Supplemental Digital Content is available in the text.
Abstract
Background:
Targeted maximum likelihood estimation has been proposed for estimating marginal causal effects, and is robust to misspecification of either the treatment or outcome model. However, due perhaps to its novelty, targeted maximum likelihood estimation has not been widely used in pharmacoepidemiology. The objective of this study was to demonstrate targeted maximum likelihood estimation in a pharmacoepidemiological study with a high-dimensional covariate space, to incorporate the use of high-dimensional propensity scores into this method, and to compare the results to those of inverse probability weighting.
Methods:
We implemented the targeted maximum likelihood estimation procedure in a single-point exposure study of the use of statins and the 1-year risk of all-cause mortality postmyocardial infarction using data from the UK Clinical Practice Research Datalink. A range of known potential confounders were considered, and empirical covariates were selected using the high-dimensional propensity scores algorithm. We estimated odds ratios using targeted maximum likelihood estimation and inverse probability weighting with a variety of covariate selection strategies.
Results:
Through a real example, we demonstrated the double robustness of targeted maximum likelihood estimation. We showed that results with this method and inverse probability weighting differed when a large number of covariates were included in the treatment model.
Conclusions:
Targeted maximum likelihood can be used in high-dimensional covariate settings. In high-dimensional covariate settings, differences in results between targeted maximum likelihood and inverse probability weighted estimation are likely due to sensitivity to (near) positivity violations. Further investigations are needed to gain better understanding of the advantages and limitations of this method in pharmacoepidemiological studies.
Propensity score methods are widely used in pharmacoepidemiology to address measured confounding in situations where a large number of covariates have to be considered, especially in studies where the outcome is rare. The propensity score is defined as an individual’s probability to receive the treatment of interest given covariates.1 Correct specification of the propensity score model conditional on a sufficient set of confounding covariates is necessary for removing confounding bias in estimated treatment effects. Typically, the propensity score model includes covariates that are assumed to be associated with both the outcome and the exposure of interest. As an alternative to traditional covariate predefinition approaches, (semi-) automated procedures to select adjustment covariates, such as the high-dimensional propensity score are gaining popularity to address residual confounding.2–4
For health policy evaluation, the marginal treatment effect at the population level is often of particular interest. It is the average causal effect of the treatment on the outcome employing a counterfactual world framework: comparing the study population if everyone were treated to the study population if everyone were untreated.5 Several approaches have been developed to estimate the marginal treatment effect. For example, inverse probability weighting (IPW) can be used to estimate a marginal treatment effect.5,6 A class of doubly robust estimators has also been proposed, where the model includes a component of the efficient influence function for the parameter of interest. This component incorporates the inverse of the propensity score.7 A related estimation procedure, targeted maximum likelihood estimation (TMLE), was subsequently developed by van der Laan and colleagues8–10 and has been shown to have certain advantages over the inverse probability weighted estimator. When estimating a marginal treatment effect, correct specification of the propensity score model is crucial to obtain an unbiased estimate by inverse probability weighting. TMLE allows specification of both the treatment model and the outcome model and is doubly robust, which means that even under misspecification of one of the two models, the TMLE effect estimator is asymptotically consistent. Furthermore, it is locally efficient, meaning that the estimator has (asymptotically) the smallest standard error (SE) among a class of models when both models are correctly specified. For these reasons, TMLE is an appealing method for “confounder-adjusted” treatment effect estimation.
TMLE is a relatively new approach that has been applied to many study designs.11–13 However, perhaps due to its novelty and theoretical complexity, it has not been widely used in pharmacoepidemiologic studies involving large administrative databases. With respect to high-dimensional covariate sets, the need for data adaptive methods was one of the open problems that motivated the development of TMLE.14 However, the properties of TMLE in high-dimensional covariate settings (e.g., considering hundreds of variables from administrative databases) have not been widely investigated for the common data setting of a single-point exposure study.15–18 Our objective was therefore to illustrate the practical implementation of TMLE and demonstrate its application in frequent pharmacoepidemiological settings. More specifically, we present the implementation of TMLE in single-point exposure studies with binary exposure and outcome and estimate risk differences (RD), risk ratios (RR), and odds ratios (OR). Furthermore, we apply the TMLE procedure in a point exposure drug effect cohort study of statin use postmyocardial infarction (MI) and the 1-year risk of all-cause mortality and compare the TMLE and IPW using varying covariate sets in the treatment and outcome models.
