Abstract
Human papillomavirus (HPV) occurs in many types, some of which cause cervical, genital, and other cancers. While vaccination is available against the major cancer-causing HPV types, many others are not covered by these preventive measures. Herein, we present a bioinformatics study for the designing of multivalent peptide vaccines against multiple HPV types as an alternative strategy to the virus-like particle vaccines being used now. Our technique of rational design of peptide vaccines is expected to ensure stability of the vaccine against many cycles of mutational changes, elicit immune response, and negate autoimmune possibilities. Using the L1 capsid protein sequences, we identified several peptides for potential vaccine design for HPV 16, 18, 33, 35, 45, and 11 types. Although there are concerns about the epitope-binding affinities for the peptides identified in this process, the technique indicates possibilities of multivalent, adjuvanted, peptide vaccines against a wider range of HPV types, and tailor-made different combinations of the peptides to address frequency variations of types over different population groups as required for prophylaxis and at lower cost than are in use at the present time.
Keywords: human papillomavirus, rational design of peptide vaccines, graphical representation, protein graph radius, multivalent peptide vaccines, continuous-discontinuous epitopes
Introduction
Certain types of cancers such as liver cancer, cervical cancer, Hodgkin’s lymphoma, and others are caused by viruses, for some of which vaccines are well established. Vaccines operate by triggering immune response in an individual. It has been shown that under some circumstances immunocompetent individuals can protect themselves against primary tumor development.1 It is also established that while a healthy immune system can act as a tumor suppressor, in other cases it might facilitate tumor progression.2 Nevertheless, given the wide variety of viral cancers and progression of the disease, using the immune system to prevent cancer through antiviral vaccines could be a reasonable procedure. Some viral cancers such as those caused by human papillomavirus (HPV) and hepatitis B viruses have been effectively prevented through such techniques.
Papillomaviruses affect mammalian, avian, and reptilian hosts. Over 170 types of HPVs have been identified.3 The large variety of HPV types is believed to have evolved from the large-scale migration of human populations and appears to follow their major migration patterns.4 The virus is generally spread by sexual activities and can lie dormant for years before symptoms develop. In the majority of cases, the virus causes warts on the body, but a small fraction of the infections develops into cancers. It is estimated that over 27,000 people develop HPV cancers every year in the USA, usually detected at a fairly late stage5; an analysis of all cancers worldwide in 2002 revealed that of about 1.9 million cases of cancer attributable to infections, around 561,200 were due to HPVs,6 with an incidence ratio of 2:7 between numbers in the developed and developing countries.
The wide varieties of HPVs are causative agents for various types of cancers and other diseases. Of the 40 types of the HPVs that affect genital areas,7 HPV types 16, 18, 33, 35, and 45 are believed to be the primary agents, with HPV16 and HPV18 identified as being responsible for about 70% of such cancers. One study conducted among a large sample of patients with histologically confirmed squamous cell cervical cancers8 determined 15 HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82) classified as high risk, three (26, 53, and 66) as probable high risk, and several others (6, 11, 40, 42, 43, 44, 54, 61, 70, 72, 81) as low-risk types. Some HPV types also cause oropharyngeal cancers: 90% of HPV-positive oropharyngeal cancers are attributable to HPV16,8,9 and HPV6 and HPV11 have also been determined to cause such cancers10; these findings are important since HPV-positive oropharyngeal squamous cell cancers (OPSCCs) are reported to have doubled in the period 1984–2004, whereas HPV-negative OPSCCs decreased by 50% while overall 18% of oral and oropharyngeal cancers worldwide are HPV associated.10 Other types of HPVs are known to cause various ailments: HPV6 and 11 cause recurrent respiratory papillomatosis,11 HPV types 2, 27, and 57 can cause skin warts, while HPV 1, 2, 27, and 63 were found on clinically normal skin.12 Moreover, the proportion of various types of HPV varies across countries and continents, and thus affects the outcome of standard preventive medications.8
HPV is a DNA virus with a genome length of about 8000 base pairs comprising nine segments that code for eight proteins and one long control region. There are basically six early proteins and two late proteins, namely, L1 and L2, which are outer capsid proteins, at approximately 95:5 ratio. The L1 proteins form a pentameric assembly on the virion shell, the individual proteins being held together by disulfide bonds; these proteins form the basis for vaccines against papillomaviruses. Unlike the RNA viruses that lack proofreading mechanism, DNA viruses are capable of correcting replication errors and are much more stable to mutational changes than their RNA counterparts. Thus, DNA viruses are more conserved, and vaccines are more effective over a longer period.
The direct linkage of some types of HPV to cervical cancers has led to much research on prophylactic vaccination against the most common HPV types that are causative agents for cervical cancers.13 Antiviral vaccines have been developed against the most common of the HPV types that cause these cancers, viz., HPV16 and HPV18; Gardasil (Merck & Co., Inc.) protects against high-risk HPV types 16 and 18 and low-risk types 6 and 11; Cervarix (GSK) protects against HPV types 16 and 18 through an AS04 adjuvanted recombinant L1 protein vaccine, which was found to also protect against HPV types 31 and 45.13 The efficacy of these vaccines is very high but limited to only the selected HPVs due to type specificity, inhibiting their preventive action against a wide range of other HPV-related cancers3; the two vaccines leave out a coverage of 20%–30% of the HPV-associated cancer-causing types. This situation has been mitigated to some extent by the launch of a 9-valent Gardasil vaccine (9vHPV) that protects against HPV types covered by the older quadrivalent HPV vaccine (HPV6, 11, 16, and 18) and five additional oncogenic types (HPV31, 33, 45, 52, and 58).14 A controlled test determined that the 9vHPV was noninferior to the previous quadrivalent HPV in preventing infections and generating antibodies against the particular types of HPVs it was designed against, but was not effective against other types.14
The HPV vaccines belong to the class of recombinant virus-like particle (VLP) vaccines, which are self-assembling bionanoparticles that mimic the structure of the original virus particle exposing multiple epitopes on the surface. They do not carry any genetic material, thus providing a safer alternative to live-attenuated or inactivated vaccines. The first VLP vaccine license was granted in 1986 for the hepatitis B virus by the US Food and Drug Administration,15 and VLP vaccines for two more viruses, such as HPV and hepatitis E, have secured licenses. The fact that over the last 30 years only three such vaccines have been marketed while many new and re-emerging viruses have been recognized indicates how difficult and expensive it must be to develop VLP vaccines. Although the VLP vaccines have proved very effective in practice, they are generally beyond the reach of patients in developing countries, and also the nature of the VLP vaccine can inhibit attempts at personalized medication, which may be required in certain population groups.16
The availability of genomic data, understanding of immune responses and immunogenetic variations, new developments in bioinformatics and computer applications, and systems biology approach are fashioning new directions in vaccine development, away from the traditional techniques of live-attenuated or inactivated virus or recombinant VLP vaccines. This is a move away from the one-size-fits-all concept of mass-applicable vaccines to one of personalized vaccines where the vaccines, ideally, can be readily designed to be individual, gender-specific, or race/community/population-specific and keep costs in control.16,17 This strategy is admittedly geared at the moment to hypervariable viruses such as human immunodeficiency virus (HIV), human coronavirus (HCV), influenza, and others, to which group the HPV does not belong, but the principles certainly can apply. Rational design of vaccines based on genomic and immunogenomic information and the science of reverse vaccinology, as yet an evolving technology, could be a pointer to the future.
At the immediate level, peptide vaccines may be such an alternative strategy to HPV VLP vaccines that could be explored. The idea is to scan the viral genome for the protein antigens that can elicit immune response and then synthesize them into a peptide vaccine. A more focused approach is to precisely locate the epitope regions within these antigens and utilize them to elicit the immune response.18 The recent advancements in the technological and bioinformatics fields enable computer-based approaches for this purpose. Synthetic peptide vaccines, which are just beginning to draw wide interest at this time, are easier to construct, the bioinformatics approaches lead to shorter development times, such vaccines can be tailor-made to suit individual applications and avoid autoimmune threats, and overall are cheaper to design and market.18 It is interesting to note that the US National Institute of Health projects portal, ClinicalTrials.gov (https://clinicaltrials.gov/), reports, as of mid April 2016, 562 projects under trial for peptide vaccines, 106 projects for VLP vaccines, and 74 projects for synthetic peptide vaccines.
We have carried out computer-based studies to identify target regions in surface proteins of influenza and rotavirus.19–21 Our interest in applying these techniques to HPV is to see whether it is feasible to design peptide vaccines against the disease that afflicts many more people in the developing nations where the economic burden is much more acute than in the developed nations. There is also the advantage with peptide vaccines that, in theory, multivalent vaccines can be more easily designed and marketed at more affordable prices than multivalent VLP vaccines against a wide range of HPV types. This makes it possible to tailor-make different combinations of the peptides to address frequency variations of types over different population groups and therefore reach a wider percentage of affected people to suit individual communities in those countries where the populations are higher and much less homogeneous than in the Western nations.
