Abstract
The study of bacterial ion channels has provided fundamental insights into the structural basis of neuronal signaling. However, the native role of ion channels in bacteria has remained elusive. Here we show that ion channels conduct long-range electrical signals within bacterial biofilm communities through spatially propagating waves of potassium. These waves result from a positive feedback loop, in which a metabolic trigger induces release of intracellular potassium, which in turn depolarizes neighboring cells. Propagating through the biofilm, this wave of depolarization coordinates metabolic states among cells in the interior and periphery of the biofilm. Deletion of the potassium channel abolishes this response. As predicted by a mathematical model, we further show that spatial propagation can be hindered by specific genetic perturbations to potassium channel gating. Together, these results demonstrate a function for ion channels in bacterial biofilms, and provide a prokaryotic paradigm for active, long-range electrical signaling in cellular communities.
Introduction
Communication through electrical signaling is prevalent among biological systems. One of the most familiar examples is the action potential in neurons that is mediated by ion channels. For many years, the study of bacterial ion channels has provided fundamental insights into the structural basis of such neuronal signaling1,2. In particular, the prokaryotic potassium ion channel KcsA provided the first structural information on ion selectivity and conductance3. More recently, it has been shown that bacteria possess many important classes of other ion channels such as sodium channels4, chloride channels5, calcium-gated potassium channels6 and ionotropic glutamate receptors7 similar to those found in neurons. However, the native role of these ion channels in bacteria has largely remained unclear8,9. Pioneering efforts to uncover ion channel function in bacteria have identified roles in the extreme acid resistance response5 and in osmoregulation10, yet ion-specific channels do not appear to be solely responsible for these cellular processes. It thus remains unclear whether ion channels can support other unique functions in prokaryotes. We hypothesized that studying bacteria in their native context, the biofilm community, may reveal new clues about the function of ion channels in bacteria.
Bacterial biofilms are organized communities containing billions of densely packed cells. Such communities can exhibit fascinating macroscopic spatial coordination11–16. However, it remains unclear how microscopic bacteria could communicate effectively over large distances. To investigate this question, we studied a Bacillus subtilis microbial community that was recently reported to undergo metabolic oscillations triggered by nutrient limitation17. The oscillatory dynamics resulted from long-range metabolic codependence between cells in the interior and periphery of the biofilm (Figure 1a)17. Specifically, interior and peripheral cells compete for glutamate, while sharing ammonium. As a result, biofilm growth halts periodically, increasing nutrient availability for the sheltered interior cells. Interestingly, glutamate (Glu−) and ammonium (NH4+) are both charged metabolites, whose respective uptake and retention is known to depend on the transmembrane electrical potential and proton motive force (PMF)18,19. Therefore, we wondered whether metabolic coordination among distant cells within the biofilm might also involve a form of electrochemical signaling.
Figure 1.
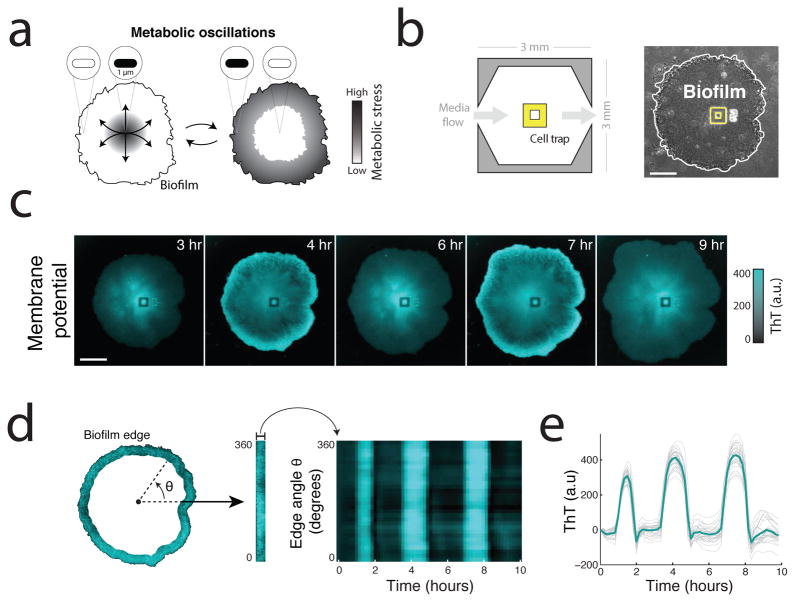
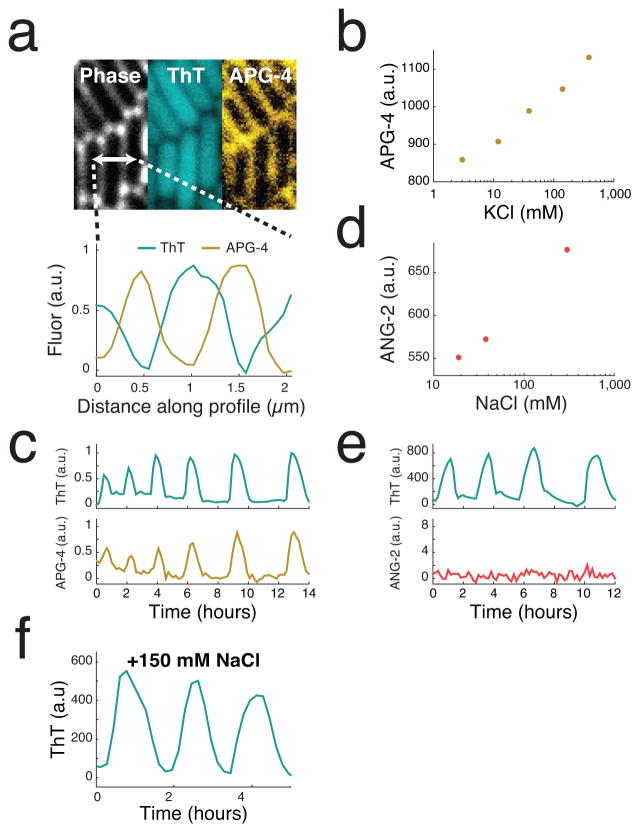
Biofilms produce synchronized oscillations in membrane potential. (a) Biofilms generate collective metabolic oscillations resulting from long-range metabolic interactions between interior and peripheral cells17. It remains unclear how microscopic bacteria are capable of communicating over such macroscopic distances within biofilm communities. (b) Schematic of the microfluidic device used throughout this study (left). Phase contrast image of a biofilm growing in the microfluidic device with the cell trap highlighted in yellow (right). Scale bar indicates 100 μm. (c) Global oscillations in membrane potential, as reported by Thioflavin T (ThT), within the biofilm community. ThT is positively charged but not known to be actively transported, so it can be retained in cells due to their inside-negative membrane potential. ThT fluorescence increases when the inside of the cell becomes more negative, and thus ThT is inversely related to the membrane potential. Scale bar indicates 0.15 mm. Representative images shown are drawn from over 75 independent biofilms. (d) Membrane potential oscillations are highly synchronized even between the most distant regions of the biofilm. To analyze synchronization, the edge region of the biofilm was identified and straightened (left) then plotted over time (right). (e) Time traces of the heatmap shown in d. Indicated in bold is the mean of 30 traces.
