Abstract
To date, more than 30 genes have been linked to monogenic diabetes. Candidate gene and genome-wide association studies have identified > 50 susceptibility loci for common type 1 diabetes (T1D) and approximately 100 susceptibility loci for type 2 diabetes (T2D). About 1–5% of all cases of diabetes result from single-gene mutations and are called monogenic diabetes. Here, we review the pathophysiological basis of the role of monogenic diabetes genes that have also been found to be associated with common T1D and/or T2D. Variants of approximately one-third of monogenic diabetes genes are associated with T2D, but not T1D. Two of the T2D-associated monogenic diabetes genes—potassium inward-rectifying channel, subfamily J, member 11 (KCNJ11), which controls glucose-stimulated insulin secretion in the β-cell; and peroxisome proliferator-activated receptor γ (PPARG), which impacts multiple tissue targets in relation to inflammation and insulin sensitivity—have been developed as major antidiabetic drug targets. Another monogenic diabetes gene, the preproinsulin gene (INS), is unique in that INS mutations can cause hyperinsulinemia, hyperproinsulinemia, neonatal diabetes mellitus, one type of maturity-onset diabetes of the young (MODY10), and autoantibody-negative T1D. Dominant heterozygous INS mutations are the second most common cause of permanent neonatal diabetes. Moreover, INS gene variants are strongly associated with common T1D (type 1a), but inconsistently with T2D. Variants of the monogenic diabetes gene Gli-similar 3 (GLIS3) are associated with both T1D and T2D. GLIS3 is a key transcription factor in insulin production and β-cell differentiation during embryonic development, which perturbation forms the basis of monogenic diabetes as well as its association with T1D. GLIS3 is also required for compensatory β-cell proliferation in adults; impairment of this function predisposes to T2D. Thus, monogenic forms of diabetes are invaluable “human models” that have contributed to our understanding of the pathophysiological basis of common T1D and T2D.
Introduction
-
Monogenic Diabetes Genes Associated With T2D, But Not T1D
KCNJ11 and ABCC8
Glucokinase
Solute carrier family 2, member 2
HNF1A and HNF4A
Hepatocyte nuclear factor 1 homeobox B
Pancreatic and duodenal homeobox 1
Paired box 4
NEUROD1/BETA2
Wolfram syndrome 1
Peroxisome proliferator-activated receptor γ
-
Monogenic Diabetes Genes Associated With Both T1D and T2D
Preproinsulin gene, INS
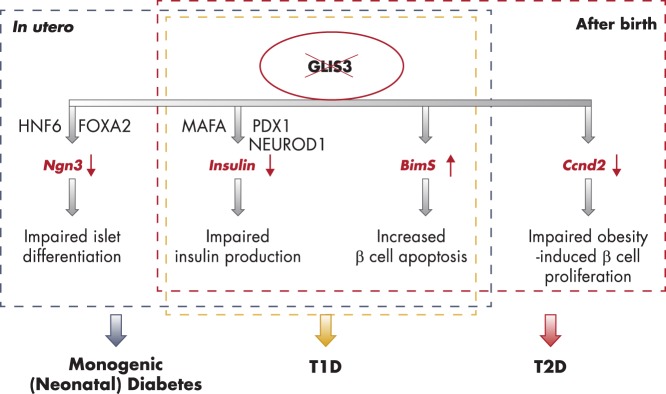
Gli-similar 3
Concluding Remarks
I. Introduction
Type 1 diabetes (type 1a; hereafter referred to as T1D) and type 2 diabetes (T2D) are of multifactorial etiology, in which genetic predisposition plays a key role. Monogenic diabetes is relatively rare. Neonatal diabetes mellitus (NDM) and maturity-onset diabetes of the young (MODY) are two major forms of monogenic diabetes, which usually result from mutations of genes for transcription factors or other proteins that regulate endocrine pancreas development or function. Mutations in a number of monogenic diabetes genes may cause NDM and MODY (Figure 1). NDM presents within the first 6 months of life and can persist throughout life (called permanent NDM [PNDM]), or it can be transient and disappear during infancy, often reappearing later in life (called transient NDM [TNDM]) (1, 2). In contrast to classic T1D (called common T1D in this article), NDM is only rarely associated with high-risk human leukocyte antigen (HLA) haplotypes or the presence of islet autoimmune antibodies (1, 3); eg, mutations of the NDM gene signal transducer and activator of transcription 3 have recently been reported to be associated with early-onset autoimmune disorders including T1D in rare cases (4). MODY is an early-onset (presenting usually before the age of 25 years) monogenic diabetes caused by mutations in a number of islet-related genes and is inherited in an autosomal dominant manner. Mutations in hepatocyte nuclear factor 1 homeobox A (HNF1A) (HNF1A-MODY or -MODY3) and glucokinase (GCK-MODY or MODY2) underlie the two most common forms of MODY (5, 6). Of note, MODY is a heterogeneous condition. The clinical phenotype can vary substantially among patients with the same MODY genotype, even among members in the same MODY family. The effect of gene-gene and gene-environment (eg, diet and exercise) interactions may underlie the heterogeneities (5).
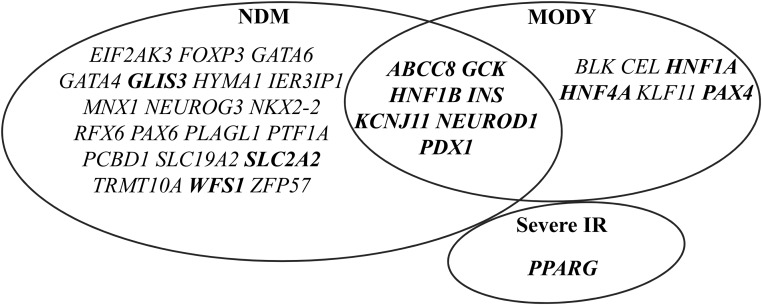
Figure 1.
Monogenic diabetes genes associated with NDM and/or MODY. Over 30 monogenic diabetes genes have been identified to cause NDM or MODY. Mutations in seven of them, including ABCC8, GCK, HNF1B, INS, KCNJ11, NEUROD1, and PDX1, may lead to both NDM and MODY. Bolded genes are discussed in this article.
To date, more than 30 genes have been linked to monogenic diabetes and related syndromes (1, 2, 5). More than 50 and 90 susceptibility loci, respectively, are found to be associated with T1D and T2D (7–11) (Figure 2). In particular, the HLA class II region of the major histocompatibility complex confers major genetic risk for T1D. Transcription factor 7-like 2 (TCF7L2) rs7903146 is one of the most robust susceptibility variants for T2D identified to date. Although T1D and T2D share some clinical and pathological similarities, GWAS indicate that their susceptibility loci are mostly distinct, and only a limited number of them have been found to be associated with both T1D and T2D (12–15). These include: INS, GLIS3, RAS guanyl nucleotide-releasing protein 1 (RASGRP1), cordon-bleu WH2 repeat protein (COBL), renalase (RNLS), and breast cancer antiestrogen resistance 1 (BCAR1). Most of the loci associated with T1D and T2D (7–10) will not be discussed further in this article because they are not known to cause monogenic diabetes. Monogenic diabetes represents unique in vivo models of disordered developmental and functional biology of pancreatic β-cells being played out, not in animals, but in people. Importantly, before the development of GWAS, monogenic diabetes genes provided the genetic landscape toward the identification of new candidate genes for the common forms of T1D and T2D. In the GWAS and post-GWAS era, the different types of monogenic diabetes continue to provide pathophysiological insight for the interpretation of GWAS data. In this context, we will focus on the mechanistic defects that underlie monogenic diabetes genes that have also been found to be associated with common T1D and/or T2D (Table 1 and Figure 2).
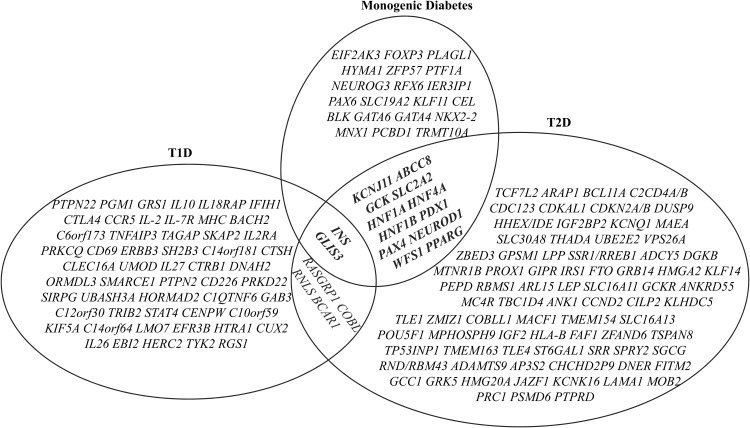
Figure 2.
Venn diagram of intersection between the loci/genes associated with T1D or T2D and known monogenic diabetes. Over 50 susceptibility loci for common T1D and approximately 100 susceptibility loci for T2D have been identified by GWAS and candidate gene association studies. INS, GLIS3, RASGRP, COBL, RNLS, and BCAR1 are associated with both T1D and T2D. About one-third of the known monogenic diabetes genes are associated with T2D. INS and GLIS3 are the two known monogenic diabetes genes whose variants are associated with both T1D and T2D.
Table 1.
Monogenic Diabetes Genes Associated With Common T1D and/or T2D
| Gene Name | Major Function | Monogenic Diabetes or Syndromes | Mutations or Variants Associated With Common T1D and/or T2D | Refs. |
|---|---|---|---|---|
| Monogenic Diabetes Genes Associated With T2D | ||||
| KCNJ11 | Encodes pore-forming inwardly-rectifying potassium channel subunits (Kir6.2) | PNDM (most common cause) and TNDM, CHI, MODY | E23K | 42–46 |
| ABCC8 | Encodes regulatory SUR1 subunits | PNDM and TNDM, CHI, MODY | A1369S, 1273AGA, R1420H | 46, 47, 52 |
| GCK | A key glucose-phosphorylating enzyme; a glucose sensor | GCK-MODY (MODY2), PNDM, CHI | rs1799884 (G/A), rs4607517 (A/G), 3′UTR SNP, chr7:44184184-G/A | 75, 78, 79 |
| SLC2A2 | Encodes GLUT2, a high-capacity facilitative glucose transporter | FBS | SNPs rs5393 (AA) and rs5394 (CC) in the promoter region and SNPs rs5400 (T110I) and rs5404 (T198T) | 93–100 |
| HNF1A/TCF1 | TF; regulator of pancreatic β-cell differentiation | HNF1A-MODY (MODY3), most common cause of MODY, CHI | G319S, c.1522G>A (p.E508K) | 114, 118, 119 |
| HNF4A | Key TF for early fetal development | HNF4A MODY (MODY1), CHI | SNPs rs2144908, rs3818247 and rs884614, rs4810424, rs1884613 | 121–124, 274 |
| HNF1B/TCF2 | TF; required for the generation of pancreatic and endocrine progenitors | RCAD syndrome, or MODY5; TNDM and PNDM (rare) | SNP rs757210 A, rs4430796 A, and rs7501939 C | 141, 144 |
| PDX1 | TF; required for pancreas development, β-cell differentiation and the maintenance of mature β-cell function | PNDM, MODY4 | C18R, Q59L, D76N, R197H, G212R, P239Q, InsCCG243, p.Gly218Alafs*12 | 163–165, 167 |
| PAX4 | Islet TF that functions mainly as a transcription repressor | MODY9 | R121W, R133W, R37W, rs10229583 G | 180, 181, 187 |
| NEUROD1/BETA2 | TF; required for the development of the endocrine pancreas; transactivates the INS gene | MODY6 and PNDM | R111L and 206 + C; A45T variant at rs1801262 (inconsistent) | 204–208 |
| WFS1 | A transmembrane protein; a negative regulator of ER stress | WFS1, sometimes referred to as DIDMOAD | R456 and H611, SNPs at rs10010131, rs6446482; variants rs10010131 G, 1801213 G, and rs734312 A | 223–225 |
| PPARG | TF; master regulator of adipogenesis, energy balance, lipid biosynthesis, and insulin sensitivity; cellular target of TZDs | Monogenic diabetes | Pro12Ala variant (rs1801282), SNP at rs4684847 | 240–243, 250 |
| Monogenic Diabetes Genes Associated With Both Common T1D and T2D | ||||
| INS | Predominant glucose-lowering hormone | PNDM (2nd most common cause), TNDM, MODY10 | Class I alleles of INS VNTR associated with T1D; Class III alleles of INS VNTR inconsistently associated with T2D | 273, 274, 276–281 |
| GLIS3 | TF; regulator of islet development, insulin gene transcription, and obesity-induced compensatory β-cell proliferation | Neonatal diabetes syndrome associated with congenital hypothyroidism, and polycystic kidneys | rs7020673 G associated with T1D; rs7034200 A and rs7041847 A associated with T2D | 78, 214, 289, 291, 292, 295–308 |
Abbreviation: TF, transcription factor.
II. Monogenic Diabetes Genes Associated With T2D, But Not T1D
GWAS revealed that approximately one-third of monogenic diabetes genes are associated with common T2D, but not T1D. The 12 monogenic diabetes genes in this category are: potassium inwardly-rectifying channel, subfamily J, member 11 (KCNJ11), ATP-binding cassette transporter subfamily C member 8 (ABCC8), GCK, solute carrier family 2, member 2 (SLC2A2), HNF1A, HNF4A, HNF1B, pancreatic and duodenal homeobox 1 (PDX1), paired box 4 (PAX4), NEUROD1/BETA2, Wolfram syndrome 1 (WFS1), and peroxisome proliferator-activated receptor γ (PPARG).
A. KCNJ11 and ABCC8
The ATP-sensitive potassium (KATP) channel is composed of four pore-forming inwardly-rectifying potassium channel subunits (Kir6.2), encoded by KCNJ11, and four regulatory sulfonylurea receptor 1 (SUR1) subunits, encoded by ABCC8. An exquisitely regulated KATP channel controls insulin secretion by coupling β-cell metabolism to calcium entry. ATP and MgADP are thought to display opposing roles in closing or opening the KATP channel. High blood glucose is taken up and metabolized by the β-cell that leads to elevated intracellular ATP level and closure of the KATP channel. The reduction in K+ efflux gives rise to membrane depolarization, resulting in opening of the voltage-gated Ca2+ channel, triggering electrical activity, Ca2+ influx, and insulin secretion (Figure 3A). Inactivating mutations in KCNJ11 or ABCC8 result in congenital hyperinsulinism (CHI), whereas activating mutations in either gene that interferes with the ATP sensitivity of the KATP channel can lead to NDM (16). Monogenic mutations in the KATP channel components have taught us how insulin secretion is regulated and how insulin secretagogues such as sulfonylureas work.
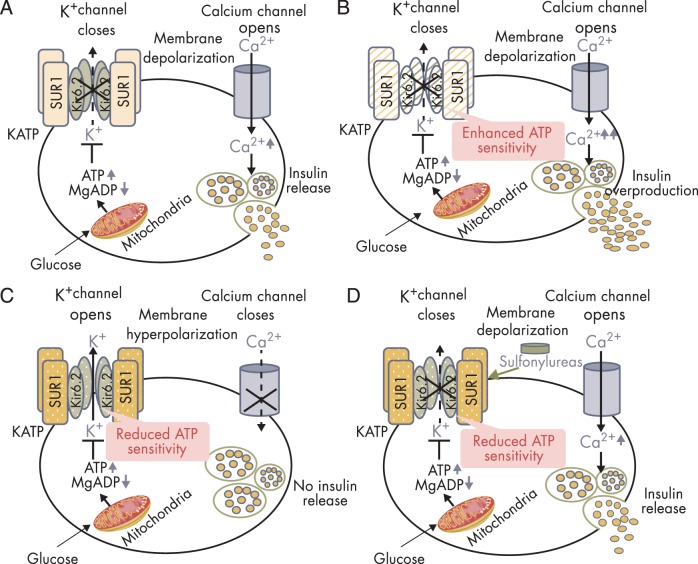
Figure 3.
Insulin secretion in normal and KATP channel mutant pancreatic β-cells. A, Glucose-stimulated insulin secretion in normal β-cells. The KATP channel is composed of four Kir6.2 subunits encoded by KCNJ11 and four SUR1 subunits encoded by ABCC8. High glucose in the circulation leads to increased glucose uptake into pancreatic β-cells. Increased intracellular glucose is metabolized via glycolytic and mitochondrial metabolism, leading to an increase in ATP production and a fall in MgADP. This results in KATP channel closure, membrane depolarization, opening of voltage-gated Ca2+ channels, Ca2+ influx, and insulin release. B, Insulin overproduction in the β-cell with KATP channel-inactivating mutations. Loss-of-function mutations in KATP channel enhances ATP binding to the channel, leading to KATP channel closure, membrane depolarization, insulin oversecretion and CHI. C, Impaired insulin secretion in the β-cell with KATP channel activating mutations. Gain-of-function mutations in KATP channel impair ATP binding to the channel Kir6.2 mutant (shown in olive green, encoded by KCNJ11), or enhance the binding of Mg-nucleotide to SUR1 mutant (shown in amber, encoded by ABCC8) leading to KATP channel opening, membrane hyperpolarization, impaired insulin release, and NDM. D, Oral sulfonylureas stimulate insulin secretion in patients with KATP channel-activating mutations. Sulfonylureas bind to the ABCC8-encoded SUR1 subunit of the KATP channel to effect channel closure independent of ATP and enable insulin secretion.
1. Inactivating mutations in the KCNJ11 and ABCC8 genes result in CHI
We will briefly review the molecular basis of CHI because it will shed light on the role of the KCNJ11 and ABCC8 genes in the regulation of insulin secretion. Loss-of-function mutations in the KCNJ11 and ABCC8 genes are the most common cause of CHI (17). Most of the disease-causing mutations in the KATP channel genes are recessively inherited, leading to medically unresponsive CHI, which frequently requires near total pancreatectomy to relieve the recurrent severe hypoglycemia. Mechanistically, these recessive mutations adversely affect the biogenesis and turnover of KATP channels, resulting in defective trafficking of channels to the plasma membrane and altered open-state frequency (18–20). Increasing numbers of mutations within the nucleotide-binding domain 2, a hotspot for dominantly acting mutations in the ABCC8 gene, have been identified in CHI patients. These include E1506K, G1479R, R1539Q, L1390R, L1431F, Q1459E, A1508P, A1537V, and R1420H (21–23). An in-frame heterozygous deletion (I284del) in the KCNJ11 gene (21) and some compound heterozygous mutations in the ABCC8 gene (24) have also been reported in CHI. These inactivating mutations cause hyperinsulinemia predominately through reduced KATP channel activity in pancreatic β-cells, which leads to abnormal membrane polarization, activation of voltage-gated calcium channels, and increased Ca2+ concentration causing insulin hypersecretion (Figure 3B).
Dominant KATP channel mutations causing CHI may predispose to the development of diabetes in adulthood. Nonpancreatectomized KATP-CHI patients eventually enter clinical remission and may progress to diabetes in later life (23–27), whereas those who are treated by partial pancreatectomy often develop late-onset diabetes that may require insulin therapy (17).
Diazoxide is one of the primary medications used to treat CHI (28). In pancreatic β-cells, diazoxide, a K+ channel opener, binds to SUR1 subunits to increase KATP channel activity, promoting K+ efflux and cell membrane hyperpolarization to block insulin release. Octreotide (a somatostatin analog) can be used for diazoxide-unresponsive CHI patients (28). The mechanism underlying the inhibition of insulin secretion by octreotide has not been fully elucidated. Octreotide inhibits Ca2+ entry into pancreatic β-cells via voltage-dependent calcium channels, leading to suppression of insulin secretion (29, 30). In addition, octreotide may hyperpolarize β-cells by activating KATP channel and thus inhibits insulin secretion (31).
2. Activating mutations in the KCNJ11 and ABCC8 genes lead to PNDM
Gain-of-function mutations in the KATP channel genes impair ATP binding to the channel, leading to KATP channel opening, membrane hyperpolarization, impaired insulin release, and NDM (Figure 3C). Activating mutations of KCNJ11 were first identified over a decade ago in patients with PNDM and account for approximately 30% of cases (32). Subsequently, gain-of-function mutations of ABCC8 were reported (33, 34); mutations of these two genes have been identified in > 40% of PNDM patients (35). Mutations in KCNJ11 and ABCC8 may also lead to TNDM. More than 100 different mutations in the KCNJ11 and ABCC8 genes have been found in patients with PNDM (36). All but one are missense mutations that cause single amino acid substitutions; the only exception to date is an in-frame 15-bp deletion in the KCNJ11 gene (37). Most of the patients with mutations in the KCNJ11 and ABCC8 genes present with isolated neonatal diabetes; a few patients display severe developmental delay, epilepsy, and NDM (called DEND syndrome) (38). In addition to causing NDM, mutations of KCNJ11 and ABCC8 have been recently linked to MODY (39, 40).
Subtle differences in the primary structure of the ABCC8-encoded SUR1 protein can produce diametrically opposite phenotypes, as illustrated by the fact that, depending on the specific amino acid substitution, mutations at the same amino acid residue may cause either CHI or NDM. For example, an E1506K substitution in the nucleotide-binding domain 2 of SUR1 leads to CHI (23, 27), whereas two other mutations at the same residue (E1506D, E1506G) were found to cause NDM (41). KATP channels are activated by Mg-nucleotides (via SUR1) and blocked by ATP (via Kir6.2). All of these mutations reduce channel activation by MgADP. Different sensitivities to ATP inhibition could be the basis of these opposing clinical phenotypes. For example, E1506D and E1506G mutant KATP channels display a markedly impaired sensitivity to ATP, resulting in hyperactive channels associated with impaired insulin secretion and PNDM. In contrast, compared to wild-type channels, the E1506K mutant channels are more sensitive to ATP inhibition, producing underactive channels that result in insulin oversecretion and CHI (41).
