Abstract
Sleep deprivation is a public health epidemic that causes wide-ranging deleterious consequences, including impaired memory and cognition. Protein synthesis in hippocampal neurons promotes memory and cognition. The kinase complex mammalian target of rapamycin complex 1 (mTORC1) stimulates protein synthesis by phosphorylating and inhibiting the eukaryotic translation initiation factor 4E binding protein 2 (4EBP2). We investigated the involvement of the mTORC1-4EBP2 axis in the molecular mechanisms mediating the cognitive deficits caused by sleep deprivation in mice. Using an in vivo protein translation assay, we found that loss of sleep impaired protein synthesis in the hippocampus. Five hours of sleep loss attenuated both mTORC1-mediated phosphorylation of 4EBP2 and the interaction between eukaryotic initiation factor 4E (eIF4E) and eukaryotic initiation factor 4G (eIF4G) in the hippocampi of sleep-deprived mice. Increasing the abundance of 4EBP2 in hippocampal excitatory neurons prior to sleep deprivation increased the abundance of phosphorylated 4EBP2, restored the amount of eIF4E-eIF4G interaction and hippocampal protein synthesis to that seen in mice that were not sleep-deprived, and prevented the hippocampus-dependent memory deficits associated with sleep loss. These findings collectively demonstrate that 4EBP2-regulated protein synthesis is a critical mediator of the memory deficits caused by sleep deprivation.
Introduction
The consolidation of long-term hippocampus-dependent memory requires protein synthesis (1–3). The rate-limiting step for de novo protein production occurs at translation initiation, which is regulated by the insulin signaling pathway, including 5′ adenosine monophosphate-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) (4–6). mTOR is a serine/threonine kinase that forms a complex called mTOR complex 1 (mTORC1) when associated with the regulatory-associated protein of mTOR (Raptor) (6–11). This complex releases the brake on protein synthesis initiation by mediating the phosphorylation of the translation repressor protein eukaryotic initiation factor 4E (eIF4E)-binding protein (4EBP). The predominant isoform of 4EBP in the brain is 4EBP2 (12, 13). Phosphorylation of 4EBP2 prevents its binding to eIF4E, freeing eIF4E to stimulate translation. mTORC1 also promotes protein synthesis by activating the p70S6 kinase (S6K) pathway (8–10).
Several studies have suggested that the mTOR signaling pathway is affected by sleep and sleep deprivation in various animal models. Sleep promotes the phosphorylation of 4EBP in the brains of cats (14), and periods of sleep have also been correlated with increased protein synthesis as measured by incorporation of radioactive leucine in the brains of rats (15) and non- human primates (16). In mice, we previously found that 5 hours of sleep deprivation leads to reduced abundance of mTOR in the hippocampus (17). Hippocampus-dependent memory is particularly sensitive to sleep loss (18–22). Here, we investigated whether sleep deprivation impairs protein synthesis in vivo and whether impaired protein synthesis causes the memory impairments associated with sleep deprivation.
Results
Sleep deprivation impairs hippocampal protein synthesis in vivo
We first assessed whether hippocampal protein synthesis is reduced by sleep deprivation using a modified non-radioactive surface sensing of translation (SUnSET) assay that tags nascent proteins with puromycin, also known as RiboPuroMycylation (23). Male C57BL6/J mice received one injection of puromycin (25 μg/μl) into the left lateral ventricle, and mice were then immediately sleep deprived for 5 hours while control mice were left undisturbed. Immediately after the sleep deprivation period, hippocampi were harvested, and the abundance of proteins tagged with puromycin was measured by Western blotting. Mice that had undergone 5 hours of sleep deprivation had a significantly reduced amount of puromycin-tagged proteins in the hippocampus compared to mice that had undisturbed sleep for those 5 hours (Fig. 1A).
Fig. 1. Sleep deprivation impairs hippocampal protein synthesis in vivo.
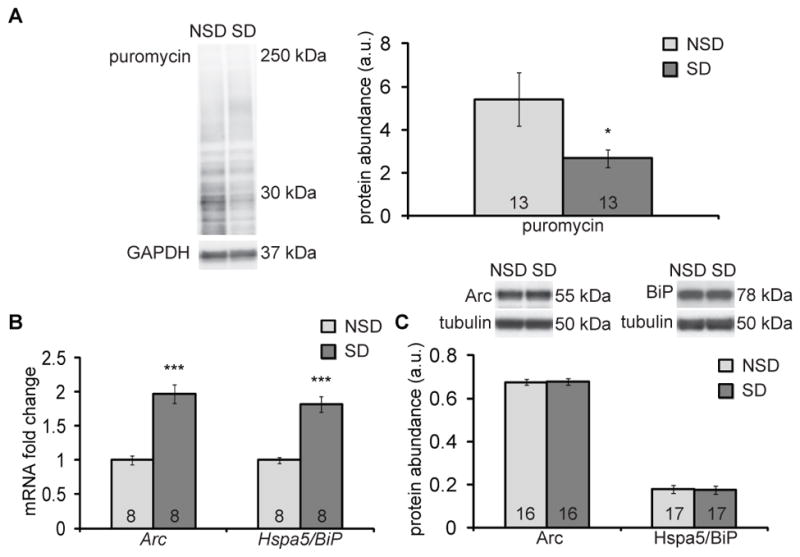
(A) Western blotting analysis for puromycin, as a proxy for protein synthesis, in the hippocampus from non-sleep-deprived (NSD) or sleep-deprived (SD) male C57BL6/J mice injected intracerebroventricularly with puromycin. Puromycin signal was normalized to the loading control, GAPDH, and quantified (right panel). Data are means ± SEM of 13 mice in each condition (* p = 0.047, t test). (B) Gene expression of Arc and Hspa5/BiP in hippocampal extracts from SD mice relative to each in those from NSD mice, assessed by qPCR. Expression was normalized to that of housekeeping gene Tuba4a. Data are means ± SEM of 8 mice in each condition (*** p < 0.001, t test). (C) Representative Western blots and quantitation of Arc and Hspa5/BiP abundance in hippocampal extracts. Abundance was normalized to the β-tubulin loading control. Data are means ± SEM of 16 or 17 mice in each condition. All blots are representative of at least 3 independent assays. a.u., arbitrary units. N, number of mice analyzed, is noted in each bar.
Our in vivo translation assay results were further corroborated when we compared the abundance of gene transcript to that of protein encoded by each of two genes in the hippocampus. The abundance of both Arc and BiP mRNA significantly increased approximately two-fold after sleep deprivation (Fig. 1B), as previously shown (17), but we observed no commensurate increase in respective protein abundance (Fig. 1C). This disassociation between gene expression and protein abundance is consistent with a reduction of protein synthesis in the hippocampus after sleep deprivation.
