Abstract
Detailed population-level description of the human immune system has recently become achievable. We used a “systems-level” approach to establish a resource of cellular immune profiles of 670 healthy individuals. We report a high level of inter-individual variation, with low longitudinal variation, at the level of cellular subset composition of the immune system. Despite the profound effects of antigen exposure on individual antigen-specific clones, the cellular subset structure proved highly elastic, with transient vaccination-induced changes being followed by a return to the unique baseline of the individual. Strikingly, the largest influence on immunological variation identified was cohabitation, with a 50% reduction in immunological variation between individuals who share an environment (parents) compared to the wider population. These results identify local environmental conditions are a key shaper of the human immune system.
Introduction
Enormous progress has been made in understanding the cellular and molecular components of the immune system. The key tool in this progression has been the use of mouse models, especially genetically modified models that have allowed the functional dissection of the myriad of interconnecting components. Despite this progress in mouse models, or, perhaps because of it, it has been convincingly argued that the focus of immunology should return to the human context1, including a comprehensive analysis of the full spectrum of immunological diversity, and the causes thereof.
Several recent studies have embraced this call for investigating human immunological diversity, identifying genetic factors as accounting for ~25-50% of measured immunological variation 2, 3, 4. A recent twin-study indicated that at least half of the immune trait variance is explained by non-genetic factors 3. Similarly the mean heritability of immune traits reported by a quantitative trail locus (QTL) study in healthy Sardinians was 41% 2. The ImmVar project, which tested for gene expression QTLs in circulating human immune cells, estimated that approximately 22% of the variance in gene expression is explained by genetic factors 4. Together these studies suggest that the immunoprofile of the healthy population is governed in a large part by non-genetic factors. As these non-genetic factors predominate, and arguably are more amenable to clinical manipulation, it is critical for us to identify and quantify the key factors that shape the human immune landscape.
Using hypothesis-based approaches, several non-genetic factors have already been identified which influence the landscape of the human immune system. Chronic infections, in particular latent herpesvirus infection, are associated with a panoply of immunological changes and discordance for CMV seropositivity in monozygotic twin pairs resulted in weaker pairwise correlations for many immune parameters 3. Another important non-genetic impact on the immune system is ageing, with potent effects on both the innate and adaptive arms of the immune response 5. Within the adaptive system, ageing is associated with a decline in naïve T cells 6. In mice, this is due to thymic involution, however, in humans, loss of naïve CD4+ T cells is primarily driven by a failure of peripheral replication of naïve cells 7, 8, again demonstrating the importance of direct assessment of human immunology rather than relying entirely on the mouse (and, indeed, far too often on a single inbred strain).
In the present study, we profiled the immune system of 670 healthy human volunteers, aged 2 to 86 years old, to provide a description of the population-level heterogeneity present in the cellular composition of the circulating immune system. Through the targeted recruitment of sub-cohorts with longitudinal sampling before and after severe immunological challenge, we were able to determine that the immunological diversity between individuals is highly robust, with an elastic return to the unique steady state of the individual following immunological challenge. We report that co-parenting profoundly reduces the immunological variation between two individuals, suggesting that environmental influences can drive convergence as well as diversity within our immunological profile.
Results
Elasticity of the human cellular immune system
To investigate diversity in the composition of the human immune system, we developed an immune phenotyping platform quantifying 54 distinct immunological parameters by flow cytometry and serum analysis, with a focus on cellular subsets within the adaptive immune system. Following optimization, we recruited 638 healthy Belgian individuals for immune profiling, ranging from 2 to 86 years of age and free from self-reported gastrointestinal, autoimmune or inflammatory disease (Supplementary Table 1). Of these, 140 individuals were recruited as 70 pairs (co-parents of children). 177 individuals were sampled at multiple time-points (average of 6 months between sampling) to allow the measurement of longitudinal variation. Within this longitudinal cohort we targeted individuals who were planning to travel with a high risk of developing gastroenteritis and obtained before and after travel samples for 50 individuals. In total, 921 samples from 638 individuals were assessed over a period of 3 years.
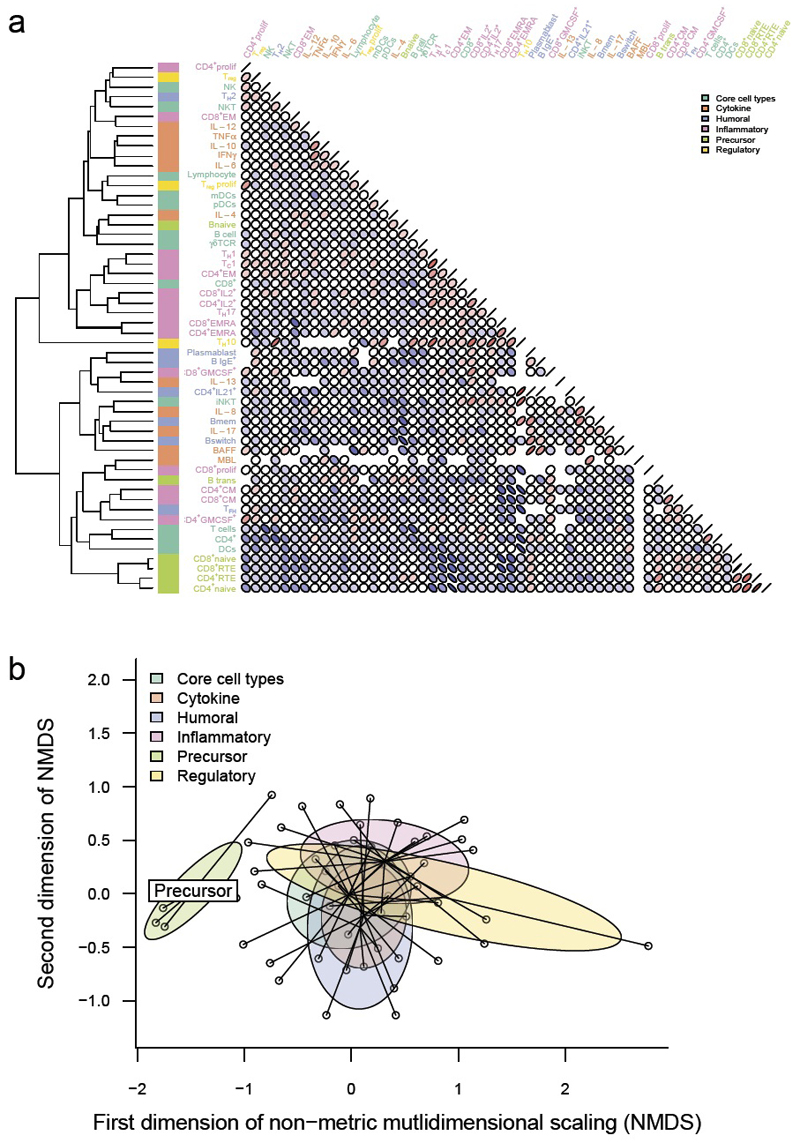
As an exploratory analysis of the dataset, we examined the degree and structure of the variation. Substantial variation was observed in all of the immunological parameters measured (Supplementary Table 2). To determine whether there were underlying patterns within the variation, we performed unsupervised hierarchical clustering (Fig. 1a). The strongest clustering was observed between parameters manually annotated as “precursor” cell types, with recent thymic emigrant (RTE) CD4 T cells, RTE CD8 T cells, naïve CD4 T cells and naïve CD8 T cells forming a single cluster (Fig. 1). This cluster was robust, being identified through iterative re-clustering (Supplementary Fig. 1). Parameters within the other manually annotated groups (humoral, inflammatory, regulatory, core cell types, cytokines) were distributed throughout the hierarchy. Using multi-dimensional scaling (MDS), we identified, in a data-driven manner, co-correlations between precursor populations (Fig. 1b). As precursor parameters separated from the rest of the parameters along the first dimension, this represents the largest source of variability between immune parameters, suggesting more co-ordinated biological control of these particular subsets. These data demonstrate that there is a high degree of variation in the immunological profiles of healthy individuals, with the largest component of the variation being a co-regulated change in the frequency of naïve or precursor cell types. Activated cell types and products, by contrast, demonstrated only minor co-regulation (with several biologically-relevant exceptions, such as between TH1 and TC1 cells).