3. KCNJ11 and ABCC8 gene variants associated with T2D
A common variant involving a glutamate (E) to lysine (K) substitution at position 23 (E23K) in KCNJ11 has been found to be associated with increased risk of T2D (42–46). Moreover, a defect in insulin secretion was found to occur in all euglycemic E23K carriers examined (45, 46). Interestingly, population studies revealed that the KCNJ11 E23K variant has a strong allelic association with an ABCC8 coding variant S1369A. It is challenging to clearly decipher which of the two variants is the responsible gene locus. Functional analyses suggested that a combination of the two variants might underlie the increased T2D risk (46, 47). Furthermore, impaired glucose tolerance (IGT) individuals with KCNJ11 E23K are not as well protected by 1-year metformin treatment in comparison with homozygous E23E IGT individuals who receive the same treatment. A similar relative resistance to metformin-mediated protection was also observed in IGT patients with ABCC8 S1369A. The authors speculated that the diabetes-protective effect of metformin may interact with the impact of these variants on insulin regulation (47). In vitro experiments indicate that metformin can confer a direct beneficial effect on pancreatic β-cells; this first-line medication for T2D treatment was shown to restore the secretory function in isolated islets that has been impaired by chronic exposure to elevated free fatty acids or glucose (48); it also appears to preserve β-cell viability by inhibiting mitochondrial permeability transition (49).
Laukkanen et al (50) found that the silent 1273AGA variant allele of the ABCC8 gene was associated with increased risk of T2D. Importantly, this silent polymorphism was in linkage disequilibrium with three promoter polymorphisms (G-2886A, G-1561A, and A-1273G) of the ABCC8 gene; together they formed a high-risk haplotype conferring a significant 2-fold risk of T2D. Individuals with this high-risk haplotype and the KCNJ11 E23K allele displayed a 6-fold risk for the conversion to diabetes in IGT patients, whereas the KCNJ11 E23K variant alone did not predict conversion to diabetes (50); in fact, it might even be protective (51). Recently, Baier et al (52) reported that ABCC8 R1420H homozygotes eventually developed diabetes, despite being hyperinsulinemic during infancy. Furthermore, the R1420H variant was associated with higher birth weights and a 2-fold increased risk for T2D in 3.3% of an American Indian population. Their increased birth weights are suggestive of insulin oversecretion due to insulin being a major growth factor in utero. However, the physiological mechanisms underlying the reversal from hyperinsulinemia to diabetes remain elusive. The possible culprits include increased β-cell apoptosis due to elevated intracellular calcium concentration and the accompanying insulin resistance secondary to increased body weight (17, 25, 27).
In a subgroup analysis of the UK Prospective Diabetes Study (UKPDS), Gloyn et al (53) found that the presence of KCNJ11 E23K did not predict failure to sulfonylurea treatment at 1 year. Subsequently, Sesti et al (54) reported that, compared with KCNJ11 E23E homozygotes, a higher proportion of E23K carriers failed sulfonylurea-metformin combination therapy. They further showed that islets isolated from E23K carriers displayed a compromised insulin response to glibenclamide stimulation.
4. Molecular mechanisms and translational significance of mutations or variations in KCNJ11 and ABCC8 and diabetes
Mechanistically, KCNJ11 activating mutations associated with PNDM decrease ATP sensitivity by reducing ATP binding to Kir6.2 or prolonging the open state of the channel and indirectly influencing ATP sensitivity (55) (Figure 3C). Most mutations identified to date lie either in the predicted ATP-binding site or in the regions associated with channel opening and closing. Activating mutations in the KCNJ11 gene decrease the sensitivity of the KATP channel to ATP, thereby compromising the insulin secretory response. In contrast, mutations in the ABCC8 gene enhance the binding of Mg-nucleotide to nucleotide-binding domains of SUR1 or alter the intrinsic gating, leading to membrane hyperpolarization and decreased insulin secretion (56).
The details of the mechanism that underlies the association of common variants in the KCNJ11 and ABCC8 genes and T2D are still being worked out. KCNJ11 E23K is thought to exert divergent effects on activatory and inhibitory modulators. It facilitates nucleoside diphosphate-mediated channel opening, whereas it inhibits ATP-triggered channel closure, synergistically causing overactivity of KATP channels in the pancreatic β-cell and inhibition of insulin release, predisposing to T2D (57). Compared to the wild-type allele, E23K also displays significantly enhanced KATP activity in response to long-chain acyl coenzyme A, leading to impaired insulin and glucagon-like peptide-1 (GLP-1) secretion and increased glucagon release (58). A recent report showed that carriers of the KCNJ11 variant display reduced Ca2+ sensitivity of exocytosis and decreased insulin release (59). As noted above, the KCNJ11 E23K variant is strongly associated with ABCC8 S1369A. The K23/A1369 variant was found to exhibit increased KATP channel MgATPase activity in comparison with the nonrisk E23/S1369 haplotype, providing a plausible mechanism by which the E23K/S1369A haplotype increases susceptibility to T2D (60).
The identification of mutations of KCNJ11 and ABCC8 in PNDM patients has important implications for therapy because most of these patients (>90%) can be switched from insulin injections to oral sulfonylureas; patients with KATP channel defects respond to this class of agents, which enable insulin secretion by binding to the ABCC8-encoded SUR1 subunit of the KATP channel to effect channel closure independent of ATP (61) (Figure 3D). An earlier age at initiation of sulfonylurea treatment is associated with improved response to the therapy in KCNJ11-related NDM patients (62).
The therapeutic efficacy of oral sulfonylureas in T2D patients with KCNJ11 E23K mutation remains controversial. KCNJ11 E23K carriers were reported to display a marginally inferior therapeutic response in one study (63), whereas another study found that they displayed a better response to sulfonylureas (64). It is important to point out that the use of sulfonylureas did not increase the risk of hypoglycemia in KCNJ11 E23K carriers with T2D in a third study (65).
B. Glucokinase
The metabolic enzyme GCK is predominantly expressed in hepatocytes and pancreatic β-cells. GCK catalyzes the phosphorylation of glucose to glucose-6-phosphate and is involved in the first step of both glycolysis and glycogen synthesis. In pancreatic β-cells, GCK is induced by glucose and considered as the glucose sensor. Mutations in GCK lead to reduced glycolysis and ATP levels and impaired insulin secretion (5, 6). Homozygous Gck-deficient mice die perinatally of severe hyperglycemia, whereas mutant Gck heterozygotes develop mild early-onset diabetes resembling GCK-MODY in humans (66–68).
1. GCK mutations underlie MODY2
Heterozygous inactivating mutations in GCK cause GCK-MODY (MODY2), which is characterized by mild fasting hyperglycemia, asymptomatic at birth and often detected later in life during routine screening. Mutations of GCK account for 30–50% of cases of MODY in different populations (6). An inactivating mutation in GCK increases the glucose threshold needed to trigger insulin secretion, although insulin secretion remains regulated with an elevated threshold that responds to postprandial glucose excursions (69). Therefore, MODY2 patients, unlike other forms of diabetes, present with mild fasting hyperglycemia and can be diagnosed at any age (70). Homozygous inactivating GCK mutations lead to PNDM presenting at birth. Although over 600 mutations affecting different parts of the GCK gene have been reported, they are all relatively rare mutations (66). The prevalence of GCK-MODY was found to be 0.5–1% among women with gestational diabetes mellitus (GDM) in recent studies of multiethnic cohorts. It was proposed that a combination of body mass index <25 kg/m2 and fasting glucose ≥99 mg/dL differentiates GCK-MODY from common GDM (71, 72). Differentiating the two conditions may have therapeutic implications. In contrast to women with simple GDM, GCK-MODY mothers and their offspring have a low prevalence of diabetic complications despite lifetime hyperglycemia; they rarely require pharmacological treatment outside pregnancy (70, 73). It is important to note that activating GCK mutations are another cause of CHI (17, 66), which may presage late-onset diabetes. GCK-CHI causes β-cell replication early on but subsequently leads to extensive β-cell apoptosis associated with DNA double-strand breaks and activation of the tumor suppressor p53. The β-cell membrane depolarization associated with KATP channel inactivation (like inactivating mutations of the KCNJ11 and ABCC8 genes) is thought to mediate this deleterious effect in GCK-CHI patients (74).
2. Variations in the GCK gene and T2D
In a large-scale GWAS and cohort meta-analysis, a common variant rs1799884 G/A of GCK was found to be associated with glycated hemoglobin A1C levels, fasting and 2-hour glucose concentrations, as well as T2D (75). The GCK rs1799884 A allele was inconsistently associated with altered fasting glucose in Chinese populations (76, 77). In another GWAS meta-analysis, GCK rs4607517 A/G was confirmed to be associated with fasting blood glucose levels and T2D (78). In Pima Indians, a 3′ untranslated region (UTR) single nucleotide polymorphism (SNP), chr7:44184184-G/A in the GCK gene, is associated with the rate of carbohydrate oxidation postabsorptively, energy expenditure, and T2D risk, which is in agreement with the role of GCK in hepatic glycolysis and energy metabolism. However, this variant was not associated with hepatic glucose production during insulin stimulation, despite a previous report demonstrating that a GCK variant in the promoter region (-258 G/A) was associated with hepatic insulin resistance (79).
3. Molecular mechanisms that underlie the association of variations in the GCK gene and T2D
The activity of GCK can be regulated by glucose-6-phosphatase, catalytic 2 (G6PC2) in the β-cell and GCK regulatory protein (GKRP, or GCKR) in the liver. In the β-cell, G6PC2 catalyzes glucose-6-phosphate dephosphorylation, thereby functionally opposing the action of GCK. Genetic variants of either gene may affect the balance between GCK and G6PC2 activities, resulting in impaired pulsatile insulin secretion, disrupting proper insulin signaling between the pancreas and insulin-sensitive tissues, and predisposing to T2D development (80).
In the liver, the activity and subcellular localization of GCK are allosterically regulated by GKRP. GCK is sequestered to the nucleus as an inactive complex with GKRP at low glucose concentrations. It is rapidly dissociated from the GCK-GKRP complex and translocated to the cytoplasm at high glucose concentrations, which triggers glucose disposal (81, 82). It has been proposed that SNP rs780094 at GKRP may perturb the GKRP/GCK system and thus modulate the effect of SNP rs1799884 at GCK, resulting in altered glucose metabolism (77). Additional functional studies are needed to prove or disprove this hypothesis.
C. Solute carrier family 2, member 2
The SLC2A2 gene encodes glucose transporter (GLUT) 2, a high-capacity facilitative glucose transporter expressed predominantly in the liver, kidney, intestine, and pancreatic β-cells. Ablation of Slc2a2 in mice leads to severe diabetes and lethality usually at 2–3 weeks of age because of an insulin secretion defect (83). It is well established that GLUT2 is the predominant glucose transporter in rodent β-cells. A recent study suggested that GLUT2 may also mediate insulin secretion in human β-cells (84), although GLUT1, not GLUT2, is thought to be the major glucose transporter in human β-cells (85).
1. Mutations in SLC2A2 lead to Fanconi-Bickel syndrome
Homozygous patients with inactivating mutations of SLC2A2 develop Fanconi-Bickel syndrome (FBS; also known as glycogen storage disease type XI), characterized by a renal Fanconi syndrome with glycosuria, galactosuria, aminoaciduria, proteinuria and phosphaturia, short stature, rickets, poor growth, hepatomegaly, and galactose intolerance (86). Some FBS patients may present with postprandial hyperglycemia and diabetes or IGT. A large number of homozygous mutations (84, 87–90) or compound heterozygous mutations (91) in the SLC2A2 gene have been identified to underlie FBS.
Mutations in SLC2A2 were recently noted to be associated with defective insulin secretion in humans (84). Five of 104 (5%) NDM patients with unknown etiology (excluding the common genetic causes) were identified to carry homozygous SLC2A2 mutations. Four of these five patients first presented with isolated diabetes and later developed other features of FBS, suggesting that GLUT2 may play a role in human insulin secretion. Notably, some compound heterozygous mutations in the SLC2A2 gene, eg, c.457_462delCTTATA (p.153_4delLI) and c.1250C>G (p.P417R), present an unusually mild clinical course, such as modest glycosuria and tubular proteinuria. Some hallmark clinical signs of FBS, like hepatomegaly and short stature, are often absent in these patients (91). Therefore, it would be prudent to include the SLC2A2 gene for genetic screening in patients with isolated glycosuria.
2. SLC2A2 gene mutations or variants and T2D
The SNP rs5400 in the SLC2A2 gene, which causes a GLUT2 T110I substitution, was first reported to be associated with T2D in Pima Native Americans (92); subsequently, other SLC2A2 gene variants and their association with T2D have been studied extensively, although conflicting conclusions remain. To date, variants of the SLC2A2 gene are associated with increased risk for T2D in some (93, 94), but not most (95–98), studies. In the Finnish Diabetes Prevention Study, SNPs rs5393 (AA) and rs5394 (CC) in the promoter region as well as SNPs rs5400 (T110I) and rs5404 (GG, T198T) in the exon of the SLC2A2 gene have been identified to predict the conversion to diabetes in obese subjects with IGT, and rs5393 (AA) increased the risk of T2D by 3-fold. Interestingly, the rs3758947 GG of ABCC8 presents an additive effect with the rs5393 AA of SLC2A2 to predict the conversion to diabetes (99), indicating that individuals with risk SNPs in the network of genes regulating insulin secretion (SLC2A2 and ABCC8) are particularly at high risk for T2D.
Kilpeläinen et al (100) found that moderate-to-vigorous physical activity could lower the risk of conversion to T2D in the carriers of the common homozygous genotype of rs5393 (AA), rs5394 (CC), or rs5404 (GG, T198T) of SLC2A2 and rs3758947 (GG) of ABCC8, an observation that implies that improvements in insulin sensitivity by lifestyle changes help preserve the insulin secretory function of β-cells.
3. Molecular mechanisms that underlie the association of mutations or variations in the SLC2A2 gene and diabetes
In vitro experiments conducted by Michau et al (101) revealed that none of four select FBS-associated SLC2A2 mutants could transport glucose to a significant level. The G20D and S242R substitution mutants are associated with loss of protein expression at the plasma membrane, whereas the P417L and W444R mutants lose their transport capacity despite adequate membrane targeting, indicating the crucial role of the two amino acids (417 Pro and 444 Trp) for GLUT2 transport function in humans (101).
A recent report showed no difference in insulin secretion during an oral glucose tolerance test in individuals with or without the risk allele at SLC2A2 rs5400 (T110I), suggesting that neither β-cell mass nor insulin secretion is significantly affected (102). In agreement with this, this GLUT2 T110I variant displays similar kinetics for glucose transport as wild-type GLUT2 (101). Additionally, this variant is in strong linkage disequilibrium with two other variants (SNPs rs5406 and rs6785803) located in the promoter region of the SLC2A2 gene (103), which raises the possibility that the functional consequence of the rs5400 (T110I) variant could be related to the difference in the transcription of the SLC2A2 gene, rather than the effect of the gene product of variant rs5400, ie, GLUT2 T110I itself. Interestingly, the variant rs5400 (T110I) is associated with habitual consumption of sugars (104), which may reflect glucose sensing in the portal vein (105) and/or the hypothalamus (106), the two organs that also harbor GLUT2-dependent glucose sensors. Although GLUT2 has been identified in the paraventricular nucleus, lateral hypothalamic area, and arcuate nucleus, it is unclear whether the high Km of GLUT2 would allow the putative transporter to function normally in these regions of the brain with normal blood-brain barrier because of the local low glucose concentrations in these brain regions (104). In contrast, GLUT2 is likely operative in parts of the brain that lack a blood-brain barrier, ie, where the transporter is exposed to physiological levels of circulating blood glucose. Indeed, the circumventricular organ, area postrema, is one such brain area that has also been found to harbor SLC2A2 mRNA (107). These observations suggest that a GLUT2-mediated glucose-sensing mechanism could regulate food intake in select parts of the brain and thus be associated with the risk of T2D.
D. HNF1A and HNF4A
HNF1A, also known as transcription factor 1 (TCF1), is first expressed in the yolk sac and in the liver bud. Later on to adulthood, HNF1A is expressed throughout organogenesis of the liver, kidney and pancreas. Hnf1a-deficient mice develop hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. A fraction of the mice lacking Hnf1a die shortly after weaning due to a progressive wasting syndrome (108). Mice with Hnf1a ablation also develop diabetes with a profound defect in glucose or arginine-stimulated insulin secretion (109).
HNF4A is first expressed in the primary endoderm in mice at embryonic day 4.5 (e4.5) and is restricted to the visceral endoderm from e5.5 to e8.5. Thereafter, it is expressed in the liver diverticulum, the hindgut, the mesonephric tubules of the kidney, and the developing pancreas, stomach, and intestine (110). Hnf4a-deficient mice display defective gastrulation and die around e9 (111). In hepatocytes, HNF4A controls the transcription of HNF1A, whereas in pancreatic islets and exocrine cells, HNF1A controls the transcription of HNF4A through an alternate tissue-specific HNF4A promoter (112).
1. Mutations in HNF1A and HNF4A lead to MODY3 and MODY1, respectively
Mutations in the HNF4A gene lead to the first subtype of MODY identified (HNF4A-MODY [MODY1]), whereas mutations in the HNF1A gene cause HNF1A-MODY (MODY3). These two types of MODY display very similar clinical characteristics. The patients develop young-onset diabetes in the second to fourth decade without features of insulin resistance or β-cell autoimmunity. Pancreatic β-cell dysfunction can be observed before the onset of diabetes in the carriers with HNF1A mutations. Glycosuria due to a low renal threshold for glucose is observed in HNF1A-MODY patients, and in nondiabetic HNF1A mutant carriers, but not in HNF4A-MODY patients (113). Interestingly, inactivating mutations in HNF1A and HNF4A can cause CHI associated with increased body weight in early life, which may improve with age (114). Both HNF1A and HNF4A MODY patients respond to low-dose sulfonylureas (12.5% or less of the maximum licensed dose) (5, 6).
Mutations in HNF1A (MODY3) are the most common cause of MODY, which underlie 52% of monogenic diabetes in a large UK series (115). Interestingly, although patients display inherent β-cell defects, they develop diabetes relatively late in life—63% at age 25 years and 94% at age 50 (116). The reason that MODY3 presents relatively late in life is caused by the capacity of HNF1A to control the transcription of sodium-glucose cotransporter 2 (SGLT2), a renal glycosuria threshold-controlling gene. Therefore, individuals with HNF1A mutations display a drastically decreased expression of SGLT2, thereby reducing glucose reabsorption through the proximal tubule (117). This defective HNF1A-mediated down-regulation of SGLT2 helps maintain relative euglycemia in MODY3 individuals before the appearance of overt hyperglycemia and diagnosis of diabetes much later in life; ie, HNF1A mutations function like SGLT2 inhibitors, and the belated appearance of overt diabetes in affected individuals could have predicted the development of this new class of hypoglycemic agent for the treatment of diabetes.
2. HNF1A gene variants and T2D
A private mutation (G319S) in HNF1A has been reported in Canadian Oji-Cree populations to be associated with early-onset T2D, lower body mass, and a higher postchallenge plasma glucose (118). Using whole-exome sequencing, MacArthur and colleagues (119) identified a single low-frequency variant (c.1522G>A [p.E508K]) in HNF1A, which is associated with T2D prevalence in Latino populations. The variant allele p.E508K is located in the HNF1A transactivation domain. Functional studies revealed that, despite the preservation of its DNA binding activity, the p.E508K variant protein displays compromised transactivation activities on the promoters of SLC2A2 and HNF4A in β-cells. Of note, a recent report showed that HNF1A variant (rs2650000) predisposes to latent autoimmune diabetes in adults and adult-onset T1D (120).
3. HNF4A gene variants and T2D
Multiple SNPs in the alternate upstream promoter (P2) region of HNF4A have been found to be associated with increased risk of T2D. Initial studies showed that SNPs at rs2144908, rs3818247, and rs884614 in the P2 region of the HNF4A gene were associated with T2D risk in Finnish (121) and Ashkenazi (122) populations. Subsequently, Weedon et al (123) reported that the risk haplotype of rs4810424 and rs2144908 in the P2 region of the HNF4A gene was associated with T2D in UK populations in both case-control and family-based studies. Recently, Barroso et al (124) found that HNF4A P2 SNPs rs1884613 and rs2144908 were associated with T2D in the Ashkenazi, but not in UK, populations. These findings suggest that P2 SNPs in the HNF4A gene confer varying risk effects in different populations.
4. Molecular mechanisms and pathophysiology that underlie the association of mutations in the HNF1A and HNF4A genes and diabetes
To dissect the molecular mechanisms underlying MODY3, Bjorkhaug et al (125) investigated the functional properties of HNF1A mutations. They reported that two mutant HNF1A proteins (R171X and R263C) manifest reduced or absent DNA binding activity. Five mutants (R131W, R171X, P379fsdelCT, S445fsdelAG, and Q466X) display defective nuclear translocation. Most of the MODY3-associated mutants exhibit reduced transactivational activity. These loss-of-function HNF1A mutants underlie the β-cell dysfunction in MODY3 (125). Recently, Nowak et al (126) reported that circulating ghrelin levels were higher in HNF1A-MODY patients than those in common T1D or T2D patients. Hnf1a-deficient mice develop diabetes caused by a defect in glucose-stimulated insulin secretion (GSIS) (109). It is intriguing that a recent study showed that serum ghrelin levels were also increased in Hnf1a-deficient mice, and treatment with a ghrelin receptor antagonist restored glucose homeostasis in Hnf1α-null mice (127). Ghrelin suppresses glucose-induced insulin secretion, but not basal insulin secretion. It counteracts cAMP-protein kinase A signaling, a well-established pathway of GSIS, by interacting with the GH secretagogue receptor GHS-R1a on β-cells. The suppression of the cAMP pathway results in activation of delayed outward K+ (Kv) channels and inhibition of Ca2+ signaling, causing decreased insulin release (128).
