Abstract
Metal ion-catalyzed oxidation of hydrazine and its derivatives leads to the formation of the hydrazyl radical and subsequently to oxy-radicals in the presence of molecular oxygen. Here we have examined the role of Cu2+-catalyzed oxidation of hydralazine in the induction of DNA damage. Neither 5, 5-dimethyl-1-pyrroline-N-oxide (DMPO) nor dimethyl sulfoxide (DMSO) were effective in inhibiting hydralazine-Cu2+-induced DNA damage. Singlet oxygen did not appear to participate in this DNA cleavage. The one-electron oxidation of hydralazine also leads to the formation of DNA radicals as confirmed by immuno-spin trapping with 5, 5-dimethyl-1-pyrroline-N-oxide. Electron spin resonance (ESR) and spin trapping studies further confirmed the formation of DNA radicals; predominantly 2′-deoxyadenosine radical adducts were detected, while some radicals were also detected with other nucleosides. Our results suggest that free hydroxyl radicals may not be the main damaging species causing DNA cleavage, and possibly, Cu-peroxide complexes, formed from Cu+-H2O2, areresponsible for this hydralazine-Cu2+-induced DNA cleavage.
Keywords: Hydralazine, Free Radicals, Oxy-radicals, ESR, Spin Trapping, DNA Damage, DNA Radicals, Immuno-spin Trapping
INTRODUCTION
Hydrazine and its derivatives constitute an important class of compounds as they are environmental pollutants, and are used in medicine. Aromatic hydrazines (e.g., agaritine) are found in edible mushrooms (Agaricus bisporus).Isoniazid is used for the treatment of tuberculosis.1 Iproniazid is a potent antidepressant; however, it is not currently in use due to its liver toxicity.2 Procarbazine is a chemotherapeutic agent used in the treatment of Hodgkin’s disease and melanoma.3 Hydralazine is a potent arterial vasodilator and plays an important role in the management of hypertension and congestive heart failure.4 Hydrazines in general are toxic, and induce a variety of toxic insults. These include liver toxicity, carcinogenicity and mutagenicity.1, 2, 4–7 Hydralazine, the least toxic of the hydrazine derivatives, induces DNA damage,8 causes severe forms of systemic lupus erythematosus,9–11 and has been shown to increase the incidence of lung tumors in mice.4–6 More recently, hydralazine has been shown to inhibit DNA Methyltransferase 1 by preventing transfer of a methyl group to DNA in several cancer silencing genes/tumor suppressor genes.9,12 It has, therefore, become an important treatment combination in cancer therapy.9,12 Furthermore, hydralazine has also been found to inhibit iron-containing prolyl hydroxylase enzymes, which are important for the induction of hypoxia induced factor (HIF) and vascular endothelial growth factor. HIF1α is a critical target in cancer chemotherapy as it is believed to be involved in tumor progression.10
Because of the significance of hydrazine derivatives as environmental pollutants and food contaminants and their utility in medicine, a large volume of research has been carried out to elucidate the mechanisms of toxicity of these compounds.1–10 Previous studies from our laboratory and others have clearly shown that hydrazine derivatives generate free radicals upon oxidation, catalyzed by metal ions, cytochrome P-450 and peroxidases, including prostaglandin synthase.13–22 In addition, certain substituted hydrazines have been proposed to alkylate DNA by nucleophilic attack rather than via free radical generation.21, 22 Irrespective of mechanisms (free radicals or nucleophilic attack),the consequences of such DNA alkylation may lead to carcinogenic events.
We have previously shown that hydralazine undergoes one-electron oxidation by both metal ions (Cu2+ and Fe3+ ions) and enzymatically (horseradish peroxidase and prostaglandin synthase) to form the hydralazyl radical (HY.),which then further decomposes to form various products.14 Reilly and Aust23 have reported that hydralazine in the presence of peroxidase/H2O2 induces single-strand DNA damage; however, the presence of a donor such as chlorpromazine is required for damage to occur. Studies of Yamamoto and Kawanishi24 have shown that the oxidation products of hydralazine and Cu2+ induce DNA damage, predominantly at guanine residues, while Fe-catalyzed oxidation of hydralazine induces damage at every nucleotide in DNA.
Copper is an essential trace element that is widely distributed throughout the body and forms the essential redox–active reaction center in a variety of metalloenzymes, including ceruloplasmin, superoxide dismutase, cytochrome oxidase, dopamine β-hydroxylase, tyrosinase and lysyl oxidase. Nevertheless, the body’s total daily requirement for copper estimated at 1–2 mg25 for adult humans is much below that for iron, which is estimated at 4–6 gm. Serum levels of copper are significantly elevated in anemia, during pregnancy and during oral contraceptives.26 It is interesting to note that copper metabolism is significantly altered in tumors and that serum concentrations are correlated with tumor incidence, progression and recurrence in a number of human tumors.27–29 Serum ceruloplasmin levels increase by a factor of 4–8 during cancer progression.30 Elevated levels of copper plays an important role in tumor angiogenesis and seems to be necessary for endothelial cell activation. Cu2+-dependent DNA fragmentation has been reported to be much more extensive than Fe3+ than iron,31 it is important and relevant to examine the toxicity of hydralazine in the presence of copper as this combination may be more potent for the DNA damage and consequently, for the formation of tumors than the single agents alone.
In this report, we have examined the consequences of Cu2+-catalyzed oxidation of hydralazine by assessing the induction of DNA cleavage in supercoiled pHOT1 DNA and the formation of DNA radicals in calf thymus DNA. Furthermore, we have investigated the Cu2+ ion: hydralazine stoichiometry that induces maximum DNA damage. Our results indicate that reactive species formed from the oxidation of hydralazine by Cu2+ induce significant DNA cleavage in pHOT1 DNA, and this nicking is dependent upon the ratio of Cu2+ ions to hydralazine. Using ESR spin-trapping techniques, we found that hydralazine-Cu2+ complexes formed significant amounts of 2′-deoxyadenosine radical spin-adducts while some radical adducts of other bases were also detected. We also found that in calf thymus DNA, formation of oxy-radicals leads to the generation of DNA radicals, which were trapped by DMPO and were easily detected by the immuno-spin trapping technique (IST). Again, the maximum DNA radical formation was dependent upon the ratio of Cu2+ ions to hydralazine, and this ratio was similar to those found for DNA cleavage.
Methods and Materials
Hydralazine, calf thymus DNA, 2,2,6,6-tetramethyl-piperidine (TEMP) and Rose Bengal were obtained from Sigma Aldrich (St. Louis, MO). DMPO was purchased from Dojindo (Rockville, MD) and used without any further purification. DNA was dissolved in 100 mM Chelex-treated PBS buffer (pH 7.4), and all incubations were carried out in this buffer. Supercoiled pHOT1 DNA was obtained from Topgen (Port Orange, FL). Copper (II) sulfate (Sigma Aldrich, St. Louis, MO) was dissolved in doubly distilled water prior to use. A fresh solution of hydralazine was prepared before use. 2′-Deoxyadenosine, 2′-deoxycytidine, 2′-deoxyguanosine and thymidine were obtained from MP Biomedicals (Irvine, CA).