Sleep deprivation reduces mTORC1 activity by increasing AMPK signaling in the hippocampus, which results in reduced 4EBP2 phosphorylation
Through bioinformatic analysis of microarray data, we previously showed that the group of genes for which expression is altered after sleep deprivation is enriched in genes that encode proteins in the insulin signaling pathway; these include adenosine monophosphate-activated kinase (AMPK) and mTOR (17). mTOR and its multiple downstream effectors serve as a nexus for numerous cellular functions, including protein synthesis (8–10, 17). AMPK negatively regulates mTORC1 indirectly by phosphorylating and activating the tuberous sclerosis complex (TSC) (24) or directly by phosphorylating Raptor (25) (Fig. 2). To determine whether the insulin signaling pathway impairs protein synthesis after sleep deprivation, we deprived mice of sleep for 5 hours and probed the abundance of several insulin signaling pathway proteins in hippocampal extracts. We found that 5 hours of sleep deprivation significantly increased the phosphorylation of AMPKα in the hippocampus (Fig. 3A).
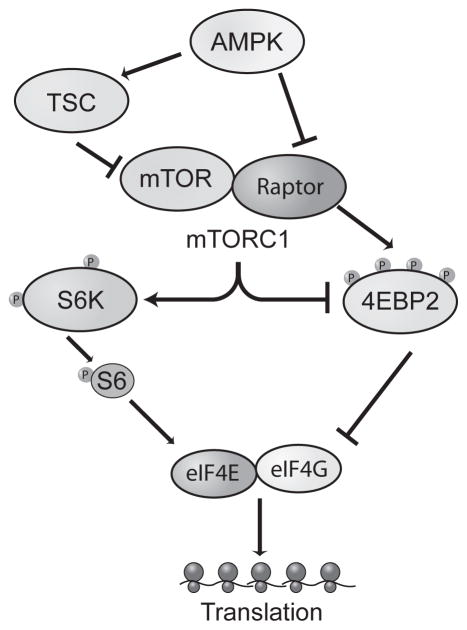
Fig. 2. Schematic of the signaling pathways regulating the initation of protein synthesis.
The insulin signaling pathway, which includes AMPK and mTOR, is one pathway that regulates protein synthesis initiation. When AMPK is activated, it inhibits mTORC1 activity either through phosphorylation of TSC or Raptor. The binding of mTOR and Raptor is necessary for mTORC1 formation and subsequent phosphorylation of p70 S6 kinase (S6K) or 4EBP2. Phosphorylation (activation) of S6K facilitates translation initiation by its phosphorylation of S6 ribosomal protein (S6). When 4EBP2 is phosphorylated (inhibited), eIF4E is able to form a complex with eukaryotic initiation factor 4G (eIF4G) to start cap-dependent translation.
Fig. 3. Sleep deprivation affects AMPK-mTORC1-4EBP2 signaling pathway in the hippocampus.
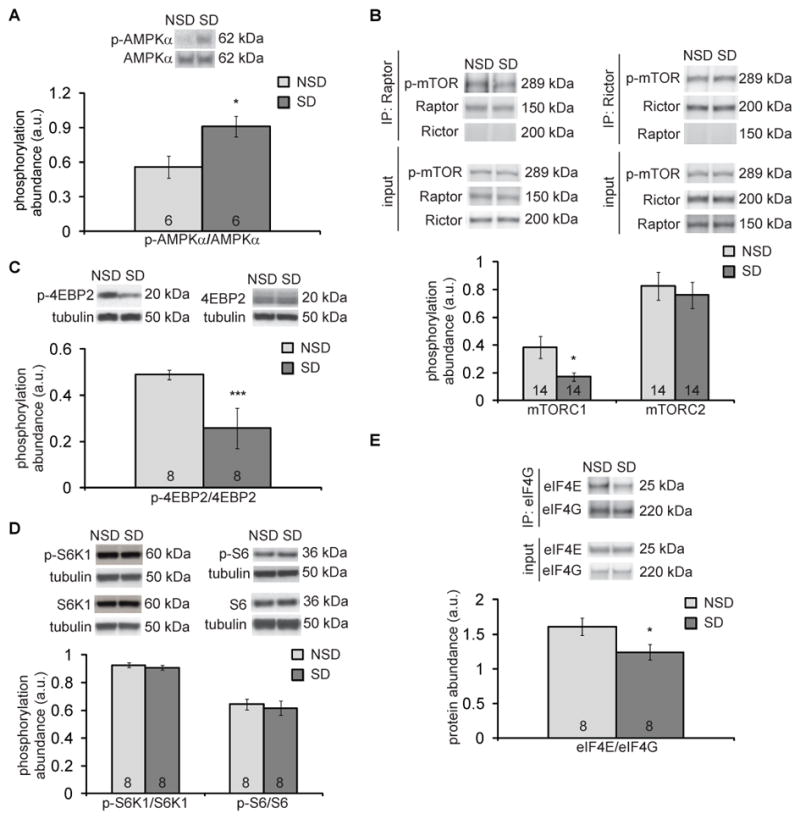
(A) Representative Western blots and quantitation of the ratios of phosphorylated to total AMPKα in hippocampal extracts from non-sleep-deprived (NSD) mice and sleep-deprived (SD) mice. Data are means ± SEM of 6 mice per condition (* p = 0.032, t test). (B) Representative Western blots and quantitation of hippocampal abundance of p-mTOR (Ser2481) after immunoprecipitation with raptor (left) or rictor (right). Negative control blots for rictor in the raptor IP (left, below raptor blot) and raptor in the rictor IP (right, below rictor blot) are shown. Control input blots for p-mTOR (Ser2481), raptor, and rictor are also shown. Data are means ± SEM of 14 mice per condition (* p = 0.042, t test). (C and D) Representative Western blots and quantitation of phosphorylated 4EBP2 (p-4EBP2) (C) or phosphorylated S6K1 (p-S6K1) and phosphorylated S6 ribosomal protein (p-S6) (D) relative to their respective total protein abundance from hippocampus of NSD and post-5 hours of SD mice. β-tubulin served as a loading control. Data are means ± SEM of 8 mice per condition (*** p < 0.005, t test). (E) Representative Western blots and quantitation of hippocampal abundance of eIF4E after immunoprecipitation with eIF4G from NSD and SD mice. Control input blots for eIF4E and eIF4G are also shown. Data are means ± SEM of 8 mice per condition (* p = 0.047, t test). All blots are representative of at least 3 independent assays. a.u., arbitrary units. N, number of mice analyzed, is noted in each bar.
