Abstract
The muscular dystrophies are a heterogeneous group of over 40 disorders that are characterised by muscle weakness and wasting. The most common are Duchenne muscular dystrophy and Becker muscular dystrophy, which result from mutations within the gene encoding dystrophin; myotonic dystrophy type 1, which results from an expanded trinucleotide repeat in the myotonic dystrophy protein kinase gene; and facioscapulohumeral dystrophy, which is associated with contractions in the subtelomeric region of human chromosome 1. Currently the only treatments involve clinical management of symptoms, although several promising experimental strategies are emerging. These include gene therapy using adeno-associated viral, lentiviral and adenoviral vectors and nonviral vectors, such as plasmid DNA. Exon-skipping and cell-based therapies have also shown promise in the effective treatment and regeneration of dystrophic muscle. The availability of numerous animal models for Duchenne muscular dystrophy has enabled extensive testing of a wide range of therapeutic approaches for this type of disorder. Consequently, we focus here on the therapeutic developments for Duchenne muscular dystrophy as a model of the types of approaches being considered for various types of dystrophy. We discuss the advantages and limitations of each therapeutic strategy, as well as prospects and recent successes in the context of future clinical applications.
Muscular dystrophy is a class of inherited disorders characterised by muscle weakness and wasting. Over 40 forms of muscular dystrophy have been identified, based on underlying genetic and molecular etiology, clinical manifestation and prognosis (Ref. 1). The most severe form, Duchenne muscular dystrophy (DMD, MIM 310200), and the milder Becker muscular dystrophy (BMD) are the most common lethal genetic disorders of children, with DMD affecting ~1 in 3500 newborn males (Ref. 2). The second and third most common muscular dystrophies are myotonic dystrophy (DM1) and facioscapulohumeral muscular dystrophy (FSHD) (Refs 3, 4).
The molecular pathology of various muscular dystrophies is diverse because of the heterogeneity of the defective proteins involved. Understanding the role of these proteins and their interactions will be a crucial aspect of the search for therapeutic targets. Many muscular dystrophies result from defects in muscle-membrane-associated proteins that help maintain the structural integrity of muscle fibres (Refs 5, 6). DMD is caused by mutations in the dystrophin gene (official symbol, DMD for human and Dmd for mouse), most of which result in translational frameshifts and failure to express a functional protein (Ref. 7). The four major domains of dystrophin (N-terminal, central rod, cysteine-rich and C-terminal) mediate a link between the subsarcolemmal cytoskeleton and a complex series of protein–protein interactions at the sarcolemma (reviewed in Ref. 8) (Fig. 1). The N-terminal and central rod domains interact with filamentous actin. The rod domain contains 24 repeats that are homologous to those in spectrin, providing a flexible and elastic region connecting the end domains that are crucial for dystrophin function. The third and fourth are the cysteine-rich and C-terminal domains, which contain many of the protein-interaction domains essential for signalling and assembly of the dystrophin–glycoprotein complex (DGC) (Fig. 1). The most critical binding site in dystrophin is the dystroglycan-binding domain (DgBD), which is made up of a WW domain (Ref. 9) at the end of the rod domain and the adjacent cysteine-rich domain. Inactivation of the DgBD renders dystrophin nonfunctional (reviewed in Ref. 8).
Figure 1. Dystrophin and the dystrophin–glycoprotein complex.
(a) Comparison of the structural domains in full-length dystrophin with those of highly functional miniaturised versions, which have been engineered in response to the limited packaging capacity of delivery vectors for gene therapy. Full-length dystrophin contains four major domains: an N-terminal (NT) cytoskeletal-binding domain, a rod domain composed of 24 spectrin-like repeats (1–24) and four hinge regions (H1–H4), a cysteine-rich (CR) and a C-terminal (CT) domain. The two known actin-binding domains (ABD-1 and ABD-2) are within the N-terminal and rod domains, respectively. The dystroglycan-binding domain (DgBD) is composed of a WW motif (in hinge 4) connected to two EF-hand-like motifs and a ZZ domain (the two EF-hand and ZZ regions comprise the so-called cysteine-rich ‘CR’ domain). Towards the C-terminus is a syntrophin-binding domain (SBD) and a dystrobrevin-binding domain (DbBD). Minidystrophin and microdystrophin retain most of the necessary regions for the signalling and structural roles of dystrophin, as well as the ability to assemble members of the dystrophin–glycoprotein complex (DGC) at the plasma membrane. (b) Important interactions among members of the DGC, many of which are defective in various muscular dystrophies. Within striated muscle fibres, dystrophin binds to cytoskeletal proteins such as filamentous actin (F-actin) at its N-terminal domain. The rod domain encodes a second actin-binding domain in spectrin-like repeats 11–17, and repeats 16–17 also participate in binding to neuronal nitric oxide synthase (nNOS). Although localisation of nNOS has been found to require the presence of both repeats 16–17 and syntrophin, the precise three-dimensional structure of the dystrophin–nNOS complex is speculative (Ref. 78). The DgBD anchors β-dystroglycan to dystrophin, and might help to assemble other proteins. The CT domain binds to and localises members of the syntrophin (Syn) and dystrobrevin (Db) protein families. Studies suggest that up to four syntrophins could attach to the DGC at any one time, two to dystrophin and two to dystrobrevin (Ref. 180). At present it is unclear how many of these syntrophins are attached to nNOS, as it has been shown that syntrophin can also bind to other proteins, such as sodium channels and aquaporin-4 (Refs 181, 182). In addition to dystrophin, dystrobrevin is known to bind to another member of the DGC, which has yet to be identified. Abbreviations: Dg, dystroglycan; Sg, sarcoglycan; SSPN, sarcospan.
Dystrophin is thought to have a primarily structural role, linking the cytoskeleton to the extracellular matrix via the DGC (Fig. 1b) (Refs 10, 11). This linkage transduces the forces of contraction to the extracellular matrix to protect myofibres from contraction-induced injury (Ref. 12). The absence of dystrophin results in membrane instability and repeated tears in the sarcolemma with calcium entry into the muscle cell (Refs 13, 14). Stretch-activated calcium channels might have a role in this process, because their blockade reduces membrane permeability and loss of force in dystrophic muscle following eccentric contractions (Ref. 15). The resulting cascade of events forces muscle fibres to undergo cycles of degeneration and regeneration until repair capacity is no longer sufficient, and muscle fibres are replaced by adipose and fibrous connective tissue (Ref. 2). In BMD, mutations typically maintain the mRNA reading frame but lead to reduced expression, or expression of truncated forms of dystrophin in striated muscle (Refs 7, 16). Mutations in other components of the DGC result in a number of other muscular dystrophies. For example, sarcoglycanopathy results in several limb-girdle muscular dystrophies, and integrin or laminin deficiencies result in congenital muscular dystrophy (Refs 17, 18).
Despite tremendous effort and major advances in our understanding of the molecular basis for the muscular dystrophies, no cure has been found. Symptom management and prolonging mobility is therefore the primary focus of clinical interventions (Refs 19, 20). Many hopes rest on recent advances in gene and cell therapies to prevent muscle degeneration and potentially reverse dystrophy-related damage. The goal of gene therapy is to deliver a functional copy of the gene, or repair the damaged gene, such that it produces sufficient product to halt the dystrophic phenotype. Methods of gene delivery include both viral and nonviral vectors. Current cell therapies involve transplantation of stem or progenitor cells with skeletal myogenic potential that can fuse with existing myofibres or form new ones. To avoid the host-versus-graft immune response, a patient’s own cells that have been corrected for the genetic defect could be used. Upon transplantation, these cells would ideally engraft and regenerate the muscle, as well as repopulate the muscle stem cell (satellite cell) niche.
Mammalian preclinical testing often uses the X-linked muscular dystrophy Dmdmdx mouse model of DMD (hereafter mdx), or the canine cxmd model (Ref. 21). In this review, many of the therapies have been developed and will be discussed with respect to models of DMD; however, these approaches are applicable to a wide range of muscle and genetic disorders.
Gene therapy for muscular dystrophies
Approaches
Gene therapy approaches for treatment of DMD must either develop methods to deliver dystrophin, or repair the locus within a patient’s genome (Fig. 2). As dystrophin encodes a very large 14 kb mRNA (the gene itself spans about 2.1 Mb), therapeutic delivery is a significant challenge (Refs 22, 23). However, the observation of large genomic deletions in some very mildly affected BMD patients (Refs 24, 25) prompted construction of highly functional ‘mini’ and ‘micro’ versions of dystrophin to facilitate gene transfer using viral vectors such as adeno-associated virus, which have a limited carrying capacity (Refs 26, 27, 28, 29). Internally deleted dystrophins, illustrated in Fig. 1a, retain their N-terminal actin-binding and C-terminal dystroglycan-binding domains, which are thought to contain most of the necessary regions for dystrophin’s role in signalling, structural support and assembly of dystrophin-associated proteins at the cell membrane (Refs 26, 30) (Fig. 1). Although inclusion of actin- and dystroglycan-binding domains is crucial, meticulous design of the deletions in the rod domain is also essential for maintaining functionality and rescuing the dystrophic phenotype (Ref. 26).
Figure 2. Strategies for treating the muscular dystrophies.
