ABSTRACT
In recent years, photosynthetic autotrophic cyanobacteria have attracted interest for biotechnological applications for sustainable production of valuable metabolites. Although biosafety issues can have a great impact on public acceptance of cyanobacterial biotechnology, biosafety of genetically modified cyanobacteria has remained largely unexplored. We set out to incorporate biocontainment systems in the model cyanobacterium Synechocystis sp. PCC 6803. Plasmid-encoded safeguards were constructed using the nonspecific nuclease NucA from Anabaena combined with different metal-ion inducible promoters. In this manner, conditional lethality was dependent on intracellular DNA degradation for regulated autokilling as well as preclusion of horizontal gene transfer. In cells carrying the suicide switch comprising the nucA gene fused to a variant of the copM promoter, efficient inducible autokilling was elicited. Parallel to nuclease-based safeguards, cyanobacterial toxin/antitoxin (TA) modules were examined in biosafety switches. Rewiring of Synechocystis TA pairs ssr1114/slr0664 and slr6101/slr6100 for conditional lethality using metal-ion responsive promoters resulted in reduced growth, rather than cell killing, suggesting cells could cope with elevated toxin levels. Overall, promoter properties and translation efficiency influenced the efficacy of biocontainment systems. Several metal-ion promoters were tested in the context of safeguards, and selected promoters, including a nrsB variant, were characterized by beta-galactosidase reporter assay.
KEY WORDS: Synechocystis, Cyanobacteria, Biocontainment, Biosafety, Nuclease nucA, Toxin/antitoxin
Summary: Biosafety of biotechnologically important microalgae was addressed by suicide switch implementation in cyanobacterium Synechocystis sp. PCC 6803. This is the first report of biocontainment safeguards in cyanobacteria.
INTRODUCTION
Cyanobacteria display high metabolic versatility and are acquiring interest for biotechnological applications, as they are photosynthetic autotrophs that require little more than water, sunlight and CO2 for their growth and sustainable production of valuable metabolites. Engineered cyanobacteria can be used for generating a number of metabolites ranging from nutrients for human consumption to bioplastics and biofuels (Lai and Lan, 2015; Lau et al., 2015). Even though biosafety issues can have a fundamental impact on public opinion and acceptance of cyanobacterial biotechnology, biosafety of genetically modified cyanobacteria has remained largely unexplored. While modified cyanobacteria can be efficiently contained within closed photobioreactors, there is always a risk of accidental release, and uncertainty about possible negative ecological effects in case the engineered bacteria survive and establish themselves in the environment. Further risk is presented by cultivation of cyanobacteria in open reactors, raceways and ponds. Cyanobacteria have also been studied for bioremediation (Lau et al., 2015; Pereira et al., 2011), which would require monitoring of their persistence in the environment. The possibility of horizontal transfer of recombinant genetic material from genetically modified cyanobacteria to endogenous population is additionally concerning, considering cyanobacteria are widely present in nature (Li et al., 2001; Vioque, 2007) and some strains are naturally transformable (Vioque, 2007).
To reduce the likelihood of environmental persistence of engineered organisms, they can be equipped with biological containment mechanisms. In passive containment systems, specific gene defects can be engineered to make cell viability dependent on addition of exogenous supplement. In active containment systems, cells can be equipped with safeguards or ‘kill switches’ that can cause cell death in a controlled suicide process. Such microbes are intended to survive normally in the absence of kill switch induction, but in the environment, engineered cells can be destroyed by induced lethality, while leaving the surrounding endogenous microbes intact. Different lethal genes have been employed in conditional cell-killing systems in various bacteria. For example, cell death has been achieved by proteins that destroy the cell membrane (Bej et al., 1988; Schweder et al., 1992), the cell wall (Morita et al., 2001), that degrade RNA (Munthali et al., 1996; Torres et al., 2003; Wright et al., 2015), or destroy cellular DNA (Ahrenholtz et al., 1994; Balan and Schenberg, 2005; Torres et al., 2003). The genes encoding these toxic proteins have been put under the control of different types of promoters, such as those inducible by increased temperature (Ahrenholtz et al., 1994), starvation (Schweder et al., 1992), the presence of specific chemicals (Bej et al., 1988; Jensen et al., 1993; Munthali et al., 1996) or growth supplements (Balan and Schenberg, 2005; Schweder et al., 1992). Employing more than one containment system in a single cell has been shown to increase the efficacy of biological safeguards (Torres et al., 2003).
While biological containment systems have been implemented in various organisms (Moe-Behrens et al., 2013; Wright et al., 2013), none have so far been developed for cyanobacteria. Even though induced lysis has been carried out in cyanobacteria for the purpose of recovering biofuel from biomass (Liu and Curtiss, 2009; Miyake et al., 2014), metabolite extraction does not require complete lysis-induced death, while total killing is necessary for biosafety purposes.
In this work, we addressed biosafety of genetically modified cyanobacteria through construction of biological containment systems in the model cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis). Two strategies were employed. In the first, conditional lethality was dependent on intracellular degradation of DNA, to accomplish not only regulated killing of cells but also destruction of cellular genetic material, which could otherwise be available for horizontal gene transfer. To that end, we made use of non-specific DNA/RNA nuclease NucA and its inhibitor NuiA from the cyanobacterium Anabaena sp. PCC 7120. By using metal-ion inducible promoters to trigger nuclease expression, we were able to elicit efficient cell killing upon inducer addition. The most efficient promoter was a PcopM variant. In the second approach, Synechocystis toxin-antitoxin (TA) systems ssr1114/slr0664 and slr6101/slr6100 were rewired for conditional lethality by using metal-ion inducible promoters. In different kill switch variants with toxins Slr0664 or Slr6100 (which encode RelE-like ribonucleases), reduced growth of bacteria rather than efficient cell killing was observed, suggesting bacteria were able to cope with the cellular damage inflicted by the toxins. Finally, as the choice of promoters used in cyanobacterial conditional suicide systems was crucial, several metal-ion promoters were tested in the context of kill switches, and selected promoters were characterized in detail by beta-galactosidase reporter assay.
RESULTS
Nuclease-based cyanobacterial kill switch
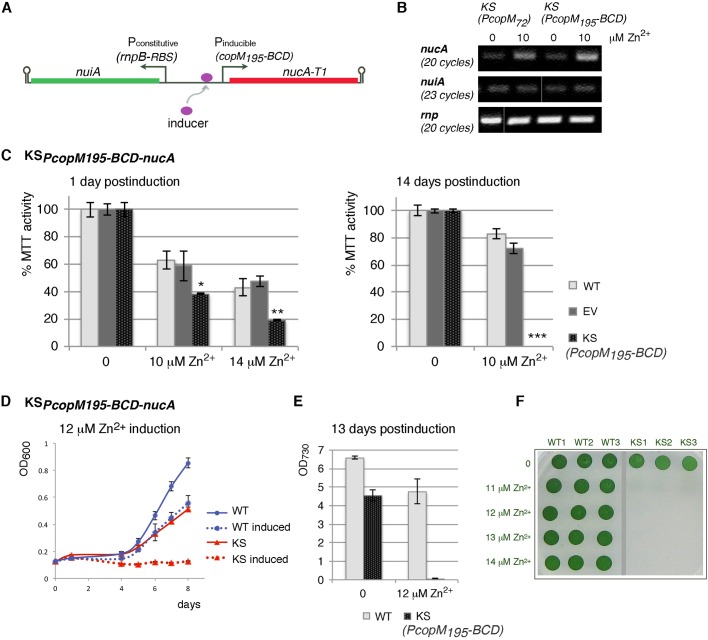
In order to construct biosafety mechanisms in cyanobacteria, we took advantage of the cyanobacterial non-specific DNA/RNA nuclease NucA and its inhibitor NuiA from Anabaena. While Synechocystis sp. PCC 6803 does not contain a NucA homolog, nucleases of this type are present in several bacterial species and are believed to have evolved to serve for nutritional purposes and sometimes as bacteriocides (Meiss et al., 1998; Muro-Pastor et al., 1992). We envisioned that by rewiring the nuclease/inhibitor pair for conditional expression, cell survival could be achieved specifically in the photobioreactor, while upon accidental release into the environment, the rewired nuclease would prevail over the inhibitor, thereby killing the cells. To create such a mechanism, the nuclease gene was placed under an inducible promoter to allow induction upon exposure to environmental inducer (Fig. 1A). The coding sequence of nucA was shortened by 69 nucleotides encoding the signal peptide (Muro-Pastor et al., 1992) in order to achieve intracellular localization of the nuclease by preventing its export to the periplasm. To protect cells from possible leaky nuclease production in the bioreactor in absence of inducer, the nuclease inhibitor gene was fused to a weak constitutive promoter (Fig. 1A).
