Abstract
To date no models exist to study MnSOD deficiency in human cells. To address this deficiency, we created a SOD2-null human cell line that is completely devoid of detectable MnSOD protein expression and enzyme activity. We utilized the CRISPR/Cas9 system to generate biallelic SOD2 disruption in HEK293T cells. These SOD2-null cells exhibit impaired clonogenic activity, which was rescued by either treatment with GC4419, a pharmacological small-molecule mimic of SOD, or growth in hypoxia. The phenotype of these cells is primarily characterized by impaired mitochondrial bioenergetics. The SOD2-null cells displayed perturbations in their mitochondrial ultrastructure and preferred glycolysis as opposed to oxidative phosphorylation to generate ATP. The activities of mitochondrial complex I and II were both significantly impaired by the absence of MnSOD activity, presumably from disruption of the Fe/S centers in NADH dehydrogenase and succinate dehydrogenase subunit B by the aberrant redox state in the mitochondrial matrix of SOD2-null cells. By creating this model we provide a novel tool with which to study the consequences of lack of MnSOD activity in human cells.
Keywords: superoxide dismutase, mitochondria, respiration, electron transport, iron metabolism
Graphical Abstract
Introduction
Manganese superoxide dismutase (MnSOD), encoded by the nuclear SOD2 gene, is a mitochondrial enzyme. MnSOD enzyme activity is essential for protecting respiring cells from oxidative damage due to excessive superoxide by converting superoxide to hydrogen peroxide. Mitochondria are major sites of reactive oxygen species production, and MnSOD is the primary antioxidant enzyme in the mitochondrial matrix responsible for protecting mitochondria from superoxide generated as a byproduct during oxidative respiration. As such, MnSOD expression is indispensable for aerobic life [1–3]. In whole animal knockout models, homozygous constitutive Sod2 knockout mice die by 2–3 weeks after birth from cardiomyopathy and neurodegenerative disease [4, 5]. Conditional Sod2 knockout models with deletion of Sod2 targeted to specific cell types has enabled the study of Sod2 deletion without the neonatal lethality observed in constitutive knockouts. Several conditional Sod2 knockout models have been reported to date including thymic T cells [6], hematopoietic stem cells [7], hepatocytes [8], epithelial keratinocytes [9], and mammary epithelial cells [10]. Extensive work has been done to characterize the phenotypic effects of lack of Sod2 expression in each of these models, which is summarized in Table 1. For example, deletion of Sod2 from thymic T cells resulted in immunodeficient mice with increased susceptibility to influenza infection in part due to defective proliferation and maturation of T cells lacking MnSOD. In a different mouse model where Sod2 was deleted in hematopoietic stem cells, a significant disruption of systemic iron homeostasis and red blood cell anemia was observed. In these mice an increase in superoxide levels led to a disruption in the specific activity of metabolic enzymes including ferrochelatase and aconitase, as well as a disruption in expression of genes responsible for maintaining iron homeostasis. These mice lived for about 76 weeks and died with severe iron overloading and apparently depleted marrow. Interestingly, no tumors were observed in these mice over their lifetime. Deletion of Sod2 in mouse liver revealed subtle global changes in the redox biology pathways in hepatocytes and demonstrated the remarkable potential of hepatocytes to maintain normal homeostasis in the complete absence of detectable MnSOD activity. Whether these livers would be able to withstand an oxidative challenge is still unclear, as the Sod2 deficient livers were not stressed with carbon tetrachloride or acetaminophen to induce a liver specific oxidative stress in those studies. Finally, when Sod2 was deleted from mammary epithelial cells it had no discernable effect on their biology or physiology and mice lacking mammary MnSOD were able to lactate normally and nurse multiple healthy litters. The diverse phenotypes resulting from tissue specific deletion of Sod2 suggest that loss of MnSOD activity confers varying degrees of toxicity depending on the cell type in which it is absent. These models have been useful to study the effects of MnSOD deficiency in mammalian cells; however, despite the existence of these Sod2-null mouse models, a model to study the effects of SOD2 deletion in human cells has, to our knowledge, not yet been established.
Table 1.
Summary of Sod2 knockout mouse models and their resulting phenotypes generated to date.
| Knockout Target | Phenotype |
|---|---|
|
Homozygous whole animal knockout |
Neonatal lethality [4, 5], reduced succinate dehydrogenase and aconitase activities [5], dilated cardiomyopathy [5], impairment of mitochondrial function in organs with high demand for oxidative metabolism including heart, liver, skeletal muscle, and hematopoietic cells [4, 5]. |
|
Heterozygous whole animal knockout |
Increased superoxide production and mitochondrial injury [46], increased oxidative stress-induced mitochondria-mediated apoptosis [25]. |
| Liver | No gross histological abnormalities [47], spontaneous oxidative damage (i.e. lipid peroxidation) in liver not detected. |
|
Mammary gland |
Embryonic knockout: maturation arrest, hyperplastic epithelium [48], Post-natal knockout: no apparent abnormalities associated with loss of Sod2 [10]. |
|
Skeletal muscle |
Reduced aerobic exercise capacity and decreased mitochondrial enzyme function; damage to glycolytic muscle fibers [49]. |
|
Hematopoietic stem cells |
Inactivation of metabolic enzymes and iron homeostasis, improper heme formation and impaired erythrocyte development [7]. |
| T-cells | Increase in superoxide and mitochondria-mediated apoptosis, reduced mitochondrial enzyme function [6]. |
Several methods exist to study the effect of gene-knockdown or deletion in addition to expensive knockout animal models, including using RNA interference (RNAi) or engineered DNA binding proteins such as zinc finger proteins and the TAL system [11–13]. The aforementioned methods are associated with several inherent limitations such as cost, difficulty in attaining stable knockdown of targeted genes and off-targeting effects. The advent of the CRISPR/Cas9 gene-editing system, the origins of which lie in the bacterial immune defense system [14], has allowed efficient and specific gene-editing in a wide variety of organisms, including yeast, zebrafish, mice, and humans, using a simple approach in which a unique guide RNA (gRNA) is designed to target disruption of the desired gene [15–17]. The repurposing of the CRISPR/Cas9 system to enable genome-wide gene editing has revolutionized the study of targeted gene deletion allowing for the generation of cells with permanent deletion of the gene of interest.
Creating deletion mutants of genes involved in maintaining cell viability can prove to be fundamentally difficult. Though previous models have been generated to characterize the effects of Sod2 deficiency in mice and other organisms, including drosophila and yeast [2, 18], to date no model exists to study lack of MnSOD expression in human cells. Here we report the use of the CRISPR/Cas9 system to generate a SOD2-null human cell line that is completely deficient in MnSOD protein expression and enzyme activity. We show that deletion of SOD2 in HEK293T cells resulted in a profound decrease in growth potential of SOD2-null cells due to mitochondrial dysfunction as a consequence of the accumulation of superoxide. These cells have many phenotypic characteristics in common with existing models thus validating the system, and provide for the first time a stable human cell line with which to study the effects of chronic MnSOD deficiency.