DNA Cleavage Assay with pHOT1
DNA cleavage in the presence of hydralazine and Cu2+ ion was analyzed using pHOT1 supercoiled DNA according to previously published methods.32 Briefly, a 20 μl assay mixture contained 0.5μg of pHOT1 supercoiled DNA and varying concentrations of hydralazine. The reaction was initiated by adding Cu2+ and incubating the reaction mixture for 30 min at room temperature. The reaction was terminated by adding 2 μl of 0.5 M EDTA. The reaction products were loaded onto 1% agarose gel for the DNA separation in Tris-acetate-EDTA buffer (pH 8.0). The DNA was stained with ethidium bromide and photographed. For the effects of scavengers on DNA damage, scavengers were preincubated for 5–10 min before adding Cu2+ ions and carrying out gel electrophoresis as before. Quantitation of various forms of DNA cleavage products was carried using an Image J soft-ware.
Oxygen Consumption with Hydralazine
Oxygen uptake assays were carried out with hydralazine and Cu2+ in the presence or absence of DNA with an YSI Model 52 Oxygen Monitor. Typically, hydralazine, dissolved in 100 mM Chelex-treated PBS buffer (pH 7.4), was equilibrated for 10 min at room temperature before adding Cu2+ and monitoring O2 consumption. When the effects of DNA on oxygen consumption were examined, DNA was first incubated with Cu2+ (200 μg/ml calf thymus DNA and 50 μM Cu2+), the reaction was initiated by adding hydralazine, and O2 consumption was measured. In some experiments, DNA-hydralazine complexes were preformed15 before the addition of Cu2+, and no significant differences in O2 consumption were observed. The effects of scavengers were examined by preincubating DNA-metal ion mixtures with the scavenger for 10 min before adding hydralazine.
ESR Studies
ESR studies for the detection of singlet oxygen33 and DNA radicals were carried out as described previously.34,35 For the detection of singlet oxygen, TEMP (100 mM) was used in the presence of hydralazine (125μM) and Cu2+ (25μM) in Chelex-treated PBS buffer (pH 7.4). Rose Bengal (20μM) was used as the positive control in the presence of light (5 min exposure).36 For the spin trapping studies, reaction mixtures contained in Chelex-treated PBS buffer (pH 7.4) individual 2′-deoxyribonucleosides (5.0 mM), hydralazine (125 μM), and DMPO (100 mM), and the reaction was initiated by adding 25 μM Cu2+. The mixture was transferred to ESR flat cells and the spectra were recorded with an ELEXSYS E 500 ESR spectrometer (Brucker Biospin, Billerica, MA) equipped with an ER4122SHQ cavity operating at 9.76 GHz at room temperature. The ESR settings were as follows: scan range 100 G; modulation frequency 100 KHz; modulation amplitude 1.0 G; microwave power 20 mW; receiver gain 2 × 104; time constant 250 ms and the conversion time 250 ms. ESR spectra were simulated as described previously.37
Formation of DNA-Nitrone Adduct with Hydralazine and Cu2+
Calf thymus DNA (0.5 mg-1.0 mg/ml) was mixed with various concentrations of hydralazine and the reaction was initiated by adding Cu2+ followed by the addition of DMPO; the reaction mixtures were then incubated for 30 min at room temperature. Reactions were terminated by adding EDTA solution (0.5 M), and DNA was precipitated in ethanol. DNA was collected by centrifugation and washed with 75% ethanol, dissolved in the Chelex-treated PBS, and analyzed for DNA-nitrone adducts.
Immuno-Spin Trapping Analysis of DNA-Nitrone Adducts
For the detection of DNA-nitrone adducts formed in calf thymus DNA by hydralazine, a standard ELISA procedure in a 96-well plate (Greiner-Bio-One) was used as described previously.38,39 Briefly, DNA samples were diluted to 5 μg/ml in the Chelex-treated PBS buffer and incubated with Reacti-bind DNA coating solution (Thermo Scientific, Rockford, IL) at 37°C for 3 hrs. The plates were washed once with a washing buffer (the Chelax-treated PBS buffer containing 0.05% nonfat dry milk and 0.1% Tween-20) and blocked with a blocking buffer (the Chelax-treated PBS buffer containing 3% nonfat dry milk) for 18 hrs. at 4°C. The plates were then washed once with the washing buffer and incubated with rabbit anti-DMPO serum (diluted 1:10,000) at 37°C for 1 hr. The plates were washed three times with the washing buffer, 100 μl of the secondary antibody (horseradish peroxidase-conjugated, diluted 1:5000 in the washing buffer) was added, and the plates were incubated at 37°C for 1 hr. Following three washes as before, the antigen-antibody complexes were developed with Immobilon Chemiluminescence substrate (Millipore Corp, Billerica, MA), and the light emitted was recorded as arbitrary light units using X-Fluor software (Tecan US, Research Triangle Park, NC).
RESULTS
We used supercoiled pHOT1 DNA (which contained 80–85% supercoiled DNA, according to the manufacturer and confirmed in our experiments) to examine the induction of DNA cleavage by hydralazine (25 μM–1000 μM) in the presence of 25 μM Cu2+ ions. Copper ion alone induced some DNA cleavage (Figure 1A, lane 1), which was significantly enhanced in the presence of hydralazine (Figure 1A). While higher concentrations of hydralazine also induced some DNA cleavage (e.g., 35 ± 5% at 1000 μM), complete DNA cleavage was observed at 125–250 μM hydralazine. At these concentrations of hydralazine, formation of both open circular and linear DNA was observed. Furthermore, as little as 10 μM Cu2+ was sufficient to induce this damage (data not shown).
Figure 1.
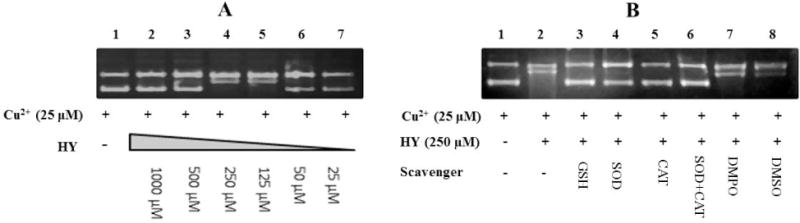
Hydralazine-mediated cleavage (A) in pHOT1 DNA following oxidation with Cu2+ (25 μM). The DNA cleavage was examined as described in the Methods section. Lane 1: DNA in the presence of Cu2+ alone; Lane 2: Hydralazine (1000 μM) and Cu2+; Lane 3: Hydralazine (500 μM) and Cu2+; Lane 4: Hydralazine (250 μM) and Cu2+; Lane 5: Hydralazine (125 μM) and Cu2+; Lane 6: Hydralazine (50 μM) and Cu2+; and Lane 7: Hydralazine (25 μM) and Cu2+. (B) Effects of scavengers on the DNA cleavage induced by hydralazine (250 μM) and Cu2+ (25 μM). Scavengers were incubated with hydralazine-DNA complexes for 5–10 min before adding Cu2+. Lane 1: control DNA; Lane 2: hydralazine in the presence of Cu2+; Lane 3: reduced GSH (1000 μM); lane 4: SOD (50 U/incubation); Lane 5: catalase (2500 U/incubation); Lane 6: SOD and catalase; Lane 7: DMPO (100 mM) and Lane 8: DMSO (1 mM).