Oscillations in membrane potential
In order to monitor long-range electrical fluctuations in the bacterial community as a function of space and time, we grew biofilms in an unconventionally large microfluidic device (Figure 1b and Supplementary Information: Microfluidics). To measure electrical signaling, we used the fluorescent cationic dye Thioflavin T (ThT) to quantify membrane potential within the biofilm. ThT is positively charged and can be retained in cells because of their inside-negative electrical membrane potential. Thus, cells with a negative membrane potential will retain more ThT, allowing it to act as a Nernstian voltage indicator20,21. We confirmed that ThT faithfully reports the membrane potential by comparing it to DiSC3(5), an established reporter of membrane potential in bacteria (Extended Data Figure 1a)22. We find that ThT has an approximately three fold higher sensitivity to changes in membrane potential compared to DiSC3(5) (Extended Data Figure 1a, inset). Furthermore, we exposed cells to minor changes in external pH, which is known to alter membrane potential23, and indeed observed the expected changes in ThT (Extended Data Figure 1b). Therefore, ThT accurately reports on changes in membrane potential for bacteria residing in biofilms.
We next investigated changes in membrane potential during metabolic oscillations. In particular, quantitative measurements of ThT fluorescence showed global and self-sustained oscillations consistent with the reported period of the metabolic oscillations (Figure 1c, Supplementary Video 1 and Extended Data Figure 1c)17. Furthermore, oscillations in ThT could be quenched by supplementation of the media with glutamine, which bypasses the need for glutamate and ammonium (Extended Data Figure 1d). These data show a connection between metabolic oscillations and membrane potential. Strikingly, oscillations in membrane potential were synchronized among even the most distant regions of the biofilm community (Figure 1d,e). We wondered whether active electrochemical signaling could be responsible for this long-range synchronization.
Active propagation of potassium signal
Changes in membrane potential involve the movement of charged species across the cellular membrane. We suspected potassium, since it is the most abundant cation in all living cells24 and has been implicated to play a role in biofilm formation25,26. B. subtilis uses active potassium transport mechanisms to concentrate intracellular potassium at approximately 300 mM27–29. This intracellular concentration is nearly 40 times the external media concentration. Consequently, sudden release of this potassium gradient would increase extracellular potassium concentration and generate a change in the membrane potential. Accordingly, we used a fluorescent chemical potassium dye, Asante Potassium Green-4 (APG-430), to measure the extracellular concentration of potassium in the biofilm (Figure 2a and Extended Data Figure 2a,b). We observed global oscillations in APG-4 that correlated with membrane potential, which suggests that the membrane potential oscillations could involve the release of potassium (Figure 2b,c and Supplementary Video 2). In agreement with this finding, oscillations in extracellular potassium extended beyond the biofilm to flood the surrounding growth media (Extended Data Figure 2c). We also measured the dynamics of sodium, another ion commonly utilized by cells to modulate membrane potential, and observed no oscillations (Extended Data Figure 2d–f). Together, these data suggest that potassium plays a role in the synchronized oscillations in membrane potential.
Figure 2.