Materials and Methods
Cells and Plasmid Transfection
HEK293T cells were maintained in DMEM supplemented with 10% fetal bovine serum. Cells were maintained in a humidified 37°C incubator with 5% CO2 and were routinely subcultured before reaching confluence by detachment with TrypLE Express (Invitrogen, Carlsbad, CA). The pD130-GFP expression vector containing expression cassettes for green fluorescent protein (GFP), Cas9 endonuclease, and a CRISPR chimeric cDNA with the gRNA moiety we designed to target exon 3 of SOD2 [ATATCAATCATAGCAT TTTC] was custom synthesized (DNA2.0, Menlo Park, CA). HEK293T cells were transiently transfected by calcium phosphate precipitation. Five days after transfection GFP+ cells were sorted by flow cytometry and colonies were selected from single clones.
Western analysis
Total protein was extracted from HEK293T clones with standard RIPA buffer plus protease inhibitors and quantified by Bradford assay. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes and probed with primary antibodies detecting CuZnSOD and MnSOD (Millipore, Billerica, MA) and beta-tubulin (University of Iowa Hybridoma Core, Iowa City, IA).
SOD enzyme activity assays
CuZnSOD and MnSOD enzyme activities were analyzed using gel zymography as previously described [19]. Briefly, 150µg total protein per sample was loaded on a 12% native acrylamide gel and separated by electrophoresis. The gel was first stained in the dark with a nitroblue tetrazolium (NBT) solution and subsequently with a riboflavin-TEMED solution for 20 minutes at room temperature then exposed to light to catalyze the production of superoxide. SOD activity is indicated by the presence of achromatic bands. MnSOD activity was specifically visualized by inclusion of sodium cyanide in the staining solution to inhibit CuZnSOD.
Cell Proliferation and Clonogenic Assays
For cell proliferation assays HEK293T cells from wild-type or SOD2-null clones were plated at a density of 1×103 cells per well in 24-well plates in complete medium. Cells were counted daily for 7 days. For clonogenic assays, HEK293T wild-type cells or SOD2-null clones were seeded at a density of 200 cells per well in 6 well plates. For hypoxia experiments, cells were placed at either 21% or 4% O2 in a humidified 37°C incubator and colonies were allowed to form for 10 days. For experiments involving the SOD mimetic GC4419 (Galera Therapeutics Inc., Malvern, PA), cells were seeded at a density of 100 cells per well in 6 well plates and were treated every other day for 10 days with 5, 10, or 20 µM GC4419 diluted in sodium bicarbonate. Cells were fixed in ice-cold methanol and stained with 0.5% crystal violet (w/v) in 25% methanol. Colonies containing >50 cells were scored.
Measurement of dihydroethidium and MitoSOX red reactive species
Cells were stained with either 10 µM dihydroethidium (DHE; Molecular Probes, Eugene, OR) or 2 µM MitoSOX red (Life Technologies, Grand Island, NY). Briefly, cells were washed once in PBS and stained in PBS for 10 minutes 37°C. The mean fluorescence intensity (MFI) was analyzed for 10,000 cells per sample.
Mitochondrial bioenergetics
The Seahorse XF96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA) was used to detect rapid, real time changes in cellular respiration and glycolysis rate to measure oxygen consumption and extracellular acidification rates. Cells were seeded in 100 µl complete DMEM at a density of 2 ×104 cells/well in XF24 24-well cell culture plates. Eight replicates per cell type were assayed in each experiment using Seahorse technology. On the day of the experiment, cells were washed and incubated in XF Base medium for 1 h at 37°C. After recording basal respiration, successive injections of oligomycin (1 µM), FCCP (1 µM) and rotenone (1 mM)/Antimycin A (5 µM) were carried out to determine OCR and ECAR. All measurement were normalized to total protein content per sample.
Succinate dehydrogenase histochemistry
Culture medium was aspirated and cells were washed twice with PBS. Cell culture dishes were allowed to air dry at room temperature. A solution containing 0.55 mM NBT and 0.05 M sodium succinate was applied and the dishes were incubated overnight at 37°C and washed again with PBS before images were captured.
Measurement of mitochondrial electron transport chain activities
Complex I was measured as the rate of rotenone-inhibitable NADH oxidation in the presence or absence of 200 µg rotenone. Complex II was assayed as previously described [20] and measured as the rate of dichloroindophenol (DCIP) reduction by Coenzyme Q in the presence and absence of succinate. Whole cell pellets were freeze-thawed, resuspended in 20mM potassium phosphate buffer and sonicated for 20s. Protein concentration was measured by the Lowry method.
TEM
Cells were collected by scraping, washed with cold PBS, and fixed with 2.5% glutaraldehyde in 0.1 M Na-cacodylate buffer. Fixed cells were incubated with 1% osmium tetroxide for 2 h and processed for embedding. The embedding procedure consisted of incubation with distilled water for 1 min, then 20 min in 2.5% uranyl acetate, 15 min in 50% ethanol, 15 min in 70% ethanol, 30 min in 95% ethanol, 2×30 min in 100% ethanol, 1 h in ethanol and epon (2:1), 1 h in ethanol and epon (1:2), and overnight in 100% epon in a 60°C oven. Ultrathin sections of 70 nm were placed on copper mesh grids, and stained with 5% uranyl acetate and lead citrate. Samples were visualized using a JEOL 1230 transmission electron microscope. All microscopy imaging was performed using The University of Iowa Central Microscopy Research Core Facility.
Results
CRISPR/Cas9 mediated gene editing of SOD2 caused efficient and complete deletion of MnSOD expression in human cells
The CRISPR/Cas9 expression system caused efficient genetic disruption of SOD2 in human cells that resulted in complete loss of detectable MnSOD protein expression and enzyme activity. Thoughtful design of the gRNA component targeting DNA double-strand breaks induced by Cas9 expression in the CRISPR/Cas9 system allows for specific editing of a genetic locus with minimal off-target effects. To disrupt MnSOD expression in HEK293T cells, we designed a single gRNA to target the third exon of SOD2.