Materials and Methods
Database
In this work, L1 being the major component of the capsid assembly and the focus of HPV vaccine development, we restrict our approach to consideration of the L1 protein only and postpone consideration of the other capsid assembly component, the L2 protein, to a future work. We have downloaded 222 sequences of the L1 capsid gene of various HPV types from GenBank, updated till March 22, 2016. Table 1 lists the number of sequences of the different HPV types forming the dataset we have used for the purposes of the present exercise. Annotated HPV data are also available through the specialized Papillomavirus Episteme database maintained by the National Institute for Allergic and Infectious Diseases. Given the high incidence of papilloma-virus infections, one could expect more data to accumulate over time.
Table 1.
Number of HPV complete L1 sequences used in our work.
| HPV TYPE | NO OF SEQS |
|---|---|
| Type 11 | 17 |
| Type 16 | 102 |
| Type 18 | 28 |
| Type 33 | 7 |
| Type 35 | 8 |
| Type 45 | 60 |
| Total | 222 |
Note: Downloaded from GenBank; updated till March 22, 2016.
Method of analysis
The method of analysis has been described in detail in our previous papers, especially in the study by Ghosh et al.20 Initially, we scan a library of sequences of the designated protein to determine segments that are unchanged or least changed among the various strains, and then, couple these with average solvent accessibility (ASA) profiles of the sequences to select those segments that are most conserved and having highest solvent accessibility profile. We then determine epitope regions within these segments with acceptable binding affinity to human leukocyte antigens and finally select those peptides that poses no autoimmune threat; in the process, the 3D crystal structure of the surface protein is also utilized to ensure that the selected peptide regions are not covered against solvent accessibility due to neighboring proteins that together constitute the quaternary structure.
As an optional step, the process may start with observations from graphical representations of DNA/RNA sequences in a 2D Cartesian coordinate system to determine similarities between the selected gene sequences or conserved segments within the selected sequences. In this system, a sequence is represented by a point for each nucleotide in succession plotted in a series of steps with one step in the negative x-direction for an adenine, positive y-direction for a cytosine, in the positive x-direction for a guanine, and in the negative y-direction for a thymine.22 This generates a plot of the sequence that reflects the base distribution in the sequence. For a quantitative measure, we take the weighted center of mass (µx, µy) of the points and the distance of the centre of mass (c.m.) from the center as a graph radius gR, which is considered as the characteristic of the sequence.23 For protein sequences, this concept is extended to a rectangular coordinate system of 20 dimensions, where each amino acid (aa) is associated with a particular direction and the protein sequence is plotted by taking successive steps in the directions dictated by the aa sequence.24 The plot is in abstract space, but we can again define a weighted center of mass and a protein graph radius, pR, just as for gR. The special property of gR and pR is that in each case when two sequences result in the same graph radius, albeit with some adjustments for pR as specified in the study by Ghosh et al.20, the sequences are found to be identical.25 This property has been used in several instances previously19,20,26 and is used in the current work to determine protein sequence segment varieties by determining the number of instances where the pR’s are identical (see more details below).
To determine the target peptides that can be considered to elicit immune response, the steps in the process are, briefly:
Global view of L1 gene sequences to determine similar and dissimilar sequences
2D graphical representation plots are generated for the L1 gene sequences of different HPV types and visually inspected to determine those that appear similar and can be clubbed together for analysis.
Identification of protein conserved segments
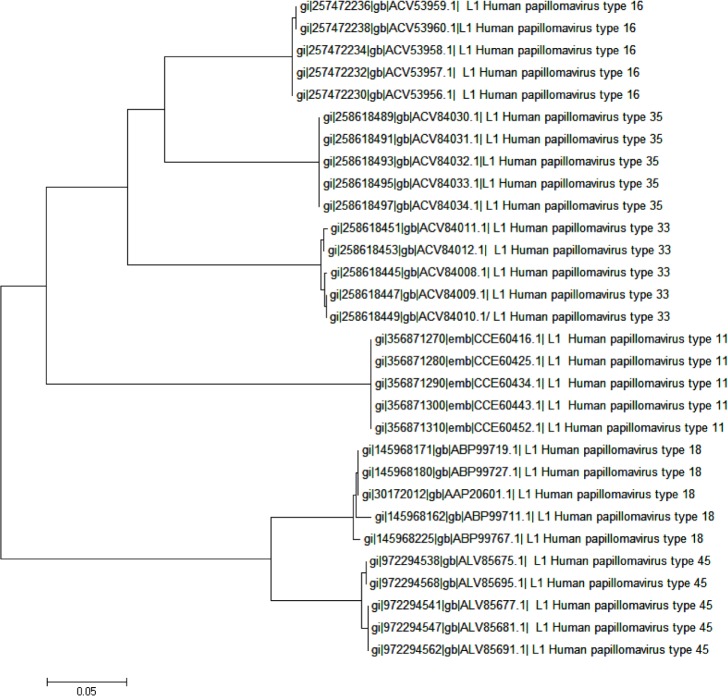
Viruses generally have high mutation rates, more for RNA viruses than for DNA viruses. The mutation rates for the dsDNA HPVs are very low, but nevertheless, mutations do occur leading to the various strains that populate the papillomavirus database – higher differences between types and smaller differences within each type; a phylogenetic tree of the protein sequences of the primary HPV types we are interested in this work, with five strains from each type, brings out this feature explicitly (Fig. 1). Now, given the strict type specificity of the neutralizing antibodies and the mutations in aa composition, deletions, and insertions that may take place,27 this implies that in specific cases neutralizing antibodies raised against one strain may not be viable against another strain, although the success of the current HPV vaccines show that this may happen seldom. Our interest is to identify those peptide segments that change the least between strains to reduce the possibility of such instances and examine whether one or more of such segments could form the basis for peptide vaccines.
Figure 1.
Phylogenetic tree of the protein sequences of five strains each of the six HPV types 11, 16, 18, 33, 35, and 45. Neighbor-joining method used, software MEGA5.22.
The methodology we follow for the purpose is to map the protein sequence using a window of 12–14 aas and sliding in increments of one aa at a time; ie, compute pR for the segment with aa nos. 1–12, say, for a chosen window size of 12 aa, then another pR for the stretch between aa nos. 2–13 and so on, until the end of the aa sequence is reached. At each stage, the peptide graph radius, pR, is computed and stored (see Refs. 19 and 20 for details). This is then repeated for each protein sequence of the selected group of sequences and then the entire lot is scanned at each mapped aa position to determine the number of different pR-values, among all the sequences. Since each pR value represents a particular peptide sequence, the number of values of the pR at each aa position map out a protein variety profile of the aas covered by the sliding window. The minima of the profile indicate regions where variety, and by inference changes in aas, are relatively least, indicating most conserved regions.
The size of the window covering a number of aas needs careful consideration. In our previous analysis of the neuraminidase protein,19 we had considered peptide lengths of 8, 10, and 12 aas; for rotavirus where mutational changes occur very frequently, it was considered prudent to consider window sizes of 12–14 aas20; 14 residue peptides were found to be more potent antigenically compared with smaller peptides,28 and we had used that size for the analysis of influenza hemagglutinin.21 It may be noted that while peptide lengths of 6–20 aas are used in scanning for B-cell epitopes, peptides of 10+ residues may contain overlapping linear B-cell epitopes29; cytotoxic T-cell epitopes have a limited length (8–11 residues), whereas for helper T-cells, longer peptide lengths are used.30 Considering these issues and the fact that the HPV viruses are dsDNA type, which therefore are comparatively more stable, we fixed window length for this exercise at 12 aas.
Solvent accessibility profile
The protein sequences are next mapped for solvent accessibility by subjecting them to an ASA server such as ITASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) or SABLE (http://sable.cchmc.org/) or any other suitable web-based server. Taking an average over 12 aas of solvent accessibility index at each aa position, and averaging over several sequences for each HPV type, this is mapped out to reveal the most solvent accessible regions by the maxima of the graph.
First cut peptide segments
Mapping both the graphs, namely, peptide variability profile and solvent accessibility profile, together, we scan for those regions where the protein variability is least and solvent accessibility is among the highest. These are the first list of conserved solvent accessible segments of the protein.
Confirmation from 3D structures and second cut choice
To confirm that the regions identified in the previous step are indeed surface situated, we examine the identified regions in a protein 3D structure, eg, as shown through a software such as Cn3D4.3 available at NIH website.31 Marking out the identified segments on one protein of the 3D structure will reveal how much of the segments lie on the surface. If composite structures of this protein with the other proteins are available, then we can further fine-tune the selection process to identify those segments that really lie on an exposed surface of the capsid protein and not get covered by a neighboring protein of the virion.