Major advantages and limitations of viral vectors, exon skipping, plasmid DNA and cell therapies. Each virus-based delivery system (a) has particular strengths, such as high striated muscle tropism (adeno-associated virus), ability to integrate into the host genome (lentivirus), and packaging capacity [helper-dependent (hd)-adenovirus]. However, viral systems must carefully address safety concerns such as insertional mutagenesis, where provirus integration into the genome might alter the structure or expression of nearby genes, and immunogenicity of capsid proteins. Exon skipping (b) is a promising new approach in which mutations can be bypassed using antisense oligonucleotides of various chemistries that modify splicing of pre-mRNAs. Shown is the phosphorodiamidate morpholino oligomer (PMO), which incorporates morpholine rings linked by phosphorodiamidate groups instead of the ribose rings linked by phosphodiester groups found in RNA. R, cell-penetrating moiety; B, RNA nucleobases. (c) Plasmid DNA is a straightforward approach and can potentially deliver full-length dystrophin. Current studies are focused on lowering toxicity of transfection reagents as well as improving delivery efficiency. (d) Cell-based therapies might be able to regenerate muscle by replacing muscle fibres lost during the course of progressive muscle-wasting conditions such as Duchenne muscular dystrophy. Transplanted cells derived from the patient (autologous setting) require genetic modification to include a functional copy of the defective gene, whereas donor cells (allogeneic setting) are at risk for immune rejection. Image shows differentiating muscle cells in culture, with immunofluorescent staining for myosin heavy chain (red). Most therapies must be adapted for systemic delivery and targeted specifically to striated muscle using muscle-specific promoters to regulate expression of the delivered transgene.
It has long been considered in the field of gene therapy that expression of a delivered transgene not normally expressed in a host could incite an immune response. Evidence from immunological studies indicates that dystrophin could act as a neoantigen in this manner (Refs 31, 32, 33). If the dystrophin neoantigen is released from degenerating muscles and absorbed by antigen-presenting cells, this could stimulate both cytotoxic and humoral immune responses, presenting a significant challenge for gene transfer in DMD (reviewed in Ref. 34).
A possible alternative is to use utrophin, a highly similar protein to dystrophin in both structure and properties (Ref. 35). Utrophin has been suggested to have a comparable functional role in muscle, with the potential to compensate for dystrophin (Refs 36, 37). Although it is primarily expressed at the neuromuscular junctions in adult muscle (Refs 38, 39), it is speculated that elevated expression of utrophin in some DMD patients partially compensates for lack of dystrophin (Refs 40, 41). Delivering and upregulating endogenous utrophin are therefore potential therapies for DMD. Full-length utrophin transferred via adenoviral vector ameliorates the dystrophic phenotype in the limb muscles of mdx mice (Ref. 42). In addition, mini-utrophins significantly improve the pathophysiology of dystrophic mdx and dystrophin and utrophin−/− (mdx:utrn−/−) double-knockout transgenic animals, as well as dystrophic dogs (Refs 43, 44, 45). Recently, microutrophins have been shown to alleviate a wide range of histopathological features of the mdx:utrn−/− model when delivered systemically using recombinant adeno-associated viral vectors (Ref. 46). Assessment of muscle morphology, mass, fibre size, and contractile properties suggests that microutrophin might work similarly to microdystrophin. These results raise the possibility of delivering micro-utrophins to dystrophic muscle as a way of ameliorating pathology, without eliciting a cellular immune response against exogenous dystrophin.
Adeno-associated virus
Recombinant vectors derived from adeno-associated virus (rAAV) are one of the most promising methods for delivery of genes to striated muscle (Fig. 2a). The wild-type virus is a nonpathogenic single-stranded DNA parvovirus that requires a helper virus to replicate (Refs 47, 48). The recombinant form carrying a desired transgene can be produced at high titres in the absence of helper virus, and it can infect both dividing and nondividing cells (Ref. 49). Although its small size and range of target tissues facilitates dissemination, the limited packaging capacity (less than 5 kb) precludes delivery of full-length dystrophin. However rAAV has been used to successfully deliver microdystrophin systemically to all striated muscle (Ref. 50).
At least nine AAV serotypes have been identified in primates, referred to as AAV1–AAV9 (Ref. 51). The different serotypes display various tropisms in vivo (Refs 52, 53, 54, 55). rAAV genomes persist as nonintegrated episomes following infection of cells (Refs 56, 57), except at very high doses, where low levels of integration have been found in cultured cells, liver and muscle (Refs 58, 59). Stable gene expression following rAAV injection into muscle has been reported for up to 2 years in mice and more than 7 years in dogs and rhesus monkeys (Refs 60, 61). However, the mostly episomal DNA transferred by rAAV in muscle will almost certainly be lost over an extended period of time because of natural muscle turnover during exercise.
AAV vectors have been widely used for gene therapy studies because they appeared unable to elicit a host cellular immune response in numerous studies. A Phase I human trial for haemophilia b showed that rAAV2 was safe, with no adverse events reported among patients who received intramuscular injections of up to 1014 vector genomes (Ref. 61). The authors also reported that pre-existing antibodies to AAV2 in patients had no effect on myofibre transduction. However, in some cases, such as in dystrophic muscles from the mdx mouse or the sarcoglycan-deficient hamster, an inflammatory response has been observed following delivery of rAAV vectors that express an immunogenic protein, such as E. coli β-galactosidase, under the control of a ubiquitously active promoter, such as CMV (Refs 50, 62). Use of a muscle-specific promoter can often block this response (Refs 50, 63). Other proteins delivered to the mdx mouse, such as human dystrophin, have not elicited a cellular immune response (Refs 26, 29, 50). Nonetheless, as investigators have expanded studies to larger animals and started to use higher doses, it is becoming clear that most, if not all, AAV serotypes can elicit a cellular immune response. Recent studies have found such responses to both rAAV2 and rAAV6 in dogs (Ref. 64), and similar observations have been made in humans (Refs 65, 66, 67). Nonetheless, the immune response is often mild, and studies in dystrophic dogs have shown that it can be blocked with brief immunosuppression (Ref. 64). Transient immune suppression has also been applied with success in nonhuman primates (Ref. 68).
Systemic delivery of rAAV will be a critical aspect of administration, given the need to target all striated muscle, including the heart and diaphragm, for an effective DMD therapy. Targeting skeletal muscle alone will strain the dystrophic heart and aggravate cardiomyopathy (Ref. 69). A major breakthrough for muscular dystrophy gene therapy came with the discovery that several serotypes of AAV could transduce muscle tissue body-wide following intravascular delivery. Initial studies of systemic delivery used vectors derived from AAV6 (Ref. 50), but subsequent studies have also had success with AAV1, AAV8 and AAV9 (Refs 70, 71, 72). Feasibility of the systemic approach coupled with early intervention is validated by successful transduction of all skeletal muscle in newborn dogs in the absence of immune suppression (Ref. 73).
The intimate association between myofibres and capillaries allows extravasated vectors to come into close contact with the surface of muscle cells. Thus, AAV serotypes readily transduce muscle following intravascular delivery. The mechanisms responsible for vector extravasation remain unclear, and few cell surface receptors have been identified for AAV1, AAV6, AAV8 or AAV9. Also, little is known of the intracellular events following vector uptake by myofibres that enable vector decapsidation and gene expression (reviewed in Ref. 74). Identification of these mechanisms could well lead to modified delivery protocols to enhance body-wide muscle transduction using lower vector doses.
Split-vector approaches for full-length dystrophin delivery by rAAVs
Restoration of the major functions of dystrophin is key to alleviating deficits in DMD, and internally truncated dystrophins lose some functionality (Refs 26, 75). For example, although most mini-and microdystrophins restore the components of the DGC to the sarcolemma, only recently has a truncation been found that supports sarcolemmal localisation of neuronal nitric oxide synthase (nNOS) (Refs 76, 77, 78). nNOS is involved in production of the signalling molecule nitric oxide (NO), which is important for maintaining vasodilation and adequate blood flow in skeletal muscle during activity (Refs 79, 80). Although reconstitution of dystrophin as represented in the internally truncated forms might be the crucial factor for stabilising muscle fibres, delivery of mini- or microdystrophins that fail to localise nNOS could lead to a mild BMD phenotype, with the associated susceptibility to fatigue. It is also unknown whether other properties of myofibres are affected by large deletions in dystrophin, such as the structure of myotendinous junctions (Refs 81, 82). It is therefore unclear whether some of the milder BMD phenotypes that result from truncated dystrophins would benefit from delivery of a designer micro- or minidystrophin, or whether an alternative therapy would be necessary to relieve symptoms.
Since rAAV packaging capacity is limited to less than 5 kb, microdystrophin is the only dystrophin variant small enough to package into a single vector. However, one possible strategy to deliver full-length dystrophin is to package fragments of the cDNA into separate vectors. Transduction of target tissues with more than one vector, each carrying a dystrophin cDNA fragment, could allow production of larger dystrophins if the parts of each vector could be brought together (Refs 83, 84, 85). Two approaches for achieving this goal are showing promise in vivo. In the first, portions of introns carrying appropriate splicing signals are incorporated into the gene fragments, such that the cellular RNA splicing mechanisms can be harnessed to reconstruct a larger gene from two smaller halves (Refs 86, 87). Another approach is to package partially overlapping fragments into the different vectors, such that a larger dystrophin expression cassette can be reconstructed via homologous recombination in transduced tissues (Refs 86, 88). A recently developed hybrid system outperforms splicing and homologous recombination split-vector systems alone, and improves efficiency of minidystrophin delivery to muscle (Ref. 89).
