Abstract
TLR3 has been implicated in the pathogenesis of several viral infections, including SIV- and HIV-1-induced inflammation and AIDS. However the molecular mechanisms of these TLR3-mediated effects are not known, and it is not known whether HIV interacts with cellular TLR3 to affect disease process. Here we investigate the effects of TLR3 ligands on HIV-1 transactivation using both primary human macrophages and cells containing integrated copies of the HIV-1 promoter. We demonstrate that TLR3 activation induced upregulation of transcription factors such as c-Jun, CCAAT/enhancer-binding protein alpha (CEBPA), signal transducer and activator of transcription (STAT)-1, STAT-2, RELB, and nuclear factor kappa-B1 (NFκB1), most of which are known to regulate the HIV promoter activity. We also demonstrate that TLR3 activation increased HIV-1 transactivation via the c-Jun N-terminal kinase (JNK) and NFκB pathways. This was associated with epigenetics modifications, including decreased histone deacetylase activity, increased histone acetyl transferase (HAT) activity, and increased acetylation of histones H3 and H4 at lysine residues in the nucleosome-0 and nucleosome-1 of the HIV-1 promoter. However, prolonged TLR3 activation decreased HIV-1 transactivation, decreased HAT activity and Tat transcription, and suppressed viral replication. Overall, data suggests TLR3 can acts as viral sensor to mediate viral transactivation, cellular signaling, innate immune response, and inflammation in HIV-infected humans. Our study provides novel insights into the molecular basis for these TLR3-mediated effects.
Keywords: TLR3, HIV, viral transactivation, LTR, NFκB, JNK, STAT1, Histone acetylation, Epigenetics, Human monocyte-derived macrophages, TZM-bl, U38 cells
1. Introduction
Toll-like receptors (TLRs) are pattern-recognition receptors that detect invading pathogens via their pathogen associated molecular patterns (PAMP), and the resulting TLR/PAMP interaction triggers the innate immune system [1, 2]. Thus, TLRs play a major role in innate immunity. Thirteen different TLRs have been discovered so far, of which 11 (TLR1 to TLR11) are expressed in human cells [3, 4]. TLR1, 2, 4, 5, 6, and 10 are mostly expressed on the outer cell membranes, whereas TLR3, 7, 8, and 9 are mostly expressed on endosomal membranes [3, 4]. However, the localization of TLRs vary amongst cell types; in many cells TLR3 is located intracellularly on endosomes, but in human astrocytes TLR3 is expressed both intracellularly and on the cell surface [5]. TLR3 can be activated by viral double-stranded RNA (dsRNA) [6, 7], viral single-stranded RNA (ssRNA) [8], endogenous viral mRNA [9], and polyinosinic-polycytidylic acid (PIC), a synthetic analogue of viral dsRNA [10]. It has been demonstrated that viral dsRNA is a replication intermediate of several ssRNA viruses and that dsRNA are produced during replication of ssRNA viruses, such as the respiratory syncytial virus, encephalomyocarditis virus, and West Nile virus, and activate TLR3 [11–13].
Ligand-induced TLR activation can be beneficial since the TLR-ligand complex initiates a quick and protective innate immune response against invading pathogens [1, 2]. However, it can also be detrimental because uncontrolled and unregulated TLR activation can result in excessive inflammation, cell injury, and tissue damage, including damage to CNS tissues [14–16]. TLR3 ligands increase the expression of inflammatory chemokines in macaques and human macrophages [17, 18] and human muscle cells [19]. TLR3 has been implicated in the pathogenesis of several viral infections, including simian immunodeficiency virus (SIV) and HIV/AIDS. Following SIV infection of macaques, there is increased TLR3 expression in lymph nodes and this is associated with increased expression of inflammatory chemokines and interferons [20].
There is evidence that TLR3 ligands inhibit SIV replication in macaque cells [17, 20], inhibit HIV-1 replication in macrophages and microglia [18, 21, 22], and culture supernatant from an endothelial cell line treated with PIC also suppressed HIV-1 replication [23]. However it has been shown that TLR3 stimulates the SIV promoter [20]; and humans with chronic, untreated HIV-1 infections showed increased TLRs expression in their peripheral blood mononuclear cells (PBMC), including increased TLR2, TRL3, TLR4, TLR6, TLR7, and TLR8 [24]. Larger increases in TLR3 levels were observed in PBMC from AIDS patients and this correlated with higher plasma viral load [24]. It is not known what are the mechanisms through which TLR3 activation decrease HIV-1 infection, whereas high viral load in AIDS patients correlate with increased TLR3 expression. In the present study, we demonstrate, using TZM-bl and monocytic (U38) cells containing the HIV-1 promoter, as well as primary human macrophages, that TLR3 activation induced upregulation of transcription factors such as c-Jun, CCAAT/enhancer-binding protein alpha (CEBPA), signal transducer and activator of transcription (STAT)-1, STAT-2, RELB, and nuclear factor kappa-B (NFκB1), most of which are known to regulate the HIV promoter activity [25–27]. We also demonstrate that TLR3 activation increased HIV-1 transactivation via the c-Jun N-terminal kinases (JNK) and NFκB pathways, and this was associated with increased LTR and Tat expression, and epigenetics modifications. However, prolonged TLR3 activation decreased HIV-1 transactivation, decreased transcription of HIV-1 Tat and LTR, and decreased viral replication.
2. Materials and Methods
2.1. PIC and TZM-bl cells
Lyophilized PIC (Catalog #31852-29-6) was purchased from Invivogen (San Diego, CA), reconstituted in endotoxin-free physiological water provided by the manufacturer, incubated for 10 minutes (min) at 65 to 70°C, and 1 hour (h) at room temperature to enable optimal annealing. PIC aliquots were stored at −20°C. TZM-bl cells were obtained from the NIH AIDS Research and Reference Program. These cells stably express large amounts of CD4 and CCR5 and contain integrated copies of the luciferase and β-galactosidase genes under the control of the HIV-1 promoter. This sensitive indicator cell line was used to measure the HIV-1 LTR promoter activity following PIC treatment. TZM-bl cells were cultured in Dulbecco’s Modified Eagles Media (DMEM) containing 10% fetal bovine serum (FBS), 100 units penicillin, and 0.1 mg/ml streptomycin. Media was changed every 2 days and cells were passaged twice every week. For treatment with PIC or pharmacological inhibitors, confluent cells were maintained in DMEM containing 1% FBS, 100 units penicillin, and 0.1 mg/ml streptomycin.
2.2. U38 cells
Monocytic U38 cells (a derivative of U937 cells that contains stably integrated, silent copies of the HIV-1 LTR promoter linked to the chloramphenicol acetyltransferase (CAT) gene) were obtained from the NIH AIDS Research and Reference Program. This sensitive indicator cell line was used to measure the HIV-1 LTR CAT activity in response to treatment with the TLR3 ligand PIC. Cells were cultured in RPMI 1640 media (Life Technologies) containing 10% FBS and 1% penicillin-streptomycin. Media was changed every 2 days and cells were passaged twice every week. For treatment with PIC or pharmacological inhibitors, cells were maintained in RPMI 1640 media containing 1% FBS and 1% penicillin-streptomycin.
2.3. Luciferase assay
The luciferase reporter assay was performed using the Luciferase Assay System (Promega, Madison, WI) according to the manufacturer’s protocol. Briefly, TZM-bl cells were treated with PIC (1, 5, 10, 15, 25, or 50 μg/ml), with or without pharmacological inhibitors (Table 1) for 48 h, or treated with PIC (50 μg/ml) for 2, 4, or 6 days. Each experimental condition was tested in triplicate. Following treatment, cells were harvested, washed with PBS, and lysed using the lysis reagent provided with the Luciferase Assay Reporter System. Cell lysates (20 μl containing 50 μg proteins) were mixed with 100 μl Luciferase Assay Reagent and luciferase activity was measured using SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA).
Table 1.
Pharmacological inhibitors used in the study
| Inhibitor name | Catalog#/ Abbreviation | Mechanism of action | Manufacturer |
|---|---|---|---|
| TLR3/dsRNA Complex Inhibitor | TLR3.CI | Acts as a direct, competitive and high affinity inhibitor of dsRNA binding to TLR3. Selectively antagonizes TLR3 signaling without affecting other TLRs. | Calbiochem-EMD Millipore |
| NFκB Activation Inhibitor | 481406 | Inhibitor of NFκB transcriptional activation | Calbiochem-EMD Millipore |
| SR 11302 | SR11302 | Inhibitor of AP-1 transcriptional activity | Tocris Biosciences |
| JNK Inhibitor II | 420119 | Selective and reversible inhibitor of JNK1, JNK2, and JNK3. | Calbiochem-EMD Millipore |
| JNK Inhibitor V | 420129 | Reversible and ATP-competitive inhibitor of JNK1, JNK2, and JNK3. | Calbiochem-EMD Millipore |
| JNK Inhibitor III | 420130 | Specifically disrupts c-Jun/JNK complex formation and subsequent phosphorylation and activation of c-Jun by JNK | Calbiochem-EMD Millipore |
| IRAK-1/4 inhibitor | IRAK-1/4 | Inhibitor of IRAK-1 and IRAK-4 | Sigma-Aldrich |
| 5Z-7-Oxozeaenol | 5ZO | ATP-competitive irreversible inhibitor of TAK1 (MEKK7), MKK7, MEK1, and ERK2. Has no activity against other MAPKs | Sigma-Aldrich |
Abbreviations: TLR3: Toll-like receptor-3; JNK: c-Jun N-terminal kinase; dsRNA: double-stranded RNA; AP-1: activator protein-1; IRAK: Interleukin-1 Receptor-Associated Kinase; NFκB: nuclear factor kappaB; TAK1: Transforming growth factor β activated kinase-1; MEKK7: MAP Kinase Kinase Kinase-7; MKK7: MAP Kinase Kinase-7; MEK1: MAP Kinase Kinase-1; ERK2: extracellular-signal-regulated kinase-2 / MAP Kinase-1
2.4. CAT assay
For the CAT assay, U38 cells were treated with PIC (1, 5, 10, 15, 25, or 50 μg/ml), with or without pharmacological inhibitors (Table 1) for 48 h, or treated with PIC (50 μg/ml) for 2, 4, or 6 days. Each experimental condition was tested in triplicate. Following treatment, cells were washed with PBS, lysed using the lysis buffer provided with the CAT ELISA kit (Roche Diagnostics Indianapolis, IN), and protein levels in cell lysates quantified using the bicinchoninic acid assay (BCA) assay as previously described [28, 29]. The amount of CAT enzyme in each sample was quantified using the CAT-ELISA kit (Roche) according to the manufacturer’s protocol. Briefly, 150 μg of each protein lysate in a 200 μl total volume were added to wells of a microplate module (in triplicate), incubated at 37 °C for 1 h, washed 5 times using the washing buffer provided with kit, incubated at 37 °C for 1 h with 200 μl of Anti-CAT-digoxigenin antibody, washed again 5 times, incubated at 37 °C for 1 h with 200 μl of anti-digoxigenin-peroxidase, and washed 5 times. The reactions were developed by incubating each microplate module for 10 to 40 min with a peroxidase substrate, and the samples’ absorbance measured at 405 nm in a SpectraMax M5 microplate reader. CAT enzymes standards were used to generate a calibration curve; and this curve was used to quantify CAT concentrations in each sample.
2.5. Pharmacological inhibitors
The IRAK-1/4 inhibitor (#I5409) and the MKK7 inhibitor 5Z-7-Oxozeaenol (#O9890) were purchased from Sigma (St. Louis, MO), and all other inhibitors were purchased from Calbiochem-EMD Millipore (Danvers, MA). For all experiments using pharmacological inhibitors, cells were treated with the inhibitor for 30 min, followed by treatment with PIC for up to 48 h in the presence of the inhibitor. Controls included untreated cells and cells treated only with inhibitor (without PIC). The concentrations of each inhibitor were based on their manufacturer recommended IC50. The TLR3/dsRNA Complex Inhibitor (TLR3.CI, EMD#614310), a thiophenecarboxamidopropionate compound that acts as a direct, competitive and high affinity inhibitor of dsRNA binding to TLR3, was used at concentrations of 5, 10, 20, 30, 40, 50, and 100 μM; the NFκB inhibitor (EMD#481406) at 20 nM and 30 nM, the AP-1 inhibitor (EMD#SR11302) at 2 μM and 10 μM, JNK inhibitor II (EMD#420119) at 10 μM and 20 μM, JNK Inhibitor III (EMD#420130) at 5 μM and 10 μM, JNK inhibitor V (EMD#420129) at 10 μM and 20 μM. The IRAK-1/4 and MKK7 inhibitors were used at 5 μM and 10 μM. The manufacturer’s names, catalog numbers, and mechanisms of action of each inhibitor are summarized in Table-1.
