Abstract
Background
Co-infection of HIV with HBV is common in West Africa but little information is available on the effects of HBV on short-term therapy for HIV patients. A 28 day longitudinal study was conducted to examine short-term antiretroviral therapy (ART) outcomes in HIV infected individuals with HBV co-infection.
Methods
Plasma from 18 HIV infected individuals co-infected with HBV and matched controls with only HIV infection were obtained at initiation, and 7 and 28 days after ART. HIV-1 viral load changes were monitored. Clinical and demographic data were also obtained from patient folders, and HIV-1 drug resistance mutation and subtype analysis performed.
Results
The presence of HBV co-infection did not significantly affect HIV-1 viral load changes within 7 or 28 days. The CD4+ counts on the other hand of patients significantly affected the magnitude of HIV-1 viral load decline after 7 days (ρ = −0.441, p = 0.040), while the pre-ART HIV-1 VL (ρ = 0.844, p = <0.001) and sex (U = 19.0, p = 0.020) also determined HIV-1 viral load outcomes after 28 days of ART. Even though the geometric sensitivity score of HIV-1 strains were influenced by the HIV-1 subtypes (U = 56.00; p = 0.036), it was not a confounder for ART outcomes.
Conclusions
There may be the need to consider the confounder effects of sex, pre-ART CD4+, and pre-ART HIV-1 viral load in the discourse on HIV and HBV co-infection.
Keywords: Human immunodeficiency virus, Hepatitis B virus, Co-infection, Antiretroviral therapy, Drug resistance, Short-term therapy
Background
In West Africa, there is a high prevalence of hepatitis B virus (HBV) infection [1], and this is also reflected in similar high numbers of individuals with HIV also co-infected with HBV in the sub-region [2–7].
There are conflicting reports on the effects of HBV on the response to antiretroviral therapy (ART). Even though some studies have shown no significant effects of HBV infections on CD4+ recovery and/or suppression of HIV-1 plasma viral load (HIV-1 VL) during ART, [8–12] others have seen some effects [4, 5, 13–15]. Apart from having higher HIV-1 VL, individuals with HIV and HBV co-infection in Taiwan on ART were found to have more frequent virological failure compared to those with HIV alone [13]. In patients co-infected with HIV and HBV in Nigeria, the presence of baseline hepatitis B e antigen (HBeAg) was associated with a slower virological response (≤400 copies/ml) as compared to those without HBeAg at week 24 [4]. However, this effect of HBeAg did not reflect in viral suppression after 48 weeks of ART. CD4+ count recovery during ART has been shown to be initially blunted in individuals with co-infection as compared to those with HIV infection alone [15]. Also, large studies done in Africa and Europe have reported impaired CD4+ outcomes due to HBV co-infection after varying periods of ART [5, 14]. These seeming discrepancies in outcomes from the various studies may be attributed to different clinical factors. Since in vitro studies have shown that HBV protein X acts as a trans-activator for HIV infections [16, 17], there is the need to understand the effects of the interaction between HIV and HBV on short and long term ART outcomes, bearing in mind demographical, epidemiological, clinical and viral factors.
In the absence of long term HIV-1 VL data and challenges with adherence after the first few weeks of ART in developing countries, changes in HIV-1 VL after 7 days (HIV-1 VL0–7) and 28 days (HIV-1 VL0–28) of ART may be useful in predicting long-term virological outcome [18–23]. In a previous study we showed that the presence of HBeAg in HIV infected patients with HIV and HBV co-infection resulted in more severe immune suppression [7]. The objective of this study was therefore to use the same study population to determine the possible effects of co-infection with HBV on short-term ART outcomes in HIV infected patients.
Methods
Patients
Briefly, pre-ART plasma samples for a cohort of 138 patients were screened for HBsAg using a third generation ELISA, Surase B 96 (TMB), General Biologicals Corp, Hsin Chu, Taiwan, and the Core HBsAg rapid test, Core Diagnostics, Birmingham, UK. All positive samples were tested for hepatitis B e antigen and antibody (HBeAg and anti-HBe) using the EASE BN-96 (TMB) and IgM antibodies to hepatitis B core antigen (anti-HBc IgM) using the ANTICORASE MB-96 (TMB) kits respectively (General Biologicals, Taiwan). Controls for all HIV patients with HBV co-infection (n = 18) were chosen based on a baseline CD4+ > or < 100 cells/μl. HIV mono-infected patients were matched (1:1) to co-infected patients on CD4+ cell count. All patients were screened with a one-step anti-HIV 1&2 test (SD Bioline, Premier Medical Corporation Ltd., India) and were shown to be HIV-1 infected. Demographic and clinical information was obtained from patient hospital records and adherence measured by self-reporting and pill count. The various host factors examined as possible confounders in this study included age, sex, pre-ART CD4+, and WHO clinical disease staging, while the viral factors were HIV-1 subtypes and drug resistance mutations (HIV-1 DRMs).
Ethics and consent
The study was approved by the University of Ghana Medical School Ethics and Protocol Review Committee and written informed consent was obtained from patients before enrolment in the study.
HIV-1 RNA quantitation and determination of early decay
Aliquots of plasma frozen in volumes of 600 μl at −85 °C were used in HIV-1 VL determination using the COBAS Amplicor Monitor v1.5 tests and the COBAS Amplicor analyzer (Roche Diagnostics GmbH, Mannheim, Germany). To ensure that marginal differences did not affect viral load analysis, all plasma samples were frozen in aliquots to avoid multiple freeze-thaws, and new frozen plasma vials at −85 °C were mainly used in determining viral loads. HIV-1 VL was determined in duplicates at baseline, days 7 and 28 to reduce errors inherent within the Amplicor Monitor v1.5 assay. Since the lower limits of the standard monitor assay was 400 copies/ml, viral load values <400 copies/ml were considered as 400 copies/ml.