We next examined the effects of various scavengers of oxy-radicals on the hydralazine-Cu2+-complex-induced DNA cleavage. As shown in Figure 1B, 1 mM GSH (an OH radical scavenger, lane 3), SOD (a scavenger of superoxide anion radical, 50 U/incubation; lane 4), catalase (a scavenger of hydrogen peroxide, 2500 U/ incubation; lane 5) or a combination of SOD and catalase (lane 6) significantly (75 ± 10%) inhibited DNA cleavage. These observations indicate the participation of both superoxide anion radical (O2.−) and hydrogen peroxide (H2O2) in the DNA damage induced by the Cu2+-hydralazine complex (lane 2). It is interesting that DMPO (lane 7), and DMSO (lane 8) both scavengers of free .OH, were not effective in inhibiting this DNA cleavage. This would suggest that the damaging radical (free .OH) if formed is in close proximity to the DNA binding site (s) of hydralazine or the hydralazine-Cu-DNA complex that DMSO/DMPO is unable to inhibit it. Alternatively, it is also possible that the species that induce DNA cleavage are not free .OH and are derived from copper-peroxide complexes.
Scavengers of free .OH were not very effective in preventing DNA cleavage from hydralazine in the presence of Cu2+.Since singlet O2 has been implicated in Cu-H2O2-mediated DNA damage,33 experiments were carried out to determine whether1O2 was formed from hydralazine-Cu2+ under our experimental conditions. In order to confirm this, we used TEMP as a probe. TEMP has been successfully used for the detection of singlet O233,36 from the Cu2+-H2O2 system. As a positive control we used Rose Bengal, a dye that has been shown to generate1O2 in the presence of light.36 Figure 2 shows that Rose Bengal generated large amounts of the TEMP nitroxide (note the difference in receiver gain), the oxidation product of TEMP by 1O2. In contrast, the hydrlazine-Cu2+ system generated very little1O2 under the conditions where significant DNA cleavage was observed. These observations would suggest that 1O2 did not participate significantly in inducing DNA cleavage from the hydralazine-Cu2+ system under our experimental conditions.
Figure 2.
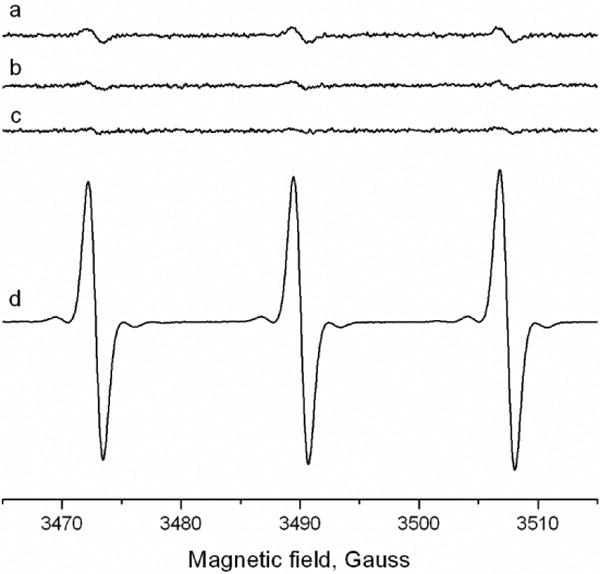
Detection of 1O2 from hydralazine (125 uM) in the presence of Cu2+ (25uM) and TEMP (100 mM). a: complete System; b: Hydralazine alone; c: Cu2+ alone and d: in the presence of Rose Bengal (20 uM) and light for 5 min. No signal for the nitroxide was detected in the absence of light with Rose Bengal. The ESR studies were carried out as described in the Methods section; a–c: conversion time = time constant = 163 ms. d: 81 ms.
Oxygen Consumption of Hydralazine in the Presence of Cu2+
The ratio dependence for the maximum DNA cleavage would suggest that at these ratios the Cu2+ ion may undergo recycling for the maximum formation of oxy-radicals. In order to confirm this, we examined O2 consumption at various metal ion-to-hydralazine ratios. The maximum O2 consumption was obtained at a ratio of 1: 2.5–5 of Cu2+: hydralazine (Figure 3A, Tracing 1 and 2), while at higher ratios, significantly decreased O2 was consumed (Figure 3A, Tracings 3–5). These observations are similar to our cleavage data where complete DNA degradation was observed (e.g., 125–250 μM hydralazine). This consumption of O2 was inhibited by the presence of SOD (250U/ml), while catalase (6,000 U/ml) was only partially effective (Figure 3B). GSH (1000μM) was also very effective in inhibiting O2 consumption, while DMPO had no effect (Figure 3B). These observations implicate O2−. as the intermediate in the formation of H2O2 and other reactive intermediates from hydralazine and Cu2+.
Figure 3.
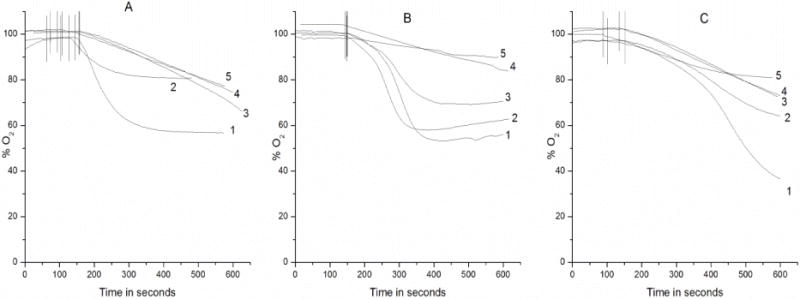
Oxygen consumption (A) with different concentrations of hydralazine in the presence of Cu2+ (50 μM). The oxygen consumption was carried out as described in the Methods section. Tracing 1: 125 μM hydralazine; Tracing 2: 50 μM hydralazine; Tracing 3: 250 μM hydralazine; Tracing 4: 500 μM hydralazine; and Tracing 5: 1000 μM hydralazine. (B) Effects of oxy-radical scavengers on oxygen consumption from 125 μM hydralazine and 50 μM Cu2+. Tracing 1: control; Tracing 2: 100 mM DMPO; Tracing 3: catalase (6000 U/ml); Tracing 4: SOD (500 U/ml); and Tracing 5: 1000 μM GSH. (C) Effect of DNA (200 μg/ml) on the oxygen consumption by various concentrations of hydralazine in the presence of Cu2+ (50 μM). Tracing 1: 250 μM hydralazine; Tracing 2: 125 μM hydralazine; Tracing 3: 500 μM hydralazine; Tracing 4: 1000 μM hydralazine and Tracing 5: 50 μM hydralazine. DNA was added to the hydralazine solutions and incubated for 10 min before adding Cu2+ and measuring the O2 consumption.
The effect of DNA on O2 consumption was also examined, and the data is presented in Figure 2C. Addition of DNA increased O2 consumption from hydralazine and Cu2+, and the maximum O2 was consumed at a Cu2+:hydralazine ratio of 1:5. Again, these observations are consistent with our cleavage reactions suggesting that Cu2+ is accessible for cycling to generate the maximum reactive intermediates.