2.6. RNA extraction
Following treatment, cells were harvested and total RNA was extracted using the Trizol reagent (Life Technologies-Ambion, Austin, TX) according to the manufacturer’s protocol. The RNA was further cleaned using Total RNA cleanup kit (Qiagen, Valencia, CA); RNA yield and quality were checked using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and for all samples absorbance ratio of 260/280 was ≥2.
2.7. TaqMan transcription factor array
Confluent TZM-bl cells in 6-well plates were treated for 48 h with 50 μg/ml high molecular weight PIC (Invivogen), harvested and RNA extracted as described above; controls consisted of RNA from untreated cells. Gene array experiments were performed using the TaqMan Human Transcription Factors array (non-HOX), 96-well plate format (PN 4418784, Applied Biosystems, Foster City, CA), which contained 92 transcription factors-associated genes and 4 endogenous control genes. Gene array experiments were performed according to the manufacturer’s instructions. Briefly, cDNA was generated using 1 μg total RNA, oligo(dT) primers, and Verso cDNA kit (Thermo Fisher Scientific, Pittsburgh, PA), according to the manufacturer’s protocol. The combined mixture of cDNA (5 μl containing 50 ng cDNA) and Taqman Fast Universal PCR Master Mix (5 μl) were added to each well. Gene amplification was quantified in real-time using StepOnePlus Real-Time PCR Systems (Applied Biosystems) and the Applied Biosystems StepOne software v2.0. Cycling conditions were as follows: 50 °C for 2 min and 95 °C for 20 seconds (sec), followed by 40 cycles of 95 °C, 3 sec and 60 °C, 30 sec. Data was analyzed using the delta-delta Ct method; untreated controls and GAPDH were used as endogenous controls to generate a fold change for each gene. Genes with a minimum of 1.5 fold change were considered for further analysis.
2.8. Real-time PCR
Each experimental condition was tested in triplicate and for each replicate sample, cDNA was generated from 1 μg RNA in a 20 μl reaction volume, using the Verso cDNA kit (Thermo Fisher), according to the manufacturer’s instructions. Reverse transcription was carried out for 30 min at 42 °C. The cDNA obtained was diluted at a 1:10 ratio in nuclease free water; a 10 μl reaction mixture containing 2 μl of each diluted cDNA sample, 5μl master mix (containing the polymerase enzyme, dNTPs and MgCl2), 0.5μl primer probe mixture [containing 900 nM of each forward and reverse primer and 250 nM Taqman minor groove binder (MGB) probe], and 2.5 μl nuclease free water was used for quantitative real-time PCR using StepOnePlus™ (Applied Biosystems) or LightCycler® 480 II (Roche) Real-Time PCR Systems. GAPDH real-time PCR was used as internal control, at the same primer-probe ratio as the target genes (900 nM of each primer and 250 nM Taqman MGB probe); and each sample was tested in triplicate, using the software monocolor hydrolysis probe universal cycling conditions (95 °C for 10 min, followed by 45 cycles of 95 °C for 10 sec, 60 °C for 30 sec, and 72 °C for 1 sec). Gene expression levels were quantified using the cycle threshold (CT) method as described in the software user manual. The mean CT values for both target and endogenous control gene (GAPDH) were determined, and fold change in each target gene was normalized to the sample’s GAPDH or to control samples. All PCR reagents, primers, and probes were obtained from Applied Biosystems; and primers’ IDs were as follows: CEBGA (Hs00269972_s1), CEBPG (Hs01922818_s1), JUN (Hs99999141_s1), STAT1 (Hs00234829_m1), RELB (Hs00232399_m1), IL-6 (Hs00985639), and GAPDH (Hs99999905_m1). For HIV-1 LTR and Tat real-time PCR, primers and probes were custom ordered from Applied Biosystems, and their sequences were as follows: LTR forward primer: 5′-GCCTCAATAAAGCTTGCCTTGA-3′; LTR reverse primer: 5′-TCCACACTGACTAAAAGGGTCTGA-3′; LTR probe: 5′-FAM-GCGAGTGCCCGTCTGTTGTGTGACTCTGGTAACTAGCTCGC-MGB-3′. Tat forward primer-5′-GGAGGAGGGTTGCTTTGATAGAG-3′; Tat reverse primer-5′-AAAGCCTTAGGCATCTCCTATGG-3′; Tat probe: 5′-FAM-CTTCGTCGCTGTCTCCGCTTCTTCC-MGB-3′.
2.9. Monocyte-derived macrophages
Human monocytes were obtained from HIV-1-, HIV-2-, and hepatitis B-seronegative donor leukopak and separated by countercurrent centrifugal elutriation as previously described [30]. To obtain monocyte-derived macrophages (MDM), freshly elutriated monocytes (2 million cells per well in 6-well plates) were cultured for 7 days in DMEM containing 2 mM L-glutamine (Invitrogen), 10% heat-inactivated human serum, 100 μg/ml gentamicin, and 10 μg/ml ciprofloxacin (Sigma) in the presence of 1,000 U/ml macrophage colony-stimulating factor (MCSF) as we have previously described [30]. All reagents were prescreened for endotoxin (<10 pg/ml, Associates of Cape Cod, Woods Hole, MA) and mycoplasma contamination (Gen-probe II, Gen-probe, San Diego, CA).
2.10. HIV-1 infection of MDM
The HIV-1ADA strain used, a clade-B, M-tropic viral isolate, was propagated in MDM or in phytohemagglutinin-stimulated human PBMC in T-75 flasks as previously described [31, 32] and titrated using previously published procedures [31, 32]. For experiments assessing the effects of HIV-1 infection and PIC on MDM HAT activity, Tat, and LTR transcription, MDM were, with or without PIC (10, 25, or 50 μg/ml), cultured for 2 h (the 2 h time point) or 4 h (all other time points) in media without MSCF containing HIV-1ADA, at a multiplicity of infection (MOI) of 0.01. Each experimental condition was tested in triplicate; after viral exposure cells were washed 3 times with serum-free media to remove free virions, harvested (2 h and 4 h time points) or cultured again for up to 120 h in media with or without PIC (10, 25, or 50 μg/ml), and harvested by centrifugation. For the 120 h time point, media was changed (replaced with fresh media) once at 48 h. For experiments assessing HAT activity, nuclear proteins were purified from MDM and the HAT activity in each sample was quantified as described section 2.12. For experiments assessing IL-6 and transcription factors mRNA levels, as well as Tat and LTR transcription, infected and non-infected MDM, without or with PIC (10, 25, and 50 μg/ml) treatment (continuous), were harvested at 2 to 120 h p.i. RNA extracted as described in section 2.6, and real-time PCR performed as described in section 2.8.
For experiments quantifying viral reverse transcriptase (RT) activity, MDM (in triplicate) were cultured for 4 h in media without MCSF containing HIV-1ADA, at MOI of 0.01. After 4 h viral exposure, cells were washed 3 times with serum-free media to remove free virions and cultured for up to 18 days; the culture media was changed (replaced with fresh media) every 2 or 3 days. Similarly, media from each sample was collected every 2 or 3 days, and cryopreserved at −80 °C for analysis. For experiments testing the effects of LTR3 ligands, MDM were treated with 10, 25, or 50 μg/ml PIC for 30 min prior to viral exposure and further cultured with or without PIC in the media: for temporary treatment with TLR3 ligands (T), after the 4 h viral exposure in the presence of PIC, MDM were washed 3 times with serum-free media and cultured for up to 18 days in media without PIC. For continuous treatment with TLR3 ligands (C), MDM were exposed to HIV-1 for 4 h in the presence of PIC, washed as above, and cultured for up to 18 days in media containing PIC (10, 25, or 50 μg/ml). To determine the levels of productive viral replication, 10 μl of each supernatant sample, collected at day-5, day-7, day-9, day-12, day-15, and day-18 post-infection (p.i.), was used in a RT assay to quantify RT enzyme levels. The RT assay method has been described in detail in previous publications [31, 33].
2.11. Protein extraction and Western blot analysis
Protein extraction, quantification, and western blot analysis were performed as previously described [28, 29]. Briefly, TZM-bl or U38 cells were stimulated with PIC (50 μg/ml) for 5 min to 2 h and cells were lysed using the mammalian cell lysis buffer CelLytic M (Sigma). For histone proteins Western blot experiments, U38 cells were treated with PIC (10 to 75 μg/ml) for 48 h, lysed, and nuclear proteins extracted using Epigentek nuclear extraction kit (Epigentek, Farmingdale, NY). Protein levels in each sample was quantified using the BCA assay as previously described [28, 29]. Each protein sample (35 μg) was fractionated in a 10% sodium dodecyl sulfate-polyacrylamide (SDS) gel for analysis with JNK, phopho-JNK, c-Jun, and phospho-c-Jun antibodies, or in a 4–20% gradient SDS gel for analysis with histone and acetyl-histone antibodies. Proteins on SDS gels were transferred onto nitrocellulose membranes, blocked for 1 h with SuperBlock T-20 (Pierce, Rockford, IL), and blotted 2 h or overnight with monoclonal antibodies to JNK, phopho-JNK, c-Jun, phospho-c-Jun, Histone H3, Histone H4, acetyl Histone H3 (Lys9), and acetyl Histone H4 (Lys8) (Cell Signaling Technology, Danvers, MA) at 1:1,000 dilution. Membranes were then washed, blotted 1 h with horseradish peroxidase-conjugated secondary antibody, washed again, and visualized using the enhanced chemiluminescence system (Pierce) and gel doc instrument (Syngene, Frederick, MD). After each western blot experiment, the membranes were stripped using Restore Western Blot Stripping Buffer (Pierce) and re-blotted with β-actin antibody (Abcam, Cambridge, MA) to confirm equal loading. Densitometry analysis was performed using the GeneTools gel analysis software (Syngene).
2.12. Histone acetyltransferase (HAT) assay
U38 and TZM-bl cells were treated with PIC (25 to 75 μg/ml) for 2, 4, or 6 days, with each experimental condition tested in triplicate. Human MDM (in triplicate) were infected as described in section 2.10, with or without PIC (50 μg/ml) treatment. Following infection and cellular treatment, nuclear extracts were prepared using the Epigentek nuclear extraction kit (Epigentek, Farmingdale, NY) and total HAT activity in nuclear extracts quantified using the EpiQuik™ HAT Activity / Inhibition Assay Kit (Epigentek), according to the manufacturer’s instructions. Briefly,20 μg of nuclear extracts diluted in HAT assay buffer (30 μl total volume) were added in triplicate to 8-wells microplate modules coated with HAT substrate and containing acetyl–coenzyme-A and incubated at 37 °C for 30 to 60 min. Samples were then aspirated from each well and all wells washed three times using a wash buffer provided with the kit. Wells were then incubated for 60 min with the HAT capture antibody, on an orbital shaker (100 rpm) at room temperature. Wells were washed four times, incubated for 30 min with the HAT detection antibody, and washed again five times. Wells were then incubated for 10 min with the developing solution (100 μl), 50 μl stop solution was added to each well to stop color development, and absorbance was measured at 450 nm using a SpectraMax M5 microplate reader. HAT standards were used to generate a calibration curve; and this curve was used quantify HAT activity in each sample, according to the manufacturer’s instructions.
2.13. Histone deacetylase (HDAC) assay
U38 cells were treated with PIC (25 to 75 μg/ml) for 48 h, with each experimental condition tested in triplicate. Following cell treatment, nuclear extracts were prepared using the Epigentek nuclear extraction kit and total HDAC in 20 μg nuclear proteins were quantified using the EpiQuick™ HDAC Activity/Inhibition Assay Kit (Epigentek), according to the manufacturer’s instructions. Briefly, 20 μg nuclear proteins in HDAC assay buffer (30 μl total volume) was added in triplicate to 8-wells microplate modules coated with biotinylated HDAC substrate and incubated for 60 min at 37 °C to allow HDAC (nuclear extracts) – substrate binding. Samples were then aspirated from each well and all wells washed three times using 150 μl of wash buffer provided with the kit. Wells were then incubated with the HDAC capture antibody for 60 min on an orbital shaker (100 rpm) at room temperature, washed four times as above, incubated for 30 min with the HDAC detection antibody, and washed again five times. Wells were then incubated for 10 min with the developing solution (100 μl); 50 μl stop solution was added to each well to stop color development, and absorbance was measured at 450nm using a SpectraMax M5 microplate reader. HDAC standards were used to generate a calibration curve; and this curve was used to quantify HADC activity in each sample, according to the manufacturer’s instructions.