All HIV-1 VL analysis were done with the log values and plasma viral decay was measured as HIV-1 VL0–7 and HIV-1 VL0–28. Since HIV-1 VL change within 6 days after ART (HIV-1 VL0–6) <0.96 log or ≥1.68 log indicated a likelihood for poor and good response good response to ART respectively [21], the proportions of patients who had <0.96 log and ≥1.68 log were also determined. The proportion of patients with HIV-1 VL0–28 of ≥2.58 log which may reflect on the possibility of having sustained HIV-1 VL suppression was also done for patients with day 28 plasma samples [22].
Primary HIV-1 mutations
Since HIV-1 DRM may be subtype related [24], the possible confounder role of primary HIV-1 DRMs and subtype on short-term ART was also investigated. The High Pure Viral RNA Kit (Roche Diagnostics GmbH, Mannheim, Germany) was used for the isolation of HIV-1 RNA in plasma. Partial polymerase genes (1065–1372 bp) were amplified with a nested PCR using six primers and a modified version of a protocol published previously [25]. Reverse transcription was done using the Titan One Tube RT-PCR system (Roche Diagnostics GmbH, Mannheim, Germany). The original protocol was modified to obtain polymerase (pol) gene fragment spanning all the protease (PR) and up to 230 amino acids in the reverse transcriptase gene (RT). The Techne TC-3000 thermocycler (Techne Incorporated, Burlington, NJ, USA), was used for all RT-PCR reactions. The RT-PCR products were cleaned with the High Pure PCR Product Purification Kit (Roche Diagnostics GmbH, Mannheim, Germany). The Big Dye Terminator Cycle sequencing kit v3.1 and the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems International Incorporated, Foster City, USA) were used for forward and reverse sequencing. In addition, a third sequencing primer (RT-sec-1-S: 5′ CAA AAA TTG GGC CTG AAA ATC CAT A 3′) was used to ensure that the region towards the end of the nucleotides needed for the first 227 amino acids in the RT region were obtained. The ViroSeq HIV-1 Genotyping System v2 (Celera Diagnostics, Foster City, Calif, USA) was used as an alternative method to sequence the pol genes of HIV-1 strains which were difficult to amplify with the in-house assay [25].
Phylogenetic relationships were determined using a partial pol region containing 867 nucleotides and a RT region covering with 691 nucleotides and methods as described previously [26]. This was done to ensure that subtyping of the RT sequences was truly representative of the pol region. Sequences used for phylogeny and in the FASTA format were submitted to the Stanford University database (http://www.HIVdb.stanford.edu) for interpretation of resistance and assignment of subtype. Mutation scores were used to derive genotypic sensitivity scores (GSS) based on specific antiretroviral drugs used by patients, and this was done using comments from the Stanford database. If the mutation site was dimorphic then the total score was divided into two and the value used as the GSS. The GenBank accession numbers of the sequences used in this study are HQ170612–HQ170613, HQ170615, HQ170617–HQ170625, HQ529236–HQ529243, HQ529245, HQ529247, HQ529249, HQ529251, HQ529253–HQ529260, HQ529262 and HQ529263.
Analysis of data
The various variables included age, gender, pre-ART CD4+, WHO clinical disease staging, HIV-1 subtypes, GSS, ARVs, patient’s adherence, pre-ART HIV-1 VL and changes within the first 28 days of ART, and co-infection status of patients. For analysis involving HIV-1 subtypes, the CFR02_AG strains and non-CRF02_AG were considered as separate groups.
The Mann–Whitney U and Kruskal–Wallis tests were used to compare the effects of HBV co-infection on HIV-1 VL0–7 and HIV-1 VL0–28. In order to assess the possibility of viral and host factors as confounders for HIV-1 VL0–7 and HIV-1 VL0–28 outcomes due to HBV co-infection, the Mann–Whitney U and Spearman’s correlation tests was used to determine the effects of the other study variables on HIV-1 VL0–7 and HIV-1 VL0–28 for all study subjects. Furthermore, since an HIV-1 VL decline <0.96 log or ≥1.68 log and ≥2.58 log may determine long term outcomes for patients on ART by day 7 and 28 respectively [18, 22], a stepwise bivariate logistic regression for HIV-1 VL decline <0.96 log or ≥1.68 log and ≥2.58 log was also done with all variables significantly affecting ART outcomes (pre-ART HIV-1 VL, pre-ART CD4+, and sex). All statistical analysis were done with SPSS Version 17 software (SPSS Inc., Chicago, Illinois) and the two-tailed p-values of <0.05 considered as significant.
Results
A description of the demographic and clinical characteristics of HIV infected patients with HBV co-infection, and those with HIV infection alone is shown in Table 1. Generally, patients had low CD4+ counts with majority having WHO clinical stage 3.
Table 1.