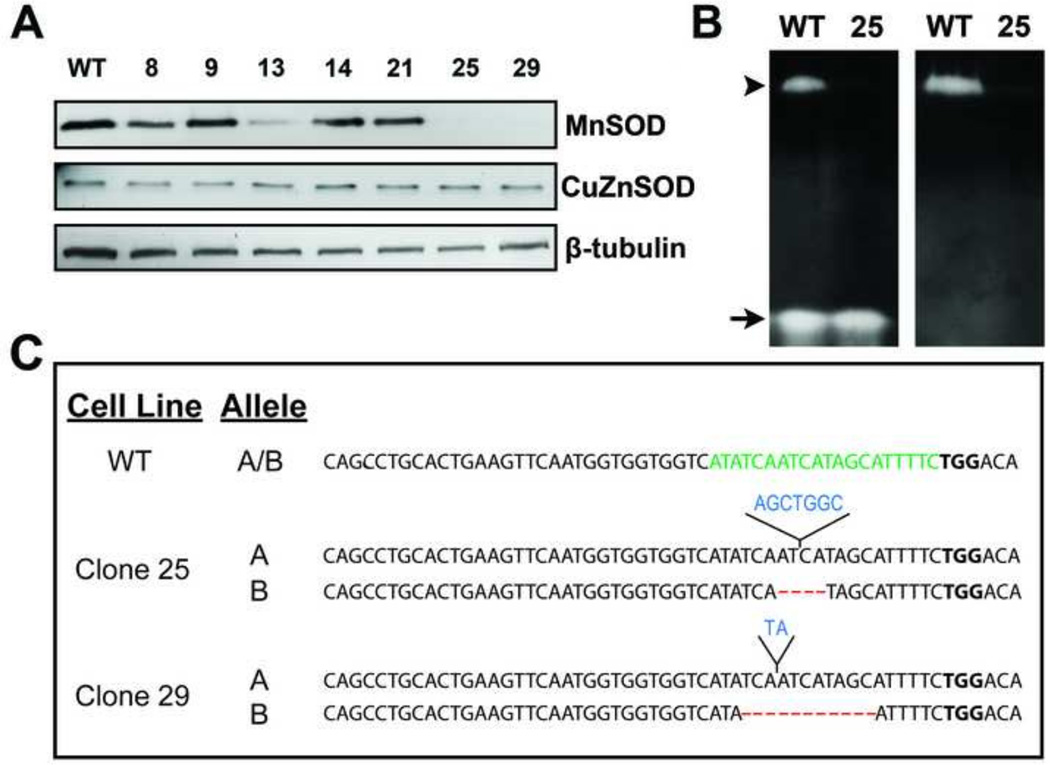
After transient expression of the CRISPR/Cas9 construct targeting SOD2 in HEK293T cells followed by selection and cloning, we performed western blot analysis to identify clones without detectable MnSOD expression. As shown in Figure 1A, two clones (clones #25 and 29), out of a total of thirty that were assayed, show no detectable MnSOD expression. To evaluate whether lack of MnSOD protein correlated with absence of enzyme activity, an SOD activity gel assay was performed on clone 25. Results from the activity gel show no detectable MnSOD enzyme activity in this clone whereas CuZnSOD activity was unaffected by deletion of SOD2 (Figure 1B).
Figure 1. CRISPR/Cas9-targeted disruption of human SOD2 results in cells lacking detectable MnSOD expression.
(A) Whole cell lysates were subjected to western blot analysis for MnSOD and CuZnSOD expression in selected clones after CRISPR/Cas9-mediated disruption of SOD2.
(B) Analysis of MnSOD (arrowhead) and CuZnSOD (arrow) activity of wild-type (WT) and SOD2 knockout clone number 25. The appearance of achromatic bands indicates the presence of SOD activity. Right panel: Sodium cyanide was included during native gel staining to inhibit CuZnSOD.
(C) Unique bi-allelic indels were detected by sequencing analysis in each knock-out clone (clone 25: 7 bp insertion, 4 bp deletion; clone 29: 2 bp insertion, 11 bp deletion). A 200 bp area encompassing the gRNA targeting site was PCR-amplified from genomic DNA, cloned, multiple clones sequenced, and compared to the reference sequence for human SOD2 (RefSeq Accession NG008729.1)
Nucleotide insertions are marked in blue and deletions are marked by red dashes. PAM motif is marked in bold and gRNA target sequence is highlighted in green.
The CRISPR/Cas9 system can produce both insertions and/or deletions (indels) in the genomic target site via the non-homologous end-joining DNA repair pathway. To determine the nature of the genetic lesions caused by expression of the SOD2 targeted CRISPR/Cas9 construct, clones lacking MnSOD protein expression and activity were subjected to DNA sequencing analysis. Genomic DNA was purified from the SOD2-null cells, PCR-amplified and cloned into a sequencing vector to verify the nature of indels present. As protein expression and activity were completely abolished in the SOD2-null cells, we expected to detect bi-allelic mutations. Both clones that were selected for further characterization were confirmed to have unique indels with bi-allelic mutations (Figure 1C). Indels consisting of an insertion of 7 nucleotides and a deletion of 4 nucleotides were detected in clone 25, whereas indels consisting of an insertion of 2 nucleotides and a deletion of 11 nucleotides were detected in clone 29. Each of these indels resulted in a frameshift of the coding sequence of SOD2 and complete loss of MnSOD protein expression.
Genetic disruption of SOD2 abrogates clonogenic potential of SOD2-null cells
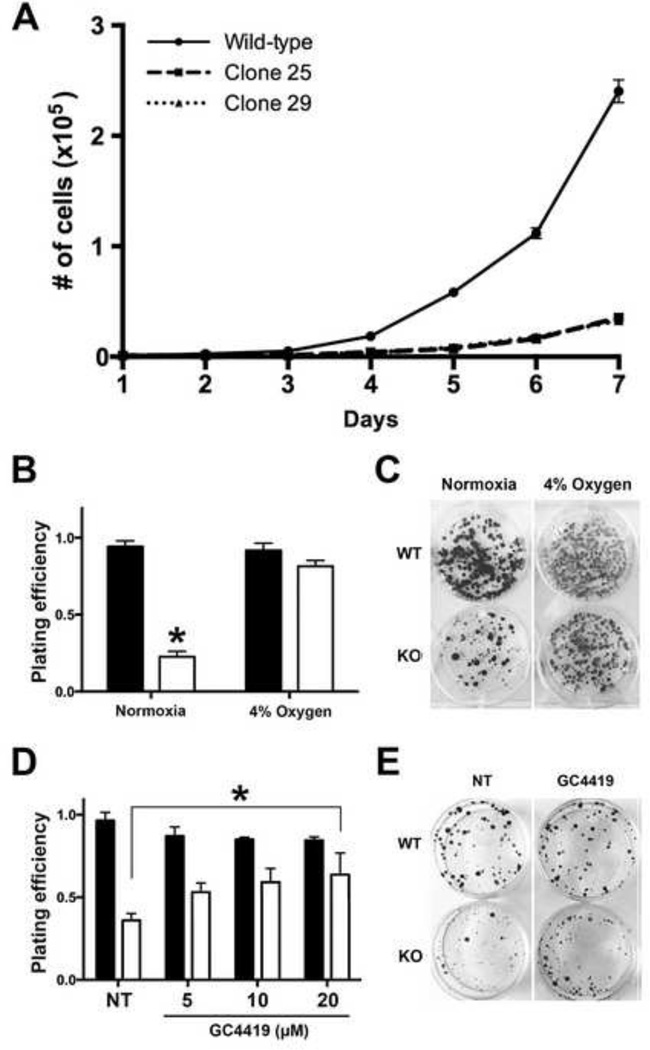
To further characterize the SOD2-null clones, we examined the growth characteristics of these cells compared to wild-type HEK293T cells. Since previous research has shown MnSOD is indispensable for cell survival, we expected the SOD2-null cells to exhibit reduced proliferative potential. As expected, the growth rate of SOD2-null cells was significantly impaired compared to wild-type cells (Figure 2A) Moreover, a greater than 5-fold reduction in clonogenic activity was observed in SOD2-null compared to wild-type HEK293T cells (Figure 2B&C; normoxia). Given the central role of MnSOD in protecting the mitochondrial function of respiring cells, we hypothesized the observed reduction in clonogenic potential of SOD2-null cells could be due to oxygen toxicity leading to impaired cell growth. To investigate this hypothesis, we repeated the clonogenic assay in a reduced oxygen (4% O2) environment. The clonogenic potential of SOD2-null cells was restored by incubation in hypoxic conditions supporting the hypothesis that lack of SOD2 expression contributes to impairment of cell growth in normoxia (Figure 2B&C; 4% O2). The clonogenic data were quantified and the data are shown in Figure 2B (right panel). This finding is particularly relevant to the hypoxic fraction of tumor cells given the heterogeneous nature of tumor blood flow. Our data supports the concept that tumor cells located within highly hypoxic areas would be less likely to be affected by loss of SOD2 expression alone. To determine whether a pharmaceutical SOD mimic could rescue cell growth and survival in the SOD2 knockout cells, wild-type and SOD2-null HEK293T cells were treated with increasing doses of GC4419, a highly specific small molecule pharmacological mimetic of SOD currently being used in clinical trials. The growth defect observed in cells without SOD2 expression was abrogated by treatment with the mimetic in a dose-dependent manner as clonogenic potential of SOD2-null cells was restored to the wild-type level with 20 µM GC4419 without significant toxicity to WT cells (Figure 2D&E). Taken together, these results demonstrate that CRISPR/Cas9-mediated gene editing of SOD2 and disruption of MnSOD expression results in impaired growth phenotype of human HEK293T cells that can be rescued by incubation in a reduced oxygen environment or by treatment with a SOD mimic.