Determination of epitope segments
Next, we consider which of the segments from the second-stage choice contains epitope regions to elicit necessary immune response. For this, we use two epitope-prediction tools, viz., the Immune Epitope Database (IEDB) and Analysis Resource server32–35 and ABCpred server,36,37 which, as mentioned earlier,21 have given good results. These tools indicate binding affinities of T-cell epitopes and B-cell epitopes (with 66% accuracy), major histocompatibility complex (MHC) I, and also of discontinuous epitopes through Ellipro.
Autoimmune test
Finally, we test the peptides that have remained after the third stage above to ensure that they do not possess any autoimmune threats and are unique peptides by themselves. Each peptide segment is subjected to a protein–protein BLAST to ensure that the selected peptides are not homologous to human proteins to cause autoimmune threats. These segments then become our recommendations for peptide vaccines subject to final verification in wet laboratories. That autoimmune threat can be a potential hazard has been explored in one of our earlier reports.38
Results and Discussions
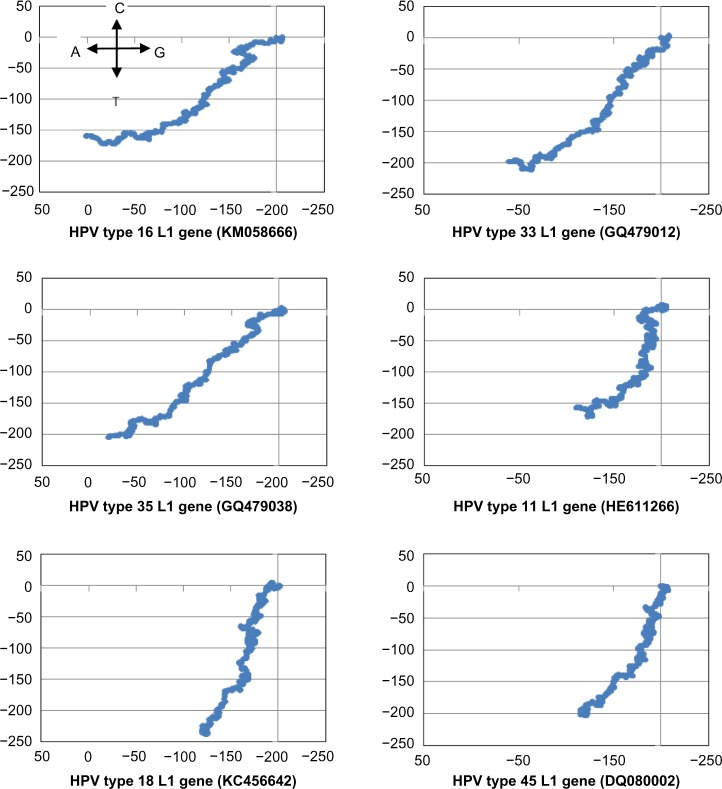
Our initial attempt at determining a common motif of conserved peptide sequences in the L1 gene of HPV, of all types available in our database as given in Table 1, met with little success. While signals for low variability were observed, these were heavily influenced by the preponderance of HPV16 data in our database; in fact, detailed inspection of the peptide stretches corresponding to the signal regions showed little homology between HPV16 and HPV18 sequences. This is observable from the nucleotide plots also (Fig. 2), which shows close similarity between HPV16, HPV33, and HPV35, and between HPV18 and HPV45, but the two groups have distinctly different plots, indicating significant differences in base distribution.
Figure 2.
2D graphical representations of five high-risk cancer-causing HPV L1 gene sequences (HPV16, 18, 33, 35, and 45) and one of low-risk type (HPV11). The assignments of the nucleotides to the four cardinal directions for the DNA walk are shown in the plot for HPV16 and are the same for all the other plots.
In our previous analyses for vaccine targets, we had considered large numbers of sequences of only one primary type, viz., the neuraminidase19 and hemagglutinin21 of influenza A genomes, or VP7 proteins of rotavirus.20 In the present instance, we have to consider different types of the HPV where the similarities within sequences of each type are greater than similarities between sequences of different types; we note that, as remarked by Chen et al.39, if the difference of the L1 gene of a cloned sequence from any other characterized type becomes 10% or more, a new type is claimed to have been established, else these are all strains of the same type group. The phylogenetic tree (Fig. 1) shows explicitly that the strains of each type belong to individual groups and the different types form separate clades: the HPV18 and 45 belong to one clade and HPV16, 33, and 35 to another. The 2D graphs (Fig. 2) also show at a glance the close similarities between HPV16, 33, and 35, and also between HPV18 and 45, and indications of where the two groups are different. Thus, our earlier experience could not be repeated here, which has to be considered as intertype differences. However, there can be large differences within a type too: eg, HPV type 16 from Iran (KM058666) and African type 2 variant (AF472509) are quite different, but because these are dsDNA types of viruses, we expect they will be regionally, ie, within the population groups, stable. Considering the preponderance of data, only the former variety of HPV16 and related sequences were used for our analyses.
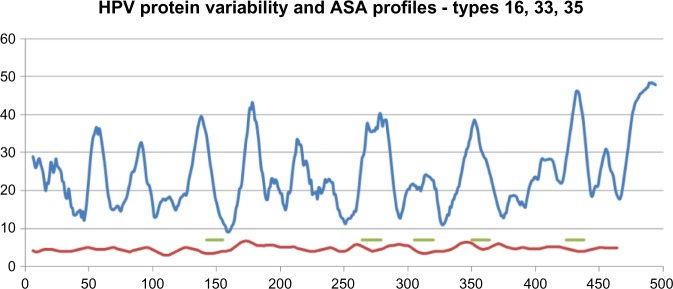
The analyses for the three high-risk types, HPV16, 33, and 35, were done together using the pR technique mentioned in the “Materials and methods” section with a 12 aa window, and the pR-values at each aa position were scanned to determine the number of different values. These numbers were then smoothed over a range of 12 positions, and a running average computed over the complete sequence. Next, ASA profiles were determined for several samples of the selected HPV types using the SABLE server, which has been found to be quite reliable. Taking an average of the solvent accessibility numbers determined residuewise for each sequence from SABLE, and then taking the running average of 12 aa, we get a smoothed out solvent accessibility profile whose peaks indicate highest solvent accessibility around that residue position. By our logic, the regions where the lowest variety numbers for aa stretches match with the highest values of the ASA profiles represent potentially the best situated highly conserved surface accessible peptide segments in the protein sequence. Figure 3 shows the plots of these two profiles of the high-risk HPV16, 33, and 35 types, where the five regions so identified are marked by short horizontal lines for further analysis.
Figure 3.
Protein variability (red) and ASA (blue) profiles of HPV types 16, 33, and 35 to determine conserved surface-exposed regions. The regions so identified are marked by short horizontal lines (green). The x-axis numbers refer to aa positions on the protein sequence; the numbers on the y-axis refer to protein segment variability numbers for the variability profile and index of surface exposure for the ASA profile.
To identify in terms of aas the regions we had determined in the varieties vs ASA profiles, we scanned the aa sequences of the HPV16, 33, and 35 of those regions. The three HPV types were found to have very close similarities in the peptide stretches, extending, in fact, to several aas beyond the 12 aa windows. aa variety vs ASA profile curves were also drawn similarly for HPV18 and HPV45 and appropriate peptide segments were identified. Table 2 lists the peptides for all identified segments for the five high-risk HPV types, and also of the low-risk HPV11 determined by a similar method.
Table 2.