Figure 1.
Data-driven analysis of immunological variation reveals biologically meaningful co-correlations between individual immune parameters. (a) On the complete dataset of 638 individuals (most recent sample only), a dendrogram of immune parameters was generated by hierarchical clustering on Euclidean distances of Spearman correlations between each parameter (left). Correlation plots using pairwise Spearman correlation coefficients between each two immunological parameters are shown (right). Coefficients are shown by the angle of eclipse (left-leaning, negative; right-leaning, positive) and colour (blue, negative; red, positive). Manually annotated thematic groups of immune parameters are shown by the colour bar next to the dendrogram. (b) Non-metric multidimensional scaling of pairwise Spearman correlations. The dataset is reduced from 54 immune parameters (54 dimensions) to a 2 dimensional representation, an exploratory approach to investigate the presence or absence of inter-relatedness between immune parameters. Each immune parameter is a point and the thematic groups are highlighted.
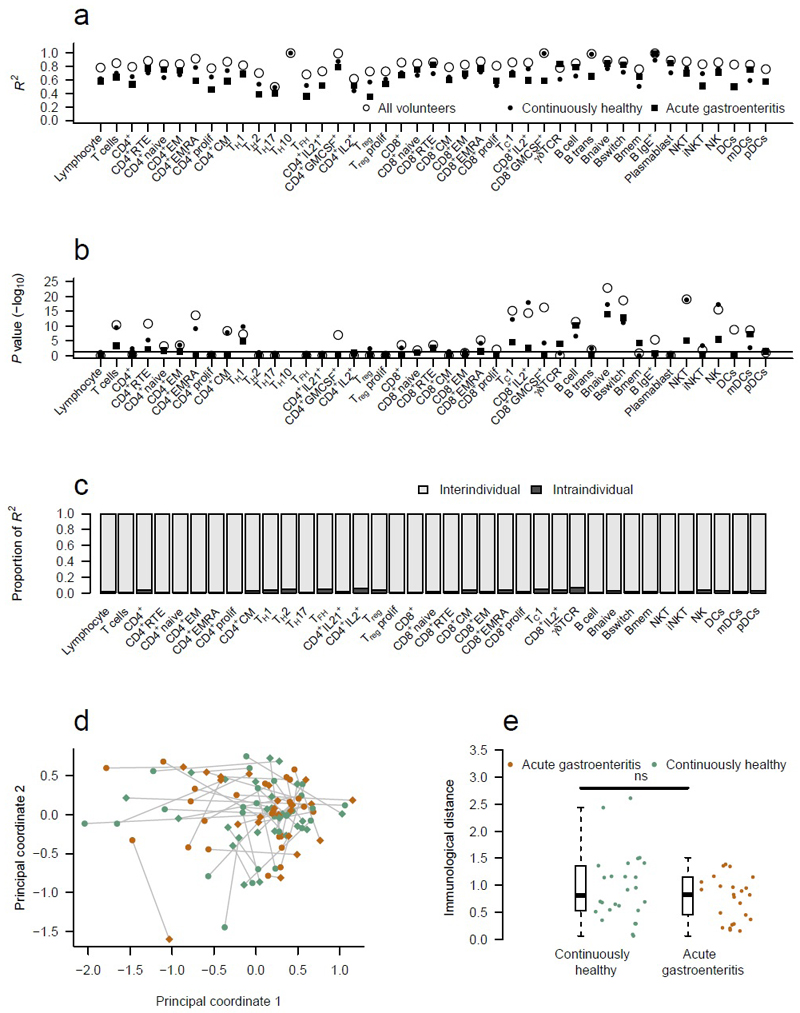
To determine whether immunological variation represented a dynamic process of change within individuals or a spectrum of different stable equilibria between individuals, we used the longitudinal sub-cohort of 177 individuals to ANOVA model with 2 independent variables – the volunteer’s unique identifier and the sample time-point (Fig. 2). Each immune parameter (Supplementary Table 2) was used as the response variable in these models (Fig. 2). The majority of the variation in each parameter was explained by a model of stable intra-individual immune profiles over longitudinal sampling (Fig. 2a), with a median R2 of 0.84 across the parameters (range, 0.5-1.0) and 60% of models maintaining statistical significance after correction for multiple testing (Fig. 2b). Variation between repeat samples of individuals, by contrast, contributed very little to the observed total variation, with a median proportion of R2 of 0.017 (range, 0.004-0.066), whereas intra-individual variation explained a much larger effect with a median proportion of R2 of 0.983 (range, 0.934-0.996) (Fig. 2c). Thus, of the total variation observed within our dataset, the majority (84%) can be explained by a model that includes both inter-individual and intra-individual variation, with just 1.4% attributable to the intra-individual variation between sample time-points. This stability in the cellular immune profile of individuals over an extended sampling period is consistent with that observed by other flow cytometry-based studies 2, 3, 9, and is consistent with a diverse set of stable equilibria observed between individuals.
Figure 2.
The human immune system is robustly maintained in multiple stable equilibriums. 177 individuals were sampled at least twice, allowing a dissection of inter- versus intra-individual variation. (a) Linear models were made for each immune parameter based on the multiple samples from each individual (ANOVA model: immune parameter ~ subject identifier + visit number). Open circles represent models built using all individuals (n=638) with multiple visits (177 individuals with up to 3 repeat visits; 921 visits in total). The filled circles represent models using only individuals who were continuously healthy between visits (152 individuals), filled squares represent models using only individuals who experienced acute gastroenteritis between their samples (24 individuals). For each cohort, R2 values and (b) -log10 of Bonferroni adjusted P values are shown for the linear models. (c) Proportion of the R2 values from all volunteers attributable to either inter-individual differences or intra-individual differences. (d) Multidimensional scaling of the pre- and post-travel study visits. Each individual is represented twice; their first and second visits depicted with a dot or diamond respectively and linked by a grey line indicating immunological distance (50 individuals; 100 visits). Continuously healthy individuals (n=26) are shown in aqua, individuals with intervening acute gastroenteritis (n=24) are shown in orange. (e) Quantification of the immunological distance between the first and second visits for continuously healthy individuals (n=26) versus individuals with intervening acute gastroenteritis (n=24). A two-tailed Mann-Whitney test was used to compare the immunological distances.