mTOR forms at least two functionally distinct complexes that are mutually exclusive (26). mTORC1 is formed when mTOR is associated with Raptor, whereas mTORC2 is formed when rapamycin-insensitive companion of mTOR (Rictor) is associated with mTOR. We investigated whether 5 hours of sleep deprivation reduced mTORC1 or mTORC2 activity. To measure mTORC-specific activity, we examined the autophosphorylation of mTOR at Ser2481 (27). We immunoprecipitated either Raptor or Rictor from hippocampal lysates and measured the amount of autophosphorylated mTOR that was bound to each. Blots from control pulldowns using rabbit IgG were negative for the proteins examined (fig. S2). We found that 5 hours of sleep deprivation specifically decreased the amount of autophosphorylated mTOR that associated with Raptor (mTORC1) by 55% but not Rictor (mTORC2) (Fig. 3B). One possible mechanism by which increased AMPK activity would reduce mTOR activity is through phosphorylation of TSC2 (Fig. 2). We measured the phosphorylation of TSC2 at the AMPK phosphorylation site (Ser1387) and did not find any changes in TSC2 phosphorylation in hippocampus extracts from sleep-deprived mice (fig. S3).
This reduction in mTORC1 but not mTORC2 activity led us to examine the downstream effectors of mTORC1-mediated translation, 4EBP2 and S6K (8). Sleep-deprived mice had significantly decreased amount of phosphorylated 4EBP2 in the hippocampus (Fig. 3C, fig. S4A). When mice were allowed to sleep for 2.5 hours after being sleep-deprived, a period of time sufficient for functional recovery from sleep deprivation (28, 29), the amount of phosphorylated 4EBP2 was no longer significantly reduced (fig. S4B). In contrast, the abundance of phosphorylated S6K1 and phosphorylated S6 remained unchanged in the hippocampus from sleep-deprived mice (Fig. 3D). The phosphorylation of eukaryotic initiation factor 2α (eIF2α) also did not change in sleep-deprived mice (fig. S5).
Reduced mTORC1 activity and increased 4EBP2 phosphorylation observed in the hippocampus of sleep-deprived mice would suggest that eIF4E binding to eukaryotic initiation factor 4G (eIF4G) would be attenuated (Fig 2). We immunoprecipitated eIF4G and measured the amount of bound eIF4E. We found that 5 hours of sleep deprivation in mice decreased the eIF4E-eIF4G association in the hippocampus (Fig. 3E). Thus, sleep deprivation attenuates protein synthesis initiation by specifically affecting the AMPK-mTORC1-4EBP2 signaling pathway, resulting in decreased eIF4E-eIF4G association in the hippocampus.
Viral expression of 4EBP2 in the hippocampus prevents deficits in protein synthesis and memory impairment caused by sleep deprivation
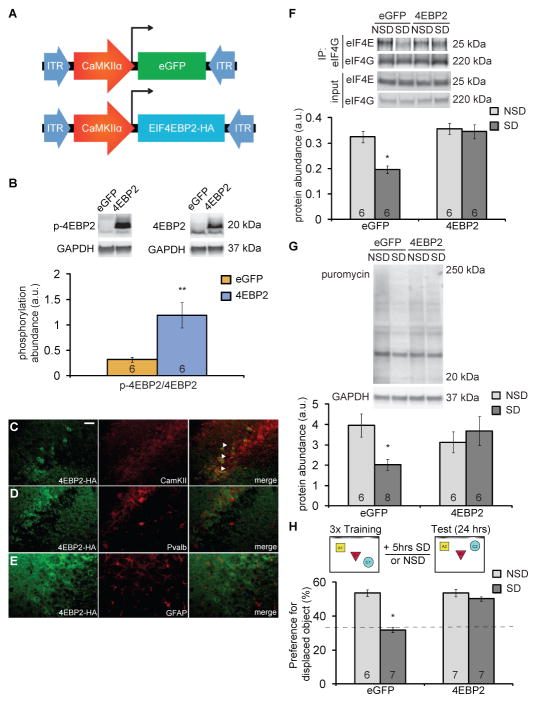
Given the decrease in 4EBP2 phosphorylation after 5 hours of sleep deprivation, we examined whether the viral expression of 4EBP2 might rescue memory deficits caused by sleep deprivation. We expressed 4EBP2 in excitatory hippocampal neurons of adult male C57BL/6J mice using a CaMKIIα promoter fragment in adeno-associated viruses (AAVs) (Fig. 4A). Three weeks after AAV vector injection into the hippocampus, phosphorylated 4EBP2 was significantly increased four-fold compared to total 4EBP2 in the hippocampus of mice injected with the 4EBP2 AAV (Fig. 4B). We also confirmed that viral expression was limited to excitatory neurons by assessing co-labeling of CaMKIIα and HA-tagged 4EBP2 (4EBP2-HA) in hippocampal cells in situ (Fig. 4C). Viral expression of 4EBP2-HA did not co-label with parvalbumin (Fig. 4D) or glial fibrillary acidic protein (GFAP, Fig. 4E), demonstrating that 4EBP2-HA was not expressed in parvalbumin-positive inhibitory neurons or glial cells.
Fig. 4. Viral expression of 4EBP2 in the hippocampus prevents deficits in protein synthesis and memory impairment caused by sleep deprivation.
