Abstract
RNA folds to form complex structures vital to many cellular functions. Proteins facilitate RNA folding at both the secondary and tertiary structure levels. An absolute prerequisite for understanding RNA folding and ribonucleoprotein (RNP) assembly reactions is a complete understanding of the RNA structure at each stage of the folding or assembly process. Here we provide a guide for comprehensive and high-throughput analysis of RNA secondary and tertiary structure using SHAPE and hydroxyl radical footprinting. As an example of the strong and sometimes surprising conclusions that can emerge from high-throughput analysis of RNA folding and RNP assembly, we summarize the structure of the bI3 group I intron RNA in four distinct states. Dramatic structural rearrangements occur in both secondary and tertiary structure as the RNA folds from the free state to the active, six-component, RNP complex. As high-throughput and high-resolution approaches are applied broadly to large protein-RNA complexes, other proteins previously viewed as making simple contributions to RNA folding are also likely to be found to exert multifaceted, long-range, cooperative, and non-additive effects on RNA folding. These protein-induced contributions add another level of control, and potential regulatory function, in RNP complexes.
1. Introduction
RNA is actively involved in diverse cellular processes, including protein synthesis, gene regulation, and maintenance of genome stability (Gesteland et al., 2006). In many critical examples, RNA function is directly related to its ability to fold back on itself to form complex higher order structures, or in some cases, the ability to achieve multiple structures. For example, during mRNA processing, catalytic RNAs such as group I and group II introns, RNase P, small ribozymes, and some riboswitches fold into highly ordered three-dimensional structures to cleave RNA (Torres-Larios et al., 2006; Scott, 2007; Cochrane and Strobel, 2008). Other RNAs, especially riboswitches, sample multiple stable conformations that then allow these RNAs to function as metabolite sensors that regulate gene expression (Winkler and Breaker, 2005; Edwards et al., 2007)
Many RNAs, especially larger RNAs like catalytic introns and the ribosome, also recruit proteins to facilitate fast and stable folding in the cell. The proteins that facilitate RNA folding are generally classified into two categories, chaperones and cofactors (Herschlag, 1995; Weeks, 1997; Rajkowitsch et al., 2007). Chaperone proteins interact transiently with RNA and function to allow an RNA to re-fold and achieve a thermodynamically favored structure. Cofactor proteins are defined by their ability to bind stably to an RNA to create a structurally well defined ribonucleoprotein (RNP) complex. Cofactors often stabilize a specific, active, tertiary structure. The mechanisms by which proteins facilitate RNA folding can be complex and emerging work has begun to emphasize that some proteins do not fit clearly into the traditional definitions of chaperones and cofactors.
An absolute prerequisite for understanding RNA folding and ribonucleoprotein assembly reactions is a complete understanding of the RNA structure at each stage of the predominant folding or assembly pathway. Information regarding the final, stable, structure of an RNA or RNA-protein complex can be obtained by high resolution NMR or by crystallography or, in a few cases, by molecular modeling. However, for most initial and intermediate states, RNA secondary and tertiary structure must be inferred from either comparative sequence analysis or chemical probing approaches.
The most successful method for determining an RNA secondary structure has been by phylogenetic comparative sequence analysis (Michel and Westhof, 1990; Gutell et al., 2002). However, covariation analysis only demonstrates that a pairing is likely to exist and not that a physical pairing occurs (Woese et al., 1980). Covariation analysis also does not reveal whether a specific physical pairing actually exists in any given RNA state or folding intermediate.
Alternatively, RNA structure information can be inferred experimentally by treating an RNA with small molecule or enzyme reagents that are sensitive to local RNA structure. Two broad classes of experiments have proven especially useful. First, many chemical and enzymatic reagents react with partial selectivity towards single stranded nucleotides. Reactivity patterns obtained using these reagents provide information useful for inferring base pairing interactions (Ehresmann et al., 1987; Brunel and Romby, 2000). Second, Fenton chemistry can be used to generate short lived hydroxyl radicals in solution, which then react with the RNA backbone in a way that is sensitive to solvent accessibility (Tullius and Greenbaum, 2005; Tullius et al., 1987). Identification of RNA backbone positions that are protected from the hydroxyl radical reagent can provide strong evidence for higher order tertiary interactions (Latham and Cech, 1989).
Conventional approaches for analyzing RNA base pairing or tertiary interactions often lead to a view of an RNA structure that is incomplete in key features. Conventional secondary structure-selective chemical and enzymatic RNA mapping reagents tend to have a narrow dynamic range and typically yield information for one-half, or less, of the nucleotides in an RNA. In addition, both secondary structure-sensitive and hydroxyl radical-mediated solvent accessibility experiments have been analyzed predominantly by sequencing gel electrophoresis. The effort required to analyze long, intact, RNAs by this approach is very large. A useful approach can be to focus on short RNAs, on simplified models of larger RNAs, or on small region within a large RNA. The challenge in focusing on a segment of a large RNA is that long-range and unanticipated interactions, critical for understanding how an RNA functions or how proteins affect an RNA folding process, will be missed. These challenges can be addressed by new chemistries and high-throughput analysis approaches.
Two methods that, in principle, yield comprehensive single nucleotide resolution information about RNA secondary and tertiary structure are SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) (Merino et al., 2005; Wilkinson et al., 2005; Wilkinson et al., 2006) and the aforementioned hydroxyl radical footprinting (Tullius et al., 1987; Latham and Cech, 1989; Brenowitz et al., 2002; Tullius and Greenbaum, 2005). SHAPE chemistry interrogates local nucleotide flexibility, while hydroxyl radical footprinting assesses solvent accessibility and, thus, the global RNA fold. Both techniques are adaptable to a variety of reaction conditions, including the presence of proteins. Additionally, both techniques are insensitive to nucleotide identity and can potentially provide structural information at nearly every nucleotide position in an RNA.
When used in tandem, SHAPE and hydroxyl radical footprinting comprehensively probe multiple levels of RNA structure and provide an impressively detailed view of an RNA or RNP folding state or assembly intermediate. Combining these sensitive structural probes with an experimental read-out using capillary electrophoresis makes it possible to characterize 350–600 nucleotides in a single high-throughput experiment. Thus, large RNAs and complex RNP structures can be studied with facility and in detail. These developments in high-throughput in-solution RNA structure analysis are revealing unanticipated, and complex, mechanisms for how RNAs fold and interact with proteins to achieve a final active structure. In this review, we will focus on the bI3 group I intron RNA and its RNP complexes as an example of how these technological advances are leading to new insights into RNA folding and RNP assembly reactions.
