Abstract
The cellular process responsible for providing energy for most life on Earth, namely photosynthetic light-harvesting, requires the cooperation of hundreds of proteins across an organelle, involving length and time scales spanning several orders of magnitude over quantum and classical regimes. Simulation and visualization of this fundamental energy conversion process pose many unique methodological and computational challenges. We present, in two accompanying movies, light-harvesting in the photosynthetic apparatus found in purple bacteria, the so-called chromatophore. The movies are the culmination of three decades of modeling efforts, featuring the collaboration of theoretical, experimental, and computational scientists. We describe the techniques that were used to build, simulate, analyze, and visualize the structures shown in the movies, and we highlight cases where scientific needs spurred the development of new parallel algorithms that efficiently harness GPU accelerators and petascale computers.
Keywords: Photosynthesis, Parallel molecular dynamics, Parallel ray tracing, GPU Computing
1. Introduction
Solar energy, directly or indirectly, powers almost all life on Earth through photosynthesis. Nature has evolved devices that utilize quantum mechanics for conversion of light energy into stable chemical energy, which can be stored and consumed by living cells to power physiological processes [1]. Studies of natural light-harvesting systems provide insights for the engineering of bio-hybrid and artificial solar devices and may contribute substantially to the current 15 TW power demand of our civilization by better harvesting Earth’s 120,000 TW average solar irradiance [2].
Light-harvesting in biological cells poses substantial computational challenges, since it involves many overlapping time and length scales, ranging from electronic excitation transfer on a picosecond timescale [3] to diffusion events on a millisecond timescale [4]. In the case of purple photosynthetic bacteria, the photosynthetic apparatus is the so-called chromatophore, now structurally known at atomic detail [5, 4]; the chromatophore is a spherical membrane of 50 nm inner diameter, comprising over a hundred proteins and ~3,000 bacteriochlorophylls for photon absorption [6] (see Fig. 1). The study of the chromatophore and its component proteins has been the subject of simulation efforts for three decades, driving software development and new methodologies in collaboration with experimental studies (see Sec. 3 and Table 1 below).
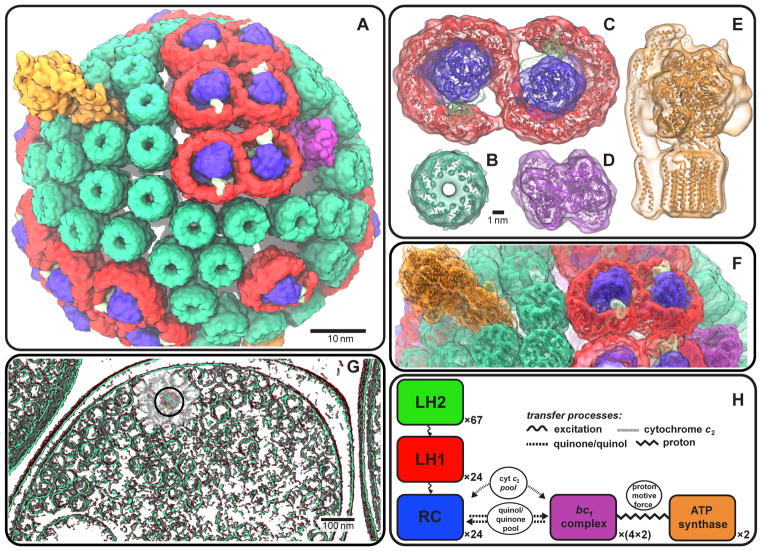
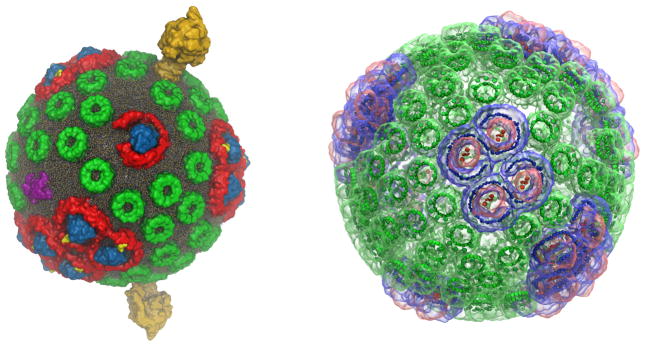
Figure 1.
The chromatophore from the purple bacterium Rba. sphaeroides (A) harvests solar energy for ATP production via a network of hundreds of cooperating proteins [4] (see accompanying movies). In order of energy utilization, these proteins are: the LH2 (B) and LH1-RC-PufX (C) complexes for light capture, electronic excitation transfer, and charge separation; the bc1 complex (D) for generation of a proton gradient across the vesicle membrane; and ATP synthase (E) for using the resultant proton gradient for ATP production (B, C, D: top view, i.e. normal to the membrane; E: side view). A minimal subunit of the four aforementioned primary protein groups is shown in (F) (proteins in cartoon representation; same colors as in B-E). The chromatophores densely populate the cell under low-light conditions, occasionally forming vesicular connections with one another (G) (the chromatophore shown in (A) is indicated by a circle). The energy conversion processes are summarized schematically in (H) (see Sec. 2 for details; image reproduced from [8]).
Table 1.
Representative Computational Milestones. (See reviews [6, 3] for a comprehensive list.) First author given for publications.
| System and Computational Achievement | Publication |
|---|---|
| magnetic field dependence of primary photochemical reactions in the RC | Werner, 1978 [22] |
| time varying electrostatic properties of the RC | Treutlein, 1988 [23] |
| structure solution of the LH2 complex | Koepke, 1996 [24] |
| excitation transfer in a photosynthetic unit consisting of LH2 & LH1-RC complexes | Ritz, 1998 [25] |
| dynamic disorder/thermal effects on excitonic properties in LH2 based on polaron models | Damjanović, 2002 [26] |
| structural modeling/excitonic properties of a chromatophore containing LH2 & LH1-RC | Sener, 2007 [5] |
| open quantum dynamics/thermal disorder in LH1 and LH2 based on hierarchy equations | Strümpfer, 2012 [27] |
| simulation of a 20M atom lamellar chromatophore | Chandler, 2014 [14] |
| integration of energy and electron transfer processes across a spherical chromatophore | Cartron, 2014 [4] |
The combination of experimental imaging, state-of-the-art molecular dynamics simulation, and high-fidelity visualization provides researchers with a powerful “computational microscope” that permits viewing of the dynamics of molecular processes with atomic detail. The simulation, analysis, and visualization of large photosynthetic membranes are replete with computational challenges due to the large size of these systems, the necessity for both quantum and classical mechanical modeling approaches, and due to the variety of timescales involved in individual molecular processes. The high performance provided by GPU-accelerated petascale computing platforms allows high quality visualization and rendering techniques to be routinely employed for the study of large photosynthetic systems such as the chromatophore. Below, we discuss recent VMD [7] developments and performance enhancements that enabled the visualizations described herein, and outline future technological opportunities we plan to pursue.