(A) Schematics of the pAAV9- CamKIIα0.4-eGFP or pAAV9-CamKIIα0.4-eIF4EBP2-HA vectors used to express eGFP or 4EBP2 in hippocampal excitatory neurons in mice. (B) Representative Western blots and quantitation of the ratio between phosphorylated 4EBP2 and total 4EBP2 in the hippocampus of mice injected with either the eGFP or the 4EBP2 vector. Abundance was normalized to GAPDH loading control. Data are means ± SD from 6 mice in each condition. (** p < 0.005, t test). (C–E) Immunofluorescence for 4EBP2-HA (green), CaMKII (C, red), parvalbumin (D, red) or GFAP (E, red) in the CA3 region of the hippocampus from 4EBP2-HA-injected mice. Arrows, colocalization. Scale bar, 50μm. Images represent 4 mice per condition, 3–5 images per mouse. (F) Representative Western blots and quantitation of hippocampal abundance of eIF4E after immunoprecipitation with eIF4G from eGFP- and 4EBP2-HA-injected mice that were either not sleep deprived (NSD) or sleep deprived (SD). Control input blots for eIF4E and eIF4G are also shown. Data are means ± SEM of 6 mice per condition (sleep deprivation effect: F(1,20) = 9.856, ** p = 0.0052; virus effect: F(1,20) = 16.53, *** p = 0.0006; interaction effect: F(1,20) = 6.988, * p = 0.0156; two-way ANOVA). (G) Representative Western blots and quantitation of proteins labeled with puromycin from hippocampi of NSD and SD mice 3 weeks after being injected with eGFP or 4EBP2. Abundance was normalized to GAPDH loading control. Data are means ± SEM of 6 or 8 mice in each condition. (sleep deprivation effect: F(1,22) = 1.585, p = 0.221; virus effect: F(1,22) = 0.577, p = 0.456; interaction effect: F(1,22) = 5.201, * p = 0.033; two-way ANOVA). (H) Performance of mice expressing eGFP or 4EBP2 in a hippocampus-dependent object-place recognition task (schematic, top) when either sleep-deprived for 5 hours immediately after training or left undisturbed. Data are means ± SEM of 6 or 7 mice in each condition. (eGFP, * p < 0.013; 4EBP2, p = 0.149; Wilcoxon rank sums test). Dotted line indicates chance performance. All blots are representative of at least 3 independent assays. a.u., arbitrary units. N, number of mice analyzed, is noted in each bar.
To determine whether viral expression of 4EBP2 rescues the reduced interaction between eIF4E and eIF4G following sleep deprivation, we immunoprecipitated eIF4G and examined eIF4E abundance bound to eIF4G in undisturbed and sleep-deprived mice injected with control eGFP AAV or 4EBP2 AAV. We found that expression of 4EBP2 in the hippocampus rescued the abundance of eIF4E bound to eIF4G after sleep deprivation (Fig. 4F). Using the in vivo translation assay described above, we measured the amount of puromycin incorporation into proteins in the hippocampus after 5 hours of sleep deprivation in mice virally expressing 4EBP2 or eGFP (control) for 3 weeks. AAV-induced increase of 4EBP2 expression in the hippocampus prevented the reduced protein synthesis that was caused by sleep deprivation (Fig. 4G). Three weeks of 4EBP2 viral expression were sufficient to restore protein synthesis amounts in the hippocampus of sleep-deprived mice. Therefore, we performed subsequent behavioral experiments at this time point after virus injection.
We previously showed that 5 hours of sleep deprivation causes hippocampus-dependent memory deficits (21, 28, 30). Here, we investigated whether restoring protein synthesis is sufficient to prevent the memory deficits associated with sleep deprivation. We trained mice expressing 4EBP2 AAV or eGFP AAV in the hippocampus in an object-place recognition task (Fig. 4H). This spatial task exploits the preference for novelty in mice (31, 32). During training, mice explored and learned the location of three objects within an arena. Immediately after training, mice expressing 4EBP2 virus or control eGFP virus were either sleep-deprived for 5 hours or left undisturbed. The next day, mice were tested for object-location memory. Consistent with previous studies, mice expressing eGFP that were left undisturbed preferentially explored the displaced object, which demonstrates consolidated memory (30). Sleep-deprived mice expressing eGFP did not exhibit increased exploration of the displaced object, demonstrating a failure to consolidate memory of it. Mice expressing 4EBP2 AAV preferentially explored the displaced object, regardless of whether they were sleep-deprived or not (Fig. 4H), indicating that 4EBP2 AAV mice were resistant to memory deficits caused by sleep deprivation. Collectively, these data demonstrate that increased 4EBP2 expression in hippocampal excitatory neurons significantly increased phosphorylated 4EBP2 abundance, which restored protein synthesis in the hippocampus and prevented memory deficits caused by sleep deprivation.
Discussion
Previous work suggested that sleep deprivation affects protein synthesis signaling pathways, specifically the insulin signaling pathway (17). In this study, we found that hippocampus-dependent memory impairments caused by sleep deprivation are because of impaired protein synthesis initiation in the hippocampus (summarized in fig. S1). Here, we found a specific subset of the insulin signaling pathway – AMPK-mTOR-4EBP2 signaling – was affected by sleep deprivation, which was sufficient to cause hippocampus-dependent memory deficits in mice. Furthermore, we rescued these memory deficits when we restored protein synthesis in the hippocampus of these mice with 4EBP2 AAV infection, which increased 4EBP2 phosphorylation in hippocampal excitatory neurons. These findings suggest that sleep deprivation impairs memory by reducing protein synthesis.
Our study found that sleep deprivation impairs protein synthesis necessary for memory consolidation. Our work supports the hypothesis that one of the functions of sleep is to facilitate protein synthesis (33, 34). We demonstrated that sleep deprivation impaired protein synthesis in the hippocampus using a non-radioactive in vivo translation assay, which was predicted by our previous findings that 5 hours of sleep deprivation lowers the abundance of mTOR in the hippocampus (17). This is consistent with previous findings that rapamycin administration inhibiting mTORC1 impairs sleep-dependent cortical plasticity (14). Gronli and colleagues have also shown that translation factors are affected by sleep deprivation (35). Conversely, periods of sleep have been correlated with increased protein synthesis as measured by a radioactive translation assay in other organisms (15, 16). Furthermore, sleep promotes the phosphorylation of 4EBP2 (14), which is in line with our findings that phosphorylated 4EBP2 is reduced with sleep deprivation. Our study demonstrates the functional importance of the impact of sleep deprivation on protein synthesis as we show that rescuing protein synthesis is sufficient to prevent these sleep deprivation-induced memory impairments. Our rescue experiments demonstrate that it is the impact of sleep deprivation on protein synthesis in the hippocampus that is critical for spatial memory impairments. In future experiments, it would be interesting to see if short periods of sleep deprivation impact other behaviors by attenuating protein synthesis in other brain regions.
We found that sleep deprivation specifically attenuated mTORC1 activity, which reduced the abundance of phosphorylated 4EBP2, but not of phosphorylated S6K, in the mouse hippocampus. This result, while surprising, is not unique to sleep deprivation. Neural stem cell self-renewal is also regulated by the specific targeting of mTORC1 to 4EBP2, and not S6K (36). One possible explanation for the specificity of the impact of sleep deprivation on the 4EBP2 pathway but not S6K signaling may be due to the action of specific protein phosphatases. Our previously published microarray data revealed several phosphatase genes with increased expression in the hippocampus after sleep deprivation (17). It would be interesting to examine in the future the role of phosphatases after sleep deprivation to discern the mechanism by which sleep deprivation selectively affects the phosphorylation of 4EBP2. Another possible hypothesis that explains why sleep deprivation specifically affects 4EBP2 activity may be due to its association with Raptor.