2. Theory
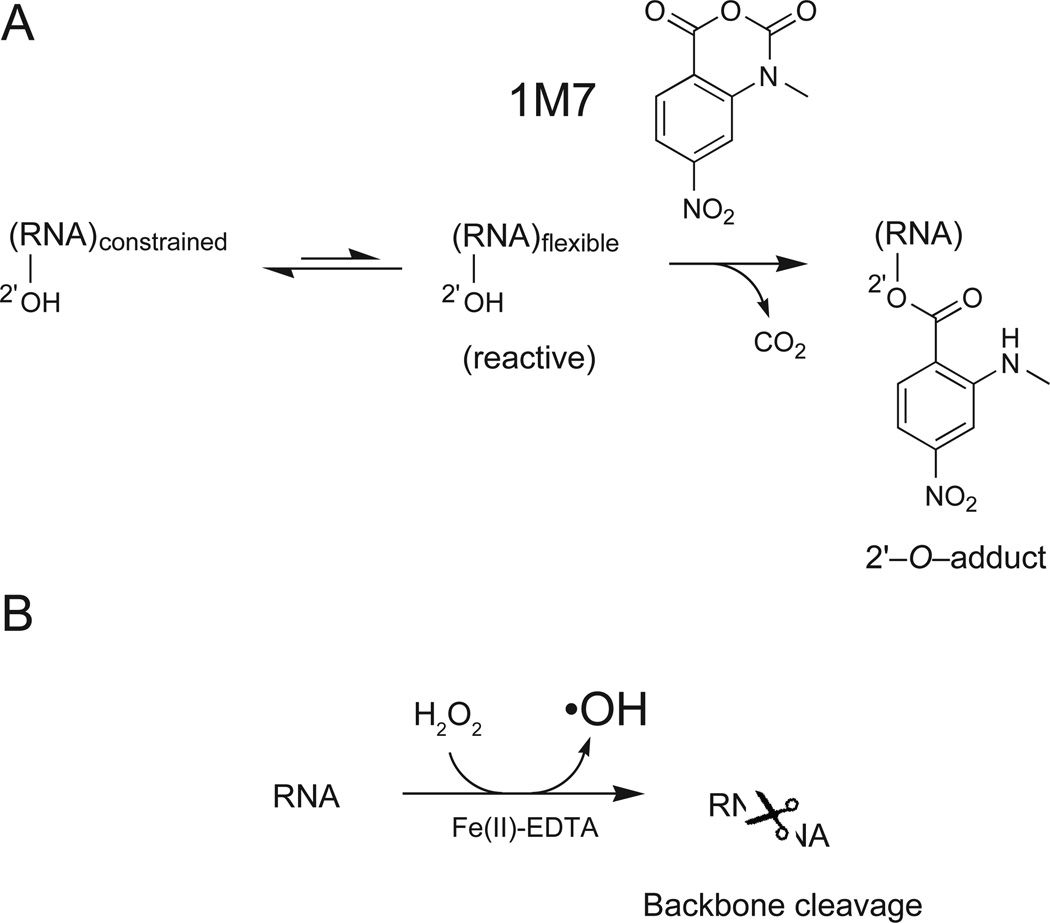
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry is a robust and comprehensive method for analyzing local nucleotide dynamics in RNA (Merino et al., 2005; Wilkinson et al., 2005; Wilkinson et al., 2006). In a SHAPE reaction, the RNA is treated with an electrophilic reagent like NMIA or 1M7 (Merino et al., 2005; Mortimer and Weeks, 2007) that selectively reacts with the ribose 2′-hydroxyl group at conformationally flexible nucleotides (Figure 1A). SHAPE reactivity reports local nucleotide flexibility because unconstrained nucleotides (like those in single stranded structures) are more likely to sample relatively rare conformations that make the 2′-hydroxyl more nucleophilic and promote the reaction that forms 2′-O-adducts. In contrast, nucleotides constrained by base pairing or other interactions react poorly with SHAPE reagents. SHAPE chemistry is not strongly sensitive to solvent accessibility and, for example, conformationally dynamic but solvent inaccessible nucleotides are generally still reactive (Merino et al., 2005; Gherghe et al., 2008). SHAPE reagents both modify flexible sites in RNA and also undergo self-inactivating hydrolysis. As a practical matter, this auto-inactivation means that reagent reactivity does not need to be quenched, as long as the 2'-O-adduct forming reaction is allowed to proceed to completion (Merino et al., 2005). These bulky RNA adducts can be detected as stops to primer extension.
Figure 1.
Schemes for (A) RNA SHAPE chemistry and (B) RNA hydroxyl radical footprinting chemistries.
The current gold standard for mapping solvent accessibility at the RNA backbone is hydroxyl radical footprinting. Short-lived hydroxyl radicals (•OH) are generated in situ in a reaction between Fe2+ and H2O2 (Tullius et al., 1987; Latham and Cech, 1989; Brenowitz et al., 2002; Tullius and Greenbaum, 2005). The iron ion is chelated by EDTA, which prevents the ion from binding directly to RNA. Hydroxyl radicals are then generated at the periphery of the RNA, leading to RNA cleavage (Figure 1B). Cleavage occurs preferentially at RNA positions accessible to solvent, although other factors also contribute to reactivity (Lu et al., 1990; Balasubramanian et al., 1998). Backbone cleavage sites can also be detected by primer extension.
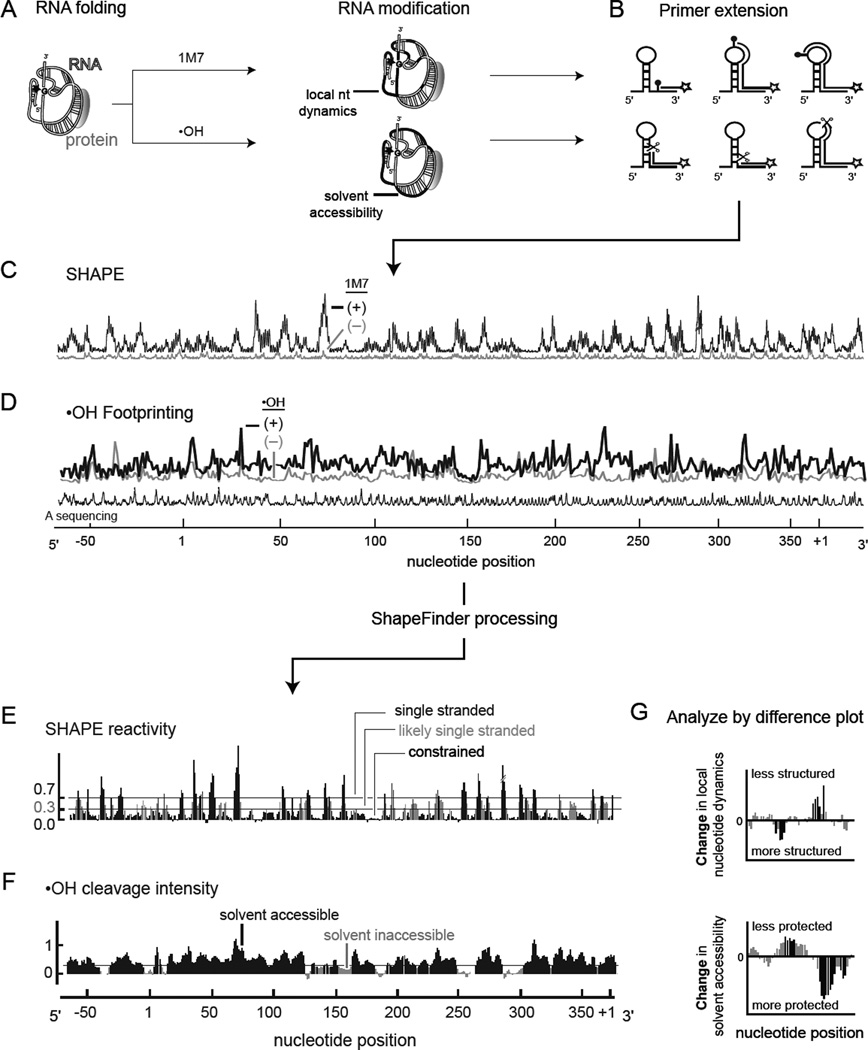
Both SHAPE and the hydroxyl radical experiment can be converted to high-throughput formats by detecting sites of 2'-O-adduct formation or of backbone cleavage, respectively, by reverse transcriptase-mediated primer extension, resolved by capillary electrophoresis (Figure 2A–D). In addition to the RNA modification reaction, a control no-reagent experiment is used to assess background. These reactions are compared to dideoxy sequencing markers to assign nucleotide positions. After data processing, each nucleotide is characterized by a quantitative SHAPE reactivity or hydroxyl radical cleavage intensity that directly reflects the underlying RNA structure at single nucleotide resolution (Figure 2E,F). In the SHAPE experiment, single stranded nucleotides are reactive while base paired nucleotides are generally unreactive (Figure 2E). Among other applications, SHAPE data can be converted to pseudo-free energy change terms to constrain the output of a thermodynamic structure prediction program. This blended approach generally produces highly accurate RNA secondary structure models (Wilkinson et al., 2008; Deigan et al., 2009). For the hydroxyl radical experiment, inspection of the final reactivity versus position histogram makes it possible to identify compact tertiary interactions as solvent inaccessible elements in the context of a large RNA (Figure 2F).