Lentivirus
Retrovirus-derived lentiviral vectors can be used to stably integrate a transgene such as dystrophin into the genome of target cells (Ref. 90). However, integration into the host genome can potentially cause insertional mutagenesis, where viral promoter and enhancer elements can activate nearby proto-oncogenes (Refs 91, 92, 93). Development of self-inactivating (SIN) lentiviral vectors may address this issue. With SIN lentiviral vectors, the viral promoter and enhancer elements are removed prior to integration into the host genome, thus preventing viral activation of proto-oncogenes at sites of integration.
Unlike vectors derived from onco-retroviruses such as Moloney murine leukaemia virus, which are commonly used for gene transfer (reviewed in Ref. 94), lentiviral vectors can infect a wide range of dividing and nondividing cells, including hepatocytes, skeletal and cardiac muscle cells, and neurons (Refs 95, 96, 97). Lentiviral vectors have low immunogenicity but the delivered transgene is still a potential neoantigen (Ref. 98). They have a larger carrying capacity (~9 kb) than rAAVs, and have been used to stably transduce myogenic cells with minidystrophin (Ref. 90), whereas rAAVs can only deliver the smaller microdystrophin (Ref. 26).
Permanent gene transfer by lentiviral vectors is especially advantageous for DMD treatment. Satellite cells within skeletal muscle are an ideal target, and lentiviral reconstitution of dystrophin into the satellite cell niche in vivo would provide a source of dystrophin during ongoing cycles of regeneration. Lentiviral vectors are also a useful tool for transducing autologously derived cells with dystrophin and other genes ex vivo before expansion and transplantation (Refs 99, 100).
Adenovirus
Vectors derived from adenovirus are attractive for gene transfer because of their large carrying capacity, high titre production and ability to infect nondividing cells. Successive modifications have improved carrying capacity, reduced immunogenicity and prolonged transgene expression. The latest generation of adenovirus vectors are helper-dependent or ‘gutted’ (hdAd) and are entirely devoid of viral genes. They thus require a helper virus to supply genes needed for vector replication and packaging during vector production (Refs 101, 102, 103). The presence of a helper virus renders production technically challenging, and additional purification steps are required to remove it from the final vector preparations (Ref. 104). Helper-dependent adenovirus has been used to stably transduce muscle with dystrophin and has improved muscle function in mdx mice (Refs 105, 106).
Despite improvements, many hurdles to effective hdAd-vector-based therapy remain. Adenovirus vectors are five times larger than AAVs, reducing their ability to exit capillaries and infect muscle (Ref. 107). Also, most intravascularly administered adenovirus vectors are taken up by the liver (Ref. 108). Coupled with low transduction efficiency in adult skeletal muscle, systemic therapeutic administration could require extremely high doses of hdAd (Refs 105, 109). Such high doses resulted in acute inflammation and a cytotoxic T cell response, which mediated lethal acute toxicity in nonhuman primates (Refs 110, 111, 112) and one patient (Ref. 113). As with AAV, delivered transgenes exist as episomes, which might be lost over time and require repeated doses. However, readministration with the same serotype is problematic given the development of neutralising antibodies to both the hdAd vector and the transgene (Refs 33, 114). Use of hdAd vectors has not progressed to clinical trials for these reasons, and much of the current work is focused on overcoming the immunological issues.
Nonviral gene transfer
Nonviral gene transfer methods involve administration of plasmid DNA (pDNA) (Ref. 115), usually in complex with synthetic compounds (Fig. 2c). Although the approach is conceptually straightforward, less expensive compared with viral vector production, and avoids the inherent risks of viral vector administration, preclinical testing has not yet demonstrated a feasible approach for system-wide treatment of DMD. Upon intramuscular and intravascular injection, naked dystrophin pDNA is expressed in only 5% of target myofibres (Ref. 116). Brief blood flow occlusion achieves targeting of the diaphragm and improves transgene expression in limb muscles following intravascular administration (Refs 116, 117). Use of cationic polymers and lipids also improves in vivo tissue transfection after systemic administration (Ref. 118). However, the possibility of acute toxicity of plasmid DNA and cationic polymer/lipid complexes must be resolved, especially considering potentially high-dose systemic administration (Refs 119, 120). The size of the plasmids encoding full-length dystrophin may also impede DNA transfer through the vasculature (Refs 116, 118). Although additional methods have improved transfection of striated muscle, clinically safe and efficient body-wide dystrophin expression in both skeletal and cardiac muscle has not yet been achieved. Still, a recent Phase I trial testing naked dystrophin pDNA by intramuscular injection resulted in low levels of dystrophin expression (Ref. 121), giving hope that development of a safe method to boost gene transfer will lead to a new DMD therapy.
Exon skipping
Exogenous delivery of dystrophin has shown promise for treatment of DMD, but limitations such as vector capacity and delivery, potential immune response and appropriate transgene expression, still need to be overcome. Exon skipping provides an alternative approach that allows direct manipulation of dystrophin transcripts, such that expression of the endogenous gene can potentially be restored in vivo (Fig. 2b).
In the various forms of muscular dystrophy, as well as other genetic diseases, mutations give rise to premature stop codons, and splice-site mutations or deletions/duplications can shift the reading frame of a transcript or cause aberrant splicing, resulting in little or no functional protein. Exon skipping addresses these disorders at the post-transcriptional level, by using the process of transcript maturation to remove problematic exons (Fig. 3). Sites within the newly synthesised pre-mRNA that signal splicing at the start of the exon are blocked, and the whole exon containing the stop codon or frameshift mutation, along with its flanking introns, are thereby removed. In the case of dystrophin, multiexon deletion in the rod domain has been found in mild and asymptomatic BMD (Ref. 7). Thus exons in this domain with a stop codon or frameshift mutation may be removed using exon skipping. Translation then generates an in-frame, albeit truncated, gene product that retains many critical functions. Studies of rare, dystrophin-positive ‘revertant’ fibres in some DMD patients first indicated that this might be a feasible therapeutic approach (Ref. 122). The revertant fibres contained a restored dystrophin reading frame, although it remains unclear whether this was a result of secondary mutations or spontaneous alternative splicing (Ref. 123).
Figure 3. The general strategy for oligonucleotide-mediated exon skipping.
(a) A hypothetical stop codon mutation in exon 4 is translated into a nonfunctional truncated protein. (b) Normal splicing results in translation of full-length protein. (c) The antisense oligonucleotide (AON) blocks the splicing site for exon 4, resulting in its removal during the processing of the pre-mRNA into a mature mRNA transcript. In the case of the mdx mouse model of Duchenne muscular dystrophy, AONs have been used to target the stop codon mutation in exon 23, leading to removal of exon 23 from the pre-mRNA and joining of exons 22 and 24 (Ref. 131). Similarly, in Duchenne and Becker muscular dystrophies, targeting of dystrophin in this manner could lead to removal of exons containing stop codon or frameshift mutations. The translated protein, although internally deleted, will ideally retain critical functional domains. In myotonic dystrophy type 1 (DM1), chloride channels have been targeted to remove an exon normally expressed only in the neonatal isoform, but which is retained in DM1 muscles, to prevent generation of a dysfunctional protein in adult muscle (Ref. 3). Recent studies have demonstrated that the specific sequence targeted within a pre-mRNA is critical to the success of an AON (Ref. 143).
Synthetic antisense oligonucleotides (AONs) were developed for specific exons within dystrophin that can block transcript splicing sites in vitro (Refs 124, 125, 126). Most of the early work on AONs for therapeutic benefit was in the form of 2’-O-methyl-phosphorothioate AONs (2’OMeAONs) (Fig. 4), which are more stable than nonmethylated RNA (Ref. 127). 2’OMeAONs restore the reading frame of dystrophin in primary muscle cells from mdx mice (Ref. 128). This was also achieved in cells from cxmd dogs (Ref. 129), human cells derived from DMD patients (Ref. 130) and in vivo, in mdx mice by intramuscular administration (Ref. 131). Other muscular disorders have also successfully used exon skipping to correct abnormal gene expression. In myotonic dystrophy, AONs have been used to correct aberrant splicing in chloride channel 1 gene (CLCN1) pre-mRNA and the associated myotonia (Ref. 3), demonstrating the broad applications of this tool.