In mammals, the HNF4A gene encodes up to nine distinct isoforms, resulting from the use of alternate promoters and alternative splicing. Promoter P1 initiates transcripts that contain exon 1A (isoforms A1–A6) encoding activation function 1 (AF-1) domain, whereas promoter P2 initiates transcripts that contain exon 1D (isoforms A7–A9) lacking AF-1. In pancreatic β-cells, the P1 promoter exhibits stronger transcriptional activities than the P2 promoter (129). Pancreatic β-cell-specific Hnf4a deletion leads to hyperinsulinemia in fasted and fed mice but paradoxically is also associated with IGT. Hnf4a mutant β-cells exhibit a defective response to stimulation by glucose and sulfonylureas. Experiments in mice showed that the expression of the potassium channel subunit Kir6.2 (encoded by KCNJ11), a downstream target of Hnf4a, is reduced by 60% in Hnf4a mutant β-cells (130).
Studies in rodents further showed that both maternal suboptimal nutrition and aging lead to progressive epigenetic silencing at the enhancer region in the Hnf4a gene, disrupting the P2 promoter-enhancer interaction and resulting in a permanent reduction in Hnf4a expression (131). It is unclear whether the variants in HNF4A predispose to T2D via a similar mechanism in people.
E. Hepatocyte nuclear factor 1 homeobox B
HNF1B, also known as transcription factor 2 (TCF2), is first expressed in the visceral endoderm at the onset of gastrulation. Later on to adulthood, it is expressed throughout organogenesis of the liver, kidney and pancreas (132). In mice, HNF1B-positive progenitors were shown to be equally capable of giving rise to pancreatic duct, acinar, and endocrine cells from e11.5 to e13.5. Global HNF1β-deficient mice die soon after implantation (132, 133). Pancreatic β-cell-specific (RIP-Cre) Hnf1b deletion resulted in IGT with reduced insulin secretion (134). HNF1B is required for the proliferation and survival of pancreatic multipotent progenitor cells. Hnf1b deficiency in pancreatic progenitors leads to severe pancreatic hypoplasia and perinatal lethality (135). HNF1B is also a key regulator of duct morphogenesis, acinar cell differentiation, and maintenance of acinar cell identity. It is indispensable for the generation of endocrine precursors, likely by directly regulating the pancreatic islet lineage-defining transcription factor neurogenin 3 (Neurog3, or Ngn3) expression (135). The sequential activation of Hnf1b, Hnf6, and Pdx1 is believed to direct the differentiation of endodermal cells into pancreatic progenitors (136).
1. Mutations in HNF1B lead to RCAD syndrome, or MODY5
In addition to causing PNDM or TNDM in very rare cases (137), heterozygous mutations in HNF1B cause renal cysts and diabetes (RCAD syndrome, or MODY5). HNF1B whole-gene deletions account for approximately 50% of MODY5 cases. To date, more than 50 missense mutations or deletions have been identified in MODY5 patients. Various phenotypes related to these mutations have been described, including urogenital malformations, hyperuricemia, young-onset gout, and pancreatic hypoplasia or atrophy. In a literature meta-analysis, renal cysts and diabetes occurred in 89.6 and 45% of the patients with RCAD syndrome, respectively. However, the concurrence of renal cysts and diabetes was observed in only 27.5% of the patients, which could delay the clinical diagnosis and timely genetic testing (138). Patients with HNF1B mutations display abnormal β-cell development and reduced insulin section (135, 136) that is usually resistant to sulfonylureas; most of them require early insulin therapy (139, 140).
2. HNF1B gene variants and T2D
Winckler et al (141) found that SNP rs757210 A in intron 2 of HNF1B showed a strong association with T2D in multiple populations. Epidemiological studies have demonstrated an inverse association between T2D and the risk of prostate cancer (142, 143). In accordance with this finding, Gudmundsson et al (144) found that a common variant rs4430796 A in HNF1B was positively associated with prostate cancer and negatively associated with T2D risk. It has been proposed that with time, metabolic status in T2D patients might switch from hyperinsulinemia to endogenous insulin deficiency, thus alleviating the pro-oncogenic effect of insulin in the prostate.
3. Molecular mechanisms and pathophysiology that underlie the association of mutations in the HNF1B gene and diabetes
HNF1A and HNF1B share a highly conserved DNA-binding domain and a more divergent C-terminal transactivation domain. HNF1B may act either as a homodimer or as a heterodimer with HNF1A (145). Diverse mechanisms have been uncovered that may underlie the development of HNF1B-MODY. For example, frameshift mutations R112fsdel (an 8-bp deletion in exon 2) or P472fsins (an 8-bp insertion in the exon 7) in the HNF1B gene were found to lead to severe pancreatic hypoplasia with underdeveloped and disorganized acini in human fetuses. The HNF1B R112fsdel mutation causes a frameshift and a truncated protein lacking part of the POU-specific domain (POUS), which is essential for DNA binding specificity, and the entire POU homeodomain (POUH), as well as the C-terminal transactivation domain, whereas the HNF1B P472fsins mutation leads to a premature stop codon and the insertion of 35 novel amino acids at the C-terminus of the transactivation domain. Functional studies showed that the truncated R112fsdel protein, retaining the N-terminal dimerization domain, functions as a weak dominant-negative mutant, through the formation of nonfunctional heterodimers with wild-type HNF1B protein. In contrast, the P472fsins mutant exhibits a decreased transactivation capacity, whereas it retains the normal DNA binding activity; however, it does not interfere with the function of the wild-type HNF1B protein (146).
GLUT2, a potential direct target of HNF1B (147), was undetectable in islet β-cells of human fetuses with R112fsdel or P472fsins mutation (146). Similarly, overexpression of the HNF1B P159L mutant (disrupting its DNA binding domain) led to significantly reduced expression of GLUT2 in Min6 mouse insulinoma cells (148). As discussed earlier, Slc2a2-deficient mice develop severe diabetes and lethality with impaired glucose sensing by pancreatic β-cells (83). Forced overexpression of a naturally occurring dominant-negative form of human HNF1A (P291fsinsC) disrupts the formation of homo- and heterodimers between wild-type HNF1A and HNF1B, leading to reduced expression of GLUT2 and E-cadherin (149). In addition, β-catenin and E-cadherin expression was drastically reduced in the islet and acinar compartments of human fetuses with R112fsdel or P472fsins mutation. Wnt/β-catenin is required for acinar lineage specification and differentiation (150–152). These findings suggest that perturbation in the Wnt/β-catenin signaling pathway may underlie the severe pancreas hypoplasia frequently observed in HNF1B mutant individuals; early loss of GLUT2 in β-cells may cause early-onset diabetes in MODY5 patients (146).
F. Pancreatic and duodenal homeobox 1
PDX1 (also known as insulin promoter factor 1 [IPF1]) is a homeodomain transcription factor expressed throughout the pancreatic epithelium at e8.5 in mice; from e15.5 onward, its expression becomes mainly restricted to β-cells and a small subpopulation of δ and PP (pancreatic polypeptide) cells (153). PDX1 is essential for pancreas development, β-cell differentiation, and the maintenance of mature β-cell function by regulating the expression of key islet-specific genes such as Ngn3, insulin, Gck, and Slc2a2 (153–155). Mice lacking Pdx1 fail to form a pancreas (156). Pancreatic β-cell-specific Pdx1 deficiency in mice leads to reduced expression of insulin and Slc2a2, causing maturity-onset diabetes (155). A large fraction of Pdx1-deleted β-cells rapidly acquires ultrastructural and physiological features of α-cells, suggesting that Pdx1 maintains β-cell identity and function via repressing an α-cell lineage (157). Mutations or variants in the PDX1 gene cause PNDM and MODY4 and are also linked to common T2D and ketosis-prone diabetes (KPD).
1. PDX1 mutations underlie PNDM and MODY4
Stoffers et al (158) first reported that a homozygous single cytosine deletion within codon 63 (Pro63fsdelC) of the human PDX1 gene was responsible for a previously described PNDM syndrome that is also associated with pancreatic exocrine insufficiency (159). Two substitution mutants, E164D and E178K in the human PDX1 gene, also individually lead to PNDM and pancreatic exocrine insufficiency (160), whereas the hypomorphic mutation (E178G) in PDX1 is responsible for PNDM but is not associated with clinical signs of exocrine insufficiency (161). In the first family pedigree, Stoffers et al (162) further identified that the heterozygous Pro63fsdelC mutation in PDX1 causes MODY4. Family members developed diabetes in six generations, with an average age at onset of 35 years (range, 17 to 67 years). Six of eight affected heterozygotes, lacking ketosis or other indications of severe insulin deficiency, were treated with diet or oral hypoglycemic agents. Notably, patients afflicted with PDX1 mutations (eg, D76N) may concurrently carry other MODY gene mutations (eg, S315fsinsA in HNF1α/MODY3). Not surprisingly, carriers of mutations in both genes (PDX1 and HNF1α) display more severe insulin deficiency than those who carry the mutation in only one of the two genes (163).
2. Mutations or variations in the PDX1 gene and T2D
Shortly after PDX1 mutations were identified in patients with PNDM and MODY4, other mutations in the PDX1 gene were also found to be linked to common T2D. Candidate gene association studies showed that PDX1 substitution mutations (C18R, Q59L, D76N, R197H, G212R, and P239Q) and an in-frame proline insertion (InsCCG243) predispose to common T2D (163–165). Of note, approximately 5% of the examined T2D patients and control nondiabetic individuals carried low-frequency PDX1 variants that were not associated with diabetic phenotype, suggesting that PDX1 mutations or variants are a very rare cause of diabetes (166). Consistent with this observation, whole-genome sequencing of 2630 Icelanders showed that a novel rare (0.20%) frameshift variant (p.Gly218Alafs*12) in PDX1 is associated with high risk of T2D (167). Furthermore, the same study observed seven previously reported variants (six missense variants and one in-frame insertion) in PDX1, but none were associated with diabetes (167). In addition, PDX1 variants have also been linked to KPD (see Section II.G for details).
3. Molecular mechanisms that underlie the association of mutations in the PDX1 gene and diabetes
As noted above, PDX1 variants associated with enhanced T2D risk are rare, despite the fact that PDX1 mutations cause PNDM and MODY4. Pro63fsdelC point deletion in PDX1 leads to a frame shift, causing a premature truncated protein with 59 aberrant codons that is lacking transactivation domain at its C terminus. The mutant protein fails to transactivate the INS gene transcription, leading to PNDM (158). The C-terminal domain of PDX1 is required to maintain all endocrine lineages in the pancreas. Expression of a single C-terminal truncated form of PDX1 results in mild hyperglycemia at birth and progresses to overt diabetes by the down-regulation of Ngn3 (154). Schwitzgebel et al (160) reported that PDX1 E178K displayed reduced transactivation activity on the INS promoter due to a decrease in PDX1 steady-state protein levels resulting from impaired protein stability, whereas substitution variant PDX1 E178G displays reduced transactivation activity via an unclear mechanism, leading to PNDM (161).
A number of PDX1 mutations (eg, C18R, Q59L, D76N, R197H, and InsCCG243) lead to reduced binding of the mutant protein to the INS gene promoter and decreased INS gene transcription in response to hyperglycemia in pancreatic β-cells, which may underlie the substantial defect in insulin secretion detected in these patients (163–165). D76N and Q59L mutations in PDX1 have also been linked to decreased GSIS in nondiabetic subjects (164).
In mice, Pdx1 haplodeficiency enhances β-cell susceptibility to endoplasmic reticulum (ER) stress-associated apoptosis and leads to a failure of β-cell compensation for high-fat diet (HFD)-induced insulin resistance (168). Mitochondrial dysfunction is an important contributor to β-cell failure, insulin resistance, and diabetes (169, 170). Mitochondrial autophagy or mitophagy is required for the selective elimination of dysfunctional or damaged mitochondria to maintain cellular respiration. Recently, Pdx1 has been shown to direct autophagosome-lysosome fusion during mitophagy and regulate mitochondrial function in pancreatic β-cells (171). Although it has not been directly examined, it is possible that these mechanisms may also underlie the association of PDX1 mutations/variants with diabetes.
G. Paired box 4
PAX4, a paired-homeodomain transcription factor, functions mainly as a transcription repressor (172). Pax4 mRNA is initially detected in the ventral spinal cord and pancreatic bud at e9.5 in mice; its expression is later restricted to pancreatic β- and δ-cells, reaching a maximal level at e13.5 to e15.5, and thereafter declining to low levels. PAX4 is also found to be expressed in adult rodent and human islets (173). Pax4-deficient mice develop severe hyperglycemia and die within 2 days of birth. The β-cells are replaced by a proportional increase in glucagon-positive α-cells; somatostatin-expressing δ-cells are also missing in Pax4-deficient mice. Thus, Pax4 deficiency is characterized by a favoring of an α-cell fate at the expense of β-/δ-cell lineages (174). The aristaless-related homeobox-encoding gene, Arx, and Pax4 are coexpressed in the same proendocrine cell during pancreas genesis. Arx-deficient mice develop early-onset hypoglycemia, dehydration, and die 2 days after birth. In the absence of Arx, endocrine progenitor cells assume β-/δ-cell destinies (175). At the molecular level, ARX and PAX4 directly bind to the respective promoter or enhancer and drastically repress each other's transcription (176).
1. Mutations in PAX4 lead to MODY9
To determine whether PAX4 mutations contribute to MODY in the Thai population, Plengvidhya et al (177) screened PAX4 coding sequences in 46 MODY probands who did not have mutations in other known MODY genes. They identified two possible pathogenic mutations of PAX4, R164W and IVS7–1G>A (a G to A substitution at splice acceptor of intron 7), in patients with MODY (PAX4-MODY, or MODY9, a very rare type of MODY), but not in nondiabetic controls and healthy subjects. Subsequently, PAX4 R192H (178) and a 39-base heterozygous deletion in exon 3 (C.374–412 del 39) in PAX4 (179) have also been reported in MODY9 patients.
2. PAX4 gene mutations or variants and T2D and KPD
In a Japanese population, a missense mutation of PAX4 R121W was found to be associated with T2D. The R121W mutation was located in the C terminus of the paired domain and was thought to affect its transcriptional activity due to a lack of DNA binding (180). In a GWAS meta-analysis, Ma et al (181) reported that the rs10229583 G risk variant near PAX4 was associated with elevated fasting glucose, impaired β-cell function, and early-onset T2D in a Chinese population. In newly diagnosed T2D patients, PAX4 variant rs6467136 correlated with a positive therapeutic response to rosiglitazone, but not repaglinide. PAX4 variant rs6467136 GA+AA carriers exhibited greater responses to rosiglitazone therapy than GG homozygotes, with greater decrement of 2-hour glucose level and amelioration of insulin resistance as well as higher cumulative attainment rates of target fasting and 2-hour glucose levels (182).
In addition to the association of PAX4 variants with T2D, one candidate gene association study (183) and a linkage analysis (184) reported that PAX4 variants including +1,168 C/A of PAX4 (rs712701) were associated with T1D, but other studies failed to find an association (185, 186), which suggests that PAX4 variants are unlikely to play an important role in the development of T1D.
Mauvais-Jarvis et al (187) first reported that PAX4 homozygous R133W and heterozygous R37W mutations may be associated with KPD. KPD is a heterogeneous group of atypical diabetic syndromes characterized by recurrent severe β-cell dysfunction and a variable clinical course (188, 189). KPD patients can be classified into four subgroups, as defined by: 1) the presence or absence of autoantibodies (that are associated with common T1D), ie, A+ or A−; and 2) the presence or absence of β-cell function, ie, β+ or β−. The A-β subgroup of KPD patients, characterized by β-cell failure without evidence of autoimmunity, often has a strong family history of diabetes (188, 189). Balasubramanyam and colleagues (190) systematically sequenced seven genes associated with monogenic diabetes in an A-β patient subgroup and found that about 30% of them harbor variants of HNF1A, PDX1, or PAX4 genes, suggesting that these variants may underlie the β-cell dysfunction in a fraction of patients with A-β KPD.
3. Molecular mechanisms and pathophysiology that underlie the association of mutations in the PAX4 gene and diabetes
Our knowledge of the molecular basis of PAX4 mutations causing diabetes remains incomplete. PAX4 predominantly represses the glucagon promoter activity in α-cells and weakly inhibits insulin promoter activity in β-cells (172). PAX4 mutations are located either in the paired domain (R37W), in the homeodomain (R164W), or between the paired domain and homeodomain (R133W). These mutations impair its transcriptional repressor activity by interfering with its capacity to bind DNA, or the binding specificity or functional stability, or its interaction with other proteins. PAX4 mutants fail to repress glucagon transcription, leading to hyperglucagonemia (187). PAX4 mutations may cause insulin deficiency, possibly through the disruption of β-cell development or impairment of GSIS in β-cells. Some PAX4 mutations (eg, R121W, IVS7-1G>A) seem to increase the susceptibility of β-cells to apoptosis upon cytokine or high glucose exposure (177, 191). Conversely, forced expression of wild-type PAX4 has been shown to protect cytokine-induced β-cell death in isolated human islets (191). In general, most MODY patients are treated with sulfonylureas, metformin, or insulin (5). A few recent studies have also documented the efficacy of GLP-1 receptor agonists (192, 193) and dipeptidyl peptidase-IV inhibitors (194, 195) in these patients. Although it has not been directly studied, it is possible that the GLP-1 receptor agonists and dipeptidyl peptidase-IV inhibitors, through their glucagon-inhibiting actions (196), are especially efficacious in the treatment of patients with PAX4 mutations.
H. NEUROD1/BETA2
The basic helix-loop-helix (HLH) transcription factor NEUROD1, also called BETA2, plays a crucial role in pancreatic β-cell maturation and maintenance. NEUROD1 is detected in the mouse embryo as early as e9.5 in glucagon-positive cells and is mostly restricted to β-cells at birth. Neurod1-deficient mice fail to develop mature islets due to enhanced apoptosis, leading to NDM and lethality within the first few days of life (197). Pancreatic β-cell-specific Neurod1-deficient mice exhibit severe glucose intolerance with greatly reduced insulin secretion. Islets lacking Neurod1 respond poorly to glucose and display a glucose metabolic profile similar to immature β-cells, characterized by increased expression of glycolytic genes, elevated levels of lactate dehydrogenase and basal oxygen consumption, and overexpression of neuropeptide Y. In sum, NEUROD1 is required for the development and maintenance of fully functional mature β-cells (198).
1. Mutations in the NEUROD1 gene lead to MODY6 and PNDM
Heterozygous loss-of-function mutations in NEUROD1 produce a very rare type of MODY (NEUROD1-MODY, or MODY6), with six families reported to date. A missense mutation E110K in NEUROD1 was first reported in an Icelandic MODY6 family (199). Subsequently, missense mutations S159P, H241Q, and R103P in NEUROD1 were identified in Chinese (200), Czech (201), and Polish (202) MODY6 families, respectively.
Recently, Rubio-Cabezas et al (203) reported that two recessive homozygous mutations in NEUROD1, a single base pair duplication (c.364dupG) and a 2-bp CT deletion (c.427_428del), led to a syndrome of PNDM and neurological abnormalities. Both mutations introduced a frameshift to produce a premature truncated protein lacking the activation domain at the C terminus.
2. NEUROD1 gene variants and T2D
Malecki et al (204) reported that a missense mutation R111L and a cytosine residue insertion (206 +C) in the NEUROD1 gene were associated with the risk of T2D. A number of studies showed that a common A45T variant at rs1801262 in NEUROD1 was inconsistently associated with T2D (204–208). Recent meta-analyses revealed that this polymorphism is associated with T2D in European descent Caucasians, but not in individuals of Chinese and Japanese descent, or others of East Asian descent (207). Of note, the A45T variant in NEUROD1 is associated with T1D in several small population studies (208–210), but not in a larger population study (211).
3. Molecular mechanisms that underlie mutations in the NEUROD1 gene and diabetes
NEUROD1 forms a heterodimer with the ubiquitous HLH protein E47 to transactivate insulin gene expression by binding to a critical E-box motif on the promoter (212), synergistically with MAFA, PDX1 (213), and GLIS3 (214). The mutation R111L in the NEUROD1 gene is located in the proximal basic portion of the basic HLH domain, which mediates DNA binding. This mutation abolishes E-box binding activity of NEUROD1 and drastically compromises insulin gene transcription in pancreatic β-cells. The 206 +C insert mutation produces a frameshift and synthesis of a nonsense peptide from amino acid 205 to 242, followed by a stop codon. The mutant peptide has lost its C-terminal transactivation domain, which normally enables interaction with the coactivators cAMP response element-binding protein and p300. Thus, these NEUROD1 mutants impair DNA binding and/or the transactivation activity on the INS gene as well as other target genes in pancreatic β-cells leading to T2D (204).
I. Wolfram syndrome 1
WFS1 encodes a 100-kDa transmembrane glycoprotein that is localized in the ER and secretory granules. Unfolded protein response (UPR) is a cellular stress response induced by the accumulation of unfolded or misfolded proteins within the ER lumen. WFS1 is a component of the inositol requiring 1 and PKR-like ER kinase UPR signaling pathway and is important in the maintenance of ER homeostasis. The maintenance of ER integrity by PKR-like ER kinase is crucial to normal β-cell function because large amounts of insulin are produced by these cells (215). WFS1 has been implicated in the activation of the UPR to mitigate ER stress (216). This ER protein is induced by ER stress and during glucose or potassium chloride-stimulated insulin secretion in β-cells. Wfs1-deficient β-cells display increased susceptibility to ER stress-mediated apoptosis (217).