Figure 2.
High-throughput RNA structure mapping. (A) The RNA is folded and modified with a SHAPE reagent (1M7 or NMIA) or cleaved by hydroxyl radical chemistry. (B) RNA modifications or cleavages yield stops to primer extension, which are detected using color-coded, fluorescently labeled primers. (C,D) Resulting cDNAs are resolved by automated capillary electrophoresis. (E,F) After quantifying the net reactivity at each nucleotide position, the data are normalized on a scale spanning 0 to ~2, where 1.0 is defined as the average intensity at highly reactive positions. (G) Changes in local nucleotide dynamics and solvent accessibility that distinguish any two RNA folding or RNP assembly states are readily detected at single nucleotide resolution by difference plot analysis.
3. Practice
Both SHAPE and hydroxyl radical footprinting advantageously yield a comprehensive and single nucleotide resolution view of an RNA state in a concise set of experiments. In outline, both SHAPE and hydroxyl radical footprinting involve folding the RNA, adding protein (if appropriate), incubating the RNA with either the SHAPE electrophile or the hydroxyl radical reagents, recovering the RNA, and mapping the sites of modified or cleaved nucleotides by primer extension (Figure 2). This experimental overview is based on the analysis of the structure of the free bI3 group I intron RNA and its complexes with protein facilitators.
3.1 RNA Folding
SHAPE and hydroxyl radical footprinting experiments are most commonly performed with RNAs that have been generated by in vitro transcription (Latham and Cech, 1989; Tullius and Greenbaum, 2005; Merino et al., 2005; Wilkinson et al., 2006; Duncan and Weeks, 2008). These RNAs require purification by denaturing gel electrophoresis and then must be renatured to achieve a biologically relevant conformation. RNA folding reactions involving the renatured RNA can then be initiated in several ways, including a change in ionic strength or addition of a protein or small molecule ligand. This protocol describes a general method in which an RNA is first heated and snap cooled in a low ionic strength buffer to eliminate multimeric forms. RNA folding is initiated by adding monovalent and divalent ions to this solution. The RNA is folded in a single step and then separated into (+) and (–) reagent reactions for either SHAPE and hydroxyl radical footprinting.
-
Add 4 pmol RNA in 10 µL sterile water to a 0.65 mL (Eppendorf) reaction tube.
NOTE: This amount of RNA is based on using 2 pmol per (+) and (–) reagent reaction. The amount of RNA needed for complete analysis may vary with the RNA. The useful range is 0.5–8 pmol per reaction.
Heat the RNA to 95 °C for 1 min. Place the RNA on ice for 1 min.
Add 24 µL 5× folding buffer [5× = 200 mM MOPS (pH 8.0), 400 mM potassium acetate (KOAc) (pH 8.0), 100 mM MgCl2]. Adjust final volume to 108 µL with water.
-
Incubate at 37 °C for 10 min. This is the “free RNA reaction mix”.
NOTE: For protein binding experiments, add proteins and incubate at appropriate, concentrations, temperature, and time for the system. Adjustment of the volume of added water will be necessary to ensure a final 30 µL reaction volume in the chemical probing step.
3.2 Chemical Probing of RNA Secondary and Tertiary Structure
Modification and cleavage reactions should take place under conditions that yield single hits over the length of RNA to be analyzed by primer extension (~1 modification per 300 nucleotides). For SHAPE, too high or low a concentration of reagent results in a steep signal decay in the fluorescence trace or in a low signal to noise ratio in the (+) reagent lane, respectively. For hydroxyl radical cleavage, excessive cleavage yields a complex cleavage pattern that is biased towards short RNA fragments. For both reactions, fresh reagents are imperative.
3.2.1 SHAPE
-
Aliquot 3 µL 50 mM 1M7 (or NMIA) in DMSO and 3 µL neat DMSO into two 0.65 mL reaction tubes. These will be the (+) and (–) reagent reactions. We strongly recommend 1M7 for routine SHAPE analysis of RNA. 1M7 can be synthesized as described (Mortimer and Weeks, 2007); alternatively, the NMIA reagent can be used.
NOTE: The optimal concentration of 1M7 can vary with RNA length. The useful range is 0.5–8 mM; 5 mM final is a good starting concentration. For longer RNAs, or very AU rich RNAs, use the lower end of these 1M7 concentrations.
Add 27 µL of the free RNA reaction mix to 3 µL 1M7 in DMSO and an additional 27 µL to the tube containing 3 µL neat DMSO (final volume = 30 µL).
-
Allow reaction mixtures to react for 5 hydrolysis half lives. This time is 70 sec for 1M7 and 35 min for NMIA (Merino et al., 2005; Mortimer and Weeks, 2007). No additional quench step is necessary.
NOTE: For protein binding experiments, remove proteins by proteolysis and phenol-chloroform extraction (Duncan and Weeks, 2008) before proceeding to step 4.
Adjust volume to 96 µL with water and recover RNA by ethanol precipitation: add 1 µL 3 M NaCl, 1 µL 20 mg/mL glycogen, 2 µL 0.5 M EDTA (pH 8.0), and 400 µL ethanol to both (+) and (–) reactions, mix, and incubate at −80 °C for 30 min. Sediment the RNA by spinning at maximum speed in a microfuge at 4 °C for 30 min.
Redissolve RNA in 10 µL sterile water.
3.2.2 Hydroxyl Radical Footprinting
NOTE: Fresh solutions are imperative for efficient cleavage reactions. Weigh out ammonium iron (II) sulfate hexahydrate [(NH4)2 Fe(SO4)2•6H2O] and ascorbic acid (sodium salt) into dry reaction tubes; wait to make the solutions until just before addition to the RNA.
-
1
Prepare a solution of 7.5 mM iron(II)/11.25 mM EDTA (pH 8.0).
-
2
Prepare a 0.3% H2O2 solution in water.
-
3
Prepare a 150 mM sodium ascorbate solution in water.
-
4
Transfer two 27 µL aliquots of the free RNA reaction mix to two sterile 0.65 mL reaction tubes. One tube will be used for the (+) •OH reaction and the other for the (−) •OH reaction.