ESR Spin Trapping Studies
In order to detect free radicals formed in DNA following oxidation of hydralazine, spin trapping studies with DMPO were carried out with nucleosides and calf thymus DNA (native and sonicated DNA). Typically, the reaction mixtures contained 5 mM individual nucleosides, dissolved in the Chelex-treated PBS buffer (pH 7.4), 125μM hydralazine and 100 mM DMPO. The reactions were initiated by adding 25 μM Cu2+ and recording the spectra.
Incubation of hydralazine (0.5 mM or more) with Cu2+ (25 μM) rapidly formed DMPO- adducts as previously reported;14 however, when the hydralazine concentration was decreased to 125 μM (ratio of 5:1 with Cu2+), the DMPO-adduct (94%) formed had the following coupling constants aN = 14.14 G; aHβ = 11.3 G and aHγ = 1.24 G (Figure 3A). These coupling constants are similar to those described for DMPO adducts of superoxide anion radicals.40 When SOD (500 U/ml) was included in the reaction mixtures, very little DMPO-adduct was detected, indicating that the DMPO-adducts trapped were derived from the trapping of superoxide anion radicals (Figure 3B). Formation of DMPO adducts required the presence of both hydralazine and Cu2.
Additionally, when the incubations were carried out for more than 20 min, a small amount (<10%) of another DMPO-adduct with coupling constants of aN = 15.4 G and aH = 22.86 G was also trapped. The identity of this radical is not known but it appears to be derived from trapping of a carbon-centered radical with DMPO. Finkelstein et al.40 have described the appearance over time of a DMPO-adduct with aN = 15.31 and aH = 22.0 during trapping of superoxide anion radical and have indicated that it was formed from the decomposition of DMPO. These coupling constants are similar to those of the DMPO-adduct trapped in our system and thus may represent this decomposition product of DMPO.
Inclusion of 2′-deoxyadenosine in the reaction mixtures containing hydralazine and Cu2+ also resulted in the formation of several different DMPO-adducts, including the 2′-deoxyadenosine radical adduct (Figure 4Ac). This radical adduct (40%) had the following coupling constants: aN1 = 15.42 G; aN2 =2.58G and aH = 19.95 G. These coupling constants are similar to those described for the N-centered exocyclic NH2 of the 2′-deoxyadenosine moiety.34,41 DMPO-OH (53%), DMPO-decomposition products and some nitroxide radical were also present. Simulated spectra of these radical adducts with their coupling constants are shown in Figure 4.
Figure 4.
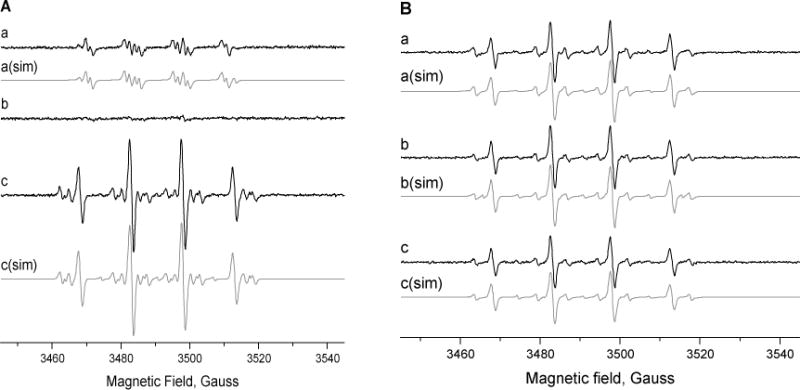
Detection of free radicals in the presence of DMPO (100 mM). Panel A, (a) hydralazine (125 μM) and Cu2+ (25 μM); (b) + SOD (500 U/ml); and (c) + 2′-deoxyadenosine (5 mM). Panel B, Radical adducts obtained from (a) 2′-deoxyguanosine (5 mM), (b) 2′-deoxycytidine (5mM) and (c) thymidine (5 mM) from hydralazine and Cu2+. These studies were carried out at pH 7.4 as described in the Methods section.
When incubations were carried out with other nucleosides, various DMPO adducts were detected, these coupling constants are in agreement with those previously published,34,41 and are summarized in Table-1. It should be noted that these radical adducts were minor products (less than 10% of the total adducts formed) as DMPO-OH was the major adduct detected.
Table 1.
Parameters of DMPO-adducts formed from nucleosides following treatment with hydralazine (125 μM) and Cu2+(25 μM) at room temperature.
| Substrate | Hyperfine Coupling Constants, G | Assignment | ||
|---|---|---|---|---|
| aN | aH | aN | ||
| 2′-deoxyadenosine | 15.42 | 19.95 | 2.58 | N-centered, exocyclic NH2 |
|
| ||||
| 2′-deoxycytidine | 15.38 | 18.95 | 2.02 | N-centered |
|
| ||||
| Thymidine | 15.88 | 19.5 | 2.71 | N-centered |
|
| ||||
| 2′-deoxyguanosine | 15.6 | 20.27 | 2.56 | N-centered, N3 position |
ESR-Spin trapping studies were carried out as described in the Methods Section.
It is interesting to note that when the spin-trapping experiments were carried out under conditions such as those described by Yamamoto and Kawanishi,24 no radicals originating from nucleosides were trapped with DMPO. More importantly, when incubations were carried out at 37°C, only exocyclic NH2 from 2′-deoxyadenosine was detected with DMPO, suggesting that this radical (or the DMPO-adduct) was significantly more stable than those formed from other nucleosides. When calf thymus DNA or sonicated calf thymus DNA was used, no radical was detected with DMPO.
Formation of DNA-Nitrone Adducts
Because oxy-radicals are formed from hydralazine-Cu2+-DNA complexes, it is possible that this oxy-radical formation would also lead to the formation of DNA radical (s) in-spite of our negative ESR spin-trapping results. This possibility was investigated by examining the formation of stable DNA-nitrone adducts by a highly sensitive immuno-spin trapping method.38,39 Data presented in Figure 5 clearly show the formation of stable nitrone adducts in calf thymus DNA. Interestingly, at high hydralazine concentrations, very few DNA-nitrone adducts were formed, and the highest amounts of adducts were detected at a Cu2+:hydralazine ratio of 1:5, a ratio at which the maximum O2 consumption was also observed in the presence of DNA (Figure 3C, Tracing 1). The time course indicated that this DNA-nitrone adduct formation was rapid and reached a maximum within 5–10 minutes (data not shown). Furthermore, when we examined the dependence of this adduct formation on DMPO concentrations, the highest amounts of adducts were formed at 50–100 mM, and no significant adduct formation was observed at 1–10 mM of DMPO.
Figure 5.