We extend our previous light harvesting visualization work presented in the Supercomputing 2014 Visualization Showcase (SC14) [8] with a second more recent short-form movie that includes additional chromatophore structure and simulation data, and with new discussion of challenges posed by other photosynthetic systems currently under study. In the two accompanying movies, all the primary energy conversion events in the chromatophore, from light absorption to ATP synthesis, are shown in a contiguous structural narrative that seamlessly connects cell-scale organization to atomic-scale function. The full length long-form movie1 presented at SC14 shows the individual steps of photosynthetic energy conversion in the chromatophore at the detail of individual electronic processes [8]. A more recent short-form movie2 displays a molecular dynamics simulation based on the structural model displayed in the first movie [8, 4] and adds visualization details such as lipids and their dynamics.
The structural models and supra-molecular organization shown in the movies were experimentally determined by atomic force microscopy, cryo-electron microscopy, electron tomography, crystallography, optical spectroscopy, mass spectroscopy, and proteomics data (see [4] and references therein). The movies represent molecular dynamics (MD) simulation and modeling of the entire chromatophore as well as of its constituent protein complexes made possible by petascale systems such as Blue Waters, demonstrating, for the first time, the complete physiological sequence of a basic photosynthetic apparatus.
In the following, first, the energy conversion processes in the chromatophore are outlined as illustrated in the movies; second, the computational challenges and representative milestones for modeling the chromatophore function are recounted; last, the visualization techniques are discussed that enable the rendering of protein function across multiple scales.
2. Visualization of Chromatophore Function
The accompanying movies present a narrative of the photosynthetic function of the chromatophore of purple bacteria as a clockwork of interlocked processes for the purpose of energy utilization, culminating in ATP synthesis, as described in [4]. Purple bacteria experience, in their habitat, often low illumination levels, such as ~1% of full sunlight, and have developed an adaptation to light starvation by overpopulating the cell interior with chromatophores (Fig. 1G). The primary step of light-absorption is carried out by pigments (bacteriochlorophylls and carotenoids) contained in the light harvesting complexes 1 and 2 (LH1 and LH2) that form a network delivering the absorbed light energy in the form of electronic excitation energy to the reaction center (RC) [9]. The RC initiates a sequence of charge transfer events, ultimately transferring two electrons and two protons to a mobile charge carrier, quinone, converting it to quinol. The quinol diffuses through the chromatophore membrane to the bc1-complex, which processes quinols for generating a proton gradient across the membrane [10]. The soluble charge carrier cytochrome c2 returns electrons back from the bc1-complex to the RC, thereby completing a circuit. The proton gradient drives ATP synthesis at the ATP synthase [11]. (See Fig. 1H for a schematic of the energy conversion processes.)
3. Computational Challenges
The simulation of chromatophore components shown in the movies has been the subject of nearly 40 publications by the senior author over the last three decades [6] (see Table 1), driving development efforts for simulation and visualization software packages NAMD [12] and VMD [7]. The chromatophore model shown in Fig. 1A was developed using VMD [13] by combining crystallographic structures of constituent proteins with supramolecular organization data [4]. Molecular dynamics (MD) simulation of a spherical chromatophore as shown in Fig. 1A contains approximately 100 million atoms, including lipids and water. A lamellar chromatophore patch containing 20 million atoms has already been simulated [14], determining excitation transfer properties of the pigment network. Current simulation efforts involve chromatophore models that contain not only the primary light harvesting complexes (LH2 and LH1-RC), but also bc1 and ATP synthase complexes [4] responsible for energy conversion, as well as diffusible charge carriers [14].
A 100 million-atom NAMD simulation was specified by the National Science Foundation in 2006 as a petascale target application, and later the chromatophore (based on a model comprising only LH2 and LH1-RC complexes [5]) specifically was made an acceptance test for the Blue Waters supercomputer, launching a multi-year effort both to prepare the model and to improve NAMD performance and parallel scaling. The extreme size of the chromatophore structure to be modeled required the development of new molecular structure and trajectory file formats, new model building tools and algorithms, techniques for parallelization of existing analysis scripts, and new visualization techniques and rendering approaches within VMD [15, 16, 17]. At SC11, Mei et al. [18] showed the utility of memory usage optimization and multithreading in NAMD to scale the 100 million atom simulation to the full Jaguar PF Cray XT5 (224,076 cores). Building on that work, at SC12 Sun et al. [19] explored techniques for improving fine-grained communication on the Cray Gemini network, demonstrating scaling to all 300,000 cores of ORNL Titan (before GPUs were installed on all nodes). Finally, at SC14 Phillips et al. [20] described the mapping of NAMD algorithms and data structures to toroidal network topologies, efficiently scaling simulation of a 224-million-atom system to 16,384 nodes of both Blue Waters and ORNL Titan (with GPUs).