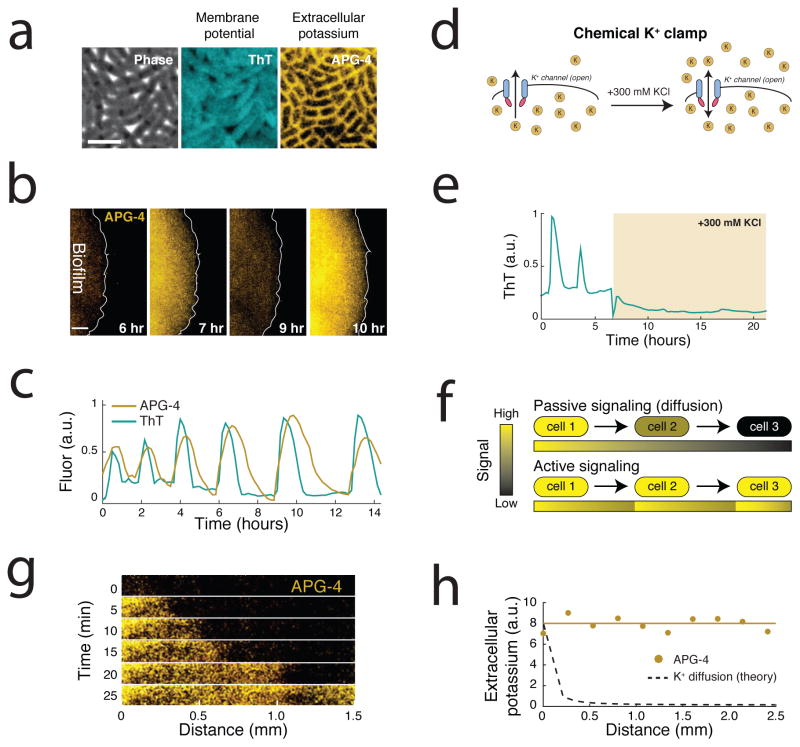
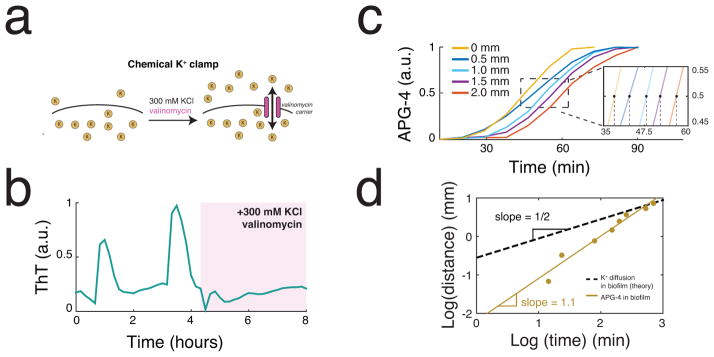
Potassium release is involved in active signal propagation within the biofilm. (a) An extracellular fluorescent chemical dye (APG-4) reports the concentration of potassium in the media (Extended Data Figure 2a,b). For comparison, the same cells are shown stained with ThT, which is inversely related to the membrane potential. These images depict cells at the peak of the ThT oscillation cycle. Representative images are selected from 6 independent experiments. Scale bar indicates 2 μm. (b) Global oscillations in extracellular potassium throughout the biofilm. A white line indicates the edge of the biofilm. Representative images are selected from 6 independent experiments. Scale bar indicates 0.2 mm. (c) Oscillations in membrane potential and extracellular potassium are synchronized, suggesting that potassium release is involved in global membrane potential oscillations. ThT is inversely related to the membrane potential. Representative traces are taken from the experiment shown in b. (d) A chemical potassium clamp (300 mM KCl, matching the intracellular concentration28) prevents the formation of potassium electrochemical gradients across the cellular membrane. (e) Clamping net potassium flux quenches oscillations in membrane potential. Representative trace is selected from 3 independent experiments. (f) Illustration of the differences between passive signaling (diffusion) and active signaling. When cells passively respond to a signal, the range that the signal can propagate is limited due to the decay of signal amplitude. In contrast, when cells actively respond by amplifying the signal, propagation can extend over greater distances. (g) We measured propagation of extracellular potassium by measuring APG-4 in time and along a length of approximately 1.5 mm within the biofilm. (h) Extracellular potassium amplitude is relatively constant as the signal propagates, in contrast to the predicted amplitude decay of a passive signal. Representative data selected from 6 independent experiments. The diffusion line is calculated using the 2D diffusion equation and the diffusion coefficient for potassium within biofilms (Supplementary Information).
Furthermore, we directly tested that oscillations in membrane potential were driven by flow of potassium across the cell membrane. Specifically, we clamped net potassium flux across the cell membrane by supplementing the growth media with 300 mM KCl (matching the intracellular K concentration) (Figure 2d). When we applied this chemical potassium clamp, oscillations in membrane potential abruptly halted (Figure 2e). Applying this clamp together with valinomycin, a potassium ionophore that acts as potassium-specific carrier in the cellular membrane31, yielded a similar quenching of oscillations (Extended Data Figure 3a,b). Therefore, changes in the electrochemical potential for potassium appear to be required for the observed oscillations in membrane potential.
Next, we determined whether cells could actively propagate the extracellular potassium signal through the biofilm to sustain long-range communication. While diffusive signals decay over space and time, active signaling processes can amplify the signal avoiding such decay (Figure 2f). To discriminate which of these processes may be operating in the biofilm, we observed the propagation of the extracellular potassium signal (Figure 2g). Results show that the signal travels at a constant rate of propagation (Extended Data Figure 3c,d). Furthermore, the amplitude of the signal does not decay with distance traveled, in contrast to what is predicted for passive potassium diffusion (Figure 2h). These findings are consistent with a process in which cells actively propagate the potassium signal. Together, these results suggest that the biofilm synchronizes global oscillations in membrane potential by an active signaling process involving potassium ions.
Potassium ion channel-mediated signaling
Motivated by our findings, we explored the role of ion channels in the observed potassium signaling. We focused on YugO, the only experimentally described potassium channel in B. subtilis, which is also reported to be important for biofilm formation32. Potassium flux through YugO is gated by an intracellular TrkA domain, known to be regulated by the metabolic state of the cell33–35. Accordingly, we hypothesized that metabolic limitation could form the initial trigger for YugO activation. Specifically, since glutamate limitation is known to drive the underlying metabolic oscillations17, we anticipated that transient removal of glutamate should initiate potassium release. To test this, we transiently deprived cells of glutamate and measured extracellular potassium in both wild type and yugO deletion strains (Supplementary Information: Strains). As expected, we observed extracellular potassium release from wild-type, but not the yugO deletion strain (Figure 3a). These findings indicate that glutamate limitation can trigger the potassium signal by activating the YugO potassium channel.
Figure 3.