Conserved surface-exposed peptides of HPV types 16, 18, 33, 35, 45, and 11.
| REGION | HPV | START POSITION | PEPTIDE |
|---|---|---|---|
| High risk types | |||
| 1 | Type 45 | 75 | YFRVVPNGAGNKQAV |
| Type 18 | 113 | VPAGGGNKQDIPKVS | |
| Type 16 | 142 | DNRECISMDYKQTQLCL | |
| Type 33 | 143 | DNRECLSMDYKQTQLCL | |
| Type 35 | 142 | DNRECISMDYKQTQLCL | |
| 2 | Type 45 | 157 | ESAHAATAVITQDVR |
| Type 18 | 189 | DTESSPAATSNVSED | |
| Type 16 | 264 | AGTVGENVPDDLYIKGS | |
| Type 33 | 264 | AGTLGEAVPDDLYIKGS | |
| Type 35 | 264 | AGTVGETVPADLYIKGT | |
| 3 | Type 45 | 194 | EHWAKGTLCKPAQLQ |
| Type 18 | 233 | GTACKSRPLSQGDCP | |
| Type 16 | 307 | FNKPYWLQRAQGHNNGI | |
| Type 33 | 307 | FNKPYWLQRAQGHNNGI | |
| Type 35 | 305 | FNKPYWLQRAQGHNNGI | |
| 4 | Type 45 | 301 | DLYIKGTSANMRETP |
| Type 18 | 336 | LYIKGTGMRASPGSC | |
| Type 16 | 354 | TYKNTNFKEYLRHGEEY | |
| Type 33 | 353 | TYKNGNFKEYIRHVEEY | |
| Type 35 | 352 | TYKNDNFKEYLRHGEEY | |
| 5 | Type 16 | 425 | AIACQKHTPPAPKEDPL |
| Type 18 | 405 | ICASTQSPVPGQYDA | |
| Type 33 | 424 | AITCQKTVPPKEKEDPL | |
| Type 35 | 423 | AVTCQKPSAPKPKDDPL | |
| Low risk type | |||
| 1 | Type 11 | 139 | DNRVNVGMDYKQTQLCM |
| 2 | Type 11 | 261 | AGTVGEPVPDDLLVKGG |
| 3 | Type 11 | 304 | FNKPYWLQKAQGHNNGI |
| 4 | Type 11 | 350 | TYTNSDYKEYMRHVEEF |
| 5 | Type 11 | 421 | AITCQKPTPEKEKQDPY |
Notes: These are listed regionwise to show peptide segment similarities. The segments marked in red are covered by neighboring proteins/sequence overlaps. Since HPV33 crystal structure was not available, overlap regions could not be ascertained, but can be expected to be similar to those for HPV35; similar considerations applied to HPV45 compared with HPV18.
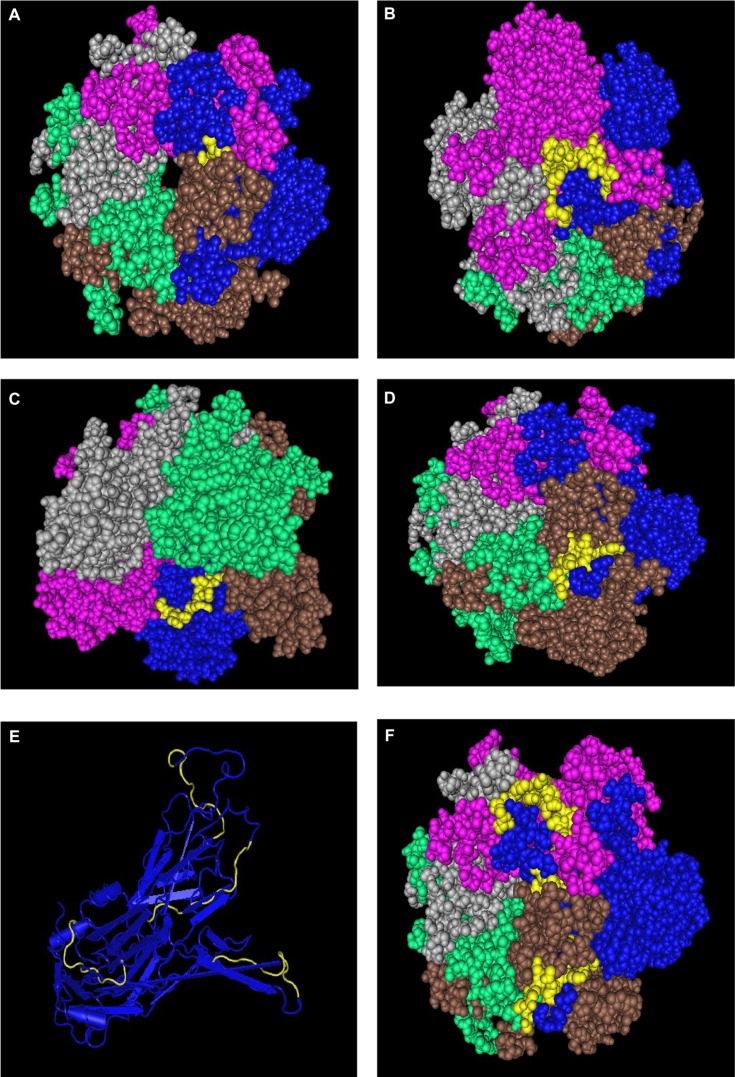
Since the HPV virion capsomers are composed of five L1 proteins, and one L2 each, it is possible that a surface accessible antigen in one protein may get covered by the folds of a neighboring protein. Step three of our search for vaccine target regions is to verify the surface accessibility of the identified regions through protein crystal structure. For this exercise, we used 2R5H for HPV16, 2R5I for HPV18, 2R5J for HPV35, and 2R5K for HPV11 available from the Protein Data Bank and individually checked for each of the regions in space-fill models; since no structure was available for HPV33 and HPV45, detailed comparison had to be omitted here. Figures 4–7 (HPV16, HPV18, HPV35, and HPV11, respectively) give snapshots of the identified peptides highlighted in yellow in one of the proteins of the pentameric structure of the capsid assembly. In the case of region 1 of HPV16, which was surface accessible for a solitary HPV protein, this peptide stretch is almost entirely covered by the adjacent protein and, therefore, has to be rejected as a vaccine target candidate. Region 2 was found to be entirely viable. In the case of regions 3 and 4, the original 12 aa stretches were found to be surface situated, but 5 aas at the 5′-end were found to be covered by the neighboring residues, ie, even if these extended segments belong to a group of conserved residues, the parts beyond the original 12aa stretches are not viable as candidate vaccine targets. Interestingly, the two regions, 3 and 4, are on two sides of the protein, but both are surface accessible. These segments that are not available for consideration as potential vaccine targets are marked in red in Table 2. Region 5 listed in the table could not be tested in a similar manner due to the nonavailability of these stretches in the protein crystals.
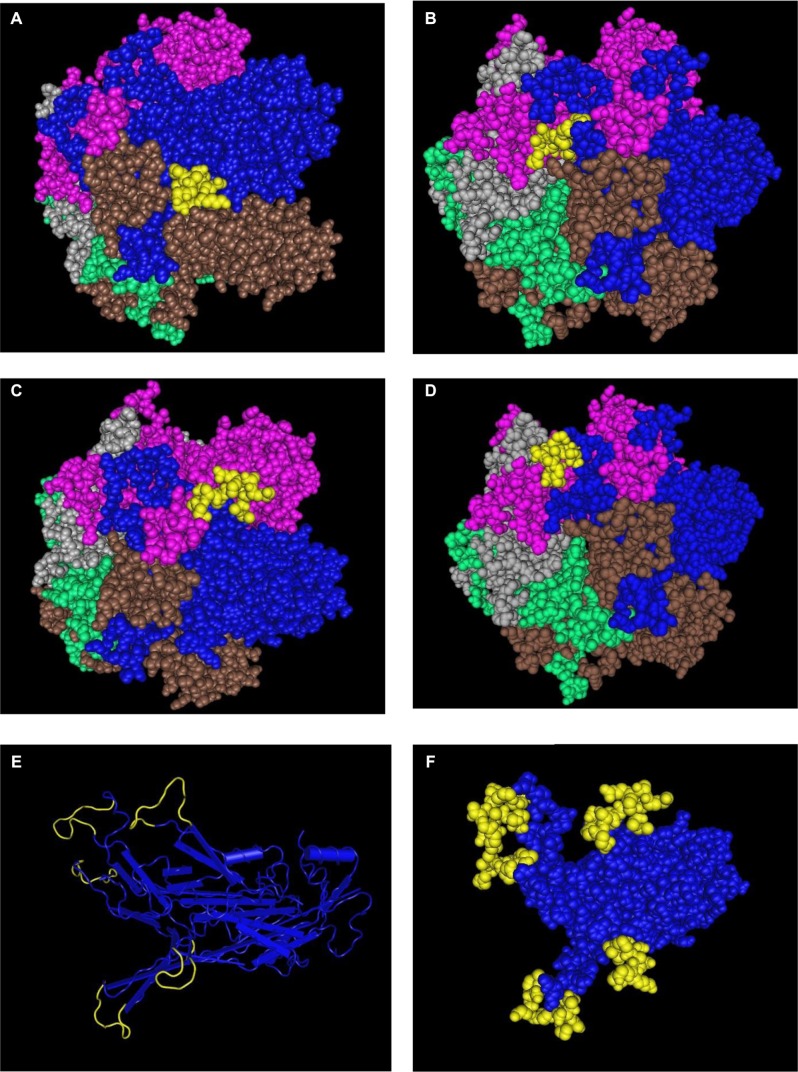
Figure 4.
Display in space-fill rendering of HPV16 L1 protein pentameric structure (2R5H). The conserved surface-exposed segments identified by comparison of protein variability and ASA profile analysis, as shown in Figure 2 and given in Table 2, are highlighted here in yellow using the protein shown in blue as a template. Peptide stretches shown are in terms of peptide stretch starting position numbers: (A) 142, (B) 264, (C) 307, and (D) 354. (E) Worm view of the protein with all four peptides. (F) A view of the three peptides on the template in the pentameric structure.