HIGH-DIMENSIONAL PROPENSITY SCORE
The high-dimensional propensity score procedure is a (semi)-automatic algorithm that identifies and selects a high-dimensional set of covariates from clinical databases or health claims databases to generate an estimated propensity score.2 This algorithm identifies the most prevalent codes within each provided data source and creates three covariates for each identified code. Subsequently, covariates are selected according to the magnitude of confounding bias induced by each covariate. These selected covariates along with some predefined variables are used to estimate the propensity score. The high-dimensional propensity score algorithm has been validated by comparing the results with those expected from randomized trials.19,20 Given the availability of large claims databases in pharmacoepidemiologic studies, the high-dimensional propensity score has been implemented very frequently in practice since its development.4,21,22 Moreover, the possibility of incorporating the procedure into other propensity score based methods has become an interesting research area.
TARGETED MAXIMUM LIKELIHOOD ESTIMATION
We illustrate TMLE in a point treatment study where for each subject, we observe a binary outcome Y, binary treatment indicator variable A, and W, which is a vector including all important confounders for the effect of A on Y. TMLE can be used to estimate the proportion of individuals experiencing a certain event (outcome) if everyone were treated in a target population and the proportion with such an event if everyone were untreated. We will refer to these two quantities as our parameters of interest. The marginal causal effect (i.e., RD, RR, or OR) can then be computed by utilizing (e.g., subtracting or dividing) the two corresponding parameter estimates.
The targeted parameter of interest can be summarized as a function of the counterfactual outcomes, i.e., 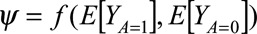 . Here,
. Here, 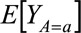 refers to the expected outcome Y in a counterfactual world where every individual in the study population receives treatment A = a. Since Y is a binary outcome variable, this average corresponds to the proportion of individuals in the study population who would develop outcome Y = 1 under treatment A = a.
refers to the expected outcome Y in a counterfactual world where every individual in the study population receives treatment A = a. Since Y is a binary outcome variable, this average corresponds to the proportion of individuals in the study population who would develop outcome Y = 1 under treatment A = a.
Let 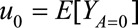 and
and 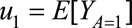 , so that any statistic that is a function of these two quantities can be subsequently computed. In particular, the common effect measures of risk such as RD, RR, and OR can be parameterized as follows:
, so that any statistic that is a function of these two quantities can be subsequently computed. In particular, the common effect measures of risk such as RD, RR, and OR can be parameterized as follows:
where 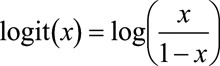 .
.
TMLE is implemented in four steps. In the first step, an initial probability of outcome Y is estimated as a function of treatment A and covariates W. In the second step, a probability of treatment A is estimated as a function of covariates W. In the third step, the initial probability of outcome Y is updated via the fluctuation parameters, which can be estimated by a parametric model that includes a clever covariate to address residual bias. The clever covariate is usually derived from the efficient influence function for the parameter of interest.9,10,23 The magnitude of the fluctuation parameter reflects the strength of the association of a function of the propensity score with the (nonrandom) signal in the residuals. When there is little signal in the residuals, the fluctuation parameter will be close to 0. In the final step, the vector of parameters of interest (i.e., u0 and u1) is estimated by the G-computation formula,24 using the updated probabilities of the counterfactual outcomes. A detailed description of the TMLE implementation is provided in eAppendix 1 (http://links.lww.com/EDE/B45). SEs, confidence intervals, and null-hypothesis testing in TMLE framework are shown in eAppendix 2 (http://links.lww.com/EDE/B45).