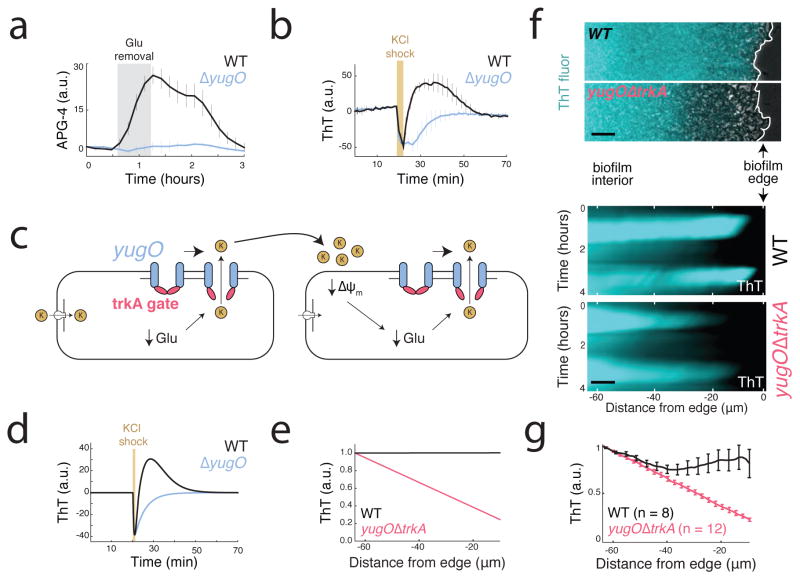
The molecular mechanism of signal propagation involves potassium channel gating. (a) yugO is a potassium channel in B. subtilis that is gated intracellularly by a trkA domain, which is regulated by the metabolic state of the cell33–35. Withdrawing glutamate (the sole nitrogen source in MSgg media) induces an increase in extracellular potassium (APG-4) for wild-type but not the yugO deletion strain. Error bars indicate the mean +/− std for 3 independent biofilms each. (b) An external potassium shock (300 mM KCl) induces a short-term membrane potential depolarization in both wild-type and yugO deletion strains. However, in the wild-type this initial depolarization was followed by hyperpolarization, which is not observed in the yugO deletion strain (mean +/− std for 12 traces drawn from 3 biofilms each). ThT is inversely related to the membrane potential. (c) Proposed toy model for potassium signaling. The initial trigger for potassium release is metabolic stress caused by glutamate limitation. External potassium depolarizes neighboring cells, producing further nitrogen limitation by limiting glutamate uptake, and thus produces further metabolic stress. This cycle results in cell-cell propagation of the potassium signal. (d) A minimal conductance-based model describing the dynamics of the cell’s membrane potential in terms of a single potassium channel and a leak current. Consistent with our experimental results, this simple model exhibits transient depolarization followed by hyperpolarization in response to local increases in extracellular potassium concentration. (e) The model predicts that manipulating channel gating and conductance will result in decaying amplitude in the spatial propagation of membrane potential oscillations. (f) Maximum intensity projection of membrane potential change illustrating attenuated communication within the biofilm in a yugOΔtrkA deletion compared to wild type biofilms (top). Heatmap of oscillations taken from wild type and yugOΔtrkA mutant biofilms (bottom). Representative images are taken from 3 independent experiments in which WT and yugOΔtrkA biofilms are compared head-to-head. Scale bars indicate 8 μm. (g) Quantification of normalized pulse amplitude from wild type (n = 8) and yugOΔtrkA (n = 12) mutant biofilms (mean +/− sem).
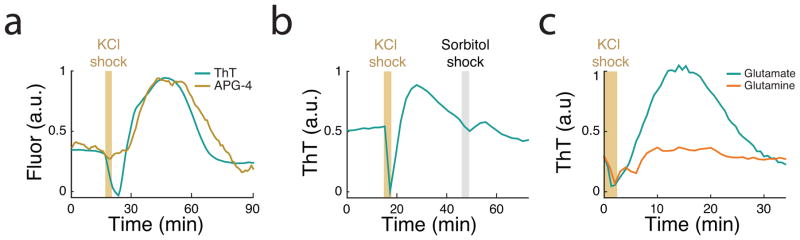
Next, we investigated whether YugO also plays a role in the active propagation of the potassium signal. To test this, we measured the response of wild-type and yugO deletion strains to transient bursts of external potassium (300 mM KCl). As expected, potassium exposure first resulted in a short-term membrane potential depolarization in both strains. However, in the wild-type this initial depolarization was followed by hyperpolarization, which was not observed in the yugO deletion strain (Figure 3b). This period of hyperpolarization was accompanied by an increase in extracellular potassium (Extended Data Figure 4a). Together, these data indicate that potassium exposure triggers a release of intracellular potassium through YugO. Exposure to an equivalent concentration of sorbitol (an uncharged solute) did not elicit a response, ruling out osmotic effects (Extended Data Figure 4b). Therefore, YugO appears to play a role in propagating the extracellular potassium signal within the biofilm.
Mathematical modeling of electrical signaling
Our data thus points to a mechanism where metabolically stressed cells release intracellular potassium, and the resulting elevated extracellular potassium imposes further metabolic stress onto neighboring cells (Figure 3c). In B. subtilis, glutamate is co-transported with 2 protons by the GltP transporter and this process depends on the proton motive force (PMF)36. Potassium mediated depolarization of the membrane potential can transiently reduce the electrical component of the PMF23, and thereby lower glutamate uptake and intracellular ammonium retention19,36. Therefore, potassium mediated signaling could propagate metabolic stress onto distant cells (Figure 3c, right). Accordingly, hyperpolarization triggered by YugO activation may represent a cellular response to enhance glutamate uptake or ammonium retention. This notion is supported by our finding that the response to extracellular potassium can be abolished by growing cells in glutamine, an uncharged metabolite and preferred nitrogen source that bypasses the need for glutamate and ammonium37 (Extended Data Figure 4c). This result further supports the specific link between potassium mediated electrical signaling and metabolic stress.
To determine whether the proposed potassium channel based mechanism is sufficient to account for the observed propagating pulses of electrical activity, we turned to mathematical modeling. Specifically, we considered a minimal conductance-based toy model describing the dynamics of the cell’s membrane potential in terms of a single potassium channel and a leak current (Supplementary Information: Mathematical Model). Consistent with our experimental results, this simple model exhibits transient depolarization followed by hyperpolarization in response to local increases in extracellular potassium concentration (Figure 3d). Furthermore, the model shows long-range propagation of these excitations without decay in the amplitude of membrane potential oscillations (Figure 3e). Therefore, the proposed mechanism is mathematically sufficient to qualitatively account for the observed membrane potential dynamics and active propagation in space.