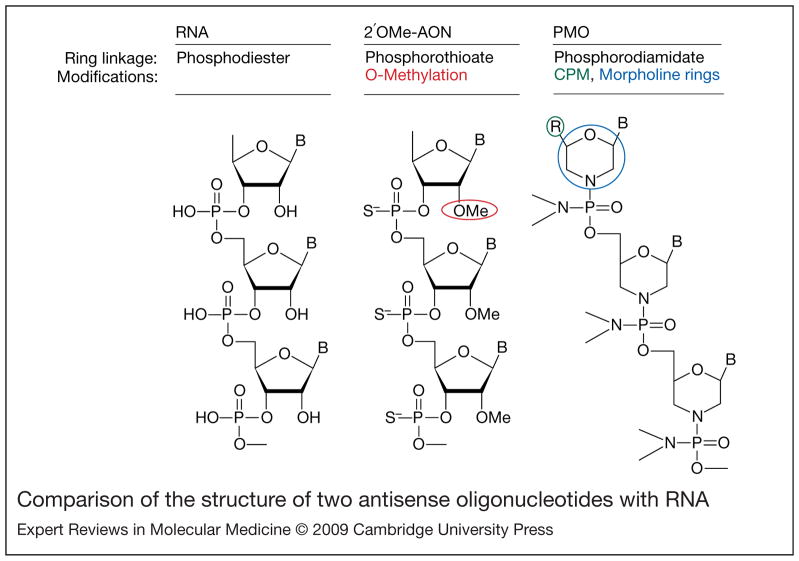
Figure 4. Comparison of the structure of two antisense oligonucleotides with RNA.
Antisense oligonucleotides (AONs) are designed to be complementary to specific sequences within mRNA transcripts, and have various chemical modifications that distinguish them from RNA. Specific modifications confer resistance to degradation and ability to penetrate cell membranes. RNA (left panel) contains phosphodiester linkages between each ribose ring, whereas 2′O-methylated (2′OMe) AONs (middle panel) have phosphorothioate linkages (the oxygen is replaced with a sulphur atom). Phosphorodiamidate morpholino oligomers (PMOs, right panel) have morpholine rings instead of ribose rings, often contain conjugated cell-penetrating moieties (CPM, R group in structure) to facilitate cell entry and subunits are linked by phosphorodiamidates (the oxygen is replaced with a nitrogen group). B, RNA nucleobases.
AON chemistry
AON modifications have undergone many iterations, resulting in various chemistries affecting efficiency and delivery to cells in vitro and in vivo (Ref. 132). Morpholino oligomers show promise for in vivo applications (Ref. 133). Antisense morpholino oligomers, commonly called PMOs (for phosphorodiamidate morpholino oligomers), resemble 2′OMeAONs, but have several key differences, as shown in Fig. 4. PMOs incorporate morpholine rings in place of the ribose sugar rings of RNA and are nonionic, which avoids nonspecific electrostatic interactions within the cell. Replacement of the phosphodiester linkage with phosphorodiamidate imparts resistance to nuclease degradation (Ref. 132). Because PMOs are non-ionic, and therefore inefficient at penetrating cell membranes (Ref. 134), they are commonly conjugated to cell-penetrating cationic moieties (Fig. 4) (Refs 135, 136). Peptide nucleic acids represent another type of chemistry developed and tested in vivo (Ref. 137). Aside from sequence specificity and efficient exon skipping, clinical considerations for AON chemistries must include the potential for toxicity or induction of a host immune response, biostability, penetration into striated muscle cells and the cost of large-scale synthesis for systemic delivery.
Administration
System-wide targeting of morpholino oligomers to skeletal muscle was initially achieved in the mdx mouse with no observed immune response or toxicity, but also without targeting of the heart (Refs 138, 139) – outcomes observed previously with 2′OMeAONs (Ref. 140). Recently, several groups have successfully targeted skeletal muscle system-wide, as well as the heart in the mdx mouse, upon incorporation of peptide or chemical moieties to facilitate cell uptake (Refs 136, 141, 142).
AON design itself must be optimised for the target sequence within a given mRNA, because of the secondary structure in single-stranded RNA and splice signal sites (Ref. 143). Indeed, this might explain why multiple AONs, or an oligo ‘cocktail’, seem to be more efficient than a single AON at targeting a single site (Ref. 144). Oligo cocktails might be important during the development of a treatment for DMD, to achieve multiexon skipping and thereby cover the majority of DMD mutations with a single treatment.
Another systemic approach for AONs uses rAAV vectors for intravascular delivery. rAAVs can be used to deliver DNA encoding an AON linked to a small nuclear RNA sequence for nuclear targeting of the transcript for modification of dystrophin (Ref. 145). Treatments show therapeutic improvement in the mdx mouse, with reduced serum creatine kinase and improved skeletal muscle function (Refs 146, 147). rAAV vector tropism also allows certain serotypes to reach the heart (Ref. 147). Another advantage of rAAV vector delivery of AONs is the persistence of rAAV episomes (Ref. 74). Longer-term in vivo AON expression from episomes would be preferred over more frequent, lifelong treatment with PMOs. For rAAV AONs, however, potential immune responses as well as scalability of vector production must also be considered.
AON safety
Although it is debatable which AON chemistry will be the most effective (Refs 142, 144, 148), it must be determined whether the most effective are actually safe for human use. The various modifications require careful investigation of potentially harmful degradation products or codelivery agents, immune responses, and nonspecific actions. Owing to inherent resistance to degradation, some morpholinos might be especially harmful if they have low specificity for RNA targets, and at high doses these effects could be amplified (Ref. 133).
Cell therapy for muscular dystrophies
Approaches
Cell transplantation as a therapeutic tool is a promising avenue for treatment of muscular dystrophies (Fig. 2d). Transplanted cells must be able to fuse with existing myofibres or form new ones, and transplanted cell nuclei within those myofibres must express the missing gene product. In addition to regeneration of myofibres, a major goal of stem cell therapies is reconstitution of the satellite cell niche, which might promote future functional regeneration of the muscle.
Cell transplantation might either be allogeneic (donor-derived) or autologous (patient-derived). Although allogeneic transplantation from a wild-type donor does not require genetic manipulation to reintroduce functional dystrophin, the risk for graft rejection remains. Autologous transplantation requires genetic manipulation, potentially by using lentiviral vectors to permanently reintroduce the defective gene. Inherent risks in using lentiviral vectors apply, but the risk of immunogenic graft rejection is much lower. With either approach, expansion and culturing of the cell population will probably be necessary, requiring careful control of conditions to preserve muscle engraftment ability. For a more extensive review of approaches and immune issues associated with cell therapy, see (Ref. 149). Developing systemic routes of administration in cell transplantation will be an important issue, as will identifying an adequate cell source for scalable autologous transplantation in humans. Ultimately, the most promising approach for muscular dystrophy cell therapy will be the use of accessible patient-derived myogenic precursors capable of efficient engraftment into skeletal muscle following ex vivo expansion and genetic correction. A variety of cell types have been investigated for their potential to fulfil these criteria.
Cell types
The most widely studied cell type for muscular dystrophy cell therapy is the satellite cell. Transplanted satellite cells from wild-type donors engraft into skeletal muscle (Ref. 150), but massive cell death, limited migration from injection sites and immune rejection of allogeneic cells is observed (Refs 151, 152, 153), even with genetic matching (Ref. 31). Isolation and autologous transplantation of satellite cells avoids host-versus-graft immune rejection. However, in later stages of muscle degeneration, fewer myogenic progenitors can be isolated, and expansion of cells in culture significantly reduces their engraftment capacity (Refs 154, 155). Many recent approaches aim to improve muscle precursor cell isolation with the rationale of avoiding in vitro expansion to determine therapeutic potential (Ref. 152). However, immune responses and the cost of antibodies commonly used in these isolation techniques prevent direct clinical applications in humans (Ref. 156). Nevertheless, these studies could potentially lead to new discoveries on how to maintain myogenic cell populations in a particular progenitor state in vitro (Refs 157, 158), and how to increase transplantation engraftment success. A further complication of these studies is that myogenic progenitors derived from muscle are typically a heterogeneous population, displaying unique properties depending on isolation methods and culture conditions (Ref. 159). It is also unclear whether satellite cells themselves might derive from a satellite cell precursor within muscle or from a circulating progenitor (Ref. 158).
Muscle-derived progenitors that can engraft in muscle after intravascular delivery have also been found; this is a crucial step to making cell-based therapies a feasible treatment for DMD. Such progenitor cells include muscle side-population cells (Refs 99, 160) and cells isolated based on expression of specific markers, such as CD34, Sca1 (Ref. 161) and CD133 (Refs 153, 162). Bone-marrow-derived progenitors, which show migration and engraftment in muscle and modest recruitment to muscle from the circulation (Ref. 163), can be enriched and expanded quickly with specific sets of factors in vitro for therapeutic applications (Ref. 164). The mesoangioblast (Ref. 165) has shown impressive levels of engraftment and functional recovery in a mouse model of limb girdle muscular dystrophy (Ref. 165) and in dystrophic dogs after arterial delivery (Ref. 166). Mesoangioblasts have been suggested to derive from the pericyte, which is a microvessel-associated cell type (Ref. 100). To understand the clinical relevance and limitations of mesoangioblasts or pericytes in muscular dystrophy, it will be crucial to develop reproducible isolation and expansion methods for them and to better characterise their skeletal muscle regenerative capacity.
Conversion of alternative cell types into myogenic precursors has recently become a plausible approach. Promising cell types that can be induced into the myogenic lineage include embryonic stem (ES) cells (Refs 167, 168), embryonic and postnatal-derived fibroblasts (Ref. 169), and induced pluripotent stem (iPS) cells derived from adult fibroblasts (Refs 170, 171). Although ES cells have shown promise in development of cell therapies, some mouse and human ES cell lines might be predisposed to chromosomal abnormalities that could limit their application in cell replacement therapy (Refs 172, 173). iPS cells are generated by lentiviral-mediated expression of a defined set of transcription factors. Once derived, they display characteristics that are remarkably similar to ES cells, including expression of ES cell markers, teratoma formation and the ability to contribute to generation of chimaeric mice following injection into a blastocyst. Use of iPS cells avoids the ethical concerns and risk of immunorejection with human ES cells, and has shown promising therapeutic potential as a pluripotent stem cell source (Ref. 174). Patient-specific iPS cells could be derived from highly accessible and expandable fibroblast populations, but the process of reprogramming and induction into the myogenic lineage must be streamlined for clinical feasibility, and carefully controlled to avoid the formation of tumours.