Description of study population
| Variablesa | Co-infected (n = 18) |
HIV infections alone (n = 18) |
|---|---|---|
| Demographic data | ||
| Sex, female (%, CI) | 11 (61.1) | 11 (61.1) |
| Age (median, IQR) | 36.5 (33.8–41.5) | 35.5 (30.8–45.2) |
| Clinical parameters | ||
| CD4+ (median/IQR) | 137 (35–197) | 125 (50–182) |
| WHO disease stage | ||
| 2 (n = 3) | 2 (11.1 %) | 1 (5.6 %) |
| 3 (n = 38) | 15 (83.3 %) | 13 (72.2 %) |
| 4 (n = 5) | 1 (5.6 %) | 4 (22.2 %) |
| ARV combination | ||
| d4T + 3TC + NVP (n = 8) | 4 (22.2 %) | 4 (22.2 %) |
| d4T + 3TC + EFV (n = 11) | 4 (22.2 %) | 3 (16.7 %) |
| CBV + NVP (n = 15) | 5 (27.8 %) | 6 (33.3 %) |
| CBV + EFV (n = 12) | 5 (27.8 %) | 5 (27.8 %) |
| Hepatitis virus markers | ||
| HBeAg | 10 (55.6 %) | – |
| Anti-HBe | 8 (44.4 %) | – |
| Anti-HBc IgM | 0 (0.0 %) | – |
aFor variable with n number of patients indicated, the percentages were calculated based on those values
Polymerase gene polymorphisms
A total of 34 samples were amplified successfully using the two PCR methods and 30 were subtyped using phylogenetic analysis (Fig. 1). The relatively shorter sequences (KAF23, KAF56, KAF104 and KAF109) were subtyped solely using the Stanford database drug resistance program (http://www.HIVdb.stanford.edu). Majority of the subtypes were CRF02_AG, with 5 subtype G, 1 CRF06_cpx, 1 subtype A, and 1 CRF01_AE (Fig. 1), but the Stanford database drug resistance program failed to correctly infer subtypes A3 and CRF06_cpx (Table 2). The topologies of the 867 pol and 691 RT phylogenetic trees were similar thus confirming subtypes assigned to the RT region. There were no major drug resistance mutations but seven HIV-1 stains had some level of drug resistance (Table 2). The V108I mutation seen in KAF41, KAF71 and KAF104 reduces NVP and EFV susceptibilities by about twofold. The V118I mutation which is a polymorphic accessory NRTI-resistance mutation that occurs in combination with multiple TAMs, was seen in the only subtype A3 identified in the study (KAF33). Finally, the V179E mutation which causes low level resistance in NVP and EFV was seen in KAF69 and KAF71, but as a polymorphic site in KAF41 (V179EV) (Table 2). The GSS were influenced by the HIV-1 subtypes (U = 56.00; p = 0.036).
Fig. 1.
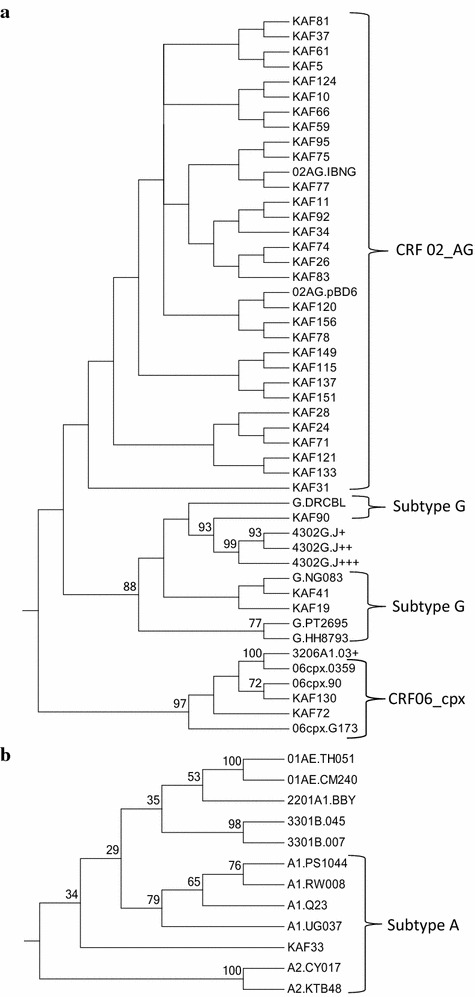
Phylogenetic trees using partial pol genes of HIV-1; a CRF02_AG, CRF06_cpx, and Subtype G clusters, and b Subtype A cluster. The reference subtype [accession number] include: A1 (PS1044 [DQ676872]; Q23 [AF004885]; RW008 [AB253421]; UG037 [AB253429]), A2 (KTB48 [AF286238]; CY017 [AF286237]), B (HXB2 [K03455]; 00T36 [AY423387]; BK132 [AY173951]; 1058 [AY331295]), C (BR025 [U52953]; ETH2220 [U46016]; IN21068 [AF067155]; SK164B1 [AY772699]), D (ELI [K03454]; 4412HAL [AY371157]; A280 [AY253311]; UG114 [U88824]), F1 (VI850 [AF077336]; BR020 [AF005494]; FIN9363 [AF075703]; MP411 [AJ249238]), F2 (0016BBY [AY371158]; MP255 [AJ249236]; MP257 [AJ249237]; 53657 [AF377956]), G (DRCBL [AF084936]; HH8793 [AF061641]; NG083 [U88826]; PT2695 [AY612637]), H (VI991 [AF190127]; VI997 [AF190128]; 056 [AF005496]), J (KTB147 [EF614151]; 7887 [AF082394]; 7022 [AF082395]), K (EQTB11C [AJ249235]; MP535 [AJ249239]), 01_AE (CM240. [U54771]; TH051. [AB220944]), 02_AG (pBD6 [AY271690]; IBNG. [L39106]), 06_cpx (90 [AF064699]; 0359 [AY535659]; G173 [AB286851]), 09_cpx (00IC [AJ866553]; 2911 [AY093605]), 22_01A1 (BBY [AY371159]), 28_BF 12313 [DQ085872]; 12609 [DQ085873]; 12817 [DQ085874]), 29_BF (16704 [DQ085876]; 11948 [DQ085871]), 32_06A1 (03+ [AY535660]), 33_01B (007 [DQ366659]; 045 [DQ366662]), 34_01B (2478 [EF165541]), 43_02G (J+ [EU697904]; J++ [EU697907]; J+++ [EU697909]), O (ANT70 [L20587])
Table 2.