-
5
For the (+) •OH reaction, sequentially add 1 µL each of the iron(II)-EDTA, hydrogen peroxide, and ascorbate solutions to the reaction solution.
-
6
Repeat step 5 for the (−) •OH reaction, substituting 1 µL water for the iron(II)-EDTA solution.
-
7
Quench the reactions after 5 min at 37 °C with 10.5 µL 75% glycerol (v/v).
NOTE: For protein binding experiments, remove proteins by proteolysis and phenol chloroform extraction (Duncan and Weeks, 2008) before proceeding to step 8.
-
8–9
Recover RNA as in steps (4–5) for the SHAPE reaction procedure.
3.3 Primer Extension to Map Modification and Cleavage Sites
Long RNA regions can be analyzed in a single experiment by performing each primer extension using a primer labeled with a color-coded fluorophore. The resulting cDNA products [from the (+) and (−) reagent reactions plus one or two dideoxy sequencing reactions] are combined and resolved in one multifluor run by automated capillary electrophoresis. In a single read, quantitative RNA structural information at single nucleotide resolution can be routinely obtained for 350–600 nts. Analysis is simplified by choosing fluorescent dyes with similar electrophoretic mobilities. The dyes 6-FAM, TET, HEX, and NED require very little correction for fluorophore-induced mobility shifts. This correction is readily performed using the ShapeFinder software (Vasa et al., 2008). Primer extension protocols are identical for RNA analyzed by SHAPE or by hydroxyl radical footprinting.
Add 3 µL of 0.3 µM fluorescently-labeled primer to the RNA solutions from (+) and (–) reagent reactions.
For sequencing reactions, add 3 µL 0.3 µM fluorescently-labeled primer to 1–2 pmol of RNA in 8 µL sterile water.
For all reactions, anneal the primer to the RNA by heating at 65 °C for 5 min, reducing the temperature to 37 °C for 1 min, and placing on ice.
-
Add 6 µL of Superscript III enzyme mix (the solution is a 4:1:1 mixture of 5× first strand buffer (from Invitrogen), 100 mM DTT, and a solution that is 10 mM in each dNTP) to the (+) and (−) reagent reactions and to the sequencing reactions. Also add 1 µL of 5 mM of a selected ddNTP to each sequencing reaction.
NOTE: For AU rich RNAs, better extension can be achieved by decreasing the deoxynucleotide concentration used for sequencing. For example, if using ddTTP to sequence at A, reduce the dTTP concentration in the reverse transcriptase enzyme mix (in step 4) to 5 mM.
Add 1 µL SuperScript III Reverse Transcriptase (Invitrogen) to each tube. Mix well and incubate at 37 °C for 5 min, 52 °C for 20 min, 60 °C for 5 min, and then place on ice.
Quench the primer extension reactions by adding 4 µL of a 1:1 mixture of 4 M NaOAc and 100 mM EDTA (pH 8.0) and place on ice.
Combine 22 µL from each (+) and (−) reaction and 1 or 2 sequencing reactions into a 1.5 mL reaction tube and recover cDNA products by ethanol precipitation (240 µL 100% ethanol). Incubate at -80 °C for 15 min. Sediment the cDNA by spinning at maximum speed in a microfuge at 4 °C for 15 min.
Wash with 800 µL 70% ethanol to remove excess salt (which can lead to poor resolution during capillary electrophoresis).
Repeat step 8 and dry pellet by vacuum for 10 min.
Resuspend in 10 µL deionized formamide.
3.4 cDNA Analysis by Capillary Electrophoresis
Load each 10 µL sample into separate input wells on a capillary electrophoresis DNA sequencing instrument and run.
Export raw traces into ShapeFinder (Vasa et al., 2008).
3.5 Data Analysis
Data analysis using ShapeFinder has been outlined in detail elsewhere (Vasa et al., 2008; Wilkinson et al., 2008). Most tools are straightforward to use. The Align and Integrate tool in ShapeFinder performs a whole trace Gaussian integration to quantify the intensity of every peak in the (+) and (−) reagent lanes. This tool works best for data in which traces have been adjusted to remove fluorescence background and in which peaks corresponding to the same positions have been aligned to overlap consistently.
Analyze SHAPE and hydroxyl radical data using ShapeFinder, as described (Wilkinson et al., 2008; Duncan and Weeks, 2008).
-
ShapeFinder is available at: http://bioinfo.unc.edu/Downloads/index.html.
In brief, adjust the fluorescent baseline (window of 40 pixels), perform a mobility shift to account for the effect of different dyes on cDNA mobility, correct for signal decay, and scale the (+) and (−) reagent traces to make them equal to each other in regions in which reagent-induced reactivities are low. Integrate all peaks in the (+) and (−) reagent traces.
Subtract the (−) from the (+) reagent reactivities to create a table of net reactivity as a function of nucleotide position.
3.6 Normalization
A consistent approach to normalization is important for comparing data sets and for making accurate secondary structure predictions. We normalize SHAPE and hydroxyl radical data to a scale starting at zero (no reactivity) and in which 1.0 is defined as the average intensity of highly reactive positions.
Identify the (usually small number of) highly reactive outliers.
For large data sets (>300 measurements), use a model-free box plot analysis (Deigan et al., 2009). Reactivities greater than 1.5 times the interquartile range are outliers (Chernick and Friis, 2003). If the box plot approach is used for small RNA datasets (less than ~100 nts), the maximum number of outliers should be capped at 5%. After eliminating outliers, compute the mean of the next (highest) 10% of intensities and divide by this value.
For smaller datasets or for RNAs with a relatively small number of reactive peaks, we use a "2%–8%" rule (Duncan and Weeks, 2008; Wilkinson et al., 2008). The most reactive 2% of peaks are taken to be outliers. Then, calculate the mean reactivity for the next 8% most reactive nucleotides and divide all intensities by this value.
Using either a box plot or the 2%–8% rule to identify outliers, the resulting reactivities typically span a scale from 0 to ~2.
For hydroxyl radical cleavage data, it is usually helpful to algebraically smooth the data over a three nucleotide window to account for the structurally heterogeneous reactivity of this reagent.
3.7 SHAPE-Directed RNA Structure Prediction
SHAPE reactivities can be converted to pseudo-free energy change constraints to enable highly accurate prediction of RNA secondary structure (Deigan et al., 2009). SHAPE energies combined with conventional thermodynamic parameters typically yield secondary structure models in which 95% of all accepted base pairs are predicted correctly, for RNAs that do not contain pseudoknots (Deigan et al., 2009).
Use RNAstructure, available at: http://rna.urmc.rochester.edu/rnastructure.html
Set the maximum base-pairing distance to 600 nts.
Input SHAPE data as pseudo-free energy change terms using 2.6 and −0.8 for the slope and intercept parameters, respectively.
4. Examples and Interpretation
4.1 High-throughput Structure Analysis of Protein-Assisted RNA Folding of the bI3 RNA
High-throughput structure probing yields a wealth of structural information about an RNA in an efficiently performed set of experiments. The methods outlined here typically yield in-solution structural information for ≥95% of the nucleotides in intact, full length RNAs (Wilkinson et al., 2008; Duncan and Weeks, 2008; Deigan et al., 2009). Because all regions in an RNA can be probed comprehensively, many fewer structural assumptions have to be made about unprobed regions of an RNA. In addition, we generally find that resolving chemical modification or backbone cleavage information by capillary electrophoresis and quantification using the tools in ShapeFinder yield reactivity information that is of much higher quality than we are able to obtain using sequencing gel approaches. Because the high-throughput data are so quantitative, the RNA structural changes that differentiate two states can be identified rapidly by simply subtracting one reactivity profile from another (Figure 2G). As an example of the impact comprehensive structural information has for developing models for RNA structure and RNP assembly, we probed the bI3 group I intron complex at four distinct folding stages using both SHAPE and hydroxyl radical footprinting.