The model also predicts that reduced efficiency of the potassium channel function could lead to degradation in long-range communication (Figure 3e). Since a complete yugO deletion interferes with development of large biofilms32, we constructed a strain in which we deleted the TrkA gating domain, leaving only the ion channel portion of YugO intact (Supplementary Information: Strains). Similarly truncated bacterial potassium channels have been shown to have altered gating and ion conductance33,38 Indeed, the yugOΔtrkA mutant biofilms exhibited a reduced propagation of membrane potential oscillations (Figure 3f and Supplementary Video 3). Specifically, in contrast to wild-type, the yugOΔtrkA mutant shows decay in the signal amplitude from the interior of the biofilm to the cells at the periphery, which is also consistent with model predictions (Figure 3g). Thus, YugO channel gating appears to promote efficient electrical communication between distant cells.
Discussion
Our findings suggest that bacteria use potassium ion channel mediated electrical signals to coordinate metabolism within the biofilm. The ensuing “bucket brigade” of potassium release allows cells to rapidly communicate their metabolic state, taking advantage of a link between membrane potential and metabolic activity. This form of electrical communication can thus enhance the previously described long-range metabolic codependence in biofilms17. Specifically, the wave of depolarization triggered by metabolically stressed interior cells would limit the ability of cells in the biofilm periphery to take up glutamate or retain ammonium, thereby allowing interior cells more access to these nutrients. This also provides a possible explanation for the observation that the yugO deletion strain has a defect in biofilm development32. Interestingly, owing to the rapid diffusivity of potassium ions in aqueous environments, it is also conceivable that even physically disconnected biofilms could be capable of synchronizing their metabolic oscillations by a similar exchange of potassium ions.
The role of ion channel mediated electrical communication has long been appreciated39. While cation channels are found in all organisms8,9 and potassium is the dominant intracellular cation24, electrical signaling is commonly viewed to be a property of neurons. However, several recent studies have suggested that in addition to traditional cell-to-cell communication systems such as quorum sensing40, bacteria may use electron flux41–43 to communicate. The here described study of electrical coordination of metabolism in microbial communities may in turn hold some general insights that extend beyond bacteria. For example, the connection between neuronal signaling and metabolic activity (neurometabolism) is an active area of research44,45. Furthermore, depletion of glutamate, the most common excitatory neurotransmitter46, also forms the initial trigger for these collective metabolic oscillations synchronized by potassium. Therefore, it is intriguing to think not only about the structural similarities between bacterial and human potassium ion channels1,2, but also their possible functional similarities with respect to long-range electrical communication.
Methods
Strains
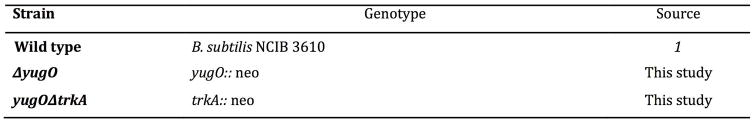
All experiments were done using Bacillus subtilis NCIB 3610. The wild type strain was a gift from Wade Winkler (University of Maryland)47, and all other strains were derived from it and are listed in Extended Data Figure 5. To make deletion strains, we used polymerase chain reaction (PCR) to amplify the desired region from the wild type strain. The PCR product is then put within the pER449 vector (gift from Wade Winkler). For the trkA mutant, we deleted the C-terminal portion of yugO (AA’s 117–328), leaving only the N-terminal ion channel portion of yugO (AA’s 1–116). We identified the trkA region using Pfam (http://pfam.xfam.org/). All constructs were confirmed by direct sequencing and then integrated into the chromosome of the wild type strain by a standard one-step transformation procedure48. Finally, chromosomal integrations were confirmed by colony PCR using the corresponding primers.
Growth conditions
The biofilms were grown in MSgg medium16 which contains 5 mM potassium phosphate buffer (pH 7.0), 100 mM MOPS buffer (pH 7.0, adjusted using NaOH), 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 100 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine HCl, 0.5% (v/v) glycerol and 0.5% (w/v) monosodium glutamate. The MSgg medium was made from stock solutions on the day of the experiment, and the stock solution for glutamate was made new weekly.
Microfluidics
We followed methods similar to a previous study17. Briefly, we used the CellASIC ONIX Microfluidic Platform and the Y04D microfluidic plate (EMD Millipore). We used a pump pressure of 1 psi with only one media inlet open, which corresponds to a flow speed of ~16 μm/s. On the day before the experiment, cells from −80°C glycerol stock were streaked onto an LB agar plate and incubated at 37°C overnight. The next morning, a single colony was picked from the plate and inoculated into 3 ml of LB broth and incubated in a 37°C shaker. After 2.5 hours of incubation, the cell culture was centrifuged at 2100 rcf for 1 min, and the cell pellet was re-suspended in MSgg and immediately loaded into microfluidics. After loading, cells in the microfluidic chamber were incubated at 37°C for 90 min, and then the temperature was kept at 30°C for the rest of the experiment.
Time-Lapse Microscopy
The growth of the biofilms was recorded using phase contrast microscopy. The microscopes used were Olympus IX83 and DeltaVision PersonalDV. To image entire biofilms, 10X objectives were used in most of the experiments. Biofilm phase contrast and fluorescence images were usually taken every 10 min, except in Figure 2g where images were taken every 5 min. To generate Figure 3b and Extended Data Figures 4a–c, where high temporal resolution was required, images were taken every minute. Whenever fluorescence images were recorded, we used the minimum exposure time that still provided a good signal-to-noise ratio (for example, we typically used 20 ms exposure for ThT and 100 ms exposure for APG-4).