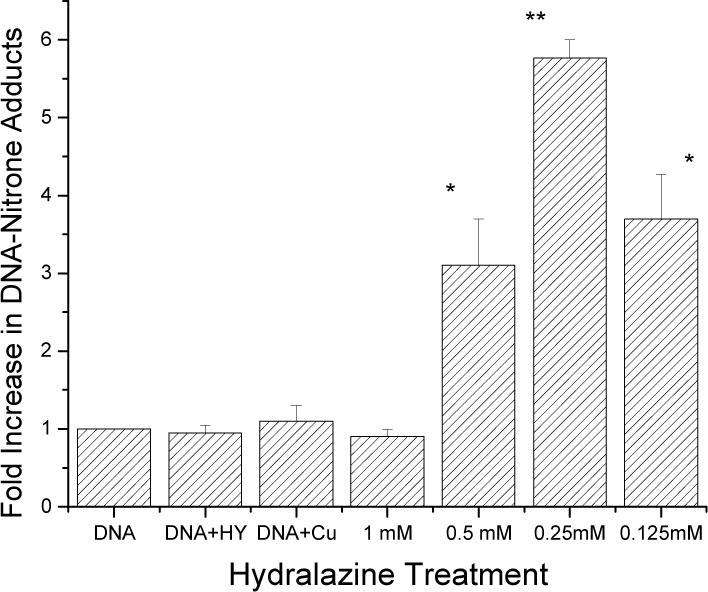
Immunological detection of the DNA radicals formed in calf thymus DNA (0.5 mg/ml) by various concentrations of hydralazine in the presence of Cu2+ (50 μM) and DMPO (100 mM). The detection of these DNA radicals was carried out as detailed in the Methods section. Starred values were significantly different (*<0.05; ** <.005, n = 3) from DNA alone or DNA in the presence of hydralazine and DNA in the presence of Cu2+. The significance was determined by Student’s t-test.
DISCUSSION
Hydrazine and its derivatives react with metal ions (Cu2+ and Fe3+) to generate hydrazyl radical, which then reacts with molecular O2 to generate superoxide anion radicals, H2O2 and, finally,.OH.13–15 Our previous studies13–15 as well as this study shows that hydralazyl radical, superoxide anion radicals, and .OH are formed from hydralazine under aerobic conditions at pH 7.4 during one-electron oxidation of hydralazine. In this study, hydralazine in the presence of Cu2+ ions induced significant DNA cleavage in the supercoiled pHOT1 DNA. Furthermore, we found that the DNA damage was O2.− and H2O2 dependent as both SOD and catalase were effective in preventing DNA damage from hydralazine and Cu2+ ions. While it has been suggested that hydroxyl radicals, once formed, will react with any species present in high concentrations in a diffusion-control manner, two typical hydroxyl radical scavengers (DMSO and DMPO) exhibited no significant inhibition of the DNA damage induced by hydralazine-Cu2+ complexes in our hands. There are a number of reports in which the addition of .OH scavengers (ethanol, mannitol, sodium formate and DMSO) conferred only partial protection or had no effect on DNA damage.33, 42–47 Because of the small or insignificant protection against DNA damage by hydroxyl radical scavengers, it has been proposed that the active species causing DNA damage are more likely copper-peroxide complexes (Scheme 1) with reactivity similar to hydroxyl radical and/or singlet oxygen rather than a free hydroxyl radical.44,47 In this study, we found no inhibition of Cu2+-hydralazine-induced DNA damage by DMSO and DMPO as observed by others.44,47 These observations lead us to suggest that free hydroxyl radicals are not the main DNA damaging species, and participation of Cu+-peroxo species (Scheme 1) in inducing DNA cleavage cannot be ruled out this time.
Scheme 1.
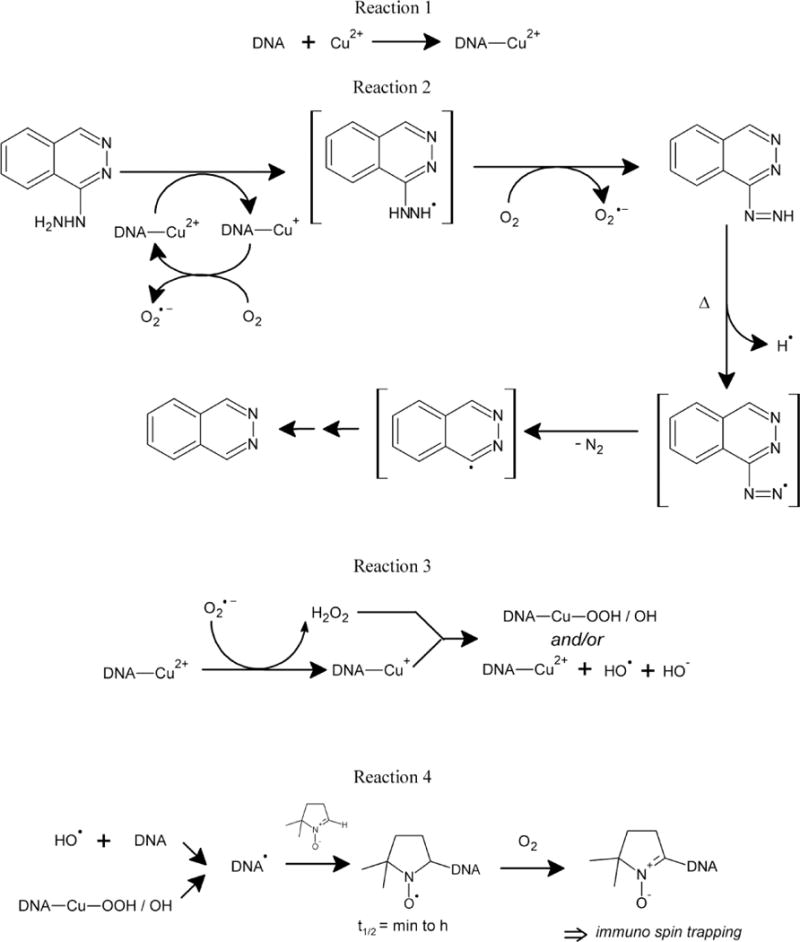
Structure of hydralazine, and DNA nitrone, and proposed formation of various free radical species from hydralazine that may induce DNA cleavage following oxidation by Cu2+ ion.
Reduced glutathione was extremely effective in preventing DNA cleavage. This would suggest that GSH rapidly traps any Cu+ formed, resulting in the formation of a stable Cu+–GSH complex and preventing the formation of reactive species. It is of interest to note that studies reported from our laboratory have shown that Cu+–GSH complexes are stable and do not undergo oxidation to generate Cu2+. Using the spin-trapping technique, we have shown that these Cu+–GSH complexes do not form DMPO-OH spin adducts in the presence of H2O2.48 Concentrations of GSH in cells and in vivo are extremely high, and it would appear that reactive species generated from hydralazine-Cu complexes may be ineffective in causing any DNA damage. However, it should be noted that many reactive species formed from various toxic drugs (e.g., cis-platin, Adriamycin) induce significant damage at submicromolar concentrations in vivo. Furthermore, it is now well established that oxidative stress causes a significant decrease in reduced GSH in cells and in vivo.49,50 Repeated use of hydralazine in the presence of elevated levels of copper ions in vivo would result in depletion of GSH, causing increased toxicity of hydralazine.