Figure 2. Deletion of MnSOD activity results in growth inhibition of HEK293T cells that is restored by incubation in hypoxia or by treatment with a SOD mimic.
(A) Growth curve of wild-type (WT) and SOD2-null (KO) HEK293T clones.
(B) Clonogenic assay of 200 WT (closed bars) or KO (open bars) cells grown in normoxia or 4% oxygen for 10 days. Incubation in 4% oxygen restores clonogenic activity of KO cells. *P<0.01 compared to WT.
(C) Photograph of 6-well plates from representative experiments in 2B is shown.
(D) Clonogenic assay of WT and KO cells supplemented with 5, 10, or 20 µM, as indicated, of the SOD mimetic GC4419. 100 cells were plated and colonies were allowed to form for 10 days. Treatment with 20 µM GC4419 restores clonogenic activity of KO cells to wild-type levels. *P<0.01 compared to untreated WT cells.
(E) Photograph of representative experiment in which cells were treated with 20 µM GC4419 is shown. *P < 0.01 compared to WT cells.
Elevated intracellular ROS contributes to mitochondrial dysfunction in SOD2-null cells
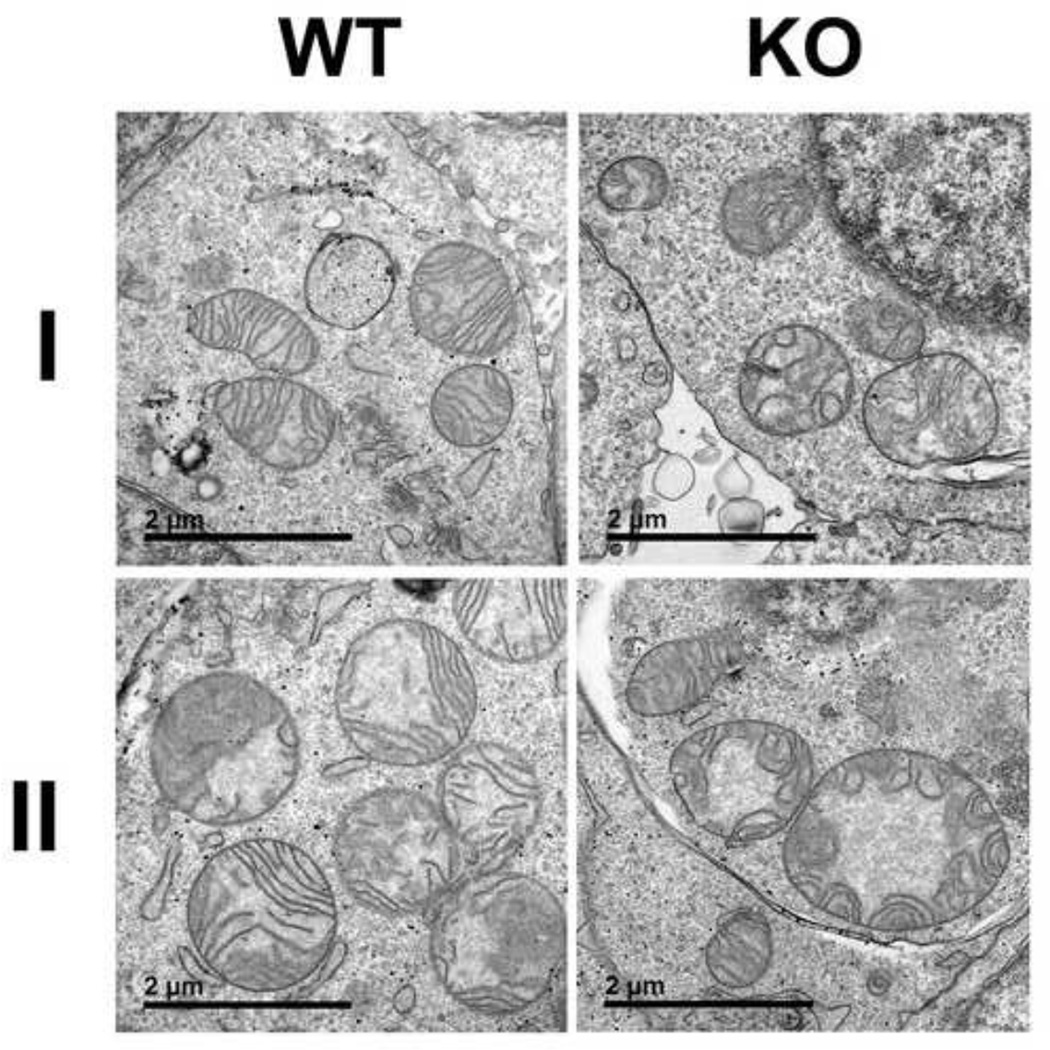
Due to its enzyme activity and its location in the mitochondrial matrix, MnSOD is the cell’s primary defense against superoxide generated by one electron reduction of oxygen as a by-product of oxidative phosphorylation. Since mitochondria are the main site of superoxide production in most normal human cell types, MnSOD activity is required to prevent superoxide-induced mitochondrial injury and subsequent dysfunction. We examined the mitochondrial ultrastructure of both SOD2-null and wild-type cells by transmission electron microscopy to determine if lack of SOD2 expression causes any observable defects in mitochondrial morphology. As shown in Figure 3, representative TEMs from SOD2-null cells show mitochondria with abnormal shape and distribution of cristae (right panels) compared to mitochondria visualized in wild-type cells (left panel). These results demonstrate that lack of MnSOD expression perturbs mitochondrial structure, a finding consistent with studies from MnSOD deficient mouse cells.