SOFTWARE FOR IMPLEMENTING TMLE
R packages for implementing TMLE in both single-point exposure and longitudinal study are available and easily implemented.23,25 We programmed a SAS macro for a general implementation of TMLE in binary point exposure and outcome study (see eAppendix 3; http://links.lww.com/EDE/B45).
EXAMPLE: STATIN USE POSTMYOCARDIAL INFARCTION AND THE 1-YEAR RISK OF ALL-CAUSE MORTALITY
Data Sources
We constructed a retrospective population-based cohort study to examine statin use and the 1-year risk of all-cause mortality following an acute MI using data extracted from the Clinical Practice Research Datalink (CPRD), a UK clinical database of general practitioner records. These data were linked to the Hospital Episode Statistics (HES) database, containing detailed hospitalization records.
Study Population
The study population consisted of patients aged 18 years and older with a recorded MI (using ICD-10 codes in HES data or Read codes in the CPRD) between 1 April 1998 and 31 March 2012. Only the first eligible MI was included for each patient. Due to our use of linked databases, diagnoses of MI recorded within 30 days of the original event were considered part of the original event. Cohort entry was defined as 30 days after the date of either hospital discharge (HES-identified events) or MI diagnosis (CPRD-identified events). The study was restricted to patients with at least 1 year of CPRD and HES history before their MI to ensure sufficient observation time to assess potential confounders. In addition, this study used a new user design, excluding patients with a statin prescription in the 365 days before the date of hospital admission or diagnosis of MI.26 Patients were followed until death from any cause, departure from the CPRD or HES, end of follow-up (1 year), or end of the study period (31 March 2012), whichever occurred first. Statin exposure was assessed during the 30 days between hospital discharge or general practitioner record of MI diagnosis and cohort entry. Patients who received a statin prescription during this period were considered exposed, and those who did not were considered unexposed. To minimize misclassification of exposure, patients were excluded if there were other hospitalizations (other than MI) during this period for which the length of stay was 14 days or more. The outcome of interest, all-cause mortality, was defined as any death recorded in either the CPRD or HES during the 1-year follow-up period. This study was approved by the Independent Scientific Advisory Committee for Medicines and Healthcare Products Regulatory Agency database research (protocol number 14_018) and the Research Ethics Board of the Jewish General Hospital in Montreal, Canada.
Potential Confounders
A range of known potential confounders was included in the propensity score model or the outcome model in the analyses. These confounders, measured in the year before MI, include demographic characteristics (e.g., age, sex), time variables (e.g., year of cohort entry), clinical characteristics (e.g., smoking, alcohol use, and obesity), comorbidities (e.g., diabetes mellitus, atrial fibrillation, coronary artery disease [recorded >30 days before the index MI], acute coronary syndrome, cerebrovascular disease, congestive heart failure, chronic obstructive pulmonary disease, hypertension, hypercholesterolemia, peripheral vascular disease, previous coronary revascularization, previous stroke, previous MI [recorded >30 days before the index MI]), and previous medications prescribed (e.g., aspirin, angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, beta-blockers, calcium-channel blockers, diuretics, fibrates, and nonsteroidal anti-inflammatory drugs). We also adjusted for the number of prescriptions issued and the number of hospitalizations in the previous year, two proxies for overall health. In addition, 400 variables empirically selected by the HDPS algorithm were also included to minimize the unmeasured confounding. A total of 32,792 subjects were included in the study cohort, of which 2,978 died during the year of follow-up. There were 3,518 subjects censored within the 1 year of follow-up.
