kudo T. Molecular generation of an Escherichia coli strain producing only the meso-isomer of 2,3-butanediol. J Ferment Bioeng. 1997;84:185–189. doi: 10.1016/S0922-338X(97)82052-1. [DOI] [Google Scholar]
- 12.Nicholson WL. The Bacillus subtilisydjL (bdhA) gene encodes acetoin reductase 2,3-butanediol dehydrogenase. Appl Environ Microb. 2008;74:6832–6838. doi: 10.1128/AEM.00881-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi G, Kang Y, Li L, Xiao A, Zhang S, Wen Z, Xu D, Chen S. Deletion of meso-2, 3-butanediol dehydrogenase gene budC for enhanced D-2, 3-butanediol production in Bacillus licheniformis. Biotechnol Biofuels. 2014;7:16. doi: 10.1186/1754-6834-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Li S, Liu L, Wu J. Acetoin production enhanced by manipulating carbon flux in a newly isolated Bacillus amyloliquefaciens. Bioresour Technol. 2013;130:256–260. doi: 10.1016/j.biortech.2012.10.036. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Zhang L, Rao B, Han Y, Chu J, Zhu J, Shen Y, Wei D. Enhanced acetoin production by Serratia marcescens H32 using statistical optimization and a two-stage agitation speed control strategy. Biotechnol Bioproc E. 2012;17:598–605. doi: 10.1007/s12257-011-0587-4. [DOI] [Google Scholar]
- 16.Ji XJ, Huang H, Du J, Zhu JG, Ren LJ, Hu N, Li S. Enhanced 2,3-butanediol production by Klebsiella oxytoca using a two-stage agitation speed control strategy. Bioresour Technol. 2009;100:3410–3414. doi: 10.1016/j.biortech.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 17.Thanh TN, Jurgen B, Bauch M, Liebeke M, Lalk M, Ehrenreich A, Evers S, Maurer KH, Antelmann H, Ernst F, et al. Regulation of acetoin and 2,3-butanediol utilization in Bacillus licheniformis. Appl Microbiol Biot. 2010;87:2227–2235. doi: 10.1007/s00253-010-2681-5. [DOI] [PubMed] [Google Scholar]
- 18.Fu J, Wang Z, Chen T, Liu W, Shi T, Wang G, Tang Y-J, Zhao X. NADH plays the vital role for chiral pure D-(−)-2,3-butanediol production in Bacillus subtilis under limited oxygen conditions. Biotechnol Bioeng. 2014;111:2126–2131. doi: 10.1002/bit.25265. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Zhang L, Li K, Wang Y, Gao C, Han B, Ma C, Xu P. A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol Biofuels. 2013;6:123. doi: 10.1186/1754-6834-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Tao F, Xu P. Glycerol dehydrogenase plays a dual role in glycerol metabolism and 2,3-butanediol formation in Klebsiella pneumoniae. J Biol Chem. 2014;289:6080–6090. doi: 10.1074/jbc.M113.525535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yangtse W, Zhou Y, Lei Y, Qiu Y, Wei X, Ji Z, Qi G, Yong Y, Chen L, Chen S. Genome sequence of Bacillus licheniformis WX-02. J Bacteriol. 2012;194:3561–3562. doi: 10.1128/JB.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Xu Q, Zhan S, Li Y, Lin H, Sun S, Sha L, Hu K, Guan X, Shen Y. A new NAD(H)-dependent meso-2,3-butanediol dehydrogenase from an industrially potential strain Serratia marcescens H30. Appl Microbiol Biot. 2014;98:1175–1184. doi: 10.1007/s00253-013-4959-x. [DOI] [PubMed] [Google Scholar]
- 23.Yamada-Onodera K, Yamamoto H, Emoto E, Kawahara N, Tani Y. Characterisation of glycerol dehydrogenase from a methylotrophic yeast, Hansenula polymorpha Dl-1, and its gene cloning. Acta Biotechnol. 2002;22:337–353. doi: 10.1002/1521-3846(200207)22:3/4<337::AID-ABIO337>3.0.CO;2-6. [DOI] [Google Scholar]
- 24.Xiao Z, Xu P. Acetoin metabolism in bacteria. Crit Rev Microbiol. 2007;33:127–140. doi: 10.1080/10408410701364604. [DOI] [PubMed] [Google Scholar]
- 25.Huang M, Oppermann-Sanio FB, Steinbuchel A. Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J Bacteriol. 1999;181:3837–3841. doi: 10.1128/jb.181.12.3837-3841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Fu J, Zhang X, Chen T. Metabolic engineering of Bacillus subtilis for enhanced production of acetoin. Biotechnol Lett. 2012;34:1877–1885. doi: 10.1007/s10529-012-0981-9. [DOI] [PubMed] [Google Scholar]