HIV-1 subtype and drug resistance mutations in patients
| Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| KAF | Subtype | Mutationsa | GSSa | HIV-1 VL | KAF | Subtype | Mutationsa | GSSa | HIV-1 VL |
| 151 | CRF02_AG | – | 0 | 5.38 | 075 | CRF02_AG | – | 0 | 5.08 |
| 083 | CRF02_AG | – | 0 | 5.74 | 124 | CRF02_AG | – | 0 | 5.46 |
| 092 | CRF02_AG | – | 0 | 5.87 | 023 | CRF01_AE | na | na | 5.49 |
| 028 | CRF02_AG | – | 0 | 5.26 | 130 | CRF02_AG/G | – | 0 | 5.73 |
| 090 | G | – | 0 | 4.76 | 033 | A/CRF01_AE | V118I | 0 | 6.02 |
| 056 | G | na | na | 5.33 | 081 | CRF02_AG | – | 0 | 5.39 |
| 071 | CRF02_AG | V108I, V179E | 25 | 5.16 | 078 | CRF02_AG | E138A | 0 | 3.52 |
| 120 | CRF02_AG | – | 0 | 5.65 | 104 | G | V108I | 15 | 5.18 |
| 026 | CRF02_AG | – | 0 | 5.76 | 149 | CRF02_AG | – | 0 | 4.65 |
| 115 | CRF02_AG | – | 0 | 5.09 | 074 | CRF02_AG | – | 0 | 5.73 |
| 031 | CRF02_AG | V90I | 0 | 4.87 | 137 | A/CRF02_AG | – | 0 | 5.28 |
| 061 | CRF02_AG | – | 0 | 5.86 | 010 | CRF02_AG | – | 0 | 6.33 |
| 077 | CRF02_AG | – | 0 | 5.52 | 064 | na | na | na | 6.45 |
| 041 | G | V108IV, E138A, V179EV | 10 | 4.20 | 121 | CRF02_AG | – | 0 | 6.27 |
| 156 | CRF02_AG | – | 0 | 5.54 | 037 | CRF02_AG | – | 0 | 5.49 |
| 011 | CRF02_AG | V90I | 0 | 5.31 | 095 | CRF02_AG | – | 0 | 6.15 |
| 109 | CRF02_AG | – | 0 | 6.08 | 019 | G | – | 0 | 6.16 |
| 200 | na | na | na | na | 024 | CRF02_AG | – | 0 | 4.46 |
KAF, patients ID; Cases, individuals infected with HIV and HBV; Controls, those infected with only HIV; Mutationsa/GSSa, HIV-1 drug resistance mutations/genotypic sensitivity scores (GSS) based on specific ARVs used by patients as determined by the Stanford database (analyzed on July 8, 2014); HIV-1 VL, pre-HAART HIV-1 plasma viral load. Subtypes were determined by the Stanford database
na not available
Outcomes after short-term HAART
A total of 35, 22 and 20 HIV-1 VL quantitation results were available for pre-ART, day 7 and day 28 plasma samples respectively. The presence of HBV co-infection or any HBV marker did not significantly affect HIV-1 VL0–7 and HIV-1 VL0–28 (Table 3). Apart from sex, pre-ART CD4+, and pre-ART HIV-1 VL, all the other variables did not influence HIV-1 VL0–7 and HIV-1 VL0–28 (Table 4). The CD4+ of patients significantly affected the magnitude of HIV-1 VL0–7 (ρ = −0.441, p = 0.040), while the pre-ART HIV-1 VL also determined HIV-1 VL0–28 outcomes (ρ = 0.844, p = <0.001). HIV-1 VL decline <0.96 log or ≥1.68 log and ≥2.58 log was generally not affected by majority of the variables. Patients who had ≥1.68 log decline in HIV-1 VL were marginally likely to have higher CD4+ counts than those who did not achieve a 1.68 log decline (p = 0.065). The pre-ART HIV-1 VL was significantly higher in those who had achieved an HIV-1 VL decline of ≥2.58 log after 28 days of ART (p = 0.006).
Table 3.
HIV-1 plasma viral loads in HIV infected patients with HBV infection
| Groups | Mean rank | Mann–Whitney U | p value |
|---|---|---|---|
| Baseline | |||
| HIV only (n = 18) | 19.56 | 125.00 | 0.355 |
| Co-infected (n = 17) | 16.37 | ||
| Day 7 | |||
| HIV only (n = 11) | 12.14 | 53.50 | 0.646 |
| Co-infected (n = 11) | 10.86 | ||
| Day 28 | |||
| HIV only (n = 10) | 11.70 | 38.00 | 0.287 |
| Co-infected (n = 10) | 9.30 | ||
| VL0–7 | |||
| HIV only (n = 11) | 10.00 | 44.00 | 0.287 |
| Co-infected (n = 11) | 13.00 | ||
| VL0–28 | |||
| HIV only (n = 10) | 12.20 | 33.00 | 0.199 |
| Co-infected (n = 10) | 8.80 | ||
Table 4.