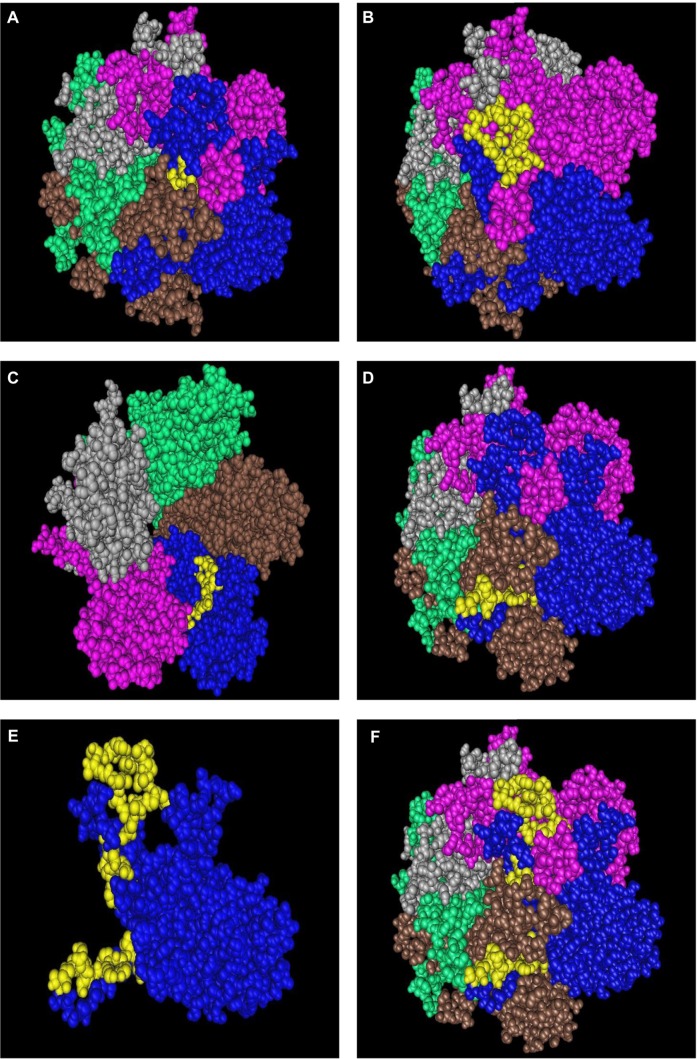
Figure 5.
Display in space-fill rendering of HPV18 L1 protein pentameric structure (2R5I). The conserved surface-exposed segments identified by comparison of protein variability and ASA profile analysis as given in Table 2 are highlighted here in yellow using the protein shown in blue as a template. Peptide stretches shown are in terms of peptide stretch starting position numbers: (A) 113, (B) 189, (C) 233, and (D) 336. (E) Worm view of the protein with all five peptides. (F) A space-fill view of the peptides on the template protein in the pentameric structure.
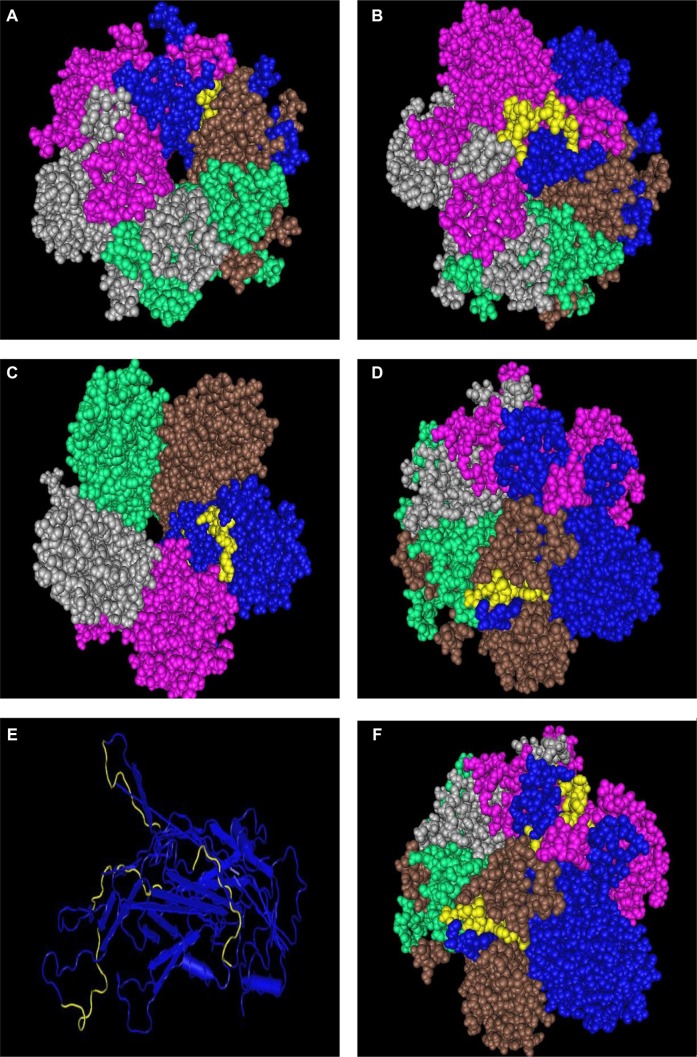
Figure 6.
Display in space-fill rendering of HPV35 L1 protein pentameric structure (2R5J). The conserved surface-exposed segments identified by comparison of protein variability and ASA profile analysis, as shown in Figure 2 and given in Table 2, are highlighted here in yellow using the protein shown in blue as a template. Peptide stretches shown are in terms of peptide stretch starting position numbers: (A) 142, (B) 264, (C) 305, and (D) 352. (E) The template protein with three of the four peptides highlighted in yellow. The fourth peptide at the back is not visible in this space fill model. (F) Another view of the three peptides on the template in the pentameric structure.
Figure 7.
Display in space-fill rendering of HPV11 L1 protein pentameric structure (2R5K). The conserved surface-exposed segments identified by comparison of protein variability and ASA profile analysis, as shown in Figure 2 and given in Table 2, are highlighted here in yellow using the protein shown in blue as a template. Peptide stretches shown are in terms of peptide stretch starting position numbers: (A) 142, (B) 264, (C) 305, and (D) 352. (E) The worm view of the protein with the four peptides highlighted in yellow. (F) Another view of the three peptides on the template in the pentameric structure.
The next stage is to determine if the identified regions have epitope potential to enable immune response. For this, we used the IEDB server to test for T-cell MHC II epitope regions using two alleles, namely, HLA-DRB1–01–01 and HLA-DRB1-04-01. Table 3 shows that all the regions we had identified passed the MHC II test, although in some instances one or the other allele gave results that were just outside the permissible range for generating immune response. However, none of the regions of any of the three HPV types failed in both alleles. Likewise, we tested for B-cell humoral response through the ABCpred server, which has been known to predict epitope regions for B-cell response with about 66% accuracy. As can be seen from the results extracted in Table 4, we find that in this instance all our regions passed this test with high acceptability ranks, except for region 4 for HPV35 whose score was outside the critical range.
Table 3.
Extract of IEDB results of regions of interest.