Estimation of the Treatment Effect
We used an intention-to-treat analysis to compare the 1-year risk of all-cause mortality among patients who initiated statin treatment and patients who did not. We conducted complete-case analysis that excluded subjects who were lost to follow-up within 1 year, assuming noninformative censoring. The data dimensions for the high-dimensional propensity scores algorithm were general practice data (diagnoses, referrals, immunizations, and laboratory tests), HES diagnosis data, HES procedure data, and CPRD medication data, with codes recorded in the year before MI assessed for potential inclusion. Using the HDPS macro,27 the 200 most prevalent codes were identified in each dimension and 400 empirical covariates were selected. Assuming missing at random, multiple imputation by chained equations was used to impute missing smoking and obesity data using the predefined confounding variables, treatment, outcome, and the 400 empirical selected covariates.28,29 TMLE and inverse probability weighted estimators with different propensity score models were used to estimate the marginal ORs of 1-year all-cause mortality associated with statin use.
Statistical Modeling
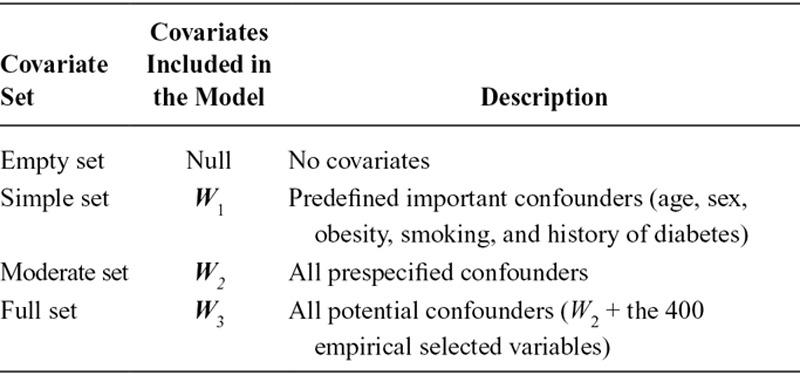
To explore the performance of TMLE in a high-dimensional data setting and compare with inverse probability weighting, we used a varying number of covariates (Table 1) in the propensity score in both approaches and for the outcome model in TMLE. The simple covariate set (W1) included some important confounders, such as age, sex, obesity, smoking, and a history of diabetes. The moderate covariate set (W2) included all the prespecified potential confounders. The full covariate set (W3) included all the potential confounders and the 400 empirically selected variables. These covariates were included as main terms in the propensity score model as well as the outcome model in TMLE using logistic regression. Interactions between covariates were not considered. Inverse probability weighted estimators were constructed using four nested covariate sets in the propensity score models. The weight for each subject is equal to the inverse of the estimated probability of having received his or her own treatment conditional on the covariate set. This weight is stabilized by multiplying by the marginal probability of treatment in the study population. Weighted unadjusted logistic regressions for the outcome were fit using generalized estimating equations, providing marginal estimates as well as robust variance estimates. These inverse probability weighted estimators yielded compatible results to the marginal estimates obtained using TMLE, as noncollapsibility of the OR was not an issue.28 For TMLE, 16 models were composed from the permutation of the covariate sets in Table 1 for the treatment and the outcome models. Although we recognize that some of these models were not the best choices in practice, we believe that these 16 models should address the consequence of different incorrect model specifications, and possibly reflect the double robustness property of TMLE. Without any adjustment in either the propensity score model or the outcome model, the inverse probability weighted estimator and TMLE both provide crude estimates (without confounder control).
TABLE 1.