Fig. 1.
Synechocystis sp. PCC 6803 carrying the plasmid-encoded nuclease suicide switch KSPcopM195-BCD-nucA displays efficient induced autokilling. (A) Diagrammatical representation of the suicide switch. The nuclease gene is under the inducible promoter PcopM195-BCD (Pinducible) that enables triggering of cell death upon exposure to inducer. To protect cells from possible leaky nuclease expression in the absence of inducer, the nuclease inhibitor is produced from the weak constitutive promoter PrnpB-RBS (Pconstitutive). T1, degradation tag; arrow indicates transcriptional start site; hairpin indicates transcription terminator. (B) RT-PCR analysis of nucA and nuiA mRNA levels in kill switch Synechocystis cells KSPcopM72-nucA and KSPcopM195-BCD-nucA 1 h after 10 µM Zn2+ induction. For detection of the less abundant nuiA, more PCR cycles were needed. rnp, loading control. (C) MTT assay showing a drop in viability of suicide switch Synechocystis cells KSPcopM195-BCD-nucA (KS) 1 day or 14 days after induction with Zn2+ (mean % MTT activity±s.e.m., n=3). WT, wild type; EV, empty vector control. Asterisks indicate significant differences (KS compared with WT) at *(1 day, 10 µM Zn2+); **(1 day, 14 µM Zn2+); ***(14 days, 10 µM Zn2+) (t-test, P<0.05). Differences between WT and EV were not statistically significant. (D) Growth of wild-type Synechocystis (WT) and KSPcopM195-BCD-nucA (KS) cells was measured by plate reader after induction with 12 µM Zn2+ (mean OD600±s.e.m.). (E) Cell density (OD730) of cultures in multiwell plates from (D) was determined manually on the last day of growth (OD730±s.e.m.). By assessing culture density at OD730 at the end of multiwell plate experiment, we validated that the efficiency of killing determined by OD600 (plate reader) and OD730 were comparable. Kill switch KSPcopM195-BCD-nucA cells showed complete killing upon 12 µM Zn2+ induction. (F) Triplicate aliquots from cultures in (D) were plated on BG11 solid media at day 13. Complete killing was observed for KSPcopM195-BCD-nucA cells (triplicates KS1-3) at or above 12 µM Zn2+, while some survivors were seen at 11 µM Zn2+. WT1-3, wild-type triplicates.
Genetic elements used in suicide switch construction
The choice of promoters was crucial for creating a successful suicide mechanism. In particular, for the fusion with the toxic nuclease, we expected that low leakiness and high promoter inducibility would be needed, with the former necessary to preclude any negative effects on growth in absence of inducer. For potential future biotechnological use, the cost of promoter inducer was also a factor. Even though numerous tight and highly responsive promoters are well characterized in Escherichia coli (e.g. Ptrc, Plac), these promoters do not work comparably in cyanobacteria, likely due to RNA polymerase differences (Heidorn et al., 2011). Some have been shown to lose their inducer responsiveness in cyanobacteria and become constitutive (Guerrero et al., 2012; Heidorn et al., 2011). Current cyanobacterial molecular biology toolbox offers only a limited number of well-characterized promoters to carry out expression of a gene of interest in Synechocystis. Several of the most inducible and tightly regulated cyanobacterial promoters belong to the group of native metal-ion responsive promoters. Examples include promoters preceding two operons involved in copper response, the copBAC operon (Giner-Lamia et al., 2012) and the copMRS operon (Giner-Lamia et al., 2015, 2012), the nickel-response operon nrsBACD (Blasi et al., 2012; Lopez-Maury et al., 2002; Peca et al., 2008), the metallothionein gene smtA (Turner et al., 1996), the plastocyanin gene petE (Briggs et al., 1990), the cytochrome c6 gene petJ (Kuchmina et al., 2012) and the coaT gene (Guerrero et al., 2012; Peca et al., 2008). These promoters react to very low (micromolar) concentrations of metal ions and typically respond to several metal ions, showing variation in response depending on the metal ion used. As inducers, metal ions are cost-efficient for larger culture volumes. For construction of inducible biosafety circuits in cyanobacteria, we selected three metal-ion responsive promoters: PcopM, PcopB, and PnrsB (Table 1).
Table 1.
Kill switch constructs and their activity
Initial attempts at cloning the suicide switch by trying to use different metal-ion responsive promoters to drive nuclease expression were repeatedly unsuccessful. Either no transformants were obtained or nuclease-disabling mutations were observed, probably due to leaky expression and cellular toxicity of the nuclease. Cloning was feasible only when degradation tags (specific amino acid sequences recognized by cytoplasmic proteases) were used to curtail the cellular half-life of the nuclease. Tags T1 (RPAANDENYAAAV) or T2 (RPAANDENYALAA) (Huang et al., 2010; Landry et al., 2013) were cloned as C-terminal fusions to the nuclease. T1 has been shown to cause weaker reporter protein destabilization in cyanobacteria than T2 (Huang et al., 2010; Landry et al., 2013).
Native PcopM promoter in nuclease-based suicide switch
We first constructed a plasmid-encoded nuclease-based kill switch (KSPcopM72-nucA) in which nucA was fused to promoter PcopM (Fig. S1A; Table 1). In Synechocystis, this promoter regulates the copMRS operon, which is involved in copper response (Giner-Lamia et al., 2015). Under native conditions, PcopM responds robustly to Cu2+. As little as 1 µM Cu2+ triggers about 26-fold increase in CopM protein levels, and 3 µM Cu2+ increases copM mRNA levels 75-fold (Giner-Lamia et al., 2015, 2012). Other metal ions (Zn2+, Cd2+, and Ni2+) trigger a poorer PcopM response (Giner-Lamia et al., 2012). The coding sequence of nucA (with T1 degradation tag) was fused to the 72-nt long PcopM72 promoter sequence up to the start codon, containing two direct repeats that are likely involved in transcriptional regulation (Giner-Lamia et al., 2012). The nuiA gene (encoding the inhibitor) was feebly expressed (Fig. 1B) from the weak constitutive promoter PrnpB (promoter of ribozyme RNase P subunit B) fused to a ribosome binding site (RBS) (Huang et al., 2010). After induction of Synechocystis KSPcopM72-nucA culture (106 cells/ml) with 2 µM Cu2+, no significant killing was observed (Fig. S1B). Rather, slower growth was recorded in comparison to wild-type (WT) cells, as measured by optical density (OD730). Some reduced growth was also seen with uninduced KSPcopM72-nucA cells, possibly due to the use of standard BG11 growth medium, which already contains small amounts of Cu2+ (0.32 μM) (Rippka, 1988). However, the choice of this medium was intentional in order to mimic conditions of bioreactor cultivation for biotechnological purposes.
Although metal ions are required for essential cellular processes, they become cytotoxic after exceeding specific critical concentrations. Synechocystis has a rather low tolerance for Cu2+ (Blasi et al., 2012) and in our experiments, substantial general toxicity toward WT Synechocystis cells is observed at Cu2+-ion concentrations above 4 µM, thereby limiting usefulness of Cu2+ as promoter inducer. On the other hand, Synechocystis tolerance to Zn2+ is relatively high, with IC50 (half growth inhibitory concentration) between 8 and 16 µM Zn2+ (Blasi et al., 2012). When KSPcopM72-nucA culture was induced with 10 µM Zn2+, a substantial, although incomplete killing of suicide switch cells was observed in comparison to WT cells, which retained normal growth (Fig. S1B). Moreover, uninduced KSPcopM72-nucA cells did not display significant growth inhibition in spite of the presence of 0.77 μM Zn2+ (Rippka, 1988) in standard BG11 medium.
Bicistronic design in PcopM-nuclease-based suicide switch
In order to improve efficiency of this suicide system, two construct modifications were made (Fig. S1C). First, a larger sequence segment (195 nt) preceding the copMRS operon was included in the suicide switch to increase the likelihood that all the important regulatory sequences were contained in the promoter fragment. Second, 26 nucleotides of the native sequence (from the copM transcriptional start site to the start codon) were replaced by bicistronic design (BCD) (Mutalik et al., 2013) (Fig. S1C). BCD is a genetic element that standardizes the mRNA secondary structure over the RBS–CDS (ribosome binding site – coding sequence) junction, thereby diminishing variation in ribosome binding and expression when combined with different genes of interest. It allows reliable initiation of translation of random genes (Mutalik et al., 2013). With the resulting modified construct (KSPcopM195-BCD-nucA), complete killing of Synechocystis cells was achieved upon addition of Zn2+ ions, as expected for a functional kill switch (Fig. 1, Table 1).