Image analysis
Fiji/ImageJ (National Institutes of Health) and MATLAB (MathWorks) were used for image analysis. We generated custom scripts and used the image analysis toolbox to perform image segmentation on biofilm phase contrast images. To measure biofilm growth rate, we identified the biofilm area in each frame by segmenting the images and took the derivative of biofilm radius over time. We identified the radius by assuming circular growth of the colony and taking the length from the center of the cell trap to the biofilm edge. To generate membrane potential curves, we measured the fluorescence of ThT within the biofilm using the ImageJ “Plot Z-axis Profile” command and performed subsequent analysis, such as normalization and subtracting of baseline signal, in MATLAB.
Experimental reproducibility
Data shown in the main figures were drawn from a minimum of 3 independent experiments and often many more. For example, we analyzed ThT oscillations (represented in Figure 1c–e) in over 75 biofilms. In cases where only a single representative trace is shown, we analyzed multiple regions within the biofilm to ensure accuracy of the analysis. In experiments comparing wild-type and a mutant strain (yugO or yugOΔtrkA), we always performed head-to-head experiments (separate chambers in the same microfluidic device) on the same day using the same media to eliminate possible artifacts.
Theoretical estimate of potassium diffusion within biofilms
We used the diffusion coefficient of potassium in water (19.7×10−6cm2/s)49 and reduced it to 70% in accordance with a reference on diffusion in biofilms50, yielding the value of the diffusion coefficient (13.8×10−6cm2/s) used in the mathematical model as well as the theoretical curves plotted alongside our experimental data. To estimate the rate of potassium propagation by diffusion, we used the formula for 2D mean squared displacement (MSD):
Where r is the displacement, D is the diffusion coefficient, and t is time. We used this relationship to generate the curve shown in Extended Data Figure 3d. We directly compared the log-log slope of the experimental data (slope = 1.1, R2 = 0.96) to that expected for diffusion (slope = 0.5) to further confirm that the experimental data cannot be explained by simple diffusion.
To estimate the decay of amplitude by diffusion, we used the formula for the concentration profile of 2D diffusion:
Where C is the concentration of potassium at displacement r and time t, M is a constant related to the initial pulse amplitude of potassium that we matched to the initial experimental pulse amplitude of APG-4, and D is the diffusion coefficient. We used this relationship to generate the curve in Figure 2h.
Dyes and Concentrations
Thioflavin T (ThT) and DiSC3(5) were used at 10 μM. We used ThT and DiSC3(5) to track relative changes in the membrane potential, where the fluorescence of ThT increases when the cell becomes more inside negative (hyperpolarizes). We found the sensitivity of ThT to be significantly higher than that of DiSC3(5), where sensitivity is defined as the ratio between the amplitude of oscillation and its error (Extended Data Figure 1a). Furthermore, under our experimental conditions, DiSC3(5) appears to be absorbed by the PDMS in the microfluidic device. This hinders quantitative analysis (lower sensitivity) and also greatly increases the time required for the dye to diffuse into the biofilm.
APG-4 (TEFLabs) was used at 2 μM. We used the membrane impermeable TMA+ salt form to track the extracellular concentration of potassium. We verified that APG-4 does not significantly diffuse into cells (Extended Data Figure 2a). We also verified that APG-4 could measure extracellular potassium in MSgg media within our microfluidic device (Extended Data Figure 2b).
We used ANG-2 (TEFLabs) at 2 μM. We used the membrane impermeable TMA+ salt form to track the extracellular concentration of sodium (Extended Data Figure 3c, d).
Extended Data
Extended Data Figure 1.
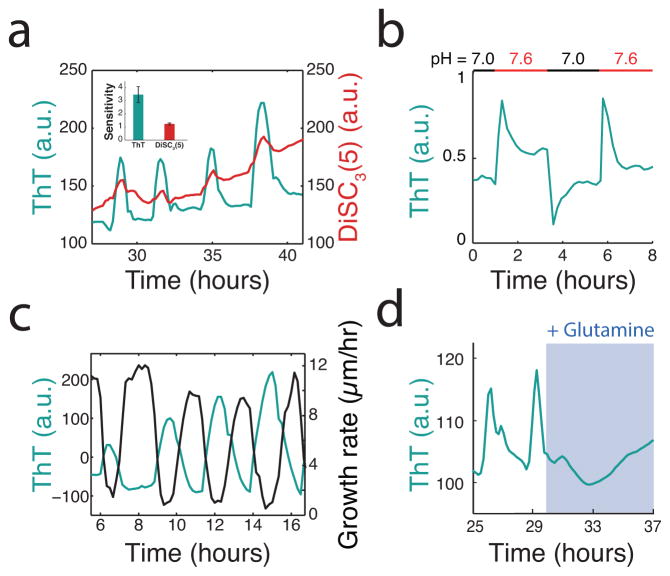
Thioflavin T (ThT) is a fluorescent reporter that is inversely related to the membrane potential. (a) ThT and DiSC3(5), an established reporter of membrane potential in bacteria22, both oscillate within biofilms. ThT has an approximately three fold higher sensitivity to changes in membrane potential compared to DiSC3(5). Sensitivity is defined as the ratio between peak height and error in peak height. Representative trace is selected from 3 independent biofilms. (b) The cellular ThT fluorescence depends on the external pH, where higher pH results in greater membrane potential as expected23. ThT itself is insensitive to these pH changes and the traces are background subtracted to eliminate possible artifacts. Representative trace is selected from 3 independent biofilms. (c) Oscillations in ThT and growth rate are inversely correlated, linking membrane potential oscillations to the metabolic cycle which produces periodic growth pauses17. Growth rate is calculated by taking the derivative of biofilm radius over time (Supplementary Information). Representative trace is selected from over 75 independent biofilms. (d) Replacing glutamate with 0.2% glutamine, which eliminates the need to take up glutamate or retain ammonium, quenches ThT oscillations. This further suggests that ThT oscillations are specific to the metabolic cycle involving glutamate and ammonium. Representative trace is selected from 3 independent experiments.