Potential Covariates to be Adjusted in the Study of Statins and Mortality Using Targeted Maximum Likelihood Estimation and Inverse Probability Weighting
RESULTS
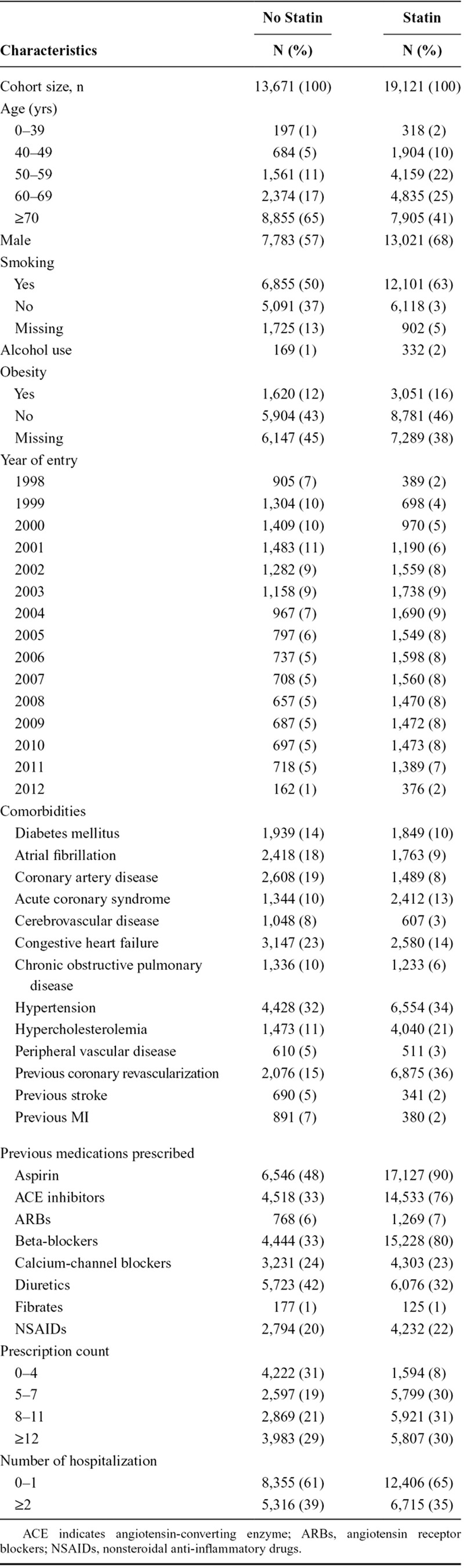
Baseline cohort characteristics are presented in Table 2. Patients who received a statin tended to be younger, entered the cohort later, had more prescriptions overall, and were more likely to have been prescribed an angiotensin-converting enzyme inhibitor, aspirin, or beta-blocker.
TABLE 2.
Baseline Characteristics of Post MI Patients With or Without Statins Use
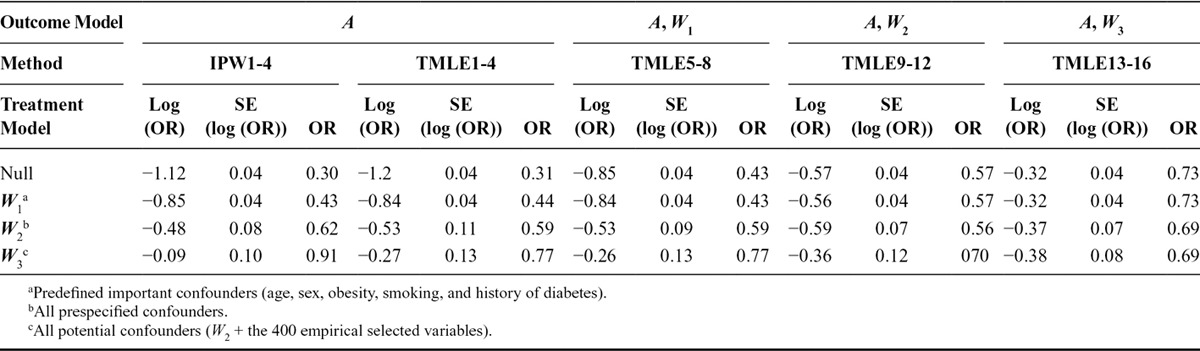
Results for the 16 models are given in Table 3. Adjusting for an increasing number of covariates from W1 to W3, the inverse probability weighted estimator and TMLE (with no covariates included in the outcome model) provided estimates that were far from the crude estimate (log OR: −1.12). With only a few important covariates (simple set W1) included in the propensity score model, the inverse probability weighted estimator and TMLE produced equivalent results for effect estimation (−0.84) and precision (the SE for both was 0.04). However, given the limited adjustment for confounding, the estimates were most likely biased. Although the inverse probability weighted estimator and the analogous TMLE used the same propensity score model, the estimates diverged when W2 and W3 were used; TMLE resulted in a stronger treatment effect as well as a larger SE compared with the inverse probability weighted estimator. TMLE with no adjustment in the propensity score model and adjustment for W1 in the outcome model provided similar results to those obtained with adjustment of W1 via the treatment model. Adjusting for W2 in the outcome model in the TMLE gave estimates close to −0.56 except for the full treatment model. Adjusting for all the measured confounders and the 400 empirical selected variables in the TMLE outcome model, the estimates (i.e., close to −0.32) were all similar regardless of the treatment model specification. The treatment model including no covariates or a few important confounders (W1) yielded the highest precision (SE, 0.04).