Effect of host and HIV-1 viral factors on day 7 and 28 changes in HIV-1 plasma viral load
| Variables | Categories | HIV-1 VL0–7 (N = 30) | HIV-1 VL0–28 (N = 22) | ||||
|---|---|---|---|---|---|---|---|
| Mean rank | U/χ2 (df) | p value | Mean rank | U/χ2 (df) | p value | ||
| HBV status | HIV alone | 10.00 (n = 11) | 44.0 | 0.300 | 12.20 (n = 10) | 33.0 | 0.218 |
| Co-infection | 13.00 (n = 11) | 8.80 (n = 10) | |||||
| Sex | Male | 12.86 (n = 7) | 43.0 | 0.503 | 15.35 (n = 9) | 19.0 | 0.020 |
| Female | 10.87 (n = 15) | 8.29 (n = 11) | |||||
| WHO clinical stage | Stage 1 | 4.25 (n = 2) | 2.794 (2) | 0.247 | 9.00 (n = 1) | 0.068 (2) | 0.966 |
| Stage 2 | 12.09 (n = 17) | 10.56 (n = 16) | |||||
| Stage 3 | 13.00 (n = 3) | 10.67 (n = 3) | |||||
| Antiretroviral drugs | d4T + 3TC + EFV | 11.00 (n = 2) | 0.737 (3) | 0.864 | 10.50 (n = 2) | 3.182 (3) | 0.364 |
| d4T + 3TC + NVP | 12.83 (n = 6) | 9.75 (n = 4) | |||||
| CBV + EFV | 12.25 (n = 8) | 13.13 (n = 8) | |||||
| CBV + NVP | 10.06 (n = 8) | 7.50 (n = 6) | |||||
| Mutations | Present | 5.75 (n = 2) | 8.5 | 0.292 | 9.5 (n = 4) | 26.0 | 0.878 |
| Absent | 10.75 (n = 17) | 10.75 (n = 16) | |||||
| HIV-1 subtype | 02AG | 10.41 (n = 17) | 24 | 0.410 | 9.5 (n = 4) | 37 | 0.898 |
| Non 02AG | 13.50 (n = 4) | 10.75 (n = 16) | |||||
Significant p values are in italics
U, Mann–Whitney U test; χ2 (df), Kruskal–Wallis H test; Co-infection, HIV and HBV co-infection; VL 0–7 HIV-1 plasma viral load changes after 7 days of therapy; VL 0–28 HIV-1 plasma viral load changes after 28 days of therapy
A bivariate logistic regression using an HIV-1 VL decline <0.96 log revealed that CD4+ was marginally significant (p = 0.067). Also, pre-ART HIV-1 VL was marginally significant after a decline of ≥2.58 after 28 days of ART (p = 0.051).
Discussion
Relatively few studies have evaluated the usefulness of early HIV-1 plasma viral decay, HIV-1 VL0–7 or HIV-1 VL0–28, in determining long term outcomes after initiation of ART or in determining the efficacy of ART regimens [18–22, 27–30]. One of such studies showed that HIV-1 VL0–6 was as good as using the decay constant in determining the likelihood for poor and good response to ART [21]. Furthermore, in a fairly large population of ART naïve adults, HIV-1 VL0–7 was significantly associated with HIV-1 VL after 48 weeks of ART but not after 24 weeks [18]. These studies therefore help to provide early warning indicators for long term outcomes of patients on ART. Even though a few patients in this study were estimated to have a reduced adherence this did not affect the short term outcomes of this study as has been seen elsewhere [22]. Thus with the relatively small population of individuals co-infected with HIV and HBV, adherence was not an issue in the analysis of the data. Admittedly, the lack of continuous HIV-1 VL data for all 36 patients may have created gaps in the analysis of data. In spite of this limitation, it is necessary to mention that the whole study population was considered in determining the effects of the different study variables on short-term ART outcomes.
In sub-Saharan Africa, individuals with HIV infections may report to ART clinics late and therefore not start ART when they are due [31]. This may reflect in low CD4+ counts which are indicators for treatment failure [32]. This is consistent with the parameters of the study population in which majority of the patients were classified as being in WHO clinical stage 3, and CD4+ affected outcomes after 7 days of ART. The HIV-1 VL0–7 has been shown to have a strong correlation with the phase 1 constant of HIV-1 decay [21, 23], and this correlation was weaker when calculated 3, 4 or 10 days after onset of ART but may be similar when done at day 6 [21]. The choice of day 7 HIV-1 VL as an indicator and the fact that CD4+ does affect its outcome, therefore suggests that those who are severely immunocompromised are more likely to have poorer outcomes during ART. The blunting of CD4+ in individuals with HIV and co-infected with HBV at different periods of ART [5, 33], may therefore affect HIV-1 VL outcomes at a point in time as shown by others [4]. This may partly have accounted for the observation that HBV co-infection may be associated with more frequent virological failure [13]. Pre-ART HIV-1 VL has been observed as one of the factors affecting long term outcomes in patients on ART [22]. Since this was also observed after 28 days on ART in this study, the use of outcomes after 7 and 28 days as representing long term outcomes to some extent is justified. Thus, in spite of the relatively small study population in this preliminary study, the results to a large extent are consistent with that which is observed in long term studies with specific respect to the effects of CD4+.
The studies cited earlier showing somewhat differences in ART outcomes were conducted in patients from different geographical regions. We postulated that since there is emerging data suggesting that HIV-1 subtype may influence the choice of antiretroviral drugs (ARVs) [24] then there is a possibility that the co-circulation of different subtypes may occur within different geographical areas. The presence of primary HIV-1 resistance which may be different in different subtypes may therefore serve as a confounder in looking at long term outcomes. Even though the GSS did not influence HIV-1 VL0–7 and HIV-1 VL0–28 significantly, the fact that those with HIV-1 subtype which were non CRF 02_AG strains had significantly higher GSS brings to the fore the hypothesis that regional subtype differences needs to be factored into the discussion of the effects of HBV on outcomes of ART. The possibility of primary HIV-1 DRMs influencing some virological outcomes in HBV co-infection in different geographical areas cannot be ruled out. This assertion is buttressed by the fact that primary HIV-1 DRMs are likely to result in treatment failure after 12 months of ART [34].
