Abstract
The spliceosome is the dynamic RNA-protein machine responsible for faithfully splicing introns from precursor messenger RNAs (pre-mRNAs). Many of the dynamic processes required for the proper assembly, catalytic activation, and disassembly of the spliceosome as it acts on its pre-mRNA substrate remain poorly understood, a challenge that persists for many biomolecular machines. Here, we developed a fluorescence-based Single Molecule Cluster Analysis (SiMCAn) tool to dissect the manifold conformational dynamics of a pre-mRNA through the splicing cycle. By clustering common dynamic behaviors derived from selectively blocked splicing reactions, SiMCAn was able to identify signature conformations and dynamic behaviors of multiple ATP-dependent intermediates. In addition, it identified a conformation adopted late in splicing by a 3′ splice site mutant, invoking a mechanism for substrate proofreading. SiMCAn presents a novel framework for interpreting complex single molecule behaviors that should prove widely useful for the comprehensive analysis of a plethora of dynamic cellular machines.
Introduction
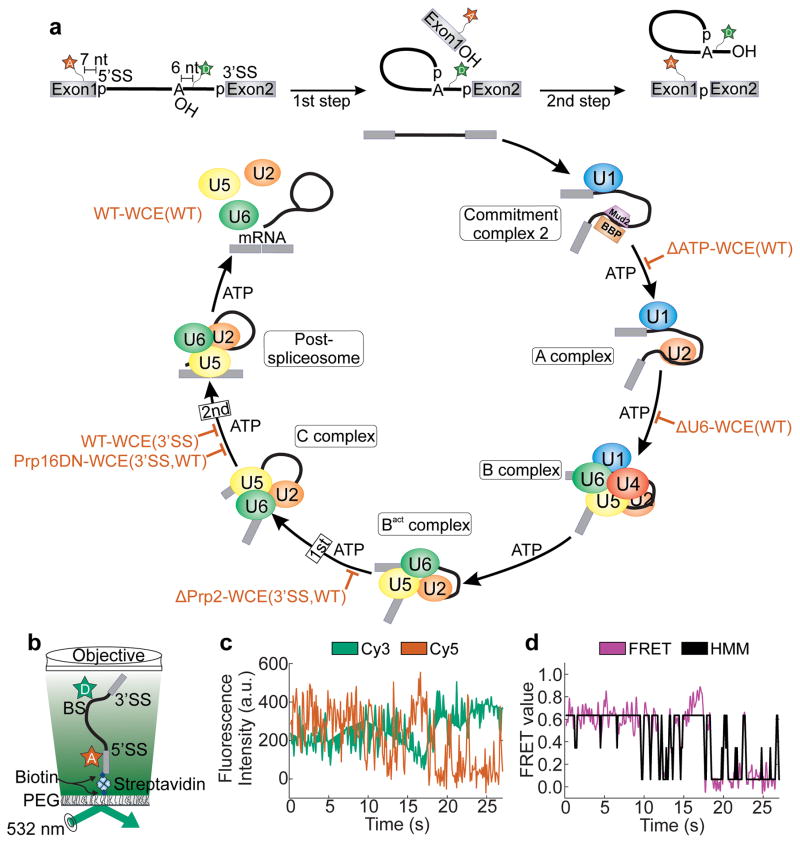
Conformational dynamics play a key role in every aspect of RNA biology, such as in RNA transcription, splicing and translation1–3. The quantitative measurement and interpretation of these dynamics are of great importance for an understanding of the common principles underlying the biological function of RNA2–4. Single molecule fluorescence approaches have recently emerged as a powerful toolset to dissect the structural dynamics that form the foundation of biomolecular machines functioning at the nanometer scale5–9. For example, single molecule fluorescence energy transfer (smFRET) has been implemented to dissect spliceosome dynamics5,6,10. The spliceosome is a multi-megadalton ribonucleoprotein (RNP) complex essential for the faithful removal of introns from eukaryotic precursor messenger RNAs (pre-mRNAs) during the two chemical steps of splicing (Fig. 1a)11. The architectural reorganization of the pre-mRNA substrate required to accommodate these two catalytic steps in a single active site are thought to be accompanied by substantial rearrangements that ensure substrate proofreading12–15. To explore these rearrangements, we have labeled the efficiently splicing yeast pre-mRNA Ubc46,16 with the FRET pair Cy5 and Cy3 seven nucleotides upstream of the 5′ splice site (5’SS) and six nucleotides downstream of the branch point (BP), respectively. This approach yields a substrate capable of detecting changes in intron conformation as a result of 5’SS and BP (un)docking (Fig. 1a, b) that we previously used to show that one of several DExD/H-box ATPases, Prp2, unlocks intrinsic conformational dynamics in the isolated spliceosomal Bact complex, setting the stage for first-step catalysis through a biased Brownian ratcheting mechanism5.
Figure 1.
smFRET analysis of pre-mRNA splicing using the hidden Markov model. (a) The fluorescent substrate used to monitor pre-mRNA dynamics contains Cy5 and Cy3 fluorophores seven nucleotides upstream of the 5’SS and six nucleotides downstream of the BP, respectively. The spliceosome assembly and catalysis pathway is thought to progress in a stepwise manner requiring ATP at several steps of assembly. The biochemical and genetic stalls utilized in this study are indicated by red blocks. (b) Prism-based TIRFM setup for smFRET. (c) Raw single molecule time trace showing the anti-correlated donor (green) and acceptor (red) intensities. (d) The corresponding FRET trace (magenta) and the HMM trace as assigned by vbFRET (black).
Despite years of utilization, the quantitative methods available for an in-depth dissection of the dynamics observed in smFRET studies are still limited. In particular, the multi-state, mostly asynchronous and often heterogeneous kinetics of many molecular machines, such as the spliceosome, render the current state-of-the-art analysis of individual state transitions as independent stochastic events insufficient for an in-depth understanding of the underlying biological function. To extract additional information, several recent studies have analyzed common smFRET metrics more thoroughly, specifically FRET probability histograms and state-to-state transition kinetics7. For example, it has been demonstrated that in certain favorable cases interstate dynamics can be extracted from histograms through an analysis of photon arrival times and lifetimes17. In addition, state-to-state transition kinetics have been extracted utilizing clustering algorithms to identify distinct kinetic behaviors18,19. All of these approaches have focused on small datasets with 2–3 FRET states and limited dynamics. Unfortunately, they are limited when more complex systems with multiple states and complex kinetic networks are examined under non-equilibrium conditions.