First, induction of KSPcopM195-BCD-nucA cells with 10 µM Zn2+ markedly increased nucA transcript levels within an hour of induction (comparably to the increase seen in KSPcopM72-nucA cells) (Fig. 1B). This correlated with a sharp decrease in the number of viable, metabolically active cells, as quantified by the MTT assay. This assay measures the conversion of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] into formazan crystals by active reductases in living cells (Li and Song, 2007) (Fig. 1C). Within a day of adding inducer to log-phase cultures, MTT-reducing activity of KSPcopM195-BCD-nucA culture significantly decreased (P<0.05 compared with WT) to 38%±0.7% at 10 µM Zn2+ and 19%±0.7% at 14 µM Zn2+. Activity of WT cells and cells carrying an empty vector (EV) also decreased (to 63%±6.7% (WT) and 59%±11.1% (EV) at 10 µM Zn2+; and to 43%±6.4% (WT) and 47%±3.8% (EV) at 14 µM Zn2+), reflecting general toxic effects of zinc ions. However, over a 14-day period WT and EV cultures were mostly able to recover, while KSPcopM195-BCD-nucA cells completely lost ability to reduce MTT, indicating they were no longer viable (Fig. 1C). Growth curves corroborated MTT results. When bacterial cultures (106 cells/ml) were induced with 12 µM Zn2+, the suicide switch culture showed no growth even after prolonged incubation (Fig. 1D,E). Plating of this culture on solid media at day 13 revealed no survivors (Fig. 1F). In contrast, WT cells remained viable, albeit displaying a small retardation of growth (Fig. 1D). Some growth hindrance was seen for suicide switch cells without Zn2+-induction (Fig. 1D), reflecting weak leakiness of nuclease expression (Fig. 1B). Together, these results indicate that the kill switch KSPcopM195-BCD-nucA is functional.
As noted, the kill switch KSPcopM195-BCD-nucA differed from the kill switch KSPcopM72-nucA twofold: in the length of the PcopM-promoter fragment, important for nucA transcription, and in the use of the BCD element (versus the native RBS), important for translation initiation. Induction of these two switches resulted in different killing efficiencies (Fig. 1D, Fig. S1B), even though upregulation of nucA transcript levels was comparable for both kill switch cells (Fig. 1B). This suggests that translation initiation efficiency (influenced by BCD) contributed to the improved killing observed for KSPcopM195-BCD-nucA cells.
Determining optimal inducer concentration for autodestruction
Up to 11 µM Zn2+, survivors of induced KSPcopM195-BCD-nucA cells could readily be observed. Complete killing was typically achieved at concentrations of 12 to 14 µM Zn2+ and culture density of 106 cells/ml (Fig. 1E). Concentrations above 14 µM Zn2+ were not useful due to nonselective Zn2+-toxicity to WT cells. Nonetheless, at higher culture density of suicide switch cells, the killing efficiency with 12 to 14 µM Zn2+ was reduced, suggesting that inducer may not have reached all cells or that it may have been getting removed from the cells. In agreement with this is our observation that WT cells also respond variably to Zn2+ at different culture densities (data not shown). Therefore, for real-life biotechnological applications, attention to inducer concentration as well as cell density will be necessary. Notably, an occasional biological replicate of KSPcopM195-BCD-nucA cells started growing even at 12-14 µM Zn2+ at 106 cells/ml. It is possible that selective pressure imposed by induced nuclease expression may have led to suicide switch instability.
Promoter PnrsB in nuclease-based suicide device
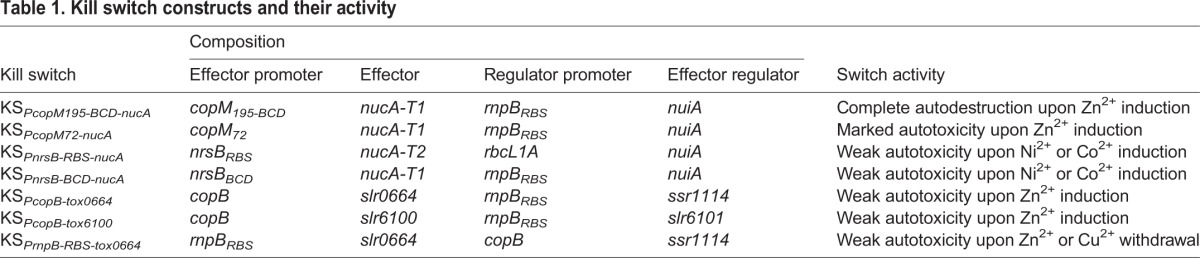
We also assessed the PnrsB promoter, which regulates the expression of the nrsBACD operon involved in cellular response to Ni2+ (Garcia-Dominguez et al., 2000; Lopez-Maury et al., 2002). PnrsB is induced by Ni2+ and Co2+ ions and exhibits low leakiness (Garcia-Dominguez et al., 2000; Lopez-Maury et al., 2002). The BG11 growth medium does not include Ni2+, but it contains 0.17 μM Co2+ (Rippka, 1988). To construct the suicide switch, nucA-T2 (gene encoding NucA with T2 degradation tag) was fused to the sequence PnrsB-RBS, which is an intergenic segment preceding the nrsB gene, in which a strong RBS (Heidorn et al., 2011) replaced the native RBS (Table 1; Fig. S2A,B). The strong RBS was used to improve chances of strong nuclease expression. The nuclease inhibitor in the switch was moderately expressed from the constitutive rbcL1A promoter (Fig. 2A) (Huang et al., 2010). The resulting construct (KSPnrsBRBS-nucA) caused only moderate autotoxicity rather than efficient killing when induced with 4 µM Co2+ (Fig. 2A), precluding its usefulness for biotechnological purposes. At Co2+ concentrations higher than 4 µM, strong general toxicity to WT cells was observed, in line with the published IC50 of 8 µM for Co2+ (Blasi et al., 2012).
Fig. 2.
Characterization of PnrsB-RBS promoter element in Synechocystis and its use in the nuclease-based suicide switch KSPnrsBRBS-nucA. (A) Diagrammatical representation of the KSPnrsBRBS-nucA suicide switch (top panel). Nuclease expression is driven by the inducible promoter PnrsB-RBS and expression of nuclease inhibitor by the moderate constitutive promoter PrbcL1A. T2, degradation tag; ; arrow indicates transcriptional start site; hairpin indicates transcription terminator. (Bottom panel) Growth of wild-type (WT) and KSPnrsBRBS-nucA (KS) Synechocystis cells was recorded by a plate reader after induction with 4 µM Co2+ (mean OD600±s.d.) (B) Evaluation of promoter activity by beta-galactosidase reporter analysis, represented as mean standardized MU (μmol ONP min−1 μg−1)±s.e.m. Promoter PnrsB-RBS (induced with different metal ions) and constitutive promoters PrnpB-RBS1 and PrbcL1A were analyzed. EV, empty vector control.
Interestingly, although PnrsB has been identified as a Ni2+-promoter that responds most robustly to Ni2+ ions, in the suicide switch context cobalt-triggered autotoxicity measured by the MTT assay exceeded that observed with Ni2+ induction (Fig. S2D,E), even though 4 µM Ni2+ did markedly induce nucA mRNA levels in KSPnrsBRBS-nucA cells (Fig. S2D). To understand the characteristics of the PnrsB-RBS genetic element, we set out to analyze it using the beta-galactosidase reporter assay (Miller, 1972). In agreement with the kill switch observations, PnrsB-RBS fused to lacZ responded well to both ions, with a more pronounced reaction to Co2+ than Ni2+ (Fig. 2B). At 8 µM Co2+, a 105-fold induction was observed, significantly higher than that with 10 µM Ni2+ (26-fold), or even 27 µM Ni2+ (80-fold promoter induction, accompanied by strong general toxicity to cells). Such inducer specificity differs from that of native PnrsB (Garcia-Dominguez et al., 2000; Lopez-Maury et al., 2002; Peca et al., 2007). It is not clear what causes the difference in specificity. It is possible that the difference may reflect sequence modifications in the RBS region (e.g. perhaps causing secondary structure changes). In the PnrsB-RBS fusion constructs, the sequence of the nrsB-regulator (nrsR) and sensor (nrsS) (Lopez-Maury et al., 2002) were not included (promoter response relied on endogenous NrsR and NrsS), which also may have influenced promoter activity.