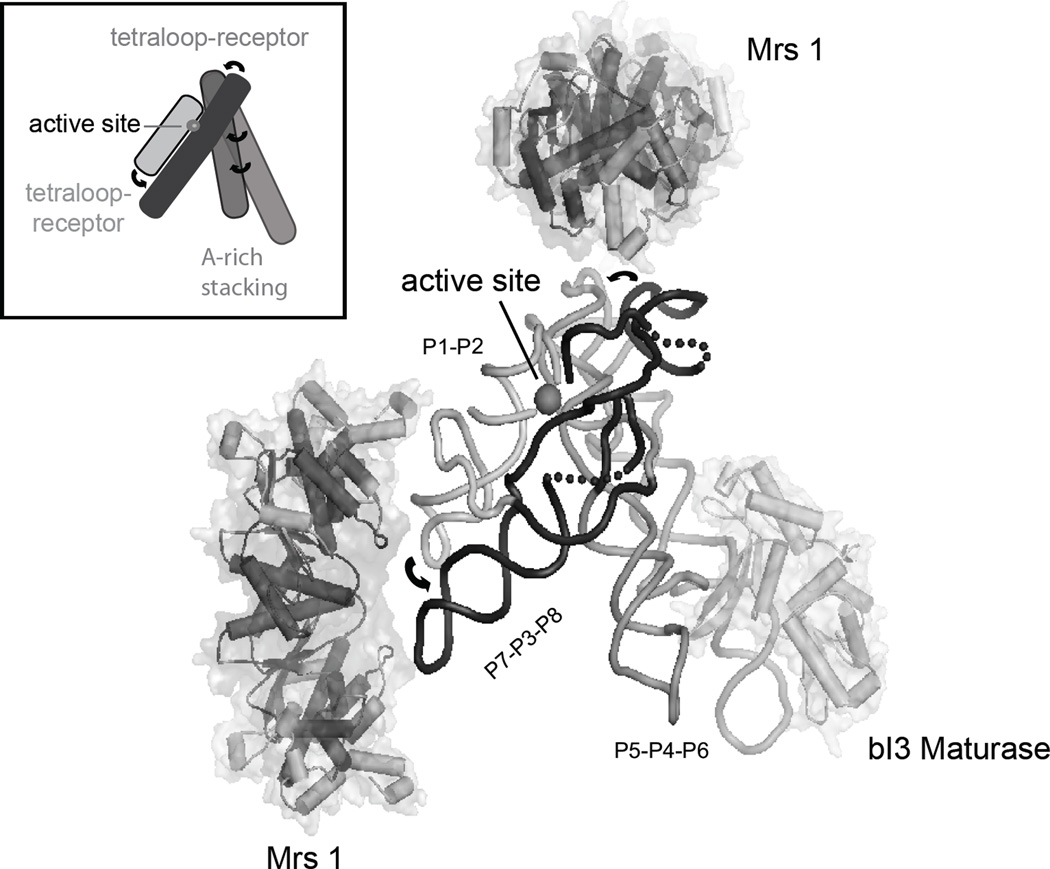
The Saccharomyces cerevisiae bI3 group I intron is a good model for complex proteinmediated RNA folding processes. It is a large (~370 nt) catalytic RNA composed of two coaxially stacked domains that interact through extensive tertiary interactions to form a highly structured catalytic core that then docks with a third helical domain to form a sophisticated active site (Cech, 1990; Michel and Westhof, 1990; Woodson, 2005; Vicens and Cech, 2006) (inset, Figure 3). The intron RNA requires binding by two proteins, the bI3 maturase and Mrs1, to splice in vivo (Kreike et al., 1987; Lazowska et al., 1989) or in vitro (Bassi et al., 2002; Bassi and Weeks, 2003; Longo et al., 2005). The active complex consists of six subunits: the intron RNA, a single monomer of the bI3 maturase protein, and two dimers of the Mrs1 protein (Figure 3). The complete bI3 RNP is ~420 kDa and splices with a kcat of 0.3 min (Bassi et al., 2002). Each protein binds the free RNA independently and each shows modest cooperativity relative to binding of the second protein (Bassi et al., 2002; Bassi and Weeks, 2003).
Figure 3.
Architecture of the six-component bI3 RNP. Tertiary interactions, including tetraloop-receptor motifs and A-minor interactions (arrows), mediate long-range interactions and stabilize a compact and tight RNA structure. The splice site is highlighted by a gray sphere; the three group I intron domains are labeled. For clarity, the P7.1 and P9.1 helices are replaced by a dashed backbone. This model summarized results from high-throughput SHAPE and hydroxyl radical footprinting, interpreted in the context of a molecular model for the bI3 group I intron.
The maturase protein binds in a peripheral helix (termed the P5c helix) in the P5-P4-P6 domain. The maturase binds across consecutive RNA minor-grooves with a Kd of 1.0 nM (Bassi and Weeks, 2003; Longo et al., 2005). Maturase binding stabilizes tertiary structure folding in the entire P5-P4-P6 domain which then stabilizes critical interactions in the catalytic active site, which lie over 50 Å away (Figure 3). Mrs1 is a dimer and two dimers bind cooperatively to the bI3 RNA with a K1/2 of 11 nM and a Hill coefficient of ~2 (Bassi and Weeks, 2003). Until the development of high-throughput RNA footprinting, the RNA binding sites for Mrs1 remained unknown.
The use of high throughput chemical probing methods has, in conjunction with three-dimensional structure modeling, enabled the development of a model for the free bI3 RNA, identified the RNA binding site for Mrs1, made it possible to visualize the global three-dimensional architecture of this RNP complex, and revealed new “non-hierarchical” contributions of protein-facilitated RNA folding (Duncan and Weeks, 2008; Duncan and Weeks, 2009; and see Figure 3).
4.2 Identification of a misfolded free RNA state using SHAPE
When studying RNA-protein interactions, often the initial secondary structure of an RNA is taken to be similar to that seen in the final complex. For example, it would be easy to assume that, prior to protein binding, the free bI3 RNA folds into the same secondary structure as that established by comparative sequence analysis. Both the bI3 maturase and Mrs1 proteins have characteristics of protein cofactors and, therefore, might have only a limited effect on the bI3 RNA secondary structure.