Having established that the relationship between immune subsets within an individual is highly stable over time, we sought to determine whether this stability was elastic or fragile. Within our dataset, we included a sub-cohort of fifty individuals who were sampled before and after travel to a developing nation, where there is an elevated risk of acquiring gastrointestinal infection. Of this sub-cohort, 24 individuals developed acute gastroenteritis whilst abroad and 26 did not (Supplementary Fig. 2a). Of these cases, 22 were classified as moderate or classic gastroenteritis with a median of 2 days of diarrhoea. Individuals were requested not to use antibiotics during this gastrointestinal challenge unless clinically indicated (87.5% did not use antibiotics), to allow a natural immune response to take course. Given the activation of the immune system during infection, as well as the importance of the gut microbiome 10, this experimental design allowed the determination of whether a combined immunological and microbiological disturbance would act as a “reset” on the immunological landscape, with individuals stabilising at an alternative equilibria point after the resolution of infection. To test this hypothesis, we repeated our earlier analysis of intra-individual variation by segregating the population into individuals that were continuously healthy during the sampling period and individuals that experienced acute gastrointestinal challenge between samples. No substantive effect was seen for gastrointestinal interlude on any single immunological parameter, with the modest changes in R2 (Fig. 2a) being driven by reduced numbers in subsets, as indicated by the accompanying reductions in P values (Fig. 2b). Notably TH17 cells, the cell type with the most compelling microbiome-interaction evidence 11, were unaffected in this analysis (Fig. 2a,b and Supplementary Fig. 3). Having failed to identify single immune parameters altered by immunological perturbation, we performed multi-dimensional scaling on paired samples to test whether severe gastrointestinal infection had an effect via the cumulative effect of minor changes on multiple parameters (Fig. 2d). In this analysis, multi-dimensions (representing each of 54 immune parameters) are reduced to 2 dimensions. Samples (either different visits or different individuals) that are closer together are more similar; those further apart are more dissimilar. The individuals affected by the diarrhoeal immunological insult did not separate from the rest of the data and the “immunological distance” between longitudinal samples was no greater than that of individuals who were continuously healthy (Fig. 2e). Even among the subset of patients with the longest duration of gastroenteritis (≥4 days) no increase in immunological distance was observed (Supplementary Fig. 2). Together our analyses demonstrate that not only can the human immune system exist in a diverse set of stable equilibria, but that these equilibria are maintained following immunological and microbiological disturbances, with each individual returning back to the original steady state following the resolution of infection. This result does not exclude functional or numerical changes within the response clones (expansion, conversion to a memory phenotype), but instead refers to the overall cell subset structure of the immune system. One possible explanation for this is that the number of antigen-specific memory cells remaining following immunological challenge are few in number 12, 13, 14, and the intrinsic biases in the individual that make up the prior immune status also apply to newly expanded clones. Thus, while individual responding T cell and B cell clones may change markedly as a response to activation, the functional landscape in which they assimilate remains intact.
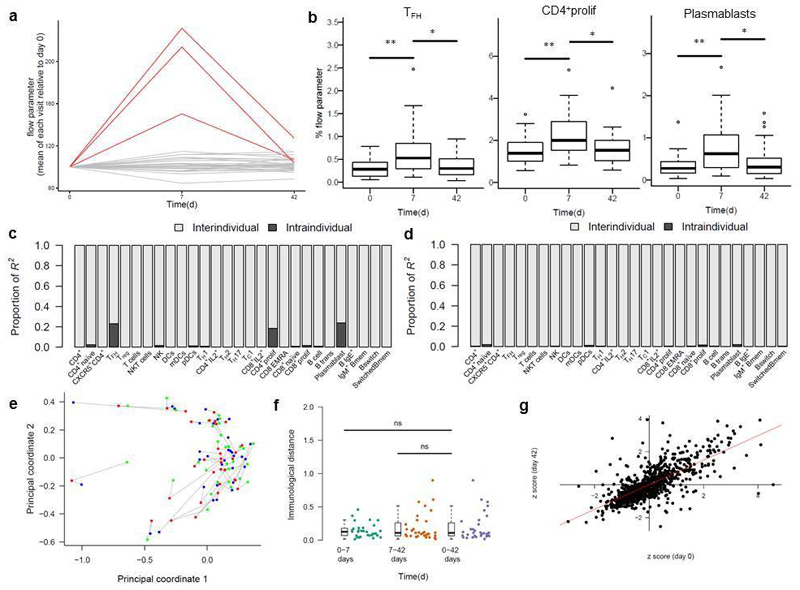
The real world context of gastrointestinal infection results in several study limitations, with the inability to take a peak-infection sample and variation in the sampling schedule and infection. To overcome these intrinsic caveats, we initiated an independent cohort to assess how a defined immunological stimulus, influenza vaccination, impacts on the immune landscape. 32 healthy English individuals, between 53 and 64 years of age, were recruited during the 2014-2015 winter influenza vaccination season. Volunteers were sampled prior to intramuscular vaccination with the standard seasonal inactivated influenza vaccine, and at 7 and 42 days post-vaccination. Samples were then phenotyped on a parallel immune phenotyping platform, which replicated the variation structure and substructure present in the Belgian cohort (Supplementary Fig. 4). Analysis of individual immune parameters indicated that most parameters remained unchanged throughout the study (Fig. 3a and Supplementary Fig. 5). The exceptions were circulating follicular T helper-like cells (cTFH), proliferating CD4+ T cells (Ki67+) and plasmablasts, all of which demonstrated marked increases at day 7, before returning to baseline at day 42 (Fig. 3b). To determine whether the immunological challenge of vaccination disrupted the immune landscape of the volunteers, we built a longitudinal ANOVA model. Between day 0 and day 7, almost the entire variation could be accounted for by inter-individual variation, with the exception of the three vaccination response parameters, where sample time-point (intra-individual variation)(Fig. 3c). With the resolution of the vaccine response (assessing day 0 and 42 time-points), even the vaccination response parameters showed no substantial time-point (intra-individual) variation (Fig. 3d), demonstrating that even perturbed parameters rebounded to pre-challenge settings. This is consistent with previous systems vaccinology papers that describe an alteration in the gene expression profile of peripheral blood samples in the first two-weeks following vaccination, followed by a return to baseline state 12, 15, 16. To demonstrate the global robustness of the immunological landscape, we used multidimensional scaling. This analysis indicated clustering of samples by individual rather than time-point (Fig. 3e), with low immunological distances between samples (Fig. 3f). To test whether the return to baseline was a population level process or whether each individual returned to their own unique baseline, we calculated a Z-score for each immunological parameter for each individual at day 0 and day 7. A strong correlation (R2 0.74, P<2x10-16) was observed (Fig. 3g), indicating that individuals regained their relative inter-individual differences after vaccination. In all, using an independent immunological challenge, these results show significant stability of human immune cell subsets, and indicate an elastic ability to respond to antigen before returning to a baseline state.