Although transcriptional induction of KSPnrsBRBS-nucA was pronounced (Fig. S2D), autotoxicity was not efficient, possibly due to issues with translation. To optimize chances of efficient translation initiation, bicistronic design (BCD) was added to the promoter sequence to create PnrsB-BCD (Fig. S2C). Nonetheless, inclusion of bicistronic design did not improve killing efficiency, as tested with a kill switch, in which nucA-T1 was expressed from PnrsB-BCD and nuiA from weak PrnpB promoter (Table 1). Therefore, other factors may have played a role in low kill switch efficiency.
Toxin/antitoxin-based cyanobacterial kill switch
In the second approach, molecular safeguards were created in Synechocystis sp. PCC 6803 by making use of cyanobacterial toxin-antitoxin (TA) systems. TAs are widespread prokaryotic genetic elements comprising a stable toxic protein and a less stable neutralizing antidote involved in various cellular functions ranging from plasmid addiction to stress response (Otsuka, 2016). Two Synechocystis TA systems were employed in safeguard constructs: the chromosomally encoded pair ssr1114/slr0664, for which lethal toxicity had been shown in the heterologous host E. coli (Ning et al., 2011; Ye and Ning, 2010); and the plasmid-encoded putative TA pair slr6101/slr6100, which was selected based on bioinformatics data (Makarova et al., 2009) (TADB database, last accessed January 15, 2016, http://202.120.12.135/TADB2/index.php) and our preliminary toxicity evaluation in E. coli (unpublished data). Both toxin genes (slr0664 and slr6100) encode proteins with homology to RNA interferase RelE. Similar to nuclease-based kill switch design, we employed metal-ion inducible promoters to rewire Synechocystis TA pairs for conditional expression.
Use of cop promoters in toxin/antitoxin-based kill switch
We first attempted to construct a TA-based kill switch by using the copM promoter, which proved optimal in the nuclease-based switch. However, our repeated cloning efforts resulted in constructs with toxin-disabling mutations, suggesting occurrence of promoter leakage and selection pressure.
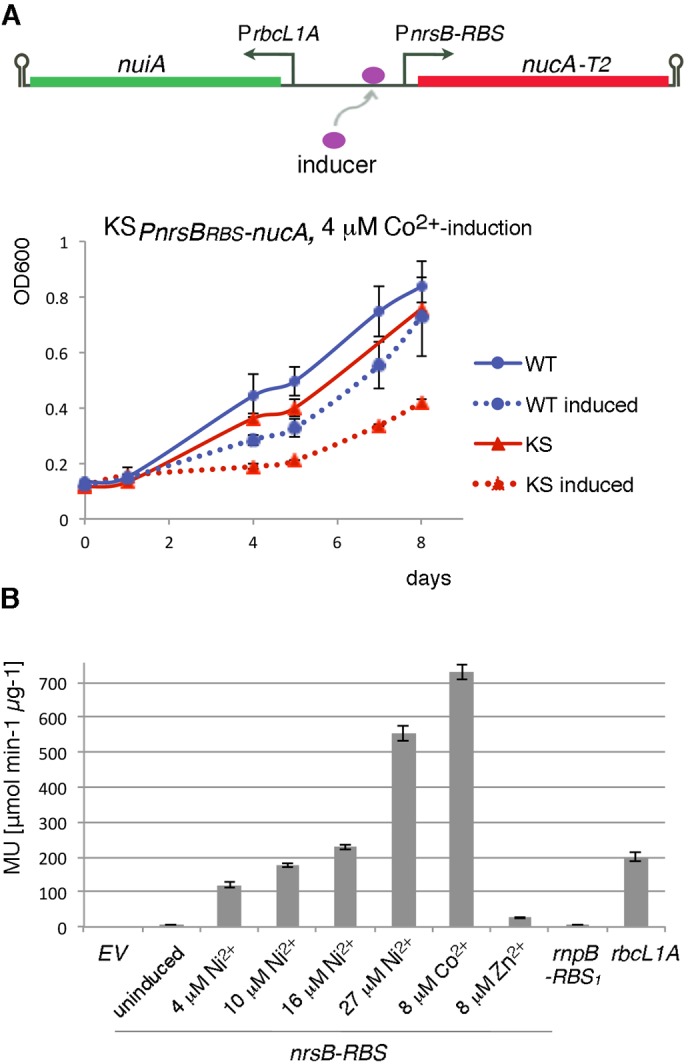
Construction was successful with PcopB, another promoter involved in cellular response to copper (Giner-Lamia et al., 2012). It responds strongly to Cu2+, with 14-fold induction at 3 µM Cu2+, moderately to Zn2+, and is fairly tight when uninduced (Giner-Lamia et al., 2012). Due to higher Synechocystis tolerance to Zn2+, this metal ion was used for induction. Kill switches containing the toxin (either slr0664 or slr6100) fused to copB (Fig. S3), with antitoxins (ssr1114 or slr6101, respectively) expressed from the weak PrnpB-RBS triggered cellular autotoxicity (Fig. 3, Table 1). Nevertheless, neither suicide switch caused efficient killing. Due to similar performances, only slr0664 kill switch KSPcopB-tox0664 is shown (Fig. 3).
Fig. 3.
Induced autotoxicity of Synechocystis sp. PCC 6803 cells carrying the plasmid-encoded toxin/antitoxin suicide switch KSPcopB-tox0664. (A) Diagrammatical representation of the KSPcopB-tox0664 suicide switch. Toxin expression is driven by the inducible promoter PcopB and expression of antitoxin by the weak constitutive promoter PrnpB-RBS. Arrow indicates transcriptional start site; hairpin indicates transcription terminator. (B) MTT assay showing a drop in viability of suicide switch Synechocystis cells KSPcopB-tox0664 (KS) 1 day after induction with 10 µM Zn2+ (mean % MTT activity±s.e.m., n=3). WT, wild type; EV, empty vector control. Asterisk indicates significant difference at 10 µM Zn2+ (KS compared with WT, t-test, P<0.05). Differences between WT and EV were not statistically significant. (Inset) Increase in toxin mRNA levels is detected by RT-PCR in Synechocystis KSPcopB-tox0664 cells 1 h after induction with 10 µM Zn2+; rnp: loading control. (C) Growth of wild-type Synechocystis (WT) and KSPcopB-tox0664 (KS) cells was recorded by a plate reader after induction with 10 µM Zn2+ (mean OD600±s.e.m., n=3).
A day after induction of log-phase KSPcopB-tox0664 culture with 10 µM Zn2+, MTT assay revealed a significant, 77%±3.0% drop in metabolically viable cells (Fig. 3B), which is comparable to that seen with the efficient nuclease switch KSPcopM195-BCD-nucA (Fig. 1C). The drop correlated with increased toxin slr0664 mRNA levels (Fig. 3B). However, following induction, the cells carrying the toxin kill switch continued to grow with reduced rate (Fig. 3C), unlike the nuclease cells KSPcopM195-BCD-nucA that died off (Fig. 1D). One explanation for different responses of the nuclease and the toxin safeguards may be that the damage inflicted by the toxin ribonuclease activity may be easier for cells to cope with than the damage imposed by NucA, which is DNA and RNA nuclease. It is also possible that the induced toxin protein levels in the kill switch cells were low (e.g. due to translation and degradation rates). One explanation may be the existence of endogenous Ssr1114 antitoxin in Synechocystis, which may be neutralizing the induced toxin. As antitoxin crosstalk has been described (Smith et al., 2012), it is also conceivable that other endogenous antitoxins may be suppressing the toxin. Consequently, in order for TA-based kill switch to be efficient, the toxin levels would have to be heavily induced to inflict substantial cellular damage as well as override possible neutralizing molecules. Toxin induction with promoter PcopB does not appear to have reached that intensity.
TA-safeguard with antitoxin induction during growth in culture medium
Another possible safeguard mechanism involves keeping both the toxin and the antitoxin expressed in the bioreactor, with the antitoxin induced by inducer that is scarce in the environment, and the toxin being continually expressed. Consequently, in the environment, the toxin is expected to prevail over the antitoxin. Potentially good inducers for this purpose may be zinc ions, considering that publicly available data on metal ion content in riverine environments (e.g. for Slovenia: http://www.arso.gov.si) indicate that zinc concentrations tend to be low (around 0.08 µM).