Extended Data Figure 2.
A fluorescent reporter of extracellular potassium (APG-4) indicates that potassium plays a role in membrane potential oscillations. (a) High-resolution images showing the intracellular localization of ThT and primarily extracellular localization of APG-4 (left). Quantification of ThT and APG-4 along the 2μm profile indicated in the phase image indicates that APG-4 does not significantly diffuse into the cell (right). Representative images are selected from 6 independent experiments. (b) Induction curve for APG-4 generated using externally supplemented KCl. The experiment was repeated twice. (c) Oscillations in extracellular potassium in the surrounding cell-free region during biofilm oscillations. These oscillations occurred during the experiment shown in Figure 2bc and the pulses are synchronized between the biofilm and the surrounding cell-free region. Representative trace is selected from 6 independent experiments. (d) Induction curve for ANG-2 generated using externally supplemented NaCl. Experiment was repeated twice. (e) Simultaneous measurement of ThT and ANG-2 within the biofilm indicates a lack of oscillations in extracellular sodium. Representative trace selected from 3 independent biofilms. (f) Furthermore, perturbing extracellular sodium concentrations in the media had no detectable effect on membrane potential oscillations. Experiment was repeated twice.
Extended Data Figure 3.
(a) A chemical potassium clamp (300 mM KCl, matching the intracellular concentration28, and 30 μM valinomycin) prevents the formation of potassium electrochemical gradients across the cellular membrane. Valinomycin is an antibiotic that creates potassium-specific carriers in the cellular membrane31. (b) Clamping net potassium flux quenches oscillations in membrane potential. (c) Propagation of extracellular potassium is estimated by tracking the halfmaximal position of the pulse over time. Representative traces are shown for a single pulse selected from one of 6 independent experiments. (d) Propagation of extracellular potassium is relatively constant over time in contrast to diffusion that is expected to decay. The diffusion line is calculated using the mean squared displacement (MSD) and the diffusion coefficient for potassium in biofilms (Supplementary Information). Slopes are calculated from the same representative data shown in c.
Extended Data Figure 4.
External potassium affects the metabolic state of the cell. (a) A potassium shock (300 mM KCl) produces an initial ThT decrease (depolarization) followed by a sustained ThT increases (hyperpolarization). ThT is inversely related to the membrane potential. A corresponding pulse in APG-4 during this ThT increase suggests that hyperpolarization is due to release of potassium. APG-4 signal due to the external potassium shock itself was subtracted using the cell-free background near the biofilm. Representative trace is selected from 3 independent experiments. (b) ThT spikes in response to external potassium shock (300 mM KCl) but not an equivalent shock of 300 mM sorbitol, an uncharged solute. Representative trace is selected from 3 independent experiments. (c) The hyperpolarization response occurs when cells are grown in glutamate but not when glutamate is replaced by 0.2% glutamine, which bypasses the need to take up glutamate or retain ammonium. Representative trace is selected from 4 independent biofilms.
Extended Data Figure 5.
List of strains used in this study.
Extended Data Figure 6.
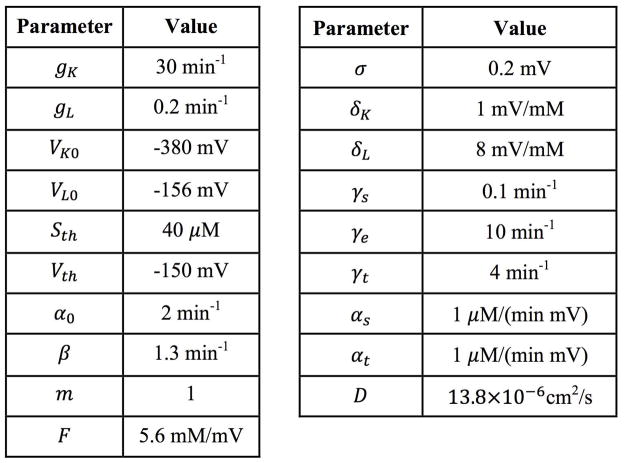
Parameter values used in the model.
Supplementary Material
Acknowledgments
We would like to thank Katherine Suel, Roy Wollman, and Tolga Cağatay for comments during the writing of the manuscript and Christopher Piggott for cloning help. A.P. is a Simons Foundation Fellow of the Helen Hay Whitney Foundation. J.G.O. is supported by the Ministerio de Economia y Competitividad (Spain) and FEDER, under project FIS2012-37655-C02-01, and by the ICREA Academia Programme. This research was funded by the National Institutes of Health, National Institute of General Medical Sciences Grant R01 GM088428 and the National Science Foundation Grant MCB-1450867 (both to G.M.S.). This work was also supported by the San Diego Center for Systems Biology (NIH Grant P50 GM085764).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions G.M.S., A.P., J.L., M.A., and J.G.O. designed the research, A.P. and J.L. performed the experiments, J.L. and A.P. performed the data analysis, J.G.O. performed the mathematical modeling, S.L. made the bacteria strains, G.M.S., A.P., J.L., and J.G.O. wrote the manuscript. All authors discussed the manuscript.