Figure 3.
Immunologial equilibria demonstrate elasticity following influenza vaccination. In a parallel cohort of 32 English individuals, volunteers were sampled prior to vaccination, with follow-up samples at day 7 and day 42 post-vaccination. (a) Samples were phenotyped, normalised to the day 0 value and assessed for change using paired t-tests. Unchanged variables are shown in grey, significantly modified variables are shown in red, with (b) boxplots for each significant immune parameter. Each parameter is labeled on the graph with an (uncorrected, two tailed) paired t-test P value. Boxes show median and interquartile ranges (IQRs), whiskers extend to 1.5 x IQR. (c) A linear model was made for each immune parameter based on the multiple samples from each individual (ANOVA model: immune parameter ~ subject identifier + visit number). For each parameter the proportion of the R2 values from all volunteers attributable to either inter-individual differences or intra-individual differences was assessed for day 0 and 7 or (d) day 0 and 42. (e) Multidimensional scaling of the vaccination time-points. Each individual is represented at day 0 (green), 7 (red) and 42 (blue) and linked by a grey line indicating immunological distance (32 individuals; 96 visits). (f) Quantification of the immunological distance between day 0 and 7, 7 and 42, and 0 and 42 for each volunteer (n=32), with paired t-test. (g) For each volunteer, a Z-score was calculated for each parameter at day 0 and 42, indicating standard deviations from the mean value. Correlation analysis indicates the line of best fit. *, p<0.001; **,p<0.0001.
Age and cohabitation affect the immunological landscape
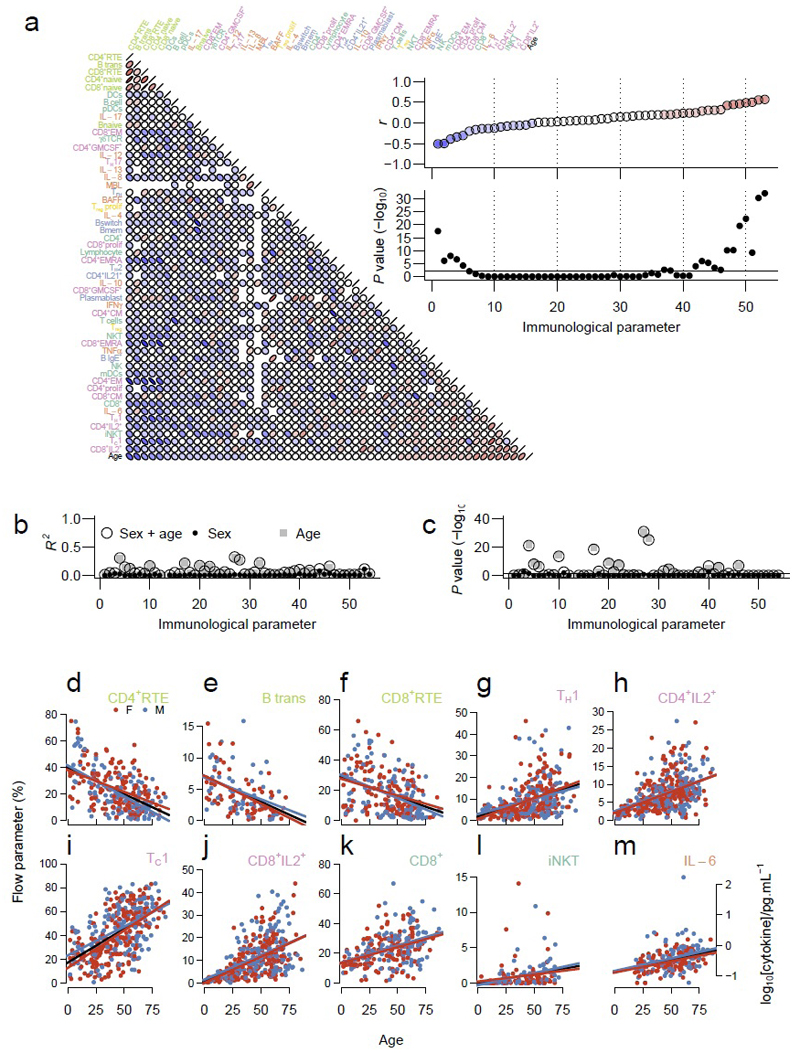
Having established the diversity of elastic stable equilibria in the human immune system, we sought to determine the underlying biological drivers. As the variability was greatest in the precursor populations, we first investigated the effect of age on immune profile. Our dataset includes individuals ranging from 2 to 86 years of age, with substantial numbers of both children (<18 years, n=40) and older persons (>65 years, n=54). Strong relationships, both positive and negative, were observed between immune parameters and age (Fig. 4a-c). Using a threshold of adjusted p<0.01 and r<-0.35 or r>0.35 (see Supplementary Fig. 6 and Supplementary Table 2), three immune parameters had a negative relationship with age and seven had a positive relationship with age. CD4+ RTE (Fig. 4b), transitional B cells (Fig. 4c) and CD8+ RTE (Fig. 4d) were all reduced in a linear fashion as individuals aged, consistent with an age-dependent reduction in thymus and bone-marrow activity. We observed a positive relationship of several inflammatory populations with age, namely TH1 cells (Figure 4E), CD4+IL-2+ cells (Fig. 4f), TC1 cells (Fig. 4g), CD8+IL-2+ cells (Fig. 4h) and iNKT cells (Fig. 4j), and an age-associated increase in CD8+ T cells (Fig. 4i). Despite finding significantly more Th1-associated cells with age, serum IFN-γ did not reach our threshold for correlation coefficient (r=0.18 [95% CI 0.06-0.29]; adjusted P=5.2x10-3). We did, however, identify IL-6, another pro-inflammatory cytokine as significantly increased with age (Fig. 4k). Together these data demonstrate that age is a major contributor to the immune profile of healthy individuals, with a successive downregulation of precursor populations and an upregulation of TH1-associated inflammatory populations with age. Interestingly, the data-points from two extremes of age (<18 and >65 years old), do not have larger variances than the central ages. This observation suggests, at least for these immune parameters, that paediatric and geriatric immunology are not “special cases” with different rules, but rather follow a set of continuous influences.
Figure 4.