Previous work in Aplysia has shown that Raptor and 4EBP2 signaling are tightly regulated (37). We restored the amount of protein synthesis in sleep-deprived mice to that seen in non-sleep-deprived mice using 4EBP2 AAV infection into the hippocampus. Thus, increasing the abundance of 4EBP2 may increase the association between Raptor and 4EBP2. It is important to note that 4EBP2 undergoes an alternate post-translational modification other than phosphorylation. 4EBP2 deamidation occurs when 2 C-terminal asparagines spontaneously convert to aspartates (38). Sleep deprivation may increase 4EBP2 deamidation in the hippocampus, and increasing 4EBP2 expression would increase the Raptor-4EBP2 association. This increase in Raptor-4EBP2 interaction, with reduced 4EBP2 sequestration of eIF4E, would ultimately decrease translation repression (39). Our finding that 4EBP2 AAV infection rescued the abundance of eIF4E bound to eIF4G supports this hypothesis. These results suggest that sleep deprivation may be affecting Raptor, due to its known interaction with 4EBP2. Additionally, we did not find changes to TSC2 phosphorylation as a result of sleep deprivation. This suggests that AMPK signaling inhibits mTORC1 activity through Raptor phosphorylation. Future experiments could probe the involvement of Raptor in the insulin signaling pathway and its changes after sleep deprivation.
Interestingly, we did not see increased protein synthesis or enhanced memory in mice expressing the 4EBP2 virus that were not sleep-deprived. We speculate that we observed a ceiling effect, where depressed translation could be rescued by 4EBP2 expression, but translation could not be facilitated beyond physiological abundance by 4EBP2 expression. The specificity of restoring memory in sleep deprived mice clearly demonstrated the importance of AMPK-mTORC1-4EBP2 activity for protein synthesis and subsequent memory impairment after sleep deprivation.
We showed that rescuing 4EBP2 expression and subsequent protein synthesis was sufficient to prevent the memory deficits associated with sleep deprivation. We have previously shown that cyclic AMP (cAMP) signaling is also impaired with sleep deprivation, leading to cognitive deficits (28, 30). Though it is possible that, with sleep deprivation, cAMP signaling and mTOR signaling occur in parallel and independent of one another, there are points of interaction between the cAMP and mTOR signaling pathways. One way in which sleep deprivation affects both cAMP and mTOR signaling may be through phosphodiesterase 4D (PDE4D), a protein that degrades cAMP. PDE4D can bind to RAS homolog enriched in brain (Rheb), inhibiting Rheb from activating mTORC1 (40). Thus, one possible explanation is that sleep deprivation reduces cAMP, which would elicit more PDE4D interaction with Rheb and thereby decrease the interaction between Rheb and mTOR (40). This would ultimately lead to reduced mTORC1-mediated translation. Previously, we have shown that sleep deprivation specifically increases PDE4A5, not PDE4D (28, 30). It would be very interesting to examine in future experiments if PDE4A5 interacts with Rheb in a similar fashion as PDE4D, as this has yet to be determined (41).
The spatial, contextual, and declarative memories that are dependent on the hippocampus are particularly susceptible to the effects of insufficient sleep (18, 19, 42). The object-based location memory task we used to measure the impact of sleep deprivation on spatial memory in mice is specifically dependent on the hippocampus. Therefore, we focused our studies on this region of the brain. Others have demonstrated that sleep deprivation also increases AMPK activity in the hypothalamus and the basal forebrain (43, 44). Further, they have shown that this increase in AMPK activity is accompanied by decreased ATP abundance (44). Protein synthesis is an energy-rich process, and the insulin signaling pathway is central to regulating whether protein synthesis occurs given the metabolic resources available to the cell (24). Increased AMPK activity with sleep deprivation suggests ATP depletion in the hippocampus (38), which would lead to reductions in protein synthesis and impairments in memory consolidation. Indeed, many of the other molecular and cellular consequences of sleep deprivation in the hippocampus including decreased cAMP signaling (22, 28, 30), and increased adenosine (22, 45, 46), could be due to reduced abundance of ATP. Thus, our results support the hypothesis that one of the functions of sleep is cellular energy restoration (47).
Materials and Methods
Animal subjects
Male C57BL/6J mice were obtained at 6 to 8 weeks of age from The Jackson Laboratories for these experiments. Mice were housed individually on a 12 hour light/dark schedule with lights on at 7:00 A.M. Food and water were available ad libitum. All experiments were conducted according to the National Institutes of Health guidelines for animal care and use and were approved by the University of Pennsylvania’s Institutional Animal Care and Use Committee.
Sleep deprivation
Mice were sleep deprived for 5 hours using the gentle handling method (17, 28, 31, 48–50). Briefly, this technique is comprised of manual cage tapping, cage jostling, and nestlet disturbance. Prior to sleep deprivation, each animal was handled daily for 3–6 days using the same interventions used during sleep deprivation for 1–2 minutes. NSD mice were left undisturbed in their home cages.
Western blotting and ELISA
Mice were sacrificed by cervical dislocation at the conclusion of sleep deprivation experiments. The hippocampus was rapidly dissected and flash frozen on dry ice. Total protein lysates were prepared by homogenizing the hippocampus in buffer containing 50 mM tris-HCl pH 7.5, 150 mM KCl, 1 mM EDTA, 1 mM DTT, as well as protease (Roche) and phosphatase inhibitors (ThermoScientific). For Western blotting, samples were normalized on the basis of total protein content measured by Bradford assay (Bio-Rad). Proteins were separated by 4–20% tris-glycine SDS-PAGE (Bio-Rad) and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked in 5% BSA-TBST or 5% milk + 0.5% BSA-TBST and incubated overnight at 4°C in primary antibody. Antibody concentrations were at manufacturers’ recommendations (p-4EBP, 4EBP2, AMPKα, Bip, eIF4E, eIF4G1, p-mTOR, mTOR, p-S6, S6, p-S6 Kinase, Rictor, p-TSC2, TSC2 [1:1000], Cell Signaling; p-AMPKα, S6K [1:2000], Cell Signaling; p-eIF2α [1:1000], Invitrogen; Raptor [1:2000], Bethyl Laboratories; Arc [1:2000], Synaptic Systems; puromycin 12D10 [1:5000] from 5 mg/ml stock, Millipore).