The authors declare no competing financial interest.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 2001 [Google Scholar]
- 2.MacKinnon R. Potassium channels and the atomic basis of selective ion conduction. Bioscience Reports. 2004;24:75–100. doi: 10.1007/s10540-004-7190-2. [DOI] [PubMed] [Google Scholar]
- 3.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 4.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 5.Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419:715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 7.Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999;402:817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- 8.Kuo MMC, Haynes WJ, Loukin SH, Kung C, Saimi Y. Prokaryotic K+ channels: From crystal structures to diversity. FEMS Microbiology Reviews. 2005;29:961–985. doi: 10.1016/j.femsre.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Saimi Y, Loukin SH, Zhou XL, Martinac B, Kung C. Ion channels in microbes. Methods Enzymol. 1998;294:507–524. doi: 10.1016/s0076-6879(99)94030-2. [DOI] [PubMed] [Google Scholar]
- 10.Martinac B, Buechner M, Delcour AH, Adler J, Kung C. Pressure-sensitive ion channel in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84:2297–2301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 12.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 13.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asally M, et al. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proceedings of the National Academy of Sciences. 2012 doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilking JN, et al. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2013;110:848–52. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne S, et al. Temporal control of self-organized pattern formation without morphogen gradients in bacteria. Mol Syst Biol. 2013;9 doi: 10.1038/msb.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. Metabolic codependence gives rise to collective oscillations within microbial communities. Nature. 2015;523 doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolner B, Ubbink-Kok T, Poolman B, Konings WN. Characterization of the proton/glutamate symport protein of Bacillus subtilis and its functional expression in Escherichia coli. J Bacteriol. 1995;177:2863–2869. doi: 10.1128/jb.177.10.2863-2869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boogerd FC, et al. AmtB-mediated NH3 transport in prokaryotes must be active and as a consequence regulation of transport by GlnK is mandatory to limit futile cycling of NH4 +/NH3. FEBS Lett. 2011;585:23–28. doi: 10.1016/j.febslet.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 20.Kralj JM, Hochbaum DR, Douglass AD, Cohen AE. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science. 2011;333:345–8. doi: 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- 21.Lo CJ, Leake MC, Pilizota T, Berry RM. Nonequivalence of membrane voltage and ion-gradient as driving forces for the bacterial flagellar motor at low load. Biophys J. 2007;93:294–302. doi: 10.1529/biophysj.106.095265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A. 2010;107:12281–6. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epstein W. The Roles and Regulation of Potassium in Bacteria. Progress in Nucleic Acid Research and Molecular Biology. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 25.Lopez D, Fischbach Ma, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2009;106:280–5. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinsinger RF, Kearns DB, Hale M, Fall R. Genetic requirements for potassium ion-dependent colony spreading in Bacillus subtilis. J Bacteriol. 2005;187:8462–8469. doi: 10.1128/JB.187.24.8462-8469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira-Pires RS, Szollosi A, Morais-Cabral JH. The structure of the KtrAB potassium transporter. Nature. 2013;496:323–8. doi: 10.1038/nature12055. [DOI] [PubMed] [Google Scholar]
- 28.Holtmann G, Bakker EP, Uozumi N, Bremer E. KtrAB and KtrCD: Two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol. 2003;185:1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whatmore aM, Chudek Ja, Reed RH. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J Gen Microbiol. 1990;136:2527–35. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]
- 30.Rimmele TS, Chatton JY. A Novel Optical Intracellular Imaging Approach for Potassium Dynamics in Astrocytes. PLoS One. 2014 doi: 10.1371/journal.pone.0109243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolin Y, Eisenbach M. Voltage clamp effects on bacterial chemotaxis. J Bacteriol. 1984;159:605–610. doi: 10.1128/jb.159.2.605-610.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundberg ME, Becker EC, Choe S. MstX and a putative potassium channel facilitate biofilm formation in Bacillus subtilis. PLoS One. 2013;8:e60993. doi: 10.1371/journal.pone.0060993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao Y, et al. Gating of the TrkH ion channel by its associated RCK protein TrkA. Nature. 2013;496:317–22. doi: 10.1038/nature12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roosild TP, Miller S, Booth IR, Choe S. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell. 2002;109:781–791. doi: 10.1016/s0092-8674(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 35.Schlosser A, Hamann A, Bossemeyer D, Schneider E, Bakker EP. NAD+ binding to the Escherichia coli K+-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Mol Microbiol. 1993;9 doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 36.Tolner B, et al. Characterization of the proton / glutamate symport protein of Bacillus subtilis and its functional expression in Escherichia coli. Updated information and services can be found at : These include : Characterization of the Proton / Glutamate Symport Prote. 1995;177 doi: 10.1128/jb.177.10.2863-2869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher SH. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol. 1999;32:223–32. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- 38.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its applications to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 41.Kato S, Hashimoto K, Watanabe K. Iron-Oxide Minerals Affect Extracellular Electron-Transfer Paths of Geobacter spp. Microbes Environ. 2013;28:141–148. doi: 10.1264/jsme2.ME12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeffer C, et al. Filamentous bacteria transport electrons over centimetre distances. Nature. 2012 doi: 10.1038/nature11586. [DOI] [PubMed] [Google Scholar]
- 43.Masi E, et al. Electrical spiking in bacterial biofilms. J R Soc Interface. 2014 doi: 10.1098/rsif.2014.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan JW, et al. Neurometabolism in human epilepsy. Epilepsia. 2008;49:31–41. doi: 10.1111/j.1528-1167.2008.01508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petroff OAC, Errante LD, Rothman DL, Kim JH, Spencer DD. Glutamate-glutamine cycling in the epileptic human hippocampus. Epilepsia. 2002;43:703–710. doi: 10.1046/j.1528-1157.2002.38901.x. [DOI] [PubMed] [Google Scholar]
- 46.Meldrum BS. Glutamate as a neurotransmitter in the brain: review of physiology and pathology. J Nutr. 2000;130:1007S–15S. doi: 10.1093/jn/130.4.1007S. [DOI] [PubMed] [Google Scholar]
- 47.Irnov I, Winkler WC. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol Microbiol. 2010;76:559–575. doi: 10.1111/j.1365-2958.2010.07131.x. [DOI] [PubMed] [Google Scholar]
- 48.Jarmer H, Berka R, Knudsen S, Saxild HH. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol Lett. 2002;206:197–200. doi: 10.1111/j.1574-6968.2002.tb11009.x. [DOI] [PubMed] [Google Scholar]
- 49.Horvath AL. Handbook of aqueous electrolyte solutions: physical properties, estimation and correlation methods. 1986 [Google Scholar]
- 50.Stewart PS. Diffusion in biofilms. Journal of Bacteriology. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.