2.13. Chromatin immunoprecipitation (ChIP) assays
ChIP assays for acetylated H3 and H4 were performed using the ChromaFlash™ Chromatin Extraction Kit and ChromaFlash™ One-Step ChIP kit (Epigentek), according to the manufacturer’s instructions. Briefly, U38 cells were treated with PIC (50 μg/ml), with or without TLR3.CI (20 μM), for 48 h. Each experimental condition was tested in triplicate; controls consisted of untreated cells and cells treated only with TRL3.CI (20 μM) for 48 h. Following treatment, cells were pelleted by centrifugation (2500 rpm for 10 min at 4 °C), resuspended in ice-cold PBS containing 0.5 mM phenylmethylsulfonyl fluoride, and pelleted again by centrifugation as above. Cellular chromatin was then sheared using a closed system ultrasonic cell disruptor (Microson™, Qsonica LLC, Newtown, CT). Sheared samples were centrifuged at 15,000 rpm for 10 min at 4 °C and chromatin aliquots (supernatants) were immunoprecipitated with acetylated H3 and H4 antibodies (Active Motif, Carlsbad, CA) using One-Step ChIP kit (Epigentek), according to the manufacturer’s instructions. An aliquot (10%) of the sheared chromatin was used as “input” DNA control; and additional controls included chromatin samples immunoprecipitated with isotype-matched control IgG. Immunoprecipitated samples were amplified by PCR using LTR-specific primers (Table 3). PCR cycle was as follows: 94 °C, 3 min initial denaturation; followed by 30 cycles of 94 °C, 20 sec, 55 °C, 20 sec, and 72 °C, 8 sec; with a final hold at 10 °C. Amplified samples were analyzed by agarose gel electrophoresis using 2.5% agarose, and images captured using the G-BOX gel doc system (Syngene). Densitometry was performed to quantify changes in the level of acetylation at each region analyzed using the gel analysis software (GeneTools software, Syngene).
Table 3.
PCR primers used in ChiP analysis
| Primer | Nucleotide position | Primer sequences | Size of the amplified product (bp) | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Nu-0 | −453/−146 | GAAGGGCTAATTTGGTCCCA | GATGCAGCTCTCGGGCCTG | 307 |
| NFR | −176/+61 | CGAGAGCTGCATCCGGAGTA | AGCTTTATTGAGGCTTAAGC | 239 |
| Nu-1 | +96/+301 | AGTAGTGTGTGCCCGTCTGT | TTGGCGTACTCACCAGTCGC | 205 |
Abbreviations: Nu-0: nucleosome 0; Nu-1: nucleosome 1; NFR: nucleosome-free region; bp: base-pairs
2.14. Statistical analysis
Data were analyzed by t-test (two-tailed) for two-group comparisons, one-way ANOVA followed by Tukey’s multiple-comparisons tests, or two-way ANOVA followed by Bonferroni multiple-comparisons tests using GraphPad Prism 5.0b. (GraphPad Software, La Jolla, CA). Threshold of significance level was 0.05. Data are presented as means ± standard error of the mean.
3. Results
3.1. TLR3 ligands induce HIV-1 transactivation
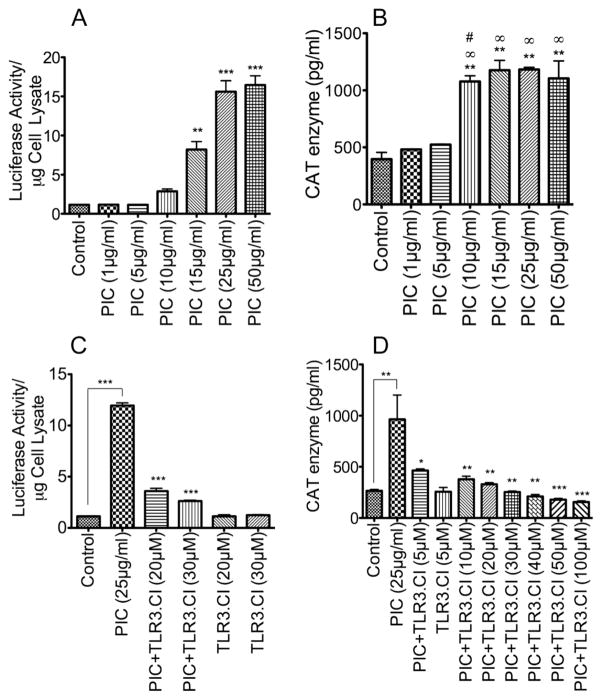
There is evidence that HIV-1 infection in humans is associated with increased TLR3 mRNA and protein in PBMC and that this correlates with high viral loads [24]. To determine whether TLR3 directly influences HIV-1 transactivation and replication, we evaluated the effects of TLR3 ligands on HIV-1 LTR promoter activity using TZM-bl cells and monocytic U38 cells containing integrated copies of the HIV-1 LTR. Dose-dependent experiments showed that exposure of TZM-bl and U38 cells to the TLR3 ligand PIC for 48 h resulted in increased HIV-1 promoter activity, as demonstrated by increased luciferase activity in TZM-bl cells (Figure 1A) and increased CAT enzymatic activity in U38 cells (Figure 1B). A significant increase in HIV-1 promoter activity was observed with 10 to 50 μg/ml PIC. Treatment with PIC at 10 to 50 μg/ml increased the luciferase activity in TZM-bl cells by 2.5- to 14-fold compared to untreated controls or cells treated with 1 or 5 μg/ml PIC (Figure 1A). Similarly, exposure of U38 cells to 10 to 50 μg/ml PIC increased the CAT enzymatic activity by 2 to 3-fold compared to untreated controls or cells treated with 1 or 5 μg/ml PIC (Figure 1B). The inhibitor of TLR3/double strand RNA complex (TLR3.CI) dose-dependently diminished PIC-induced HIV-1 transactivation (Figure 1C, D).
Figure 1. TLR3 ligands increased HIV-1 LTR activity in both TZM-bl and U38 cells.
Exposure of TZM-bl (A, C) or U38 cells (B, D) to PIC for 48 h increased HIV-1 LTR activity in a dose-dependent manner, with maximal LTR activity observed with 25 to 50 μg/ml PIC in TZM-bl cells (A), and from 15 μg/ml PIC in U38 cells (B). The TLR3/dsRNA complex inhibitor (TLR3.CI) dose-dependently blocked PIC-induced transactivation of HIV-1 LTR (C, D). Data shown are representative data from 3 independent experiments. Each experimental condition was tested in triplicate and values shown are mean ± SEM of each triplicates sample. For panel A, **P<0.01, ***P<0.001, and p-values are in comparison to untreated controls. For panel B, **P<0.01 compared to untreated control, ∞P<0.01 compared to cells treated with 1 μg/ml PIC, #P<0.05 compared to cells treated with 5 μg/ml PIC. For panel C, ***P<0.001, and p-values for inhibitor-treated samples are in comparison to LTR activity in PIC-treated cells. For panel D, *P<0.05, **P<0.01, ***P<0.001, p-values for inhibitor-treated samples are in comparison to LTR activity in PIC-treated cells.
3.2. TLR3 ligands upregulate transcription factors known to increase HIV-1 transactivation
Transcription factors play a critical role in the regulation of HIV gene transcription and viral replication. To determine which transcriptional regulators are involved in TLR3-mediated increase in HIV-1 transactivation, we performed gene array experiments using Taqman transcription factor arrays (Applied Biosystems). Exposure of TZM-bl cells to PIC (50 μg/ml), induced differential expression of transcription factors, with 21 transcription factors upregulated by 1.5- to 3.6-fold and 6 transcription factors downregulated by at least 1.5-fold compared to untreated controls (Table 2). Transcription factors upregulated following cells exposure to PIC included JUN, CEBPA, CEBPG, NFκB1, RELB, STAT1, and STAT2 (Table 2). Transcription factors downregulated included early growth response-1 (EGR1), nuclear factor of activated T-cells (NFATC)-2, NFATC4, and FBJ Murine Osteosarcoma Viral Oncogene Homolog (FOS) (Table 2). Transcription factors of the NFκB family, c-Jun/activator protein-1 (AP1), and CEBP family have been shown to regulate and induce HIV-1 transactivation [25–27].
Table 2.
Differentially expressed transcription factors in PIC-exposed TZM-bl cells
| Gene Symbol | Gene Name | Fold change PIC vs. Control |
|---|---|---|
| JUN | c-Jun | 3.59 |
| CEBPA | CCAAT/enhancer-binding protein alpha | 2.91 |
| HNF4A | Hepatocyte nuclear factor 4 alpha | 2.58 |
| NFKB1 | Nuclear factor NF-kappa-B p105 subunit | 2.53 |
| ID1 | Inhibitor of DNA binding 1 | 2.39 |
| RELB | v-Rel avian reticuloendotheliosis viral oncogene homolog B | 2.38 |
| STAT2 | Signal transducer and activator of transcription 2 | 2.14 |
| CTNNB1 | Catenin beta-1 | 2.01 |
| NFAT5 | Nuclear factor of activated T-cells 5 | 1.93 |
| STAT1 | Signal transducer and activator of transcription 1 | 1.86 |
| TGIF1 | TGFB-induced factor homeobox 1 | 1.77 |
| PGK1 | Phosphoglycerate kinase 1 | 1.65 |
| ETS1 | V-Ets avian erythroblastosis virus E26 oncogene homolog1 | 1.63 |
| HIF1A | Hypoxia inducible factor 1, alpha | 1.62 |
| CEBPG | CCAAT/enhancer-binding protein gamma | 1.61 |
| FOXO1 | Forkhead box O1 | 1.60 |
| B2M | Beta-2-microglobulin | 1.57 |
| SMAD9 | Mothers against decapentaplegic homolog 9 | 1.54 |
| GTF2B | Transcription initiation factor IIB | 1.54 |
| TP53 | Tumor protein p53 | 1.53 |
| GATA2 | GATA binding protein 2 | 1.51 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 0.74 |
| HAND2 | heart and neural crest derivatives expressed 2 | 0.45 |
| NFATC4 | Nuclear factor of activated T-cells, cytoplasmic 4 | 0.44 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic 2 | 0.41 |
| EGR1 | Early growth response 1 | 0.18 |
| GATA1 | GATA binding protein 1 | 0.003 |
3.3. Validation of the signaling array data in TZM-bl and U38 cells
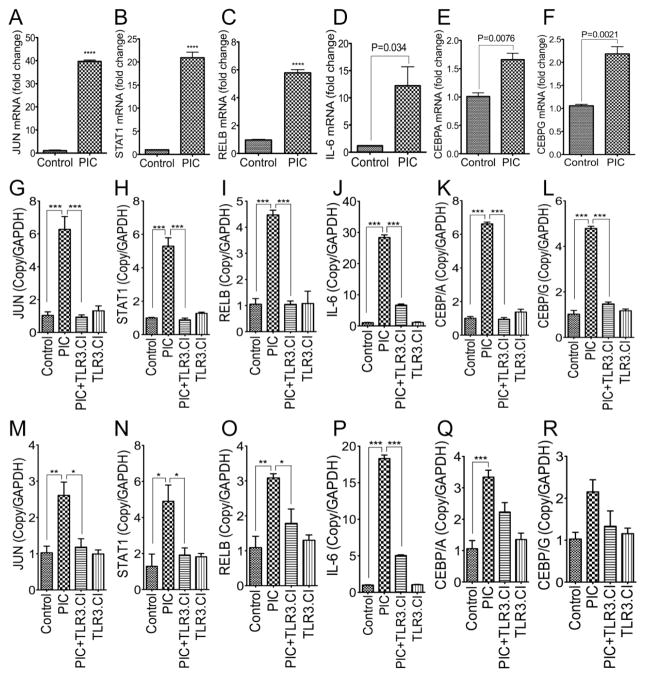
To validate our transcription factors array results, we performed quantitative real-time PCR using RNA from untreated TZM-bl cells and TZM-bl cells treated for 48 h with the TLR3 ligand PIC (50 μg/ml). Exposure of TZM-bl cells to TLR3 ligands for 48 h increased mRNA for JUN, STAT1, and RELB by 37-, 20-, and 6-fold respectively (Figure 2A–C, P<0.0001); increased mRNA of IL-6 by 10-fold (Figure 2D, P=0.034), CEBPA by 1.6-fold (Figure 2E P=0.0076), and CEBPG by 2-fold (Figure 2F, P=0.0021). To further validate these results, we performed additional experiments using TZM-bl cells and U38 cells treated with PIC (50 μg/ml), with or without TLR3.CI (20 μM) for 48 h; with controls consisting of untreated cells and cells treated only TLR3.CI. Compared to untreated controls, treatment of TZM-bl cells with PIC increased mRNA for c-Jun, STAT1, RELB, IL-6, CEBPA, and CEBPG by 6-, 5.3-, 4.2-, 28-, 6.5-, and 4.7-fold respectively (Figure 2G–L). TLR3.CI blocked PIC-induced transcriptional upregulation of c-Jun, STAT1, RELB, IL-6, CEBPA, and CEBPG in TZM-bl cells (Figure 2G–L). Similarly, treatment of U38 cells with PIC increased mRNA for JUN, STAT1, RELB, IL-6, CEBPA, and CEBPG by 2- to 18.3-fold (Figure 2M–R). TLR3.CI inhibited PIC-induced transcriptional upregulation of IL-6 (Figure 2P, P<0.001); significantly diminished PIC-induced transcriptional upregulation of c-Jun, STAT1, and RELB (Figure 2M–O, P<0.05); and also diminished PIC-induced transcriptional upregulation of CEBPA and CEBPG (Figure 2Q, R) in U38 cells.