We present here a method that utilizes hierarchical clustering as a means to group, sort, and identify commonalities of smFRET trajectories fit using Hidden Markov Modeling (HMM, Fig. 1c, d). We termed this tool Single Molecule Cluster Analysis (SiMCAn) and used it to characterize the pre-mRNA dynamics associated with the assembly and catalytic steps of the yeast spliceosome. SiMCAn reduces every single molecule trajectory, regardless of its number of states, to an easily comparable unit of information that we coin the FRET Similarity Matrix (FSM). By leveraging hierarchical clustering techniques, we identified common dynamic behaviors across 10,680 different Ubc4 pre-mRNA molecules. Importantly, we accomplished an unbiased, model-free identification of commonalities and differences between splicing complexes through a second level of clustering based on the abundance of dynamic behaviors exhibited by defined functional intermediates. Applying SiMCAn thus allowed us to efficiently assign pre-mRNA FRET states and transitions to specific splicing complexes, including a heretofore undescribed low-FRET conformation adopted late in splicing by a 3′ splice site mutant. These results establish SiMCAn as an effective bioinformatics tool to characterize complex smFRET behavior of dynamic cellular machines.
Results
Hierarchical clustering of complex smFRET behaviors
State-to-state transitions in single molecule trajectories report on the accessibility of conformational states and their ability to interconvert. HMMs are the most commonly utilized tools for identifying state-to-state transitions in smFRET trajectories (Fig. 1c, d). HMM fits create challenges, however, when comparing trajectories with different states and kinetic properties across a variety of experimental conditions (Supplementary Note 1). These challenges can be addressed by fitting all data with a single HMM model so that consistent state values are used across all trajectories6,7. Such an approach effectively imposes a single, preordained kinetic model on all molecules and experimental conditions, which may not be appropriate for highly complex systems such as the spliceosome.
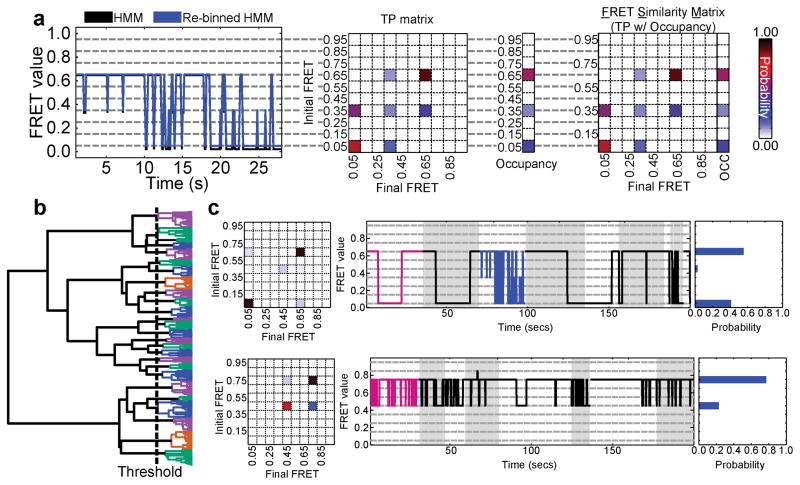
SiMCAn introduces a solution for sorting and identifying commonalities among large numbers of HMM-fitted smFRET trajectories by first binning each FRET state into one of ten evenly spaced FRET values (0.05–0.95, with increments of 0.10) (Fig. 2a). This binning enables the direct comparison across a large dataset with FRET values that together evenly span the viable FRET range and are commensurate with typical signal-to-noise ratios. The resulting HMMs are used to construct transition probability (TP) matrices that describe the FRET states as well as the kinetics of transition between them (Fig. 2a). Each TP matrix is then combined with the occupancies of the individual FRET states to create an FSM (Fig. 2a). The Euclidean (ordinary) distance between FSM provides a suitable weighted, information-rich metric by which to compare thousands of HMM-fitted smFRET trajectories using hierarchical clustering analysis (Supplementary Note 1), an agglomerative clustering technique that aims to group data of similar characteristics without the need for a preconceived experimental model or hypothesis20,21 (Online Methods). The result of this clustering is a hierarchical tree, where each leaf on the tree represents the dynamics of an individual molecule, while branch points indicate a split in dynamic behavior of the group of molecules at a given level of coarseness (Fig. 2b). The number of clusters is determined using an iterative measurement of the inter-cluster distances and a modified k-means algorithm22. Henceforth, each cluster will be represented using the average TP matrix, a random collection of traces, and the probability distribution of FRET states within the cluster (Fig. 2c).
Figure 2.
Workflow of SiMCAn to sort and cluster single-molecule derived HMMs for common dynamic behaviors. (a) Assigned FRET trace before (black) and after (blue) reassignment to the closest of 10 evenly spaced states (0.05–0.95, increment of 0.10, gray dashed lines). Transition probability (TP) matrix corresponding to the re-binned FRET trace in left panel and occupancy values for each of the ten FRET values for the molecule in left panel. FRET Similarity Matrix (FSM), which contains the transition probability (TP) matrix and FRET occupancies that describe the FRET states and transition kinetics between them for the molecule in panel a. (b) Hierarchical tree as a result of hierarchical clustering analysis using all 6,079 dynamic molecules. Each colored branch describes a set of molecules that shares common FRET transition probabilities. The dashed line indicates the threshold of 25 clusters used to describe the data. Static molecules were identified and analyzed by SiMCAn separately. (c) Cluster description for two of the 25 dynamic clusters of the full splicing dataset. Each representation shows the TP matrix of the cluster, the trace closest to the cluster center (magenta) and up to 200 s of random (black) traces from the cluster, and the probability of FRET states within the cluster. The highlighted blue trace in the top right panel indicates the example trace used in panel a. Grey and white backgrounds demarcate individual trajectories in (c).
Validation of SiMCAn using simulated datasets
To evaluate whether SiMCAn is able to correctly identify and segregate HMM-fitted trajectories with known FRET states, we applied it first to a simulated dataset containing 1,500 trajectories that reversibly transition from a 0.15 to a 0.45 FRET state and an equal number of trajectories that transition from the same 0.15 FRET state to a 0.85 state instead (Supplementary Fig. 1a), with average rate constants of k0.15→0.45 = 0.54 s−1, k0.45→0.15 = 0.54 s−1 and k0.15→0.85 = 0.54 s−1, k0.85→0.15 = 0.54 s−1, respectively. Utilizing the inter-cluster distances and modified k-means algorithm, SiMCAn properly identified and separated these two molecular behaviors (Supplementary Fig. 1b), demonstrating that FSMs can be clustered and distinguished based on the identity of their FRET states. A second and more important feature of SiMCAn is the ability to segregate HMMs based on differing kinetics. We analyzed a second set of 3,000 simulated HMMs possessing two FRET states of 0.15 and 0.75, with half designed to have identical interconversion rate constants of 0.54 s−1, whereas the other half transitioned much more slowly with rate constants of 0.15 s−1 (Supplementary Fig. 1c). SiMCAn identified two clusters with distinct transition rate constants between the two states (Supplementary Fig. 1d). These results demonstrate SiMCAn’s ability to differentiate HMM-fitted FRET trajectories based on their FRET states and kinetics.