1. Mutations in WFS1 lead to Wolfram syndrome 1
Recessive mutations in the WFS1 gene cause pancreatic β-cell death, resulting in a monogenic form of diabetes known as Wolfram syndrome 1 (WFS1), which is sometimes referred to as DIDMOAD (diabetes insipidus, diabetes mellitus, optic atrophy, and deafness). It is noteworthy that, in certain populations, eg, a Lebanese population, patients with some WFS1 mutations may present with isolated early-onset diabetes, contributing to a substantial proportion of patients who are often mislabeled as having “T1D” (218). There is currently no effective treatment for the syndrome, which usually ends in premature death. With respect to the optic atrophy, a recent report showed that average retinal thickness is significantly lower in WFS1 patients, compared with age-matched T1D patients and healthy individuals. Moreover, a strong negative correlation exists between average retinal thickness and both age at examination and duration of diabetes in WFS1 patients. It has therefore been proposed that retinal thinning is a marker of disease progression in these patients (219). To date, over 180 mutations in the WFS1 gene, most commonly in exon 8, have been identified (220). A WFS1-like syndrome, WFS2, is caused by homozygous mutation in the CDGSH iron sulfur domain 2 (CISD2) gene. CISD2 encodes a highly conserved zinc-finger protein, ER intermembrane small (ERIS), which plays a crucial role in calcium homeostasis (221). WFS1 and WFS2 share some common clinical features such as diabetes mellitus, hearing loss, and optic atrophy or neuropathy. They also display distinct characteristics, eg, diabetes insipidus is a specific feature for WFS1, whereas defective platelet aggregation resulting in bleeding peptic ulcer appears to be specific for WFS2 (222).
2. WFS1 gene variants and T2D
Minton et al (223) reported the first evidence that variation in the WFS1 gene may influence susceptibility to T2D. In a parent-offspring trios study, R456 and H611 in WFS1 were found to be overtransmitted to affected offspring from heterozygous parents in diabetic families. In a further cohort study, the H611 allele and the R456-H611 haplotype were present more frequently in T2D individuals than in control subjects.
After genotyping 1536 SNPs in 84 genes regulating pancreatic β-cell development, function, and survival, Sandhu et al (224) found that rs10010131 (minor allele frequency [MAF] = 40%) and rs6446482 (MAF = 41%) in the WFS1 gene were strongly associated with T2D risk in UK and other populations. The frequency at the rs10010131 risk allele is 60%, and it was estimated that the population attributable fraction of rs10010131 is 9%, explaining 0.3% of the excess familial risk of T2D.
Cheurfa et al (225) examined the association of allelic variations in the WFS1 gene with insulin secretion and risk of T2D in the prospective Data from Epidemiological Study on the Insulin Resistance Syndrome (DESIR) study. They found that the three variants rs10010131 G, 1801213 G, and rs734312 A were significantly associated with plasma glucose, glycated hemoglobin A1C levels, and insulin secretion at baseline and throughout the study in individuals with T2D or at risk of developing diabetes, but not in those who remained euglycemic at the end of the follow-up. Carriers of the combined GGA haplotype for these SNPs had a 26% increased risk for developing diabetes compared with carriers of the ACG haplotype. These associations between the three variants and T2D were replicated in a large case-control, cross-sectional study. These findings suggested that variants of the WFS1 gene affect insulin secretion and are associated with an increased risk of T2D. However, variants of WFS1 including rs10010131G were found not to be associated with T2D in a well-designed fine-mapping study, raising the argument that low frequency variants in WFS1 may or may not have significant impact on T2D risk in the examined populations (226).
Awata et al (227) reported that variant R456H in the WFS1 gene was associated with an increased risk of common T1D in a small candidate gene association study, which needs validation in larger populations.
3. Molecular mechanisms and pathophysiology that underlie the association of mutations in the WFS1 gene and diabetes
In WFS1 patients, loss-of-function mutations in the WFS1 gene lead to deficiency of wolframin protein and an inadequate activation of the UPR in response to accumulation of unfolded proteins within the ER lumen, which induces ER stress and results in apoptotic cell death. These mechanisms are believed to underlie progressive neurodegeneration and endocrine dysfunction in these patients (216, 228). Interestingly, ER stress-mediated diabetes in WFS1 was diagnosed earlier in life than the common type of autoimmune-mediated T1D. WFS1 mutations induce ER stress, which in turn mediates the decline of β-cell mass and diabetes in WFS1 patients. Poor glycemic control-induced oxidative stress is thought to further aggravate the progression of neurodegeneration in this disease (216). The development of therapeutic measures that alleviate ER stress and oxidative stress could benefit WFS1 patients. Calpain, a family member of calcium-dependent, nonlysosomal cysteine proteases, may be a plausible therapeutic target. This enzyme is thought to be a mechanistic link between the ER and β-cell death in WFS1 (229). Loss-of-function mutations of WFS1 increase cytoplasmic calcium levels, leading to calpain activation, which causes ER stress-induced β-cell death (230). Treatment with the ryanodine receptor inhibitor dantrolene was shown to reduce calcium leakage from the ER to cytosol, lowering cytosolic calcium levels and suppressing calpain activation; it also prevents cell death in pancreatic β-cells and neural progenitor cells. The protective effect of dantrolene suggests that modulating calcium levels by suppressing calpain activation could be one approach to treat WFS1 and other diseases associated with excess ER stress (229).
In addition to its role in the UPR, WFS1 may have other β-cell-specific functions. WFS1 localizes not only to the ER but also to secretory granules in pancreatic β-cells. Compared with wild-type β-cells, a 32% reduction in granular acidification was observed in Wfs1-deficient β-cells (231). This aberration may account for the impaired proinsulin processing and an increased circulating proinsulin level in Wfs1-null β-cells. Electron microscopy showed that Wfs1 deficiency leads to a reduction in plasma membrane-attached secretory granules in the β-cell, indicating that WFS1 may play a role in the intracellular distribution of secretory granules, especially the docking of insulin granules to the plasma membrane. These perturbations may cause the profoundly impaired GSIS in Wfs1-deficient mice, providing new insights into the molecular basis of β-cell dysfunction in WFS1 patients (231).
J. Peroxisome proliferator-activated receptor γ
The PPARG gene, predominantly expressed in white and brown adipose tissue, is a master regulator of adipogenesis as well as a potent modulator of whole-body energy balance, lipid biosynthesis, and insulin sensitivity (232). PPARG exists in two splice isoforms, PPARG1 and PPARG2, with the latter carrying an additional 30 amino acids at its N terminus. PPARG is a nuclear hormone receptor and a target of thiazolidinediones (TZDs), a commonly used class of antidiabetic medication despite concerns on side effects of weight gain, fluid retention, bone loss, and heart failure. PPARG activation by TZDs leads to a marked improvement of whole-body insulin sensitivity and glucose metabolism (232). Recently, fibroblast growth factor (FGF) 21 and FGF1 are thought to act locally in adipose tissue to mediate the pharmacological actions as well as the side effects of TZDs (233, 234). Fgf21-deficient mice are resistant to the insulin-sensitizing effects of TZDs as well as the associated weight gain and fluid retention (234). In contrast, FGF21 administration in mice produces strong antidiabetic effects along with bone loss (235). In addition, obesity can activate cyclin-dependent kinase 5 in adipose tissue, resulting in the phosphorylation of PPARG at serine 273. This modification underlies the dysregulation of a number of genes associated with obesity and insulin resistance in adipose tissue. PPARG ligands such as TZDs can block this phosphorylation and confer their antidiabetic therapeutic effects (236, 237).
1. Mutations in PPARG lead to severe insulin resistance and diabetes
Barroso et al (238) first reported the identification of two heterozygous mutations, P467L and V290M, in the ligand-binding domain of PPARG in three subjects with severe insulin resistance who presented with diabetes and hypertension at an unusually early age. Although these mutations are uncommon, detected in just three of 85 severely insulin-resistant individuals and none in 314 controls, they represent a unique model of monogenic diabetes through decreased insulin sensitivity, without overtly defective β-cell function.
2. PPARG gene variants and T2D
Several variants in the PPARG gene have been identified, with the P12A variant (rs1801282), first identified by Yen et al (239), being the most extensively examined in epidemiological studies. Ethnic heterogeneity has been observed between the PPARG P12A and the magnitude of the association with T2D (240–242). In a large meta-analysis that involved 32 849 T2D cases and 47 456 controls, which included a population-based cohort, case-control, cross-sectional, and GWAS analyses, Gouda et al (243) found that the PPARG P12A polymorphism was associated with a reduction in T2D risk, with a combined overall odds ratio of 0.86.
Of note, PPARG SNP rs1801282 (P12A C>G) was found to be associated with T1D in one report (244) but not in another (245), suggesting that this T2D locus may or may not play a role in T1D genetic susceptibility.
3. Molecular mechanisms and pathophysiology that underlie the association of mutations or variations in the PPARG gene and diabetes
The P467L mutation discussed above (238) lies in an amphipathic α-helix at the C terminus of the PPARG ligand-binding domain, a motif that is critical for mediating ligand-dependent transactivation and coactivator recruitment. The mutant displays a severely impaired binding capacity to its ligand and coactivator. The V290M receptor mutation is located proximally within the ligand-binding domain. Both PPARG mutants inhibit wild-type receptor function in a dominant-negative manner, which may lead to silencing of basal gene transcription (238). Notably, a recent study highlights that motif-altering SNPs in target genes of PPARG cause differential PPARG binding to the DNA, modulating the recruitment of cooperating factors including CCAAT/enhancer binding protein, and thus determine individual disease risk and the response to TZD treatment (246).
Peroxisome proliferator-activated receptor α (PPARA) is abundantly expressed in the liver, heart, and muscle, where it plays important roles in the regulation of fatty acid oxidation, lipoprotein metabolism, and glucose homeostasis (247). In the Quebec family study, Bossé et al (248) examined the interactions and combined effects of PPARG2 A12 and PPARA V162 on glucose and insulin homeostasis. Carriers of the PPARA V162 allele were found to display higher insulin and C-peptide levels in response to a glucose challenge. The PPARG2 A12 allele reduced plasma insulin or C-peptide levels only on a PPARA V162 genetic background. The PPARG2 A12 allele appeared to attenuate the deleterious effect of the PPARA V162 allele and improve insulin sensitivity (248), an observation that is consistent with the fact that obese subjects carrying the PPARG2 A12 allele exhibited greater insulin sensitivity as measured by euglycemic-hyperinsulinemic clamp (249).
Recently, Claussnitzer et al (250) identified the T2D risk SNP rs4684847 as a cis-regulatory variant located at 6.5 kb upstream of the PPARG2-specific promoter. Functional studies revealed that paired related homeobox 1 (PRRX1) may act as a repressor of PPARG2 via its enhanced binding at the rs4684847 C risk allele. Consistent with the role of PPARG2 for insulin sensitivity rather than insulin secretion, this enhanced binding activity was observed only in insulin target cells (adipocytes, hepatocytes, and myocytes), but not in pancreatic β-cells or kidney cells. These findings support the interpretation that the rs4684847 C risk allele-mediated enhanced binding of PRRX1 represses the expression of PPARG2, leading to reduced insulin sensitivity and impaired glucose homeostasis.
III. Monogenic Diabetes Genes Associated With Both T1D and T2D
Among the list of monogenic diabetes genes, variants of the human preproinsulin gene (INS) are strongly associated with common T1D, but inconsistently with T2D. Interestingly, GWAS and meta-analyses showed that variants of GLIS3, a NDM gene, are associated with susceptibility to both common T1D and T2D.
A. Preproinsulin gene, INS
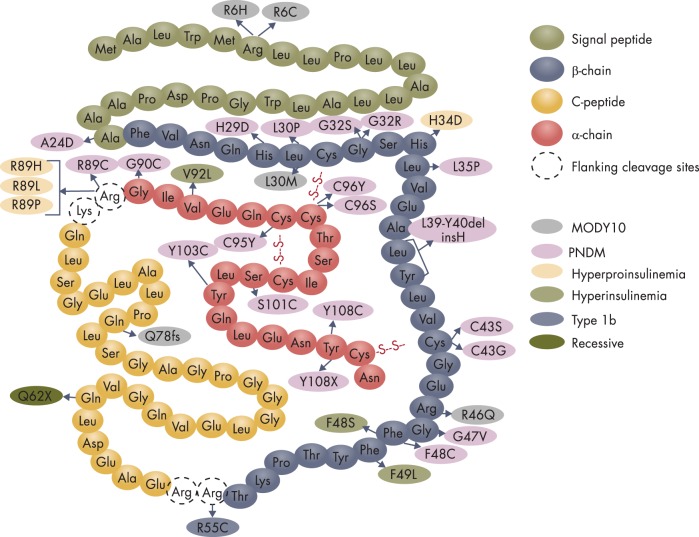
INS is unique among monogenic diabetes genes in that INS mutations can cause hyperinsulinemia, hyperproinsulinemia, NDM, MODY10, as well as autoantibody-negative T1D (type 1b) (Figure 4). Moreover, variants of the INS gene are also strongly associated with common T1D (type 1a), but inconsistently with T2D.
Figure 4.
Diagram depicting the human preproinsulin molecule with mutations that are currently known to cause diabetes and hyperinsulinemia or hyperproinsulinemia. The amino acid residues in the signal peptide, the β-chain, the C-peptide, and the α-chain are shown in different colors. The dashed circles indicate the basic residues that are the cleavage sites for conversion from proinsulin to insulin. The mutations underlying different types of diabetes and hyperinsulinemia or hyperproinsulinemia are presented in oval shapes filled in distinct colors as indicated. We note that some translation initiation codon mutations (eg, 3G>T, 3G>A), in-frame deletion mutants in the coding region, and mutations in the promoter or intron regions of the INS gene have been left out of this diagram. In addition, several INS substitution mutants, eg, A23S, A23T, L68M, and G84R, that were initially reported to occur in patients with diabetes have been left out of this diagram because subsequent studies showed that they are functional incidental variants. (See Section III.A for details.)
1. Mutations in the INS gene lead to hyperinsulinemia and hyperproinsulinemia
Mutations of the INS gene that produce structurally defective insulins can cause hyperinsulinemia and late-onset diabetes (Figure 4). These mutated insulins display drastically reduced bioactivity and receptor binding capacity, and often prolonged half-life (251). For example, the INS mutations F49L (252), F48S (253, 254), and V92L (255) have been identified in such patients. It is important to note that not all individuals who harbor heterozygous INS gene mutations necessarily develop diabetes because affected individuals who produce both normal and mutant insulins may compensate for the defect by overproducing the normal insulin. In addition, other factors such as aging, body weight gain, and insulin resistance may also play a role.
Insulin gene mutations affecting the conversion of proinsulin to insulin cause hyperproinsulinemia with or without mild diabetes. Mutations at the junction of C-peptide and α-chain (R89H, R89L, R89P; Figure 4) (251) disrupt the cleavage site of the prohormone convertase 2, and the mutated insulins can be cleaved only at the junction of C-peptide and β-chain by prohormone convertase 1/3. Another mutation at β-chain (H34D) was also shown to cause hyperproinsulinemia via an unclear mechanism. H34D proinsulin has a 4- to 5-fold higher receptor binding activity than wild type, and it has been suggested that it may bind to insulin receptors in the trans-Golgi and be secreted constitutively (256, 257).
2. Mutations in the INS gene lead to PNDM, MODY10, and type 1b diabetes
In 2007, Støy et al (258) first reported that mutations of the INS gene are a novel cause of PNDM. They identified 10 heterozygous mutations in INS among 16 PNDM families. These patients develop diabetes at a median age of 9 weeks, usually with diabetic ketoacidosis or severe hyperglycemia that needs immediate insulin therapy. To date, heterozygous autosomal dominantly inherited mutations in the INS gene are a common cause of PNDM (∼12%), second only to mutations in KCNJ11 (35). These dominant heterozygous INS mutations lead to preproinsulin misfolding and accumulation in the ER of the β-cell, causing ER stress and β-cell apoptosis (see Section II.A.5 for details).
Some INS mutations outside the exonic regions can also cause diabetes. For example, mutations in the intron regions of the INS gene such as heterozygous c.188–31G>A (259) and homozygous c.187+241G>A (260) cause PNDM. The intronic c.188–31G>A mutation introduces an ectopic splice site leading to the insertion of 29 nucleotides from the intronic sequence into the mature mRNA, which results in an aberrant transcript, producing misfolded proteins to induce ER stress and β-cell death (259). It is interesting that this c.188–31G>A mutation was also reported to cause MODY in one family (261). The other intronic c.187+241G>A mutation introduces a donor 5′ splice site, producing a truncated 482-bp INS mRNA product that includes a 237-bp intronic sequence. In addition, this mutation also produces an aberrant transcript with an insertion of a 79-nucleotide pseudoexon following exon 2 using a native 3′ acceptor site. These two mutant transcripts have been predicted to undergo nonsense and nonstop-mediated mRNA decay, respectively (260). Notably, a number of single amino acid substitutions in insulin, such as A23S, A23T, L68M, and G84R, that had been initially reported as possible deleterious pathogenic mutants in subjects with diabetes (35, 262, 263) could represent instances of ascertainment bias (264); the same “mutated” insulin molecules were subsequently shown to be nonpathogenic incidental variants (35, 265–267).
Molven et al (268) extended the phenotype of the INS gene mutations to include rare cases of MODY (MODY10) and type 1b diabetes. They reported the INS mutation c.137G>A (R46Q) in a MODY10 family and the INS mutation c.163C>T (R55C) in a type 1b diabetes family who presented with ketoacidosis and insulin dependency. The R46Q mutation disrupts a critical hydrogen bond formation, which impairs the stability of the insulin molecule. The R55C substitution is thought to perturb insulin biosynthesis via the introduction of an unpaired cysteine, as noted in the Akita mouse harboring a C96Y mutation in the ins2 gene (269, 270), or it may affect proteolytic processing of proinsulin to insulin (268). It is significant that the C96Y mutation has also been found in rare patients with PNDM (271). A novel heterozygous single nucleotide deletion (c.233delA) leading to a frameshift mutation (Q78fs) in the INS gene was recently identified in a MODY family. This mutation produces an aberrant “proinsulin” that is devoid of the native structures of C-peptide and α-chain (261).
Although most of the diabetogenic mutations in the INS gene occur in a dominant heterozygous manner, Garin et al (272) identified 10 different recessive INS mutations that cause PNDM or TNDM by interfering with the efficiency of insulin biosynthesis. These include four homozygous mutations involving the coding region: c.184C > T (p.Q62X), c.3G > T (p.0?), c.3G > A (p.0?), and a deletion (c.-370-? 186+?del); five homozygous mutations in the promoter or 3′ UTR regions: c.-331C > A, c.-331C > G, c.-218A > C, and c.-366_-343del, and c.*59A > G; and one compound heterozygote for two regulatory region mutations, c.-331C > G and c.-332C > G. Interestingly, patients with recessive INS mutations were diagnosed earlier (median, 1 week vs 10 weeks) and presented with a lower birth weight (−3.2 vs −2.0 SD score) than those with dominant heterozygous INS mutations. A plausible explanation of these observations is that two distinct disease mechanisms are involved in pathogenesis. In babies with recessive INS mutations, disrupted insulin synthesis occurs as soon as the β-cell starts to produce insulin during early fetal development. In contrast, in patients with dominant heterozygous INS mutations, the mutant preproinsulin misfolds and accumulates in the ER of the β-cell, causing increasingly severe ER stress that with time ultimately disrupts insulin production by the β-cell (267).
3. INS gene variants associated with T1D and possible mechanisms
Prior to GWAS, the INS gene on chromosome 11p15 was one of a few loci identified to be associated with susceptibility to T1D (273). The genetic risk conferred by the INS locus maps to a DNA region with a variable number of tandem repeats (VNTRs) of a 14- to 15-bp sequence approximately 0.5 kb upstream of the INS gene. The class I alleles of INS VNTR (26 to 63 repeats, frequency ∼70% in Caucasians) are associated with T1D, whereas the class III alleles (140 to more than 200 repeats, frequency ∼30% in Caucasians) seem to confer protection against T1D. Although these classes of VNTR alleles have little effect on insulin transcript levels in the pancreas, class I alleles are associated with lower levels of insulin mRNA and protein in the thymus, which may compromise negative selection and allow more autoreactive T cells to escape into the periphery, increasing susceptibility to T1D, whereas the higher insulin levels associated with class III alleles may enhance negative selection for autoreactive T cells, protecting against T1D (274). In support of this hypothesis, recent GWAS and other studies highlight INS as the second strongest risk locus (right after HLA) for T1D (273), insulin and its precursors being major initiating autoantigens in human T1D (275). It is unclear whether any missense mutation in the INS gene underlies common T1D, although the INS mutation c.163C>T (R55C) has been linked to type 1b diabetes (268).
4. INS gene variants associated with T2D
Although the class III alleles of INS VNTR may render protection against T1D, association studies suggested that they are inconsistently associated with the risk of T2D, probably due to population admixture (276–281). In Caucasians, class III INS VNTR appeared to confer a modest increase in risk of T2D (276), but the observation could not be replicated in a large family-based association study (279). The latter study did, however, report a marked excess transmission of class III INS VNTR from fathers to diabetic probands, suggesting the potential contribution of imprinted genes in the risk of T2D. In the Pima population, significant linkage disequilibrium was observed between INS VNTR (-23Hph1, an A/T SNP within the Hph1 restriction endonuclease site) and birth weight, but not T2D (280).