Figure 2. Real-time PCR validation of TLR3 ligands-induced differential expression of transcription factors and IL-6 in TZM-bl and U38 cells.
Quantitative real-time PCR confirmed that exposure of TZM-bl cells to TLR3 ligands induce transcriptional upregulation of JUN (A, G), STAT1 (B, H), RELB (C, I), IL-6 (D, J), CEBPA (E, K), and CEBPG (F, L). Exposure of U38 cells to TLR3 ligands induced transcriptional upregulation of JUN (M), STAT1 (N), RELB (O), IL-6 (P), CEBPA (Q), and CEBPG (R). A TLR3/dsRNA complex inhibitor (TLR3.CI) blocked TLR3 ligand-induced upregulation of JUN (G, M), STAT1 (H, N), RELB (I, O), IL-6 (J, P), CEBPA (K, Q), and CEBPG (L, R). Control represents untreated TZM-bl cells (A–L), and untreated U38 cells (M–R). PIC represents TZM-bl or U38 cells treated for 48h with 50 μg/ml poly I:C. Additional controls included cells treated only with the TLR3.CI. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
3.4. Validation of the signaling array data in primary human macrophages
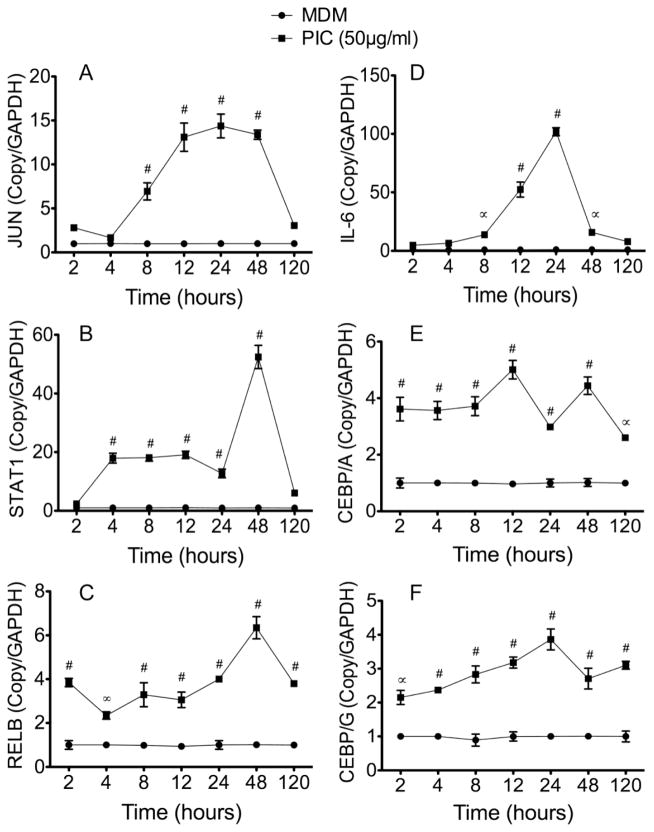
To determine whether our signaling array results validated using TZM-bl and U38 cells could be reproduced in primary human cells, we performed additional real-time PCR experiments using HIV-1-infected and non-infected human MDM treated with PIC (50 μg/ml) for 2 to 120 h. Compared to untreated cells, exposure of human MDM to 50 μg/ml PIC for 2 to 120 h increased mRNA for JUN by 1.6 to 14-fold (Figure 3A), STAT1 by 2 to 52-fold (Figure 3B), RELB by 2 to 6-fold (Figure 3C), IL-6 by 4.8 to 102-fold (Figure 3D), CEBPA by 2.6 to 5-fold (Figure 3E), and CEBPG by 2 to 3.8-fold (Figure 3F). Similarly, exposure of human MDM to lower concentrations of PIC (10 or 25 μg/ml) for 2 to 120 h significantly increased JUN, STAT1, RELB, IL-6, CEBPA, and CEBPG mRNA (see Figure 1 in Data In Brief appendix).
Figure 3. TLR3 ligands induced upregulation of transcription factors and IL-6 in human macrophages.
Primary human MDM in triplicate were treated with PIC (50 μg/ml) for 2 to 120 h, mRNA extracted, and analyzed by quantitative real-time PCR. Data showed that TLR3 ligands induced transcriptional upregulation of JUN (A), STAT1 (B), RELB (C), IL-6 (D), CEBPA (E), and CEBPG (F). *P<0.05, ∞P<0.001. ∝P<0.001, #P<0.0001, compared to untreated controls (MDM).
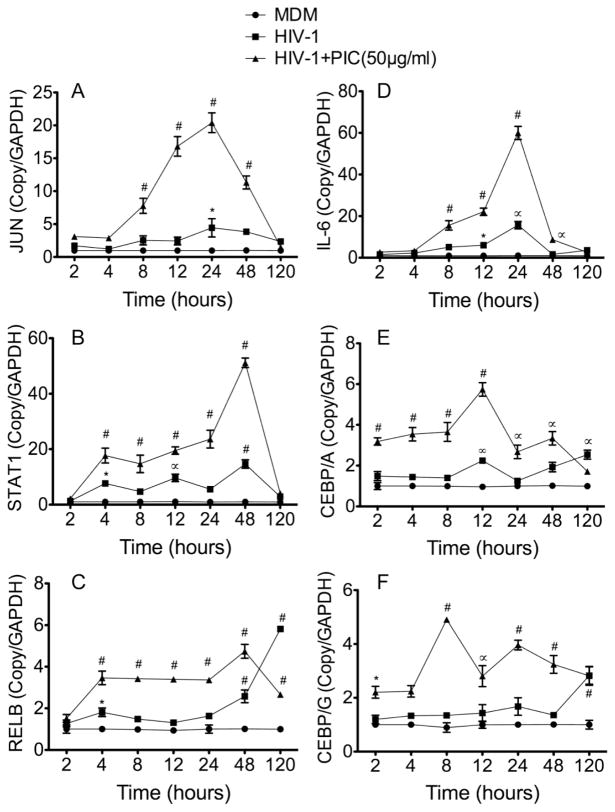
Analysis of HIV-1-infected MDM at 2 to 120 h p.i. showed that, compared to non-infected MDM, HIV-1 infection resulted in increased mRNA for JUN by 1.2 to 4.4-fold (Figure 4A), STAT1 by 1.3 to 14.6-fold (Figure 4B), RELB by 1.2 to 5.8-fold (Figure 4C), IL-6 by 1.5 to 15.6-fold (Figure 4D), CEBPA by 1.2 to 2.2-fold (Figure 4E), and CEBPG by 1.2 to 2.8-fold (Figure 4F). PIC potentiated viral-induced upregulation of these transcription factors. Compared to infected MDM not treated with PIC, exposure of HIV-1-infected MDM to PIC (50 μg/ml) for 2 to 120 h increased mRNA for JUN by 1.5 to 20-fold (Figure 4A), STAT1 by 2 to 51-fold (Figure 4B), RELB by 1.5 to 4.7-fold (Figure 4C), IL-6 by 2 to 60-fold (Figure 4D), CEBPA by 1.7 to 5.7-fold (Figure 4E), and CEBPG by 2 to 4.9-fold (Figure 4F). Similarly, exposure of infected MDM to lower concentrations of PIC (10 or 25 μg/ml) for 2 to 120 h significantly increased JUN, STAT1, RELB, IL-6, CEBPA, and CEBPG mRNA compared to infected MDM not treated with PIC (see Figure 2 in Data In Brief appendix). However, this potentiating effect of PIC was not seen at longer time points (120 h).
Figure 4. TLR3 ligands induced upregulation of transcription factors and IL-6 in HIV-infected macrophages.
Primary human MDM were infected with HIV-1ADA (MOI: 0.01), in triplicate, with or without treatment with PIC (50 μg/ml), for 2 to 120 h, mRNA extracted, and analyzed by quantitative real-time PCR. Data showed that HIV-1 infection induced transcriptional upregulation of JUN (A), STAT1 (B), RELB (C), IL-6 (D), CEBPA (E), and CEBPG (F), and TLR3 ligands further increased this transcriptional upregulation. *P<0.05, ∞P<0.001. ∝P<0.001, #P<0.0001. P-values of infected MDM (HIV-1) are in comparison to non-infected controls (MDM), and P-values of PIC-treated infected MDM (HIV-1+PIC (50 μg/ml)) are in comparison to infected MDM.
PIC-induced expression of these transcription factors was generally dose-dependent in both infected and non-infected MDM, with expression in MDM treated with 50 μg/ml PIC greater than levels in cells treated with 25 μg/ml PIC, and the levels of transcription factors in MDM treated with 25 μg/ml PIC were greater than levels in cells treated with 10 μg/ml PIC (Figures 3 and 4; Figures 1 and 2 of the Data In Brief appendix).
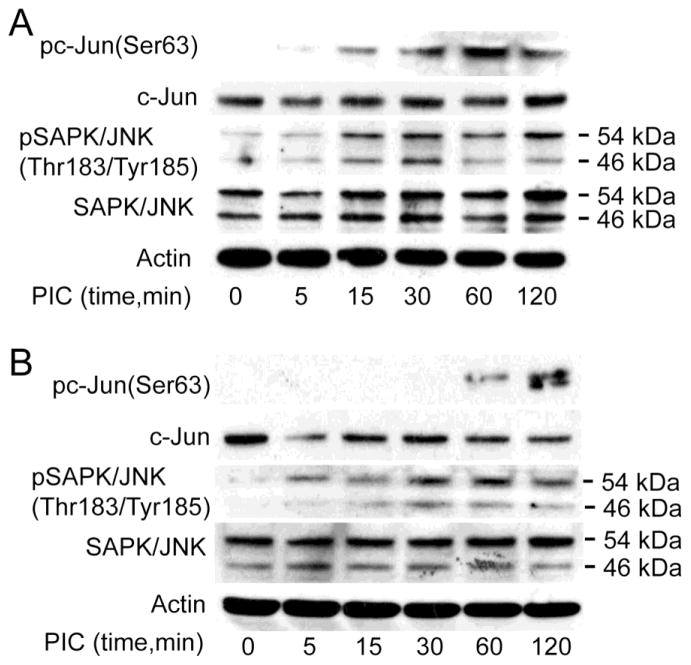
3.5. TLR3 ligands phosphorylate c-Jun and JNK
Our studies showed that TLR3 ligands induced transcriptional upregulation of JUN and transcription factors associated with JNK pathways in TZM-bl cells, U38 cells, and primary human MDM (Table 2, Figures 2 to 4). To determine whether TLR3 ligands effect c-Jun or JNK expression and activation, we analyzed the expression and phosphorylation of c-Jun and JNK in protein lysates from TZM-bl and U38 cells treated with PIC (50 μg/ml) for 5 min to 2 h. Exposure of TZM-bl cells to PIC induced the phosphorylation of c-Jun at serine-63 (Ser63) and phosphorylation of SAPK/JNK at Thr183/Tyr185, with maximal phosphorylation occurring between 30 min and 2 h (Figure 5A). Similarly, exposure of U38 cells to PIC induced the phosphorylation of c-Jun at Ser63 and SAPK/JNK at Thr183/Tyr185 with maximal phosphorylation occurring between 30 min and 2 h (Figure 5B).