Afterwards, they were washed and incubated with appropriate horseradish peroxidase conjugated goat anti-mouse or anti-rabbit IgG ([1:5,000], Santa Cruz) at room temperature for 1 hour. Blot were washed and exposed via chemiluminescence using ImageQuant LAS 4000 (GE) and quantified using ImageJ. The density of signal was normalized to GAPDH ([1:1000], Santa Cruz) or β-tubulin ([1:10,000], Sigma). For immunoprecipitations, hippocampal lysates were pre-cleared using protein A/G agarose beads (Thermoscientific/Pierce) while rotating for 1 hour at 4°C. Antibody was added to the cell lysates and incubated while rotating overnight at 4°C. Protein A/G agarose beads were then added to the lysates and incubated while rotating overnight at 4°C. Lysates were centrifuged and agarose beads were washed four times in lysis buffer. Immunoprecipitates were analyzed by Western blot. Negative control IgG pulldowns were analyzed in lysates that were treated with rabbit IgG (Bethyl Laboratories). ELISA analysis of hippocampal lysates were conducted in accordance to the manufacturer’s guidelines (Cell Signaling).
Quantitative PCR
RNA preparation, cDNA synthesis, and quantitative real-time RT-PCR (qPCR) analyses were performed as previously described (17, 51). Briefly, RNA concentration and purity were quantified by NanoDrop spectrophotometry (Thermoscientific), and cDNA were generated using the RETROscript kit (Ambion) with 1 μg of RNA as template. Three technical replicates were measured by qPCR using ABI Prism 7000. Data were normalized to Actg1, Hprt, and Tuba4a. Fold change was calculated from the delta Ct values with corrections for housekeeping gene expression for each sample.
In vivo translation assay
Proteins were labeled using an adaptation of the SUnSET protocol (52–54), also known as RiboPuroMycylation (23). A week before sleep deprivation experiments to allow for post-operative recovery, mice were anesthetized with isoflurane and mounted onto a stereotaxic apparatus. Intracerebroventricular (ICV) cannulae were implanted unilaterally at −0.5 mm anterioposterior and +1.0 mm mediolateral from bregma. At the start of sleep deprivation experiments, 25 μg in 1 μl of puromycin (Sigma) was infused into the ventricle over 5 minutes. At the end of sleep deprivation experiments, the contralateral hippocampus was dissected and flash frozen. Proteins labeled with puromycin were identified by Western blot analysis from hippocampal lysates.
Viral constructs and surgeries
The eIF4EBP2 gene construct was generated using Geneart (Millipore) with an HA peptide tag at the C-terminal end. A 0.4 kb CamKIIα promoter fragment was used to restrict expression to excitatory neurons. The pAAV9-CamKIIα0.4-eIF4EBP2-HA recombinant, along with control pAAV9-CamKIIα0.4-eGFP, were constructed and packaged by the University of Pennsylvania Vector Core. Viral titers ranged from 2.4 × 1012 to 1.47 × 1013 genome copy per μl. Mice were anesthetized with isoflurane and mounted onto a stereotaxic apparatus. Depending on the titer, approximately 1 μl of the virus suspension was injected bilaterally at −1.9 mm anterioposterior, 1.5 mm mediolateral, and −1.5 mm dorsoventral from bregma into the hippocampi of mice using a 33 ga beveled NanoFil needle, a microsyringe pump, and controller (WPI). Three weeks after surgery, mice were tested in the object-place recognition task and hippocampal in vivo translation was assayed.
Perfusion and immunohistochemistry
Mice were transcardially perfused with 10 ml of PBS prior to infusion of 4% paraformaldehyde (PFA) in PBS. Perfused brains were post-fixed for 24 hours in 4% PFA at 4°C before immersion in 30% sucrose. After cryoprotection with 30% sucrose, 30 μm coronal sections were cut. To validate expression of 4EBP2 virus in the hippocampus, immunohistochemical analyses were performed on mouse brain sections. Sections were washed 3 times in PBS for 5 minutes each, and then incubated in 0.3% H2O2 in PBS for 30 minutes. Afterwards, sections were washed 3 times in PBS for 10 minutes each. They were blocked in 5% of appropriate serum (Normal Goat Serum; Normal Donkey Serum) in 0.1% Triton in PBS for 30 minutes. Sections were incubated in primary antibodies (anti-HA rat monoclonal antibody, [1:500], Roche; anti-CaMKII goat polyclonal, [1:50] Santa Cruz; anti-parvalbumin rabbit polyclonal, [1:1000], Abcam; anti-GFAP mouse polyclonal, [1:500], FMD Millipore) in 1% serum in 1% Triton-PBS at room temperature overnight. Sections were washed 3 times in PBS for 5 minutes each. They were incubated in appropriate secondary antibodies [1:1000] (goat anti-rat IgG, Alexa Fluor 488; donkey anti-goat IgG, Alexa Fluor 555; donkey anti-rabbit IgG, Alexa Fluor 555; goat anti-mouse IgG, Alexa Fluor 555) in PBS for 4 hours at room temperature in the dark. They were washed 6 times over 60 minutes in PBS and mounted onto slides using 0.7% gelatin in H2O. Slides were allowed to dry overnight and then cover slipped with Permafluor. Sections were imaged with an epiflourescent microscope to determine specificity of viral 4EBP2 expression in the hippocampus.
Object-place recognition task
This hippocampus-dependent memory task was conducted as previously described (30, 31, 55, 56). Sixteen days after viral surgeries, mice were handled for 2 minutes each day, for 5 consecutive days leading up to experimentation. At the beginning of the light phase (zeitgeber time 0), mice were placed in the empty box for 6 minutes for habituation. After a 3 minute intertrial interval, mice were placed in the box with 3 different objects (a 100 ml glass bottle, a white cylinder, and a metallic rectangular tower) for 3 consecutive 6-minute training sessions. Each training session was separated by a 3-minute interval during which the animals were returned to the holding cages. Directly after training, mice were either sleep deprived or left undisturbed. Twenty-four hours following the training session, mice were re-introduced to the spatial context in a single 6-minute test session with 1 of the 3 objects moved to a novel location. Objects and locations were balanced between treatment groups. Video of behavior was recorded and scored off-line by an experimenter that was blind to treatment. Exploration of the objects was defined as the amount of time mice were oriented toward an object within close proximity or touching it.