Future prospects for experimental muscular dystrophy therapies
Despite current limitations, many recent developments point to the promise of viral-vector-mediated gene therapy. Increased understanding of viral transduction mechanisms have allowed generation of hybrid vectors that target desired tissues with high efficiency and long-term expression (Refs 74, 175, 176). We envision that further tailoring with hybrid or chimaeric vectors could lead to more exclusive tissue-specific tropism, thereby improving safety while retaining adequate carrying capacity for most defective genes. In addition, modulation of transgene promoters and enhancers could improve muscle-specific expression for virtually all gene-transfer methods (Ref. 177). Immune responses remain a critical issue, and evasion will be an important aspect of viral-mediated therapies (Ref. 178). A promising approach for DMD treatment entails multisite intravascular delivery of rAAV vectors carrying microutrophin cassettes. Transient immune suppression could be used to block cellular immunity against the rAAV vector, whereas use of utrophin cDNAs could avoid such responses against dystrophin. Development of optimal methods for delivery and immune suppression, as well as design of microutrophin cDNA and transcriptional regulation of the cassette, are active areas of study that hold promise for clinical intervention. Exon skipping is a rapidly developing potential therapy, and further modifications to the approach might make it possible to target a large number of the mutations and deletions observed in DMD and BMD (Ref. 179). However, further testing is necessary to determine optimal chemistries for the safety and feasibility of systemic delivery. Questions also remain as to the functionality of the truncated proteins that would arise from some of these induced splicing events (Ref. 26).
Summary and conclusions
Because ongoing clinical trials are currently focused mostly on safety, intramuscular delivery is the chosen delivery route. Local administration might be of therapeutic benefit to individual muscles or small groups of muscles; however, striated muscle must eventually be targeted systemically for an effective therapy. Combinations of local, regional and systemic routes of administration might also be viable therapeutic options to achieve system-wide targeting of striated muscle. We should consider gene transfer, gene repair, and aspects of regenerative medicine to be complementary approaches – not at all mutually exclusive – that could potentially be most effective in a wide array of muscular dystrophies and other disorders when used in conjunction. Although underestimation of challenges to gene therapy initially led to some disappointing clinical trials, there has been a recent resurgence in interest as new and exciting discoveries in the field have broken though barriers to viral, nonviral and cell-mediated therapies, bringing us a few steps closer to safe and effective treatments for the muscular dystrophies.
Acknowledgments
Funding
We thank members of the Chamberlain lab for helpful discussions and the peer reviewers for their thoughtful comments. Our research is supported by grants from the National Institutes of Health (R37AR040864 and P01NS46788) and the Muscular Dystrophy Association (USA).
References
- 1.Emery AEH, editor. The Muscular Dystrophies. Oxford University Press; Oxford: 2001. [Google Scholar]
- 2.Emery AE, Muntoni F. Duchenne Muscular Dystrophy. 3. Oxford University Press; Oxford: 2003. [Google Scholar]
- 3.Wheeler TM, et al. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. Journal of Clinical Investigation. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tawil R, et al. Facioscapulohumeral dystrophy: A distinct regional myopathy with a novel molecular pathogenesis. Annals of Neurology. 1998;43:279–282. doi: 10.1002/ana.410430303. [DOI] [PubMed] [Google Scholar]
- 5.Bansal D, Campbell KP. Dysferlin and the plasma membrane repair in muscular dystrophy. Trends in Cell Biology. 2004;14:206–213. doi: 10.1016/j.tcb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Davies KE, Nowak KJ. Molecular mechanisms of muscular dystrophies: old and new players. Nature Reviews Molecular Cell Biology. 2006;7:762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- 7.Monaco AP, et al. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 8.Abmayr S, Chamberlain JS. The structure and function of dystrophin. In: Winder SJ, editor. The Molecular Mechanisms in Muscular Dystrophy. Landes Biosciences; Georgetown: 2006. pp. 14–34. [Google Scholar]
- 9.Huang X, et al. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nature Structural and Molecular Biology. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida M, Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. Journal of Biochemistry. 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- 11.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. Journal of Cell Biology. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrof BJ, et al. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner PR, et al. Increased protein degradation results from elevated free calcium levels found in muscle from mdx mice. Nature. 1988;335:735–738. doi: 10.1038/335735a0. [DOI] [PubMed] [Google Scholar]
- 14.Batchelor CL, Winder SJ. Sparks, signals and shock absorbers: how dystrophin loss causes muscular dystrophy. Trends in Cell Biology. 2006;16:198–205. doi: 10.1016/j.tcb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Whitehead NP, et al. Streptomycin reduces stretch-induced membrane permeability in muscles from mdx mice. Neuromuscular Disorders. 2006;16:845–854. doi: 10.1016/j.nmd.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Baumbach LL, et al. Molecular and clinical correlations of deletions leading to Duchenne and Becker muscular dystrophies. Neurology. 1989;39:465–474. doi: 10.1212/wnl.39.4.465. [DOI] [PubMed] [Google Scholar]
- 17.Ozawa E, et al. From dystrophinopathy to sarcoglycanopathy: Evolution of a concept of muscular dystrophy. Muscle & Nerve. 1998;21:421–438. doi: 10.1002/(sici)1097-4598(199804)21:4<421::aid-mus1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 18.Campbell KP. Three muscular dystrophies: Loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 19.Manzur A, et al. Glucocorticoid corticosteroids for Duchenne muscular dystrophy. Cochrane Database of Systematic Reviews. 2008;1:CD003725. doi: 10.1002/14651858.CD14003725. [DOI] [PubMed] [Google Scholar]
- 20.Chamberlain JS, Rando TA, editors. Duchenne Muscular Dystrophy: Advances in Therapeutics. Taylor & Francis Group; New York: 2006. [Google Scholar]
- 21.Banks GB, Chamberlain JS, Krauss RS. Chapter 9, The value of mammalian models for Duchenne muscular dystrophy in developing therapeutic strategies. Current Topics in Developmental Biology. 2008;84:431–453. doi: 10.1016/S0070-2153(08)00609-1. [DOI] [PubMed] [Google Scholar]
- 22.Chamberlain J, Caskey CT. Duchenne Muscular Dystrophy. In: Appel SH, editor. Current Neurology. Vol. 10. Yearbook Medical Publishers; Chicago: 1990. pp. 65–103. [Google Scholar]
- 23.Chamberlain JS. Gene therapy of muscular dystrophy. Human Molecular Genetics. 2002;11:2355–2362. doi: 10.1093/hmg/11.20.2355. [DOI] [PubMed] [Google Scholar]
- 24.England SB. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 25.Matsumura K, et al. Immunohistochemical analysis of dystrophin-associated proteins in Becker/Duchenne muscular dystrophy with huge in-frame deletions in the NH2-terminal and rod domains of dystrophin. Journal of Clinical Investigation. 1994;93:99–105. doi: 10.1172/JCI116989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper SQ, et al. Modular flexibility of dystrophin: Implications for gene therapy of Duchenne muscular dystrophy. Nature Medicine. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 27.Phelps SF, et al. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Human Molecular Genetics. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto M. Micro-dystrophin cDNA ameliorates dystrophic phenotypes when introduced into mdx mice as a transgene. Biochemical and Biophysical Research Communications. 2002;293:1265–1272. doi: 10.1016/S0006-291X(02)00362-5. [DOI] [PubMed] [Google Scholar]
- 29.Wang B, Li J, Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishikawa-Sakurai M, et al. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to {beta}-dystroglycan. Human Molecular Genetics. 2004;13:693–702. doi: 10.1093/hmg/ddh087. [DOI] [PubMed] [Google Scholar]
- 31.Huard J, et al. Human myoblast transplantation between immunohistocompatible donors and recipients produces immune reactions. Transplant Proceedings. 1992;24:3049–3051. [PubMed] [Google Scholar]
- 32.Ohtsuka Y, et al. Dystrophin acts as a transplantation rejection antigen indystrophin-deficient mice: implication for gene therapy. Journal of Immunology. 1998;160:4635–4640. [PubMed] [Google Scholar]
- 33.Gilchrist SC, et al. Immune response to full-length dystrophin delivered to DMD muscle by a high-capacity adenoviral vector. Molecular Therapy. 2002;6:359–368. doi: 10.1006/mthe.2002.0675. [DOI] [PubMed] [Google Scholar]
- 34.Wells DJ, Ferrer A, Wells KE. Immunological hurdles in the path to gene therapy for Duchenne muscular dystrophy. Expert Reviews in Molecular Medicine. 2002;4:1–23. doi: 10.1017/S146239940200515X. [DOI] [PubMed] [Google Scholar]
- 35.Tinsley JM, et al. Primary structure of dystrophin-related protein. Nature. 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 36.Matsumura K, et al. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 37.Winder SJ, et al. Utrophin actin binding domain: analysis of actin binding and cellular targeting. Journal of Cell Science. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen TM. Localization of the DMDL gene-encoded dystrophin-related protein using a panel of nineteen monoclonal antibodies: presence at neuromuscular junctions, in the sarcolemma of dystrophic skeletal muscle, in vascular and other smooth muscles, and in proliferating brain cell lines. Journal of Cell Biology. 1991;115:1695–1700. doi: 10.1083/jcb.115.6.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khurana TS, et al. Immunolocalization and developmental expression of dystrophin related protein in skeletal muscle. Neuromuscular Disorders. 1991;1:185–194. doi: 10.1016/0960-8966(91)90023-l. [DOI] [PubMed] [Google Scholar]