- 27.Chen C, Wei D, Shi J, Wang M, Hao J. Mechanism of 2,3-butanediol stereoisomer formation in Klebsiella pneumoniae. Appl Microbiol Biot. 2014;98:4603–4613. doi: 10.1007/s00253-014-5526-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Xu Q, Peng X, Xu B, Wu Y, Yang Y, Sun S, Hu K, Shen Y. Cloning, expression and characterization of glycerol dehydrogenase involved in 2,3-butanediol formation in Serratia marcescens H30. J Ind Microbiol Biot. 2014;41:1319–1327. doi: 10.1007/s10295-014-1472-x. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Q, Wu J, Wu M. Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance. Process Biochem. 2014;49:1223–1230. doi: 10.1016/j.procbio.2014.05.005. [DOI] [Google Scholar]
- 31.Wei X, Ji Z, Chen S. Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-gamma-glutamic acid. Appl Biochem Biotechnol. 2010;160:1332–1340. doi: 10.1007/s12010-009-8681-1. [DOI] [PubMed] [Google Scholar]
- 32.Qiu Y, Xiao F, Wei X, Wen Z, Chen S. Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biot. 2014;98:8895–8903. doi: 10.1007/s00253-014-5978-y. [DOI] [PubMed] [Google Scholar]
- 33.Liang C, Huo Y, Qi G, Wei X, Wang Q, Chen S. Enhancement of l-valine production in Bacillus licheniformis by blocking three branched pathways. Biotechnol Lett. 2015;37:1243–1248. doi: 10.1007/s10529-015-1783-7. [DOI] [PubMed] [Google Scholar]
- 34.Tian G, Fu J, Wei X, Ji Z, Ma X, Qi G, Chen S. Enhanced expression of pgdS gene for high production of poly-γ-glutamic aicd with lower molecular weight in Bacillus licheniformis WX-02. J Chem Technol Biot. 2014;89:1825–1832. doi: 10.1002/jctb.4261. [DOI] [Google Scholar]
- 35.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Moes J, Griot M, Keller J, Heinzle E, Dunn IJ, Bourne JR. A microbial culture with oxygen-sensitive product distribution as a potential tool for characterizing bioreactor oxygen-transport. Biotechnol Bioeng. 1985;27:482–489. doi: 10.1002/bit.260270413. [DOI] [PubMed] [Google Scholar]
- 37.Biswas R, Yamaoka M, Nakayama H, Kondo T, Yoshida K, Bisaria VS, Kondo A. Enhanced production of 2,3-butanediol by engineered Bacillus subtilis. Appl Microbiol Biot. 2012;94:651–658. doi: 10.1007/s00253-011-3774-5. [DOI] [PubMed] [Google Scholar]
- 38.Yang T, Rao Z, Zhang X, Lin Q, Xia H, Xu Z, Yang S. Production of 2,3-butanediol from glucose by GRAS microorganism Bacillus amyloliquefaciens. J Basic Microbiol. 2011;51:650–658. doi: 10.1002/jobm.201100033. [DOI] [PubMed] [Google Scholar]
- 39.Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST. Enhanced 2,3-butanediol production from biodiesel-derived glycerol by engineering of cofactor regeneration and manipulating carbon flux in Bacillus amyloliquefaciens. Microb Cell Fact. 2015;14:122. doi: 10.1186/s12934-015-0317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilegaonkar S, Bhosale S, Kshirsagar D, Kapadi A. Production of 2,3-butanediol from glucose by Bacillus licheniformis. World J Microb Biot. 1992;8:378–381. doi: 10.1007/BF01198748. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Chen T, Zhao X, Chamu J. Metabolic engineering of thermophilic Bacillus licheniformis for chiral pure D-2,3-butanediol production. Biotechnol Bioeng. 2012;109:1610–1621. doi: 10.1002/bit.24427. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Li K, Wang K, Chen C, Gao C, Ma C, Xu P. Efficient production of 2,3-butanediol from corn stover hydrolysate by using a thermophilic Bacillus licheniformis strain. Bioresour Technol. 2014;170:256–261. doi: 10.1016/j.biortech.2014.07.101. [DOI] [PubMed] [Google Scholar]
- 43.Jurchescu IM, Hamann J, Zhou X, Ortmann T, Kuenz A, Prusse U, Lang S. Enhanced 2,3-butanediol production in fed-batch cultures of free and immobilized Bacillus licheniformis DSM 8785. Appl Microbiol Biot. 2013;97:6715–6723. doi: 10.1007/s00253-013-4981-z. [DOI] [PubMed] [Google Scholar]
- 44.Giovannini PP, Mantovani M, Grandini A, Medici A, Pedrini P. New acetoin reductases from Bacillus stearothermophilus: meso- and 2R,3R-butanediol as fermentation products. J Mol Catal B Enzym. 2011;69:15–20. doi: 10.1016/j.molcatb.2010.12.004. [DOI] [Google Scholar]