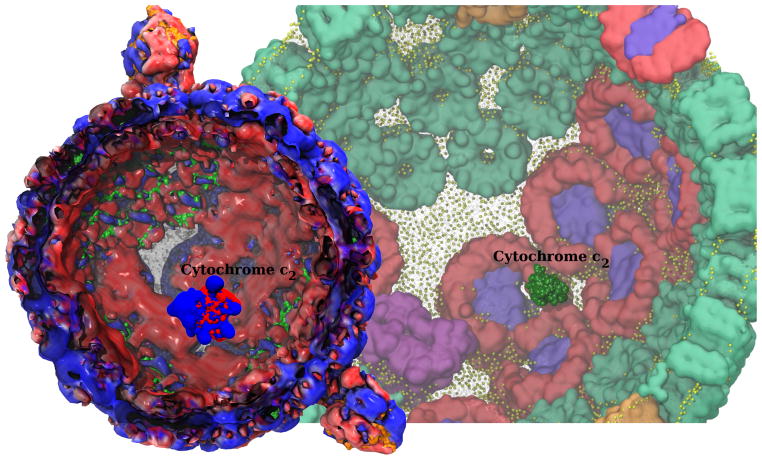
The chromatophore has now been simulated for 100 ns on ORNL Titan. Electrostatic potentials derived from the all-atom structure, employing the Adaptive Poisson Boltzmann Solver (APBS) [21], provide unforeseen insights into the synergy between the structural arrangement of the light harvesting proteins and their respective functions. Figure 2 presents a visualization of the electrostatic potential isosurfaces in concert with the underlying atomic structure, leading to the discovery of regular binding patterns on the extremely heterogeneous chromatophore surface. Indeed, these patterns are recognized by transport proteins for their optimal activity. The visualization of such patterns was highly memory intensive since every trajectory frame contains 100-million atoms and volumetric potential maps totaling 10-million grid points. In the near future, the pathways of protein transport spanning the entire chromatophore will be simulated and visualized to capture, for the very first time in atomic detail, a light-absorbing organelle in action.
Figure 2.
A snapshot of single electron carrier cytochrome c2 diffusing within the inner cavity of the chromatophore, transporting charge between the bc1 complex and LH1, visualized in all-atom structural detail. The electrostatic potential characterizing the chromatophore inner surface shows red (negative) patches that provide binding sites for cytochrome c2 blue (positive) binding surface.
4. Visualization Techniques
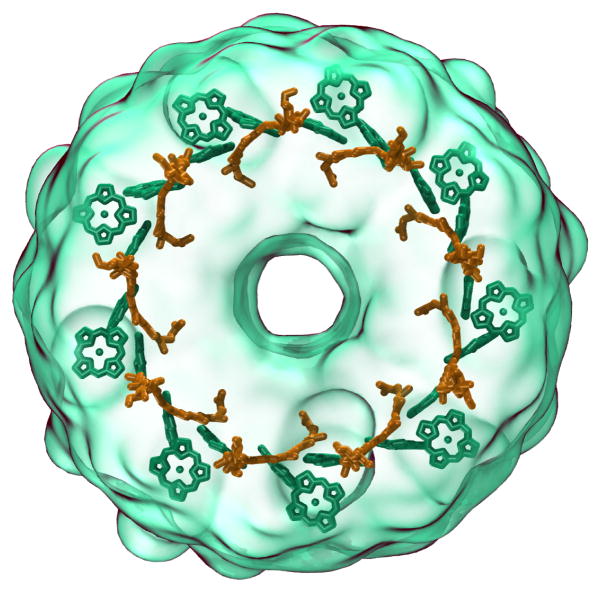
All of the molecular structure and cellular tomography renderings in the movies were produced by VMD [7], using combinations of many common molecular graphics structural representation techniques such as ball-stick, ribbon, secondary structure, and surface representations, but on a much larger-scale biomolecular complex than is typically visualized by such methods [15, 16, 17]. The fast GPU-accelerated “QuickSurf” molecular surface representation in VMD [28] was used extensively throughout the movies, enabling emphasis to be placed on overall chromatophore architecture or on atomic detail as needed in different contexts. In many of the movie clips, QuickSurf representations were used in combination with variants of the VMD “GlassBubble” material (a set of shading parameters) with a viewing-angle-modulated transparency effect that yields surfaces that are entirely transparent when surface normals align with the view direction, varying smoothly and becoming fully-opaque when the boundary of the surface is seen edge-on. The transparent materials were used to combine transparent QuickSurf representations that illustrate overall architecture and context with secondary structure and ribbon representations that emphasize the protein backbone of key components, as shown in Fig. 3, and Fig. 1, in panels B, C, D and E.
Figure 3.
Use of surface representations with angle-modulated transparency allows structure details to be viewed clearly while retaining context, exemplified in this rendering of an LH2 complex (outer transparent green surface) with its interior chlorophyll (green) and carotenoid (orange) structures exhibiting mutually orthogonal orientations.
The movies are composed of a series of short clips and transitions that were produced using the VMD ViewChangeRender plugin, hereafter referred to as “VCR”. The VCR plugin allows researchers to create a list of visualization “viewpoints” that combine the camera view orientation, the molecular representations and their settings, and the associated simulation trajectory frame or other time series data. Individual movie clips are generated using previously defined viewpoints and associated transition times that allow the VCR plugin to interpolate viewing orientations, graphical representation properties, and simulation timestep indices as it renders individual frames. The VCR plugin provided the production team with live dry-run movie visualization using the VMD OpenGL renderer, and final production rendering was performed using the built-in GPU-accelerated ray tracing engine described below.
The complexity of the chromatophore movie required that the VCR plugin be extended significantly beyond its original capabilities to enable a large team of researchers to work together to create movie content, render movie clips, and assemble clips into complete movies for final editing. Support for the script-driven definition of movie clips, 3-D structure animations, and complex scene transitions was added, along with export of a human-readable annotated “edit list” for use in the final video editing process. The VCR plugin was also extended to allow rendering of large sequences of clips and to support batch mode parallel rendering on petascale computing systems, using built-in VMD scripting interfaces for parallel execution and load balancing on distributed memory parallel computers [16, 29].
5. GPU-Accelerated Ray Tracing Engine
High quality image rendering was performed using a new GPU-accelerated variant of the Tachyon ray tracing (RT) engine that is built into VMD [30, 16, 17], based on the CUDA GPU computing toolkit and the OptiX RT framework [31]. Embedded RT engines allow VMD to efficiently render molecular geometry in-situ directly from in-memory data structures without disk I/O [32], while enabling the use of high quality rendering techniques such as shadow filtering and ambient occlusion lighting [17]. The built-in RT engines have been significantly extended with new rendering capabilities including depth-of-field focal blur effects, improved algorithms for stochastic sampling, single-pass stereoscopic rendering, and several new camera projection modes that support image and movie renderings for display in planetariums and panoramic theaters.