It is noteworthy that DNA cleavage studies, oxygen consumption studies, ESR spin trapping studies and immunological detection of DNA-DMPO adducts all show a very similar stoichiometry for Cu2+ ion with hydralazine for the maximum oxy-radicals formation, DNA cleavage, oxygen uptake and nitrone formation at a ratio of 5–10. The reason for this is not clear at the present; however, it would suggest that hydralazine at these concentrations (e.g. 125–250μM) is extremely effective in cycling Cu2+ ions for Fenton-type chemistry and facilitates the maximum formation of oxy-radicals. We found that this ratio of hydralazine to Cu2+ ions of between 5–10 is extremely effective in inducing DNA damage when this ratio is maintained throughout the range of concentrations e.g., 50 μM hydralazine and 10 μM Cu2+ or 1mM hydralazine and 100 μM Cu2+ (data not shown). It is possible, then, that hydralazine, at higher concentrations, acts as an antioxidant inhibiting cycling of copper ions and formation of oxy-radicals, and consequently, decreasing O2 consumption, DNA cleavage and DNA radical formation, as observed in this study. This would be similar to ascorbic acid, which can act both as an antioxidant and as a pro-oxidant, causing significant DNA damage in the presence of Fe or Cu ions.51 It should be noted that hydralazine concentrations greater than 100 μM are extremely difficult to achieve in patients undergoing treatment with hydralazine even at the highest doses of hydralazine used.52 Furthermore, due to acetylation and rapid metabolism, the concentration of hydralazine is significantly lower52 than 100 μM and thus, at these lower concentrations, hydralazine is expected to act more as a pro-oxidant, inducing significant oxidatively generated damage in vivo in the presence of Cu2+.
The most important finding is that hydralazine in the presence of Cu2+ induces significant DNA radical formation, detected as DNA-DMPO nitrone adducts as shown in Scheme 1. The implications of this finding are that DNA radicals, once formed, may induce formation of 8-oxo-7,8-dihydroguanine (8-oxoGua), and DNA-aldehydes.53,54 Formation of DNA aldehydes (or 4,6-diamino-5-formidopyrimidine and/or2,6-diamino-4-hydroxy-5-formamidopyrimidine) is common to many oxidizing systems such as ionizing radiation and some chemotherapeutic agents, e.g., bleomycin.55,56 We have previously shown that hydralazine binds to DNA and induces DNA aldehyde formation in the presence of metal ions.15 As copper is distributed throughout the body, DNA damage involving copper would be relatively favored in tissues/cells where copper concentration is elevated. Furthermore, in tissues where O2 concentration is low, the oxidation of Cu (I) by H2O2 would be favored, resulting in DNA damage as studied in this paper. Repeated use of drugs such as hydralazine is expected to further enhance DNA damage, resulting in more serious adverse effects, e.g., tumor formation and possibly have implications in autoimmune disease.
Using spin trapping studies, we show here direct trapping of DMPO-nucleoside adducts from hydralazine-Cu2+ complexes. Furthermore, we clearly show that the Cu2+-hydralazine system induces adduct formation predominantly at the 2′-deoxyadenosine moiety of the DNA. Thomas et al.55 have shown that hydralazine binds to the poly dA-poly dT sequences of DNA, and it is possible that hydralazine-Cu2+ also binds to these sequences in DNA and induces radical formation in 2′-deoxyadenosine similar to that induced by the Cu2+-H2O2 system.35 Yamamoto and Kawanishi24 have presented evidence for the formation of nucleoside radicals from hydralazine-peroxidase/H2O2 and hydralazine-Cu2+ complexes. However, no nucleoside radical adducts of DMPO were trapped in their studies. Furthermore, both 2′-deoxyguanosine and 2′-deoxyadenosine were equally effective in producing radicals.24 In contrast, our studies suggest that the exocyclic -NH2 adenosine radical was predominantly formed and trapped (40%) by DMPO, and only a small amount of radicals (5%) was detected from 2′-deoxyguanosine. However, it should be noted that other nucleoside radicals may have been formed in calf thymus DNA but not trapped/detected at 37°C, as these radical adducts of DMPO were not stable at 37°C as observed with our spin-trapping studies.
Work is in progress to understand how DNA radical formation may be linked to inhibition of DNA Methyltransferase 1. In this regard, formation of 8-oxoGua has been implicated in decreasing the ability of DNA Methyltransferase 1 to methylate neighboring cytosines.58,59 Furthermore, the formation of 8-oxoGua in the CpG island significantly alters binding of the enzyme to DNA, resulting in the inhibition of methylation.60,61 It is not known at this time whether hydralazyl radical or hydralazine diazonium radical23, or the phthalazinyl radical derived from the diazonium radical (Scheme 1), can directly alkylate DNA. Furthermore, it is also not known if the diazoniun radical can generate oxy-radicals that could induce DNA damage as found in this study.
Acknowledgments
The authors thank Jean Corbett for assisting with oxygen uptake experiments and Dr. Ann Motten and Mrs. Mary Mason for critically editing the manuscript. The authors also thank Drs. Fiona Summers and Douglas da Silva Ganini for their critical review of the manuscript.
FUNDING: This research was supported [in part] by the intramural research program of the NIH, National Institute of Environmental Health Sciences; however, statements contained herein do not necessarily represent the statements, opinions or conclusions of NIEHS, NIH or the U.S. Government.