Validation of SiMCAn using purified spliceosomal complexes
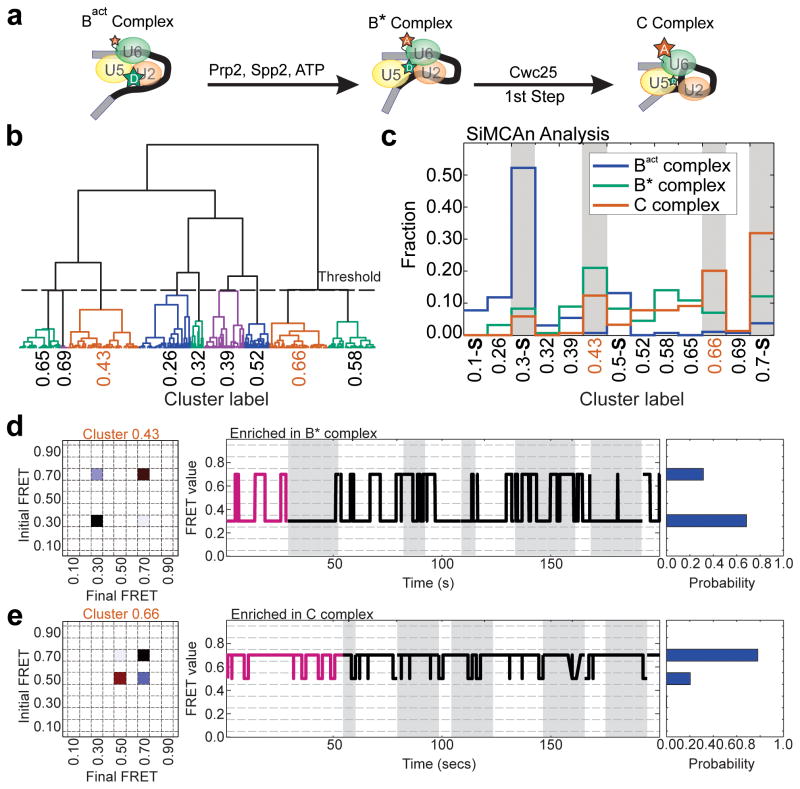
To benchmark SiMCAn against a more complex experimental dataset featuring multiple FRET states, numerous rate constants of transition, and the inherent experimental limitations (e.g., signal noise and premature photobleaching), we chose to analyze a previously published dataset collected during the Prp2-mediated conformational transition immediately prior to the first step of splicing5. Briefly, the immobilized Bact complex containing FRET-labeled Ubc4 was monitored as it progresses through the B* to the C complex upon addition of recombinant proteins Prp2, Spp2 and Cwc25 (Fig. 3a). Only upon exhaustive manual sorting were we able to identify distinct FRET state and kinetic signatures for the intermediate Bact, B* and C complexes (Supplementary Fig. 2a). Notably, SiMCAn was able to rapidly (within minutes) and correctly identify these previously only manually identified (Supplementary Fig. 2b)5 sub-populations of pre-mRNA molecules.
Figure 3.
Validation of SiMCAn using a previously analyzed dataset describing the transition from the purified Bact to the C complex5. (a) Protein requirements for the transition from the Bact complex through B* to the C complex. (b) Hierarchical tree based on hierarchical clustering analysis of the dynamic molecules re-fit with FRET states of 0.1, 0.3, 0.5, and 0.75. Static molecules were identified and analyzed by SiMCAn separately. (c) Cluster occupancy bar graph showing the fraction of molecules from each experimental condition that occupy the nine dynamic and four static clusters found using SiMCAn. Dynamic clusters were labeled by the weighted average FRET value of the molecules within the cluster (e.g., 0.2563) while static clusters are labeled by the single state they describe (e.g., 0.1-S). Grey bars highlight the most populated clusters occupied by each of the complexes. (d and e) Dynamic clusters enriched in the B* (d, cluster 0.4267) and C (e, cluster 0.6478) complexes. Each representative for the B* and C complex shows the TP matrix of the cluster (left), the closest (magenta) and several random (black) traces from the cluster (middle), and the probability of FRET states within the cluster (right).
To this end, the HMM-fitted FRET traces under Bact, B*, and C complex conditions were combined and analyzed using SiMCAn to determine if the analysis could recapitulate the manual annotation of these traces. Maximizing the inter-cluster distances while minimizing the intra-cluster distances using SiMCAn revealed 9 dynamic and 4 static clusters as best fitting the data (Fig. 3b and Supplementary Fig. 3). These clusters were combined into a single bar graph to depict the fraction of molecules that occupy each cluster, allowing for the identification of clusters most populated under each experimental condition (Fig. 3c). Reproducing our previous analysis, a cluster of molecules adopting a static low-FRET state (0.3-S) was identified as dominant under Bact conditions (Fig. 3c), whereas a static high-FRET cluster (0.7-S) was most abundant under C complex conditions (Fig. 3c). In addition, SiMCAn identified two dynamic clusters increasingly populated under B* (cluster 0.43, green) and C (cluster 0.66, red) complex conditions (Fig. 3c). Cluster 0.43 contains molecules with a short-lived high-FRET state and longer dwell times in the low-FRET state that are most abundant under B* conditions (Fig 3d). By contrast, cluster 0.66 contains molecules with a longer-lived high-FRET state featuring rapid excursions back to a mid-FRET state that are enriched upon addition of Cwc25 to form the C complex (Fig. 3e), matching our previous manual analysis5. These results demonstrate that, when applied to a complex experimental dataset, SiMCAn is able to segregate the data efficiently based on FRET states and differences in state-to-state interconversion kinetics to derive a biologically meaningful result.