Figure 5. TLR3 ligands phosphorylate c-Jun/JNK in TZM-bl and monocytic U38 cells.
(A): Exposure of TZM-bl cells to the TLR3 ligand PIC (50 μg/ml) induced phosphorylation of c-Jun at Serine-63 (Ser63), with maximal phosphorylation at 1 h, and increased the phosphorylation of SAPK/JNK at Threonine-183 / Tyrosine-185 (Thr183/Tyr185). (B): Exposure of U38 cells to the TLR3 ligand PIC (50 μg/ml) induced phosphorylation of c-Jun at Ser63, with maximal phosphorylation at 2 h, and increased the phosphorylation of SAPK/JNK at Thr183/Tyr185.
3.6. NFκB and JNK pathways mediates TLR3 ligands-induced HIV-1 transactivation
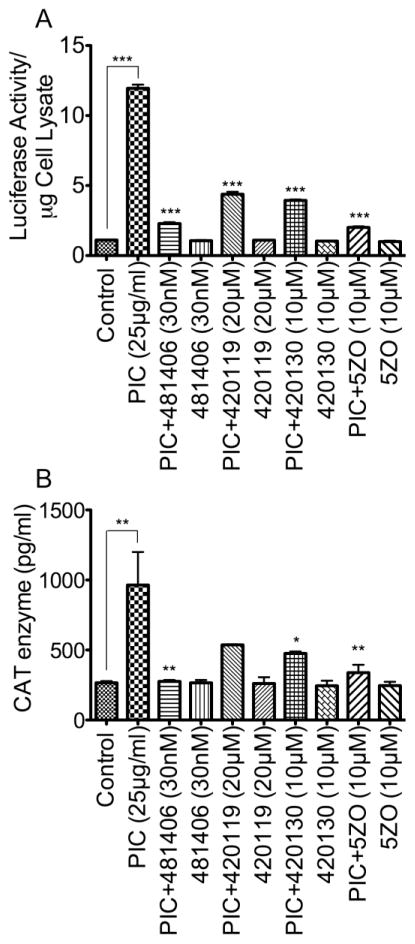
To further investigate the role and involvement of upregulated transcription factors in TLR3-mediated HIV-1 transactivation, we determined the effects of pharmacological inhibitors targeting the NFκB and MAPK/JNK pathways (Table 1) on PIC-induced HIV-1 transactivation. Exposure of TZM-bl (Figure 6A) and U38 (Figure 6B) cells to PIC (25 μg/ml for 48 h) significantly increased HIV LTR transactivation. The inhibitor of NFκB transcriptional activation (481406, 30 nM), the JNK inhibitor II (420119, 20 μM), the JNK inhibitor III (420130, 10 μM), and the MEKK7/MKK7 inhibitor (5ZO, 10 μM) significantly diminished PIC-induced transactivation of the HIV-1 LTR in TZM-bl (Figure 6A) and U38 (Figure 6B) cells. The inhibitor of NFκB transcriptional activation diminished PIC-induced transactivation of the HIV-1 LTR in TZM-bl cells by 5-fold (Figure 6A, P<0.001) and diminished PIC-induced increase in HIV-1 LTR CAT activity in U38 cells by 3-fold (Figure 6B, P<0.001). The JNK inhibitor II diminished PIC-induced increase in HIV-1 LTR luciferase activity in TZM-bl cells by 2.5 to 2.7-fold (Figure 6A, P<0.001) and diminished PIC-induced increase in HIV-1 LTR CAT activity in U38 cells by 1.6 to 1.8-fold (Figure 6B, P<0.05). The JNK inhibitor III, a compound that disrupts c-Jun/JNK complex and prevents JNK phosphorylation of c-Jun, diminished PIC-induced increase in HIV-1 LTR luciferase activity in TZM-bl cells by 2 to 3-fold (Figure 6A, P<0.001) and diminished PIC-induced increase in HIV-1 LTR CAT activity in U38 cells by 1.8 to 2-fold (Figure 6B, P<0.01). The MEKK7/MKK7 inhibitor diminished PIC-induced increase in HIV-1 LTR luciferase activity in TZM-bl cells by 5.6 to 6-fold (Figure 6A, P<0.001), and diminished PIC-induced increase in HIV-1 LTR CAT activity in U38 cells by 2.8 to 3.6-fold (Figure 6B, P<0.01). Lower concentrations of these inhibitors significantly diminished PIC-induced increase in HIV-1 LTR luciferase activity in TZM-bl cells (see Figure 3A in Data In Brief appendix), and significantly diminished PIC-induced increase in HIV-1 LTR CAT activity in U38 cells (see Figure 3B in Data In Brief appendix). Other inhibitors tested included the AP-1 inhibitor (SR11302, 2 μM and 10 μM), the JNK inhibitor V (420129, 2 μM and 10 μM), and the IRAK1/4 inhibitor (5 μM and 10 μM), and they had a lesser to no effect on PIC-induced HIV-1 transactivation in TZM-bl cells (see Figure 4A in Data In Brief appendix) and U38 cells (see Figure 4B in Data In Brief appendix).
Figure 6. NFκB and JNK pathways mediate TLR3 ligands-induced increase in HIV-1 LTR activity in both TZM-bl and U38 cells.
Exposure of TZM-bl (A) or U38 cells (B) to PIC (25 or 50 μg/ml) for 48 h increased HIV-1 LTR activity. PIC-induced HIV-1 LTR activity in TZM-bl and U38 cells could be blocked by the inhibitor of NFκB transcriptional activation (481406), the JNK inhibitor (420119), the inhibitor of c-Jun/JNK complex (420130), and the MEKK7/MKK7 inhibitor (5ZO). *P<0.05, **P<0.01, ***P<0.001; p-values for inhibitors-treated samples are in comparison to LTR activity in PIC-treated cells.
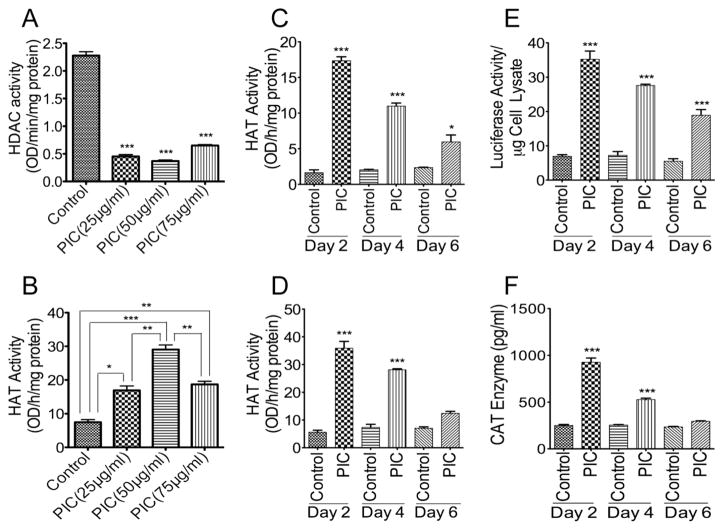
3.7. TLR3-mediated HIV-1 transactivation is associated with decreased HDAC and increased HAT activity
It is known that epigenetics regulate HIV-1 transactivation and viral replication [25–27]. To determine the epigenetic mechanisms involved in TLR3-mediated HIV-1 transactivation, we assessed the effects of TLR3 ligands on histone deacetylases (HDAC) and histone acetyl transferases (HAT) activities in U38 cells following 48 h PIC treatment. PIC concentrations from 25 to 75 μg/ml significantly decreased HDAC activity (Figure 7A) and significantly increased HAT activity, with maximal HAT activity observed with 50 μg/ml PIC (Figure 7B). To determine whether PIC-induced increase of HAT activity was time-dependent, we assessed the effects of PIC on HAT activity in TZM-bl cells and U38 cells following 2, 4, and 6 days of treatment with PIC (50 μg/ml). PIC treatment significantly increased HAT activity in TZM-bl cells (Figure 7C) and U38 cells (Figure 7D). Maximal HAT activity was observed at day-2 in TZM-bl cells (10.6-fold increase, Figure 7C) and U38 cells (6.36-fold increase, Figure 7D). HAT activity gradually decreased after day-2. At day-4, PIC increased HAT activity in TZM.bl and U38 cells by 5.37-fold and 3.8-fold, respectively; at day-6, PIC increased HAT activity in TZM-bl and U38 cells by 2.5-fold and 1.7-fold, respectively (Figure 7C,D). Similarly, PIC (50 μg/ml) treatment significantly increased HIV-1 transactivation in TZM-bl cells (Figure 7E) and U38 cells (Figure 7F), as demonstrated by increased luciferase activity and CAT levels. In a similar pattern to HAT activity, maximal PIC-induced increase of HIV-1 transactivation occurred at day-2 in both TZM-bl cells and U38 cells, decreasing gradually from day-2 to day-6 (Figure 7E,F).
Figure 7. Effects of TLR3 ligands on HIV-1 LTR activity, HDAC, and HAT enzymatic activities.
Exposure of U38 cells to 25 μg/ml, 50 μg/ml, and 75 μg/ml PIC for 48 h resulted in the inhibition of HDAC activity (A) and increased HAT activity, with maximal HAT activity observed with 50 μg/ml PIC (B). Exposure of TZM-bl cells (C) and U38 cells (D) to PIC (50 μg/ml) for 2, 4, and 6 days resulted in a significant increase in HAT activity but this PIC-induced increase in HAT activity decreased gradually from day 2 to day 6. Similarly, exposure of TZM-bl cells (E) and U38 cells (F) to PIC (50 μg/ml) for 2, 4, and 6 days resulted in significant increase in HIV-1 LTR activity, but this PIC-induced increase in HIV-1 LTR activity decreased gradually from day 2 to day 6. *P<0.05, **P<0.01, ***P<0.001. For panels A and C-F, p-values are in comparison to untreated controls.
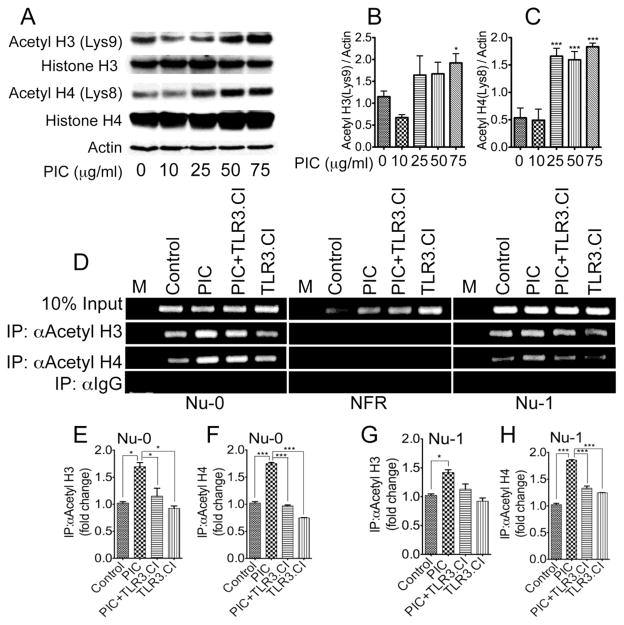
3.8. TLR3-mediated HIV-1 transactivation is associated with increased acetylation of histone H3 and H4 in the viral promoter
HIV-1 transactivation is often associated with increased acetylation of histone proteins [26]. To determine which acetylated histone might be responsible for the TLR3-mediated increase in HIV-1 transactivation and HAT activity, we performed Western blot analysis using acetyl histone antibodies on nuclear protein lysates from U38 cells treated for 48 h with PIC (10 to 75 μg/ml). PIC increased the acetylation of histone H3 at Lysine-9 (Lys9) and increased the acetylation of histone H4 at Lysine-8 (Lys8), with maximal histone acetylation observed with 25 to 75 μg/ml PIC (Figure 8A–C). We then performed ChIP assays to confirm whether PIC increased the acetylation of H3 and H4 on HIV-1 LTR chromatin and to determine on which LTR region this histone acetylation was occurring. Results showed that treatment of U38 cells with PIC (50 μg/ml) for 48 h increased the levels of acetylated H3 and acetylated H4 in the nucleosome-0 (Nu-0) (Figure 8D–F) and nucleosome-1 (Nu-1) (Figure 8D,G,H) regions of the HIV-1 promoter. No acetylated histone was detected in the nucleosome-free region (NFR). Controls included untreated cells and cells treated for 48 h with PIC (50 μg/ml) in the presence of TLR3.CI (20 μM). TLR3.CI significantly diminished PIC-induced increases in acetylated H3 and acetylated H4 at Nu-0 (Figure 8D–F), as well as PIC-induced increase in acetylated H4 at Nu-1 (Figure 8D,H).