A kill switch in which the toxin slr0664 was expressed constitutively from PrnpB-RBS and the antitoxin ssr1114 was inducible from the PcopB promoter (Fig. 4A, Table 1) was tested in Synechocystis. As expected, growth of these cells on BG11 plates containing 4 µM Zn2+ was indistinguishable from that of WT cells (Fig. 4B). In contrast, substantially less growth was observed for kill switch cells on BG11 alone. Plate colony counting and stereomicroscopic evaluation revealed that toxin expression resulted in cell growth inhibition rather than cell killing, as indicated by a marked change in colony size but similar colony counts between plates (Fig. 4C). This suggests that kill switch cells are able to cope with the damage imposed by toxin activity, and higher toxin levels may be necessary to achieve efficient killing. Thus, in order for this safeguard mechanism to be useful for biotechnological purposes, other promoter combinations would have to be tested.
Fig. 4.
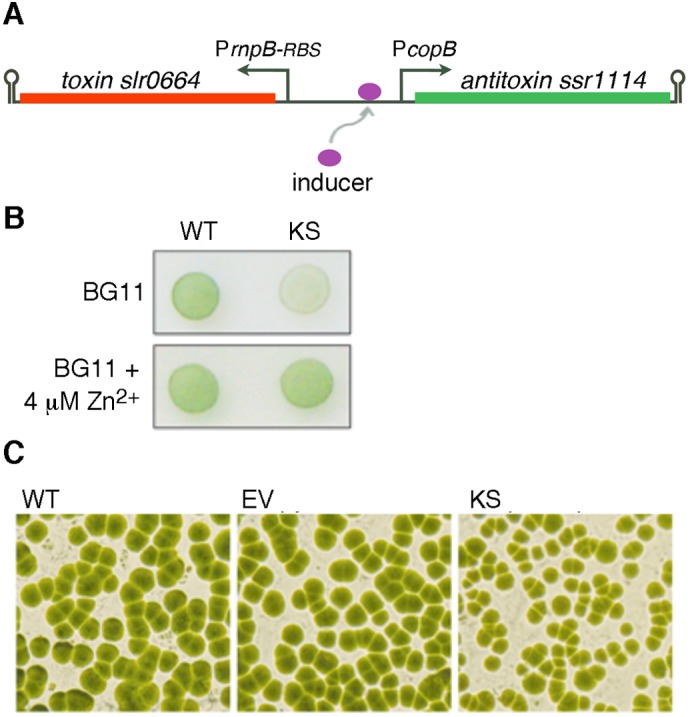
Autotoxicity triggered by withdrawal of antitoxin inducer in Synechocystis sp. PCC 6803 cells carrying a plasmid-encoded toxin/antitoxin suicide switch. (A) Diagrammatical representation of the suicide switch. Antitoxin expression is driven by the inducible promoter PcopB and expression of toxin by the constitutive promoter PrnpB-RBS. Antitoxin levels can be reduced upon withdrawal of inducer from the growth medium, allowing the toxin to prevail. Arrow indicates transcriptional start site; hairpin indicates transcription terminator. (B) Growth of wild type Synechocystis (WT) and suicide switch cells (KS) on BG11 plates with or without added Zn2+. Inducer limitation in BG11 plates results in autotoxicity of suicide switch cells. (C) Stereomicroscopic evaluation of colonies from agar plates in (B) reveals that kill switch cells (KS) exhibit similar numbers of colony forming units but different growth rates in comparison to wild-type Synechocystis (WT) and empty vector (EV) control.
DISCUSSION
In this work, a functional biocontainment mechanism was implemented in the model cyanobacterium Synechocystis sp. PCC 6803. Our biosafety suicide switch KSPcopM195-BCD-nucA was based on induction of an intracellular DNA/RNA nuclease NucA, which causes killing of cells by degrading their genetic material. The nuclease gene was driven by a variant of the metal-ion inducible promoter copM (PcopM195-BCD). The advantage of having a nuclease incorporated into a biosafety system is the destruction of recombinant DNA, so that it cannot be available for horizontal transfer. As expected for a functional suicide switch, induction of nuclease expression in KSPcopM195-BCD-nucA cells resulted in efficient programmed autokilling. This is the first report of biocontainment implementation in Synechocystis. Being a standalone circuit, it remains to be tested if this biosafety device may be transferrable to other cyanobacteria. This system can serve as a starting point for further cyanobacterial biosafety research. The biocontainment safeguard was developed under constant laboratory growth conditions. For biotechnological applications, it will need further assessment under different conditions, for example fluctuating temperatures and light intensities. Additionally, local environmental inducer levels will have to be determined.
It should be noted that inducible lysis in Synechocystis has been carried out before; for the purpose of metabolite extraction, lysis has been triggered by bacteriophage-derived lysis genes (Liu and Curtiss, 2009; Miyake et al., 2014). However, for metabolite recovery, complete lysis-induced killing is not necessary, while it is required for biosafety purposes. Thus, for recovery of biofuel-related compounds from Synechocystis, partial cell lysis was observed with green-light inducible lytic system, with about 40% cell death recorded (Miyake et al., 2014). In another metabolite-recovery lysis system (Liu and Curtiss, 2009), the use of nrsB promoter caused efficient killing in Synechocystis. Nonetheless, the two most efficient strains with bacteriophage lysis genes showed substantial growth retardation, and the cells of these strains adhered to each other and to the container walls in absence of induction, indicating probable cell wall damage caused by likely leakage of lysis enzymes (Liu and Curtiss, 2009). These traits can be disadvantageous in biotechnological applications. The benefit of using a nuclease as the toxic protein in biosafety devices is avoiding such problems of adherence in absence of inducer.
To construct a cyanobacterial biosafety device, we tested different promoters (seeking good promoter inducibility and low promoter leakiness) and translation initiation elements. The efficient KSPcopM195-BCD-nucA suicide switch was regulated by PcopM195-BCD, a variant of the metal-ion responsive promoter copM. PcopM195-BCD in the kill switch showed good Zn2+-responsiveness. Additionally, putting a bicistronic translation initiation element BCD in place of the native RBS in the region preceding the copM start codon appears to have contributed to the killing efficiency of the switch. Nonetheless, the PcopM195-BCD promoter displayed some leakiness, as determined by nucA transcript levels and a decrease in growth rate of uninduced kill switch cells in comparison to WT. This decrease suggested the expression level of the inhibitor in the kill switch may have been insufficient to completely neutralize the nuclease in absence of induction. Leakage can be a problem in suicide biosafety systems, as it causes reduced growth rate due to basal expression of the toxic gene; this can give selective growth advantage to cells with mutated suicide functions (Knudsen and Karlstrom, 1991). In agreement with this, sporadic biological replicates of kill switch KSPcopM195-BCD-nucA started growing in the presence of inducer. In addition to promoter leakiness and induction capability, the practical usefulness of kill switches can be influenced by the ability of the inducer to reach all cells and activate gene expression. At higher cell densities, reaching all cells may be a problem that can result in inconsistent induction. Furthermore, less inducer may be available per cell. Also possible in denser cultures are cell aggregation and biofilm formation that can pose a further challenge. Additionally, it has been reported that with varying cell density, promoter tightness can vary (described for a Plac promoter variant used in Synechocystis) (Guerrero et al., 2012). In denser cultures we have observed some variation in response of WT cells to the presence of metal ions, and some inconsistency in kill switch response to inducer. In biotechnological applications, all these limitations will need to be taken into consideration. Taking everything into account, we can envision that kill switch KSPcopM195-BCD-nucA is suitable to achieve a containment level where controllable reduction of genetically modified bacteria and decreased quantities of recombinant genetic material are desired, but complete elimination is not a requirement. When testing for a reduction or elimination of bacteria, it should be noted that available methods for monitoring cell survival cannot prove complete eradication of engineered strains, as there is always a possibility that the cells could be surviving in a dormant state.
Currently, cyanobacterial toolbox is lacking in really tight promoters with very high inducibility. Actually, cyanobacterial metal-ion responsive promoters are among the best available, even though their usefulness in biological safeguards is not ideal. Further work on cyanobacterial genetic elements will be required to improve our understanding of cyanobacterial promoters. We have contributed to the cyanobacterial toolbox by analyzing PnrsB-RBS (a nrsB promoter variant) with beta-galactosidase assay. Interestingly, unlike the native PnrsB, this variant exhibits better responsiveness to Co2+ than Ni2+ ions.