Figure 3. Mitochondria of SOD2-null cells display ultrastructure abnormalities.
Representative transmission electron micrographs from two independent samples (I and II) from wild-type (WT) and SOD2-null (KO) cells. Micrographs from KO cells show multiple mitochondria with abnormal cristae structure.
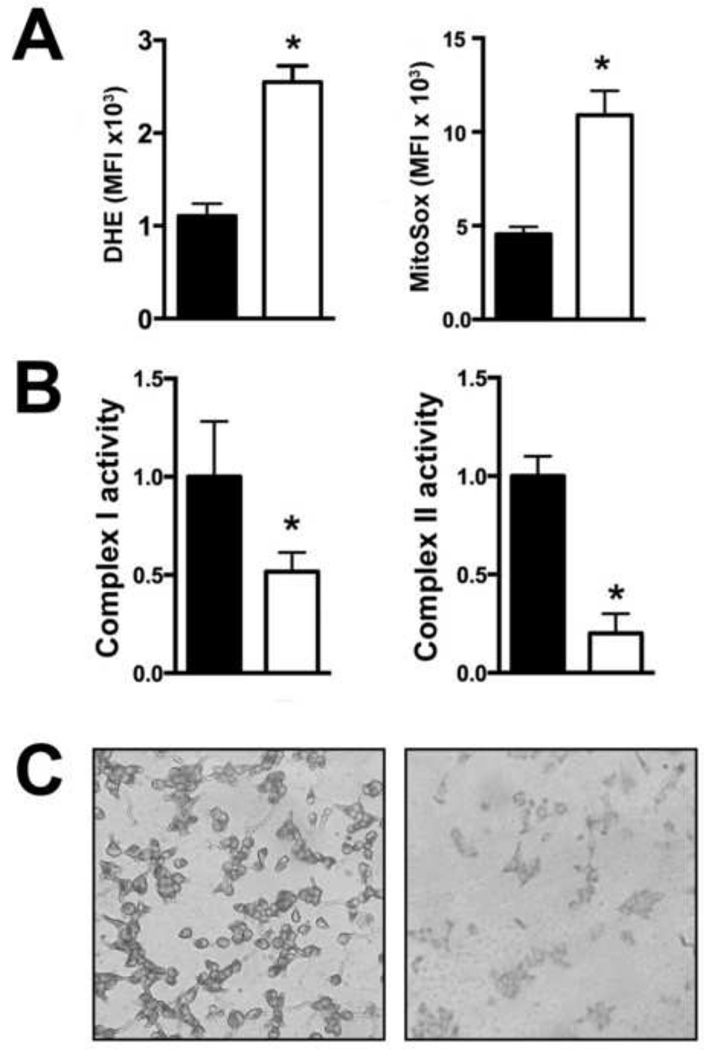
Given the abnormal appearance of SOD2-null mitochondria, we next sought to determine whether elevated levels of superoxide were detectable in these cells compared to wild-type HEK293T cells. To confirm that loss of MnSOD expression resulted in the anticipated increase in mitochondrial oxidative stress, the levels of dihydroethidium and MitoSOX red reactive species were examined and quantified by flow cytometry. We detected a 2.5-fold increase in dihydroethidium reactive species and 2-fold increase in MitoSOX red reactive species in SOD2-null cells compared to wild-type cells (Figure 4A), showing that in the absence of functional MnSOD activity human cells are subjected to increased baseline mitochondrial oxidative stress.
Figure 4. Absence of MnSOD expression results in increased mitochondrial oxidative stress and alters mitochondrial electron transport chain function.
(A) An increase in mean fluorescent intensity (MFI) of dihydroethidium (DHE) and MitoSOX red staining was detected by flow cytometry in SOD2 knock-out cells (open bars) compared to wild-type cells (closed bars). *P = 0.012 compared to wild-type.
(B) Activities of complex I (left panel) and complex II (right panel) (µmol/min/mg protein) were measured spectrophotometrically. Complex I activity was measured as the rate of rotenone-inhibitable NADH oxidation in the presence or absence of rotenone. Complex II was assayed as the rate of DCIP reduction by Coenzyme Q in the presence and absence of succinate. Activity measurements were normalized to total cellular protein. *P<.05 compared to wild-type.
(C) Increased succinate dehydrogenase activity was detected by histochemical staining in wild-type cells (left panel) compared to knock-out cells (right panel)
Increased levels of oxidative stress have been shown to cause the oxidation of mitochondrial electron transport chain (ETC) complexes containing iron-sulfur (Fe/S) clusters [21–23]. Both NADH dehydrogenase (complex I) and succinate dehydrogenase (SDH; complex II) contain Fe/S clusters that are particularly sensitive to oxidation and subsequent inactivation by increased levels of superoxide. Therefore, we hypothesized that elevated superoxide levels in SOD2-null cells contribute to functional alterations in ETC complexes. To test this hypothesis, the activities of complex I, which contains 7 Fe/S clusters, and complex II, which contains 3 Fe/S clusters, were assayed spectrophotometrically. Complex I activity was assayed via the rate of rotenone-inhibitable NADH oxidation and complex II was measured as the rate of DCIP reduction by Coenzyme Q in the presence and absence of succinate. As expected, a 2-fold reduction in complex I activity and a 5-fold reduction in complex II activity was observed in SOD2-null compared to wild-type cells (Figure 4B). Complex I and II activities were also found to be reduced in a mouse model of Sod2 deficiency, which is consistent with our results and the idea that respiratory chain enzymes with Fe/S clusters are sensitive superoxide-mediated inactivation [24, 25]. In concordance with this finding, histochemical staining of SDH activity was markedly decreased in cells devoid of MnSOD expression (Figure 4C). Altogether these results indicate MnSOD is essential for protecting ETC complexes I and II from superoxide-induced inactivation.