Figure 8. Effects of TLR3 ligands on histone proteins.
Exposure of U38 cells to PIC increased the acetylation of histone H3 at Lysine 9 (A, B) and increased the acetylation of histone H4 at Lysine 8 (A, C). ChIP assays further showed increased levels of acetylated H3 and acetylated H4 in PIC-treated U38 cells and showed that PIC-induced acetylation of H3 and H4 occurred in the nucleosome 0 (Nu-0) (D–F) and nucleosome 1 (Nu-1) (D, G, H) regions of the HIV-1 TLR. No histone acetylation was observed in the LTR nucleosome-free region (NFR). *P<0.05, ***P<0.001. For panel B and C, p-values are in comparison to untreated controls or cells treated with 10 μg/ml PIC. For panel D, M represents the molecular weight marker.
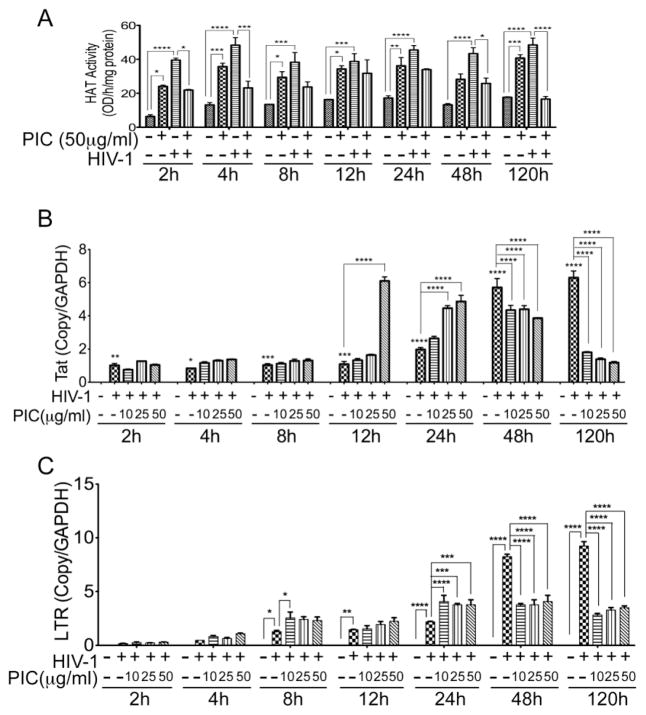
3.9. TLR3 ligands and HIV-1 increased HAT activity in human macrophages
We demonstrated that PIC-induced increase in HIV-1 transactivation was associated with increased HAT activity in TZM-bl and U38 cells. To validate these results in primary human cells, we assessed HAT activity in uninfected or HIV-1-infected human MDM treated with PIC for 2 to 120 h. Exposure of human MDM to PIC (50 μg/ml) for 2 to 120 h induced a 2- to 3.77-fold increase in HAT activity (Figure 9A). Analysis of infected MDM from 2 to 120 h p.i. showed that HIV-1 infection also increased HAT activity in MDM by 2.37- to 6.2-fold (Figure 9A). However, combined HIV-1 infection and PIC treatment of human MDM resulted in a decreased HAT activity (Figure 9A).
Figure 9. Effects of TLR3 ligands on HAT enzymatic activities, HIV-1 LTR, and Tat in human macrophages.
(A): Exposure of human MDM to PIC (50 μg/ml) for 2 to 120 h resulted in increased HAT activity. HIV-1 infection resulted in an even larger increase in HAT activity. However PIC decreased HAT activity in infected MDM. (B): Real-time PCR quantification of the HIV-1 Tat transcripts in human MDM infected with HIV-1 for 2 to 120 h, showed a gradual and significant increase in the levels of HIV-1 Tat in infected cells from 2 to 120 h. Compared to infected cells not treated with PIC, treatment of infected MDM with PIC (at 10, 25, or 50 μg/ml), did not significantly change Tat levels at 2 to 8 h p.i., but increased Tat levels at 12 and 24 h p.i. Longer PIC treatment (48 and 120 h) decreased the levels of Tat transcripts. (C): Real-time PCR quantification of the viral LTR in human MDM infected with HIV-1 for 2 to 120h, with or without PIC (at 10, 25, or 50 μg/ml), showed a gradual and significant increase in the levels of HIV-1 LTR in infected cells from 2 to 120 h. PIC treatment gradually increased LTR levels from 2 to 24 h, but decreased LTR levels at 48 and 120 h. Controls consisted of non-infected MDM, and HIV-1-infected MDM not exposed to PIC. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Data shown are representative data from 2 independent experiments, using MDM from 2 different donors, with each donor tested in triplicate.
3.10. TLR3 ligands mediates HIV-1 transactivation, Tat transcription, and HIV-1 infection of human macrophages
The transactivator of transcription, Tat, is essential for HIV transcription and viral replication. We determined the effects of HIV-1 and PIC on Tat transcription in human MDM from 2 to 120 h p.i. Real-time PCR experiments showed that HIV-1 infection of MDM increased Tat transcript levels at all time points, with higher levels of Tat transcripts at longer time points (Figure 8B). Treatment of HIV-infected MDM with PIC (at 10, 25, and 50 μg/ml) did not significantly change Tat levels at 2 to 8 h p.i. compared to infected cells not treated with PIC. At 12 h p.i., the levels of Tat transcripts increased in infected MDM exposed to 10, 25, and 50 μg/ml PIC by 1.2-fold, 1.5-fold, and 5.6-fold (Figure 9B, P<0.0001), respectively, compared to infected cells not treated with PIC. At 24 h p.i., the levels of Tat transcripts increased in infected MDM exposed to 10, 25, and 50 μg/ml PIC by 1.3-fold, 2.2-fold, and 2.4-fold, respectively (Figure 9B, P<0.0001), compared to infected cells not treated with PIC. However, longer treatment of infected MDM with TLR3 ligands decreased Tat transcription. The levels of Tat transcripts in infected MDM treated with 10, 25, and 50 μg/ml PIC decreased by 1.3- to 1.7-fold at 48 h p.i. and decreased by 3.5- to 5.3-fold at 120 h p.i. compared to infected cells not treated with PIC (Figure 9B).
The HIV-1 LTR regulates viral transcriptional activation and reverse transcription. We determined the effects of HIV-1 and PIC on LTR levels in human MDM from 2 to 120 h p.i. HIV-1 infection gradually increased LTR copies in MDM from 2 to 120 h (Figure 9C). Treatment of infected MDM with PIC (at 10, 25, or 50 μg/ml) gradually increased LTR copies from 2 to 24 h p.i, but decreased LTR levels at 48 h and 120 h (Figure 9C). No HIV-1 LTR was detected in non-infected MDM (Figure 9C) or in non-infected MDM treated with PIC (data not shown).
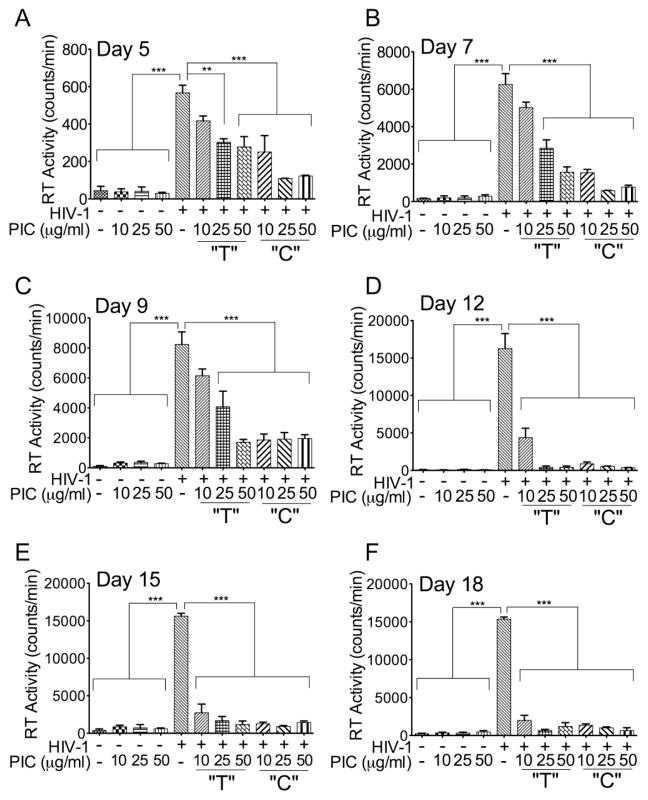
Quantification of RT activity in HIV-1-infected MDM from day-5 to day-18 p.i, also showed that PIC dose-dependently decreased RT activity (Figure 10). Both infected MDM treated only temporarily and treated continuously with PIC showed a decrease in RT activity from day-5 to day-18 p.i. (Figure 10A–F, P<0.01 or P<0.001). This PIC-induced decrease of HIV-1 infection was more pronounced in MDM treated continuously with PIC (Figure 10A–F, P<0.001).
Figure 10. Effects of TLR3 ligands on HIV-1 infection of MDM.
Human MDM were infected with the macrophage tropic HIV-1ADA, with or without PIC (10, 25, or 50 μg/ml) in the media. Culture supernatants were collected every 2 or 3 days from day-5 to day-18 post-infection (A–F) and viral replication was estimated by quantifying reverse transcriptase activity. For some experiments, infected MDM were treated with PIC only during the 4h initial viral exposure and subsequent cultures performed without PIC in the media (“T”); for other experiments, infected MDM were treated with PIC throughout the 5 to 18 days experiments (“C”). From day-5 to day-18 p.i., PIC decreased HIV-1 infection in a dose-dependent manner, with higher inhibition observed in infected MDM continuously cultured with media containing PIC (“C”). ***P<0.001, **P<0.01. Data shown are representative data from 3 independent experiments, using MDM from 3 different donors, with each donor tested in triplicate.
4. Discussion
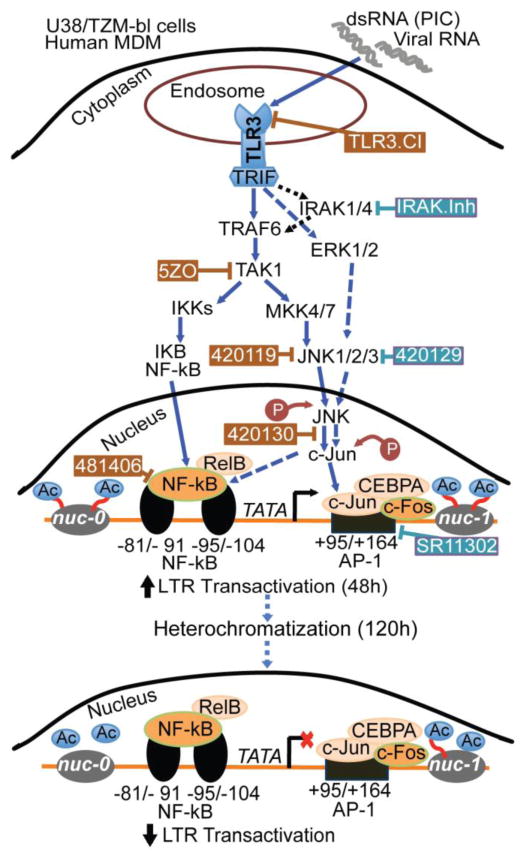
TLR3 has been implicated in the pathogenesis of viral infection; there is increased TLR3 expression in biopsy specimens and PBMC from HIV-infected patients, and this correlated with increased viral load [19, 24, 34]. Because the initial transactivation of the HIV-1 LTR promotes subsequent HIV-1 replication to increase viral load, we assessed the effects of TLR3 ligands on HIV-1 transactivation. Data using two different cell lines containing integrated copies of HIV-1 LTR showed that TLR3 ligands significantly increased the transactivation of the HIV-1 promoter and upregulated the expression of transcription factors such as RELB and NFκB1, both of which are components of the NFκB transcription factor complex [35], and JUN. Experiments in primary human macrophages confirmed these findings and showed that both HIV-1 and TLR3 ligands upregulated the expression of IL-6 and transcription factors such as STAT1, JUN, RELB, and NFκB1 in human MDM. c-Jun dimerizes with c-Fos to form AP-1 [36] and both NFκB and AP1 are HIV transcriptional activators [26]. c-Jun is activated through phosphorylation by the JNK pathways [36, 37]. The role of these transcription factors in TLR3-mediated HIV-1 transactivation were further validated by data showing that TLR3 ligands-induced HIV-1 transactivation can be blocked by pharmacological inhibitors of NFκB transcriptional activation, c-Jun activation, and JNK. These data are in agreement with previous studies showing that PIC activates NFκB in muscle cells and this is associated with increased chemokine release [19]. The inhibitor of AP-1 transcriptional activity did not have any major effect on TLR3 ligands-induced HIV-1 transactivation. This would suggest that following activation of JNK and c-Jun upstream, HIV transactivation in the nucleus occurs predominantly via JNK/c-Jun crosstalk with NFκB and the alternative NFκB pathway (Figure 11).