Statistical analyses
All blots and images are representative of at least 3 independent assays. Data represent means ± SEM. Student’s t tests were used to analyze the gene expression and biochemical data from sleep deprivation experiments. Two-way ANOVAs were used to analyze the biochemical data from the four group viral experiments. Dunnett’s test was used for post hoc comparisons. The Wilcoxon Rank Sums test and the Kruskal-Wallis test were used to analyze the behavioral data. When p < 0.05, differences were considered statistically significant.
Supplementary Material
Fig. S1: Schematic showing the mechanism of attenuated translation in the hippocampus caused by sleep deprivation.
Fig. S2: Control immunoblots of hippocampal extracts treated with rabbit IgG.
Fig. S3: The abundance of phosphorylated TSC2 does not change after 5 hours of sleep deprivation.
Fig. S4: Five hours of sleep deprivation reduces the abundance of phosphorylated 4EBP2, which rebounds after 2.5 hours of recovery sleep.
Fig. S5: Abundance of phosphorylated eIF2α does not change after 5 hours of sleep deprivation.
Acknowledgments
We would like to thank N. Grissom, T. Jongens, and N. Sonenberg for editorial input on the manuscript, P. Hernandez for technical support, and C. Hoeffer and E. Klann for fruitful discussions and advice.
Funding: This work is supported by National Institutes of Health grants F32MH099730 (to J.C.T.), R21MH102703 (to T.A.), P01AG017628 (to T.A.), K12GM081259 (to J.C.T., Principle Investigator Y. Paterson), and T32NS007413 (to L.P., PI M. Robinson). This work is also supported by the Kwanjeong Educational Foundation (to A.P.).
Footnotes
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author contributions: J.C.T., L.P., M.W., A.P., and R.H. designed and performed the sleep deprivation studies. J.C.T., E.J.D., E.v.T., and C.W.C. performed surgeries, along with the biochemistry and behavioral experiments. J.C.T. and S.G.P. performed the qPCR experiments. J.C.T, E.G., and P.P. designed the in vivo translation experiment. J.C.T. and J.H. performed the statistical analyses. J.C.T. and T.A. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The pAAV9-CamKIIα0.4-eIF4EBP2-HA virus is available by contacting T.A..
References and Notes
- 1.Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learning & memory. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham WC, Williams JM. LTP maintenance and its protein synthesis-dependence. Neurobiology of learning and memory. 2008;89:260–268. doi: 10.1016/j.nlm.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 5.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nature reviews Molecular cell biology. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buffington SA, Huang W, Costa-Mattioli M. Translational control in synaptic plasticity and cognitive dysfunction. Annual review of neuroscience. 2014;37:17–38. doi: 10.1146/annurev-neuro-071013-014100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes & development. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 9.Yang Q, Guan KL. Expanding mTOR signaling. Cell research. 2007;17:666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 10.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends in neurosciences. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert V, Hall MN. mTOR signaling in cellular and organismal energetics. Current opinion in cell biology. 2015;33:55–66. doi: 10.1016/j.ceb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, Harper ME, Tremblay ML, Sonenberg N. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nature medicine. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- 13.Banko JL, Poulin F, Hou L, DeMaria CT, Sonenberg N, Klann E. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:9581–9590. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seibt J, Dumoulin MC, Aton SJ, Coleman T, Watson A, Naidoo N, Frank MG. Protein synthesis during sleep consolidates cortical plasticity in vivo. Current biology: CB. 2012;22:676–682. doi: 10.1016/j.cub.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiology & behavior. 1990;48:749–753. doi: 10.1016/0031-9384(90)90220-x. [DOI] [PubMed] [Google Scholar]
- 16.Nakanishi H, Sun Y, Nakamura RK, Mori K, Ito M, Suda S, Namba H, Storch FI, Dang TP, Mendelson W, Mishkin M, Kennedy C, Gillin JC, Smith CB, Sokoloff L. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. The European journal of neuroscience. 1997;9:271–279. doi: 10.1111/j.1460-9568.1997.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 17.Vecsey CG, Peixoto L, Choi JH, Wimmer M, Jaganath D, Hernandez PJ, Blackwell J, Meda K, Park AJ, Hannenhalli S, Abel T. Genomic analysis of sleep deprivation reveals translational regulation in the hippocampus. Physiological genomics. 2012;44:981–991. doi: 10.1152/physiolgenomics.00084.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abel T, Havekes R, Saletin JM, Walker MP. Sleep, plasticity and memory from molecules to whole-brain networks. Current biology: CB. 2013;23:R774–788. doi: 10.1016/j.cub.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banks S, Dinges DF. Behavioral and physiological consequences of sleep restriction. Journal of clinical sleep medicine: JCSM: official publication of the American Academy of Sleep Medicine. 2007;3:519–528. [PMC free article] [PubMed] [Google Scholar]
- 20.Curcio G, Ferrara M, De Gennaro L. Sleep loss, learning capacity and academic performance. Sleep medicine reviews. 2006;10:323–337. doi: 10.1016/j.smrv.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Graves LA, Heller EA, Pack AI, Abel T. Sleep deprivation selectively impairs memory consolidation for contextual fear conditioning. Learning & memory. 2003;10:168–176. doi: 10.1101/lm.48803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havekes R, Vecsey CG, Abel T. The impact of sleep deprivation on neuronal and glial signaling pathways important for memory and synaptic plasticity. Cellular signalling. 2012;24:1251–1260. doi: 10.1016/j.cellsig.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. The Journal of cell biology. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annual review of pharmacology and toxicology. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 25.Gwinn DM, Schackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology: CB. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 27.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, Fingar DC. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. The Journal of biological chemistry. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vecsey CG, Baillie GS, Jaganath D, Havekes R, Daniels A, Wimmer M, Huang T, Brown KM, Li XY, Descalzi G, Kim SS, Chen T, Shang YZ, Zhuo M, Houslay MD, Abel T. Sleep deprivation impairs cAMP signalling in the hippocampus. Nature. 2009;461:1122–1125. doi: 10.1038/nature08488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber R, Deboer T, Tobler I. Effects of sleep deprivation on sleep and sleep EEG in three mouse strains: empirical data and simulations. Brain research. 2000;857:8–19. doi: 10.1016/s0006-8993(99)02248-9. [DOI] [PubMed] [Google Scholar]