The hypothesis that HBV protein X may cause different outcomes in ART in those with and without HBV co-infection is not suggested in this study. However, CD4+, pre-ART HIV-1 VL and sex need to be considered as possible confounders. It is unclear how the latter will affect virological outcome.
As compared to other studies in Ghana, the mutations seen confirm the absence of major HIV-1 DRM in treatment naïve patients [35, 36]. Since no transmitted HIV-1 drug resistance mutations were seen, most of the mutations were more likely to be subtype or strain related. In spite of the small study population as compared to others [35, 37, 38], it seems that the V108I mutation may be fairly common in naïve patients.
Conclusion
Pre-ART CD4+, pre-ART HIV-1 pVL, and sex may be important in determining ART outcomes in individuals on ART with HIV and HBV co-infections.
Authors’ contributions
Conceived and designed the experiments: KWCS, FZ, AAA. Analysed the data: KWCS, MA. Wrote the first draft of the manuscript: KWCS. Contributed to the writing of the manuscript: KWCS, KOD. Agree with manuscript results and conclusions: KWCS, KOD, FZ, AAA, TKA, ML, JAAM, MA. Jointly developed the structure and arguments for the paper: KWCS, KOD, JAAM. Made critical revisions and approved final version: KWCS, KOD. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to the nurses and doctors at the Fevers Unit, Korle-Bu Teaching Hospital for making the study possible. The technical support of Anna Aba Hayford and Makafui Seshie of the Clinical Virology Laboratory, School of Biomedical and Allied Health Sciences, University of Ghana, and Magda Dwidar, Sheila Mason and Laura Blair at the Retrovirus Laboratory, Department of Pediatrics, Washington University Medical School, St. Louis, Missouri, are acknowledged. Rev. Dr. Tom Ndanu of the College of Health Sciences, School of Medicine and Dentistry, is acknowledged for statistical support.
Max Arens is currently on retirement.
Funding sources
The study was partly funded by the University of Ghana staff Ph. D. research grant.
Contributor Information
Kwamena William Coleman Sagoe, Phone: (233) 21 687704, Phone: (233) 27 7408528, Email: ksagoe@ug.edu.gh.
Kwabena Obeng Duedu, Email: koduedu@gmail.com.
Francesca Ziga, Email: akorfzig@yahoo.com.
Afrakoma Adjoa Agyei, Email: afrakomaagyei@yahoo.com.
Theophilus Korku Adiku, Email: tkadiku@ug.edu.gh.
Margaret Lartey, Email: mlartey@ug.edu.gh.
Julius Abraham Addo Mingle, Email: jaamingle@gmail.com.
Max Arens, Email: max.arens@gmail.com.
References
- 1.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 2.Diop-Ndiaye H, Touré-Kane C, Etard JF, Lô G, Diaw P, Ngom-Gueye NF, Gueye PM, Ba-Fall K, Ndiaye I, Sow PS, et al. Hepatitis B, C seroprevalence and delta viruses in HIV-1 Senegalese patients at HAART initiation (retrospective study) J Med Virol. 2008;80:1332–1336. doi: 10.1002/jmv.21236. [DOI] [PubMed] [Google Scholar]
- 3.Geretti AM, Patel M, Sarfo FS, Chadwick D, Verheyen J, Fraune M, Garcia A, Phillips RO. Detection of highly prevalent hepatitis B virus coinfection among HIV-seropositive persons in Ghana. J Clin Microbiol. 2010;48:3223–3230. doi: 10.1128/JCM.02231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Idoko J, Meloni S, Muazu M, Nimzing L, Badung B, Hawkins C, Sankalé J-L, Ekong E, Murphy R, Kanki P, Thio CL. Impact of hepatitis B virus infection on human immunodeficiency virus response to antiretroviral therapy in Nigeria. Clin Infect Dis. 2009;49:1268–1273. doi: 10.1086/605675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ladep NG, Agaba PA, Agbaji O, Muazu A, Ugoagwu P, Imade G, Cooke G, McCormack S, Taylor-Robinson SD, Idoko J, Kanki P. Rates and impact of hepatitis on human immunodeficiency virus infection in a large African cohort. World J Gastroenterol. 2013;19:1602–1610. doi: 10.3748/wjg.v19.i10.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouet F, Chaix M-L, Inwoley A, Msellati P, Viho I, Combe P, Leroy V, Dabis F, Rouzioux C, DITRAME-B&A Study Group HBV and HCV prevalence and viraemia in HIV-positive and HIV-negative pregnant women in Abidjan, Côte d’Ivoire: the ANRS 1236 study. J Med Virol. 2004;74:34–40. doi: 10.1002/jmv.20143. [DOI] [PubMed] [Google Scholar]
- 7.Sagoe KWC, Agyei AA, Ziga F, Lartey M, Adiku TK, Seshi M, Arens MQ, Mingle JAA. Prevalence and impact of hepatitis B and C virus co-infections in antiretroviral treatment naïve patients with HIV infection at a major treatment center in Ghana. J Med Virol. 2012;84:6–10. doi: 10.1002/jmv.22262. [DOI] [PubMed] [Google Scholar]
- 8.Konopnicki D, Mocroft A, de Wit S, Antunes F, Ledergerber B, Katlama C, Zilmer K, Vella S, Kirk O, Lundgren JD, EuroSIDA Group Hepatitis B and HIV: prevalence, AIDS progression, response to highly active antiretroviral therapy and increased mortality in the EuroSIDA cohort. AIDS. 2005;19:593–601. doi: 10.1097/01.aids.0000163936.99401.fe. [DOI] [PubMed] [Google Scholar]