Notably, one large-scale meta-analysis showed that tyrosine hydroxylase (TH)-INS variant (rs10770141), a T1D risk allele, is protective for T2D. This variant, different from INS VNTR, is located in the promoter region of the TH gene and 11 kb upstream of the INS gene. TH-INS variant regulates the expression of TH, but not INS (282). The mechanism underlying the association between this variant of TH and diabetes has not been investigated.
5. Molecular mechanisms and pathophysiology that underlie INS mutations and diabetes
Heterozygous dominant mutations of the INS gene have been identified within all regions of the preproinsulin molecule including the signal peptide, C-peptide, and flanking cleavage sites (Figure 4) (283). The mutations within the signal peptide region likely impair one or more of the earliest events of proinsulin biosynthesis, including completion of translocation, excision of the signal peptide, initiation of β-chain folding, and disulfide bond formation (283). For example, R6C and R6H mutations in the INS gene lead to loss of the N-terminal positive charge of the 24-amino acid signal peptide, which fails to be fully translocated across the ER membrane and accumulate in a juxtanuclear cytoplasmic compartment, inducing ER stress and β-cell death (284). The preproinsulin A24D mutation that affects the last signal peptide residue likely disrupts normal signal peptide cleavage and interferes with downstream proinsulin folding and trafficking (265), leading to PNDM.
Proper disulfide pairing is a key event that determines whether a proinsulin molecule can achieve its native folded structure. Proinsulin contains six cysteine residues that form three evolutionarily conserved disulfide bonds: B7-A7, B19-A20, and A6-A11 (Figure 4). Many INS mutations introduce or eliminate a cysteine residue in the polypeptide chain, producing unpaired cysteine residues, which may alter or disrupt normal proinsulin disulfide pairing (283). For example, the mutations C43G and C96Y in the INS gene disrupt normal disulfide bonds at B19-A20 and A7-B7, respectively. The mutations R89C and G90C introduce an additional unpaired cysteine residue at the α-chain-C-peptide cleavage site (258). It is noteworthy that some mutations, such as G32S, G32R, and G47V, do not change proinsulin cysteine content, but may still disrupt the normal proinsulin folding, an observation that may be related to the fact that most of these mutations involve evolutionarily highly conserved residues (258). These mutations in the INS gene may lead to proinsulin misfolding and accumulation in the ER of the β-cell and may cause ER stress, β-cell apoptosis, and diabetes. Autophagy is a physiological process that maintains cellular homeostasis via clearance of misfolded proteins and damaged organelles (285). Accumulation of excessive amounts of misfolded proinsulin molecules per se, eg, in Akita mice, can stimulate autophagy by inhibiting mammalian target of rapamycin complex 1, alleviates ER stress, and partially prevents β-cell apoptosis. Treatment of female diabetic Akita mice with rapamycin (mammalian target of rapamycin complex 1 inhibitor) improves diabetes and further down-regulates β-cell apoptosis (286). These findings suggest that augmentation of autophagy may be a novel therapeutic approach for this type of diabetes. The challenge would be to find agents that are safe and efficacious in alleviating the excess ER stress in the β-cell.
Dominant-negative inhibition of wild-type proinsulin trafficking appears to be a feature of many of the INS mutants that cause PNDM (287). Forced expression of mouse proinsulin mutants with either cysteine residue substitution (C96Y) or noncysteine residue substitution (H29D or L35P), all known to be associated with human PNDM (Figure 4), inhibits the production of mature human insulin in the rat INS-1-derived 832/13 β-cell line, which coexpresses wild-type human insulin. On the other hand, it is noteworthy that the simultaneous introduction of missense substitutions that replace all six cysteine residues of proinsulin fails to repress the secretion of coexpressed wild-type human insulin. This suggests that pathways for proinsulin folding/processing are complicated; cysteine residues (paired or unpaired) in the INS mutants play a critical role in causing intracellular entrapment of coexpressed wild-type proinsulin, disrupting insulin production (287). In addition, INS mutants associated with PNDM facilitate the wild-type proinsulin to recruit into non-native disulfide-linked protein complexes in a cell line and in pancreatic islets of Akita mice (269, 270), which leads to disruption of normal folding of wild-type proinsulin. Although the C96Y mutation affects only one copy of the Ins2 gene, leaving one normal Ins2 gene and two normal Ins1 genes intact in these animals, β-cells from Akita mice display severe ER stress, accompanied by failure of GSIS, culminating in diabetes at an early age (269, 270). As noted earlier, heterozygous C96Y mutation was also found to cause PNDM in humans (271). Thus, it has been proposed that perturbation of the disulfide-coupled folding pathway of wild-type proinsulin initiates the molecular pathogenesis of PNDM associated with INS mutations (287).
In addition to INS mutations that cause proinsulin misfolding and severe ER stress that cripples the insulin production machinery of the β-cell, recessive INS mutations can also cause decreased insulin biosynthesis and diabetes via other mechanisms (272). These include absent or altered translation because of coding-sequence deletions or mutations, reduced insulin transcripts because of mutations in the promoter, or increased insulin mRNA instability. A few examples of INS mutations that impair insulin production directly include: two point mutations (c.3G > A and c.3G > T) at the translation initiation codon that abolish the native translation initiation for the preproinsulin protein; a 3′ UTR mutation (c.*59A > G) that abolishes the polyadenylation signal and potentially impairs mRNA stability; a 24-bp deletion (c.-366_-343del) in the INS promoter region that disrupts the promoter evolutionary conserved C1 and E1 elements, which are involved in the binding of MAFA and NEUROD1 to the INS promoter, respectively (288); and the CC dinucleotide mutations, c.-331(C > G, C > A) and c.-332C > G, that abolish a canonical binding motif for a key INS transactivator protein GLIS3 (214, 272, 289) (see discussion in the next section), producing up to 90% reduction in the INS promoter transcriptional activity.
B. Gli-similar 3
GLIS3, a member of the Krüppel-like family of transcription factors, is predominantly expressed in pancreatic β-cells, thyroid, and kidney (290, 291). Interestingly, GLIS3 is a NDM gene that is also associated with the susceptibility of both common T1D and T2D, as identified by GWAS and large- scale meta-analyses that include multiple populations.
1. Mutations in the GLIS3 gene lead to monogenic diabetes-related syndromes
About a decade ago, Senée et al (291) first reported that mutations in the GLIS3 gene cause a previously described rare syndrome characterized by PNDM, congenital hypothyroidism, polycystic kidneys, and in some cases, also with intrauterine growth retardation, facial anomalies, congenital glaucoma, and hepatic fibrosis. Six individuals from three families were affected. In one family (with neonatal diabetes, hypothyroidism [NDH]), they identified a homozygous insertion (2067insC) in GLIS3 leading to a frameshift and a truncated protein. In two other families with an incomplete syndrome, affected individuals harbor homozygous large fragment (426 and 149 Kb) deletions affecting the 5′ UTR of the GLIS3 gene. Recently, additional patients carrying homozygous partial GLIS3 gene deletions or mutations have been reported, most presenting with NDH as well as renal cysts and liver disease (292–294). It is noteworthy that Dimitri et al (293) reported a patient with compound heterozygous mutations in GLIS3 (p.Arg589Trp/exons 1–11 del) presenting with NDM, but not congenital hypothyroidism or renal cysts, who survived to adulthood.
2. GLIS3 gene variants and T1D
Barrett et al (295) first identified the association between a GLIS3 variant and common T1D in a report that combined a new GWAS and a meta-analysis of two prior studies that involved a total of 7514 T1D patients and 9045 control individuals. They found that the region of strong linkage disequilibrium at chromosome 9p24.2 with a MAF of 50.2% harbors only a single gene, GLIS3. The risk allele rs7020673 G was strongly associated with susceptibility to T1D in GWAS, case-control, and families analyses. This association was replicated recently in Pakistani T1D patients (296).
In another recent study, Winkler et al (297) found that the inclusion of 40 non-HLA gene SNPs could significantly improve the prediction accuracy of T1D, compared to that provided by HLA alone. Furthermore, they identified that HLA plus nine SNPs including the one from GLIS3 could achieve similar prediction accuracy as the total SNP set for T1D. They proposed a weighted risk model with selected SNPs that could be considered for recruitment of at-risk infants into studies of the natural history or disease prevention for T1D.
3. GLIS3 gene variants and T2D
Shortly after the report that the GLIS3 variant was strongly associated with common T1D (295), Dupuis et al (78) identified the association between a variation in GLIS3 and common T2D, which has been subsequently extensively examined in epidemiological studies among different populations.
In meta-analyses involving 46 186 nondiabetic participants in 21 GWAS and follow-up studies, Dupuis et al (78) identified the GLIS3 variant (rs7034200 A) as one of nine newly identified loci associated with fasting glucose in T2D trait, but not associated with body mass index, blood pressure, or lipid profile in participants of European descent. These findings were replicated in Danes (298), Chinese (299, 300), South Asians (301), and East Asians (302). Boesgaard et al (298) found that GLIS3 variant rs7034200 A was associated with reduced glucose-stimulated β-cell function in middle-aged Danish people. Barker et al (303) reported that GLIS3 variant rs7034200 was associated with fasting blood glucose levels in healthy children and adolescents of European origin. GLIS3 variant rs7034200 carriers showed impaired β-cell function, as indicated by homeostasis model assessment. Additionally, variant rs7041847 A in GLIS3 is associated with susceptibility to T2D in East Asians (302, 304).
4. Molecular mechanisms and pathophysiology that underlie the association of GLIS3 mutations or variations and diabetes
The gene association studies in humans are tantalizing. However, the underlying mechanisms were largely unknown until Glis3 global and β-cell-specific knockout mice were generated. Early studies in vitro identified a GLIS3 response element in the insulin gene promoter and revealed that GLIS3 physically and functionally interacts with PDX1, MAFA, and NEUROD1 to control insulin gene transcription (214). Moreover, the direct binding of GLIS3 to the insulin promoter is required for the interaction of PDX1, MAFA, and NEUROD1 with the promoter (305), indicating that GLIS3 plays a pivotal role in forming a transactivator complex to control insulin transcription. In agreement with these in vitro findings, Yang et al (306) found that tamoxifen-mediated β-cell-specific inactivation of Glis3 in adult mice down-regulates insulin expression, leading to fulminant diabetes and death. Thus, both in vitro and in vivo data demonstrated that GLIS3 is a potent transactivator of the insulin gene.
To gain insight into the role of GLIS3 in monogenic diabetes, three groups independently generated Glis3-deficient mice (289, 307, 308). Recapitulating the phenotype of NDH patients with GLIS3 mutations (291), Glis3-null mice died with severe hyperglycemia and ketoacidosis within the first days of life. The mice also developed hypothyroidism and polycystic kidney, whereas exocrine development was unaffected. Mechanistically, GLIS3 was shown to control fetal islet differentiation, synergistically with HNF6 and forkhead box A2 (FOXA2), via direct transactivation of Ngn3, the endocrine lineage-defining transcription factor (308).
Consistent with an autosomal recessive inheritance observed in humans, heterozygous Glis3-mutant mice remained euglycemic while on regular chow diet. However, when they were challenged with a HFD, adult mice developed diabetes because of impaired compensatory β-cell proliferation and mass expansion. It has been shown that GLIS3 controls β-cell proliferation in response to HFD feeding partly by direct regulation of cyclin D2 (Ccnd2) mRNA expression (306). CCND2 is essential for postnatal pancreatic β-cell growth (309, 310) and compensatory mass expansion in response to insulin resistance (311). Interestingly, recent GWAS identified that the variant rs11063069 G in CCND2 confers susceptibility risk to T2D (312), whereas the variant rs76895963 G in CCND2 reduces the risk of T2D by half and is correlated with increased CCND2 expression (167).
GLIS3 may also protect pancreatic β-cells against apoptosis. Proinflammatory cytokines, particularly IL-1β and interferon-γ (313), and the saturated nonesterified fatty acid palmitate (314), may contribute to β-cell loss in T1D and T2D, respectively. Nogueira et al (315) reported that Glis3 knockdown in INS-1 cells increased basal and inflammatory cytokine (IL-1β + interferon-γ)- or palmitate-induced β-cell apoptosis. They suggested that reduced Glis3 expression might modulate the alternative splicing of the proapoptotic BH3-only protein Bim, promoting the expression of the prodeath variant BimS and β-cell death.
These data from cell lines and mouse models provide the mechanistic basis for the genetic associations of the GLIS3 gene and NDM (291), common T1D (295, 296), and T2D (78, 298–304) (Figure 5). GLIS3 may underlie all three forms of diabetes by potently controlling insulin gene transcription and likely β-cell survival. The homozygous insertion (2067insC) in GLIS3 identified in an NDH family (291) leads to a frameshift and a truncated protein that lacks the C-terminal transactivation domain (290), which is required to activate both NGN3 (308) and INS (214, 305, 306) gene transcription. This serious developmental deficiency causes severely abnormal β-cell development in utero and marked impairment in insulin production at birth, culminating in PNDM. In addition to causing PNDM (289, 291, 307, 308), GLIS3 was also found by GWAS to be linked to common T1D (295, 296), probably through its action on insulin gene transcription and β-cell survival. Finally, GWAS showed that the GLIS3 locus is also linked to T2D (78, 298–304). Studies in adult mouse models suggest that GLIS3 plays a critical role in compensatory obesity-induced β-cell proliferation via the regulation of Ccnd2 transcription (306). Of note, the effects of the susceptibility gene variants examined in isolation on the risk of T1D or T2D are typically modest, often making it a challenge to decipher such subtle effects by current functional assays (10). The precise mechanisms underlying the GLIS3 variants and diabetes remain to be fully elucidated.
Figure 5.
Multifaceted role of GLIS3 in diabetes. In utero, GLIS3 controls islet differentiation by transactivating Ngn3, synergistically with HNF6 and forkhead box A2 (FOXA2, also known as HNF3B or transcription factor 3B [TCF-3B]). Loss-of-function mutations or ablation of Glis3 cause impaired islet differentiation and NDM. After birth, GLIS3 predominantly controls insulin gene transcription, cooperating with MAFA, PDX1, and NEUROD1. GLIS3 is also required for obesity-induced β-cell proliferation and compensatory β-cell mass expansion by transactivating Ccnd2. In addition, GLIS3 plays a protective role for β-cell survival, possibly by regulating BimS. Therefore, impairment of GLIS3 function also plays a key role in the development of T1D and T2D. This figure summarizes the pathways by which ablation, loss-of-function mutations, or functional impairment of GLIS3 cause NDM, T1D, and T2D. Pathways in the blue rectangle take place in the developing pancreas in utero and underlie monogenic NDM; pathways in the yellow square and the red rectangle pertain to the postnatal β-cell and contribute to the development of T1D and T2D, respectively.
IV. Concluding Remarks
In the past few decades, an increasing number of genes have been linked to monogenic diabetes and related syndromes. GWAS have markedly expanded the repertoire of susceptibility loci associated with common T1D or T2D. Although T1D and T2D share overlap in select clinical and pathological features, the susceptibility loci identified by GWAS indicate that the genetic basis for the two common types of diabetes is mostly distinct (7–15) (Figure 2). In agreement with this premise, candidate gene-association studies and GWAS showed that, to date, at least 12 monogenic diabetes genes are associated only with T2D, but not T1D; the INS locus appears to be an exception in that it is strongly associated with T1D but is inconsistently associated with T2D. GLIS3 is a developmental transcription factor gene that was shown to underlie a rare kind of monogenic PNDM. It is unique that GWAS showed that variants of GLIS3 are associated with both common T1D and T2D in different populations. Not unexpectedly, the T1D and T2D GLIS3 variants are unique and do not overlap, likely because they confer their diabetogenic effects via different mechanisms.
The monogenic forms of diabetes offer excellent models to unravel the developmental biology of pancreatic β-cells and the role of the implicated genes in the regulation of β-cell function. Information gained from these studies also facilitates the interpretation of GWAS data for common T1D and T2D. Because more and more rare variants are being identified in diabetic patients with advanced sequencing technologies, special caution needs be taken to avoid ascertainment bias, overinterpretation, and misidentification of risk variants for common forms of diabetes (264). A number of rare mutations of late-onset MODY genes (eg, PDX1) have been reported to be variants associated with common T2D, blurring the boundary between the two diabetes syndromes. Differentiating between an incompletely penetrant MODY mutation from a rare T2D variant is often difficult and can be a matter of semantics (264, 316).
Most of the T2D risk variants in islet transcription factors are enriched in clustered islet enhancers in human islet cells where they interact functionally with the epigenome. Individual variants may display disrupted islet enhancer activity, which may be an important mechanism for the genetic predisposition of T2D (317, 318). It remains a challenge to translate our knowledge from research in the monogenic diabetes genes into effective forms of treatment. Because many of these have been shown to occur in T1D or T2D, we are hopeful that future research into these genes will provide new insights into disease pathogenesis and treatment target identification, not only for the rare monogenic disorders, but also for the worldwide epidemic of T1D and T2D.
Acknowledgments
Research in the authors' laboratories that was discussed in this review was supported by the National Institutes of Health (P30-DK079638) for a Diabetes Endocrinology Research Center at Baylor College of Medicine (T32-HL066991), Juvenile Diabetes Foundation (Award 46-2010-752), the American Diabetes Association (Grant 1-14-MN-01), the T.T. and W.F. Chao Global Foundation, the Cindy and Frank Liu Family Foundation, the Cunningham Family Foundation, the Betty Rutherford Chair in Diabetes Research at Baylor St. Luke's Medical Center (to L.C.), the American Heart Association (Grant 13SDG17090096; to Y.Y.), and MetroHealth Medical Center, Case Western Reserve University start-up funds (to Y.Y.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ABCC8
- ATP-binding cassette transporter subfamily C member 8
- CCND2
- cyclin D2
- CHI
- congenital hyperinsulinism
- e
- embryonic day
- ER
- endoplasmic reticulum
- FBS
- Fanconi-Bickel syndrome
- FGF
- fibroblast growth factor
- GCK
- glucokinase
- GDM
- gestational diabetes mellitus
- GKRP
- GCK regulatory protein
- GLIS3
- Gli-similar 3
- GLP-1
- glucagon-like peptide-1
- GLUT
- glucose transporter
- G6PC2
- glucose-6-phosphatase, catalytic 2
- GSIS
- glucose-stimulated insulin secretion
- GWAS
- genome-wide association studies
- HFD
- high-fat diet
- HLA
- human leukocyte antigen
- HLH
- helix-loop-helix
- HNF1A
- hepatocyte nuclear factor 1 homeobox A
- IGT
- impaired glucose tolerance
- KATP
- ATP-sensitive potassium
- KCNJ11
- potassium inward-rectifying channel, subfamily J, member 11
- KPD
- ketosis-prone diabetes
- MAF
- minor allele frequency
- MODY
- maturity-onset diabetes of the young
- NDH
- neonatal diabetes, hypothyroidism
- NDM
- neonatal diabetes mellitus
- PAX4
- paired box 4
- PDX1
- pancreatic and duodenal homeobox 1
- PNDM
- permanent NDM
- PPARA
- peroxisome proliferator-activated receptor α
- PPARG
- peroxisome proliferator-activated receptor γ
- RCAD
- renal cysts and diabetes
- SGLT2
- sodium-glucose cotransporter 2
- SLC2A2
- solute carrier family 2, member 2
- SUR1
- sulfonylurea receptor 1
- T1D
- type 1 diabetes
- T2D
- type 2 diabetes
- TH
- tyrosine hydroxylase
- TNDM
- transient NDM
- TZD
- thiazolidinedione
- UPR
- unfolded protein response
- UTR
- untranslated region
- VNTR
- variable number of tandem repeats
- WFS1
- Wolfram syndrome 1.