The bioenergetic profile of SOD2-null cells is altered compared to wild-type
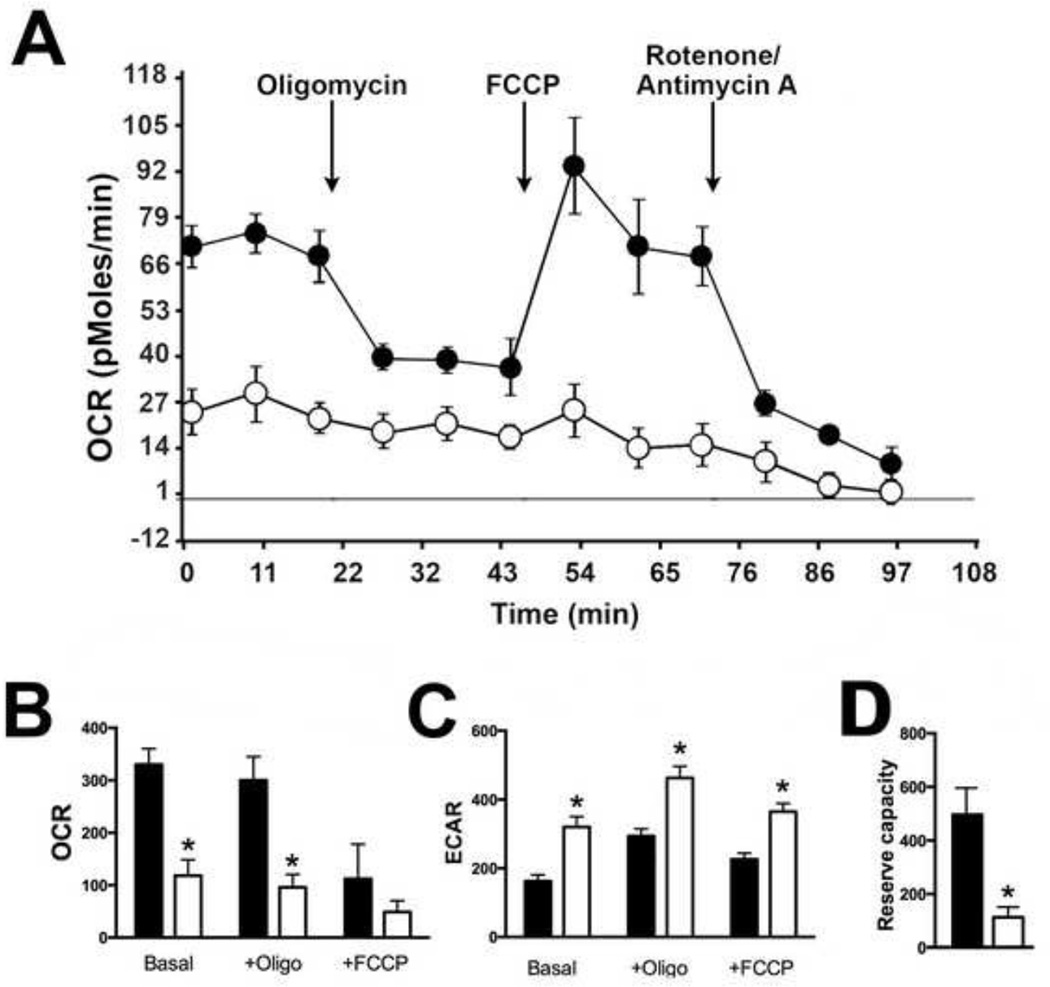
To determine whether mitochondrial dysfunction due to alterations in ETC activity had an effect on cellular respiration, we utilized Seahorse technology to conduct metabolic assays to compare the bioenergetic profiles in real-time of SOD2-null versus wild-type cells. Compared to wild-type cells, SOD2-null cells displayed a nearly 3.5-fold reduction in baseline oxygen consumption rate (OCR) indicating a significant decrease in mitochondrial respiration (Figure 5B). This was accompanied by an increase in extracellular acidification rate (ECAR) in SOD2-null cells compared to wild-type cells (Figure 5C). These findings indicate that SOD2-null cells preferentially utilize glycolysis rather than oxidative phosphorylation to make ATP. The largest difference between the bioenergetics profiles of wild-type and null cells was in the reserve respiratory capacity of the two cell types where a 5-fold reduction in reserve capacity was noted in cells devoid of MnSOD expression (Figure 5D). Taken together, these results indicate deletion of SOD2 in human cells results in significant mitochondrial dysfunction and metabolic oxidative stress.
Figure 5. SOD2-null HEK293T cells displayed significant alterations in mitochondrial bioenergetics.
(A) Real-time oxygen consumption rate (OCR; pmol/min) was measured with the Seahorse Extracellular Flux Analyzer. A representative trace from a mitochondrial stress test is shown comparing OCR of wild-type cells (closed circles) to knock-out cells (open circles) with arrows indicating sequential additions of oligomycin, FCCP, and antimycin A/rotenone.
(B) Quantification of OCR (pMol/min/µg protein) in wild-type and SOD2-null cells. *P<0.01 compared to wild-type.
(C) Quantification of extracellular acidification rate (ECAR; mpH/min/µg protein) in wild-type and SOD2-null cells. *P<0.01 compared to wild-type.
(D) A 3.5-fold decrease in mitochondrial reserve capacity was detected in knock-out cells. *P = 0.01 compared to wild-type.
Discussion
MnSOD, encoded by the nuclear SOD2 gene, localizes to the mitochondrial matrix and is the main enzyme that scavenges superoxide to prevent oxidative damage in the mitochondria. Mice with constitutive homozygous deletion of Sod2 develop several pathologies such as cardiac myopathy and motor neuron dysfunction and die soon after birth [5, 26]. As a result of their short life span, studying the long-term effects of loss of Sod2 in mice is nearly impossible without conditional Sod2 knockout models. Moreover, translating data derived from animal models in humans is often difficult, if not impossible, but the availability of appropriate human model systems is a substantial obstacle impeding research progress. Studying genetic knockouts associated with disease phenotypes, especially those pertaining to genes involved in cell proliferation and survival, such as SOD2, presents a unique set of challenges. We present for the first time herein a human cell model appropriate for studying cellular and molecular phenotypes caused by loss of MnSOD activity.
The CRISPR/Cas9 system provides the opportunity to easily and efficiently delete genes in human cell models [15–17]. We show targeting SOD2 with a single gRNA resulted in complete abrogation of MnSOD protein expression and eliminated detectable MnSOD enzyme activity. Unlike siRNA, which often results in multiple levels of success in terms of totality of knockdown and is typically transient, the mutations created in SOD2 in this study are stable and therefore heritable and provide an excellent model with which to study SOD2 deficiency in a human cell model.
SOD2 is located on 6q25.3 and loss of heterozygosity of this chromosome has been reported in several types of cancer and other diseases. For example, LOH on the long arm of chromosome 6 has been reported in melanoma [27, 28], acute lymphocytic leukemia[29, 30], as well as in breast [31–35], stomach [36], and ovarian[37] cancers. Therefore, our model would be particularly useful in the context of a Tet-inducible system to regulate expression of SOD2. The use of the CRISPR/Cas9 system to delete SOD2 would allow for customizable re-expression of SOD2 to evaluate the effect(s) of variable levels of MnSOD expression and activity in cancers and other diseases where the expression of MnSOD is altered.
Given MnSOD’s role as the major primary antioxidant protein responsible for protecting mitochondria from oxidative damage, we expected mitochondrial function to be severely impaired in SOD2-null cells. Without proper mitochondrial function due to increased oxidative stress, cellular metabolism and therefore cell growth and proliferation are impaired by lack of MnSOD expression. We found SOD2-null cells exhibited reduced clonogenic potential compared to their wild-type counterparts. This characteristic was attributable to increased oxidative stress since growth in reduced oxygen or treatment with the SOD small molecule mimetic GC4419 both restored the clonogenic potential of SOD2-null cells.