- 9.De Luca A, Bugarini R, Lepri AC, Puoti M, Girardi E, Antinori A, Poggio A, Pagano G, Tositti G, Cadeo G, et al. Coinfection with hepatitis viruses and outcome of initial antiretroviral regimens in previously naive HIV-infected subjects. Arch Intern Med. 2002;162:2125–2132. doi: 10.1001/archinte.162.18.2125. [DOI] [PubMed] [Google Scholar]
- 10.Omland LH, Weis N, Skinhøj P, Laursen A, Christensen PB, Nielsen HI, Møller A, Engsig F, Sørensen HT, Obel N. Impact of hepatitis B virus co-infection on response to highly active antiretroviral treatment and outcome in HIV-infected individuals: a nationwide cohort study. HIV Med. 2008;9:300–306. doi: 10.1111/j.1468-1293.2008.00564.x. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann CJ, Charalambous S, Martin DJ, Innes C, Churchyard GJ, Chaisson RE, Grant AD, Fielding KL, Thio CL. Hepatitis B virus infection and response to antiretroviral therapy (ART) in a South African ART program. Clin Infect Dis. 2008;47:1479–1485. doi: 10.1086/593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun HM, Mesner O, Thio CL, Bebu I, Macalino G, Agan BK, Bradley WP, Malia J, Peel SA, Jagodzinski LL, et al. HIV outcomes in Hepatitis B virus coinfected individuals on HAART. J Acquir Immune Defic Syndr. 2014;66:197–205. doi: 10.1097/QAI.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheng W-H, Chen M-Y, Hsieh S-M, Hsiao C-F, Wang J-T, Hung C-C, Chang S-C. Impact of chronic hepatitis B virus (HBV) infection on outcomes of patients infected with HIV in an area where HBV infection is hyperendemic. Clin Infect Dis. 2004;38:1471–1477. doi: 10.1086/420744. [DOI] [PubMed] [Google Scholar]
- 14.Wandeler G, Gsponer T, Bihl F, Bernasconi E, Cavassini M, Kovari H, Schmid P, Battegay M, Calmy A, Egger M, et al. Hepatitis B virus infection is associated with impaired immunological recovery during antiretroviral therapy in the Swiss HIV cohort study. J Infect Dis. 2013;208:1454–1458. doi: 10.1093/infdis/jit351. [DOI] [PubMed] [Google Scholar]
- 15.Law WP, Duncombe CJ, Mahanontharit A, Boyd MA, Ruxrungtham K, Lange JMA, Phanuphak P, Cooper DA, Dore GJ. Impact of viral hepatitis co-infection on response to antiretroviral therapy and HIV disease progression in the HIV-NAT cohort. AIDS. 2004;18:1169–1177. doi: 10.1097/00002030-200405210-00010. [DOI] [PubMed] [Google Scholar]
- 16.Twu JS, Chu K, Robinson WS. Hepatitis B virus X gene activates kappa B-like enhancer sequences in the long terminal repeat of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1989;86:5168–5172. doi: 10.1073/pnas.86.13.5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Twu JS, Rosen CA, Haseltine WA, Robinson WS. Identification of a region within the human immunodeficiency virus type 1 long terminal repeat that is essential for transactivation by the hepatitis B virus gene X. J Virol. 1989;63:2857–2860. doi: 10.1128/jvi.63.6.2857-2860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haubrich RH, Riddler SA, Ribaudo H, Direnzo G, Klingman KL, Garren KW, Butcher DL, Rooney JF, Havlir DV, Mellors JW, et al. Initial viral decay to assess the relative antiretroviral potency of protease inhibitor-sparing, nonnucleoside reverse transcriptase inhibitor-sparing, and nucleoside reverse transcriptase inhibitor-sparing regimens for first-line therapy of HIV infection. AIDS (London, England) 2011;25:2269–2278. doi: 10.1097/QAD.0b013e32834d0c20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, De Gruttola V, Fischl M, Hammer S, Richman D, Havlir D, Gulick R, Squires K, Mellors J. Patterns of plasma human immunodeficiency virus type 1 RNA response to antiretroviral therapy. J Infect Dis. 2001;183:1455–1465. doi: 10.1086/320192. [DOI] [PubMed] [Google Scholar]
- 20.Lepri AC, Miller V, Phillips AN, Rabenau H, Sabin CA, Staszewski S. The virological response to highly active antiretroviral therapy over the first 24 weeks of therapy according to the pre-therapy viral load and the weeks 4–8 viral load. AIDS. 2001;15:47–54. doi: 10.1097/00002030-200101050-00008. [DOI] [PubMed] [Google Scholar]
- 21.Polis MA, Sidorov IA, Yoder C, Jankelevich S, Metcalf J, Mueller BU, Dimitrov MA, Pizzo P, Yarchoan R, Dimitrov DS. Correlation between reduction in plasma HIV-1 RNA concentration 1 week after start of antiretroviral treatment and longer-term efficacy. Lancet. 2001;358:1760–1765. doi: 10.1016/S0140-6736(01)06802-7. [DOI] [PubMed] [Google Scholar]
- 22.Powderly WG, Saag MS, Chapman S, Yu G, Quart B, Clendeninn NJ. Predictors of optimal virological response to potent antiretroviral therapy. AIDS. 1999;13:1873–1880. doi: 10.1097/00002030-199910010-00009. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Lathey J, Ruan P, Douglas SD, Spector SA, Lindsey J, Hughes MD, Rudy BJ, Flynn PM, Team P. Relationship of plasma HIV-1 RNA dynamics to baseline factors and virological responses to highly active antiretroviral therapy in adolescents (aged 12–22 years) infected through high-risk behavior. J Infect Dis. 2004;189:593–601. doi: 10.1086/381500. [DOI] [PubMed] [Google Scholar]