| HPV TYPE | REGION | ALLELE | START | END | PEPTIDE | COMBLIB SCORE | COMBLIB RANK | Smm ALIGN ic50 | Smm ALIGN RANK |
|---|---|---|---|---|---|---|---|---|---|
| High risk types | |||||||||
| HPV16 | I | HLA-DRB1*04:01 | 145 | 159 | ECISMDYKQTQLCLI | 3611 | 29.6 | ||
| I | HLA-DRB1*01:01 | 145 | 159 | ECISMDYKQTQLCLI | 90412.08 | 81.79 | 354 | 38.82 | |
| II | HLA-DRB1*04:01 | 269 | 283 | ENVPDDLYIKGSGST | – | – | 3849 | 30.84 | |
| II | HLA-DRB1*01:01 | 269 | 283 | ENVPDDLYIKGSGST | 213.59 | 48.86 | 187 | 27.4 | |
| III | HLA-DRB1*04:01 | 307 | 321 | FNKPYWLQRAQGHNN | – | – | 1492 | 15.32 | |
| III | HLA-DRB1*01:01 | 307 | 321 | FNKPYWLQRAQGHNN | 587.21 | 55.37 | 191 | 27.75 | |
| IV | HLA-DRB1*04:01 | 354 | 368 | TYKNTNFKEYLRHGE | – | – | 8082 | 46.58 | |
| IV | HLA-DRB1*01:01 | 354 | 368 | TYKNTNFKEYLRHGE | 423.57 | 53.61 | 4625 | 83.28 | |
| V | HLA-DRB1*04:01 | 423 | 437 | SQAIACQKHTPPAPK | – | – | 2489 | 22.87 | |
| V | HLA-DRB1*01:01 | 423 | 437 | SQAIACQKHTPPAPK | 90.99 | 43.54 | 2814 | 76.58 | |
| HPV33 | I | HLA-DRB1*04:01 | 146 | 160 | ECLSMDYKQTQLCLL | – | – | 3598 | 29.52 |
| I | HLA-DRB1*01:01 | 146 | 160 | ECLSMDYKQTQLCLL | 90412.08 | 81.79 | 253 | 32.54 | |
| II | HLA-DRB1*04:01 | 269 | 283 | EAVPDDLYIKGSGTT | – | – | 4427 | 33.59 | |
| II | HLA-DRB1*01:01 | 269 | 283 | EAVPDDLYIKGSGTT | 500.93 | 54.2 | 183 | 27.06 | |
| III | HLA-DRB1*04:01 | 307 | 321 | FNKPYWLQRAQGHNN | – | – | 1492 | 15.32 | |
| III | HLA-DRB1*01:01 | 307 | 321 | FNKPYWLQRAQGHNN | 587.21 | 55.37 | 191 | 27.75 | |
| IV | HLA-DRB1*04:01 | 353 | 367 | TYKNGNFKEYIRHVE | – | – | 7313 | 44.34 | |
| IV | HLA-DRB1*01:01 | 353 | 367 | TYKNGNFKEYIRHVE | 227.95 | 49.46 | 4520 | 82.99 | |
| V | HLA-DRB1*04:01 | 422 | 436 | SQAITCQKTVPPKEK | – | – | 712 | 7.76 | |
| V | HLA-DRB1*01:01 | 422 | 436 | SQAITCQKTVPPKEK | 25205.94 | 76.18 | 2166 | 72.56 | |
| HPV35 | I | HLA-DRB1*04:01 | 145 | 159 | ECISMDYKQTQLCLI | – | – | 3611 | 29.6 |
| I | HLA-DRB1*01:01 | 145 | 159 | ECISMDYKQTQLCLI | 90412.08 | 81.79 | 354 | 38.82 | |
| II | HLA-DRB1*04:01 | 269 | 283 | ETVPADLYIKGTTGT | – | – | 2709 | 24.3 | |
| II | HLA-DRB1*01:01 | 269 | 283 | ETVPADLYIKGTTGT | 44.86 | 39.46 | 316 | 36.62 | |
| III | HLA-DRB1*04:01 | 305 | 319 | FNKPYWLQRAQGHNN | – | – | 1492 | 15.32 | |
| III | HLA-DRB1*01:01 | 305 | 319 | FNKPYWLQRAQGHNN | 587.21 | 55.37 | 191 | 27.75 | |
| IV | HLA-DRB1*04:01 | 352 | 366 | TYKNDNFKEYLRHGE | – | – | 7975 | 46.29 | |
| IV | HLA-DRB1*01:01 | 352 | 366 | TYKNDNFKEYLRHGE | 15387.68 | 73.97 | 5655 | 85.56 | |
| V | HLA-DRB1*04:01 | 421 | 435 | SQAVTCQKPSAPKPK | – | – | 2871 | 25.32 | |
| V | HLA-DRB1*01:01 | 421 | 435 | SQAVTCQKPSAPKPK | 2120.5 | 63.29 | 1603 | 67.63 | |
| HPV18 | I | HLA-DRB1*04:01 | 113 | 127 | VPAGGGNKQDIPKVS | – | – | 7042 | 43.49 |
| I | HLA-DRB1*01:01 | 113 | 127 | VPAGGGNKQDIPKVS | 1000000 | 89.54 | 5462 | 85.18 | |
| II | HLA-DRB1*04:01 | 189 | 203 | DTESSPAATSNVSED | – | – | 3369 | 28.23 | |
| II | HLA-DRB1*01:01 | 189 | 203 | DTESSPAATSNVSED | 8148.71 | 70.61 | 2278 | 73.37 | |
| III | HLA-DRB1*04:01 | 233 | 247 | GTACKSRPLSQGDCP | – | – | 5455 | 37.98 | |
| III | HLA-DRB1*01:01 | 233 | 247 | GTACKSRPLSQGDCP | 32629.66 | 77.45 | 3346 | 79.03 | |
| IV | HLA-DRB1*04:01 | 336 | 350 | LYIKGTGMRASPGSC | – | – | 1170 | 12.4 | |
| IV | HLA-DRB1*01:01 | 336 | 350 | LYIKGTGMRASPGSC | 98.96 | 44.12 | 79 | 14.64 | |
| V | HLA-DRB1*04:01 | 405 | 419 | ICASTQSPVPGQYDA | – | – | 3202 | 27.3 | |
| V | HLA-DRB1*01:01 | 405 | 419 | ICASTQSPVPGQYDA | 7271.97 | 70.12 | 1907 | 70.55 | |
| HPV45 | I | HLA-DRB1*01:01 | 75 | 89 | YFRVVPNGAGNKQAV | 0.01 | 0.01 | 113 | 19.37 |
| I | HLA-DRB1*04:01 | 75 | 89 | YFRVVPNGAGNKQAV | – | – | 837 | 9.07 | |
| II | HLA-DRB1*04:01 | 157 | 171 | ESAHAATAVITQDVR | – | – | 3466 | 28.8 | |
| II | HLA-DRB1*01:01 | 157 | 171 | ESAHAATAVITQDVR | 0.08 | 9.62 | 540 | 47.14 | |
| III | HLA-DRB1*01:01 | 194 | 208 | EHWAKGTLCKPAQLQ | 1.34 | 20.07 | 1281 | 63.69 | |
| III | HLA-DRB1*04:01 | 194 | 208 | EHWAKGTLCKPAQLQ | – | – | 9494 | 50.17 | |
| IV | HLA-DRB1*04:01 | 301 | 315 | DLYIKGTSANMRETP | – | – | 56 | 0.39 | |
| IV | HLA-DRB1*01:01 | 301 | 315 | DLYIKGTSANMRETP | 5.09 | 26.48 | 51 | 10 | |
| Low risk type | |||||||||
| HPV11 | I | HLA-DRB1*04:01 | 142 | 156 | VNVGMDYKQTQLCMV | – | – | 4913 | 35.76 |
| I | HLA-DRB1*01:01 | 142 | 156 | VNVGMDYKQTQLCMV | 791736.5 | 88.8 | 420 | 42.19 | |
| II | HLA-DRB1*04:01 | 266 | 280 | EPVPDDLLVKGGNNR | – | 8388 | 47.43 | ||
| II | HLA-DRB1*01:01 | 266 | 280 | EPVPDDLLVKGGNNR | 812.21 | 57.66 | 1166 | 61.98 | |
| III | HLA-DRB1*04:01 | 304 | 318 | FNKPYWLQKAQGHNN | 1607 | 16.29 | |||
| III | HLA-DRB1*01:01 | 304 | 318 | FNKPYWLQKAQGHNN | 247.24 | 50.05 | 200 | 28.5 | |
| IV | HLA-DRB1*04:01 | 350 | 364 | TYTNSDYKEYMRHVE | – | – | 13755 | 58.62 | |
| IV | HLA-DRB1*01:01 | 350 | 364 | TYTNSDYKEYMRHVE | 14290.32 | 73.49 | 6877 | 87.61 | |
| V | HLA-DRB1*04:01 | 419 | 433 | SQAITCQKPTPEKEK | – | – | 3608 | 29.58 | |
| V | HLA-DRB1*01:01 | 419 | 433 | SQAITCQKPTPEKEK | 811.45 | 57.66 | 1894 | 70.43 | |
Note: Binding affinity is considered high if IC50 <50, intermediate for IC50 <500, and low for IC <5000.
Table 4.