Age is a major determinant of immunological equilibria. (a) Each immune parameter was correlated with age and each other immune parameter using pairwise Spearman r values. The arrangement of the immune parameters is determined by their correlation with age. Inset panels show Spearman’s r (upper) and -log10 Bonferroni corrected P values (lower) plotted against age. (b) R2 for each immune parameter for models incorporating gender (filled circles), age (grey squares) or both gender and age (open circles) as the independent variable(s), with (c) accompanying -log10 of Bonferroni corrected P values. (d) to (m) Individual scatterplots for each of the immune parameters with significant association with age. The percentage values of the flow parameters, or log10 cytokine concentrations, are plotted against age, with women in red, men in blue and linear regression lines for the whole cohort in black. Data shown for (d) CD4+ RTE (p=3x10-18), (e) transitional B cells (p=8x10-7), (f) CD8+ RTE (p=1x10-8), (g) TH1 (p=3x10-20), (h) CD4+ IL-2+ T cells (p=5x10-23), (i) Tc1 (p=5x10-31), (j) CD8+ IL-2+ T cells (p=8x10-33), (k) CD8+ T cells (p=8x10-11) (l) iNKT cells (p=6x10-10) and (m) serum IL-6 (p=7x10-11).
There are well-defined differences in ageing-related diseases between the genders. We therefore assessed whether any of the immune parameters were associated with gender, while controlling for age. We found that gender added no explanatory power to a model already including age, with essentially no effect on the variance of each immune parameter with a median difference in R2 of 0.006 (Fig. 4b,c). The only significant effects of gender on the immune profile was an increase in CD4+ T cell numbers in women compared to men, with a median CD4+ T cell frequency of 17.8% in women and 14.6% in men (adjusted P=1.3x10-3), consistent with previous reports 17.
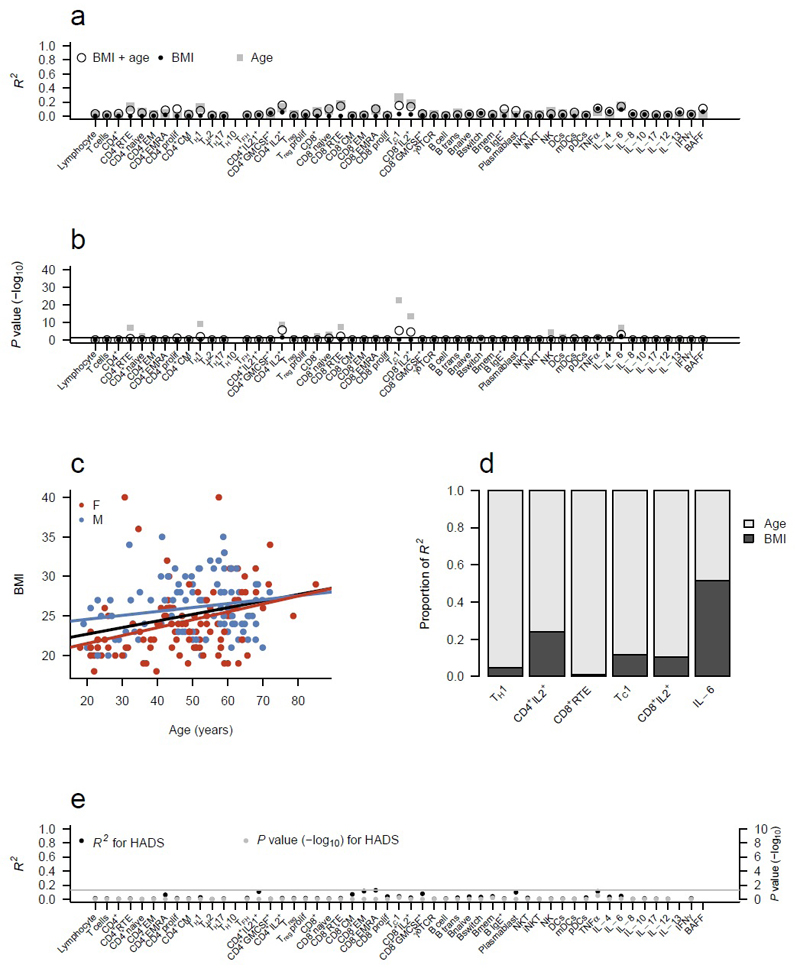
To extend the analysis on physiological influences on immune profiles, we assessed body mass index (BMI), anxiety and depression in a subset of adult volunteers (>18 years). BMI provided two significant associations with immune parameters: CD4+IL-2+ (adjusted P=0.03) and IL-6 (P=0.007), however their effect sizes were modest, with an R2 of 0.06 and 0.09, respectively (Fig. 5a,b). As an extra complexity, there is a relationship between BMI and age (Fig. 5c), with a tendency for BMI to increase with age. To control for this, we built both age and BMI into our model. For all the age-BMI immune associations, with the exception of IL-6, BMI was the minor contributor to the observed variation, with age playing a much greater role (Fig. 5d). By contrast, serum concentrations of IL-6, an adipose-associated cytokine, was equally influenced by both age and BMI (Fig. 5d). Many of the other changes associated with BMI in the literature 18, 19, 20, 21 were not observed in our dataset. This discrepancy may be due to inadequate controlling for age in prior studies (based on the relative strength of associations observed here, using age as a category rather than as a linear variable would substantially over-estimate the effect of BMI), or a relative lack of individuals at the extreme ends of the BMI scale in the current study. To investigate a potential neuro-immunological connection, we sought to determine whether anxiety and depression scores 19 altered immune parameters, however no substantial or significant effects were observed (Fig. 5e). Together, these data indicate that BMI, depression or anxiety do not substantively alter immune equilibria point of healthy individuals, beyond the association of BMI with serum IL-6 concentrations.
Figure 5.
Immunoprofile is not influenced by BMI, depression, or anxiety. (a) R2 for each immune parameter for models incorporating BMI (filled circles; 213 individuals), age (filled squares; 367 individuals) or both BMI and age (open circles; 213 individuals) as the independent variable(s), with (b) accompanying -log10 of Bonferroni corrected P values. Analysis excluded children (<18 years). (c) Relationship between BMI and age in the analysed cohort. (d) The relative R2 contributions of age and BMI to immune parameters that were significant in a model including both BMI and age (adjusted P<0.05). (e) R2 and -log10 adjusted P values for each immune parameter for a model incorporating HADS anxiety score and HADS depression score (235 individuals).