Stalling of the spliceosome leads to distinct behaviors
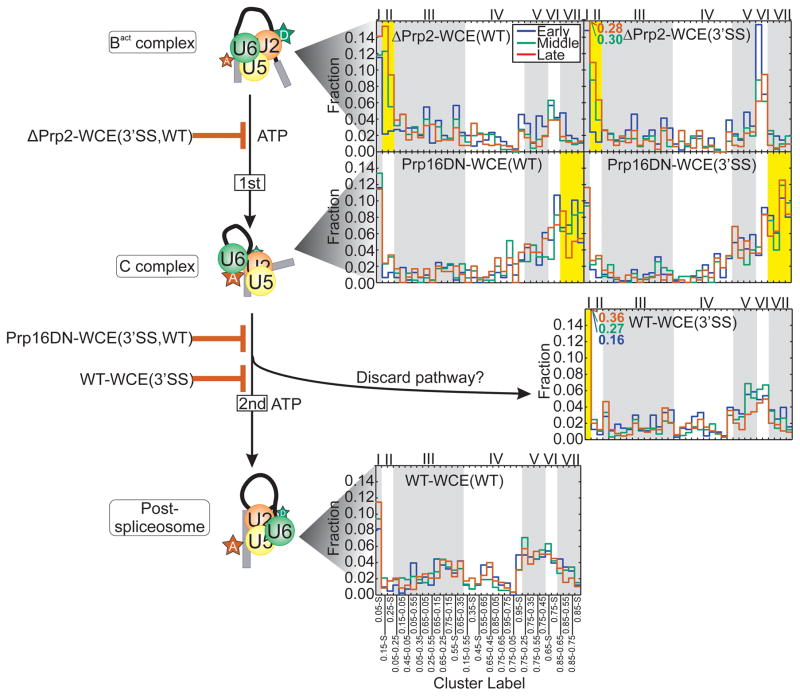
Having established that SiMCAn identifies known dynamic behaviors in simulated (Supplementary Fig. 1) and experimental, HMM-fitted smFRET trajectories (Fig. 3), we next utilized it on a new dataset enriched for specific stages of splicing through the use of biochemical and genetic stalls for which no previous behaviors are known. smFRET data were collected upon incubation of FRET-labeled wild-type (WT) Ubc4 pre-mRNA with WT yeast whole cell extract (WT-WCE), allowing for spliceosomal assembly on and splicing of the fluorescent substrate (condition WT-WCE(WT), Fig. 1a). Time courses were performed during which smFRET was recorded within time windows 0–8 min (early), 18–23 min (middle) and 33–40 min (late) after addition of WCE. To assign dynamics to particular splicing intermediates without a need for cumbersome biochemical isolation, we chose to utilize eight mutations, and combinations thereof, known to allow for efficient accumulation of specific splicing intermediates in WCE (Fig. 1a and Supplementary Table 1). Blockage and release by reconstitution were verified by bulk in vitro splicing assays in yeast WCE (Supplementary Fig. 4). smFRET data for each stall were then acquired using the same time lapse approach utilized for the WT-WCE(WT) condition. FRET probability distributions and Transition Occupancy Density Plots (TODPs) (Supplementary Fig. 5 and Supplementary Fig. 6) were utilized to broadly summarize the behavior of hundreds of molecule trajectories per condition7, confirming that the blocks lead to different ensemble and time averaged behaviors. However, this far more complex dataset is not amenable to standard analysis techniques as it includes a large number of traces, FRET states, and transition rate constants from splicing complexes stalled by mutation throughout the splicing cycle. As such, it represents an ideal application for SiMCAn.
Identifying biologically defined dynamics using SiMCAn
Application of SiMCAn to this new dataset allowed us to identify and cluster sets of molecules that share common dynamic behaviors. Each of the 10,680 smFRET trajectories was first fit with a HMM using vbFRET23, although any HMM fitting tool can be utilized that satisfies the user’s fitting preferences. Prior to clustering, 4,601 static molecules were identified and analyzed separately. Hierarchical clustering of the remaining 6,079 dynamic molecules produced a tree that was pruned to a height of 25 distinct clusters (Fig. 2b, Supplementary Fig. 7), so that each cluster represented a unique dynamic behavior (Fig. 2c, Supplementary Fig. 8). Static clusters were named by their sole FRET state (e.g., 0.05-S), whereas dynamic cluster names were assigned based on the first and second most occupied FRET states within the cluster (e.g., cluster 0.65–0.05 primarily occupies 0.65 and 0.05 FRET states). Bootstrap analysis based on the 25 SiMCAn identified clusters confirmed the ability to identify input HMMs from increasingly complex datasets, and that the SiMCAn-identified clusters for the large experimental dataset capture the molecular behavior exhaustively. (Supplementary Fig. 9a–b).
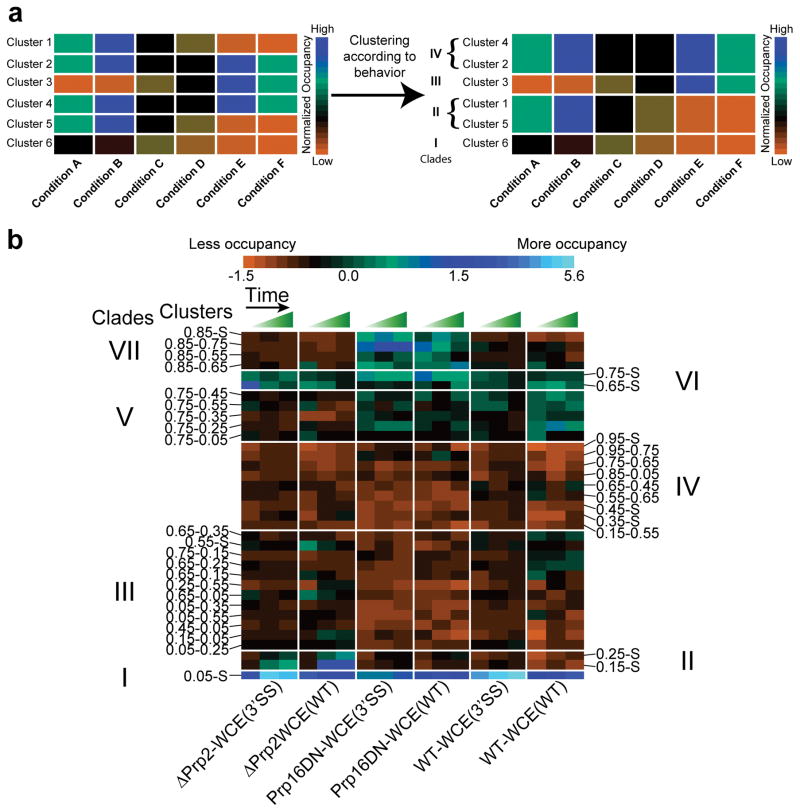
We next sought to identify clusters whose occupancies are similarly enriched or depleted for the same group of conditions, i.e., follow a similar pattern of high and low occupancies across conditions, suggesting they can be grouped into a ‘clade’ through a second round of hierarchical clustering (Fig. 4a). Upon application of this second level of SiMCAn to the full dataset, a tree height of seven clades (Supplementary Fig. 10) allowed for the identification of clusters representative of particular splicing conditions, thus most naturally capturing the changes in dynamic behavior expected to occur as the pre-mRNA progresses through the splicing cycle (Fig. 4b and Supplementary Fig. 11). A bar graph of all 35 (25 dynamic plus 10 static) clusters was also constructed, revealing the extent to which each cluster contributes to the overall dynamics for each condition (Fig. 5 and Supplementary Figs. 12, 13). Statistical analysis found that the average length of molecules within each cluster was similar, indicating that SiMCAn does not segregate by trace length (Supplementary Fig. 14 and Supplementary Table 2).