- 30.Havekes R, Bruinenberg VM, Tudor JC, Ferri SL, Baumann A, Meerlo P, Abel T. Transiently increasing cAMP levels selectively in hippocampal excitatory neurons during sleep deprivation prevents memory deficits caused by sleep loss. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:15715–15721. doi: 10.1523/JNEUROSCI.2403-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prince TM, Wimmer M, Choi J, Havekes R, Aton S, Abel T. Sleep deprivation during a specific 3-hour time window post-training impairs hippocampal synaptic plasticity and memory. Neurobiology of learning and memory. 2014;109:122–130. doi: 10.1016/j.nlm.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poplawski SG, Schoch H, Wimmer ME, Hawk JD, Walsh JL, Giese KP, Abel T. Object-location training elicits an overlapping but temporally distinct transcriptional profile from contextual fear conditioning. Neurobiology of learning and memory. 2014;116:90–95. doi: 10.1016/j.nlm.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisor JP. The sleep-deprived hippocampus: a loss in translation. Physiological genomics. 2013;45:26–27. doi: 10.1152/physiolgenomics.00156.2012. [DOI] [PubMed] [Google Scholar]
- 34.Gronli J, Soule J, Bramham CR. Sleep and protein synthesis-dependent synaptic plasticity: impacts of sleep loss and stress. Frontiers in behavioral neuroscience. 2013;7:224. doi: 10.3389/fnbeh.2013.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gronli J, Dagestad G, Milde AM, Murison R, Bramham CR. Post-transcriptional effects and interactions between chronic mild stress and acute sleep deprivation: regulation of translation factor and cytoplasmic polyadenylation element-binding protein phosphorylation. Behavioural brain research. 2012;235:251–262. doi: 10.1016/j.bbr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Hartman NW, Lin TV, Zhang L, Paquelet GE, Feliciano DM, Bordey A. mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell reports. 2013;5:433–444. doi: 10.1016/j.celrep.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Carroll M, Dyer J, Sossin WS. Serotonin increases phosphorylation of synaptic 4EBP through TOR, but eukaryotic initiation factor 4E levels do not limit somatic cap- dependent translation in aplysia neurons. Molecular and cellular biology. 2006;26:8586–8598. doi: 10.1128/MCB.00955-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bidinosti M, Ran I, Sanchez-Carbente MR, Martineau Y, Gingras AC, Gkogkas C, Raught B, Bramham CR, Sossin WS, Costa-Mattioli M, DesGroseillers L, Lacaille JC, Sonenberg N. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Molecular cell. 2010;37:797–808. doi: 10.1016/j.molcel.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graber TE, McCamphill PK, Sossin WS. A recollection of mTOR signaling in learning and memory. Learning & memory. 2013;20:518–530. doi: 10.1101/lm.027664.112. [DOI] [PubMed] [Google Scholar]
- 40.Kim HW, Ha SH, Lee MN, Huston E, Kim DH, Jang SK, Suh PG, Houslay MD, Ryu SH. Cyclic AMP controls mTOR through regulation of the dynamic interaction between Rheb and phosphodiesterase 4D. Molecular and cellular biology. 2010;30:5406–5420. doi: 10.1128/MCB.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC. Advances in targeting cyclic nucleotide phosphodiesterases. Nature reviews Drug discovery. 2014;13:290–314. doi: 10.1038/nrd4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diekelmann S, Born J. The memory function of sleep. Nature reviews Neuroscience. 2010;11:114–126. doi: 10.1038/nrn2762. [DOI] [PubMed] [Google Scholar]
- 43.Chikahisa S, Fujiki N, Kitaoka K, Shimizu N, Sei H. Central AMPK contributes to sleep homeostasis in mice. Neuropharmacology. 2009;57:369–374. doi: 10.1016/j.neuropharm.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 44.Dworak M, McCarley RW, Kim T, Kalinchuk AV, Basheer R. Sleep and brain energy levels: ATP changes during sleep. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:9007–9016. doi: 10.1523/JNEUROSCI.1423-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Florian C, Vecsey CG, Halassa MM, Haydon PG, Abel T. Astrocyte-derived adenosine and A1 receptor activity contribute to sleep loss-induced deficits in hippocampal synaptic plasticity and memory in mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:6956–6962. doi: 10.1523/JNEUROSCI.5761-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, Frank MG. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron. 2009;61:213–219. doi: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chikahisa S, Sei H. The role of ATP in sleep regulation. Frontiers in neurology. 2011;2:87. doi: 10.3389/fneur.2011.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagewoud R, Havekes R, Novati A, Keijser JN, Van der Zee EA, Meerlo P. Sleep deprivation impairs spatial working memory and reduces hippocampal AMPA receptor phosphorylation. Journal of sleep research. 2010;19:280–288. doi: 10.1111/j.1365-2869.2009.00799.x. [DOI] [PubMed] [Google Scholar]
- 49.Hagewoud R, Whitcomb SN, Heeringa AN, Havekes R, Koolhaas JM, Meerlo P. A time for learning and a time for sleep: the effect of sleep deprivation on contextual fear conditioning at different times of the day. Sleep. 2010;33:1315–1322. doi: 10.1093/sleep/33.10.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vecsey CG, Wimmer ME, Havekes R, Park AJ, Perron IJ, Meerlo P, Abel T. Daily acclimation handling does not affect hippocampal long-term potentiation or cause chronic sleep deprivation in mice. Sleep. 2013;36:601–607. doi: 10.5665/sleep.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nature methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 53.Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoeffer CA, Tang W, Wong H, Santillan A, Patterson RJ, Martinez LA, Tejada-Simon MV, Paylor R, Hamilton SL, Klann E. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron. 2008;60:832–845. doi: 10.1016/j.neuron.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliveira AM, Hawk JD, Abel T, Havekes R. Post-training reversible inactivation of the hippocampus enhances novel object recognition memory. Learning & memory. 2010;17:155–160. doi: 10.1101/lm.1625310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Havekes R, Canton DA, Park AJ, Huang T, Nie T, Day JP, Guercio LA, Grimes Q, Luczak V, Gelman IH, Baillie GS, Scott JD, Abel T. Gravin orchestrates protein kinase A and beta2-adrenergic receptor signaling critical for synaptic plasticity and memory. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:18137–18149. doi: 10.1523/JNEUROSCI.3612-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Schematic showing the mechanism of attenuated translation in the hippocampus caused by sleep deprivation.
Fig. S2: Control immunoblots of hippocampal extracts treated with rabbit IgG.
Fig. S3: The abundance of phosphorylated TSC2 does not change after 5 hours of sleep deprivation.
Fig. S4: Five hours of sleep deprivation reduces the abundance of phosphorylated 4EBP2, which rebounds after 2.5 hours of recovery sleep.
Fig. S5: Abundance of phosphorylated eIF2α does not change after 5 hours of sleep deprivation.