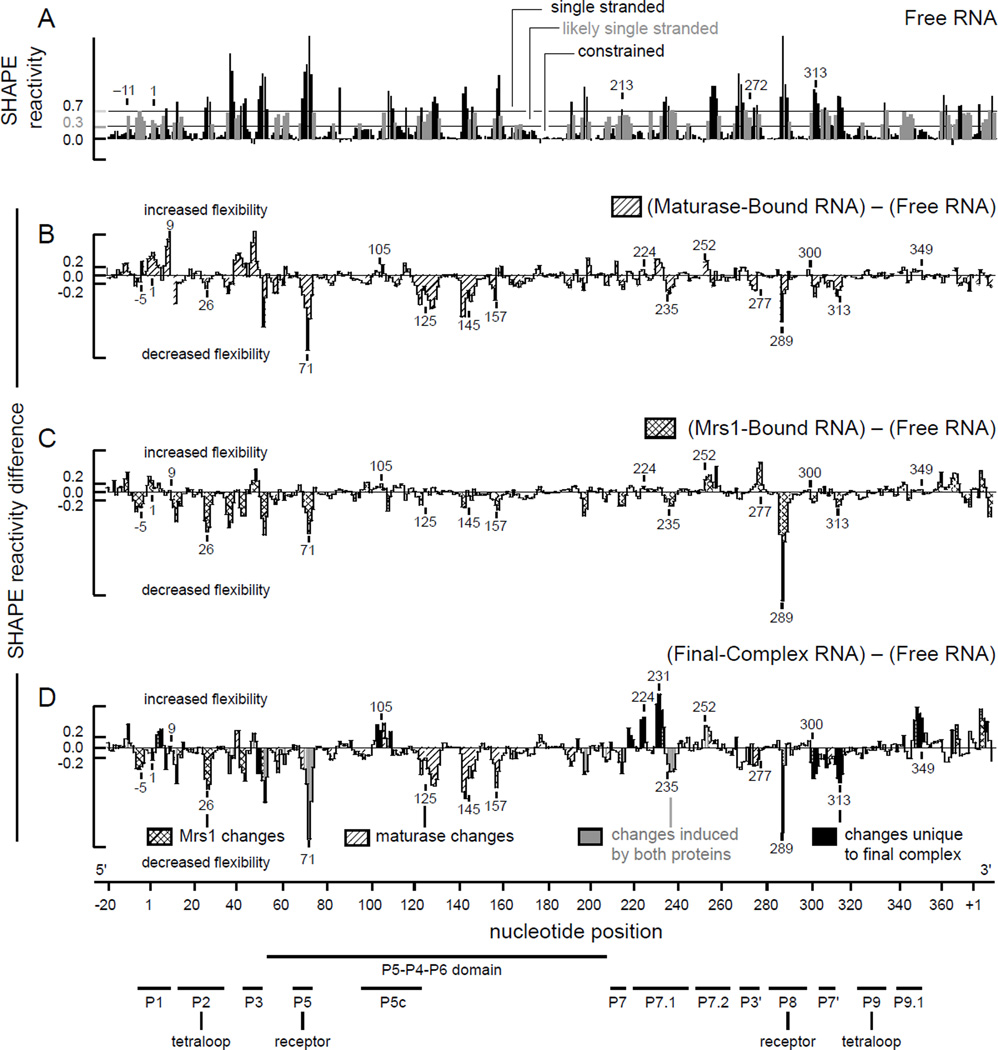
The structure of the entire free bI3 RNA was analyzed in a single experiment using high-throughput SHAPE, as outlined above. Inspection of the SHAPE reactivity data indicated that about one-half of the RNA folds into a structure consistent with the evolutionarily conserved secondary structure common to all group I introns (Michel and Westhof, 1990; Duncan and Weeks, 2008). For example, in the P5–P4–P6 domain, all base paired regions are unreactive towards SHAPE whereas the single-stranded nucleotides that link these regions are reactive (Figure 4A). In contrast, SHAPE data also indicate that about one-half of the free bI3 RNA does not fold in a manner consistent with this consensus group I intron secondary structure (Duncan and Weeks, 2008). For example, nucleotides in the P1 helix are predicted to be base paired in the active RNA state and should therefore be unreactive towards SHAPE, in contrast, these nucleotides are reactive (Figure 4A). The conserved P3 and P7 helices also show significant reactivity. In a single experiment, high-throughput SHAPE readily identified a misfolded free RNA state.
Figure 4.
Protein-induced folding of the bI3 RNA monitored by SHAPE. (A) Reactivity as a function of nucleotide position for the free RNA. Horizontal lines indicate reactivity cut-offs characteristic of base paired and single-stranded nucleotides. (B–D) Difference histograms for the maturase-bound, Mrs1-bound, and final complex RNAs. Nucleotides with significant changes in reactivity are shaded to indicate the protein that induces a given effect. Structural landmarks are highlighted below the axis.
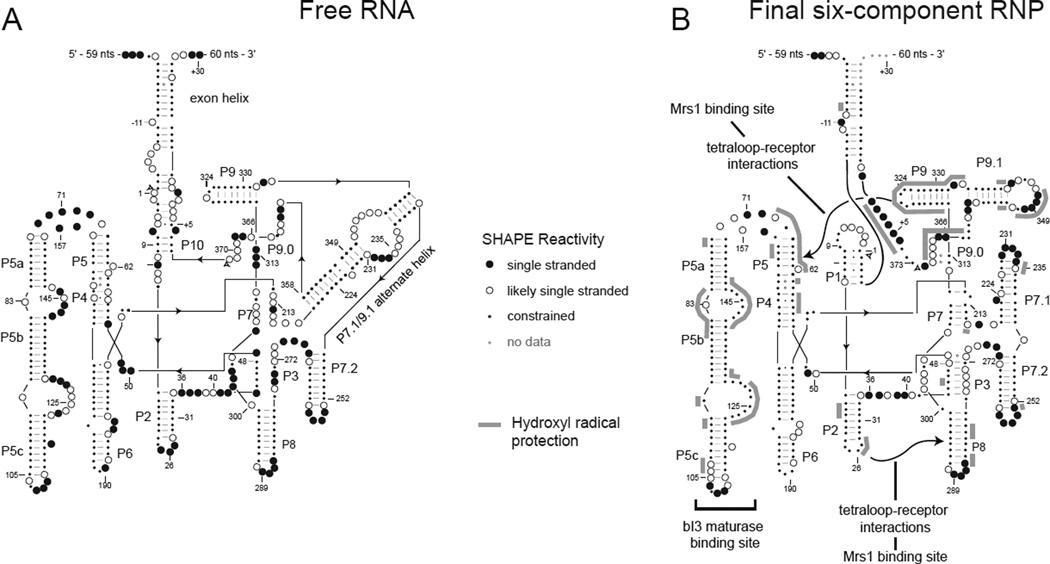
A new secondary structure model for the RNA was developed using SHAPE-directed secondary structure prediction (Duncan and Weeks, 2008; Deigan et al., 2009). This model supported the existence of canonical pairings in regions where SHAPE reactivities were consistent with the phylogenetic model, including in the P5–P4–P6 domain and in the P2, P8, P9, and P7.2 helices (Figure 6A). However, in this new model, roughly half of the RNA is folded in a conformation different from that of the phylogenetic structure, including in the P1 helix, in the P7–P3 pseudoknot, and in the peripheral P7.1 and P9.1 helices (termed the P7.1/P9.1 alternative helix). Additionally, an unanticipated helix between 5′ and 3′ exons was predicted (Figure 6A). SHAPE data are almost exactly consistent with this alternate secondary structure model.
Figure 6.
RNA folding in the six-component bI3 RNP. Secondary structural models for the (A) free RNA and (B) final six-component RNP. SHAPE reactivities are indicated by spheres. Protection from hydroxyl radical cleavage is emphasized with gray lines.