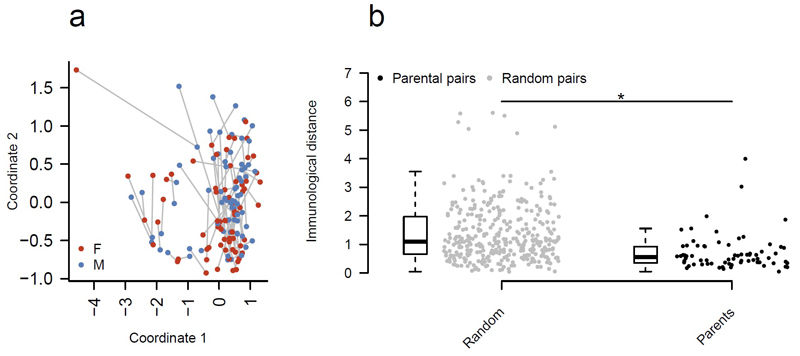
Finally, we sought to determine whether cohabitation had an impact on immune equilibria. Within our study, we sampled 70 parental pairs (adults, 18 to 65, with one or more children living at home). From these 140 individuals we could ask whether a shared environment altered immunoprofiles. Our hypothesis was that the immunoprofiles of a parental pair would be more closely related than a random in silico male:female pairing. To test this assertion, we used multi-dimensional scaling to reduce the diversity of the immune system down to two dimensions (Fig. 6a) and linked each pair with a line. We measured the distance between mother and father for all parental pairs (Fig. 6b) and compared this to the distance between randomly generated pairings (Fig. 6b). We found that there was a significant and substantial (~50%) reduction in the immune variability between genuine parental pairs compared to the randomised, null pairs, suggesting that a shared environment drives a convergence between immunoprofile. This effect was independent of age, as exclusion of the identified age-related parameters gave no difference in the result (Supplementary Fig. 7). Likewise, measurement of cytokine production following in vitro stimulations of PBMCs demonstrated significant convergence between the profiles of parents (Supplementary Fig. 8), demonstrating that the effect extends to alternative methodologies. These results demonstrate that while the immunological equilibria point is robust and stable within an individual, two individuals in a close relationship converge towards a single immunological equilibria point.
Figure 6.
Parenthood shapes the immune system towards a shared equilibria. (a) 140 individuals were identified as adult (18-65 years) biological parents with a child still living at home. The immune profile (54 parameters) was compressed using multidimensional scaling (k=2) of the correlation matrix between individuals, visualising pairwise Spearman’s correlation coefficients between each individual. The immunological distance between each male:female pair is indicated by the connecting gray line. (b) The immunological distance, as measured by multidimensional scaling, between parental pairs versus random male:female pairs. To generate the random distribution, each male was computationally paired in a random fashion with 5 females from the parental dataset. Distributions were compared using a two-tailed Mann-Whitney test. *, p=8x10-11.
Discussion
Non-genetic factors are estimated to account for ~50-75% of immunological variation between healthy individuals 2, 3, 4, yet a thorough understanding of the causative factors at play remains lacking. Through the use of a systems immunology approach and targeted sub-cohorts of healthy individuals, we were able to assess the main non-genetic factors. Of the intrinsic factors of age, BMI, sex and psychological state, an individual’s age was the most important influence on their immunological landscape. This result complements longstanding observations that immune function (response to vaccination, infection, cancer immunosurveillance) deteriorates with age. The reduction in T cell precursors may be explained by thymic involution 22, however the highly concordant decrease in transitional B cells, suggests a common root cause, such as impaired bone-marrow function 23. The age-dependent increase in TH1-associated populations is striking because TH2 and TH17 cell populations did not show an association with age, demonstrating that this effect is specific to the TH1 arm of the immune system, rather than a generic increase in T cell activation. Overall, for the 10 age-associated traits, the combined effect of age and genetics appears to account for almost the entire variability in the human population. In the current study, the median age-dependent R2 for these traits was 0.13. The median published heritability for these traits (R2 for genetic effects) 3, is 0.67. We estimate that for age-related immune parameters, ~80% of their variation would be explicable by age and genetic factors, with more variability potentially explained by age-genetic interaction. It is intriguing that the immune parameters that are age-related have higher heritability estimates than the rest of the immunoprofile 3.
The immunological effect of BMI could perhaps be best regarded as a minor acceleration in the normal immunoageing process. The striking exception to the negligible effect of BMI was the positive association with serum IL-6 concentrations. One possibility is that this IL-6 is “non-immune” in origin, as monocytes from older individuals secrete less IL-6 in response to TLR ligation 5. While the precise cellular origin of increased IL-6 in aged and high BMI humans is unknown, a plausible candidate is vascular smooth muscle cells (VSMCs). Aged VSMC from mice and non-human primates produce more IL-6 than younger controls 24, 25, while obesity increases the inflammatory phenotype of VSMCs 26. With the complex biological functions of IL-6 27, this effect could partially account for the alteration of clinical outcomes that obesity has on diseases such heart failure.
We found very little effect of gender on the immune landscape. This is at odds to the longstanding observation that autoimmune diseases are, in general, more frequent in women than men (at pre-menopausal ages) and that vaccine responses are reportedly more robust in women than men 28, although the sex difference in vaccine response varies greatly 28 29 30. Notably, the gender-based differences are more limited at the cellular level compared to the molecular level 31. The incomplete correlation between gene signature and cell type suggests that the discrepancy can be resolved by a model where high diversity in molecular expression is largely compensated for at the cellular level.
One of the most surprising results from our study was the degree to which the immune profiles of parents were more similar to one another than to unrelated pairs. This suggests that a shared environment acts in some way to bring immunoprofiles towards a convergent equilibria. Within the environment shared by parents, there is a panoply of plausible biological mechanisms. For example, an individual’s microbiome converges with those that they live with 32, even including their dogs 33. Individuals in a relationship, rather than just co-habiting, have a more similar microbiome 34, possibly via direct transmission 35, which would make the microbial convergence even stronger for parents. Within our study design, this shared environment also includes a shared vector (the child), the significance of which requires further investigation. Beyond the bacterial components of the microbiome, close proximity allows the transmission of viral pathogens, including CMV, which was found to influence twin concordance in more than half of their immune traits 3. A shared environment, including, presumably, socio-economic status, will also bring shared behaviours, a process called spousal concordance: diet (which can also influence gut microbiome 36), smoking 37, alcohol intake 38, exercise levels and even control of chronic diseases like hypertension 39 are all likely to be influenced by a partner’s attitudes towards them. It is fascinating to speculate that partner choice may also influence response to immunity and immune pathologies. Finally, we also note, but decline to comment further, that this “parenthood effect” is a far stronger influence on the immune system than acute and untreated gastroenteritis.
On-line Methods
Participant selection and ethical approval
All participants were Caucasian in origin and all sampling was conducted in Belgium, except for the vaccination cohort, who were Caucasian and sampled in England. All individuals, or their legal guardians, gave written informed consent and the study was approved by the Ethics Committee of the University Hospitals Leuven. The influenza vaccination study protocol was approved by the Health Research Authority, National Research Ethics Service committee South Central, Hampshire A, UK (REC reference:14/SC/1077). Demographic and clinical data were collected through a questionnaire. Exclusion criteria were cancer, autoimmunity or gastrointestinal complaints. Individuals that were parents of the same child were considered to be cohabiting. Individuals from the travel clinic planning a trip to developing countries in South America, Africa or Asia were considered to be at elevated risk of gastrointestinal infection. Acute gastroenteritis was defined as self-reported diarrhoea (passage of unformed stools), with or without additional symptoms of nausea, vomiting, abdominal cramps, pain, fever or blood in stools (using ROME-III criteria). Gasteroenteritis was classified as mild if no additional symptoms were reported, moderate with at least one additional symptom and classic with at least three diarrhoeal episodes per day and at least one additional symptom. Anxiety and depression was assessing using the Hospital Anxiety and Depression Scale (HADS) and the Patient Health Questionnaire (PHQ-15). For recruitment to the vaccination study potential participants were excluded if they have already received the 2014-2015 seasonal influenza vaccination, if they have had a previous adverse reaction to any vaccination, a known allergy to any components of the vaccine, were taking immune modulating medication, and women who are pregnant or breastfeeding. In total 32 healthy donors between 53 and 64 years of age were recruited from the Cambridge BioResource as part of the vaccination study during the 2014-2015 winter. Participants were administered the inactivated influenza vaccine (split virion) BP vaccine (Sanofi Pasteur) by intramuscular injection in the right deltoid.