The photosynthesis visualizations described here have been selected for future inclusion in digital film productions to be shown in a variety of large scale immersive 360° panoramic displays such as planetariums and full-dome theaters. The task of producing high quality renderings of molecular visualizations for non-planar displays poses a variety of technical problems for conventional rasterization-based image generation approaches, but it is aptly suited to ray tracing [33, 34]. To facilitate routine development and previewing of stereoscopic molecular visualizations for planetariums and panoramic theaters, the RT engines were also extended with support for rendering of omnidirectional stereoscopic images and movies that support viewing with head mounted displays (HMDs) such as Oculus Rift, Google Cardboard, and others (see Fig. 4). Stereoscopic rendering and depth-of-field were implemented conventionally for common planar camera projection modes, but they require special handling when implemented for panoramic or omnidirectional projections. Depth-of-field focal blur was implemented for non-planar projections using a spherical virtual focal surface and a radial focal distance. Support for stereoscopic rendering of panoramic and omnidirectional projections was implemented using an adaptation of the circular projection technique for stereoscopic photography described by Peleg and Ben-Ezra [35]. Our stereoscopic rendering approach employs a spherical eye convergence surface; stereoscopic eye positions are calculated independently for every primary ray or image pixel by computing eye positions as offsets orthogonal to the primary ray direction. The stereoscopic eye separation distance is modulated by the cosine of the angle from the equator of the spherical projection, yielding no stereoscopic effect at the zenith and nadir (the two poles), where the spherical projection comes to a point, thereby preventing inadvertent viewing with backward stereo near the poles. To facilitate panoramic viewing deep within the interiors of largely or fully enclosed molecular structures, the RT engine was extended by adding support for a special omnidirectional positional light that always tracks the location of the camera, and a modification of ambient occlusion lighting that ignores shadows from objects beyond a user-specified maximum occlusion distance. The RT workload associated with panoramic or omnidirectional rendering is much more demanding than for conventional perspective or orthographic projections since the fan of primary rays subtends a very large or omnidirectional field of view, since primary and secondary rays will traverse a much larger fraction of the scene geometry than conventional planar projections do.
Figure 4.
Panoramic ray tracing of the entire chromatophore structure from within its interior. Ray tracing is aptly suited to 360° panoramic and omnidirectional camera models needed for rendering of movies for projection in planetariums and fulldome theaters, and for viewing in head mounted displays such as Oculus Rift and Google Cardboard. The images shown are a panoramic planetarium dome master projection (top left), an omnidirectional equirectangular (latitude-longitude) spherical projection (top right), and an omnidirectional cube map projection (bottom).
The combined effects of the large size and geometric complexity of many of the scenes in the chromatophore movies presented significant computational challenges, even in the context of parallel rendering runs on a large supercomputer such as Blue Waters. Overall rendering performance was increased significantly by optimizing several of the performance critical components of the TachyonL-OptiX RT engine. The previous TachyonL-OptiX implementation used a single code path for all shading and surface material properties [17]. At runtime, OptiX combines application-specific RT code written in CUDA with OptiX-provided code for building and traversing spatial acceleration data structures, and for RT work distribution and load balancing [31]. The final result of runtime code generation is a so-called “megakernel” that implements the complete RT engine from all of the parts. By splitting the shading and material processing code into a set of 64 case-specific shaders using the combinatorial expansion of a set of 6 shading features as CUDA C++ template parameters, the rendering performance is significantly improved on average due to elimination of unnecessary code, by reduced branching, and by reduced GPU register consumption.
All previous VMD TachyonL-OptiX implementations used CPU-based RT acceleration structure (AS) builder algorithms that could handle large molecular geometry effectively [36, 37]. The Cray XK7 compute nodes use moderate clock rate 16-core AMD Opteron 6276 CPUs that require careful multithreading for best performance. The CPU-based OptiX AS builders are not optimized for the particular characteristics of the Opteron 6276 in the Cray XK7. Ray tracing performance tests on Blue Waters showed that the CPU-based AS builders that yielded the highest RT performance often took longer to run than the RT phase itself, negating their benefits for movies with time-varying scenes and high geometric complexity, such as the chromatophore movies. In the past, the TachyonL-OptiX RT engine would therefore select the “MedianBvh” CPU-based AS builder as the best default compromise between AS build time, GPU memory usage, and RT performance. Recent OptiX releases include a high performance GPU-accelerated “Trbvh” AS builder that now provides fast AS build times as well as high performance during the RT phase [38]. The Trbvh AS implementation in OptiX 3.8 is robust for very large molecular scenes, and is now the best performing AS builder on the Cray XK7 nodes when GPU memory capacity is not a limitation. In cases when the memory requirement for a molecular scene and its AS would exceed GPU memory the TachyonL-OptiX engine reverts to the “MedianBvh” CPU-based AS builder. Using this approach, VMD is capable of rendering scenes with over 171 million triangles, while achieving interactive RT display rates, as described below.
Performance was increased further by reusing the same OptiX context for the life of the VMD process, thereby eliminating some GPU management overhead, albeit at the cost of increased code complexity, since VMD must manage competing uses of the relatively small on-board GPU memory for RT, surface generation, and a variety of molecular structure analysis tasks. Table 2 compares the performance of the newest TachyonL-OptiX GPU RT engine with the previous implementation, rendering a 64M-atom HIV-1 movie sequence on Blue Waters [17]. For movies rendered at high-definition resolutions (1920 × 1080) or higher, the overall performance increased by up to a factor of 1.46×, greatly reducing the node-hour rendering cost of such movies and making so-called “4K” movies (3840×2160 resolution) and “8K” movies (7680 × 4320 resolution) more practical going forward.
Table 2.