Figure 4.
Clustering of clusters to identify ‘clades’ of similar behavior. (a) Illustration of the second round of clustering to group the clusters by common occupancy patterns. In this example, six clusters (Y-axis, 1–6) have been populated by six conditions (X-axis, A–F). Each cluster has an occupancy pattern across the conditions as represented by a heat map, with high occupancy shown in blue and low occupancy shown in orange. By applying a second round of SiMCAn clustering, clusters with similar occupancies across the six conditions become grouped to form a clade (labeled I–IV). (b) Performing the second round of clustering with the 35 clusters from our experimental splicing dataset reveals 7 clades (labeled I–VII) of clusters enriched in particular splicing complexes. The fraction of molecules within each cluster for each experimental condition at each time was normalized to a mean of zero with unit variance. Green and blue colors indicate increased occupancy of a particular cluster while orange indicates decreased occupancy. Rows identify the clusters and are ordered by increasing average FRET of the clade. Columns identify the cluster occupancy of each condition for the early, middle, and late time points.
Figure 5.
Cluster occupancy histogram showing the raw fraction of molecules occupying each cluster for the late assembly stages of the splicing cycle. Alternating gray and white backgrounds demarcate the clusters (bottom) comprising each of the 7 clades (top). Clusters of occupancy characteristic of a specified condition are highlighted in yellow.
Characterization of pre- and post-first step blocks
Application of SiMCAn revealed a disperse set of dynamics and cluster occupancies in the early splicing conditions ΔATP-WCE(WT) and ΔU6-WCE(WT) that stall at the Commitment Complex 2 (CC2) and A complexes, respectively (Supplementary Fig. 13). Interestingly, SiMCAn identified a time-dependent increase in clade I upon A complex formation (Supplementary Note 2). This low-FRET behavior has been proposed to be sustained upon incorporation of the U5·U4/U6 tri-snRNP during B complex formation10 (Fig. 1). In our corresponding ΔPrp2-WCE(WT) and ΔPrp2-WCE(3SS) datasets, conditions known to enrich the activated spliceosome Bact24,25, SiMCAn recognized a pair of static clusters, 0.25-S and 0.15-S, found to be overrepresented and thus grouped to form clade II (Figs. 4 and 5). These clusters represent molecules that are stalled in a static low-FRET Bact conformation prior to activation of Prp2’s ATPase activity and are similar to those previously determined5 using the isolated Bact complex lacking free extract (Fig. 3). Notably, SiMCAn was able to distinguish these clusters from the equally static, but even lower FRET cluster 0.05-S of the A complex, which is not resolvable in the FRET histograms (Supplementary Fig. 5). In addition to the static clusters of clade II, the dynamic cluster 0.05–0.25 (Supplementary Fig. 15a) is moderately enriched in these conditions relative to other conditions, suggesting that occasional excursions back into an A or B-like conformation occur.
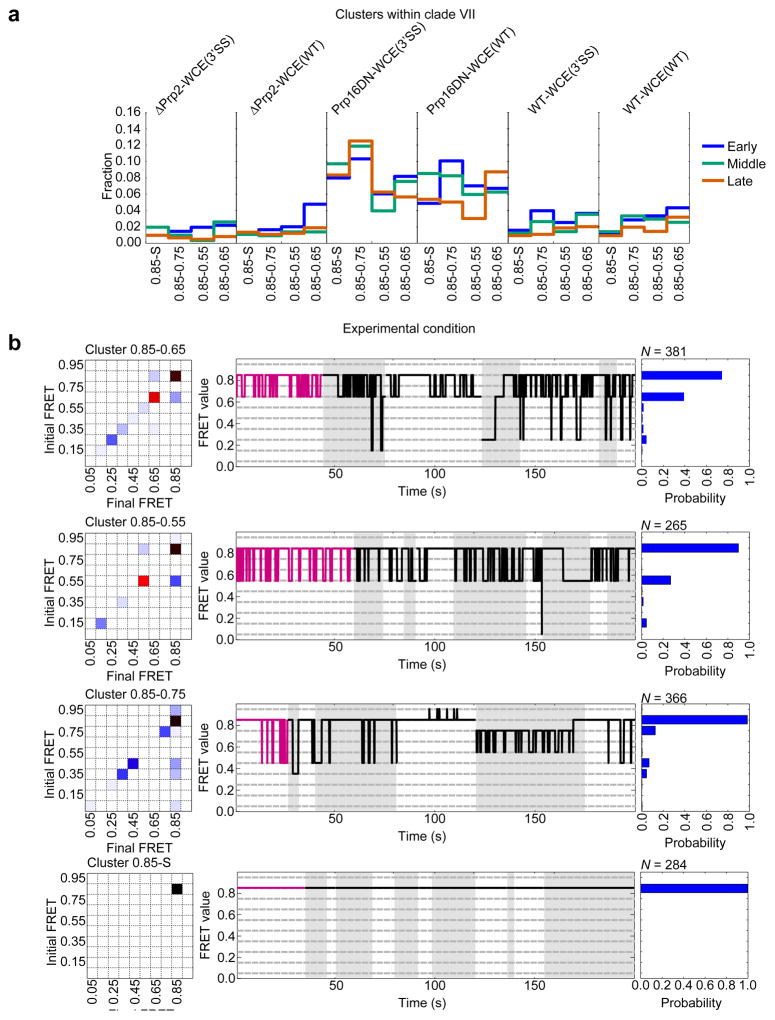
In contrast to Prp2 depletion, SiMCAn identified clade VII as particularly enriched upon addition of recombinant Prp16 dominant negative mutant ATPase (Prp16DN-WCE(WT) and Prp16DN-WCE(WT)), known to stall splicing within the post-first-step C complex5,26,27 (Figs. 4–5 and Supplementary Fig. 15b). Within this clade were a static cluster 0.85-S and three dynamic clusters, all containing the 0.85 FRET state (Fig. 6), which is distinct from the 0.75-S/0.65-S conformational state of clade VI enriched in early splicing intermediates (Supplementary Fig. 13). The dynamics of the clusters enriched at the Prp16DN stage indicate a preference for the 0.85 high-FRET state (Fig. 6b), suggesting we are enriching for and identifying molecules just before catalysis or transiently sampling the first catalytic conformation before proceeding to the 0.85-S cluster characteristic of molecules that have undergone first-step splicing. Although the ΔPrp2-WCE(3’SS) stall did show a delay in Bact complex formation (Supplementary Note 3), these observations suggest that only faithful spliceosome assembly leads to juxtaposition of the 5’SS and BP in a stable fashion, thus favoring first-step catalysis independent of the identity of the 3’SS28.