B-cell epitopes for selected regions from ABCpred server.
| RANK | SEQUENCE | START POSITION | SCORE |
|---|---|---|---|
| Type 16 | |||
| 7 | RECISMDYKQTQLCLI | 144 | 0.87 |
| 16 | AGTVGENVPDDLYIKG | 264 | 0.76 |
| 15 | YWLQRAQGHNNGICWG | 311 | 0.78 |
| 32 | SETTYKNTNFKEYLRH | 351 | 0.53 |
| 4 | AIACQKHTPPAPKEDP | 425 | 0.9 |
| Type 33 | |||
| 3 | GADNRECLSMDYKQTQ | 141 | 0.91 |
| 12 | HFFNRAGTLGEAVPDD | 259 | 0.81 |
| 14 | LGEAVPDDLYIKGSGT | 267 | 0.79 |
| 16 | SMVTSESQLFNKPYWL | 298 | 0.77 |
| 15 | YWLQRAQGHNNGICWG | 311 | 0.78 |
| 26 | TYKNGNFKEYIRHVEE | 353 | 0.56 |
| 1 | AITCQKTVPPKEKEDP | 424 | 0.93 |
| Type 35 | |||
| 8 | RECISMDYKQTQLCLI | 144 | 0.87 |
| 14 | AGTVGETVPADLYIKG | 264 | 0.81 |
| 11 | YWLQRAQGHNNGICWS | 309 | 0.84 |
| 1 | AVTCQKPSAPKPKDDP | 423 | 0.97 |
| 12 | PSAPKPKDDPLKNYTF | 429 | 0.83 |
| Type 11 | |||
| 11 | GQDNRVNVGMDYKQTQ | 137 | 0.81 |
| 14 | PVPDDLLVKGGNNRSS | 267 | 0.78 |
| 14 | SSEAQLFNKPYWLQKA | 298 | 0.78 |
| 10 | YWLQKAQGHNNGICWG | 308 | 0.83 |
| 7 | TYTNSDYKEYMRHVEE | 350 | 0.86 |
| 2 | AITCQKPTPEKEKQDP | 421 | 0.93 |
| Type 18 | |||
| 13 | RVPAGGGNKQDI | 112 | 0.69 |
| 21 | DDTESSPAATSN | 188 | 0.6 |
| 1 | TACKSRPLSQGD | 234 | 0.86 |
| 12 | DTVPQSLYIKGT | 330 | 0.7 |
| 21 | GTGMRASPGSCV | 340 | 0.6 |
| 10 | STQSPVPGQYDA | 408 | 0.72 |
| Type 45 | |||
| 12 | YFRVVPNGAGNKQAVP | 75 | 0.81 |
| 7 | AHAATAVITQDVRDNV | 159 | 0.86 |
| 8 | EHWAKGTLCKPAQLQP | 194 | 0.85 |
| 12 | TDLYIKGTSANMRETP | 300 | 0.81 |
Notes: Low rank, high score indicates high binding affinity (cut-off score = 0.51).
Both, linear and conformational epitopes, are found on the L1 protein surface, but it is well established that most of the neutralizing antibody production are through conformational epitopes.27,40 Ludmerer et al.41 showed that a distinct surface-exposed linear epitope overlaps the neighboring conformational epitope in both HPV11 and HPV16 VLPs and that neutralizing epitopes also bind to a linear epitope. Combita et al.42 experimenting with mice model confirmed the existence of linear epitopes inducing cross-neutralization, but also inferred that such cross-neutralization will not exceed 1% of the effects of the dominant conformational epitopes. Type-specific and cross-reactive linear epitopes have been determined on the L2 protein surface also,43 but for the present purposes, we restrict our analyses to the L1 protein epitopes only.
Our tests above identified linear epitopes but since the papillomavirus L1 proteins have mostly conformational epitopes, we tested for discontinuous epitopes on the aa sequences also through Ellipro available in the IEDB web server. We found that for all the HPV types we investigated, almost all the identified peptides had overlaps with the conformational epitopes. The peptides that had the overlaps are marked in Table 5. Table 5 also shows the percentile rank of binding affinity for HLA for MHC Class I predicted by the IEDB server; a number of peptide regions show low ranks, which indicate high binding affinity.
Table 5.
Final list of conserved surface-exposed peptides of HPV types 16, 18, 33, 35, 45, and 11 selected through our screening process.
| HPV | START POSITION | PEPTIDE | MHC-II | |||
|---|---|---|---|---|---|---|
| T-CELL | B-CELL RANK | DISCONTINUOUS EPITOPES | MHCI RANK | |||
| High risk types | ||||||
| HPV16 | 264 | AGTVGENVPDDLYIKGS | Y | 16 | Y | 46 |
| 307 | FNKPYWLQRAQG | Y | 15 | – | 82 | |
| 354 | TYKNTNFKEYLR | Y | 32 | Y | 97 | |
| 425 | AIACQKHTPPAPKEDPL | Y | 4 | Y | 65 | |
| HPV18 | 113 | VPAGGGNKQDIPKVS | Y | 13 | 78 | |
| 189 | DTESSPAATSNVSED | Y | 21 | * | 2.7 | |
| 233 | GTACKSRPLSQGDCP | Y | 1 | 97 | ||
| 336 | LYIKGTGMRASPGSC | Y | 12 | 100 | ||
| 405 | ICASTQSPVPGQYDA | Y | 10 | 78 | ||
| HPV33 | 143 | DNRECLSMDYKQTQLCL | Y | 3 | Y | 55 |
| 264 | AGTLGEAVPDDLYIKGS | Y | 12 | Y | 54 | |
| 307 | FNKPYWLQRAQGHNNGI | Y | 15 | – | 82 | |
| 353 | TYKNGNFKEYIRHVEEY | Y | 26 | Y | 99 | |
| 424 | AITCQKTVPPKEKEDPL | Y | 1 | Y | 77 | |
| HPV35 | 142 | DNRECISMDYKQTQLCL | Y | 8 | Y | 52 |
| 264 | AGTVGETVPADLYIKGT | Y | 14 | Y | 49 | |
| 305 | FNKPYWLQRAQGHNNGI | Y | 11 | – | 82 | |
| 352 | TYKNDNFKEYLRHGEEY | Y | – | Y | 97 | |
| 423 | AVTCQKPSAPKPKDDPL | Y | 1 | Y | 86 | |
| HPV45 | 75 | YFRVVPNGAGNKQAV | Y | 12 | – | 31 |
| 157 | ESAHAATAVITQDVR | Y | 7 | – | 21 | |
| 194 | EHWAKGTLCKPAQLQ | Y | 8 | Y | 87 | |
| 301 | DLYIKGTSANMRETP | Y | 12 | Y | 69 | |
| Low risk type | ||||||
| HPV11 | 261 | AGTVGEPVPDDLLVKGG | Y | 14 | Y | 45 |
| 304 | FNKPYWLQKAQG | Y | 10 | – | 84 | |
| 350 | TYTNSDYKEYMR | Y | 7 | Y | 100 | |
| 421 | AITCQKPTPEKEKQDPY | Y | 2 | Y | 78 | |
Notes: “Y” represents acceptable binding affinities. The discontinuous epitopes for HPV18 could not be determined due to mismatch with homologs.
The remaining test was envisaged for autoimmune possibilities. In the protein–protein BLAST analysis using the nonredundant protein sequences from GenBank, PDB, SWISSPROT, etc., the results were found to match the super papillomavirus family with no matches with any non-HPV protein.
The final list of identified peptide stretches that met all our criteria and could serve as potential peptide vaccine targets are given in Table 5. We have ended up with four possible surface accessible highly conserved peptide regions in each of the HPV16, 18, 33, 35, 45, and HPV11, with some being common to several types. Some show less binding affinities in T-cell or B-cell activity, other regions appear to have purely linear epitopes with no overlaps with any discontinuous epitopes, but by and large, every peptide segment identified in this exercise show prospects of eliciting immune response; in some instances, cross-neutralization between types seem possible. What would be required, however, are wet laboratory experiments and adjuvant combinations to validate this model of alternate strategies and improve and expand upon the possible multivalent peptide vaccine approach to the prevention of HPV-induced cancers.
Conclusion
HPV comes in many types and causes numerous cases of cervical, genital, and other cancers around the world every year, more so in developing and underdeveloped countries where economic considerations make it difficult to afford the preventive care that is almost taken for granted in the developed nations3,8,18; there is also the need to consider future trends where vaccine development will tend to be individual, gender specific, or community/population group specific.17 Partly to alleviate this condition, we have proposed an alternative strategy of multivalent peptide vaccines for conditions where VLP vaccines like Gardasil and Cervarix may not be ideal. Our exercise through bioinformatics means has yielded a set of peptides for each HPV type analyzed, which a priori appears to provide a feasible alternative. Because of the nature of the viral coat proteins, the peptides proposed in this paper may not provide the high level of efficiency that the VLP vaccines can provide, but with suitable adjuvants to enhance the neutralizing antibody production and appropriate mix of wider range of HPV types, it is hoped that the preventive actions can be augmented and a substantial fraction of the population may be afforded some relief at lower cost. For this to happen, however, it is necessary to conduct wet laboratory experiments to test the efficacy of the results of bioinformatics studies like this. Given the wide spread of papillomavirus-induced cervical, genital, and other cancers and the sufferings these cause, and the large variety of the virus, such preparations are to be strongly advocated.