Comparison of VMD parallel movie rendering performance for a 1,079 frame HIV-1 test movie on BlueWaters, after implementing several new optimizations within the TachyonL-OptiX GPU-accelerated ray tracing engine. For reference, the best previously published performance results are included for both GPU and CPU rendering [17]. Repeated timings reveal overall movie rendering performance gains ranging from a factor of 1.2× faster for presentation-sized movies (not shown in table) up to as much as 1.46× faster for high-definition 1920 × 1080 movies.
| Movie | Ray Tracing Engine | Nodes | Full-Movie Wall Clock Execution Time
|
|||
|---|---|---|---|---|---|---|
| Script Load | State Load | Geometry and Rendering | Total | |||
|
| ||||||
| HIV-1 Capsid HD 1920 × 1080 144 AO samples/pixel |
New TachyonL-OptiX (GPU) VMD 1.9.3 |
64 XK7 | 2 s | 39 s | 435 s | 476 s |
| 128 XK7 | 3 s | 62 s | 230 s | 295 s | ||
|
| ||||||
| TachyonL-OptiX (GPU) VMD 1.9.2-UltraVis’13 [17] |
64 XK7 | 2 s | 38 s | 655 s | 695 s | |
| 128 XK7 | 4 s | 74 s | 331 s | 410 s | ||
| 256 XK7 | 7 s | 110 s | 171 s | 288 s | ||
|
| ||||||
| Tachyon (CPU) VMD 1.9.2-UltraVis’13 [17] |
256 XE6 | 7 s | 160 s | 1,374 s | 1,541 s | |
| 512 XE6 | 13 s | 211 s | 808 s | 1,032 s | ||
6. Interactive Ray Tracing with Progressive Refinement
During the development of the movies, it became clear that a fully interactive RT engine for previewing visualizations with the same lighting and material shading as the final rendering would be tremendously helpful to researchers producing movie content. One of the most significant recent advances to VMD is a fully interactive version of the TachyonL-OptiX RT engine that uses a progressive refinement approach. The progressive RT engine decomposes the rendering algorithm into an iterative stochastic sampling process that combines both high interactivity with high quality rendering, enabling fully interactive viewing and manipulation of the scene, and adjustment of important rendering parameters such as the lighting and shadowing settings, depth-of-field aperture and focal point distance, and stereoscopic display modes.
The progressive renderer operates by using an accumulation buffer to gather stochastic samples, and drawing a renormalized copy of the latest accumulation buffer contents to the display as rendering continues. When the user changes the view orientation, window size, or other parameters that invalidate the previously accumulated samples, the accumulation buffer is cleared and progressive rendering restarts anew. The interactive display performance of the progressive renderer is dominated by the number of pixels that must accumulate samples during each rendering pass. The renderer computes one or more stochastic samples per pixel per rendering pass, depending on the complexity of the scene. By default, the progressive renderer uses a moving window average to estimate the current display frame rate, and it adjusts the number of samples per rendering pass up or down to try and maintain a display rate of 30 frames per second. If the user prefers more rapid image convergence over interactivity or vice-versa, the auto-tuning mechanism can be disabled and the user can set the desired behavior. When rendering direct-to-HMD, the auto-tuning mechanism adjusts for peak display update rate to prevent motion sickness associated with HMD redraw latency. VMD has achieved direct-to-HMD rendering rates limited by the HMD display hardware (75 frames per second on Oculus Rift DK2) for moderate complexity scenes containing on the order of one million atoms, with direct lighting and a small number of ambient occlusion lighting samples. The default RT parameters in VMD lead to progressive RT display rates that are up to 150× faster than the time required for a complete batch mode rendering of the same image. An image that would require 10 seconds of rendering time in batch mode can be rendered interactively using progressive RT at roughly 15 frames per second. To achieve the same final image quality as in batch rendering, progressive RT requires slightly more runtime due to intermediate display updates, but the benefits of full interactivity when making scene adjustments are difficult to overstate.
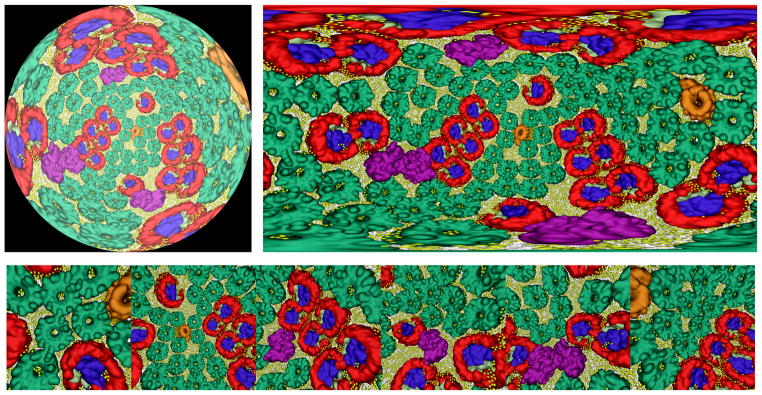
The performance of a single GPU can be insufficient for smooth interactive rendering of geometrically large scenes with complex shading requirements such as the full chromatophore model shown with membrane lipids, extensive use of transparent surfaces, and ambient occlusion lighting, as in Fig 1(f). In such cases, VMD’s TachyonL-OptiX RT engine exploits multiple GPUs within the same workstation or compute node to achieve higher rendering rates. For cases that greatly exceed the capabilities of a multi-GPU workstation or compute node, the TachyonL-OptiX supports the use of remote visualization clusters through a new set of progressive rendering APIs introduced in OptiX 3.8. Using this feature, VMD sends molecular scene geometry to the remote visualization cluster, and the TachyonL-OptiX progressive renderer is run on a visualization cluster composed of NVIDIA Visual Computing Appliance (VCA) nodes that are tightly coupled through InfiniBand. Intermediate images are streamed live from the visualization cluster back to the VMD client application using H.264 video compression, enabling remote visualization frame rates as high as 24 frames per second, even for trans-continental network traversals. The VCA nodes can be configured by VMD to use their eight GPUs to optimize the display update frame rate, or to optimize the rate of stochastic sampling and progressive image convergence. The frame-rate-optimized VCA configuration doubles image-space parallelism, halving the number of stochastic samples per display update. The image-convergence-optimized VCA configuration doubles the number of stochastic samples per display update, and halves image-space parallelism. The use of a remote 16-node (128-GPU) VCA visualization cluster has enabled interactive RT to be used for even the most challenging chromatophore visualizations. When using large VCA node counts in an image-convergence-optimized mode, the RT engine accumulates up to 64 times as many stochastic samples per rendering pass as it would with a single GPU, typically resulting in a fully converged image within a half-second to one second, even for geometrically highly complex scenes as shown in Figs. 5 and 6.