Figure 6.
Dynamic clusters of clade VII enriched in the Prp16DN-WCE conditions show repeated excursions from the 0.85 state to lower FRET states. (a) Fraction of molecules within each late assembly stage for the clusters of clade VII. (b) Cluster description for each of the four clusters within clade VII. Each representation shows the TP matrix of the cluster (left), the trace closest to the cluster center (magenta) and up to 200 s of random (black) traces from the cluster (middle), and the probability of FRET states within the cluster (right). Grey and white backgrounds demarcate individual trajectories.
A 3’SS mutant undocks late in spliceosome assembly
Finally, SiMCAn identified differences in smFRET behavior between the WT and 3’SS mutant substrates upon incubation with WT WCE containing no blocks (WT-WCE(WT) and WT-WCE(3’SS)), thus allowing for the unabated assembly towards the final step of splicing. The 3’SS mutant is known to assemble in a complex that includes the splicing factors responsible for the second step of catalysis, yet the 3’SS mutant is not amenable to splicing (Supplementary Fig. 4). Since both substrates progress through most of the splicing cycle, it is not surprising that SiMCAn revealed a similar set of pre-mRNA conformations sampled (Fig. 5). However, the 3’SS over time adopted an increasingly dominant 0.05-S cluster (Fig. 5, clade I), indicating a large separation of the 5’SS and BP not found in the Prp16DN-WCE(3’SS) dataset. This 0.05-S state is thus stabilized to a much greater extent in the 3’SS mutant than the WT substrate, supporting the appearance of a conformation in which the 5’SS and BP become greatly separated only after the first step of splicing when the mutated 3’SS is detected. Our data suggest that the 3’SS is either unable to dock into the catalytic core or is unable to remain docked in the catalytic core after the ATP-dependent action of Prp16. This deficiency in docking may be a result of second-step factors preventing docking into the second-step conformation29,30. Alternatively, this open conformation may be caused by Prp22, an ATPase known to be involved in proofreading mutant substrates during the second step of splicing (Supplementary Note 4)13,31 Taken together, our SiMCAn analysis generates the hypothesis that the lack of a proper 3’SS sequence marker leads to robust proofreading against a substrate not kinetically competent for the second step of splicing by undocking from the active site.
Discussion
We here have demonstrated the power of Single Molecule Cluster Analysis (SiMCAn) to reveal unique dynamic properties associated with specific splicing cycle intermediates that could not be identified using classical smFRET analysis (Supplementary Figs. 5, 6). Since SiMCAn does not make assumptions about the heterogeneity or completeness of the underlying biochemical reactions, it allows one to identify consistent molecular behaviors in model-free fashion (Supplementary Note 1). Through such unbiased and thorough analysis, we were able to assign dynamic FRET states to specific complexes, identify molecules transitioning between complexes, and demonstrate that the 5’SS and BP undock completely after the first step of splicing when the spliceosome encounters a 3’SS mutation (Fig. 5). SiMCAn thus can use exploratory datasets collected from complex reaction pathways to generate testable hypotheses, for example, that the spliceosome exploits similar undocked intermediates to proofread substrates along the splicing cycle, providing checkpoints that trap suboptimal substrates not meeting the criteria for cycle progression.
Single molecule FRET experiments provide a unique perspective into the dynamic behavior of complex reactions like splicing. Our experiments revealed a complex set of dynamic behaviors throughout the splicing cycle. SiMCAn was born of the necessity to classify common kinetic behaviors over a broad range of experimental states. Building hierarchical trees from disparate sets of data is the basis of most phylogenetic inference, and the methods presented here are inspired from evolutionary analysis32. The clades identified by SiMCAn allow us to define common subsets of relative dynamic behavior occurring at different biochemical blocks of the splicing cycle. Building on the phylogenetic analogy, the dynamic clades identified represent common kinetic pathways traversing the splicing cycle. We thus observed conserved pathways in the splicing cycle driven by a limited number of transitions. A limitation of investigating complex systems, such as the spliceosome, is that it does not allow us to unambiguously define conformations from FRET states. In a simpler system, like the P4-P6 subdomain of the T. thermophila group I intron, docking/undocking of the GNRA tetraloop could be assigned to specific FRET values, which enabled an unambiguous kinetic model to be developed19. Emerging approaches involving multiple probes such as the coincidence analysis of colocalization single-molecule spectroscopy (CoSMoS)33, combined with SiMCAn, are poised to resolve this ambiguity and facilitate the development of a complete kinetic model of the eukaryotic splicing cycle. Furthermore, as point detector-mediated photon counting becomes more high-throughput, these methods should introduce a substantial improvement in time resolution and allow a detailed description of shot-noise limited FRET efficiency distributions17. As single molecule techniques are applied to increasingly complex biochemical processes, SiMCAn is an approach that will make it possible to no longer limit the experimental strategy to one with a low number of states while still seeing the forest for the trees.
In summary, our results demonstrate that SiMCAn vastly improves the amount of information possible to extract from a large quantity of complex smFRET data. It is a powerful tool for the unbiased extraction of FRET states and kinetics from single molecule trajectories. By combining Hidden Markov Models with hierarchical clustering, we have utilized the strengths of both techniques to allow for the identification of biologically related dynamics. Beyond the identification of FRET states, SiMCAn helps distinguish molecules with similar FRET levels but differing rates of interconversion. By applying an additional layer of clustering based on the occupancy of behaviors across a systematic set of experimental conditions with known effects, we have created a tool for the identification of common and distinct behaviors among large numbers of single molecules. As such, SiMCAn can help generate hypotheses that drive focused experiments on isolated pathway intermediates. We anticipate that SiMCAn will be a powerful analysis tool that can be applied to any single molecule dataset, allowing for unprecedented in-depth analyses of the dynamics of complex biomolecular machines.