References
- 1. Steck AK, Winter WE. Review on monogenic diabetes. Curr Opin Endocrinol Diabetes Obes. 2011;18(4):252–258. [DOI] [PubMed] [Google Scholar]
- 2. Greeley SA, Naylor RN, Philipson LH, Bell GI. Neonatal diabetes: an expanding list of genes allows for improved diagnosis and treatment. Curr Diab Rep. 2011;11(6):519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edghill EL, Dix RJ, Flanagan SE, et al. HLA genotyping supports a nonautoimmune etiology in patients diagnosed with diabetes under the age of 6 months. Diabetes. 2006;55(6):1895–1898. [DOI] [PubMed] [Google Scholar]
- 4. Flanagan SE, Haapaniemi E, Russell MA, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014;46(8):812–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fajans SS, Bell GI. MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care. 2011;34(8):1878–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gardner DS, Tai ES. Clinical features and treatment of maturity onset diabetes of the young (MODY). Diabetes Metab Syndr Obes. 2012;5:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32(4):457–467. [DOI] [PubMed] [Google Scholar]
- 8. Grarup N, Sandholt CH, Hansen T, Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014;57(8):1528–1541. [DOI] [PubMed] [Google Scholar]
- 9. Hara K, Shojima N, Hosoe J, Kadowaki T. Genetic architecture of type 2 diabetes. Biochem Biophys Res Commun. 2014;452(2):213–220. [DOI] [PubMed] [Google Scholar]
- 10. Bonnefond A, Froguel P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 2015;21(3):357–368. [DOI] [PubMed] [Google Scholar]
- 11. Prasad RB, Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes. 2015;6(1):87–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Basile KJ, Guy VC, Schwartz S, Grant SF. Overlap of genetic susceptibility to type 1 diabetes, type 2 diabetes, and latent autoimmune diabetes in adults. Curr Diab Rep. 2014;14(11):550. [DOI] [PubMed] [Google Scholar]
- 13. Pociot F, Akolkar B, Concannon P, et al. Genetics of type 1 diabetes: what's next? Diabetes. 2010;59(7):1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fortune MD, Guo H, Burren O, et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet. 2015;47(7):839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McTaggart JS, Clark RH, Ashcroft FM. The role of the KATP channel in glucose homeostasis in health and disease: more than meets the islet. J Physiol. 2010;588:3201–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yorifuji T. Congenital hyperinsulinism: current status and future perspectives. Ann Pediatr Endocrinol Metab. 2014;19(2):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shyng SL, Ferrigni T, Shepard JB, et al. Functional analyses of novel mutations in the sulfonylurea receptor 1 associated with persistent hyperinsulinemic hypoglycemia of infancy. Diabetes. 1998;47(7):1145–1151. [DOI] [PubMed] [Google Scholar]
- 19. Cartier EA, Conti LR, Vandenberg CA, Shyng SL. Defective trafficking and function of KATP channels caused by a sulfonylurea receptor 1 mutation associated with persistent hyperinsulinemic hypoglycemia of infancy. Proc Natl Acad Sci USA. 2001;98(5):2882–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taschenberger G, Mougey A, Shen S, Lester LB, LaFranchi S, Shyng SL. Identification of a familial hyperinsulinism-causing mutation in the sulfonylurea receptor 1 that prevents normal trafficking and function of KATP channels. J Biol Chem. 2002;277(19):17139–17146. [DOI] [PubMed] [Google Scholar]
- 21. Kapoor RR, Flanagan SE, James CT, et al. Hyperinsulinaemic hypoglycaemia and diabetes mellitus due to dominant ABCC8/KCNJ11 mutations. Diabetologia. 2011;54(10):2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pinney SE, MacMullen C, Becker S, et al. Clinical characteristics and biochemical mechanisms of congenital hyperinsulinism associated with dominant KATP channel mutations. J Clin Invest. 2008;118(8):2877–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huopio H, Reimann F, Ashfield R, et al. Dominantly inherited hyperinsulinism caused by a mutation in the sulfonylurea receptor type 1. J Clin Invest. 2000;106(7):897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gussinyer M, Clemente M, Cebrián R, Yeste D, Albisu M, Carrascosa A. Glucose intolerance and diabetes are observed in the long-term follow-up of nonpancreatectomized patients with persistent hyperinsulinemic hypoglycemia of infancy due to mutations in the ABCC8 gene. Diabetes Care. 2008;31(6):1257–1259. [DOI] [PubMed] [Google Scholar]
- 25. Abdulhadi-Atwan M, Bushman J, Tornovsky-Babaey S, et al. Novel de novo mutation in sulfonylurea receptor 1 presenting as hyperinsulinism in infancy followed by overt diabetes in early adolescence. Diabetes. 2008;57(7):1935–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vieira TC, Bergamin CS, Gurgel LC, Moisés RS. Hyperinsulinemic hypoglycemia evolving to gestational diabetes and diabetes mellitus in a family carrying the inactivating ABCC8 E1506K mutation. Pediatr Diabetes. 2010;11(7):505–508. [DOI] [PubMed] [Google Scholar]
- 27. Huopio H, Otonkoski T, Vauhkonen I, Reimann F, Ashcroft FM, Laakso M. A new subtype of autosomal dominant diabetes attributable to a mutation in the gene for sulfonylurea receptor 1. Lancet. 2003;361(9354):301–307. [DOI] [PubMed] [Google Scholar]
- 28. Arnoux JB, Verkarre V, Saint-Martin C, et al. Congenital hyperinsulinism: current trends in diagnosis and therapy. Orphanet J Rare Dis. 2011;6:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsu WH, Xiang HD, Rajan AS, Kunze DL, Boyd AE., 3rd Somatostatin inhibits insulin secretion by a G-protein-mediated decrease in Ca2+ entry through voltage-dependent Ca2+ channels in the β cell. J Biol Chem. 1991;266(2):837–843. [PubMed] [Google Scholar]
- 30. Schmidt A, Hescheler J, Offermanns S, et al. Involvement of pertussis toxin-sensitive G-proteins in the hormonal inhibition of dihydropyridine-sensitive Ca2+ currents in an insulin-secreting cell line (RINm5F). J Biol Chem. 1991;266(27):18025–18033. [PubMed] [Google Scholar]
- 31. de Weille JR, Schmid-Antomarchi H, Fosset M, Lazdunski M. Regulation of ATP-sensitive K+ channels in insulinoma cells: activation by somatostatin and protein kinase C and the role of cAMP. Proc Natl Acad Sci USA. 1989;86(8):2971–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gloyn AL, Pearson ER, Antcliff JF, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–1849. [DOI] [PubMed] [Google Scholar]
- 33. Babenko AP, Polak M, Cavé H, et al. Activating mutations in the ABCC8 gene in neonatal diabetes mellitus. N Engl J Med. 2006;355(5):456–466. [DOI] [PubMed] [Google Scholar]
- 34. Proks P, Arnold AL, Bruining J, et al. A heterozygous activating mutation in the sulphonylurea receptor SUR1 (ABCC8) causes neonatal diabetes. Hum Molec Genet. 2006;15(11):1793–1800. [DOI] [PubMed] [Google Scholar]
- 35. Edghill EL, Flanagan SE, Patch AM, et al. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57(4):1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flanagan SE, Clauin S, Bellanné-Chantelot C, et al. Update of mutations in the genes encoding the pancreatic β-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat. 2009;30(2):170–180. [DOI] [PubMed] [Google Scholar]
- 37. Craig TJ, Shimomura K, Holl RW, Flanagan SE, Ellard S, Ashcroft FM. An in-frame deletion in Kir6.2 (KCNJ11) causing neonatal diabetes reveals a site of interaction between Kir6.2 and SUR1. J Clin Endocrinol Metab. 2009;94(7):2551–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edghill EL, Flanagan SE, Ellard S. Permanent neonatal diabetes due to activating mutations in ABCC8 and KCNJ11. Rev Endocr Metab Disord. 2010;11(3):193–198. [DOI] [PubMed] [Google Scholar]
- 39. Bonnefond A, Philippe J, Durand E, et al. Whole-exome sequencing and high throughput genotyping identified KCNJ11 as the thirteenth MODY gene. PLoS One. 2012;7(6):e37423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bowman P, Flanagan SE, Edghill EL, et al. Heterozygous ABCC8 mutations are a cause of MODY. Diabetologia. 2012;55(1):123–127. [DOI] [PubMed] [Google Scholar]
- 41. Männikkö R, Flanagan SE, Sim X, et al. Mutations of the same conserved glutamate residue in NBD2 of the sulfonylurea receptor 1 subunit of the KATP channel can result in either hyperinsulinism or neonatal diabetes. Diabetes. 2011;60(6):1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hani EH, Boutin P, Durand E, et al. Missense mutations in the pancreatic islet β cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of type II diabetes mellitus in Caucasians. Diabetologia. 1998;41(12):1511–1515. [DOI] [PubMed] [Google Scholar]
- 43. Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic β-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52(2):568–572. [DOI] [PubMed] [Google Scholar]
- 44. Love-Gregory L, Wasson J, Lin J, Skolnick G, Suarez B, Permutt MA. E23K single nucleotide polymorphism in the islet ATP-sensitive potassium channel gene (Kir6.2) contributes as much to the risk of type II diabetes in Caucasians as the PPARγ Pro12Ala variant. Diabetologia. 2003;46(1):136–137. [DOI] [PubMed] [Google Scholar]
- 45. Nielsen EM, Hansen L, Carstensen B, et al. The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes. 2003;52(2):573–577. [DOI] [PubMed] [Google Scholar]
- 46. Florez JC, Burtt N, de Bakker PI, et al. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53(5):1360–1368. [DOI] [PubMed] [Google Scholar]
- 47. Florez JC, Jablonski KA, Kahn SE, et al. Type 2 diabetes-associated missense polymorphisms KCNJ11 E23K and ABCC8 A1369S influence progression to diabetes and response to interventions in the Diabetes Prevention Program. Diabetes. 2007;56(2):531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Patanè G, Piro S, Rabuazzo AM, Anello M, Vigneri R, Purrello F. Metformin restores insulin secretion altered by chronic exposure to free fatty acids or high glucose: a direct metformin effect on pancreatic β-cells. Diabetes. 2000;49(5):735–740. [DOI] [PubMed] [Google Scholar]
- 49. Lablanche S, Cottet-Rousselle C, Lamarche F, et al. Protection of pancreatic INS-1 β-cells from glucose- and fructose-induced cell death by inhibiting mitochondrial permeability transition with cyclosporin A or metformin. Cell Death Dis. 2011;2:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laukkanen O, Pihlajamäki J, Lindström J, et al. Polymorphisms of the SUR1 (ABCC8) and Kir6.2 (KCNJ11) genes predict the conversion from impaired glucose tolerance to type 2 diabetes. The Finnish Diabetes Prevention Study. J Clin Endocrinol Metab. 2004;89(12):6286–6290. [DOI] [PubMed] [Google Scholar]
- 51. Lyssenko V, Almgren P, Anevski D, et al. Genetic prediction of future type 2 diabetes. PLoS Med. 2005;2(12):e345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baier LJ, Muller YL, Remedi MS, et al. ABCC8 R1420H loss-of-function variant in a Southwest American Indian community: association with increased birth weight and doubled risk of type 2 diabetes. Diabetes. 2015;64(12):4322–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gloyn AL, Hashim Y, Ashcroft SJ, et al. Association studies of variants in promoter and coding regions of β-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with type 2 diabetes mellitus (UKPDS 53). Diabet Med. 2001;18(3):206–212. [DOI] [PubMed] [Google Scholar]
- 54. Sesti G, Laratta E, Cardellini M, et al. The E23K variant of KCNJ11 encoding the pancreatic β-cell adenosine 5′-triphosphate-sensitive potassium channel subunit Kir6.2 is associated with an increased risk of secondary failure to sulfonylurea in patients with type 2 diabetes. J Clin Endocrinol Metab. 2006;91(6):2334–2339. [DOI] [PubMed] [Google Scholar]
- 55. Proks P, Antcliff JF, Lippiat J, Gloyn AL, Hattersley AT, Ashcroft FM. Molecular basis of Kir6.2 mutations associated with neonatal diabetes or neonatal diabetes plus neurological features. Proc Natl Acad Sci USA. 2004;101(50):17539–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Proks P, Shimomura K, Craig TJ, Girard CA, Ashcroft FM. Mechanism of action of a sulphonylurea receptor SUR1 mutation (F132L) that causes DEND syndrome. Hum Molec Genet. 2007;16(16):2011–2019. [DOI] [PubMed] [Google Scholar]
- 57. Schwanstecher C, Neugebauer B, Schulz M, Schwanstecher M. The common single nucleotide polymorphism E23K in K(IR)6.2 sensitizes pancreatic β-cell ATP-sensitive potassium channels toward activation through nucleoside diphosphates. Diabetes. 2002;51(suppl 3):S363–S367. [DOI] [PubMed] [Google Scholar]
- 58. Riedel MJ, Boora P, Steckley D, de Vries G, Light PE. Kir6.2 polymorphisms sensitize β-cell ATP-sensitive potassium channels to activation by acyl CoAs: a possible cellular mechanism for increased susceptibility to type 2 diabetes? Diabetes. 2003;52(10):2630–2635. [DOI] [PubMed] [Google Scholar]
- 59. Rosengren AH, Braun M, Mahdi T, et al. Reduced insulin exocytosis in human pancreatic β-cells with gene variants linked to type 2 diabetes. Diabetes. 2012;61(7):1726–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fatehi M, Raja M, Carter C, Soliman D, Holt A, Light PE. The ATP-sensitive K(+) channel ABCC8 S1369A type 2 diabetes risk variant increases MgATPase activity. Diabetes. 2012;61(1):241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pearson ER, Flechtner I, Njølstad PR, et al. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med. 2006;355(5):467–477. [DOI] [PubMed] [Google Scholar]
- 62. Thurber BW, Carmody D, Tadie EC, et al. Age at the time of sulfonylurea initiation influences treatment outcomes in KCNJ11-related neonatal diabetes. Diabetologia. 2015;58(7):1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Holstein A, Hahn M, Stumvoll M, Kovacs P. The E23K variant of KCNJ11 and the risk for severe sulfonylurea-induced hypoglycemia in patients with type 2 diabetes. Horm Metab Res. 2009;41(5):387–390. [DOI] [PubMed] [Google Scholar]
- 64. Javorsky M, Klimcakova L, Schroner Z, et al. KCNJ11 gene E23K variant and therapeutic response to sulfonylureas. Eur J Intern Med. 2012;23(3):245–249. [DOI] [PubMed] [Google Scholar]
- 65. Ragia G, Tavridou A, Petridis I, Manolopoulos VG. Association of KCNJ11 E23K gene polymorphism with hypoglycemia in sulfonylurea-treated type 2 diabetic patients. Diabetes Res Clin Pract. 2012;98(1):119–124. [DOI] [PubMed] [Google Scholar]
- 66. Osbak KK, Colclough K, Saint-Martin C, et al. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum Mutat. 2009;30(11):1512–1526. [DOI] [PubMed] [Google Scholar]
- 67. Postic C, Shiota M, Niswender KD, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274(1):305–315. [DOI] [PubMed] [Google Scholar]
- 68. Grupe A, Hultgren B, Ryan A, Ma YH, Bauer M, Stewart TA. Transgenic knockouts reveal a critical requirement for pancreatic β cell glucokinase in maintaining glucose homeostasis. Cell. 1995;83(1):69–78. [DOI] [PubMed] [Google Scholar]
- 69. Byrne MM, Sturis J, Clément K, et al. Insulin secretory abnormalities in subjects with hyperglycemia due to glucokinase mutations. J Clin Invest. 1994;93(3):1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Steele AM, Shields BM, Wensley KJ, Colclough K, Ellard S, Hattersley AT. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA. 2014;311(3):279–286. [DOI] [PubMed] [Google Scholar]
- 71. Rudland VL, Hinchcliffe M, Pinner J, et al. Identifying glucokinase monogenic diabetes in a multiethnic gestational diabetes mellitus cohort: new pregnancy screening criteria and utility of HbA1c. Diabetes Care. 2016;39(1):50–52. [DOI] [PubMed] [Google Scholar]
- 72. Chakera AJ, Spyer G, Vincent N, Ellard S, Hattersley AT, Dunne FP. The 0.1% of the population with glucokinase monogenic diabetes can be recognized by clinical characteristics in pregnancy: the Atlantic Diabetes in Pregnancy cohort. Diabetes Care. 2014;37(5):1230–1236. [DOI] [PubMed] [Google Scholar]
- 73. Stride A, Shields B, Gill-Carey O, et al. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia. 2014;57(1):54–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tornovsky-Babeay S, Dadon D, Ziv O, et al. Type 2 diabetes and congenital hyperinsulinism cause DNA double-strand breaks and p53 activity in β cells. Cell Metab. 2014;19(1):109–121. [DOI] [PubMed] [Google Scholar]
- 75. Soranzo N, Sanna S, Wheeler E, et al. Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Qi Q, Wu Y, Li H, et al. Association of GCKR rs780094, alone or in combination with GCK rs1799884, with type 2 diabetes and related traits in a Han Chinese population. Diabetologia. 2009;52(5):834–843. [DOI] [PubMed] [Google Scholar]
- 77. Tam CH, Ma RC, So WY, et al. Interaction effect of genetic polymorphisms in glucokinase (GCK) and glucokinase regulatory protein (GCKR) on metabolic traits in healthy Chinese adults and adolescents. Diabetes. 2009;58(3):765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Muller YL, Piaggi P, Hoffman D, et al. Common genetic variation in the glucokinase gene (GCK) is associated with type 2 diabetes and rates of carbohydrate oxidation and energy expenditure. Diabetologia. 2014;57(7):1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ingelsson E, Langenberg C, Hivert MF, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59(5):1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brown KS, Kalinowski SS, Megill JR, Durham SK, Mookhtiar KA. Glucokinase regulatory protein may interact with glucokinase in the hepatocyte nucleus. Diabetes. 1997;46(2):179–186. [DOI] [PubMed] [Google Scholar]
- 82. de la Iglesia N, Veiga-da-Cunha M, Van Schaftingen E, Guinovart JJ, Ferrer JC. Glucokinase regulatory protein is essential for the proper subcellular localisation of liver glucokinase. FEBS Lett. 1999;456(2):332–338. [DOI] [PubMed] [Google Scholar]
- 83. Guillam MT, Hümmler E, Schaerer E, et al. Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2. Nat Genet. 1997;17(3):327–330. [DOI] [PubMed] [Google Scholar]
- 84. Sansbury FH, Flanagan SE, Houghton JA, et al. SLC2A2 mutations can cause neonatal diabetes, suggesting GLUT2 may have a role in human insulin secretion. Diabetologia. 2012;55(9):2381–2385. [DOI] [PubMed] [Google Scholar]
- 85. De Vos A, Heimberg H, Quartier E, et al. Human and rat β cells differ in glucose transporter but not in glucokinase gene expression. J Clin Invest. 1995;96(5):2489–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Santer R, Schneppenheim R, Dombrowski A, Götze H, Steinmann B, Schaub J. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat Genet. 1997;17(3):324–326. [DOI] [PubMed] [Google Scholar]
- 87. Abbasi F, Azizi F, Javaheri M, Mosallanejad A, Ebrahim-Habibi A, Ghafouri-Fard S. Segregation of a novel homozygous 6 nucleotide deletion in GLUT2 gene in a Fanconi-Bickel syndrome family. Gene. 2015;557(1):103–105. [DOI] [PubMed] [Google Scholar]
- 88. Mihout F, Devuyst O, Bensman A, et al. Acute metabolic acidosis in a GLUT2-deficient patient with Fanconi-Bickel syndrome: new pathophysiology insights. Nephrol Dial Transplant. 2014;29(suppl 4):iv113–iv116. [DOI] [PubMed] [Google Scholar]
- 89. Mannstadt M, Magen D, Segawa H, et al. Fanconi-Bickel syndrome and autosomal recessive proximal tubulopathy with hypercalciuria (ARPTH) are allelic variants caused by GLUT2 mutations. J Clin Endocrinol Metab. 2012;97(10):E1978–E1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Al-Haggar M, Sakamoto O, Shaltout A, Al-Hawari A, Wahba Y, Abdel-Hadi D. Mutation analysis of the GLUT2 gene in three unrelated Egyptian families with Fanconi-Bickel syndrome: revisited gene atlas for renumbering. Clin Exp Nephrol. 2012;16(4):604–610. [DOI] [PubMed] [Google Scholar]
- 91. Grünert SC, Schwab KO, Pohl M, Sass JO, Santer R. Fanconi-Bickel syndrome: GLUT2 mutations associated with a mild phenotype. Mol Genet Metab. 2012;105(3):433–437. [DOI] [PubMed] [Google Scholar]
- 92. Janssen RC, Bogardus C, Takeda J, Knowler WC, Thompson DB. Linkage analysis of acute insulin secretion with GLUT2 and glucokinase in Pima Indians and the identification of a missense mutation in GLUT2. Diabetes. 1994;43(4):558–563. [DOI] [PubMed] [Google Scholar]
- 93. Alcolado JC, Baroni MG, Li SR. Association between a restriction fragment length polymorphism at the liver/islet cell (GluT 2) glucose transporter and familial type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia. 1991;34(10):734–736. [DOI] [PubMed] [Google Scholar]
- 94. Barroso I, Luan J, Middelberg RP, et al. Candidate gene association study in type 2 diabetes indicates a role for genes involved in β-cell function as well as insulin action. PLoS Biol. 2003;1(1):E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Matsutani A, Koranyi L, Cox N, Permutt MA. Polymorphisms of GLUT2 and GLUT4 genes. Use in evaluation of genetic susceptibility to NIDDM in blacks. Diabetes. 1990;39(12):1534–1542. [DOI] [PubMed] [Google Scholar]
- 96. Patel P, Bell GI, Cook JT, Turner RC, Wainscoat JS. Multiple restriction fragment length polymorphisms at the GLUT2 locus: GLUT2 haplotypes for genetic analysis of type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia. 1991;34(11):817–821. [DOI] [PubMed] [Google Scholar]
- 97. Tanizawa Y, Riggs AC, Chiu KC, et al. Variability of the pancreatic islet β cell/liver (GLUT 2) glucose transporter gene in NIDDM patients. Diabetologia. 1994;37(4):420–427. [DOI] [PubMed] [Google Scholar]
- 98. Matsubara A, Tanizawa Y, Matsutani A, Kaneko T, Kaku K. Sequence variations of the pancreatic islet/liver glucose transporter (GLUT2) gene in Japanese subjects with noninsulin dependent diabetes mellitus. J Clin Endocrinol Metab. 1995;80(11):3131–3135. [DOI] [PubMed] [Google Scholar]
- 99. Laukkanen O, Lindström J, Eriksson J, et al. Polymorphisms in the SLC2A2 (GLUT2) gene are associated with the conversion from impaired glucose tolerance to type 2 diabetes: the Finnish Diabetes Prevention Study. Diabetes. 2005;54(7):2256–2260. [DOI] [PubMed] [Google Scholar]