In this work, two toxin/antitoxin systems were tested in kill switch constructs with metal-ion inducible promoter PcopB. In both cases, elevated levels of RelE-like toxins caused reduced growth rather than efficient killing of cells. Bacteriostatic rather than bactericidal effect of TA systems has been reported, and has been overcome in other suicide systems (e.g. E. coli) by high toxin expression (Knudsen et al., 1995; Kristoffersen et al., 2000; Wright et al., 2013). In our biocontainment system, bacteriostatic effect was likely the result of insufficient toxin levels, coupled with the presence of endogenous antitoxins that could neutralize toxin activity. Our results are in agreement with those of Cheah et al. (2013), who used the metal-ion responsive promoter PnrsB for induction of E. coli-MazF toxin in Synechocystis to serve as a counter-selection marker for genetic modification. Despite high inducibility of metal-ion inducible promoters, growth arrest rather than killing was observed by Ni2+-induction of mazF in their experiments, similar to what we have observed with RelE-like toxins and PcopB.
MATERIALS AND METHODS
Strains and constructs
Cyanobacterium Synechocystis sp. PCC 6803 (WT) was used in this work. Suicide switch constructs and lacZ-fusions were constructed in plasmid pPMQAK1 (Huang et al., 2010). Cloning was carried out in Escherichia coli DH5α, and sequence-verified constructs were transferred to Synechocystis. Table S1 describes the parts used in suicide switch constructs and lacZ-fusions, the templates used for amplification, and the primers with restriction sites.
To generate suicide switch constructs, a double transcription terminator (biobrick BBa_B0015) was first inserted into the EcoRI site of pPMQAK1, introducing a NheI site. Kill switch constructs were assembled separately and then inserted between the PstI and the new NheI site. To assemble the kill switch constructs, individual parts of the switch were first amplified by PCR (genes, promoters, RBSs, transcription terminators) or synthesized (BCD bicistronic design). Primers from Sigma-Aldrich (St. Louis, MO, USA) and Platinum Pfx DNA polymerase (Fisher Scientific, Pittsburgh, PA, USA) were used for PCR. Fragment synthesis was done by GeneArt Gene Synthesis (Thermo Scientific). Next, the parts were digested with restriction nucleases (Thermo Scientific), and two parts at the time were ligated with T4 DNA ligase (Thermo Scientific). The ligated parts were amplified by PCR using outer primers. Several steps of restriction, ligation and PCR were needed to assemble all parts and amplify an entire kill switch. The final kill switch composition included an additional double transcription terminator BBa_B0015 between the SpeI and PstI sites. The last PCR product, which amplified the entire kill switch, was digested with PstI and NheI and ligated into the vector. Empty vector control plasmid (EV) was obtained by deletion of the ccdB-containing fragment (XbaI-SpeI) from pPMQAK1.
To generate lacZ-fusions, the lacZ sequence was combined with different promoters (PnrsB-RBS, PrbcL1A and PrnpB-RBS1) by using a BamHI restriction site. A double transcription terminator (biobrick BBa_B0015) was placed downstream of lacZ by using a SpeI site. The final lacZ composition was inserted into pPMQAK1 between XbaI and PstI.
Growth rate measurements and kill switch induction
Wild-type Synechocystis sp. PCC 6803 was grown in BG11 medium (Rippka, 1988), and strains carrying constructs in plasmid pPMQAK1 were grown in BG11 supplemented with antibiotics. Cultures were grown at 22°C under continuous light (∼28 µmol photons/m2/s) in flasks (gentle shaking) or in test tubes that were vortexed daily to mix bacterial suspensions. Growth was monitored by measuring optical density OD730 with Varian Cary 50 spectrophotometer (Agilent Technologies, Santa Clara, CA, USA).
For kill switch induction experiments, log-phase cultures (OD730∼0.3) were used. At least three biological replicates were performed for individual kill switch constructs. Log-phase cultures (5 ml), diluted to OD730∼0.03, were induced with ZnCl2 (12 μM final concentration), CuSO4 (2 μM final concentration) or were left untreated (uninduced controls), and growth was followed by measuring OD730. For kill switch induction in Fig. 4, BG11 plates with or without added 4 μM ZnCl2 were inoculated with 10-μl bacterial suspensions, and colonies were evaluated under a stereomicroscope with 25× magnification (Zeiss 476100-9901, Oberkochen, Germany).
Some kill switch inductions were carried out in Nunc 96-well plates (Thermo Scientific). Log-phase bacterial cultures (OD730∼0.3) growing in BG11 were used to inoculate plates. To start the growth experiment, log-phase cultures were inoculated in triplicates into wells, and inducer (for treated cells) or BG11 (for untreated cells) was added, resulting in wells containing 200 μl of bacterial suspension at cell density OD730∼0.03 [the inducers were prepared in BG11 medium and were added to bacteria to final concentrations of 10 or 12 μM (for ZnCl2) or 4 μM (for Co(NO3)2)]. Plates were incubated with gentle shaking under standard growth conditions (light, temperature), and samples were mixed daily by pipetting. Optical density (OD600) was followed by Sunrise™ absorbance plate reader (Tecan, Männedorf, Switzerland). On the last day of the experiment, optical density OD730 of KSPcopM195-BCD-nucA was measured by Varian Cary 50 spectrophotometer (Agilent Technologies), and 10-μl culture aliquots were plated on solid BG11 media.
MTT cell viability assay
MTT analysis is used to assess cell viability by measuring reduction of yellow-colored MTT [3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide] to purple formazan by active reductases in live cells (Li and Song, 2007). The level of reduction, which correlates with the number of living cells, can be quantified in 96-well plates by an absorbance plate reader. Kill switch cultures, wild-type cells and empty vector control cells were grown under standard growth conditions to log phase (OD730∼0.3). Cells were pelleted, washed 3 times with sterile dH2O, and final pellets were resuspended in BG11 medium. Aliquots of 100 μl (106 cells) were pipetted into individual wells of a 96-well plate (BRANDplates®, BrandTech Scientific, Essex, CT, USA). To these cells, the following was added: ZnCl2 (to 10 or 14 μM final concentration), NiSO4 (to 4 μM final concentration), Co(NO3)2 (to 4 μM final concentration) or H2O (uninduced control samples). Treatments were carried out in triplicates. For background calculation, wells containing BG11 without bacterial cells were used. Multiwell plates containing treated cells were incubated with gentle shaking under standard growth conditions for 24 h. At that time, 20 μl of filter-sterilized MTT solution [5 mg/ml MTT in PBS (pH 6.9)] (Sigma-Aldrich) was added to each well, after which the plate was incubated in the dark at 37°C. After 3-4 h, 100 μl MTT solvent solution (1 g SDS and 8.3 μl HCl in 10 ml H2O) was added to each well, the plate was covered with tinfoil and incubated overnight at 28°C with gentle shaking. The next day, absorbance was read at 560 nm with a reference filter of 620 nm by Sunrise™ absorbance plate reader (Tecan). Three biological replicates were performed for each suicide switch construct. For analyzing cell viability 14 days postinduction, cells were grown and induced in test tubes (4-ml cultures) instead of multiwell plates to avoid the problem of drying. Log-phase cultures were inoculated into fresh BG11 medium in triplicates at the initial concentration OD730∼0.03. After being treated with ZnCl2 (10 μM final concentration) or H2O (uninduced control), cells were left to grow under standard conditions for two weeks. On day 14, 50-μl aliquots were taken from every culture (50 μl of wild-type culture contained ∼4×106 cells), diluted with BG11 to 100 μl, and pipetted into 96-well plates for MTT analysis. MTT assay was then carried out as described above.
Statistical significance between the control and the kill switch cells in MTT assays was calculated using Student's two-tailed t-test.