Blood sampling and peripheral blood mononuclear cell isolation
Blood samples from Belgian participants were collected in Heparin tubes and rested at 22°C for four hours prior to separation of serum and peripheral blood mononuclear cells (PBMC) using lymphocyte separation medium (LSM, MP Biomedicals). PBMCs were frozen in 10% DMSO (Dimethyl sulfoxide, Sigma) and stored at -80°C for a maximum of 10 weeks. For the vaccination cohort, research nurses at the Cambridge BioResource collected blood samples into EDTA coated tubes on the day of vaccination (prior to administration of the vaccine), seven days and 42 days following vaccination. PBMC were isolated using 15mL of Histopaque-1077 (Sigma) then frozen in Foetal Bovine Serum supplemented with 10% Dimethyl sulfoxide (Sigma) overnight at -80°C, then stored in the liquid nitrogen freezer prior to analysis by flow cytometry.
Flow cytometry phenotyping
Thawed cells were stained with antibodies as listed in Supplementary Table 3. Ki67 and Foxp3 staining was performed following treatment with fixation/permeabilization buffer (eBioscience). Cytokine staining was performed following ex vivo stimulation for five hours in 50 ng/ml PMA (Phorbol 12-myristate 13-acetate, Sigma) and 500 ng/ml ionomycin (Sigma) in the presence of GolgiStop (BD Biosciences). Stimulated cells were surface stained, fixed and permeabilized with Cytofix/cytoperm (BD), prior to staining for cytokines. Additional cells were stimulated for 72 h for supernatant assessment by MSD (see below). Data was acquired on a BD FACSCantoII and analyzed with FlowJo (Tree star). The vaccination cohort data was acquired on a BD LSRFORTESSA and analysed using FlowJo (Tree star).
Serological assessment
Plasma samples collected were stored at -80°C. Circulating levels of MBL were quantified in plasma using the MBL Oligomer ELISA Kit form Bioporto® (Copenhagen, Denmark). Circulating levels of BAFF were measured using a human BAFF Quantikine ELISA (R&D Systems). Cytokine plasma concentrations were quantified by an electrochemiluminescence immunoassay format using the V-Plex™ human Pro-inflammatory panel MSD (Meso Scale Discovery. Rockville, Maryland, USA) plates. All reagents and standards were provided by each manufacturer. Samples and standards were prepared according to each manufacturer’s instructions.
Data handling
Data (phenotypic, flow cytometric and serological) were collated and stored in Microsoft Excel. All data analysis was performed using R (http://www.r-project.org version 3.1.0 40) via the RStudio IDE (http://www.rstudio.com version 0.98.1102). Figures were drawn using knitr 41, which produces pdf output via LaTeX.
ELISA and MSD data were pre-processed as follows: Any experimental value lower than the lower limit of detection for the assay was replaced with the lower limit of detection of that cytokine. After this step, all ELISA and MSD data were log10 transformed. The flow cytometry data was used as percentage as exported from FlowJo. No data was excluded from analysis. Inter-quartile range, median, number of missing values for each immune parameter (flow cytometry derived and serologically derived parameters together) are shown in Supplementary Table 2. For non-longitudinal analyses only the most recent sampling of each individual was used. The original data (phenotypic, flow cytometric and serological) is available to download as an xls or an RData file.
Statistical analysis
All sample collection, data acquisition and data processing was performed blind prior to statistical analysis. Spearman’s rank correlation coefficient was used throughout for pairwise correlation comparisons. Euclidean distances were calculated from correlation matrices (using Spearman) as pre-processing for multi-dimensional scaling (using either cmdscale in base R, or monoMDS from the vegan package 42) or hierarchical clustering (using heatmap in stats package 40). Consensus clustering was performed using the R package ConsensusClusterPlus, an R implementation of the original algorithm 43. Correlation plots were drawn using the plotcorr function in the ellipse package 44. Linear regression modelling was performed using base R function (lm), with the proportions of R2 calculated with the relaimpo package 45. Lm will provide differing statistical models depending upon the input data. For most of the modelling shown, a continuous variable (immune parameter) is modelled using a categorical variable (such as subject identifier, visit number). In this scenario, lm provides an ANOVA model. If both variables are continuous, linear regression modelling is used. Two group comparisons were made using two-tailed Mann-Whitney tests, except for the paired vaccination data, where a paired t-test was used to compare samples from the same individual at different time points. Bonferroni correction was used for all corrections for multiple testing, as implemented in base R.
Code availability
Original datasets are provided as both .xls and .RData files, split into the Belgian profiling cohort and the English vaccination cohort. The code used to produce our figures is provided as both a pdf (for easier reading) or as an R markdown file (for easier re-running or evaluation).
Supplementary Material
AOP Summary.
Large-scale flow cytometric and serological assessment of the human immune landscape by Liston and colleagues demonstrates a wide inter-individual variation that is modified by age and cohabitation.
Acknowledgements
This study was supported via funding from the European Research Council (Start Grant IMMUNO to AL and Start Grant TWILIGHT (637801) to MAL). EJC is an academic clinical fellow funded by the NIHR. MAL is supported by the Biotechnology and Biological Sciences Research Council. AG is supported by the Research Fund KU Leuven (OT/11/087) and Research Foundation Flanders (G073415N). JCL is supported by the Wellcome Trust (105920/Z/14/Z). We gratefully acknowledge the participation of all volunteers. We thank the Cambridge BioResource staff for their help with volunteer recruitment. We thank members of the Cambridge BioResource SAB and Management Committee for their support of our study and the National Institute for Health Research Cambridge Biomedical Research Centre for funding. The authors thank Anh Nuygen, Dean Franckaert, Dina Danso-Abeam and Lien Van Eyck (KUL-VIB) for technical assistance.
Footnotes
Author’s contribution
EJC analysed the data and drafted the manuscript. JD and JG performed the experiments. VL analysed the vaccination cohort. JCL, CW, IM, AN and GB designed and recruited sub-cohorts. MAL designed and supervised the vaccination study, contributed to the data analysis and the manuscript. AL designed and supervised the study and drafted the manuscript.