Figure 11. Model illustrating the signaling pathways in TLR3-induced HIV-1 transactivation.
Arrows with solid blue lines indicates activation and potential direct signaling. Dotted blue lines show cross-talk and alternative signaling pathways that may be involved in TLR3-induced HIV-1 transactivation. Dotted black lines show known pathways that do not appear to be involved in TLR3-induced HIV-1 transactivation. The ⊥ symbol indicates pharmacological inhibitors; brown symbol and brown rectangular boxes indicates compounds with inhibitory effects, while blue symbol and blue rectangular boxes indicating compounds with only partial or no inhibitory effects. “Ac” indicates acetylation.
 indicates a decrease.
indicates a decrease.
 indicates an increase.
indicates an increase.
TLR3 dimerizes to form a signaling complex [38] and ligands bind to the TLR3 dimer via its N-terminal ectodomain [39]. Ligand binding results in a conformational change and recruitment of Toll/IL-1 receptor domain-containing adapter inducing interferon (IFN)-β (TRIF), which then triggers a signaling cascade that results in expression of IFN-β or proinflammatory cytokines [7]. Our data confirm the specificity of TLR3 pathways in HIV-1 transactivation and the expression of transcriptional activators and IL-6, as we demonstrated that PIC-induced HIV-1 transactivation, increased expression of IL-6 and transcription factors such as STAT1, JUN, RELB, and CEBP could be blocked by an inhibitor of the TLR3/dsRNA complex that specifically antagonizes TLR3 signaling by blocking the binding of dsRNA to TLR3.
TLR3-mediated HIV-1 transactivation was also blocked by 5ZO, an ATP-competitive inhibitor of MEKK7, MKK7, MEK1, and ERK2. Each of these MAP kinases (MAPK) contains a cysteine residue in their ATP-binding site; thus, 5ZO may act by interfering with cysteine residues in the ATP-binding region of these MAPK. This would suggest that a region on the MAPK ATP-binding site is involved in TLR3-mediated HIV-1 transactivation. In our current study, inhibitors of IRAK1/IRAK4, downstream effectors of TLR signaling, and inhibitors of AP-1 transcriptional activity did not have any major effect on TLR3-induced HIV-1 transactivation. This suggests that TLR3-mediated HIV-1 transactivation does not involve IRAK1 and IRAK4, and TLR3 ligands do not induce HIV-1 transactivation by affecting AP1 transcriptional activity (Figure 11). Our data also showed that TLR3 ligands decreased HDAC activity, increased HAT activity, and increased acetylation of histone H3 at Lys9 and histone H4 at Lys8. These data are in agreement with literature showing that increased HIV-1 transactivation and viral replication is associated with acetylation of histone proteins and increased HAT activity; while a decrease in HIV-1 transactivation, lower viral replication, and viral latency are associated with deacetylation and increased HDAC activity [26, 40, 41]. Our current data suggests that TLR3 ligands induce HIV-1 transactivation by mechanisms that involve epigenetic modifications.
Maximal TLR3 ligands-induced HIV-1 transactivation in both U38 monocytic cells and TZM-bl cells were observed at 48 h and decreased gradually thereafter. Similarly, experiments in HIV-1-infected human MDM from 2 h to 18 days p.i. showed that PIC increased LTR transcription in the first 24 h, but subsequently decreased LTR transcription and viral replication in HIV-infected MDM, with more decrease in viral replication seen with longer culture time. This was associated with reduced levels of Tat transcripts in PIC-treated MDM. It is likely that this decline in viral transactivation after 48 h and reduced viremia in PIC-treated infected MDM involves histone proteins and epigenetic modifications, because TLR3 ligands-induced changes in HIV-1 promoter activity paralleled TLR3 ligands-induced changes in HAT activity. In fact, TLR3 ligands significantly increased HAT activity at 48 h, but HAT activity decreased gradually and was almost similar to control levels at day-6. It has been shown that short dsRNA cause promoter heterochromatization in both TZM-bl cells and human PBMC; this was associated with histone modification and resulted in decreased HIV-1 replication for up to 3 weeks p.i. [42, 43]. The TLR3 ligand PIC is a short dsRNA and may use similar mechanisms to decrease HAT activity and HIV-1 transactivation at longer time points and to decrease Tat and HIV-1 replication in human MDM. In fact, it has been shown that dsRNA can act as a small interfering RNA (siRNA) to induce the cleavage of mRNA sequence and induce transcriptional or post-transcriptional gene silencing [44–46]. Micro-RNAs, which play a critical role in RNA silencing, can also be induced by PIC and may modulate PIC antiviral effects in macrophages [17, 18, 22]. Our current study provides novel insights into the molecular basis for TLR3-mediated HIV-1 transactivation and the mechanisms of these TLR3-mediated effects are summarized in Figure 11.
There is evidence that agonists for TLR3, TLR7, TLR8, and TLR9 suppress SIV and HIV-1 infection in PBMC, macrophages, and microglia by inducing interferon and interferon-stimulated genes [17, 18, 20, 21, 47]. TLR3 agonists also decreased HIV-1 replication in a human model of ex vivo cervical tissues by enhancing interferon regulatory factor-7-mediated antiviral responses, and this was modulated by NFκB [48]. TLR3 ligands-induced activation of transcription factors such as NFκB, inflammatory cytokines such as IL-6, and increased T-cell immune responses suggests that in HIV-infected humans TLR3 can acts as viral sensor to mediate cellular signaling, innate immune response, and inflammation. Thus, dsRNA and ssRNA from circulating viruses or co-infection with other RNA viruses such as the hepatitis C virus could continuously activate TLR3 and contribute to inflammation, prolonged dysfunction of the innate immune system, and HIV pathogenesis in infected patients. Our data also showed that TLR3 ligands induce transcriptional upregulation of HIV transactivators such as NFκB, CEBP, and JUN in the first 24 to 48 h, but subsequently decreased HIV-1 transactivation and viral replication in human macrophages. TLR3 ligands could be inducing antiviral effects by effecting cellular immunity; it has been shown that stimulation of human PBMC with TLRs ligands resulted in a stronger adaptive immune response [49]. TLRs ligands enhance the ability of dendritic cells to activate cytomegalovirus and HIV specific T-cells and induce protective CD4+ T-cells responses, suggesting that TLRs ligands could be useful as adjuvants for vaccine and immunotherapy [50, 51]. In fact, a study of several TLR ligands as adjuvants for immunization of Rhesus macaques with SIV Gag proteins showed that PIC induced the most effective T-cell responses in the animals, including significantly higher Th1 responses, and this correlated with a better control of SIV replication in infected macaques [52]. Our future studies will investigate this potential role of TLR3 in HIV-induced dysfunction of the innate immunity and as therapeutic target against latent virus and viral reservoirs.
5. Conclusions
Our current data suggests that the NFκB, MAPK, and JNK pathways mediate TLR3-induced HIV-1 transactivation and that this is associated with increased HAT activity and increased acetylation of histones H3 and H4 in the viral Nu-0 and Nu-1 promoter regions. However, maximal TLR3 ligands-induced HIV-1 transactivation and HAT activity occurred at 48 h post-treatment and decreased thereafter. TLR3 ligands also increased HIV-1 LTR and Tat transcription in infected human macrophages in the first 24 h, but decreased LTR levels thereafter and this correlated with decreased levels of Tat transcripts and decreased HIV-1 replication in infected human macrophages. Evidence suggests that epigenetic changes are involved in these TLR3-mediated early HIV-1 transactivation and subsequent suppression of viral infection.
Highlights.
TLR3 activation induced transcription factors that modulate HIV promoter activity
TLR3 activation increased HIV transactivation via JNK and NFκB pathways
Epigenetics modifications modulate TLR3-induced HIV transactivation
Prolonged TLR3 activation suppress HIV transactivation and viral replication
TLR3 ligands could help target and eliminate HIV in latently infected cells
Acknowledgments
We would like to thank the NIH AIDS Reagents Program for providing TZM-bl and U38 cells, Ms. Sangya Singh for technical assistance, and Dr. Matthew Omojola for critical reading of the manuscript. This work was partly supported by grant from the National Institute of Health, National Institute of Mental Health, to G.D.K (MH081780 and MH094160).
List of abbreviations
- AP-1
Activator protein-1
- B2M
Beta-2-microglobulin
- BCA
bicinchoninic acid
- CAT
Chloramphenicol acetyltransferase
- CCR5
Chemokine (C-C motif) receptor 5
- CEBPA
CCAAT/enhancer-binding protein-alpha
- CEBPG
CCAAT/enhancer-binding protein-gamma
- CTNNB1
Catenin beta-1
- CXCR4
Chemokine (C-X-C motif) receptor 4
- dsRNA
double-stranded RNA
- ETS1
V-Ets avian erythroblastosis virus E26 oncogene homolog1
- EGR1
Early growth response 1
- ERK2
Extracellular-signal-regulated kinase-2 / MAP Kinase-1
- FBS
Fetal bovine serum
- FOXO1
Forkhead box O1
- FOS
FBJ murine osteosarcoma viral oncogene homolog
- GATA1
GATA binding protein 1
- GATA2
GATA binding protein 2
- GTF2B
Transcription initiation factor IIB
- HAT
Histone acetyl transferase
- HAND2
Heart and neural crest derivatives expressed 2
- HDAC
Histone deacetylase
- HNF4A
Hepatocyte nuclear factor 4 alpha
- HIF1A
Hypoxia inducible factor 1, alpha
- IL-6
Interleukin-6
- IRAK-1/4
Interleukin-1 receptor-associated kinases-1 and -4
- JNK
c-Jun N-terminal kinases
- LTR
Long terminal repeat
- M
Molecular weight
- MCSF
Macrophage colony stimulating factor
- MDM
Monocyte-derived macrophages
- MEK1
Mitogen-activated protein (MAP) Kinase Kinase-1
- MEKK7
MAP Kinase Kinase Kinase-7
- MKK7
MAP Kinase Kinase-7
- MOI
Multiplicity of infection
- NFATC
Nuclear factor of activated T-cells
- NFATC4
Nuclear factor of activated T-cells, cytoplasmic 4
- NFATC2
Nuclear factor of activated T-cells, cytoplasmic 2
- NFκB
Nuclear factor kappa-B
- NFR
Nucleosome-free region
- Nu-0
Nucleosome-0
- Nu-1
Nucleosome-1
- PAMP
Pathogen associated molecular patterns
- PBS
Phosphate buffered saline
- PBMC
Peripheral blood mononuclear cells
- PCR
Polymerase chain reaction
- PIC/ Poly I
C, Poly Inosinic-polycytidylic acid
- p.i
Post infection
- PGK1
Phosphoglycerate kinase 1
- RELB
V-Rel avian reticuloendotheliosis viral oncogene homolog B
- SMAD9
Mothers against decapentaplegic homolog 9
- ssRNA
single-stranded RNA
- STAT
Signal transducer and activator of transcription
- TAK1
Transforming growth factor β activated kinase-1
- TGIF1
Transforming growth factor β-induced factor homeobox 1
- TLR-3
Toll-like receptor-3
- TLR3.CI
TLR3 /double strand RNA complex inhibitor
- TP53
Tumor protein p53
- TRIF
Toll/IL-1 receptor domain-containing adapter-inducing interferon-β
- 5ZO
5Z-7-Oxozeaenol
Footnotes
Competing interests: The authors declare that they have no competing interests
Authors’ contributions: B.B. carried out immunoassays, real-time PCR, Western blot and ChIP assays, participated in the making of Figures, Tables 2 and 3, data analysis and writing the manuscript. S.M.W. carried out immunoassays, TaqMan transcription factor array, real-time PCR, and prepared Table 1. G.D.K. conceived and designed the study, participated in the making of Figures and Tables, data analysis, and wrote the manuscript. All authors read and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Biju Bhargavan, Email: bbhargavan@unmc.edu.