Figure 5.
Chromatophore scenes used to evaluate interactive ray tracing performance using a 16-node (128 GPU) NVIDIA VCA remote visualization cluster. The chromatophore scene on the left shows lipids in atomic detail, yielding a scene comprised of opaque geometry containing 1.6M spheres, 3.3M cylinders, and 3.2M triangles (8.1M objects in total). The scene on the left can be rendered at 1920 × 1080 resolution at up to 30 frames per second using a single VCA node (8 GPUs). The chromatophore scene on the right highlights light harvesting complexes and pigments combining outer transparent molecular surface representations and opaque inner surfaces, comprised of roughly 5M triangles in total. The scene on the right is extremely challenging for ray tracing due to the large number of secondary transmission rays generated due to the predominantly transparent geometry. The scene on the right can be rendered at 1024 × 1024 resolution at frame rates up to 5 frames per second using one VCA node. Both scenes benefit from greatly increased parallelism for stochastic sampling. The use of 16 VCA nodes (128 GPUs) can produce image quality levels in under one second of interactive display that would require roughly two minutes of batch rendering on a single GPU.
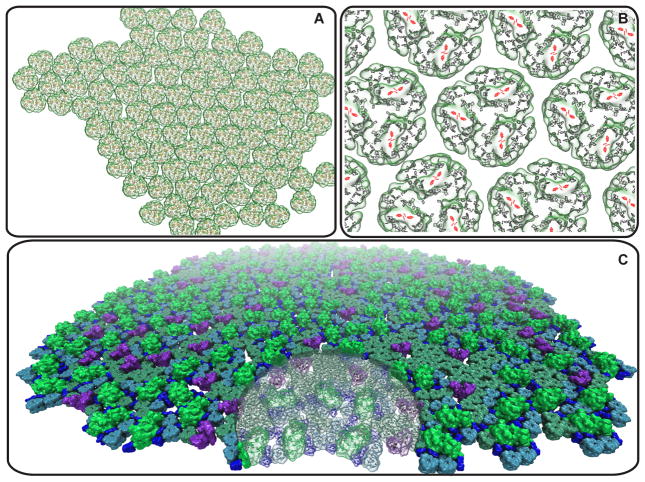
Figure 6.
Future challenges for the simulation and visualization of photosynthetic membranes. (A) Cyanobacterial membrane domain comprising 96 trimeric photosystem I complexes, containing a total of 27,648 chlorophylls, corresponding to a light harvesting capacity of about 10 chromatophores like those shown in Fig. 1. (B) Close-up of photosystem I trimers within the cyanobacterial domain shown in (A) illustrating the quasi-periodic packing pattern of the protein complexes; chlorophylls are highlighted as porphyrin rings with the reaction center shown in red. (C) Plant thylakoid membrane domain comprising 164 photosystem II dimers (green), 58 cytochrome b6 f dimers (purple), 991 LHC2 trimers (cyan and lime), 402 of which are within a supercomplex with photosystem II, and 902 CP29/CP26/CP24 complexes (blue), also in supercomplex with photosystem II. As a size comparison, the transparent disk shaped region has the same radius as the chromatophore in Fig. 1; the thylakoid domain shown has the light-harvesting capacity of about 20 chromatophores.
The interactive RT engine has been extremely valuable as a tool to help researchers that are not rendering experts to produce high quality images and movies, making it much easier to fine tune camera, lighting, and rendering parameters for complex molecular scenes. We have recently begun to use the interactive RT engine in conjunction with HMDs as a way of previewing material intended for display in planetariums and other panoramic theater venues. The task of creating effective visualizations for panoramic displays is not as intuitive as it is for conventional planar projections. Previewing with HMDs eliminates much of the need to obtain access to a real theater for evaluating visualization revisions. One limitation of the remote rendering approach is that the network latencies associated with video streaming are far too high for effective use of direct-to-HMD rendering. We will address this limitation in the future by combining progressive RT on remote GPU clusters with video streaming of omnidirectional projections, and local OpenGL rasterization for low-latency view-dependent reprojection on HMDs.
7. Parallel Production Rendering and Post Production Video Editing
Historically, MD simulation trajectories were generated at supercomputer centers and then transferred to the researcher’s home lab, where they were ultimately analyzed. However, petascale MD simulations generate terabytes of simulation trajectory output that must be analyzed, and the sheer size of such data is creating a shift in visualization and analysis practice, leading us to adapt our software to perform analytical and visualization tasks on the supercomputer where the data is generated, thereby avoiding days or weeks of off-site data transfer [16, 17, 13].
All of the VMD parallel rendering work required for production of the present chromatophore movies was performed on BlueWaters, using the Cray XK7 GPU-accelerated compute nodes which are operating in a fully graphics-enabled “GPU operation mode”. Fully graphics-enabled GPUs on Blue Waters allow the use of OpenGL for rapid turnaround of full-length preview visualizations [39]. The built-in TachyonL-OptiX GPU RT engine was used for production renderings [31, 17]. Movie frames were rendered in 16:9 aspect ratio at HD 1920 × 1080 resolution, with 12 antialiasing samples per pixel, 144 ambient occlusion shadow feeler rays per pixel, with direct lighting contributions and shadows from two directional lights; transmission rays and shadow filtering were performed for transparent geometry. One complete parallel rendering of the movie frames for the long-form movie using 96 GPU-accelerated Cray XK7 compute nodes consumed ~290 node hours with a wall-clock turnaround time of 3 hours, and produced ~7,500 frames which used ~45 GB of disk space.
Non-linear video editing was employed to compose the complete set of rendered VMD movie clips with hand-drawn figures, transitions, captions and annotations, and other materials. Cross-fade scene transitions were designed in the VMD VCR plugin using the live dry-run visualization mode, but rather than being rendered by brute force within VMD itself, the “edit list” exported by the VMD VCR plugin was used to guide generation of the transitions within the video editing software. Several clips in the movies utilize animated sequences produced within Final Cut Pro and Motion 5 to illustrate excitation migration between pigment clusters in modified Förster formalism, involving rapid delocalization and thermal equilibration of excitonic states within one LH-protein prior to transfer to neighboring proteins [5, 3, 27]. The animated illustrations were composited with VMD imagery through the use of multiple renderings, transparency, and depth layering. The final movies were exported in both 16:9 and cropped 4:3 aspect ratios for presentation on diverse display hardware.