RNA isolation and RT-PCR analysis
Exponentially growing cyanobacterial cultures (60 ml, OD730=0.3) were used for RT-PCR analysis. Aliquots of 30 ml were induced with 10 μM ZnCl2 (cultures KSPcopM72-nucA, KSPcopM195-BCD-nucA, KSPcopB-tox0664) or 4 μM NiSO4 (culture KSPnrsBRBS-nucA), and the remaining 30 ml were left untreated. Wild-type cultures served as controls for absence of nucA/nuiA expression and for basal slr0664 expression. Following a 1-h induction, the cultures were rapidly chilled, pelleted (4500 g, 10 min, 4 °C) and the cells were broken with 150-212 µm glass beads (Sigma-Aldrich). Total RNA was isolated with RNeasy Mini Kit (Qiagen, Hilden, Germany) as per manufacturer's instructions. Following removal of genomic DNA by DNase I (1 U per 1 μg RNA) (Thermo Scientific), 20 µl-reverse transcription reactions were carried out with 0.5 µg RNA (0.14 µg in case of KSPcopB-tox0664) by using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) per manufacturer's instructions. Reactions containing cDNA were diluted 1:1 with dH2O and used for PCR with DreamTaq DNA polymerase (Thermo Scientific) following manufacturer's protocol (1.5 µl cDNA was used as template for KSPcopM72-nucA and KSPcopM195-BCD-nucA; 2.5 µl for KSPcopB-tox0664; and 3 µl for KSPnrsBRBS-nucA). Control reverse transcription reactions without added reverse transcriptase were used for PCR-verification of successful (prior) removal of genomic DNA, and PCR reactions without template were used to verify absence of reagent contamination. PCR primers were: for nucA (5′-CCATCAATCAGCGTGCATTTAC-3′ and 5′-TGTTGTTGTAGGAGAGTGCATATT-3′); for nuiA (5′-GAGTGAGTCTGAATACCCATTTGA-3′ and 5′-TCTTGTCCGTGACCTGTTTG-3′); for slr0664 toxin (5′-GCAAAAAGGCGTCATTAGT-3′ and 5′-AATCGGTAGCCAGAACTTT-3′); for control housekeeping gene rnpB (Pinto et al., 2012) (5′-CGTTAGGATAGTGCCACAG-3′ and 5′-CGCTCTTACCGCACCTTTG-3′).
Beta-galactosidase assay
To measure promoter activity in Synechocystis, we made modifications to a previously described beta-galactosidase protocol (Miller, 1972). Log-phase cultures harboring reporter constructs (10 ml) were treated with NiSO4 (4, 10, 16 or 27 μM final concentration), Co(NO3)2 (8 μM final concentration), ZnCl2 (8 μM final concentration) or H2O (uninduced control samples) for 18 h and harvested by centrifugation (12,500 g, 4°C, 10 min). Pellets were resuspended in 1 ml of ice cold buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7), which was freshly supplemented with 2-mercaptoethanol (50 mM final concentration) and EDTA-free Pierce protease inhibitors (1× final concentration) (Fisher Scientific). Chloroform (100 μl) was added to the samples (use of SDS was avoided due to interference with subsequent Bradford assay). About 300 μl of 150-212 µm glass beads (Sigma-Aldrich) were added and cells were broken by vortexing (10 times for 30 s, with 1-min incubations on ice in-between). After centrifugation (10 min, 4°C, maximal speed), about 1 ml of aqueous phase was collected. Two 200-μl aliquots were used to determine total protein concentration by Bradford assay and the rest of the aqueous phase was used to measure beta-galactosidase activity. Triplicate cell aliquots (from 2-200 μl, depending on expected strength of promoter response) were diluted with buffer Z to 400 μl and incubated at 28°C for 2 min. Initial reaction time was recorded when 72 μl of ONPG [o-nitrophenyl-β-D-galactoside; 4 mg/ml solution in phosphate buffer (60 mM Na2HPO4, 40 mM NaH2PO4, pH 7)] was added to each of the three samples. Reaction in one sample (blank sample) was immediately terminated by addition of 176 μl of 1 M Na2CO3. The two other samples were terminated simultaneously when they developed sufficient yellow color, and termination time was recorded. After centrifugation (5 min, maximal speed), samples (500 μl) were diluted with Milli-Q water (500 μl), transferred to UV-Vis cuvette, and absorbance at 420 and 550 nm (A420 and A550) was measured against the blank sample by Varian Cary 50 spectrophotometer (Agilent Technologies). Promoter activity was calculated from the modified equation, which gives standardized Miller Units, (s)MU:
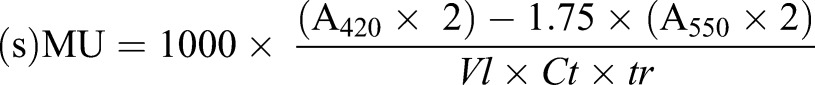 |
where Vl is the volume of cell lysate used (ml), Ct is the total protein concentration (μg/ml) and tr is the reaction time (min). Absolute unit of (s)MU is given as μmol of ONP (o-nitrophenol) product per min per μg of total protein. Three biological replicates were performed for each reporter construct.
Acknowledgements
This work was performed within the European consortium CyanoFactory funded by the European Commission (FP7). We would like to thank Peter Lindblad from Uppsala University for kindly providing us with the plasmids pPMQAK1 and pPMQAK1-Ptrc1-GFP. We are thankful to Enrique Flores from University of Seville, Centro de Investigaciones Científicas, for sending us the strain Anabaena sp. PCC 7120.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Growth and kill-induction experiments were performed by H.Č., A.T. and A.T. MTT analyses were carried out by A.T. and A.T. Reporter assays were done by J.M. RNA analyses were performed by A.V., V.V. and H.Č. Cloning was done by H. Č., J.M. and V.V. H.Č. wrote the manuscript. H. Č., M.D., A.T., A.T., A.V., J.M., V.V. contributed to designing experiments and presenting the data.
Funding
This project has received funding from the European Union Seventh Framework Programme for research, technological development and demonstration under grant agreement [No 308518, CyanoFactory].
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/suppl/doi:10.1242/bio.017129/-/DC1
References
- Ahrenholtz I., Lorenz M. G. and Wackernagel W. (1994). A conditional suicide system in Escherichia coli based on the intracellular degradation of DNA. Appl. Environ. Microbiol. 60, 3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan A. and Schenberg A. C. G. (2005). A conditional suicide system for Saccharomyces cerevisiae relying on the intracellular production of the Serratia marcescens nuclease. Yeast 22, 203-212. 10.1002/yea.1203 [DOI] [PubMed] [Google Scholar]
- Bej A. K., Perlin M. H. and Atlas R. M. (1988). Model suicide vector for containment of genetically engineered microorganisms. Appl. Environ. Microbiol. 54, 2472-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi B., Peca L., Vass I. and Kos P. B. (2012). Characterization of stress responses of heavy metal and metalloid inducible promoters in synechocystis PCC6803. J. Microbiol. Biotechnol. 22, 166-169. 10.4014/jmb.1106.06050 [DOI] [PubMed] [Google Scholar]
- Briggs L. M., Pecoraro V. L. and McIntosh L. (1990). Copper-induced expression, cloning, and regulatory studies of the plastocyanin gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 15, 633-642. 10.1007/BF00017837 [DOI] [PubMed] [Google Scholar]
- Cheah Y. E., Albers S. C. and Peebles C. A. M. (2013). A novel counter-selection method for markerless genetic modification in Synechocystis sp. PCC 6803. Biotechnol. Prog. 29, 23-30. 10.1002/btpr.1661 [DOI] [PubMed] [Google Scholar]
- Garcia-Dominguez M., Lopez-Maury L., Florencio F. J. and Reyes J. C. (2000). A gene cluster involved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182, 1507-1514. 10.1128/JB.182.6.1507-1514.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giner-Lamia J., Lopez-Maury L., Reyes J. C. and Florencio F. J. (2012). The CopRS two-component system is responsible for resistance to copper in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 159, 1806-1818. 10.1104/pp.112.200659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giner-Lamia J., Lopez-Maury L. and Florencio F. J. (2015). CopM is a novel copper-binding protein involved in copper resistance in Synechocystis sp. PCC 6803. Microbiol. Open 4, 167-185. 10.1002/mbo3.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero F., Carbonell V., Cossu M., Correddu D. and Jones P. R. (2012). Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803. PLoS ONE 7, e50470 10.1371/journal.pone.0050470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn T., Camsund D., Huang H.-H., Lindberg P., Oliveira P., Stensjo K. and Lindblad P. (2011). Synthetic biology in cyanobacteria engineering and analyzing novel functions. Methods Enzymol. 497, 539-579. 10.1016/B978-0-12-385075-1.00024-X [DOI] [PubMed] [Google Scholar]