- 100. Kilpeläinen TO, Lakka TA, Laaksonen DE, et al. Physical activity modifies the effect of SNPs in the SLC2A2 (GLUT2) and ABCC8 (SUR1) genes on the risk of developing type 2 diabetes. Physiol Genomics. 2007;31(2):264–272. [DOI] [PubMed] [Google Scholar]
- 101. Michau A, Guillemain G, Grosfeld A, et al. Mutations in SLC2A2 gene reveal hGLUT2 function in pancreatic β cell development. J Biol Chem. 2013;288(43):31080–31092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Burgdorf KS, Grarup N, Justesen JM, et al. Studies of the association of Arg72Pro of tumor suppressor protein p53 with type 2 diabetes in a combined analysis of 55,521 Europeans. PLoS One. 2011;6(1):e15813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Willer CJ, Bonnycastle LL, Conneely KN, et al. Screening of 134 single nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes replicates association with 12 SNPs in nine genes. Diabetes. 2007;56(1):256–264. [DOI] [PubMed] [Google Scholar]
- 104. Eny KM, Wolever TM, Fontaine-Bisson B, El-Sohemy A. Genetic variant in the glucose transporter type 2 is associated with higher intakes of sugars in two distinct populations. Physiol Genomics. 2008;33(3):355–360. [DOI] [PubMed] [Google Scholar]
- 105. Burcelin R, Dolci W, Thorens B. Glucose sensing by the hepatoportal sensor is GLUT2-dependent: in vivo analysis in GLUT2-null mice. Diabetes. 2000;49(10):1643–1648. [DOI] [PubMed] [Google Scholar]
- 106. Roncero I, Alvarez E, Chowen JA, et al. Expression of glucose transporter isoform GLUT-2 and glucokinase genes in human brain. J Neurochem. 2004;88(5):1203–1210. [DOI] [PubMed] [Google Scholar]
- 107. Li B, Xi X, Roane DS, Ryan DH, Martin RJ. Distribution of glucokinase, glucose transporter GLUT2, sulfonylurea receptor-1, glucagon-like peptide-1 receptor and neuropeptide Y messenger RNAs in rat brain by quantitative real time RT-PCR. Brain Res Mol Brain Res. 2003;113:139–142. [DOI] [PubMed] [Google Scholar]
- 108. Pontoglio M, Barra J, Hadchouel M, et al. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84(4):575–585. [DOI] [PubMed] [Google Scholar]
- 109. Pontoglio M, Sreenan S, Roe M, et al. Defective insulin secretion in hepatocyte nuclear factor 1α-deficient mice. J Clin Invest. 1998;101(10):2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10(2):267–282. [PubMed] [Google Scholar]
- 111. Chen WS, Manova K, Weinstein DC, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8(20):2466–2477. [DOI] [PubMed] [Google Scholar]
- 112. Boj SF, Parrizas M, Maestro MA, Ferrer J. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc Natl Acad Sci USA. 2001;98(25):14481–14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Stride A, Ellard S, Clark P, et al. β-Cell dysfunction, insulin sensitivity, and glycosuria precede diabetes in hepatocyte nuclear factor-1α mutation carriers. Diabetes Care. 2005;28(7):1751–1756. [DOI] [PubMed] [Google Scholar]
- 114. Stanescu DE, Hughes N, Kaplan B, Stanley CA, De León DD. Novel presentations of congenital hyperinsulinism due to mutations in the MODY genes: HNF1A and HNF4A. J Clin Endocrinol Metab. 2012;97(10):E2026–E2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia. 2010;53(12):2504–2508. [DOI] [PubMed] [Google Scholar]
- 116. Ellard S, Colclough K. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 α (HNF1A) and 4 α (HNF4A) in maturity-onset diabetes of the young. Hum Mutat. 2006;27(9):854–869. [DOI] [PubMed] [Google Scholar]
- 117. Pontoglio M, Prié D, Cheret C, et al. HNF1α controls renal glucose reabsorption in mouse and man. EMBO Rep. 2000;1(4):359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hegele RA, Cao H, Harris SB, Hanley AJ, Zinman B. The hepatic nuclear factor-1α G319S variant is associated with early-onset type 2 diabetes in Canadian Oji-Cree. J Clin Endocrinol Metab. 1999;84(3):1077–1082. [DOI] [PubMed] [Google Scholar]
- 119. SIGMA Type 2 Diabetes Consortium, Estrada K, Aukrust I, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311(22):2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Andersen MK, Sterner M, Forsén T, et al. Type 2 diabetes susceptibility gene variants predispose to adult-onset autoimmune diabetes. Diabetologia. 2014;57(9):1859–1868. [DOI] [PubMed] [Google Scholar]
- 121. Silander K, Mohlke KL, Scott LJ, et al. Genetic variation near the hepatocyte nuclear factor-4 α gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53(4):1141–1149. [DOI] [PubMed] [Google Scholar]
- 122. Love-Gregory LD, Wasson J, Ma J, et al. A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 α gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an Ashkenazi Jewish population. Diabetes. 2004;53(4):1134–1140. [DOI] [PubMed] [Google Scholar]
- 123. Weedon MN, Owen KR, Shields B, et al. Common variants of the hepatocyte nuclear factor-4α P2 promoter are associated with type 2 diabetes in the U.K. population. Diabetes. 2004;53(11):3002–3006. [DOI] [PubMed] [Google Scholar]
- 124. Barroso I, Luan J, Wheeler E, et al. Population-specific risk of type 2 diabetes conferred by HNF4A P2 promoter variants: a lesson for replication studies. Diabetes. 2008;57(11):3161–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bjørkhaug L, Sagen JV, Thorsby P, Søvik O, Molven A, Njølstad PR. Hepatocyte nuclear factor-1 α gene mutations and diabetes in Norway. J Clin Endocrinol Metab. 2003;88(2):920–931. [DOI] [PubMed] [Google Scholar]
- 126. Nowak N, Hohendorff J, Solecka I, et al. Circulating ghrelin level is higher in HNF1A-MODY and GCK-MODY than in polygenic forms of diabetes mellitus. Endocrine. 2015;50(3):643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brial F, Lussier CR, Belleville K, Sarret P, Boudreau F. Ghrelin inhibition restores glucose homeostasis in hepatocyte nuclear factor-1α (MODY3)-deficient mice. Diabetes. 2015;64(9):3314–3320. [DOI] [PubMed] [Google Scholar]
- 128. Dezaki K, Kakei M, Yada T. Ghrelin uses Gαi2 and activates voltage-dependent K+ channels to attenuate glucose-induced Ca2+ signaling and insulin release in islet β-cells: novel signal transduction of ghrelin. Diabetes. 2007;56(9):2319–2327. [DOI] [PubMed] [Google Scholar]
- 129. Eeckhoute J, Moerman E, Bouckenooghe T, et al. Hepatocyte nuclear factor 4 α isoforms originated from the P1 promoter are expressed in human pancreatic β-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology. 2003;144(5):1686–1694. [DOI] [PubMed] [Google Scholar]
- 130. Gupta RK, Vatamaniuk MZ, Lee CS, et al. The MODY1 gene HNF-4α regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115(4):1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sandovici I, Smith NH, Nitert MD, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci USA. 2011;108(13):5449–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Coffinier C, Thépot D, Babinet C, Yaniv M, Barra J. Essential role for the homeoprotein vHNF1/HNF1β in visceral endoderm differentiation. Development. 1999;126(21):4785–4794. [DOI] [PubMed] [Google Scholar]
- 133. Barbacci E, Reber M, Ott MO, Breillat C, Huetz F, Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126(21):4795–4805. [DOI] [PubMed] [Google Scholar]
- 134. Wang L, Coffinier C, Thomas MK, et al. Selective deletion of the Hnf1β (MODY5) gene in β-cells leads to altered gene expression and defective insulin release. Endocrinology. 2004;145(8):3941–3949. [DOI] [PubMed] [Google Scholar]
- 135. De Vas MG, Kopp JL, Heliot C, Sander M, Cereghini S, Haumaitre C. Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development. 2015;142(5):871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Poll AV, Pierreux CE, Lokmane L, et al. A vHNF1/TCF2-HNF6 cascade regulates the transcription factor network that controls generation of pancreatic precursor cells. Diabetes. 2006;55(1):61–69. [PubMed] [Google Scholar]
- 137. Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic β-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4(4):200–213. [DOI] [PubMed] [Google Scholar]
- 138. Chen YZ, Gao Q, Zhao XZ, et al. Systematic review of TCF2 anomalies in renal cysts and diabetes syndrome/maturity onset diabetes of the young type 5. Chin Med J. 2010;123(22):3326–3333. [PubMed] [Google Scholar]
- 139. Edghill EL, Stals K, Oram RA, Shepherd MH, Hattersley AT, Ellard S. HNF1B deletions in patients with young-onset diabetes but no known renal disease. Diabet Med. 2013;30(1):114–117. [DOI] [PubMed] [Google Scholar]
- 140. Haldorsen IS, Vesterhus M, Raeder H, et al. Lack of pancreatic body and tail in HNF1B mutation carriers. Diabet Med. 2008;25(7):782–787. [DOI] [PubMed] [Google Scholar]
- 141. Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56(3):685–693. [DOI] [PubMed] [Google Scholar]
- 142. Rodriguez C, Patel AV, Mondul AM, Jacobs EJ, Thun MJ, Calle EE. Diabetes and risk of prostate cancer in a prospective cohort of US men. Am J Epidemiol. 2005;161(2):147–152. [DOI] [PubMed] [Google Scholar]
- 143. Giovannucci E, Harlan DM, Archer MC, et al. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33(7):1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–983. [DOI] [PubMed] [Google Scholar]
- 145. Barbacci E, Chalkiadaki A, Masdeu C, et al. HNF1β/TCF2 mutations impair transactivation potential through altered co-regulator recruitment. Hum Molec Genet. 2004;13(24):3139–3149. [DOI] [PubMed] [Google Scholar]
- 146. Haumaitre C, Fabre M, Cormier S, Baumann C, Delezoide AL, Cereghini S. Severe pancreas hypoplasia and multicystic renal dysplasia in two human fetuses carrying novel HNF1β/MODY5 mutations. Hum Molec Genet. 2006;15(15):2363–2375. [DOI] [PubMed] [Google Scholar]
- 147. Cha JY, Kim H, Kim KS, Hur MW, Ahn Y. Identification of transacting factors responsible for the tissue-specific expression of human glucose transporter type 2 isoform gene. Cooperative role of hepatocyte nuclear factors 1α and 3β. J Biol Chem. 2000;275(24):18358–18365. [DOI] [PubMed] [Google Scholar]
- 148. Kim EK, Lee JS, Cheong HI, Chung SS, Kwak SH, Park KS. Identification and functional characterization of P159L mutation in HNF1B in a family with maturity-onset diabetes of the young 5 (MODY5). Genomics Inform. 2014;12(4):240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Yamagata K, Nammo T, Moriwaki M, et al. Overexpression of dominant-negative mutant hepatocyte nuclear fctor-1 α in pancreatic β-cells causes abnormal islet architecture with decreased expression of E-cadherin, reduced β-cell proliferation, and diabetes. Diabetes. 2002;51(1):114–123. [DOI] [PubMed] [Google Scholar]
- 150. Dessimoz J, Bonnard C, Huelsken J, Grapin-Botton A. Pancreas-specific deletion of β-catenin reveals Wnt-dependent and Wnt-independent functions during development. Curr Biol. 2005;15(18):1677–1683. [DOI] [PubMed] [Google Scholar]
- 151. Papadopoulou S, Edlund H. Attenuated Wnt signaling perturbs pancreatic growth but not pancreatic function. Diabetes. 2005;54(10):2844–2851. [DOI] [PubMed] [Google Scholar]
- 152. Murtaugh LC, Law AC, Dor Y, Melton DA. β-Catenin is essential for pancreatic acinar but not islet development. Development. 2005;132(21):4663–4674. [DOI] [PubMed] [Google Scholar]
- 153. Oliver-Krasinski JM, Stoffers DA. On the origin of the β cell. Genes Dev. 2008;22(15):1998–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Oliver-Krasinski JM, Kasner MT, Yang J, et al. The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest. 2009;119(7):1888–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12(12):1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. [DOI] [PubMed] [Google Scholar]
- 157. Gao T, McKenna B, Li C, et al. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab. 2014;19(2):259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15(1):106–110. [DOI] [PubMed] [Google Scholar]
- 159. Wright NM, Metzger DL, Borowitz SM, Clarke WL. Permanent neonatal diabetes mellitus and pancreatic exocrine insufficiency resulting from congenital pancreatic agenesis. Am J Dis Child. 1993;147(6):607–609. [DOI] [PubMed] [Google Scholar]
- 160. Schwitzgebel VM, Mamin A, Brun T, et al. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J Clin Endocrinol Metab. 2003;88(9):4398–4406. [DOI] [PubMed] [Google Scholar]
- 161. Nicolino M, Claiborn KC, Senée V, Boland A, Stoffers DA, Julier C. A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes. 2010;59(3):733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17(2):138–139. [DOI] [PubMed] [Google Scholar]
- 163. Weng J, Macfarlane WM, Lehto M, et al. Functional consequences of mutations in the MODY4 gene (IPF1) and coexistence with MODY3 mutations. Diabetologia. 2001;44(2):249–258. [DOI] [PubMed] [Google Scholar]
- 164. Hani EH, Stoffers DA, Chèvre JC, et al. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest. 1999;104(9):R41–R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Macfarlane WM, Frayling TM, Ellard S, et al. Missense mutations in the insulin promoter factor-1 gene predispose to type 2 diabetes. J Clin Invest. 1999;104(9):R33–R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Edghill EL, Khamis A, Weedon MN, et al. Sequencing PDX1 (insulin promoter factor 1) in 1788 UK individuals found 5% had a low frequency coding variant, but these variants are not associated with Type 2 diabetes. Diabet Med. 2011;28(6):681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Steinthorsdottir V, Thorleifsson G, Sulem P, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46(3):294–298. [DOI] [PubMed] [Google Scholar]
- 168. Sachdeva MM, Claiborn KC, Khoo C, et al. Pdx1 (MODY4) regulates pancreatic β cell susceptibility to ER stress. Proc Natl Acad Sci USA. 2009;106(45):19090–19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol. 2012;8(2):92–103. [DOI] [PubMed] [Google Scholar]
- 170. Sivitz WI, Yorek MA. Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid Redox Signal. 2010;12(4):537–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Soleimanpour SA, Ferrari AM, Raum JC, et al. Diabetes susceptibility genes Pdx1 and Clec16a function in a pathway regulating mitophagy in β-cells. Diabetes. 2015;64(10):3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Smith SB, Ee HC, Conners JR, German MS. Paired-homeodomain transcription factor PAX4 acts as a transcriptional repressor in early pancreatic development. Molec Cell Biol. 1999;19(12):8272–8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Brun T, Gauthier BR. A focus on the role of Pax4 in mature pancreatic islet β-cell expansion and survival in health and disease. J Mol Endocrinol. 2008;40(2):37–45. [DOI] [PubMed] [Google Scholar]
- 174. Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature. 1997;386(6623):399–402. [DOI] [PubMed] [Google Scholar]
- 175. Collombat P, Mansouri A, Hecksher-Sorensen J, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17(20):2591–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Collombat P, Hecksher-Sørensen J, Broccoli V, et al. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the α- and β-cell lineages in the mouse endocrine pancreas. Development. 2005;132(13):2969–2980. [DOI] [PubMed] [Google Scholar]
- 177. Plengvidhya N, Kooptiwut S, Songtawee N, et al. PAX4 mutations in Thais with maturity onset diabetes of the young. J Clin Endocrinol Metab. 2007;92(7):2821–2826. [DOI] [PubMed] [Google Scholar]
- 178. Kooptiwut S, Plengvidhya N, Chukijrungroat T, et al. Defective PAX4 R192H transcriptional repressor activities associated with maturity onset diabetes of the young and early onset-age of type 2 diabetes. J Diabetes Complications. 2012;26(4):343–347. [DOI] [PubMed] [Google Scholar]
- 179. Jo W, Endo M, Ishizu K, Nakamura A, Tajima T. A novel PAX4 mutation in a Japanese patient with maturity-onset diabetes of the young. Tohoku J Exp Med. 2011;223(2):113–118. [DOI] [PubMed] [Google Scholar]
- 180. Shimajiri Y, Sanke T, Furuta H, et al. A missense mutation of Pax4 gene (R121W) is associated with type 2 diabetes in Japanese. Diabetes. 2001;50(12):2864–2869. [DOI] [PubMed] [Google Scholar]
- 181. Ma RC, Hu C, Tam CH, et al. Genome-wide association study in a Chinese population identifies a susceptibility locus for type 2 diabetes at 7q32 near PAX4. Diabetologia. 2013;56(6):1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Chen M, Hu C, Zhang R, et al. Association of PAX4 genetic variants with oral antidiabetic drugs efficacy in Chinese type 2 diabetes patients. Pharmacogenomics J. 2014;14(5):488–492. [DOI] [PubMed] [Google Scholar]
- 183. Biason-Lauber A, Boehm B, Lang-Muritano M, et al. Association of childhood type 1 diabetes mellitus with a variant of PAX4: possible link to β cell regenerative capacity. Diabetologia. 2005;48(5):900–905. [DOI] [PubMed] [Google Scholar]
- 184. Holm P, Rydlander B, Luthman H, Kockum I. Interaction and association analysis of a type 1 diabetes susceptibility locus on chromosome 5q11–q13 and the 7q32 chromosomal region in Scandinavian families. Diabetes. 2004;53(6):1584–1591. [DOI] [PubMed] [Google Scholar]
- 185. Hermann R, Mantere J, Lipponen K, et al. Lack of association of PAX4 gene with type 1 diabetes in the Finnish and Hungarian populations. Diabetes. 2005;54(9):2816–2819. [DOI] [PubMed] [Google Scholar]
- 186. Zhang Y, Xiao X, Liu Y, et al. The association of the PAX4 gene with type 1 diabetes in Han Chinese. Diabetes Res Clin Pract. 2008;81(3):365–369. [DOI] [PubMed] [Google Scholar]
- 187. Mauvais-Jarvis F, Smith SB, Le May C, et al. PAX4 gene variations predispose to ketosis-prone diabetes. Hum Molec Genet. 2004;13(24):3151–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Balasubramanyam A, Nalini R, Hampe CS, Maldonado M. Syndromes of ketosis-prone diabetes mellitus. Endocr Rev. 2008;29(3):292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Patel SG, Hsu JW, Jahoor F, et al. Pathogenesis of A−β+ ketosis-prone diabetes. Diabetes. 2013;62(3):912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Haaland WC, Scaduto DI, Maldonado MR, et al. A-β-subtype of ketosis-prone diabetes is not predominantly a monogenic diabetic syndrome. Diabetes Care. 2009;32(5):873–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Brun T, Franklin I, St-Onge L, et al. The diabetes-linked transcription factor PAX4 promotes β-cell proliferation and survival in rat and human islets. J Cell Biol. 2004;167(6):1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Docena MK, Faiman C, Stanley CM, Pantalone KM. Mody-3: novel HNF1A mutation and the utility of glucagon-like peptide (GLP)-1 receptor agonist therapy. Endocr Pract. 2014;20(2):107–111. [DOI] [PubMed] [Google Scholar]
- 193. Østoft SH, Bagger JI, Hansen T, et al. Glucose-lowering effects and low risk of hypoglycemia in patients with maturity-onset diabetes of the young when treated with a GLP-1 receptor agonist: a double-blind, randomized, crossover trial. Diabetes Care. 2014;37(7):1797–1805. [DOI] [PubMed] [Google Scholar]
- 194. Lumb AN, Gallen IW. Treatment of HNF1-α MODY with the DPP-4 inhibitor sitagliptin(1). Diabet Med. 2009;26(2):189–190. [DOI] [PubMed] [Google Scholar]
- 195. Katra B, Klupa T, Skupien J, et al. Dipeptidyl peptidase-IV inhibitors are efficient adjunct therapy in HNF1A maturity-onset diabetes of the young patients–report of two cases. Diabetes Technol Ther. 2010;12(4):313–316. [DOI] [PubMed] [Google Scholar]
- 196. Lefèbvre PJ, Paquot N, Scheen AJ. Inhibiting or antagonizing glucagon: making progress in diabetes care. Diabetes Obes Metab. 2015;17(8):720–725. [DOI] [PubMed] [Google Scholar]
- 197. Naya FJ, Huang HP, Qiu Y, et al. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11(18):2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Gu C, Stein GH, Pan N, et al. Pancreatic β cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 2010;11(4):298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kristinsson SY, Thorolfsdottir ET, Talseth B, et al. MODY in Iceland is associated with mutations in HNF-1α and a novel mutation in NeuroD1. Diabetologia. 2001;44(11):2098–2103. [DOI] [PubMed] [Google Scholar]
- 200. Liu L, Furuta H, Minami A, et al. A novel mutation, Ser159Pro in the NeuroD1/BETA2 gene contributes to the development of diabetes in a Chinese potential MODY family. Mol Cell Biochem. 2007;303:115–120. [DOI] [PubMed] [Google Scholar]
- 201. Gonsorcíková L, Průhová S, Cinek O, et al. Autosomal inheritance of diabetes in two families characterized by obesity and a novel H241Q mutation in NEUROD1. Pediatr Diabetes. 2008;9:367–372. [DOI] [PubMed] [Google Scholar]
- 202. Szopa M, Ludwig-Galezowska AH, Radkowski P, et al. A family with the Arg103Pro mutation in the NEUROD1 gene detected by next-generation sequencing-clinical characteristics of mutation carriers. Eur J Med Genet. 2016;59(2):75–79. [DOI] [PubMed] [Google Scholar]
- 203. Rubio-Cabezas O, Minton JA, Kantor I, Williams D, Ellard S, Hattersley AT. Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes. 2010;59(9):2326–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Malecki MT, Jhala US, Antonellis A, et al. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet. 1999;23(3):323–328. [DOI] [PubMed] [Google Scholar]
- 205. Liu L, Jia W, Zheng T, Li M, Lu H, Xiang K. Ala45Thr variation in neuroD1 gene is associated with early-onset type 2 diabetes with or without diabetic pedigree in Chinese. Mol Cell Biochem. 2006;290:199–204. [DOI] [PubMed] [Google Scholar]
- 206. Malecki MT, Cyganek K, Klupa T, Sieradzki J. The Ala45Thr polymorphism of BETA2/NeuroD1 gene and susceptibility to type 2 diabetes mellitus in a Polish population. Acta Diabetol. 2003;40(2):109–111. [DOI] [PubMed] [Google Scholar]
- 207. Han X, Xiao J, Ren Q, Tang Y, Yang W, Ji L. Evaluation of variant A45T in NEUROD1/BETA2 for its association with type 2 diabetes mellitus. Endocrine. 2013;44(1):99–106. [DOI] [PubMed] [Google Scholar]