References
- 1.Davis MM. Immunology taught by humans. Sci Transl Med. 2012;4(117):117fs112. doi: 10.1126/scitranslmed.3003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orru V, Steri M, Sole G, Sidore C, Virdis F, Dei M, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155(1):242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, Furman D, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160(1-2):37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Jager PL, Hacohen N, Mathis D, Regev A, Stranger BE, Benoist C. ImmVar project: Insights and design considerations for future studies of “healthy” immune variation. Semin Immunol. 2015 doi: 10.1016/j.smim.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nat Rev Immunol. 2013;13(12):875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamieson BD, Douek DC, Killian S, Hultin LE, Scripture-Adams DD, Giorgi JV, et al. Generation of functional thymocytes in the human adult. Immunity. 1999;10(5):569–575. doi: 10.1016/s1074-7613(00)80056-4. [DOI] [PubMed] [Google Scholar]
- 7.den Braber I, Mugwagwa T, Vrisekoop N, Westera L, Mogling R, de Boer AB, et al. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity. 2012;36(2):288–297. doi: 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Johnson PL, Yates AJ, Goronzy JJ, Antia R. Peripheral selection rather than thymic involution explains sudden contraction in naive CD4 T-cell diversity with age. Proc Natl Acad Sci U S A. 2012;109(52):21432–21437. doi: 10.1073/pnas.1209283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caballero S, Pamer EG. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol. 2015;33:227–256. doi: 10.1146/annurev-immunol-032713-120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucas M, Day CL, Wyer JR, Cunliffe SL, Loughry A, McMichael AJ, et al. Ex vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. Journal of virology. 2004;78(13):7284–7287. doi: 10.1128/JVI.78.13.7284-7287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JD, van der Most RG, Akondy RS, Glidewell JT, Albott S, Masopust D, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Nino D, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. The Journal of infectious diseases. 2011;203(7):921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, et al. Systems biology of vaccination for seasonal influenza in humans. Nature immunology. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amadori A, Zamarchi R, De Silvestro G, Forza G, Cavatton G, Danieli GA, et al. Genetic control of the CD4/CD8 T-cell ratio in humans. Nat Med. 1995;1(12):1279–1283. doi: 10.1038/nm1295-1279. [DOI] [PubMed] [Google Scholar]
- 18.Spiegelman BM, Hotamisligil GS. Through thick and thin: wasting, obesity, and TNF alpha. Cell. 1993;73(4):625–627. doi: 10.1016/0092-8674(93)90243-j. [DOI] [PubMed] [Google Scholar]
- 19.Spielmann G, Johnston CA, O'Connor DP, Foreyt JP, Simpson RJ. Excess body mass is associated with T cell differentiation indicative of immune ageing in children. Clin Exp Immunol. 2014;176(2):246–254. doi: 10.1111/cei.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin-Romero C, Santos-Alvarez J, Goberna R, Sanchez-Margalet V. Human leptin enhances activation and proliferation of human circulating T lymphocytes. Cell Immunol. 2000;199(1):15–24. doi: 10.1006/cimm.1999.1594. [DOI] [PubMed] [Google Scholar]
- 21.Damluji AA, Ramireddy A, Al-Damluji MS, Marzouka GR, Otalvaro L, Viles-Gonzalez JF, et al. Association between anti-human heat shock protein-60 and interleukin-2 with coronary artery calcium score. Heart. 2015;101(6):436–441. doi: 10.1136/heartjnl-2014-306649. [DOI] [PubMed] [Google Scholar]
- 22.Dooley J, Liston A. Molecular control over thymic involution: from cytokines and microRNA to aging and adipose tissue. Eur J Immunol. 2012;42(5):1073–1079. doi: 10.1002/eji.201142305. [DOI] [PubMed] [Google Scholar]
- 23.Franckaert D, Schlenner SM, Heirman N, Gill J, Skogberg G, Ekwall O, et al. Premature thymic involution is independent of structural plasticity of the thymic stroma. Eur J Immunol. 2015;45(5):1535–1547. doi: 10.1002/eji.201445277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y, Shen H, Schenten D, Shan P, Lee PJ, Goldstein DR. Aging enhances the basal production of IL-6 and CCL2 in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2012;32(1):103–109. doi: 10.1161/ATVBAHA.111.236349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csiszar A, Sosnowska D, Wang M, Lakatta EG, Sonntag WE, Ungvari Z. Age-associated proinflammatory secretory phenotype in vascular smooth muscle cells from the non-human primate Macaca mulatta: reversal by resveratrol treatment. J Gerontol A Biol Sci Med Sci. 2012;67(8):811–820. doi: 10.1093/gerona/glr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van de Voorde J, Boydens C, Pauwels B, Decaluwe K. Perivascular adipose tissue, inflammation and vascular dysfunction in obesity. Curr Vasc Pharmacol. 2014;12(3):403–411. doi: 10.2174/1570161112666140423220628. [DOI] [PubMed] [Google Scholar]
- 27.Fontes JA, Rose NR, Cihakova D. The varying faces of IL-6: From cardiac protection to cardiac failure. Cytokine. 2015;74(1):62–68. doi: 10.1016/j.cyto.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giefing-Kröll C, Berger P, Lepperdinger G, Grubeck-Loebenstein B. How sex and age affect immune responses, susceptibility to infections, and response to vaccination. Aging Cell. 2015;14(3):309–321. doi: 10.1111/acel.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furman D, Hejblum BP, Simon N, Jojic V, Dekker CL, Thiebaut R, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proceedings of the National Academy of Sciences. 2014;111(2):869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, et al. Global Analyses of Human Immune Variation Reveal Baseline Predictors of Postvaccination Responses. Cell. 157(2):499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, et al. Individuality and variation in gene expression patterns in human blood. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(4):1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kort R, Caspers M, van de Graaf A, van Egmond W, Keijser B, Roeselers G. Shaping the oral microbiota through intimate kissing. Microbiome. 2014;2:41. doi: 10.1186/2049-2618-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmody RN, Gerber GK, Luevano JM, Jr, Gatti DM, Somes L, Svenson KL, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17(1):72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falba TA, Sindelar JL. Spousal concordance in health behavior change. Health Serv Res. 2008;43(1 Pt 1):96–116. doi: 10.1111/j.1475-6773.2007.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham K, Braun K. Concordance of use of alcohol and other substances among older adult couples. Addict Behav. 1999;24(6):839–856. doi: 10.1016/s0306-4603(99)00059-3. [DOI] [PubMed] [Google Scholar]
- 39.McAdams DeMarco M, Coresh J, Woodward M, Butler KR, Kao WH, Mosley TH, Jr, et al. Hypertension status, treatment, and control among spousal pairs in a middle-aged adult cohort. Am J Epidemiol. 2011;174(7):790–796. doi: 10.1093/aje/kwr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2014. [Google Scholar]
- 41.Xie Y. knitr: A General-Purpose Package for Dynamic Report Generation in R. 2015 [Google Scholar]
- 42.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. vegan: Community Ecology Package. 2015 [Google Scholar]
- 43.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52(1-2):91–118. [Google Scholar]
- 44.Murdoch D, Chow ED. ellipse: Functions for drawing ellipses and ellipse-like confidence regions. 2013 [Google Scholar]
- 45.Grömping U. Relative Importance for Linear Regression in R: The Package relaimpo. Journal of Statistical Software. 2006;17(1):1–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.