The current study describes the negative effect of loss of MnSOD activity on ETC enzyme function and the subsequent metabolic impairment exhibited by SOD2-null cells supporting the hypothesis that oxidative stress is increased in cells without functional MnSOD. Lack of MnSOD expression resulted in cells that preferentially utilize glycolysis to make ATP rather than oxidative phosphorylation as assessed in real-time by the Seahorse Extracellular Flux analyzer. At baseline, a significant difference in oxygen consumption was observed in wild-type vs. SOD-null cells indicating cells without MnSOD activity are respirationally challenged. Addition of the mitochondrial uncoupler FCCP failed to induce an increase in respiratory rate of SOD2-null cells, indicating these cells have essentially no spare reserve respiratory capacity. Reserve capacity is a measure of mitochondrial reserve energy and is oftentimes only revealed when substrate oxidation is limited by increased demand in ATP. Several factors influence reserve capacity including the ability of the cell to deliver substrate to the mitochondria and proper functioning of enzymes associated with the ETC [38]. The near total depletion of reserve capacity in SOD2-null cells indicates these cells are operating at their bioenergetic limit. Taken together, the decrease in mitochondrial complex I and II activities in SOD2-null cells and the Seahorse OCR data from the mitochondrial stress test indicate severe mitochondrial dysfunction caused by lack of MnSOD expression.
Overall our results support the hypothesis that MnSOD is essential for normal mitochondrial function due to its role in neutralizing superoxide radicals, preserving iron homeostasis, and preserving ETC function. We have created what is to our knowledge the first human cell line with targeted biallelic genetic disruption of SOD2 that is devoid of both detectable MnSOD protein expression and enzyme activity. In addition to studying the metabolic consequences arising as a result of deletion of MnSOD activity, this new model provides a precedent for the genesis of other SOD2-null human cells from other cell types including primary human stem cells. Given that decreased MnSOD expression has been observed in transformed cells and early tumors [39–41], whereas increased expression has been noted in advanced disease [42–45], the work described here provides an excellent model with which to further study the role of MnSOD expression in relevant disease pathologies, including cancer.
HIGHLIGHTS.
We describe the generation of novel human cell line to study MnSOD deficiency
SOD2-null cells exhibit growth deficiency compared to wild-type cells
O2 consumption and ETC enzyme activities were inhibited without MnSOD
SOD2-null cells exhibit significantly impaired mitochondrial function
The SOD2-null phenotype could be rescued by hypoxia or SOD mimic drugs
Acknowledgments
The authors thank Jamie Soto in the Foundation of Eagles Diabetes Research Center Core Facility for assistance with the Seahorse Extracellular Flux experiments, and Kathy Walters of the Central Microscopy Research Facility for assistance with the transmission electron microscopy. This work was supported by a grant from American Diabetes Association and from the University of Iowa Holden Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Loon AP, Pesold-Hurt B, Schatz G. A yeast mutant lacking mitochondrial manganese-superoxide dismutase is hypersensitive to oxygen. Proc Natl Acad Sci U S A. 1986;83:3820–3824. doi: 10.1073/pnas.83.11.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majima HJ, Oberley TD, Furukawa K, Mattson MP, Yen HC, Szweda LI, St Clair DK. Prevention of mitochondrial injury by manganese superoxide dismutase reveals a primary mechanism for alkaline-induced cell death. The Journal of biological chemistry. 1998;273:8217–8224. doi: 10.1074/jbc.273.14.8217. [DOI] [PubMed] [Google Scholar]
- 4.Lebovitz RM, Zhang HJ, Vogel H, Cartwright J, Dionne L, Lu NF, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. P Natl Acad Sci USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature genetics. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 6.Case AJ, McGill JL, Tygrett LT, Shirasawa T, Spitz DR, Waldschmidt TJ, Legge KL, Domann FE. Elevated mitochondrial superoxide disrupts normal T cell development, impairing adaptive immune responses to an influenza challenge. Free radical biology & medicine. 2011;50:448–458. doi: 10.1016/j.freeradbiomed.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Case AJ, Madsen JM, Motto DG, Meyerholz DK, Domann FE. Manganese superoxide dismutase depletion in murine hematopoietic stem cells perturbs iron homeostasis, globin switching, and epigenetic control in erythrocyte precursor cells. Free radical biology & medicine. 2013;56:17–27. doi: 10.1016/j.freeradbiomed.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cyr AR, Brown KE, McCormick ML, Coleman MC, Case AJ, Watts GS, Futscher BW, Spitz DR, Domann FE. Maintenance of mitochondrial genomic integrity in the absence of manganese superoxide dismutase in mouse liver hepatocytes. Redox biology. 2013;1:172–177. doi: 10.1016/j.redox.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velarde MC, Flynn JM, Day NU, Melov S, Campisi J. Mitochondrial oxidative stress caused by Sod2 deficiency promotes cellular senescence and aging phenotypes in the skin. Aging-Us. 2012;4:3–12. doi: 10.18632/aging.100423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Case AJ, Domann FE. Manganese superoxide dismutase is dispensable for post-natal development and lactation in the murine mammary gland. Free radical research. 2012;46:1361–1368. doi: 10.3109/10715762.2012.715370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 12.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nature biotechnology. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 15.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 2003;165:2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weydert CJ, Cullen JJ. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nature protocols. 2010;5:51–66. doi: 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dayal D, Martin SM, Owens KM, Aykin-Burns N, Zhu Y, Boominathan A, Pain D, Limoli CL, Goswami PC, Domann FE, Spitz DR. Mitochondrial complex II dysfunction can contribute significantly to genomic instability after exposure to ionizing radiation. Radiat Res. 2009;172:737–745. doi: 10.1667/RR1617.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazzolotto X, Gaillard J, Pantopoulos K, Hentze MW, Moulis JM. Human cytoplasmic aconitase (Iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron-responsive element binding. The Journal of biological chemistry. 1999;274:21625–21630. doi: 10.1074/jbc.274.31.21625. [DOI] [PubMed] [Google Scholar]