Online Methods
Synthesis of pre-mRNA substrates
The Ubc4 pre-mRNA substrates used in this study (Supplementary Table 3) were synthesized as previously described6. Briefly, the 135-nucleotide pre-mRNA was ligated from two fragments: a 59-nucleotide 3′ segment with 5-amino-allyl-uridine at the +6 position relative to the BP adenosine and a 76-nucleotide 5′ segment with 5-amino-allyl-uridine at the −7 position relative to the 5’SS. The 3’SS mutant had the guanines at positions 115 and 117 on the 3′ segment replaced with cytosines. The 5′ and 3′ fragments were coupled to Cy5 and Cy3 N-hydroxysuccinimidyl ester (GE Healthcare), respectively, by resuspending 4 nanomoles of RNA in 40 μl of 0.1 M sodium bicarbonate buffer, pH 9.0, and incubating for 30 min at 60 °C with the proper dye pack dissolved in DMSO. The conjugated fragments were ethanol precipitated and washed with 70% (v/v) ethanol to remove unconjugated dye. Unlabeled RNA was removed by purification on benzoylated naphthoylated DEAE (BND)-cellulose (Sigma) that was washed with 1 M NaCl containing 5% (v/v) ethanol. Fully labeled RNA fragments were eluted with 1.5 M NaCl containing 20% (v/v) ethanol and further precipitated to remove excess salt. Labeled fragments were combined with an equal molar amount of DNA splint (Supplementary Table 3) and ligated by incubating with RNA Ligase 1 (NEB) for 4 h at 37 °C as described6,16. Full length, labeled Ubc4 was then purified on a denaturing 7 M urea, 15% (w/v) polyacrylamide gel.
Preparation of yeast whole cell extract
Splicing active whole cell extract (WCE) was prepared from either yeast strain BJ2168 or a prp2-1 cef1-TAP yeast strain (ATCC 201388: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) as previously described6,34. Briefly, cells were grown in YPD medium to an OD600 of 1.6–2.0 before they were harvested and washed in AGK buffer (10 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 200 mM KCl, 10% (v/v) glycerol, 0.5 mM DTT, 0.6 mM PMSF, and 1.5 mM benzamidine). A thick slurry of cells was dripped into liquid nitrogen to form small cell pellets that could be stored at −80 °C. The frozen pellets were disrupted by manual grinding with a mortar and pestle half-submerged in liquid nitrogen for 30 min. The resulting frozen powder was thawed in an ice bath and centrifuged at 17,000 rpm in a type 45 Ti Beckman rotor. The supernatant was then centrifuged at 37,000 rpm in a Ti-70 rotor for 1 h. The clear middle layer was removed with a syringe and dialyzed for 4 h against 20 mM HEPES-KOH, pH 7.9, 0.2 mM EDTA, 0.5 mM DTT, 50 mM KCL, 20% (v/v) glycerol, 0.1 mM PMSF, and 0.25 mM benzamidine with one buffer exchange.
Accumulation of Splicing Complexes
Supplementary Table 1 describes all experimental conditions by identifying the substrate and WCE used along with the complex formed. All splicing products were confirmed via in vitro splicing assays by incubating 4 nM fluorescent Ubc4 in splicing buffer (8 mM HEPES-KOH, pH 7.0, 2 mM MgCl2, 0.08 mM EDTA, 60 mM Ki(PO4), 20 mM KCl, 8% (v/v) glycerol, 3% (w/v) PEG, 0.5 mM DTT) and 40% (v/v) WCE at 25 °C for 40 min. Products were analyzed by separation on a 7 M urea, 15% (w/v) polyacrylamide gel and scanned on a Typhoon variable mode imager (GE Healthcare, Supplementary Fig. 4). ATP depletion was performed by pre-incubating WCE with 1 mM glucose at 25 °C for 10 min prior to incubation with splicing buffer and substrate. Endogenous U6 snRNA was depleted by pre-incubation of WCE with 300 nM D1 oligodeoxynucleotide (Supplementary Table 3) in splicing buffer, 50% (v/v) WCE, and 2 mM ATP at 33 °C for 30 min prior to incubation with substrate. Knockdown of endogenous Prp2 was performed by heating prp2-1 cef1-TAP WCE to 37 °C for 40 min prior to incubation with splicing buffer, ATP, and pre-mRNA substrate. Endogenous Prp16 was inactivated using 100 nM of a Prp16 dominant-negative mutant (Prp16DN; K379A) added to the BJ2168 WCE for 10 min prior to incubation with splicing buffer, 2 mM ATP, and pre-mRNA substrate. On-slide splicing assays were performed as the in vitro splicing assays with the exception that all materials were combined prior to flowing reaction mixtures onto a substrate-coated, PEG-passivated slide using established procedures5,6.
Single Molecule FRET
Single Molecule FRET was carried out in the same manner as previously described5,6. Using a prism-based TIRF microscope8,35,36, we collected data from single molecules incubated under the desired conditions (Supplementary Table 1). Data were collected from two to three fields of view for each time period of 0–8 min (early), 18–23 min (middle), and 33–40 min (late) after addition of WCE. The donor (Cy3) near the BS adenosine was excited with a 532 nm laser for 100 seconds, followed by a direct excitation of the Cy5 acceptor near the 5’SS with a 635 nm laser for another 100 seconds, with the resulting emission recorded at 100 ms time resolution with a Princeton Instruments, I-PentaMAX intensified CCD camera. Molecules selected for further analysis by SiMCAn were required to last longer than 3 seconds before photobleaching of Cy3, show anti-correlated changes in Cy3 and Cy5 intensity, undergo single-step photobleaching, and still contain active Cy5 fluorophore at the time of its direct excitation. The FRET ratio was calculated by dividing the intensity of the acceptor emission by the total emission from both donor and acceptor. Each individual FRET trace was fitted with an individual Hidden Markov Model (HMM) with up to 10 states using vbFRET23 in Mathwork’s MATLAB environment with no assumptions about the values or distributions; in principle, any HMM-fitted trajectories can be used (generated by vbFRET, HaMMy, QuB, etc.)7. Regardless of the HMM software utilized, a certain degree of uncertainty in the number of FRET states and transitions among those states will be present in the data due to the noise associated with smFRET analysis. However, an improvement of HMM analysis techniques is not the focus of this manuscript.