The working model for the free bI3 RNA could be tested and was ultimately confirmed through SHAPE analysis of point mutations. SHAPE analysis of point mutations is proving to be a powerful approach for evaluating unconventional secondary structure models. The basic idea is to make a precise sequence change in one region of a large RNA and then evaluate the structural consequences in the RNA element that is postulated to interact with the mutated region. The second site can be located hundreds of nucleotides away in the primary sequence. For example, mutations were designed in the bI3 exon helix, in the P1 splice site helix, and in the large P7.1/P9.1 alternative helix. Single nucleotide mutations that either disrupted or stabilized the proposed alternative secondary structure or that induced small helical defects were easily detected by the single nucleotide resolution SHAPE experiment and provided strong confirmation of widespread misfolding in the free bI3 RNA state (Duncan and Weeks, 2008).
Comparing the structure of the free RNA to the protein-bound, catalytically active RNA showed extensive differences in SHAPE reactivity, confirming that dramatic structural rearrangements occurred as the RNA formed the intact RNP complex. Interestingly, a second SHAPE analysis of the bI3 RNA, after proteins were removed, showed that the RNA relaxed back to a structural state similar to that of the free RNA (Duncan and Weeks, 2008). These experiments indicate that the catalytically active secondary structure is not the thermodynamically most stable structure for the bI3 RNA. In a single experiment, high-throughput SHAPE thus made it possible to establish whether an individual large RNA folds to the structure characteristic of its phylogenetic family and to evaluate the extent of protein-induced secondary structure rearrangement.
4.3 Identification of a Protein Cofactor that Binds the Ubiquitous RNA Tetraloop-Receptor Motif
Identification of the RNA interaction site is a crucial step in understanding the role a protein plays in RNP function. High-throughput hydroxyl radical footprinting identified two tetraloop-receptor interactions as the binding sites for the Mrs1 protein on the bI3 RNA (Duncan and Weeks, 2009). The tetraloop-receptor interaction is a common long-range tertiary interaction in RNA. The motif consists of a GNRA tetraloop in which the RA (purine-adenosine) nucleotides form A-minor type hydrogen bonds in the minor groove of a receptor helix (Jaeger et al., 1994; Cate et al., 1996; Nissen et al., 2001).
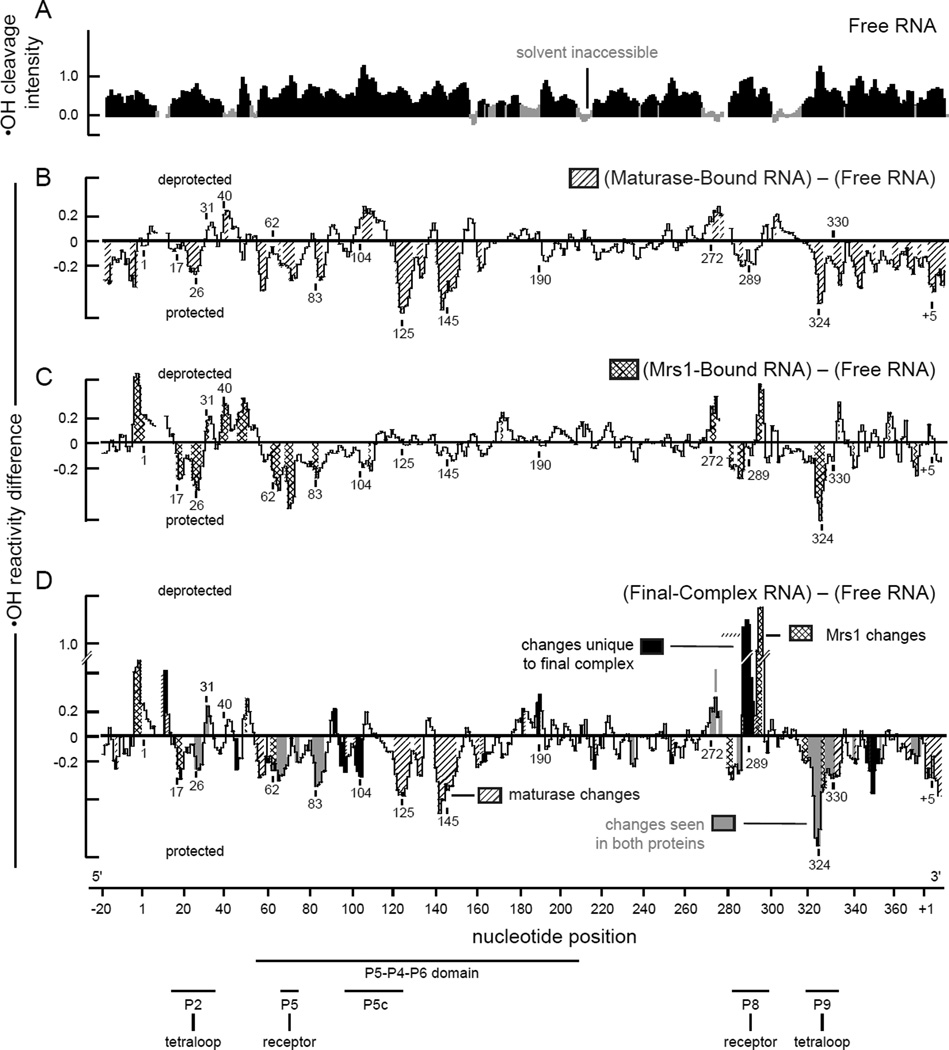
In the free bI3 RNA, only 20% of nucleotides are protected from hydroxyl radical cleavage prior to binding by the Mrs1 protein (Figure 5A). Most RNA elements expected to form tertiary contacts are solvent accessible, including the entire P5–P4–P6 domain, the GNRA tetraloops at the ends of the P2 and P9 helices, and their respective receptors in the P8 and P5 helices.
Figure 5.
Protein-mediated changes in backbone solvent accessibility in the bI3 RNA. (A) Hydroxyl radical (•OH) cleavage intensity versus nucleotide position for the free RNA. Protected nucleotides (reactivities less than one-half the mean) are emphasized in gray. (B–D) Difference histograms for the maturase-bound, Mrs1-bound, and final complex RNAs. Nucleotide regions with significant protein-induced changes in reactivity are shaded as in Figure 4. Structural landmarks are highlighted below the axis.
Upon addition of Mrs1, extensive regions in the RNA become protected from cleavage. The effects are readily quantified by creating a difference plot in which the hydroxyl radical cleavage intensities for the free RNA are subtracted from those for the Mrs1-bound RNA (Figure 5C). Most significant differences in cleavage intensity are localized at or near the two distinct GNRA tetraloop-receptor interactions in the bI3 RNA. These tetraloop-receptor interactions are the L2 loop with the P8 helix and the L9 loop with the P5 helix (see Figure 5C; indicated with arrows on the secondary structure in Figure 6B). These results provide strong evidence that Mrs1 binds to and stabilizes the two independent GNRA tetraloop-receptor interactions in the otherwise misfolded bI3 RNA.
4.4 Mechanism of Cooperative Folding of the bI3 RNP
Folding for some RNAs is strongly hierarchical, such that the secondary structure forms first, followed by tertiary structure (Brion and Westhof, 1997; Tinoco and Bustamante, 1999). However, there are substantial and accumulating examples in which RNA tertiary structure either folds in tandem with the secondary structure or induces changes to the secondary structure during folding (LeCuyer and Crothers, 1993; Gluick and Draper, 1994; Wu and Tinoco, 1998; Buchmueller et al., 2000; Zheng et al., 2001; Chauhan et al., 2005; Wilkinson et al., 2005; Chauhan and Woodson, 2008; Wang et al., 2008). A protein cofactor that stabilizes an RNA tertiary structure element might therefore also have a dramatic impact on secondary structure. The use of high-throughput SHAPE and hydroxyl radical footprinting, as applied to the bI3 RNP, has identified a new and complex non-hierarchical mechanism for protein-facilitated RNA folding. The net contributions of the bI3 maturase and Mrs1 on protein-facilitated folding of the bI3 group I intron are distinct from the conventional definitions of RNA cofactor and chaperone functions.