Shawna M. Woollard, Email: swoollar@unmc.edu.
Georgette D. Kanmogne, Email: gkanmogne@unmc.edu.
References
- 1.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochemical and biophysical research communications. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 2.Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jack CS, Arbour N, Manusow J, Montgrain V, Blain M, McCrea E, Shapiro A, Antel JP. TLR signaling tailors innate immune responses in human microglia and astrocytes. J Immunol. 2005;175:4320–4330. doi: 10.4049/jimmunol.175.7.4320. [DOI] [PubMed] [Google Scholar]
- 6.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-[kappa]B by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 7.Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser BJ, Rodrigo AV, Zlotnick A, Mukhopadhyay S, Ranjith-Kumar CT, Kao CC. Viral double-strand RNA-binding proteins can enhance innate immune signaling by toll-like Receptor 3. PLoS One. 2011;6:e25837. doi: 10.1371/journal.pone.0025837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Smith D, Bot S, Dellamary L, Bloom A, Bot A. Noncoding RNA danger motifs bridge innate and adaptive immunity and are potent adjuvants for vaccination. The Journal of Clinical Investigation. 2002;110:1175–1184. doi: 10.1172/JCI15536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA Is an Endogenous Ligand for Toll-like Receptor 3. Journal of Biological Chemistry. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 10.Wesch D, Beetz S, Oberg HH, Marget M, Krengel K, Kabelitz D. Direct Costimulatory Effect of TLR3 Ligand Poly(I:C) on Human γδ T Lymphocytes. The Journal of Immunology. 2006;176:1348–1354. doi: 10.4049/jimmunol.176.3.1348. [DOI] [PubMed] [Google Scholar]
- 11.Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- 12.Vercammen E, Staal J, Beyaert R. Sensing of Viral Infection and Activation of Innate Immunity by Toll-Like Receptor 3. Clin Microbiol Rev. 2008;21:13–25. doi: 10.1128/CMR.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the TLR3 pathway. Rev Med Virol. 2011 doi: 10.1002/rmv.680. [DOI] [PubMed] [Google Scholar]
- 14.Arroyo DS, Soria JA, Gaviglio EA, Rodriguez-Galan MC, Iribarren P. Toll-like receptors are key players in neurodegeneration. International immunopharmacology. 2011;11:1415–1421. doi: 10.1016/j.intimp.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong Y, Le Y. Toll-like receptors in inflammation of the central nervous system. International immunopharmacology. 2011;11:1407–1414. doi: 10.1016/j.intimp.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 16.Hanamsagar R, Hanke ML, Kielian T. Toll-like receptor (TLR) and inflammasome actions in the central nervous system. Trends in immunology. 2012;33:333–342. doi: 10.1016/j.it.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sang M, Liu JB, Dai M, Wu JG, Ho WZ. Toll-like receptor 3 signaling inhibits simian immunodeficiency virus replication in macrophages from rhesus macaques. Antiviral Res. 2014;112:103–112. doi: 10.1016/j.antiviral.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Wang X, Liu M, Hu Q, Song L, Ye L, Zhou D, Ho W. A critical function of toll-like receptor-3 in the induction of anti-human immunodeficiency virus activities in macrophages. Immunology. 2010;131:40–49. doi: 10.1111/j.1365-2567.2010.03270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiner B, Voss J, Wischhusen J, Dombrowski Y, Steinle A, Lochmuller H, Dalakas M, Melms A, Wiendl H. Expression of toll-like receptors by human muscle cells in vitro and in vivo: TLR3 is highly expressed in inflammatory and HIV myopathies, mediates IL-8 release and up-regulation of NKG2D-ligands. FASEB J. 2006;20:118–120. doi: 10.1096/fj.05-4342fje. [DOI] [PubMed] [Google Scholar]
- 20.Sanghavi SK, Reinhart TA. Increased expression of TLR3 in lymph nodes during simian immunodeficiency virus infection: implications for inflammation and immunodeficiency. J Immunol. 2005;175:5314–5323. doi: 10.4049/jimmunol.175.8.5314. [DOI] [PubMed] [Google Scholar]
- 21.Suh HS, Zhao ML, Choi N, Belbin TJ, Brosnan CF, Lee SC. TLR3 and TLR4 are innate antiviral immune receptors in human microglia: role of IRF3 in modulating antiviral and inflammatory response in the CNS. Virology. 2009;392:246–259. doi: 10.1016/j.virol.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaminathan G, Rossi F, Sierra LJ, Gupta A, Navas-Martin S, Martin-Garcia J. A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages. PLoS Pathog. 2012;8:e1002937. doi: 10.1371/journal.ppat.1002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Wang Y, Wang X, Ye L, Zhou Y, Persidsky Y, Ho W. Immune activation of human brain microvascular endothelial cells inhibits HIV replication in macrophages. Blood. 2013;121:2934–2942. doi: 10.1182/blood-2012-08-450353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lester RT, Yao XD, Ball TB, McKinnon LR, Kaul R, Wachihi C, Jaoko W, Plummer FA, Rosenthal KL. Toll-like receptor expression and responsiveness are increased in viraemic HIV-1 infection. AIDS. 2008;22:685–694. doi: 10.1097/QAD.0b013e3282f4de35. [DOI] [PubMed] [Google Scholar]
- 25.Quivy V, De Walque S, Van Lint C. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell Biochem. 2007;41:371–396. [PubMed] [Google Scholar]
- 26.Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B. Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology. 2009;6:118. doi: 10.1186/1742-4690-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med. 2012;2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri A, Yang B, Gendelman HE, Persidsky Y, Kanmogne GD. STAT1 signaling modulates HIV-1-induced inflammatory responses and leukocyte transmigration across the blood-brain barrier. Blood. 2008;111:2062–2072. doi: 10.1182/blood-2007-05-091207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang B, Akhter S, Chaudhuri A, Kanmogne GD. HIV-1 gp120 induces cytokine expression, leukocyte adhesion, and transmigration across the blood-brain barrier: modulatory effects of STAT1 signaling. Microvascular research. 2009;77:212–219. doi: 10.1016/j.mvr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhuri A, Duan F, Morsey B, Persidsky Y, Kanmogne GD. HIV-1 activates proinflammatory and interferon-inducible genes in human brain microvascular endothelial cells: putative mechanisms of blood-brain barrier dysfunction. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2008;28:697–711. doi: 10.1038/sj.jcbfm.9600567. [DOI] [PubMed] [Google Scholar]
- 31.Gorantla S, Che M, Gendelman HE. Isolation, propagation, and HIV-1 infection of monocyte-derived macrophages and recovery of virus from brain and cerebrospinal fluid. Methods Mol Biol. 2005;304:35–48. doi: 10.1385/1-59259-907-9:035. [DOI] [PubMed] [Google Scholar]
- 32.Schuitemaker H, Kootstra NA. Isolation, propagation, and titration of human immunodeficiency virus type 1 from peripheral blood of infected individuals. Methods Mol Biol. 2005;304:17–24. doi: 10.1385/1-59259-907-9:017. [DOI] [PubMed] [Google Scholar]
- 33.Woollard SM, Li H, Singh S, Yu F, Kanmogne GD. HIV-1 induces cytoskeletal alterations and Rac1 activation during monocyte-blood-brain barrier interactions: modulatory role of CCR5. Retrovirology. 2014;11:20. doi: 10.1186/1742-4690-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scagnolari C, Selvaggi C, Chiavuzzo L, Carbone T, Zaffiri L, d’Ettorre G, Girardi E, Turriziani O, Vullo V, Antonelli G. Expression levels of TLRs involved in viral recognition in PBMCs from HIV-1-infected patients failing antiretroviral therapy. Intervirology. 2009;52:107–114. doi: 10.1159/000218082. [DOI] [PubMed] [Google Scholar]
- 35.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 37.Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ. Selective interaction of JNK protein kinase isoforms with transcription factors. Embo J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell JK, Askins J, Hall PR, Davies DR, Segal DM. The dsRNA binding site of human Toll-like receptor 3. Proc Natl Acad Sci U S A. 2006;103:8792–8797. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C. HIV-1 tat transcriptional activity is regulated by acetylation. Embo J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lusic M, Marcello A, Cereseto A, Giacca M. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. Embo J. 2003;22:6550–6561. doi: 10.1093/emboj/cdg631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, Rao S, Kelleher AD. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008;283:23353–23363. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh A, Palanichamy JK, Ramalingam P, Kassab MA, Bhagat M, Andrabi R, Luthra K, Sinha S, Chattopadhyay P. Long-term suppression of HIV-1C virus production in human peripheral blood mononuclear cells by LTR heterochromatization with a short double-stranded RNA. J Antimicrob Chemother. 2014;69:404–415. doi: 10.1093/jac/dkt348. [DOI] [PubMed] [Google Scholar]
- 44.Montgomery MK, Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 45.Lim HG, Suzuki K, Cooper DA, Kelleher AD. Promoter-targeted siRNAs induce gene silencing of simian immunodeficiency virus (SIV) infection in vitro. Mol Ther. 2008;16:565–570. doi: 10.1038/sj.mt.6300380. [DOI] [PubMed] [Google Scholar]
- 46.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buitendijk M, Eszterhas SK, Howell AL. Toll-like receptor agonists are potent inhibitors of human immunodeficiency virus-type 1 replication in peripheral blood mononuclear cells. AIDS Res Hum Retroviruses. 2014;30:457–467. doi: 10.1089/AID.2013.0199. [DOI] [PubMed] [Google Scholar]
- 48.Rollenhage C, Macura SL, Lathrop MJ, Mackenzie TA, Doncel GF, Asin SN. Enhancing Interferon Regulatory Factor 7 Mediated Antiviral Responses and Decreasing Nuclear Factor Kappa B Expression Limit HIV-1 Replication in Cervical Tissues. PLoS One. 2015;10:e0131919. doi: 10.1371/journal.pone.0131919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biasin M, Piacentini L, Lo Caputo S, Naddeo V, Pierotti P, Borelli M, Trabattoni D, Mazzotta F, Shearer GM, Clerici M. TLR activation pathways in HIV-1-exposed seronegative individuals. J Immunol. 2010;184:2710–2717. doi: 10.4049/jimmunol.0902463. [DOI] [PubMed] [Google Scholar]
- 50.Lore K, Betts MR, Brenchley JM, Kuruppu J, Khojasteh S, Perfetto S, Roederer M, Seder RA, Koup RA. Toll-like receptor ligands modulate dendritic cells to augment cytomegalovirus- and HIV-1-specific T cell responses. J Immunol. 2003;171:4320–4328. doi: 10.4049/jimmunol.171.8.4320. [DOI] [PubMed] [Google Scholar]
- 51.Trumpfheller C, Caskey M, Nchinda G, Longhi MP, Mizenina O, Huang Y, Schlesinger SJ, Colonna M, Steinman RM. The microbial mimic poly IC induces durable and protective CD4+ T cell immunity together with a dendritic cell targeted vaccine. Proc Natl Acad Sci U S A. 2008;105:2574–2579. doi: 10.1073/pnas.0711976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park H, Adamson L, Ha T, Mullen K, Hagen SI, Nogueron A, Sylwester AW, Axthelm MK, Legasse A, Piatak M, Jr, Lifson JD, McElrath JM, Picker LJ, Seder RA. Polyinosinic-polycytidylic acid is the most effective TLR adjuvant for SIV Gag protein-induced T cell responses in nonhuman primates. J Immunol. 2013;190:4103–4115. doi: 10.4049/jimmunol.1202958. [DOI] [PMC free article] [PubMed] [Google Scholar]