Single molecule Cluster Analysis – SiMCAn
The HMM-idealized data were assigned to the closest of 10 evenly spaced FRET states (0.05–0.95, increment of 0.10 as our resolution limit). Traces of less than 3 s (30 frames) length were discarded and a transition probability (TP) matrix was constructed for each of the remaining molecule traces. Each TP matrix was then combined with the vector describing the percent of the trace that occupies each FRET state to create a FRET similarity matrix (FSM) where FSM[i,j] = [TP[i−1,j],P[n,j]] where i is from 1 to n+1 and j is from 1 to n. The FSMs were divided into categories containing static traces and dynamic traces, the dynamic traces identified and characterized by having at least one FRET transition between two FRET states. Static traces were identified automatically based on their unique signature with just a single FRET value and kept separate for the remaining analysis. Static molecules could arise due to fluorophores photobleaching prior to a transition taking place. Alternatively, formation of a particular complex may lead to a very stable, unchanging conformation that results in a single (static) FRET state. The FSMs corresponding to dynamic traces were used as input for a hierarchical clustering analysis performed by MATLAB that calculates the distance between FSMs using the Euclidean (ordinary) distance. The resulting hierarchical tree was then used to identify clusters of traces with similar behavior as identified from their FSM. The tree was pruned at a height that resulted in 25 dynamic clusters in addition to 10 static clusters as assigned by their FRET state. The height used to determine the clusters in the hierarchical tree was determined using an iterative measurement of the inter-cluster distances and a modified k-means algorithm. The specific cut-off was chosen at the first point where randomly assigned traces had a higher inter-cluster distance than the hierarchical clustering, which provided the best option among several for determining an optimal cluster selection. The resulting clusters were analyzed and labeled according to their occupancy in the FRET states. All analysis and descriptions of the clusters were performed using MATLAB. For each experimental condition, the fraction of molecules within each SiMCAn identified cluster was computed by dividing the number of molecules of that condition assigned to each cluster by the total number of molecules in that condition. The occupancy within all the clusters was used as a new similarity matrix to compute the distance between each SiMCAn cluster using Euclidean distance measurement. Clades were generated by the iterative k-means approach used in SiMCAn, with the aim to generate groups of clusters whose occupancy patterns across conditions are most alike (as measured by Euclidean distance).
Generation of the Simulated Datasets
Artificial HMMs containing the distinctions of interest were used to generate traces of 106 time step length for each of four clusters. These traces were used to generate 1,500 subtraces where the starting points were uniformly selected along the full trace and the length determined by a Poisson distribution with a lambda of 100. The resulting traces were treated exactly like experimentally acquired data fit by vbFRET for analysis by SiMCAn.
Code availability
Custom MATLAB code for SiMCAn analysis and figure generation is available upon request.
Acknowledgments
The authors wish to thank A. Price (University of California, San Francisco) for providing native gel analysis of CC2 formation using Ubc4; D.R. Semlow and J.P. Staley (University of Chicago, Illinois) for providing the dominant negative Prp16 protein expression plasmid; Nguyen N. (Josh) Vo for compiling all MATLAB scripts of SiMCAn into a GUI; as well as C. Guthrie, D.R. Semlow, J.P. Staley and A.A. Hoskins for providing valuable comments on the manuscript. The authors acknowledge funding from US National Institutes of Health grant R01GM098023 to N.G.W. and J.A., National Heart, Lung, and Blood Institute R01HL111527-01 to A.L., and the National Science Foundation through the National Evolutionary Synthesis Center (NESCent) grant NSF\#EF-0905606 to J.M.
Abbreviations
- SiMCAn
Single Molecule Cluster Analysis
Footnotes
Author Contributions
M.L.K. and R.K. performed the in vitro splicing verification assays. M.L.K. and M.R.B. performed the single molecule experiments and performed data analysis. M.L.K. expressed and purified the Prp16DN protein. M.R.B. and J.S.M. wrote and developed the MATLAB scripts for SiMCAn. M.L.K. prepared all fluorescent substrates and yeast whole cell extracts. M.R.B., J.S.M., M.L.K., J.A., A.L., and N.G.W jointly wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Pitchiaya S, Heinicke LA, Custer TC, Walter NG. Single molecule fluorescence approaches shed light on intracellular RNAs. Chem Rev. 2014;114:3224–3265. doi: 10.1021/cr400496q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustoe AM, Brooks CL, Al-Hashimi HM. Hierarchy of RNA functional dynamics. Annu Rev Biochem. 2014;83:441–466. doi: 10.1146/annurev-biochem-060713-035524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Krishnan R, et al. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nat Struct Mol Biol. 2013;20:1450–1457. doi: 10.1038/nsmb.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abelson J, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco M, Walter NG. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010;472:153–178. doi: 10.1016/S0076-6879(10)72011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter NG, Bustamante C. Introduction to single molecule imaging and mechanics: seeing and touching molecules one at a time. Chem Rev. 2014;114:3069–3071. doi: 10.1021/cr500059w. [DOI] [PubMed] [Google Scholar]
- 10.Crawford DJ, et al. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. Proc Natl Acad Sci USA. 2013;110:6783–6788. doi: 10.1073/pnas.1219305110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brody E, Abelson J. The “spliceosome”: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- 12.Egecioglu DE, Chanfreau G. Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control. RNA. 2011;17:383–389. doi: 10.1261/rna.2454711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci. 2012;37:263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 15.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Abelson J, Hadjivassiliou H, Guthrie C. Preparation of fluorescent pre-mRNA substrates for an smFRET study of pre-mRNA splicing in yeast. Methods Enzymol. 2010;472:31–40. doi: 10.1016/S0076-6879(10)72017-6. [DOI] [PubMed] [Google Scholar]
- 17.Gopich IV, Szabo A. Theory of the energy transfer efficiency and fluorescence lifetime distribution in single-molecule FRET. Proc Natl Acad Sci U S A. 2012;109:7747–7752. doi: 10.1073/pnas.1205120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller BG, et al. Complex RNA folding kinetics revealed by single-molecule FRET and hidden Markov models. J Am Chem Soc. 2014;136:4534–4543. doi: 10.1021/ja4098719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenfeld M, Pavlichin DS, Mabuchi H, Herschlag D. Single Molecule Analysis Research Tool (SMART): an integrated approach for analyzing single molecule data. PLoS One. 2012;7:e30024. doi: 10.1371/journal.pone.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruno AE, et al. Comparing chemistry to outcome: the development of a chemical distance metric, coupled with clustering and hierarchal visualization applied to macromolecular crystallography. PLoS One. 2014;9:e100782. doi: 10.1371/journal.pone.0100782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mall R, Langone R, Suykens JA. Multilevel hierarchical kernel spectral clustering for real-life large scale complex networks. PLoS One. 2014;9:e99966. doi: 10.1371/journal.pone.0099966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J Royal Stat Soc Ser B. 2001;63:411–423. [Google Scholar]
- 23.Bronson JE, et al. Learning Rates and States from Biophysical Time Series: A Bayesian Approach to Model Selection and Single-Molecule FRET Data. Biophys J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SH, Lin RJ. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warkocki Z, et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 26.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider S, Hotz HR, Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J Biol Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- 28.Rymond BC, Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 29.Ohrt T, et al. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA. 2013;19:902–915. doi: 10.1261/rna.039024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umen JG, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 31.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens SW, Abelson J. Yeast pre-mRNA splicing: methods, mechanisms, and machinery. Methods Enzymol. 2002;351:200–220. doi: 10.1016/s0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- 35.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widom JR, Dhakal S, Heinicke LA, Walter NG. Single-molecule tools for enzymology, structural biology, systems biology and nanotechnology: an update. Arch Toxicol. 2014;88:1965–1985. doi: 10.1007/s00204-014-1357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]