- Huang H. H., Camsund D., Lindblad P. and Heidorn T. (2010). Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology . Nucleic Acids Res. 38, 2577-2593. 10.1093/nar/gkq164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. B., Ramos J. L., Kaneva Z. and Molin S. (1993). A substrate-dependent biological containment system for Pseudomonas putida based on the Escherichia coli gef gene. Appl. Environ. Microbiol. 59, 3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S. M. and Karlstrom O. H. (1991). Development of efficient suicide mechanisms for biological containment of bacteria. Appl. Environ. Microbiol. 57, 85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S., Saadbye P., Hansen L. H., Collier A., Jacobsen B. L., Schlundt J. and Karlstrom O. H. (1995). Development and testing of improved suicide functions for biological containment of bacteria. Appl. Environ. Microbiol. 61, 985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen P., Jensen G. B., Gerdes K. and Piskur J. (2000). Bacterial toxin-antitoxin gene system as containment control in yeast cells. Appl. Environ. Microbiol. 66, 5524-5526. 10.1128/AEM.66.12.5524-5526.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchmina E., Wallner T., Kryazhov S., Zinchenko V. V. and Wilde A. (2012). An expression system for regulated protein production in Synechocystis sp. PCC 6803 and its application for construction of a conditional knockout of the ferrochelatase enzyme. J. Biotechnol. 162, 75-80. 10.1016/j.jbiotec.2012.06.016 [DOI] [PubMed] [Google Scholar]
- Lai M. C. and Lan E. I. (2015). Advances in metabolic engineering of cyanobacteria for photosynthetic biochemical production. Metabolites 5, 636-658. 10.3390/metabo5040636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry B. P., Stockel J. and Pakrasi H. B. (2013). Use of degradation tags to control protein levels in the Cyanobacterium Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 79, 2833-2835. 10.1128/AEM.03741-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.-S., Matsui M. and Abdullah A. A.-A. (2015). Cyanobacteria: photoautotrophic microbial factories for the sustainable synthesis of industrial products. BioMed Res. Int. 2015, 754934 10.1155/2015/754934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. and Song L. (2007). Applicability of the MTT assay for measuring viability of cyanobacteria and algae, specifically for Microcystis aeruginosa (Chroococcales, Cyanobacteria). Phycologia 46, 593-599. 10.2216/07-11.1 [DOI] [Google Scholar]
- Li P., Harding S. E. and Liu Z. (2001). Cyanobacterial exopolysaccharides: their nature and potential biotechnological applications. Biotechnol. Genet. Eng. Rev. 18, 375-404. 10.1080/02648725.2001.10648020 [DOI] [PubMed] [Google Scholar]
- Liu X. and Curtiss R. III (2009). Nickel-inducible lysis system in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 106, 21550-21554. 10.1073/pnas.0911953106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Maury L., Garcia-Dominguez M., Florencio F. J. and Reyes J. C. (2002). A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 43, 247-256. 10.1046/j.1365-2958.2002.02741.x [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I. and Koonin E. V. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4, 19 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiss G., Franke I., Gimadutdinow O., Urbanke C. and Pingoud A. (1998). Biochemical characterization of Anabaena sp. strain PCC 7120 non-specific nuclease NucA and its inhibitor NuiA. Eur. J. Biochem. 251, 924-934. 10.1046/j.1432-1327.1998.2510924.x [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Miyake K., Abe K., Ferri S., Nakajima M., Nakamura M., Yoshida W., Kojima K., Ikebukuro K. and Sode K. (2014). A green-light inducible lytic system for cyanobacterial cells. Biotechnol. Biofuels 7, 56 10.1186/1754-6834-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe-Behrens G. H. G., Davis R. and Haynes K. A. (2013). Preparing synthetic biology for the world. Front. Microbiol. 4, 5 10.3389/fmicb.2013.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Asami K., Tanji Y. and Unno H. (2001). Programmed Escherichia coli cell lysis by expression of cloned T4 phage lysis genes. Biotechnol. Prog. 17, 573-576. 10.1021/bp010018t [DOI] [PubMed] [Google Scholar]
- Munthali M. T., Timmis K. N. and Diaz E. (1996). Use of colicin e3 for biological containment of microorganisms. Appl. Environ. Microbiol. 62, 1805-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor A. M., Flores E., Herrero A. and Wolk C. P. (1992). Identification, genetic analysis and characterization of a sugar-non-specific nuclease from the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 6, 3021-3030. 10.1111/j.1365-2958.1992.tb01760.x [DOI] [PubMed] [Google Scholar]
- Mutalik V. K., Guimaraes J. C., Cambray G., Lam C., Christoffersen M. J., Mai Q.-A., Tran A. B., Paull M., Keasling J. D., Arkin A. P. et al. (2013). Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods 10, 354-360. 10.1038/nmeth.2404 [DOI] [PubMed] [Google Scholar]
- Ning D., Ye S., Liu B. and Chang J. (2011). The proteolytic activation of the relNEs (ssr1114/slr0664) toxin-antitoxin system by both proteases Lons and ClpP2s/Xs of Synechocystis sp. PCC 6803. Curr. Microbiol. 63, 496-502. 10.1007/s00284-011-0011-5 [DOI] [PubMed] [Google Scholar]
- Otsuka Y. (2016). Prokaryotic toxin-antitoxin systems: novel regulations of the toxins. Curr. Genet. [Epub ahead of print]. 10.1007/s00294-015-0557-z. [DOI] [PubMed] [Google Scholar]
- Peca L., Kos P. and Vass I. (2007). Characterization of the activity of heavy metal-responsive promoters in the cyanobacterium Synechocystis PCC 6803. Acta Biol. Hungarica 58 Suppl., 11-22. 10.1556/ABiol.58.2007.Suppl.2 [DOI] [PubMed] [Google Scholar]
- Peca L., Kos P. B., Mate Z., Farsang A. and Vass I. (2008). Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS Microbiol. Lett. 289, 258-264. 10.1111/j.1574-6968.2008.01393.x [DOI] [PubMed] [Google Scholar]
- Pereira S., Micheletti E., Zille A., Santos A., Moradas-Ferreira P., Tamagnini P. and De Philippis R. (2011). Using extracellular polymeric substances (EPS)-producing cyanobacteria for the bioremediation of heavy metals: do cations compete for the EPS functional groups and also accumulate inside the cell? Microbiology 157, 451-458. 10.1099/mic.0.041038-0 [DOI] [PubMed] [Google Scholar]
- Pinto F., Pacheco C. C., Ferreira D., Moradas-Ferreira P. and Tamagnini P. (2012). Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS ONE 7, e34983 10.1371/journal.pone.0034983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R. (1988). Isolation and purification of cyanobacteria. Methods Enzymol. 167, 3-27. 10.1016/0076-6879(88)67004-2 [DOI] [PubMed] [Google Scholar]
- Schweder T., Schmidt I., Herrmann H., Neubauer P., Hecker M. and Hofmann K. (1992). An expression vector system providing plasmid stability and conditional suicide of plasmid-containing cells. Appl. Microbiol. Biotechnol. 38, 91-93. 10.1007/BF00169425 [DOI] [PubMed] [Google Scholar]
- Smith A. B., Lopez-Villarejo J., Diago-Navarro E., Mitchenall L. A., Barendregt A., Heck A. J., Lemonnier M., Maxwell A. and Diaz-Orejas R. (2012). A common origin for the bacterial toxin-antitoxin systems parD and ccd, suggested by analyses of toxin/target and toxin/antitoxin interactions. PLoS ONE 7, e46499 10.1371/journal.pone.0046499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres B., Jaenecke S., Timmis K. N., Garcia J. L. and Diaz E. (2003). A dual lethal system to enhance containment of recombinant micro-organisms. Microbiology 149, 3595-3601. 10.1099/mic.0.26618-0 [DOI] [PubMed] [Google Scholar]
- Turner J. S., Glands P. D., Samson A. C. R. and Robinson N. J. (1996). Zn2+-sensing by the cyanobacterial metallothionein repressor SmtB: different motifs mediate metal-induced protein-DNA dissociation. Nucleic Acids Res. 24, 3714-3721. 10.1093/nar/24.19.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque A. (2007). Transformation of cyanobacteria. Adv. Exp. Med. Biol. 616, 12-22. 10.1007/978-0-387-75532-8_2 [DOI] [PubMed] [Google Scholar]
- Wright O., Stan G.-B. and Ellis T. (2013). Building-in biosafety for synthetic biology. Microbiology 159, 1221-1235. 10.1099/mic.0.066308-0 [DOI] [PubMed] [Google Scholar]
- Wright O., Delmans M., Stan G.-B. and Ellis T. (2015). GeneGuard: a modular plasmid system designed for biosafety. ACS Synth. Biol. 4, 307-316. 10.1021/sb500234s [DOI] [PubMed] [Google Scholar]
- Ye S. and Ning D. (2010). [The interaction between chromosome-encoded toxin Slr0664 and antitoxin Slr1114 of cyanobacteria Synechocystis sp. PCC6803]. Wei Sheng Wu Xue Bao 50, 743-748. [PubMed] [Google Scholar]