- 22.Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. The Journal of biological chemistry. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 23.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. The Journal of biological chemistry. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 24.Melov S, Coskun P, Patel M, Tuinstra R, Cottrell B, Jun AS, Zastawny TH, Dizdaroglu M, Goodman SI, Huang TT, Miziorko H, Epstein CJ, Wallace DC. Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc Natl Acad Sci U S A. 1999;96:846–851. doi: 10.1073/pnas.96.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, Epstein CJ, Huang TT, Richardson A. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am J Physiol Heart Circ Physiol. 2001;281:H1422–H1432. doi: 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- 26.Oh SS, Sullivan KA, Wilkinson JE, Backus C, Hayes JM, Sakowski SA, Feldman EL. Neurodegeneration and early lethality in superoxide dismutase 2-deficient mice: a comprehensive analysis of the central and peripheral nervous systems. Neuroscience. 2012;212:201–213. doi: 10.1016/j.neuroscience.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Church SL, Grant JW, Ridnour LA, Oberley LW, Swanson PE, Meltzer PS, Trent JM. Increased manganese superoxide dismutase expression suppresses the malignant phenotype of human melanoma cells. Proc Natl Acad Sci U S A. 1993;90:3113–3117. doi: 10.1073/pnas.90.7.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson GP, Coleman AB, Lugo TG. Mechanisms of human melanoma cell growth and tumor suppression by chromosome 6. Cancer research. 1996;56:1635–1641. [PubMed] [Google Scholar]
- 29.Sherratt T, Morelli C, Boyle JM, Harrison CJ. Analysis of chromosome 6 deletions in lymphoid malignancies provides evidence for a region of minimal deletion within a 2-megabase segment of 6q21. Chromosome Res. 1997;5:118–124. doi: 10.1023/a:1018418224660. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi S, Koike M, Seriu T, Bartram CR, Schrappe M, Reiter A, Park S, Taub HE, Kubonishi I, Miyoshi I, Koeffler HP. Frequent loss of heterozygosity on the long arm of chromosome 6: identification of two distinct regions of deletion in childhood acute lymphoblastic leukemia. Cancer research. 1998;58:2618–2623. [PubMed] [Google Scholar]
- 31.Theile M, Seitz S, Arnold W, Jandrig B, Frege R, Schlag PM, Haensch W, Guski H, Winzer KJ, Barrett JC, Scherneck S. A defined chromosome 6q fragment (at D6S310) harbors a putative tumor suppressor gene for breast cancer. Oncogene. 1996;13:677–685. [PubMed] [Google Scholar]
- 32.Devilee P, van Vliet M, van Sloun P, Kuipers Dijkshoorn N, Hermans J, Pearson PL, Cornelisse CJ. Allelotype of human breast carcinoma: a second major site for loss of heterozygosity is on chromosome 6q. Oncogene. 1991;6:1705–1711. [PubMed] [Google Scholar]
- 33.Fujii H, Zhou W, Gabrielson E. Detection of frequent allelic loss of 6q23-q25.2 in microdissected human breast cancer tissues. Genes Chromosomes Cancer. 1996;16:35–39. doi: 10.1002/(SICI)1098-2264(199605)16:1<35::AID-GCC5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Noviello C, Courjal F, Theillet C. Loss of heterozygosity on the long arm of chromosome 6 in breast cancer: possibly four regions of deletion. Clinical cancer research : an official journal of the American Association for Cancer Research. 1996;2:1601–1606. [PubMed] [Google Scholar]
- 35.Sheng ZM, Marchetti A, Buttitta F, Champeme MH, Campani D, Bistocchi M, Lidereau R, Callahan R. Multiple regions of chromosome 6q affected by loss of heterozygosity in primary human breast carcinomas. Br J Cancer. 1996;73:144–147. doi: 10.1038/bjc.1996.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Queimado L, Seruca R, Costa-Pereira A, Castedo S. Identification of two distinct regions of deletion at 6q in gastric carcinoma. Genes Chromosomes Cancer. 1995;14:28–34. doi: 10.1002/gcc.2870140106. [DOI] [PubMed] [Google Scholar]
- 37.Saito S, Sirahama S, Matsushima M, Suzuki M, Sagae S, Kudo R, Saito J, Noda K, Nakamura Y. Definition of a commonly deleted region in ovarian cancers to a 300-kb segment of chromosome 6q27. Cancer research. 1996;56:5586–5589. [PubMed] [Google Scholar]
- 38.Sansbury BE, Jones SP, Riggs DW, Darley-Usmar VM, Hill BG. Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chem Biol Interact. 2011;191:288–295. doi: 10.1016/j.cbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marlhens F, Nicole A, Sinet PM. Lowered level of translatable messenger RNAs for manganese superoxide dismutase in human fibroblasts transformed by SV 40. Biochemical and biophysical research communications. 1985;129:300–305. doi: 10.1016/0006-291x(85)91437-8. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, Colburn NH, Oberley LW. Decreased expression of manganese superoxide dismutase mRNA and protein after immortalization and transformation of mouse liver cells. Oncol Res. 1993;5:127–132. [PubMed] [Google Scholar]
- 41.St Clair DK, Oberley LW. Manganese superoxide dismutase expression in human cancer cells: a possible role of mRNA processing. Free Radic Res Commun. 1991;12–13(Pt 2):771–778. doi: 10.3109/10715769109145858. [DOI] [PubMed] [Google Scholar]
- 42.Tsanou E, Ioachim E, Briasoulis E, Damala K, Charchanti A, Karavasilis V, Pavlidis N, Agnantis NJ. Immunohistochemical expression of superoxide dismutase (MnSOD) anti-oxidant enzyme in invasive breast carcinoma. Histol Histopathol. 2004;19:807–813. doi: 10.14670/HH-19.807. [DOI] [PubMed] [Google Scholar]
- 43.Kamarajugadda S, Cai Q, Chen H, Nayak S, Zhu J, He M, Jin Y, Zhang Y, Ai L, Martin SS, Tan M, Lu J. Manganese superoxide dismutase promotes anoikis resistance and tumor metastasis. Cell Death Dis. 2013;4:e504. doi: 10.1038/cddis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen PM, Wu TC, Shieh SH, Wu YH, Li MC, Sheu GT, Cheng YW, Chen CY, Lee H. MnSOD promotes tumor invasion via upregulation of FoxM1-MMP2 axis and related with poor survival and relapse in lung adenocarcinomas. Mol Cancer Res. 2013;11:261–271. doi: 10.1158/1541-7786.MCR-12-0527. [DOI] [PubMed] [Google Scholar]
- 45.Hempel N, Carrico PM, Melendez JA. Manganese superoxide dismutase (Sod2) and redox-control of signaling events that drive metastasis. Anticancer Agents Med Chem. 2011;11:191–201. doi: 10.2174/187152011795255911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami K, Kondo T, Kawase M, Li Y, Sato S, Chen SF, Chan PH. Mitochondrial susceptibility to oxidative stress exacerbates cerebral infarction that follows permanent focal cerebral ischemia in mutant mice with manganese superoxide dismutase deficiency. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:205–213. doi: 10.1523/JNEUROSCI.18-01-00205.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikegami T, Suzuki Y, Shimizu T, Isono K, Koseki H, Shirasawa T. Model mice for tissue-specific deletion of the manganese superoxide dismutase (MnSOD) gene. Biochemical and biophysical research communications. 2002;296:729–736. doi: 10.1016/s0006-291x(02)00933-6. [DOI] [PubMed] [Google Scholar]
- 48.Parmar H, Melov S, Samper E, Ljung BM, Cunha GR, Benz CC. Hyperplasia, reduced E-cadherin expression, and developmental arrest in mammary glands oxidatively stressed by loss of mitochondrial superoxide dismutase. Breast. 2005;14:256–263. doi: 10.1016/j.breast.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Lustgarten MS, Jang YC, Liu Y, Muller FL, Qi W, Steinhelper M, Brooks SV, Larkin L, Shimizu T, Shirasawa T, McManus LM, Bhattacharya A, Richardson A, Van Remmen H. Conditional knockout of Mn-SOD targeted to type IIB skeletal muscle fibers increases oxidative stress and is sufficient to alter aerobic exercise capacity. American journal of physiology. Cell physiology. 2009;297:C1520–C1532. doi: 10.1152/ajpcell.00372.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]