- 208. Hansen L, Jensen JN, Urioste S, et al. NeuroD/BETA2 gene variability and diabetes: no associations to late-onset type 2 diabetes but an A45 allele may represent a susceptibility marker for type 1 diabetes among Danes. Danish Study Group of Diabetes in Childhood, and the Danish IDDM Epidemiology and Genetics Group. Diabetes. 2000;49(5):876–878. [DOI] [PubMed] [Google Scholar]
- 209. Malecki MT, Klupa T, Moczulski DK, Rogus JJ. The Ala45Thr polymorphism of BETA2/NeuroD1 gene and susceptibility to type 1 diabetes mellitus in Caucasians. Exp Clin Endocrinol Diabetes. 2003;111(5):251–254. [DOI] [PubMed] [Google Scholar]
- 210. Oliveira CS, Hauache OM, Vieira JG, et al. The Ala45Thr polymorphism of NEUROD1 is associated with type 1 diabetes in Brazilian women. Diabetes Metab. 2005;31(6):599–602. [DOI] [PubMed] [Google Scholar]
- 211. Vella A, Howson JM, Barratt BJ, et al. Lack of association of the Ala(45)Thr polymorphism and other common variants of the NeuroD gene with type 1 diabetes. Diabetes. 2004;53(4):1158–1161. [DOI] [PubMed] [Google Scholar]
- 212. Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9(8):1009–1019. [DOI] [PubMed] [Google Scholar]
- 213. Zhao L, Guo M, Matsuoka TA, et al. The islet β cell-enriched MafA activator is a key regulator of insulin gene transcription. J Biol Chem. 2005;280(12):11887–11894. [DOI] [PubMed] [Google Scholar]
- 214. Yang Y, Chang BH, Samson SL, Li MV, Chan L. The Krüppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37(8):2529–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Fonseca SG, Fukuma M, Lipson KL, et al. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic β-cells. J Biol Chem. 2005;280(47):39609–39615. [DOI] [PubMed] [Google Scholar]
- 216. Rohayem J, Ehlers C, Wiedemann B, et al. Diabetes and neurodegeneration in Wolfram syndrome: a multicenter study of phenotype and genotype. Diabetes Care. 2011;34(7):1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Fonseca SG, Ishigaki S, Oslowski CM, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120(3):744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Zalloua PA, Azar ST, Delépine M, et al. WFS1 mutations are frequent monogenic causes of juvenile-onset diabetes mellitus in Lebanon. Hum Molec Genet. 2008;17(24):4012–4021. [DOI] [PubMed] [Google Scholar]
- 219. Zmyslowska A, Fendler W, Niwald A, et al. Retinal thinning as a marker of disease progression in patients with Wolfram syndrome. Diabetes Care. 2015;38(3):e36–e37. [DOI] [PubMed] [Google Scholar]
- 220. Rigoli L, Lombardo F, Di Bella C. Wolfram syndrome and WFS1 gene. Clin Genet. 2011;79(2):103–117. [DOI] [PubMed] [Google Scholar]
- 221. Amr S, Heisey C, Zhang M, et al. A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am J Hum Genet. 2007;81(4):673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Rigoli L, Di Bella C. Wolfram syndrome 1 and Wolfram syndrome 2. Curr Opin Pediatr. 2012;24(4):512–517. [DOI] [PubMed] [Google Scholar]
- 223. Minton JA, Hattersley AT, Owen K, et al. Association studies of genetic variation in the WFS1 gene and type 2 diabetes in U.K. populations. Diabetes. 2002;51(4):1287–1290. [DOI] [PubMed] [Google Scholar]
- 224. Sandhu MS, Weedon MN, Fawcett KA, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39(8):951–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Cheurfa N, Brenner GM, Reis AF, et al. Decreased insulin secretion and increased risk of type 2 diabetes associated with allelic variations of the WFS1 gene: the Data from Epidemiological Study on the Insulin Resistance Syndrome (DESIR) prospective study. Diabetologia. 2011;54(3):554–562. [DOI] [PubMed] [Google Scholar]
- 226. Fawcett KA, Wheeler E, Morris AP, et al. Detailed investigation of the role of common and low-frequency WFS1 variants in type 2 diabetes risk. Diabetes. 2010;59(3):741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Awata T, Inoue K, Kurihara S, et al. Missense variations of the gene responsible for Wolfram syndrome (WFS1/wolframin) in Japanese: possible contribution of the Arg456His mutation to type 1 diabetes as a nonautoimmune genetic basis. Biochem Biophys Res Commun. 2000;268(2):612–616. [DOI] [PubMed] [Google Scholar]
- 228. Lemaire K, Schuit F. Integrating insulin secretion and ER stress in pancreatic β-cells. Nat Cell Biol. 2012;14(10):979–981. [DOI] [PubMed] [Google Scholar]
- 229. Lu S, Kanekura K, Hara T, et al. A calcium-dependent protease as a potential therapeutic target for Wolfram syndrome. Proc Natl Acad Sci USA. 2014;111(49):E5292–E5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Huang CJ, Gurlo T, Haataja L, et al. Calcium-activated calpain-2 is a mediator of β cell dysfunction and apoptosis in type 2 diabetes. J Biol Chem. 2010;285(1):339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Hatanaka M, Tanabe K, Yanai A, et al. Wolfram syndrome 1 gene (WFS1) product localizes to secretory granules and determines granule acidification in pancreatic β-cells. Hum Molec Genet. 2011;20(7):1274–1284. [DOI] [PubMed] [Google Scholar]
- 232. Ahmadian M, Suh JM, Hah N, et al. PPARγ signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19(5):557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Jonker JW, Suh JM, Atkins AR, et al. A PPARγ-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;485(7398):391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Dutchak PA, Katafuchi T, Bookout AL, et al. Fibroblast growth factor-21 regulates PPARγ activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148(3):556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Wei W, Dutchak PA, Wang X, et al. Fibroblast growth factor 21 promotes bone loss by potentiating the effects of peroxisome proliferator-activated receptor γ. Proc Natl Acad Sci USA. 2012;109(8):3143–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Choi JH, Banks AS, Estall JL, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARγ by Cdk5. Nature. 2010;466(7305):451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Banks AS, McAllister FE, Camporez JP, et al. An ERK/Cdk5 axis controls the diabetogenic actions of PPARγ. Nature. 2015;517(7534):391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Barroso I, Gurnell M, Crowley VE, et al. Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402(6764):880–883. [DOI] [PubMed] [Google Scholar]
- 239. Yen CJ, Beamer BA, Negri C, et al. Molecular scanning of the human peroxisome proliferator activated receptor γ (hPPAR γ) gene in diabetic Caucasians: identification of a Pro12Ala PPAR γ 2 missense mutation. Biochem Biophys Res Commun. 1997;241(2):270–274. [DOI] [PubMed] [Google Scholar]
- 240. Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80. [DOI] [PubMed] [Google Scholar]
- 241. Soriguer F, Morcillo S, Cardona F, et al. Pro12Ala polymorphism of the PPARG2 gene is associated with type 2 diabetes mellitus and peripheral insulin sensitivity in a population with a high intake of oleic acid. J Nutr. 2006;136(9):2325–2330. [DOI] [PubMed] [Google Scholar]
- 242. Stumvoll M, Häring H. The peroxisome proliferator-activated receptor-γ2 Pro12Ala polymorphism. Diabetes. 2002;51(8):2341–2347. [DOI] [PubMed] [Google Scholar]
- 243. Gouda HN, Sagoo GS, Harding AH, Yates J, Sandhu MS, Higgins JP. The association between the peroxisome proliferator-activated receptor-γ2 (PPARG2) Pro12Ala gene variant and type 2 diabetes mellitus: a HuGE review and meta-analysis. Am J Epidemiol. 2010;171(6):645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Raj SM, Howson JM, Walker NM, et al. No association of multiple type 2 diabetes loci with type 1 diabetes. Diabetologia. 2009;52(10):2109–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Qu HQ, Grant SF, Bradfield JP, et al. Association analysis of type 2 diabetes loci in type 1 diabetes. Diabetes. 2008;57(7):1983–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Soccio RE, Chen ER, Rajapurkar SR, et al. Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell. 2015;162(1):33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications–a review. Nutr J. 2014;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Bossé Y, Weisnagel SJ, Bouchard C, Després JP, Pérusse L, Vohl MC. Combined effects of PPARγ2 P12A and PPARα L162V polymorphisms on glucose and insulin homeostasis: the Québec Family Study. J Hum Genet. 2003;48(12):614–621. [DOI] [PubMed] [Google Scholar]
- 249. Koch M, Rett K, Maerker E, et al. The PPARγ2 amino acid polymorphism Pro 12 Ala is prevalent in offspring of type II diabetic patients and is associated to increased insulin sensitivity in a subgroup of obese subjects. Diabetologia. 1999;42(6):758–762. [DOI] [PubMed] [Google Scholar]
- 250. Claussnitzer M, Dankel SN, Klocke B, et al. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156:343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Steiner DF, Tager HS, Chan SJ, Nanjo K, Sanke T, Rubenstein AH. Lessons learned from molecular biology of insulin-gene mutations. Diabetes Care. 1990;13(6):600–609. [DOI] [PubMed] [Google Scholar]
- 252. Tager H, Given B, Baldwin D, et al. A structurally abnormal insulin causing human diabetes. Nature. 1979;281(5727):122–125. [DOI] [PubMed] [Google Scholar]
- 253. Haneda M, Chan SJ, Kwok SC, Rubenstein AH, Steiner DF. Studies on mutant human insulin genes: identification and sequence analysis of a gene encoding [SerB24]insulin. Proc Natl Acad Sci USA. 1983;80(20):6366–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Shoelson S, Fickova M, Haneda M, et al. Identification of a mutant human insulin predicted to contain a serine-for-phenylalanine substitution. Proc Natl Acad Sci USA. 1983;80(24):7390–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Nanjo K, Sanke T, Miyano M, et al. Diabetes due to secretion of a structurally abnormal insulin (insulin Wakayama). Clinical and functional characteristics of [LeuA3] insulin. J Clin Invest. 1986;77(2):514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Carroll RJ, Hammer RE, Chan SJ, Swift HH, Rubenstein AH, Steiner DF. A mutant human proinsulin is secreted from islets of Langerhans in increased amounts via an unregulated pathway. Proc Natl Acad Sci USA. 1988;85(23):8943–8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Schwartz GP, Burke GT, Katsoyannis PG. A superactive insulin: [B10-aspartic acid]insulin(human). Proc Natl Acad Sci USA. 1987;84(18):6408–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Støy J, Edghill EL, Flanagan SE, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci USA. 2007;104(38):15040–15044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Garin I, Perez de Nanclares G, Gastaldo E, Harries LW, Rubio-Cabezas O, Castano L. Permanent neonatal diabetes caused by creation of an ectopic splice site within the INS gene. PLoS One. 2012;7(1):e29205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Carmody D, Park SY, Ye H, et al. Continued lessons from the INS gene: an intronic mutation causing diabetes through a novel mechanism. J Med Genet. 2015;52(9):612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Dusatkova L, Dusatkova P, Vosahlo J, et al. Frameshift mutations in the insulin gene leading to prolonged molecule of insulin in two families with maturity-onset diabetes of the young. Eur J Med Genet. 2015;58(4):230–234. [DOI] [PubMed] [Google Scholar]
- 262. Bonfanti R, Colombo C, Nocerino V, et al. Insulin gene mutations as cause of diabetes in children negative for five type 1 diabetes autoantibodies. Diabetes Care. 2009;32(1):123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Oda N, Nakai A, Fujiwara K, et al. Polymorphisms of the insulin gene among Japanese subjects. Metabolism. 2001;50(6):631–634. [DOI] [PubMed] [Google Scholar]
- 264. Flannick J, Beer NL, Bick AG, et al. Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat Genet. 2013;45(11):1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Park SY, Ye H, Steiner DF, Bell GI. Mutant proinsulin proteins associated with neonatal diabetes are retained in the endoplasmic reticulum and not efficiently secreted. Biochem Biophys Res Commun. 2010;391(3):1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16(5):198–200. [DOI] [PubMed] [Google Scholar]
- 267. Støy J, Steiner DF, Park SY, Ye H, Philipson LH, Bell GI. Clinical and molecular genetics of neonatal diabetes due to mutations in the insulin gene. Rev Endocr Metab Disord. 2010;11(3):205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Molven A, Ringdal M, Nordbø AM, et al. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008;57(4):1131–1135. [DOI] [PubMed] [Google Scholar]
- 269. Wang J, Takeuchi T, Tanaka S, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic β-cell dysfunction in the Mody mouse. J Clin Invest. 1999;103(1):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, and proteotoxicity. Proc Natl Acad Sci USA. 2007;104(40):15841–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Ahamed A, Unnikrishnan AG, Pendsey SS, et al. Permanent neonatal diabetes mellitus due to a C96Y heterozygous mutation in the insulin gene. A case report. JOP. 2008;9(6):715–718. [PubMed] [Google Scholar]
- 272. Garin I, Edghill EL, Akerman I, et al. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc Natl Acad Sci USA. 2010;107(7):3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Bakay M, Pandey R, Hakonarson H. Genes involved in type 1 diabetes: an update. Genes. 2013;4(3):499–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Vafiadis P, Bennett ST, Todd JA, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet. 1997;15(3):289–292. [DOI] [PubMed] [Google Scholar]
- 275. Zhang L, Nakayama M, Eisenbarth GS. Insulin as an autoantigen in NOD/human diabetes. Curr Opin Immunol. 2008;20(1):111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Bennett ST, Todd JA. Human type 1 diabetes and the insulin gene: principles of mapping polygenes. Ann Rev Genet. 1996;30:343–370. [DOI] [PubMed] [Google Scholar]
- 277. Meigs JB, Dupuis J, Herbert AG, Liu C, Wilson PW, Cupples LA. The insulin gene variable number tandem repeat and risk of type 2 diabetes in a population-based sample of families and unrelated men and women. J Clin Endocrinol Metab. 2005;90(2):1137–1143. [DOI] [PubMed] [Google Scholar]
- 278. Hansen SK, Gjesing AP, Rasmussen SK, et al. Large-scale studies of the HphI insulin gene variable-number-of-tandem-repeats polymorphism in relation to type 2 diabetes mellitus and insulin release. Diabetologia. 2004;47(6):1079–1087. [DOI] [PubMed] [Google Scholar]
- 279. Huxtable SJ, Saker PJ, Haddad L, et al. Analysis of parent-offspring trios provides evidence for linkage and association between the insulin gene and type 2 diabetes mediated exclusively through paternally transmitted class III variable number tandem repeat alleles. Diabetes. 2000;49(1):126–130. [DOI] [PubMed] [Google Scholar]
- 280. Lindsay RS, Hanson RL, Wiedrich C, Knowler WC, Bennett PH, Baier LJ. The insulin gene variable number tandem repeat class I/III polymorphism is in linkage disequilibrium with birth weight but not type 2 diabetes in the Pima population. Diabetes. 2003;52(1):187–193. [DOI] [PubMed] [Google Scholar]
- 281. Brookes KJ. The VNTR in complex disorders: the forgotten polymorphisms? A functional way forward? Genomics. 2013;101(5):273–281. [DOI] [PubMed] [Google Scholar]
- 282. Saxena R, Elbers CC, Guo Y, et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90(3):410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Liu M, Hodish I, Haataja L, et al. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endocrinol Metab. 2010;21(11):652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Guo H, Xiong Y, Witkowski P, et al. Inefficient translocation of preproinsulin contributes to pancreatic β cell failure and late-onset diabetes. J Biol Chem. 2014;289(23):16290–16302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Fernández ÁF, López-Otín C. The functional and pathologic relevance of autophagy proteases. J Clin Invest. 2015;125(1):33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Bachar-Wikstrom E, Wikstrom JD, Ariav Y, et al. Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes. Diabetes. 2013;62(4):1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Liu M, Haataja L, Wright J, et al. Mutant INS-gene induced diabetes of youth: proinsulin cysteine residues impose dominant-negative inhibition on wild-type proinsulin transport. PLoS One. 2010;5(10):e13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Hay CW, Docherty K. Comparative analysis of insulin gene promoters: implications for diabetes research. Diabetes. 2006;55(12):3201–3213. [DOI] [PubMed] [Google Scholar]
- 289. Kang HS, Kim YS, ZeRuth G, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic β-cell development and insulin gene expression. Mol Cell Biol. 2009;29(24):6366–6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Kim YS, Nakanishi G, Lewandoski M, Jetten AM. GLIS3, a novel member of the GLIS subfamily of Krüppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003;31(19):5513–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Senée V, Chelala C, Duchatelet S, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38(6):682–687. [DOI] [PubMed] [Google Scholar]
- 292. Dimitri P, Warner JT, Minton JA, et al. Novel GLIS3 mutations demonstrate an extended multisystem phenotype. Eur J Endocrinol. 2011;164(3):437–443. [DOI] [PubMed] [Google Scholar]
- 293. Dimitri P, Habeb AM, Garbuz F, et al. Expanding the clinical spectrum associated with GLIS3 mutations. J Clin Endocrinol Metab. 2015;100(10):E1362–E1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Cao B, Gong C, Wu D, et al. Genetic analysis and follow-up of 25 neonatal diabetes mellitus patients in China. J Diabetes Res. 2016;2016:6314368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Kiani AK, John P, Bhatti A, et al. Association of 32 type 1 diabetes risk loci in Pakistani patients. Diabetes Res Clin Pract. 2015;108(1):137–142. [DOI] [PubMed] [Google Scholar]
- 297. Winkler C, Krumsiek J, Buettner F, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia. 2014;57(12):2521–2529. [DOI] [PubMed] [Google Scholar]
- 298. Boesgaard TW, Grarup N, Jørgensen T, et al. Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated β cell function in middle-aged Danish people. Diabetologia. 2010;53(8):1647–1655. [DOI] [PubMed] [Google Scholar]
- 299. Liu C, Li H, Qi L, et al. Variants in GLIS3 and CRY2 are associated with type 2 diabetes and impaired fasting glucose in Chinese Hans. PLoS One. 2011;6(6):e21464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Hu C, Zhang R, Wang C, et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS One. 2010;5(11):e15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Rees SD, Hydrie MZ, O'Hare JP, et al. Effects of 16 genetic variants on fasting glucose and type 2 diabetes in South Asians: ADCY5 and GLIS3 variants may predispose to type 2 diabetes. PLoS One. 2011;6(9):e24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44(1):67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Barker A, Sharp SJ, Timpson NJ, et al. Association of genetic loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes. 2011;60(6):1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Sakai K, Imamura M, Tanaka Y, et al. Replication study for the association of 9 East Asian GWAS-derived loci with susceptibility to type 2 diabetes in a Japanese population. PLoS One. 2013;8(9):e76317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. ZeRuth GT, Takeda Y, Jetten AM. The Krüppel-like protein Gli-similar 3 (Glis3) functions as a key regulator of insulin transcription. Mol Endocrinol. 2013;27(10):1692–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Yang Y, Chang BH, Chan L. Sustained expression of the transcription factor GLIS3 is required for normal β cell function in adults. EMBO Mol Med. 2013;5(1):92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Watanabe N, Hiramatsu K, Miyamoto R, et al. A murine model of neonatal diabetes mellitus in Glis3-deficient mice. FEBS Lett. 2009;583(12):2108–2113. [DOI] [PubMed] [Google Scholar]
- 308. Yang Y, Chang BH, Yechoor V, et al. The Krüppel-like zinc finger protein GLIS3 transactivates neurogenin 3 for proper fetal pancreatic islet differentiation in mice. Diabetologia. 2011;54(10):2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Georgia S, Bhushan A. β-Cell replication is the primary mechanism for maintaining postnatal β cell mass. J Clin Invest. 2004;114(7):963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Kushner JA, Ciemerych MA, Sicinska E, et al. Cyclins D2 and D1 are essential for postnatal pancreatic β-cell growth. Mol Cell Biol. 2005;25(9):3752–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Georgia S, Hinault C, Kawamori D, et al. Cyclin D2 is essential for the compensatory β-cell hyperplastic response to insulin resistance in rodents. Diabetes. 2010;59(4):987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and β-cell loss in type 1 diabetes. Nat Rev Endocrinol. 2009;5(4):219–226. [DOI] [PubMed] [Google Scholar]
- 314. Cnop M, Ladrière L, Igoillo-Esteve M, Moura RF, Cunha DA. Causes and cures for endoplasmic reticulum stress in lipotoxic β-cell dysfunction. Diabetes Obes Metab. 2010;12(suppl 2):76–82. [DOI] [PubMed] [Google Scholar]
- 315. Nogueira TC, Paula FM, Villate O, et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic β cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet. 2013;9(5):e1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Thomsen SK, Gloyn AL. The pancreatic β cell: recent insights from human genetics. Trends Endocrinol Metab. 2014;25(8):425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46(2):136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]