- 40.Mizuno Y, et al. Reciprocal expression of dystrophin and utrophin in muscles of Duchenne muscular dystrophy patients, female DMD-carriers and control subjects. Journal of the Neurological Sciences. 1993;119:43–52. doi: 10.1016/0022-510x(93)90190-a. [DOI] [PubMed] [Google Scholar]
- 41.Weir AP, Morgan JE, Davies KE. A-utrophin up-regulation in mdx skeletal muscle is independent of regeneration. Neuromuscular Disorders. 2004;14:19–23. doi: 10.1016/j.nmd.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Deol JR, et al. Successful compensation for dystrophin deficiency by a helper-dependent adenovirus expressing full-length utrophin. Molecular Therapy. 2007;15:1767–1774. doi: 10.1038/sj.mt.6300260. [DOI] [PubMed] [Google Scholar]
- 43.Tinsley JM. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- 44.Deconinck AE. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- 45.Cerletti M. Dystrophic phenotype of canine X-linked muscular dystrophy is mitigated by adenovirus-mediated utrophin gene transfer. Gene Therapy. 2003;10:750–757. doi: 10.1038/sj.gt.3301941. [DOI] [PubMed] [Google Scholar]
- 46.Odom GL, et al. Microutrophin delivery through rAAV6 increases lifespan and improves muscle function in dystrophic dystrophin/utrophin-deficient mice. Molecular Therapy. 2008;16:1539–1545. doi: 10.1038/mt.2008.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atchison R, Casto B, Hammon W. Adenovirus-associated defective virus particles. Science. 1965;13:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 48.Muzyczka N, Berns KI. Parvoviridae: the viruses and their replication. In: Knipe D, Howley P, editors. Fields Virology. 4. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 2327–2359. [Google Scholar]
- 49.Podsakoff G, Wong KK, Jr, Chatterjee S. Efficient gene transfer into nondividing cells by adeno-associated virus-based vectors. Journal of Virology. 1994;68:5656–5666. doi: 10.1128/jvi.68.9.5656-5666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gregorevic P, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nature Medicine. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao G, et al. Clades of adeno-associated viruses are widely disseminated in human tissues. Journal of Virology. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chao H, et al. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Molecular Therapy. 2000;2:619–623. doi: 10.1006/mthe.2000.0219. [DOI] [PubMed] [Google Scholar]
- 53.Duan D, et al. Enhancement of muscle gene delivery with pseudotyped adeno-associated virus type 5 correlates with myoblast differentiation. Journal of Virology. 2001;75:7662–7671. doi: 10.1128/JVI.75.16.7662-7671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimm D, et al. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood. 2003;102:2412–2419. doi: 10.1182/blood-2003-02-0495. [DOI] [PubMed] [Google Scholar]
- 55.Halbert CL, Allen JM, Miller AD. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. Journal of Virology. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duan D, et al. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. Journal of Virology. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnepp BC, et al. Infectious molecular clones of adeno-associated virus isolated directly from human tissues. Journal of Virology. 2009;83:1456–1464. doi: 10.1128/JVI.01686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chamberlain JR, et al. Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science. 2004;303:1198–1201. doi: 10.1126/science.1088757. [DOI] [PubMed] [Google Scholar]
- 59.Inagaki K, et al. DNA palindromes with a modest arm length of greater than or equal to 20 base pairs are a significant target for recombinant adeno-associated virus vector integration in the liver, muscles, and heart in mice. Journal of Virology. 2007;81:11290–11303. doi: 10.1128/JVI.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herzog RW, et al. Long-term correction of canine hemophilia B by gene transfer of blood coagulation factor IX mediated by adeno-associated viral vector. Nature Medicine. 1999;5:56–63. doi: 10.1038/4743. [DOI] [PubMed] [Google Scholar]
- 61.Manno CS. AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B. Blood. 2003;101:2963–2972. doi: 10.1182/blood-2002-10-3296. [DOI] [PubMed] [Google Scholar]
- 62.Yuasa K. Adeno-associated virus vector-mediated gene transfer into dystrophin-deficient skeletal muscles evokes enhanced immune response against the transgene product. Gene Therapy. 2002;9:1576–1588. doi: 10.1038/sj.gt.3301829. [DOI] [PubMed] [Google Scholar]
- 63.Hartigan-O’Connor D, et al. Immune evasion by muscle-specific gene expression in dystrophic muscle. Molecular Therapy. 2001;4:525–533. doi: 10.1006/mthe.2001.0496. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, et al. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Molecular Therapy. 2007;15:1160–1166. doi: 10.1038/sj.mt.6300161. [DOI] [PubMed] [Google Scholar]
- 65.Manno CS, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nature Medicine. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 66.Mingozzi F, High K. Immune responses to AAV in clinical trials. Current Gene Therapy. 2007;7:316–324. doi: 10.2174/156652307782151425. [DOI] [PubMed] [Google Scholar]
- 67.Mingozzi F, et al. CD8+ T-cell responses to adeno-associated virus capsid in humans. Nature Medicine. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 68.Jiang H, et al. Effects of transient immunosuppression on adeno-associated virus-mediated liver-directed gene transfer in rhesus macaques and implications for human gene therapy. Blood. 2006;108:3321–3328. doi: 10.1182/blood-2006-04-017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Townsend D, et al. Emergent dilated cardiomyopathy caused by targeted repair of dystrophic skeletal muscle. Molecular Therapy. 2008;16:832–835. doi: 10.1038/mt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mah C, et al. Sustained correction of glycogen storage disease type II using adeno-associated virus serotype 1 vectors. Gene Therapy. 2005;12:1405–1409. doi: 10.1038/sj.gt.3302550. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nature Biotechnology. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 72.Inagaki K, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Molecular Therapy. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yue Y, et al. A single intravenous injection of adeno-associated virus serotype-9 leads to whole body skeletal muscle transduction in dogs. Molecular Therapy. 2008;16:1944–1952. doi: 10.1038/mt.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schultz BR, Chamberlain JS. Recombinant adeno-associated virus transduction and integration. Molecular Therapy. 2008;16:1189–1199. doi: 10.1038/mt.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Banks GB, et al. Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain. Human Molecular Genetics. 2007;16:2105–2113. doi: 10.1093/hmg/ddm158. [DOI] [PubMed] [Google Scholar]
- 76.Wells KE, et al. Relocalization of neuronal nitric oxide synthase (nNOS) as a marker for complete restoration of the dystrophin associated protein complex in skeletal muscle. Neuromuscular Disorders. 2003;13:21–31. doi: 10.1016/s0960-8966(02)00191-8. [DOI] [PubMed] [Google Scholar]
- 77.Judge LM, Haraguchi M, Chamberlain JS. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. Journal of Cell Science. 2006;119:1537–1546. doi: 10.1242/jcs.02857. [DOI] [PubMed] [Google Scholar]
- 78.Lai Y, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. Journal of Clinical Investigation. 2009;119:624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomas GD, et al. Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15090–15095. doi: 10.1073/pnas.95.25.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kobayashi YM, et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 2008;456:511–515. doi: 10.1038/nature07414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Banks GB, et al. Molecular and cellular adaptations to chronic myotendinous strain injury in mdx mice expressing a truncated dystrophin. Human Molecular Genetics. 2008;17:3975–3986. doi: 10.1093/hmg/ddn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Banks GB, Chamberlain JS, Froehner SC. Truncated dystrophins can influence neuromuscular synapse structure. Molecular and Cellular Neuroscience. 2009;40:433–441. doi: 10.1016/j.mcn.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun L, Li J, Xiao X. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nature Medicine. 2000;6:599–602. doi: 10.1038/75087. [DOI] [PubMed] [Google Scholar]
- 84.Yan Z, et al. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakai H, Storm TA, Kay MA. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nature Biotechnology. 2000;18:527–532. doi: 10.1038/75390. [DOI] [PubMed] [Google Scholar]
- 86.Duan D, Yue Y, Engelhardt JF. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Molecular Therapy. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- 87.Lai Y, et al. Synthetic intron improves transduction efficiency of trans-splicing adeno-associated viral vectors. Human Gene Therapy. 2006;17:1036–1042. doi: 10.1089/hum.2006.17.ft-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Halbert CL, Allen JM, Miller AD. Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nature Biotechnology. 2002;20:697–701. doi: 10.1038/nbt0702-697. [DOI] [PubMed] [Google Scholar]
- 89.Ghosh A, et al. A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Molecular Therapy. 2007;16:124–130. doi: 10.1038/sj.mt.6300322. [DOI] [PubMed] [Google Scholar]