The secondary and tertiary structures of the bI3 RNA were probed in four conformational states: the free RNA, the maturase-bound and Mrs1-bound complexes, and the final sixcomponent complex (Figures 4 & 5). High-throughput SHAPE and hydroxyl radical experiments indicate that binding by the individual maturase and Mrs1 proteins primarily stabilize long-range RNA tertiary structures but, nonetheless, do not repair the misfolded secondary structure (Figures 4B, C and 5B, C). However, upon binding all protein components to form the final six-component complex, the RNA undergoes large-scale secondary structure rearrangements to form the active group I intron structure (Figures 4D & 6). Strikingly, dramatic structural rearrangements occur in RNA regions located long distances from the protein binding sites. These data imply secondary structure rearrangements in the bI3 RNA involve highly cooperative interactions and that, apparently, binding by both the maturase and Mrs1 proteins “pulls” the RNA secondary structure to the native conformation.
The cooperative rearrangement of secondary structure in the bI3 RNA supports a new, and ambiguous, classification for the bI3 maturase and Mrs1 proteins. Individually, each seems to function as a cofactor: each binds tightly to the bI3 RNA to stabilize specific long-range and higher order tertiary structures (Figures 4B,C and 5B,C). In concert, these proteins also induce additional large-scale structural rearrangements in the RNA secondary structure (Figures 4D & 5D), a role usually attributed to proteins with chaperone-like activities. However, because the active secondary structure relaxes back to the inactive structure when proteins are removed by proteolysis, these proteins must remain bound to the RNA to exert their structure rearranging functions, which is not consistent with the traditional definition of a chaperone. The detailed information provided by high-throughput RNA structure analysis has revealed unexpectedly rich and complex consequences of protein binding on RNA folding.
5. Perspectives and Conclusion
Intricate and elegant relationships are emerging between the functional roles of RNA in the cell and its ability to form multiple, complex structures, often in concert with proteins. Only by obtaining detailed structural information for almost every nucleotide in each folded state of an RNA can the relationships that govern folding, assembly, conformational changes, and function be fully understood.
Together, SHAPE and hydroxyl radical footprinting comprehensively probe many levels of RNA structure and RNA-protein interactions at single nucleotide resolution. These techniques, in combination with resolution by high-throughput capillary electrophoresis, yield quantitative, single nucleotide resolution structural information for large and complex RNAs and RNPs in a concise and efficiently performed experiments. In our experience, this level of detail consistently yields new insights and structural surprises, even in intensively studied RNAs and RNPs.
As the high-throughput and high-resolution approaches described here are applied more broadly to large RNA-protein complexes, many proteins previously thought to make simple contributions to RNA folding will also likely to be found to exert highly complex, cooperative, and non-additive effects on RNA folding and RNP function. These sophisticated protein-induced affects add another level of control, and potential regulatory function, in RNP complexes.
Acknowledgments
This research program on high-throughput RNA structure analysis is supported by the U.S. National Institutes of Health (AI068462 to K.M.W.). Experimental work on the bI3 intron RNP was supported by GM056222.
References
- Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi GS, de Oliveira DM, White MF, Weeks KM. Recruitment of intron-encoded and co-opted proteins in splicing of the bI3 group I intron RNA. Proc Natl Acad Sci USA. 2002;99:128–133. doi: 10.1073/pnas.012579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi GS, Weeks KM. Kinetic and thermodynamic framework for assembly of the six-component bI3 group I intron ribonucleoprotein catalyst. Biochemistry. 2003;42:9980–9988. doi: 10.1021/bi0346906. [DOI] [PubMed] [Google Scholar]
- Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical "footprinting". Curr Opin Struct Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- Brunel C, Romby P. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. doi: 10.1016/s0076-6879(00)18040-1. [DOI] [PubMed] [Google Scholar]
- Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nat Struct Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J Mol Biol. 2005;353:1199–1209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J Am Chem Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernick MR, Friis RH. Introductory biostatistics for the health sciences : modern applications including bootstrap. Hoboken, N.J.: Wiley-Interscience; 2003. [Google Scholar]
- Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CDS, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CDS, Weeks KM. The Mrs1 splicing factor binds the RNA tetraloop-receptor motif. 2009 doi: 10.1371/journal.pone.0008983. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Klein DJ, Ferré-D'Amaré AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland RF, Cech T, Atkins JF. The RNA world : the nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluick TC, Draper DE. Thermodynamics of folding a pseudoknotted mRNA fragment. J Mol Biol. 1994;241:246–262. doi: 10.1006/jmbi.1994.1493. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Kreike J, Schulze M, Ahne F, Lang BF. A yeast nuclear gene, MRS1, involved in mitochondrial RNA splicing: nucleotide sequence and mutational analysis of two overlapping open reading frames on opposite strands. EMBO J. 1987;6:2123–2129. doi: 10.1002/j.1460-2075.1987.tb02479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- Lazowska J, Claisse M, Gargouri A, Kotylak Z, Spyridakis A, Slonimski PP. Protein encoded by the third intron of cytochrome b gene in Saccharomyces cerevisiae is an mRNA maturase. Analysis of mitochondrial mutants, RNA transcripts proteins and evolutionary relationships. J Mol Biol. 1989;205:275–289. doi: 10.1016/0022-2836(89)90341-0. [DOI] [PubMed] [Google Scholar]
- LeCuyer KA, Crothers DM. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993;32:5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- Longo A, Leonard CW, Bassi GS, Berndt D, Krahn JM, Hall TM, Weeks KM. Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase. Nat Struct Mol Biol. 2005;12:779–787. doi: 10.1038/nsmb976. [DOI] [PubMed] [Google Scholar]
- Lu M, Guo Q, Wink DJ, Kallenbach NR. Charge dependence of Fe(II)-catalyzed DNA cleavage. Nucleic Acids Res. 1990;18:3333–3337. doi: 10.1093/nar/18.11.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. SciU.SA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- Scott WG. Ribozymes. Curr. Opin. Struct. Biol. 2007;17:280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of ribonuclease P--a universal ribozyme. Curr. Opin. Struct. Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Tullius TD, Dombroski BA, Churchill ME, Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr Opin Chem Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Wang B, Wilkinson KA, Weeks KM. Complex ligand-induced conformational changes in tRNAAsp revealed by single nucleotide resolution SHAPE chemistry. Biochemistry. 2008;47:3454–3461. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J Am Chem Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, Noller HF. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA. Structure and assembly of group I introns. Curr Opin Struct Biol. 2005;15:324–330. doi: 10.1016/j.sbi.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wu M, Tinoco I., Jr Formation of a GNRA tetraloop in P5abc can disrupt an interdomain interaction in the Tetrahymena group I ribozyme. Proc Natl Acad Sci USA. 2001;98:3695–3700. doi: 10.1073/pnas.051608598. [DOI] [PMC free article] [PubMed] [Google Scholar]