Acknowledgments
The authors express their thanks to the publishers and to Prof Chun Li, in particular, for the invitation to forward an article for publishing in this thematic issue. One of the authors (AN) thanks Ms. Anindita Basu for bringing the HPV issue to his attention. The authors acknowledges with thanks the many important comments and insights offered by the referees, which has made this study more complete.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Editor in Chief
PEER REVIEW: Seven peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2,703 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: AN. Analyzed the data: AD, SD. Wrote the first draft of the manuscript: AN. Contributed to the writing of the manuscript: AD. Agreed with manuscript results and conclusions: AN, AD, SD. Jointly developed the structure and arguments for the paper: AN, AD. Made critical revisions and approved the final version: AN. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Shankaran V, Ikeda H, Bruce AT, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 2.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 3.Ghittoni R, Accardi R, Chiocca S, Tommasino M. Role of human papillomaviruses in carcinogenesis. Ecancermedicalscience. 2015;9:526. doi: 10.3332/ecancer.2015.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Schiffman M, Herrero R, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes. PLoS One. 2011;6(5):1–16. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC CDC Report on Human Papillomavirus (HPV) 2015. [Accessed December 10, 2015]. Available at: http://www.cdc.gov/hpv/parents/cancer.html.
- 6.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–44. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 7.CDC CDC Report on Human Papillomavirus (HPV) 2015. [Accessed December 10, 2015]. Available at: http://www.cdc.gov/hpv/parents/whatishpv.html.
- 8.Muñoz N, Bosch FX, Castellsagué X, et al. Against which human papilloma-virus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111(2):278–85. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 9.Lowy DR, Schiller JT. Prophylactic human papillomavirus vaccines. J Clin Invest. 2006;116(5):1167–73. doi: 10.1172/JCI28607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller DL, Puricelli MD, Stack MS. Virology and molecular pathogenesis of human papillomavirus (HPV)-associated oropharyngeal squamous cell carcinoma. Biochem J. 2012;443(2):339–53. doi: 10.1042/BJ20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derkay CS. Recurrent respiratory papillomatosis. Laryngoscope. 2001;111:7–69. doi: 10.1097/00005537-200101000-00011. [DOI] [PubMed] [Google Scholar]
- 12.De Koning MNC, Quint KD, Bruggink SC, et al. High prevalence of cutaneous warts in elementary school children and the ubiquitous presence of wart-associated human papillomavirus on clinically normal skin. Br J Dermatol. 2015;172:196–201. doi: 10.1111/bjd.13216. [DOI] [PubMed] [Google Scholar]
- 13.Harper DM, Franco EL, Wheeler CM, et al. HPV Vaccine Study Group Sustained efficacy up to 4.5 years of a bivalent L1 virus-like particle vaccine against human papillomavirus types 16 and 18: follow-up from a randomised control trial. Lancet. 2006;367(9518):1247–55. doi: 10.1016/S0140-6736(06)68439-0. [DOI] [PubMed] [Google Scholar]
- 14.Joura EA, Giuliano AR, Iversen O-E, et al. A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N Engl J Med. 2015;372(8):711–21. doi: 10.1056/NEJMoa1405044. [DOI] [PubMed] [Google Scholar]
- 15.Chroboczek J, Szurgot I, Szolajska E. Virus-like particles as vaccine. Acta Biochim Pol. 2014;61:531–9. [PubMed] [Google Scholar]
- 16.Poland GA, Whitaker JA, Poland CM, Ovsyannikova IG, Kennedy RB. Vaccinology in the third millennium: scientific and social challenges. Curr Opin Virol. 2016;17:116–25. doi: 10.1016/j.coviro.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poland GA, Kennedy RB, Ovsyannikova IG. Vaccinomics and personalized vaccinology: is science leading us toward a new path of directed vaccine development and discovery? PLoS Pathog. 2011;7(12):e1002344. doi: 10.1371/journal.ppat.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell AW, McCluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev. 2007;6:404–141. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh A, Nandy A, Nandy P. Computational analysis and determination of a highly conserved surface exposed segment in H5N1 avian flu and H1N1 swine flu neuraminidase. BMC Struct Biol. 2010;10:6. doi: 10.1186/1472-6807-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh A, Chattopadhyay S, Chawla-Sarkar M, Nandy P, Nandy A. In silico study of rotavirus VP7 surface accessible conserved regions for antiviral drug/vaccine design. PLoS One. 2012;7(7):e40749. doi: 10.1371/journal.pone.0040749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkar T, Das S, De A, et al. H7 N9 influenza outbreak in China 2013: in silico analyses of conserved segments of the hemagglutinin as a basis for the selection of peptide vaccine targets. Comput Biol Chem. 2015;59:8–15. doi: 10.1016/j.compbiolchem.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Nandy A. A new graphical representation and analysis of DNA sequence structure: I. methodology and application to globin genes. Curr Sci. 1994;66(4):309–14. [Google Scholar]
- 23.Raychaudhury C, Nandy A. Indexing scheme and similarity measures for macromolecular sequences. J Chem Inf Comput Sci. 1999;39:243–7. doi: 10.1021/ci980077v. [DOI] [PubMed] [Google Scholar]
- 24.Nandy A, Ghosh A, Nandy P. Numerical characterization of protein sequences and application to voltage-gated sodium channel alpha subunit phylogeny. In Silico Biol. 2009;9:77–87. [PubMed] [Google Scholar]
- 25.Nandy A, Nandy P. On the uniqueness of quantitative DNA difference descriptors in 2D graphical representation models. Chem Phys Lett. 2003;368:102–7. [Google Scholar]
- 26.Ghosh A, Nandy A, Nandy P, Gute BD, Basak SC. Computational study of dispersion and extent of mutated and duplicated sequences of the H5N1 influenza neuraminidase over the period 1997–2008. J Chem Inf Model. 2009;49:2627–38. doi: 10.1021/ci9001662. [DOI] [PubMed] [Google Scholar]
- 27.Bishop B, Dasgupta J, Klein M, et al. Crystal structures of four types of human papillomavirus L1 capsid proteins. J Biol Chem. 2007;282:31803–11. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- 28.Multhoff G, Pfister K, Gehrmann M, et al. A 14-mer Hsp70 peptide stimulates natural killer (NK) cell activity. Cell Stress Chaperones. 2001;6:337–44. doi: 10.1379/1466-1268(2001)006<0337:amhpsn>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olenina LV, Nikolaeva LI, Sobolev BN, Blokhina NP, Archakov AI, Kolesanova EF. Mapping and characterization of B cell linear epitopes in the conservative regions of hepatitis C virus envelope glycoproteins. J Viral Hepat. 2002;9(3):174–82. doi: 10.1046/j.1365-2893.2002.00358.x. [DOI] [PubMed] [Google Scholar]
- 30.Moisa AA, Kolesanova EF. Ch.11, Synthetic peptide vaccines. In: Priti R, editor. Insight and Control of Infectious Disease in Global Scenario. InTech; 2012. pp. 201–28. Available at: http://www.intechopen.com/books/insight-and-control-of-infectious-disease-in-global-scenario/syntheticpeptide-Vaccines. [Google Scholar]
- 31.NIH NIH Website. Available at: http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=64261.
- 32.IEDB IEDB Server at Website. Available at: http://tools.iedb.org/mhcii/
- 33.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4(4):e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Sidney J, Kim Y, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vita R, Zarebski L, Greenbaum JA, et al. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–62. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ABCpred ABCpred Server at Website. Available at: http://www.imtech.res.in/raghava/abcpred/ABC_submission.html.
- 37.Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65:40–8. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar T, Das S, Nandy P, Bhowmick R, Nandy A. In silico study of potential autoimmune threats from rotavirus infection. Comput Biol Chem. 2014;51:51–6. doi: 10.1016/j.compbiolchem.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Schiffman M, Herrero R, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One. 2011;6(5):e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleury MJ, Touzé A, Maurel M-C, Moreau T, Coursaget P. Identification of neutralizing conformational epitopes on the human papillomavirus type 31 major capsid protein and functional implications. Protein Sci. 2009;18:1425–38. doi: 10.1002/pro.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludmerer SW, Benincasa D, Mark GE, III, Christensen ND. A neutralizing epitope of human papillomavirus type 11 is principally described by a continuous set of residues which overlap a distinct linear, surface-exposed epitope. J Virol. 1997;71:3834–9. doi: 10.1128/jvi.71.5.3834-3839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Combita A-L, Touzé A, Bousarghin L, Christensen ND, Coursaget P. Identification of two cross-neutralizing linear epitopes within the L1 major capsid protein of human papillomaviruses. J Virol. 2002;76:6480–6. doi: 10.1128/JVI.76.13.6480-6486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roden RBS, Yutzy WH, IV, Fallon R, Inglis S, Lowy DR, Schiller JT. Minor capsid protein of human genital papillomaviruses contains subdominant, cross-neutralizing epitopes. Virology. 2000;270:254–7. doi: 10.1006/viro.2000.0272. [DOI] [PubMed] [Google Scholar]