Abbreviations
- DMPO
Dimethyl-1-pyrrolidine N-oxide
- GSH
reduced glutathione
- DMSO
dimethylsulfoxide
- SOD
Superoxide dismutase
- OH
hydroxyl radical
- IST
Immuno-spin trapping
- TEMP
2,2,6,6-tetramethyl-piperidine
References
- 1.Mitchell JR, Zimmerman HJ, Ishak KG, Thorgeirsson UP, Timberell JA, Snodgrass WR, Nelson SD. Isoniazid liver injury-clinical spectrum, pathology, and probable pathogenesis. Ann Intern Med. 1976;94:181–192. doi: 10.7326/0003-4819-84-2-181. [DOI] [PubMed] [Google Scholar]
- 2.Nelson SD, Mitchell JR, Timberell JA, Snodgrass WR, Corcoran GB. Isoniazid and Iproniazid-activation of metabolites to toxic intermediates in man and rat. Science (Wash, D.C.) 1976;193:901–903. doi: 10.1126/science.7838. [DOI] [PubMed] [Google Scholar]
- 3.Martz G, Dalessandri A, Keel HJ, Bollag W. Preliminary clinical results with a new antitumor agent RO-4-6467 (NSC-77213) Cancer Chemother Rep. 1963;33:5–14. [PubMed] [Google Scholar]
- 4.De Flora S, Zanacchi P, Bennicelli C, Camoirano A, Cavanna M, Sciaba L, Faggin P, Brambilla G. In vivo and in vitro genotoxicity of three antihypertensive hydrazine derivatives (hydralazine, dihydralazine, and endralazine) Environ Mutagen. 1982;4:605–619. doi: 10.1002/em.2860040512. [DOI] [PubMed] [Google Scholar]
- 5.Toth B. Tumorigenic effects of 1-hydrazinophthalazine hydrochloride in mice. J Natl Cancer Inst. 1978;61:1363–1365. doi: 10.1093/jnci/61.5.1363. [DOI] [PubMed] [Google Scholar]
- 6.Toth B. Actual cancer-causing hydrazines, hydrazides, and hydrazones. J Cancer Res Clinc Oncol. 1980;97:97–108. doi: 10.1007/BF00409895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parodi S, De Flora S, Cavanna M, Pino A, Bennicecelli C, Brambilla G. DNA damaging activity in vivo and bacterial mutagenicity of sixteen hydrazine derivatives as related to quantitatively to their carcinogenicity. Cancer Res. 1981;41:1469–1482. [PubMed] [Google Scholar]
- 8.Freese E, Sklarow S, Fresse EB. DNA damage caused by antidepressant hydrazines and related drugs. Mutat Res. 1968;5:343–348. doi: 10.1016/0027-5107(68)90004-3. [DOI] [PubMed] [Google Scholar]
- 9.Arce C, Segura-Pacheco B, Perez-Cardenas E, Taja-Chayeb L, Candelaria M, Duenas-Gonzalez A. Hydralazine targets: From blood vessels to epigenome. J Transl Med. 2006;4:10–22. doi: 10.1186/1479-5876-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knowles HJ, Tian YM, Mole DR, Harris AL. Novel mechanism of action of hydralazine: Induction of hypoxia-inducible factor I alpha, vascular endothelial growth factor, and angiogenesis by inhibition of prolyl hydroxylases. Circ Res. 2004;95:162–169. doi: 10.1161/01.RES.0000134924.89412.70. [DOI] [PubMed] [Google Scholar]
- 11.Timbrell JA, Facchini V, Harland SJ, Mansilla-Tinoco R. Hydralazine-induced Lupus: is there a toxic metabolic pathway? Eur J Clinc Pharmacol. 1984;27:555–559. doi: 10.1007/BF00556891. [DOI] [PubMed] [Google Scholar]
- 12.Cuz-Hernandez E, Perez-Cardena E, Taja-Chayeb L, Chavez-Blanco A, Trejo-Becerril C, Trujillo J, Franco-Meddina JL, Duenas-Gonzalez A. DNA demethylating activity of hydralazine in cancer cell lines. Life Sci and Med Res. 2011:2–8. [Google Scholar]
- 13.Sinha BK, Motten AG. Oxidative metabolism of hydralazine: evidence for nitrogen-centered radicals formation. Biochim Biophys Res Commun. 1982;105:1044–1051. doi: 10.1016/0006-291x(82)91075-0. [DOI] [PubMed] [Google Scholar]
- 14.Sinha BK. Enzymatic activation of hydrazine derivatives: a spin-trapping study. J Biol Chem. 1983;25:796–801. [PubMed] [Google Scholar]
- 15.Sinha BK, Patterson MA. Free radical metabolism of hydralazine: binding and degradation of nucleic acids. Biochem Pharmacol. 1983;32:3279–3284. doi: 10.1016/0006-2952(83)90351-9. [DOI] [PubMed] [Google Scholar]
- 16.Sinha BK. Metabolic activation of Procarbazine: evidence for carbon-centered radical intermediates. Biochem Pharmacol. 1984;33:2777–2781. doi: 10.1016/0006-2952(84)90695-6. [DOI] [PubMed] [Google Scholar]
- 17.Kalyanaraman B, Sinha BK. Free radical-mediated activation of hydrazine derivatives. Envior Health Prespect. 1985;64:179–184. doi: 10.1289/ehp.8564179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha BK. Activation of hydrazine derivatives to free radicals in the perfused rat liver: A spin-trapping study. Biochim Biophys Acta. 1987;924:261–269. doi: 10.1016/0304-4165(87)90021-3. [DOI] [PubMed] [Google Scholar]
- 19.Ranguelova K, Suarez J, Magliozzo RS, Mason RP. Spin trapping investigations of peroxide- and isoniazid-induced radicals in Mycobacterium tuberculosis catalase-peroxidase. Biochemistry. 2008;47:11377–11385. doi: 10.1021/bi800952b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiala ES. Investigations into the metabolism and mode of action of the colon carcinogen, DMH. Cancer. 1975;36:2407–2412. doi: 10.1002/1097-0142(197512)36:6<2407::aid-cncr2820360620>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 21.Leite LCC, Augusto O. DNA alterations induced by the carbon-centered radical derived from the oxidation 2-phenylhydrazine. Arch Biochem Biophys. 1989;270:560–572. doi: 10.1016/0003-9861(89)90538-9. [DOI] [PubMed] [Google Scholar]
- 22.Gamberini M, Cidade MR, Valotta LA, Armelin MCS, Leite LCC. Contribution of hydrazines-derived alkyl radicals to cytotoxicity and transformation induced in normal c-myc-overexpressing mouse fibroblasts. Carcinogenesis. 1998;19:147–155. doi: 10.1093/carcin/19.1.147. [DOI] [PubMed] [Google Scholar]
- 23.Reily CA, Aust SD. Peroxide substrates stimulate the oxidation of hydralazine to metabolites which cause single-strand breaks in DNA. Chem Res Toxicol. 1997;10:328–334. doi: 10.1021/tx960189l. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto K, Kawanishi S. Free radical production and site-specific DNA damage induced by hydralazine in the presence of metal ions or peroxidase/hydrogen peroxide. Biochem Pharmacol. 1991;41:905–914. doi: 10.1016/0006-2952(91)90195-b. [DOI] [PubMed] [Google Scholar]
- 25.Copper: Environmental health criteria. Geneva: World Health Organization (WHO); 2000. [Google Scholar]
- 26.Markovitz H, Gubler CJ, Mahoney JP, Cartwright GE, Wintrobe MM. Studies on copper metabolism XIV. Copper ceruloplasmin and oxidase activity in serum of normal subject, pregnant women, and patients with infection, hepatolenticular degeneration and nephric syndrome. J Clin Invest. 1955;34:1498–1508. doi: 10.1172/JCI103201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zowczak M, Iskara M, Torlinski S, Cofta S. Analysis of serum copper and zinc concentrations in cancer patients. Biol Trace Elem Res. 2001;82:1–8. doi: 10.1385/BTER:82:1-3:001. [DOI] [PubMed] [Google Scholar]
- 28.Gupta SK, Shukla VK, Vaidya MP, Roy SK, Gupta S. Serum and tissue trace elements in colorectal cancer. J Surg Oncol. 1993;52:172–175. doi: 10.1002/jso.2930520311. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida D, Ikeda Y, Nakazawa S. Quantitative analysis of copper, zinc, and copper/zinc ratio in selected human brain tumors. J Neurooncol. 1993;16:109–115. doi: 10.1007/BF01324697. [DOI] [PubMed] [Google Scholar]