8. Conclusions and Future Work
A complete description of the energy harvesting and conversion processes in a photosynthetic pseudo-organelle is achieved for the first time through a combination of petascale simulations, theoretical modeling, and experimental collaborations, visualized in two comprehensive movies. Both the modeling and visualization tasks currently utilize the capabilities of the BlueWaters petascale computer. The movies described here required significant post production effort. A goal for future VMD development will be to provide improved mechanisms for adding arbitrary illustrative graphics to movies intended for planetariums or panoramic theaters and for stereoscopic head mounted displays, none of which are well supported by mainstream commercial video editing tools.
Simulation and visualization of cellular processes pose software and hardware challenges due to system size and complexity. Beyond the photosynthetic apparatus employed by the bacterial chromatophore, shown in Fig. 1 and the accompanying movies, the cyanobacterial and plant photosynthetic systems shown in Fig. 6 exhibit a greater diversity of proteins, a higher pigment density, and a larger surface area per functional domain when compared with the chromatophore. Future simulations will describe structure and dynamics of ever larger cellular machinery up to a whole cell and will require exascale computing, along with new simulation concepts and algorithms.
Acknowledgments
The authors acknowledge support from NSF grants MCB1157615 and PHY0822613, NIH grants 9P41GM104601 and 5R01GM098243-02, the CUDA Center of Excellence at the University of Illinois, the NCSA AVL and the CADENS project supported in part by NSF award ACI-1445176, the Blue Waters sustained-petascale computing project funded by NSF awards OCI-0725070 and ACI-1238993 and the state of Illinois, “The Computational Microscope” NSF PRAC awards OCI-0832673 and ACI-1440026, and the Oak Ridge Leadership Computing Facility at Oak Ridge National Laboratory supported by the Office of Science of the Department of Energy under Contract DEAC05- 00OR22725 (K. S.), the Biotechnology and Biological Research Council, UK, award number BB/M000265/1 (C. N. H., M. P. J.), the European Research Council, Advanced Award 338895 (C. N. H), the Leverhulme Trust, the Krebs Institute at the University of Sheffield and Project Sunshine, University of Sheffield (M. P. J), the Alexander von Humboldt Foundation for a postdoctoral fellowship (L. F. K.), and the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC 0001035 (K. S., C. N. H., C. M-C.). Cryo-electron microscopy was performed under the guidance of H. Engelhardt, J. Plitzko, W. Baumeister and with support from J. Lubieniecki, A, Rigort at the MPI of Biochemistry, Martinsried, Germany (L. F. K.).
Footnotes
References
- 1.Blankenship RE. Molecular Mechanisms of Photosynthesis. 2. Wiley Blackwell; 2014. [Google Scholar]
- 2.Blankenship RE, Tiede DM, Barber J, Brudvig GW, Fleming G, Ghirardi M, Gunner MR, Junge W, Kramer DM, Melis A, Moore TA, Moser CC, Nocera DG, Nozik AJ, Ort DR, Parson WW, Prince RC, Sayre RT. Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science. 2011;332(6031):805–809. doi: 10.1126/science.1200165. [DOI] [PubMed] [Google Scholar]
- 3.Sener M, Strümpfer J, Hsin J, Chandler D, Scheuring S, Hunter CN, Schulten K. Förster energy transfer theory as reflected in the structures of photosynthetic light harvesting systems. ChemPhysChem. 2011;12:518–531. doi: 10.1002/cphc.201000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartron ML, Olsen JD, Sener M, Jackson PJ, Brindley AA, Qian P, Dickman MJ, Leggett GJ, Schulten K, Hunter CN. Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides. Biochim Biophys Acta – Bioener. 2014;1837:1769–1780. doi: 10.1016/j.bbabio.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sener MK, Olsen JD, Hunter CN, Schulten K. Atomic level structural and functional model of a bacterial photosynthetic membrane vesicle. Proc Natl Acad Sci USA. 2007;104:15723–15728. doi: 10.1073/pnas.0706861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strümpfer J, Hsin J, Sener M, Chandler D, Schulten K. The light-harvesting apparatus in purple photosynthetic bacteria, introduction to a quantum biological device. In: Roux B, editor. Molecular Machines. Ch 2. World Scientific Press; 2011. pp. 19–48. [Google Scholar]
- 7.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 8.Sener M, Stone JE, Barragan A, Singharoy A, Teo I, Vandivort KL, Isralewitz B, Liu B, Goh BC, Phillips JC, Kourkoutis LF, Hunter CN, Schulten K. Visualization of energy conversion processes in a light harvesting organelle at atomic detail. Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, SC ’14; 2014.IEEE Press; [Google Scholar]
- 9.Strümpfer J, Schulten K. Excited state dynamics in photosynthetic reaction center and light harvesting complex 1. J Chem Phys. 2012;137:065101, 8. doi: 10.1063/1.4738953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barragan AM, Crofts AR, Schulten K, Solov’yov IA. Identification of ubiquinol binding motifs at the Qo-site of the cytochrome bc1 complex. J Phys Chem B. 2015;119:433–447. doi: 10.1021/jp510022w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinekathoefer U, Isralewitz B, Dittrich M, Schulten K. Domain motion of individual F1-ATPase β-subunits during unbiased molecular dynamics simulations. J Phys Chem. 2011;115:7267–7274. doi: 10.1021/jp2005088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone JE, McGreevy R, Isralewitz B, Schulten K. GPU-accelerated analysis and visualization of large structures solved by molecular dynamics flexible fitting. Faraday Discuss. 2014;169:265–283. doi: 10.1039/C4FD00005F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler D, Strümpfer J, Sener M, Scheuring S, Schulten K. Light harvesting by lamellar chromatophores in Rhodospirillum photometricum. Biophys J. 2014;106:2503–2510. doi: 10.1016/j.bpj.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stone JE, Vandivort KL, Schulten K. Immersive out-of-core visualization of large-size and long-timescale molecular dynamics trajectories. Lect Notes in Comp Sci. 2011;6939:1–12. [Google Scholar]