- 24.Lessells RJ, Katzenstein DK, de Oliveira T. Are subtype differences important in HIV drug resistance? Curr Opin Virol. 2012;2:636–643. doi: 10.1016/j.coviro.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steegen K, Demecheleer E, De Cabooter N, Nges D, Temmerman M, Ndumbe P, Mandaliya K, Plum J, Verhofstede C. A sensitive in-house RT-PCR genotyping system for combined detection of plasma HIV-1 and assessment of drug resistance. J Virol Methods. 2006;133:137–145. doi: 10.1016/j.jviromet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Sagoe KW, Dwidar M, Adiku TK, Arens MQ. HIV-1 CRF 02 AG polymerase genes in Southern Ghana are mosaics of different 02 AG strains and the protease gene cannot infer subtypes. Virol J. 2009;6:27. doi: 10.1186/1743-422X-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Leth F, Andrews S, Grinsztejn B, Wilkins E, Lazanas MK, Lange JMA, Montaner J, group Ns The effect of baseline CD4 cell count and HIV-1 viral load on the efficacy and safety of nevirapine or efavirenz-based first-line HAART. AIDS. 2005;19:463–471. doi: 10.1097/01.aids.0000162334.12815.5b. [DOI] [PubMed] [Google Scholar]
- 28.Louie M, Hogan C, Di Mascio M, Hurley A, Simon V, Rooney J, Ruiz N, Brun S, Sun E, Perelson AS, et al. Determining the relative efficacy of highly active antiretroviral therapy. J Infect Dis. 2003;187:896–900. doi: 10.1086/368164. [DOI] [PubMed] [Google Scholar]
- 29.Torti C, Quiros-Roldan ME, Cologni G, Nichelatti M, Ceresoli F, Pinti M, Nasi M, Cossarizza A, Lapadula G, Costarelli S, et al. Plasma HIV load and proviral DNA decreases after two standard antiretroviral regimens in HIV-positive patients naïve to antiretrovirals. Curr HIV Res. 2008;6:43–48. doi: 10.2174/157016208783571982. [DOI] [PubMed] [Google Scholar]
- 30.Wu H, Ding AA, De Gruttola V. Why are the decay rates in plasma HIV-1 different for different treatments and in different patient populations? AIDS. 1999;13:429–430. doi: 10.1097/00002030-199902250-00022. [DOI] [PubMed] [Google Scholar]
- 31.Kigozi IM, Dobkin LM, Martin JN, Geng EH, Muyindike W, Emenyonu NI, Bangsberg DR, Hahn JA. Late-disease stage at presentation to an HIV clinic in the era of free antiretroviral therapy in Sub-Saharan Africa. J Acquir Immune Defic Syndr. 1999;2009(52):280–289. doi: 10.1097/QAI.0b013e3181ab6eab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilks CF, Walker AS, Munderi P, Kityo C, Reid A, Katabira E, Goodall RL, Grosskurth H, Mugyenyi P, Hakim J, et al. A single CD4 test with 250 cells/mm3 threshold predicts viral suppression in HIV-infected adults failing first-line therapy by clinical criteria. PLoS One. 2013;8:e57580. doi: 10.1371/journal.pone.0057580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chadwick D, Ankcorn M, Sarfo F, Phillips R, Fox Z, Garcia A, Appiah L, Bedu-Addo G, Geretti AM. Outcomes of starting first-line antiretroviral therapy in hepatitis B virus/HIV-coinfected patients in Ghana. J Antimicrob Chemotherapy. 2012;67:2939–2942. doi: 10.1093/jac/dks333. [DOI] [PubMed] [Google Scholar]
- 34.Rusine J, Asiimwe-Kateera B, van de Wijgert J, Boer KR, Mukantwali E, Karita E, Gasengayire A, Jurriaans S, de Jong M, Ondoa P. Low primary and secondary HIV drug-resistance after 12 months of antiretroviral therapy in human immune-deficiency virus type 1 (HIV-1)-infected individuals from Kigali, Rwanda. PloS one. 2013;8:e64345. doi: 10.1371/journal.pone.0064345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nii-Trebi NI, Ibe S, Barnor JS, Ishikawa K, Brandful JAM, Ofori SB, Yamaoka S, Ampofo WK, Sugiura W. HIV-1 drug-resistance surveillance among treatment-experienced and -naïve patients after the implementation of antiretroviral therapy in Ghana. PLoS One. 2013;8:e71972. doi: 10.1371/journal.pone.0071972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sagoe KWC, Dwidar M, Lartey M, Boamah I, Agyei AA, Hayford AA, Mingle JAA, Arens MQ. Variability of the human immunodeficiency virus type 1 polymerase gene from treatment naïve patients in Accra, Ghana. J Clin Virol. 2007;40:163–167. doi: 10.1016/j.jcv.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 37.Delgado E, Ampofo WK, Sierra M, Torpey K, Pérez-Alvarez L, Bonney EY, Mukadi YD, Lartey M, Nyarko C, Amenyah RN, et al. High prevalence of unique recombinant forms of HIV-1 in Ghana: molecular epidemiology from an antiretroviral resistance study. J Acquir Immune Defic Syndr. 1999;2008(48):599–606. doi: 10.1097/QAI.0b013e3181806c0e. [DOI] [PubMed] [Google Scholar]
- 38.Fischetti L, Opare-Sem O, Candotti D, Sarkodie F, Lee H, Allain JP. Molecular epidemiology of HIV in Ghana: dominance of CRF02_AG. J Med Virol. 2004;73:158–166. doi: 10.1002/jmv.20070. [DOI] [PubMed] [Google Scholar]