- 90.Li S, et al. Stable transduction of myogenic cells with lentiviral vectors expressing a minidystrophin. Gene Therapy. 2005;12:1099–1108. doi: 10.1038/sj.gt.3302505. [DOI] [PubMed] [Google Scholar]
- 91.Hacein-Bey-Abina S, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. New England Journal of Medicine. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 92.Ciuffi A, et al. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Molecular Therapy. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 93.Beard BC, et al. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, or foamy virus. Human Gene Therapy. 2007;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- 94.Barquinero J, Eixarch H, Perez-Melgosa M. Retroviral vectors: new applications for an old tool. Gene Therapy. 11:S3–S9. doi: 10.1038/sj.gt.3302363. [DOI] [PubMed] [Google Scholar]
- 95.Kafri T, et al. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nature Genetics. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 96.Bonci D, et al. ‘Advanced’ generation lentiviruses as efficient vectors for cardiomyocyte gene transduction in vitro and in vivo. Gene Therapy. 2003;10:630–636. doi: 10.1038/sj.gt.3301936. [DOI] [PubMed] [Google Scholar]
- 97.Naldini L, et al. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Annoni A, et al. The immune response to lentiviral-delivered transgene is modulated in vivo by transgene-expressing antigen-presenting cells but not by CD4 + CD25+ regulatory Tcells. Blood. 2007;110:1788–1796. doi: 10.1182/blood-2006-11-059873. [DOI] [PubMed] [Google Scholar]
- 99.Bachrach E, et al. Muscle engraftment of myogenic progenitor cells following intraarterial transplantation. Muscle & Nerve. 2006;34:44–52. doi: 10.1002/mus.20560. [DOI] [PubMed] [Google Scholar]
- 100.Dellavalle A, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nature Cell Biology. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 101.Kumar-Singh R, Chamberlain JS. Encapsidated adenovirus minichromosomes allow delivery and expression of a 14 kb dystrophin cDNA to muscle cells. Human Molecular Genetics. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- 102.Clemens P, et al. In vivo muscle gene transfer of full-length dystrophin with an adenoviral vector that lacks all viral genes. Gene Therapy. 1996;3:965–972. [PubMed] [Google Scholar]
- 103.Chen HH, et al. Persistence in muscle of an adenoviral vector that lacks all viral genes. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1645–1650. doi: 10.1073/pnas.94.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hartigan-O’Connor D, et al. Generation and growth of adenoviral vectors. Methods in Enzymology. 2002;346:224–246. doi: 10.1016/s0076-6879(02)46058-2. [DOI] [PubMed] [Google Scholar]
- 105.DelloRusso C, et al. Functional correction of adult mdx mouse muscle using gutted adenoviral vectors expressing full-length dystrophin. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12979–12984. doi: 10.1073/pnas.202300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gilbert R, et al. Prolonged dystrophin expression and functional correction of mdx mouse muscle following gene transfer with a helper-dependent (gutted) adenovirus-encoding murine dystrophin. Human Molecular Genetics. 2003;12:1287–1299. doi: 10.1093/hmg/ddg141. [DOI] [PubMed] [Google Scholar]
- 107.Su LT, et al. Uniform scale-independent gene transfer to striated muscle after transvenular extravasation of vector. Circulation. 2005;112:1780–1788. doi: 10.1161/CIRCULATIONAHA.105.534008. [DOI] [PubMed] [Google Scholar]
- 108.Gao GP, Yang Y, Wilson JM. Biology of adenovirus vectors with E1 and E4 deletions for liver- directed gene therapy. Journal of Virology. 1996;70:8934–8943. doi: 10.1128/jvi.70.12.8934-8943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Acsadi G, et al. A differential efficiency of adenovirus-mediated in vivo gene transfer into skeletal muscle cells of different maturity. Human Molecular Genetics. 1994;3:579–584. doi: 10.1093/hmg/3.4.579. [DOI] [PubMed] [Google Scholar]
- 110.Zoltick PW, et al. Biology of E1-deleted adenovirus vectors in nonhuman primate muscle. Journal of Virology. 2001;75:5222–5229. doi: 10.1128/JVI.75.11.5222-5229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muruve DA, et al. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Human Gene Therapy. 1999;10:965–976. doi: 10.1089/10430349950018364. [DOI] [PubMed] [Google Scholar]
- 112.Brunetti-Pierri N, et al. Acute toxicity after high-dose systemic injection of helper-dependent adenoviral vectors into nonhuman primates. Human Gene Therapy. 2004;15:35–46. doi: 10.1089/10430340460732445. [DOI] [PubMed] [Google Scholar]
- 113.Raper SE, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Molecular Genetics and Metabolism. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 114.Morral N, et al. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12816–12821. doi: 10.1073/pnas.96.22.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wolff J, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 116.Zhang G, et al. Intraarterial delivery of naked plasmid DNA expressing full-length mouse dystrophin in the mdx mouse model of Duchenne muscular dystrophy. Human Gene Therapy. 2004;15:770–782. doi: 10.1089/1043034041648408. [DOI] [PubMed] [Google Scholar]
- 117.Hagstrom JE, et al. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Molecular Therapy. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 118.Richard P, et al. Amphiphilic block copolymers promote gene delivery in vivo to pathological skeletal muscles. Human Gene Therapy. 2005;16:1318–1324. doi: 10.1089/hum.2005.16.1318. [DOI] [PubMed] [Google Scholar]
- 119.Chollet P, et al. Side-effects of a systemic injection of linear polyethylenimine-DNA complexes. The Journal of Gene Medicine. 2002;4:84–91. doi: 10.1002/jgm.237. [DOI] [PubMed] [Google Scholar]
- 120.Trubetskoy VS, et al. Recharging cationic DNA complexes with highly charged polyanions for in vitro and in vivo gene delivery. Gene Therapy. 2003;10:261–271. doi: 10.1038/sj.gt.3301888. [DOI] [PubMed] [Google Scholar]
- 121.Romero NB, et al. Phase I study of dystrophin plasmid-based gene therapy in Duchenne/Becker muscular dystrophy. Human Gene Therapy. 2004;15:1065–1076. doi: 10.1089/hum.2004.15.1065. [DOI] [PubMed] [Google Scholar]
- 122.Sherratt TG, et al. Exon skipping and translation in patients with frameshift deletions in the dystrophin gene. American Journal of Human Genetics. 1993;53:1007–1015. [PMC free article] [PubMed] [Google Scholar]
- 123.Partridge T, Lu Q-L. The enigma of the ‘dystrophin revertant’ muscle fibre. In: Tsuchida K, Takeda S, editors. Recent advances in skeletal muscle differentiation. Research Signpost; Trivandrum: 2008. pp. 93–107. [Google Scholar]
- 124.Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takeshima Y, et al. Modulation of in vitro splicing of the upstream intron by modifying an intra-exon sequence which is deleted from the dystrophin gene in dystrophin Kobe. Journal of Clinical Investigation. 1995;95:515–520. doi: 10.1172/JCI117693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pramono ZA. Induction of exon skipping of the dystrophin transcript in lymphoblastoid cells by transfecting an antisense oligodeoxynucleotide complementary to an exon recognition sequence. Biochemical and Biophysical Research Communications. 1996;226:445–449. doi: 10.1006/bbrc.1996.1375. [DOI] [PubMed] [Google Scholar]
- 127.Shibahara S, et al. Inhibition of human immunodeficiency virus (HIV-1) replication by synthetic oligo-RNA derivatives. Nucleic Acids Research. 1989;17:239–252. doi: 10.1093/nar/17.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dunckley MG, et al. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Human Molecular Genetics. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- 129.McClorey G, et al. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Therapy. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- 130.van Deutekom JC. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Human Molecular Genetics. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 131.Mann CJ, et al. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense and Nucleic Acid Drug Development. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 133.Yokota T, et al. A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground. Archives of Neurology. 2009;66:32–38. doi: 10.1001/archneurol.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sazani P, et al. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nature Biotechnology. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 135.Lebleu B, et al. Cell penetrating peptide conjugates of steric block oligonucleotides. Advanced Drug Delivery Reviews. 2008;60:517–529. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu B, et al. Octa-guanidine morpholino restores dystrophin expression in cardiac and skeletal muscles and ameliorates pathology in dystrophic mdx mice. Molecular Therapy. 2009;17:864–871. doi: 10.1038/mt.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ivanova GD, et al. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Research. 2008;36:6418–6428. doi: 10.1093/nar/gkn671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Alter J, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nature Medicine. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 139.Fletcher S, et al. Morpholino oligomer-mediated exon skipping averts the onset of dystrophic pathology in the mdx mouse. Molecular Therapy. 2007;15:1587–1592. doi: 10.1038/sj.mt.6300245. [DOI] [PubMed] [Google Scholar]
- 140.Lu QL, et al. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jearawiriyapaisarn N, et al. Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice. Molecular Therapy. 2008;16:1624–1629. doi: 10.1038/mt.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wu B, et al. Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14814–14819. doi: 10.1073/pnas.0805676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Popplewell LJ, et al. Design of phosphorodiamidate morpholino oligomers (PMOs) for the induction of exon skipping of the human DMD gene. Molecular Therapy. 2009;17:554–561. doi: 10.1038/mt.2008.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mitrpant C, et al. By-passing the nonsense mutation in the 4 (CV) mouse model of muscular dystrophy by induced exon skipping. The Journal of Gene Medicine. 2009;11:46–56. doi: 10.1002/jgm.1265. [DOI] [PubMed] [Google Scholar]