- 30.Nasulewicz A, Mazur A, Opolski A. Role of copper in tumor angiogenesis-clinical implications. J Trace Elem in Med And Biol. 2004;18:1–8. doi: 10.1016/j.jtemb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Aruoma OI, Halliwell B, Gajewski E, Dizdarouglu M. Copper-ion-dependent damage to the bases in DNA in the presence of hydrogen peroxide. Biochem J. 1991;273:601–604. doi: 10.1042/bj2730601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinha BK, Antholine WM, Kalyanaraman B, Eliot HM. Copper ion-Dependent Oxy-Radical Mediated DNA Damage from Dihydroxy Derivative of Etoposide. Biochim Biophys Acta. 1990;1096:81–83. doi: 10.1016/0925-4439(90)90015-h. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto K, Kawanishi S. Hydroxyl free radical is not the main active species in site-specific DNA damage induced by copper (II) ion and hydrogen peroxide. J Biol Chem. 1989;264:15435–15440. [PubMed] [Google Scholar]
- 34.Bhattacharjee S, Deterding LJ, Chatterjee S, Jiang J, Ehrenshaft M, Lardinosis O, Ramirez DC, Tomer KB, Mason RP. Site-specific radical formation in DNA induced by the Cu (II)-H2O2 oxidizing system, using ESR, immuno-spin trapping, LC-MS and MS/MS. Free Radic Biol Med. 2011;50:1536–1545. doi: 10.1016/j.freeradbiomed.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee S, Chatterjee S, Jiang J, Sinha BK, Mason RP. Detection and imaging of the free radical DNA in cells-Site-specific radical formation induced by Fenton chemistry and its repair in cellular DNA as seen by electron spin resonance, immuno-spin trapping and confocal microscopy. Nucl Acid Res. 2012;40:5477–5486. doi: 10.1093/nar/gks180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura K, Ishiyama K, Ikai H, Kanno T, Sasaki K, Niwano Y, Kohno K. Reevaluation of analytical methods for photogenerated singlet oxygen. J Clin Biochem Nutr. 2011;49:87–95. doi: 10.3164/jcbn.10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dulling DR. Simulation of multiple isotropic spin traps ESR spectra. J Magn Reson Ser B. 1994;104:105–110. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- 38.Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radical in time and space with immuno-spin trapping. Free Rad Biol Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping analysis of DNA radicals. Nat Protocol. 2007;2:512–522. doi: 10.1038/nprot.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finkelstein E, Rosen GM, Rauckman EJ, Paxton J. Spin trapping of superoxide. Mol Phramacol. 1979;16:676–686. [PubMed] [Google Scholar]
- 41.Hawkins CL, Davies MJ. Hypochlorite-induced damage to nucleosides: formation of chloramine and nitrogen-centered radicals. Chem Res Toxicol. 2001;14:1071–1081. doi: 10.1021/tx010071r. [DOI] [PubMed] [Google Scholar]
- 42.Fujimoto S, Adachi Y, Ishimitsu S, Ohara A. Release of bases from deoxyribonucleic acid by ascorbic acid in the presence of Cu2+ Chem Pharm Bull. 1986;34:4848–4851. doi: 10.1248/cpb.34.4848. [DOI] [PubMed] [Google Scholar]
- 43.Sagripanti JS, Kraemer KH. Site-specific oxidative DNA damage at polyguanosine produced by copper plus hydrogen peroxide. J Biol Chem. 1989;264:1729–1734. 1989. [PubMed] [Google Scholar]
- 44.Oikawa S, Kawanishi S. Copper-mediated DNA damage by metabolites of p-dichlorobenzene. Carcinogenesis. 1996;17:2733–2739. doi: 10.1093/carcin/17.12.2733. [DOI] [PubMed] [Google Scholar]
- 45.Oikawa S, Kawanishi S. Distinct mechanisms of site-specific DNA damage induced by endogenous reductants in the presence of iron (III) and copper (II) Biochim Biophys Acta. 1998;1399:19–30. doi: 10.1016/s0167-4781(98)00092-x. [DOI] [PubMed] [Google Scholar]
- 46.Frelon S, Douki T, Favier A, Cadet J. Hydroxyl radical is not the main reactive species involved in the degradation of DNA bases by copper in the presence of hydrogen peroxide. Chem Res Toxicol. 2003;16:191–197. doi: 10.1021/tx025650q. [DOI] [PubMed] [Google Scholar]
- 47.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 48.Hanna PM, Mason RP. Direct evidence for inhibition of free radical formation from Cu(I) and hydrogen peroxide by glutathione and other potential ligands using the EPR- spin-trapping technique. Arch Biochem Biophys. 1992;295:205–213. doi: 10.1016/0003-9861(92)90507-s. [DOI] [PubMed] [Google Scholar]
- 49.Kidd PM. Glutathione: Systemic protectant against oxidative and free radical damage. Alter Med Rev. 1997;2:155–176. [Google Scholar]
- 50.Vaziri ND, Wang XQ, Oveisi F, Rad B. Induction of oxidative stress by glutathione depletion causes severe hypertension in normal rats. Hypertension. 2000;36:142–146. doi: 10.1161/01.hyp.36.1.142. [DOI] [PubMed] [Google Scholar]
- 51.Halliwell B. Vitamin C: Antioxidant or pro-oxidant in vivo? Free Radic Res. 1999;25:439–454. doi: 10.3109/10715769609149066. [DOI] [PubMed] [Google Scholar]
- 52.Shepherd AMM, Luden TM, McNay JL, Lin MS. Hydralazine kinetics after single and repeated oral doses. Clin Pharmacol Ther. 1980;28:804–811. doi: 10.1038/clpt.1980.238. [DOI] [PubMed] [Google Scholar]
- 53.Fujita S, Steenken S. Pattern of OH radical addition to uracil and methyl- and carbon substituted uracils: Electron transfer of OH adducts with N,N,N,Ntertramethylphenyldiamine and tetranitromethane. J Am Chem Soc. 1982;103:2540–2545. [Google Scholar]
- 54.Cadet J, Douki T, Ravanat JL. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc Chem Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 55.Burger RM, Peisac J, Horwitz SB. Stoichiometry of DNA strand scission and aldehyde formation by bleomycin. J Biol Chem. 1982;257:8612–8614. [PubMed] [Google Scholar]
- 56.Guteridge JM, West M, Eneff K, Floyd RA. Bleomycin-iron damage with formation of 8-hydroxydeoxyguanosine and base propenals. Implications that xanthine oxidase generates superoxide from DNA degradation products. Free Radic Res Commun. 1990;10:159–165. doi: 10.3109/10715769009149884. [DOI] [PubMed] [Google Scholar]
- 57.Thomas TJ, Seibold JR, Adams LE, Hess EV. Triplex-DNA stabilization by hydralazine and the presence of anti-(triplex DNA) antibodies in patients treated with hydralazine. Biochem J. 1995;311:183–188. doi: 10.1042/bj3110183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tudek B. Imidazole ring-opened DNA purines and their biological significance. J Biochem Mol Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 59.Turk PW, Laayoun A, Smith SS, Weitzman SA. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of DNA Methyltransferase. Carcinogenesis. 1995;16:1253–1255. doi: 10.1093/carcin/16.5.1253. [DOI] [PubMed] [Google Scholar]
- 60.Cerda S, Weitzman SA. Influences of oxygen radical injury on DNA methylation. Mutat Res. 1997;386:141–152. doi: 10.1016/s1383-5742(96)00050-6. [DOI] [PubMed] [Google Scholar]
- 61.Maltseva DV, Baykov AA, Jeltsch A, Gromova ES. Impact of 7,8-dihydro-8-oxoguanine on methylation of CpG sites by Dnmt3a. Biochemistry. 2009;48:1361–1368. doi: 10.1021/bi801947f. [DOI] [PubMed] [Google Scholar]