- 16.Stone JE, Isralewitz B, Schulten K. Early experiences scaling VMD molecular visualization and analysis jobs on BlueWaters. Extreme Scaling Workshop (XSW); 2013; 2013. pp. 43–50. [DOI] [Google Scholar]
- 17.Stone JE, Vandivort KL, Schulten K. GPU-accelerated molecular visualization on petascale supercomputing platforms. Proceedings of the 8th International Workshop on Ultrascale Visualization, UltraVis ’13; New York, NY, USA: ACM; 2013. pp. 6:1–6:8. [Google Scholar]
- 18.Mei C, Sun Y, Zheng G, Bohm EJ, Kalé LV, Phillips JC, Harrison C. Enabling and scaling biomolecular simulations of 100 million atoms on petascale machines with a multicore-optimized message-driven runtime. Proceedings of the 2011 ACM/IEEE conference on Supercomputing; Seattle, WA. 2011; pp. 61:1–61:11. [Google Scholar]
- 19.Sun Y, Zheng G, Mei C, Bohm EJ, Jones T, Kalé LV, Phillips JC. Optimizing fine-grained communication in a biomolecular simulation application on Cray XK6. Proceedings of the 2012 ACM/IEEE conference on Supercomputing; Salt Lake City, Utah. 2012; IEEE press; pp. 1–11. [Google Scholar]
- 20.Phillips JC, Sun Y, Jain N, Bohm EJ, Kalé LV. Mapping to irregular torus topologies and other techniques for petascale biomolecular simulation. Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, SC ’14; IEEE Press; 2014. pp. 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner H-J, Schulten K, Weller A. Electron transfer and spin exchange contributing to the magnetic field dependence of the primary photochemical reaction of bacterial photosynthesis. Biochim Biophys Acta. 1978;502:255–268. doi: 10.1016/0005-2728(78)90047-6. [DOI] [PubMed] [Google Scholar]
- 23.Treutlein H, Schulten K, Deisenhofer J, Michel H, Brünger A, Karplus M. Molecular dynamics simulation of the primary processes in the photosynthetic reaction center of Rhodopseudomonas viridis. In: Breton J, Verméglio A, editors. The Photosynthetic Bacterial Reaction Center: Structure and Dynamics, Vol. 149 of NATO Sci. Ser. A. Plenum; New York: 1988. pp. 139–150. [Google Scholar]
- 24.Koepke J, Hu X, Muenke C, Schulten K, Michel H. The crystal structure of the light harvesting complex II (B800-850) from Rhodospirillum molischianum. Structure. 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 25.Ritz T, Hu X, Damjanović A, Schulten K. Excitons and excitation transfer in the photosynthetic unit of purple bacteria. J Luminesc. 1998;76–77:310–321. [Google Scholar]
- 26.Damjanović A, Kosztin I, Kleinekathoefer U, Schulten K. Excitons in a photosynthetic light-harvesting system: A combined molecular dynamics, quantum chemistry and polaron model study. Phys Rev E. 2002;65:031919, 24. doi: 10.1103/PhysRevE.65.031919. [DOI] [PubMed] [Google Scholar]
- 27.Strümpfer J, Schulten K. Open quantum dynamics calculations with the hierarchy equations of motion on parallel computers. J Chem Theor Comp. 2012;8:2808–2816. doi: 10.1021/ct3003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krone M, Stone JE, Ertl T, Schulten K. Fast visualization of Gaussian density surfaces for molecular dynamics and particle system trajectories. EuroVis - Short Papers 2012. 2012:67–71. [Google Scholar]
- 29.Phillips JC, Stone JE, Vandivort KL, Armstrong TG, Wozniak JM, Wilde M, Schulten K. Petascale Tcl with NAMD, VMD, and Swift/T. SC’14 workshop on High Performance Technical Computing in Dynamic Languages, SC ’14; IEEE Press; 2014. pp. 6–17. [Google Scholar]
- 30.Stone JE. Master’s thesis. Computer Science Department, University of Missouri-Rolla; Apr, 1998. An Efficient Library for Parallel Ray Tracing and Animation. [Google Scholar]
- 31.Parker SG, Bigler J, Dietrich A, Friedrich H, Hoberock J, Luebke D, McAllister D, McGuire M, Morley K, Robison A, Stich M. OptiX: a general purpose ray tracing engine. ACM SIGGRAPH 2010 papers, SIGGRAPH ’10; New York, NY, USA: ACM; 2010. pp. 66:1–66:13. [Google Scholar]
- 32.Stone J, Underwood M. Rendering of numerical flow simulations using MPI. Second MPI Developer’s Conference, IEEE Computer Society Technical Committee on Distributed Processing; IEEE Computer Society Press; 1996. pp. 138–141. [Google Scholar]
- 33.Max NL. ATOMLLL: ATOMS with shading and highlights. SIGGRAPH Comput Graph. 1979;13(2):165–173. [Google Scholar]
- 34.Max NL. Computer graphics distortion for IMAX and OMNIMAX projection. Nicograph ’83 Proceedings. 1983:137–159. [Google Scholar]
- 35.Peleg S, Ben-Ezra M. Stereo panorama with a single camera. Computer Vision and Pattern Recognition, 1999. IEEE Computer Society Conference on; 1999. p. 401. [Google Scholar]
- 36.Stich M, Friedrich H, Dietrich A. Spatial splits in bounding volume hierarchies. Proceedings of the Conference on High Performance Graphics 2009, HPG ’09; New York, NY, USA: ACM; 2009. pp. 7–13. [Google Scholar]
- 37.Garanzha K, Pantaleoni J, McAllister D. Simpler and faster HLBVH with work queues. Proceedings of the ACM SIGGRAPH Symposium on High Performance Graphics, HPG ’11; New York, NY, USA: ACM; 2011. pp. 59–64. [Google Scholar]
- 38.Karras T, Aila T. Fast parallel construction of high-quality bounding volume hierarchies. Proceedings of the 5th High-Performance Graphics Conference, HPG ’13; New York, NY, USA: ACM; 2013. pp. 89–99. [Google Scholar]
- 39.Klein MD, Stone JE. Unlocking the full potential of the Cray XK7 accelerator. Cray User Group Conf., Cray; 2014. [Google Scholar]