- 145.De Angelis FG. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in [delta] 48–50 DMD cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goyenvalle A, et al. Rescue of Dystrophic Muscle Through U7 snRNA-Mediated Exon Skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 147.Denti MA, et al. Chimeric adeno-associated virus/antisense U1 small nuclear RNA effectively rescues dystrophin synthesis and muscle function by local treatment of mdx mice. Human Gene Therapy. 2006;17:565–574. doi: 10.1089/hum.2006.17.565. [DOI] [PubMed] [Google Scholar]
- 148.Heemskerk HA, et al. In vivo comparison of 2’-O-methyl phosphorothioate and morpholino antisense oligonucleotides for Duchenne muscular dystrophy exon skipping. The Journal of Gene Medicine. 2009;11:257–266. doi: 10.1002/jgm.1288. [DOI] [PubMed] [Google Scholar]
- 149.Skuk D, Tremblay JP. Myoblast transplantation: the current status of a potential therapeutic tool for myopathies. Journal of Muscle Research and Cell Motility. 2003;24:285–300. [PubMed] [Google Scholar]
- 150.Skuk D, et al. First test of a “high-density injection” protocol for myogenic cell transplantation throughout large volumes of muscles in a Duchenne muscular dystrophy patient: eighteen months follow-up. Neuromuscular Disorders. 2007;17:38–46. doi: 10.1016/j.nmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 151.Skuk D, et al. Ischemic central necrosis in pockets of transplanted myoblasts in nonhuman primates: implications for cell-transplantation strategies. Transplantation. 2007;84:1307–1315. doi: 10.1097/01.tp.0000288322.94252.22. [DOI] [PubMed] [Google Scholar]
- 152.Montarras D, et al. Direct Isolation of Satellite Cells for Skeletal Muscle Regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 153.Peault B, et al. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Molecular Therapy. 2007;15:867–877. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 154.Webster C, Blau H. Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: implications for cell and gene therapy. Somatic Cell and Molecular Genetics. 1990;16:557–565. doi: 10.1007/BF01233096. [DOI] [PubMed] [Google Scholar]
- 155.Fan Y, et al. Rapid death of injected myoblasts in myoblast transfer therapy. Muscle & Nerve. 1996;19:853–860. doi: 10.1002/(SICI)1097-4598(199607)19:7<853::AID-MUS7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 156.Tremblay JP, Skuk D. Another new “super muscle stem cell” leaves unaddressed the real problems of cell therapy for Duchenne muscular dystrophy. Molecular Therapy. 2008;16:1907–1909. doi: 10.1038/mt.2008.243. [DOI] [PubMed] [Google Scholar]
- 157.Deasy BM, et al. Mechanisms of muscle stem cell expansion with cytokines. Stem Cells. 2002;20:50–60. doi: 10.1634/stemcells.20-1-50. [DOI] [PubMed] [Google Scholar]
- 158.Collins CA, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 159.Cornelison DDW. Context matters: In vivo and in vitro influences on muscle satellite cell activity. Journal of Cellular Biochemistry. 2008;105:663–669. doi: 10.1002/jcb.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gussoni E, et al. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 161.Lee JY, et al. Clonal isolation of muscle-derived cells capable of enhancing muscle regeneration and bone healing. Journal of Cell Biology. 2000;150:1085–1100. doi: 10.1083/jcb.150.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Torrente Y, et al. Autologous transplantation of muscle-derived CD133+ stem cells in Duchenne muscle patients. Cell Transplantation. 2007;16:563–577. doi: 10.3727/000000007783465064. [DOI] [PubMed] [Google Scholar]
- 163.Ferrari G, et al. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 164.Dezawa M, et al. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science. 2005;309:314–317. doi: 10.1126/science.1110364. [DOI] [PubMed] [Google Scholar]
- 165.Sampaolesi M, et al. Cell therapy of {alpha}-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- 166.Sampaolesi M, et al. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- 167.Barberi T, et al. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nature Medicine. 2007;13:642–648. doi: 10.1038/nm1533. [DOI] [PubMed] [Google Scholar]
- 168.Darabi R, et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nature Medicine. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- 169.Kimura E, et al. Cell-lineage regulated myogenesis for dystrophin replacement: a novel therapeutic approach for treatment of muscular dystrophy. Human Molecular Genetics. 2008;17:2507–2517. doi: 10.1093/hmg/ddn151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 171.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 172.Rebuzzini P, et al. Chromosome number variation in three mouse embryonic stem cell lines during culture. Cytotechnology. 2008;58:17–23. doi: 10.1007/s10616-008-9164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Catalina P, et al. Human ESCs predisposition to karyotypic instability: Is a matter of culture adaptation or differential vulnerability among hESC lines due to inherent properties? Molecular Cancer. 2008;7:76. doi: 10.1186/1476-4598-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 175.Dickson G, et al. Recombinant micro-genes and dystrophin viral vectors. Neuromuscular Disorders. 2002;12:S40–S44. doi: 10.1016/s0960-8966(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 176.Goncalves MAFV, et al. Targeted chromosomal insertion of large DNA into the human genome by a fiber-modified high-capacity adenovirus-based vector system. PLoS ONE. 2008;3:3084. doi: 10.1371/journal.pone.0003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Salva MZ, et al. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Molecular Therapy. 2007;15:320–329. doi: 10.1038/sj.mt.6300027. [DOI] [PubMed] [Google Scholar]
- 178.Zaldumbide A, Hoeben RC. How not to be seen: immune-evasion strategies in gene therapy. Gene Therapy. 2007;15:239–246. doi: 10.1038/sj.gt.3303082. [DOI] [PubMed] [Google Scholar]
- 179.van Deutekom JC, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. New England Journal of Medicine. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 180.Newey SE, et al. Alternative splicing of dystrobrevin regulates the stoichiometry of syntrophin binding to the dystrophin protein complex. Current Biology. 2000;10:1295–1298. doi: 10.1016/s0960-9822(00)00760-0. [DOI] [PubMed] [Google Scholar]
- 181.Adams ME, Mueller HA, Froehner SC. In vivo requirement of the {alpha}-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. Journal of Cell Biology. 2001;155:113–122. doi: 10.1083/jcb.200106158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gee SH, et al. Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. Journal of Neuroscience. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading, resources and contacts
Publications
- Blake DJ, et al. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiological Reviews. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. A comprehensive review on dystrophin and dystrophin-associated proteins and their involvement in the pathophysiology of DMD. [DOI] [PubMed] [Google Scholar]
- Abmayr S, Chamberlain JS. The structure and function of dystrophin. In: Winder SJ, editor. The Molecular Mechanisms in Muscular Dystrophy. Landes Biosciences; Georgetown: 2006. This book chapter provides a detailed discussion of dystrophin domain structures in the context of DMD. [Google Scholar]
- Chamberlain JS, Rando TA. Duchenne muscular dystrophy: advances in therapeutics. Taylor & Francis Group; New York: 2006. A comprehensive book covering the molecular etiology of DMD, clinical management, and experimental therapies. [Google Scholar]
- Odom GL, Gregorevic P, Chamberlain JS. Viral-mediated gene therapy for the muscular dystrophies: Successes, limitations and recent advances. Biochimica et Biophysica Acta (BBA) -Molecular Basis of Disease. 2007;1772:243–262. doi: 10.1016/j.bbadis.2006.09.007. A detailed review on viral vectors in gene therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- Online Mendelian Inheritance in Man (OMIM) information on the dystrophin gene (DMD), available from the National Center for Biotechnology Information (NCBI), no. 300377: http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=300377
- The following organisations provide information to patients and researchers on neuromuscular disorders: The Muscular Dystrophy Association (MDA-USA) http://www.mda.org/
- The TREAT-NMD Neuromuscular Network. http://www.treat-nmd.eu/home.php.
- The Leiden Muscular Dystrophy pages. http://www.dmd.nl/
- The American Society for Gene Therapy (ASGT) provides information for researchers regarding development and clinical applications of gene therapy: http://www.asgt.org/
- Ongoing and recently completed trials for the muscular dystrophies relevant to discussed approaches can be accessed via the clinical trials website and searching for the national clinical trial (NCT) identifiers at: http://clinicaltrials.gov.
- rAAV-mediated gene transfer: 2006 Phase 1 Clinical Trial of rAAV2.5-CMV-Mini-Dystrophin Gene Vector in Duchenne Muscular Dystrophy (NCT00428935); 2008 Phase I Gene Transfer of rAAV1.tMCK.Human-Alpha-Sarcoglycan for Limb Girdle Muscular Dystrophy Type 2D (LGMD2D) (NCT00494195).
- Exonskipping:2007RestoringDystrophinExpressioninDuchenneMuscularDystrophy:APhaseI/IIClinicalTrialUsing AVI-4658 (NCT00159250); 2009 Clinical Study to Assess the Safety of AVI-4658 in Subjects With Duchenne Muscular Dystrophy Due to a Frame-Shift Mutation Amenable to Correction by Skipping Exon 51 (NCT00844597).