TABLE 3.
Effect Estimate of Statins Use Post MI on 1-year All-cause Mortality by Different Targeted Maximum Likelihood Estimation and Inverse Probability Weighting Modeling Approaches
Inspection of the Distribution of the Propensity Score
In contrast to the results where only a few covariates were considered, the inverse probability weighted estimator and TMLE produced different estimates in high dimensional covariate settings (including W2 or W3). With a large number of covariates included in the propensity score model and given the multivariate covariate distribution, the assumption of positivity, which requires the probability of receiving any level of treatment conditional on the covariates has to be greater than zero for each individual, may have been violated or nearly violated. This may have generated large SEs and/or biased estimates.16,31–33
Therefore, we investigated the distribution of the propensity score from the three different model specifications. The histograms of the estimated propensity score are shown in the Figure for the three treatment models. With an increasing number of covariates included in the treatment model, the distribution of the propensity score was more dispersed. The range, median, and quartiles of the propensity score are presented in Table 4 by treatment groups. When including the moderate or full set in the propensity score model, the maximum values of the propensity score were extremely close to 1 in both the treated and untreated groups, while the minimum values were extremely close to 0. The Figure and Table 4 show indications of the near positivity violation and partially explain the instability of the effect estimates after adjusting for all the measured covariates and/or 400 empirically selected variables. With the same adjustment for covariates in the treatment model, the divergent results between the inverse probability weighted estimator and TMLE that includes the exposure only in the outcome model (uninformative outcome model specification) may reflect their different utilization of the propensity score.
TABLE 4.
Distribution of Propensity Score Estimated from Three Different Models by Treatment Group
FIGURE.
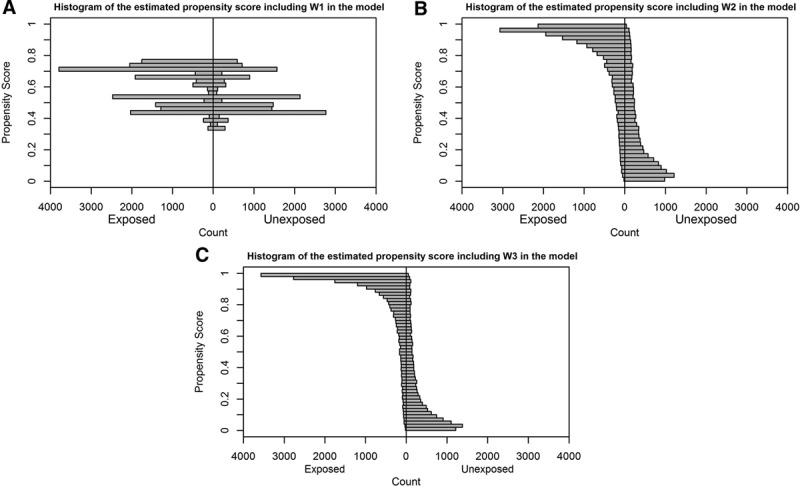
Histograms of the estimated propensity score from three different models. A, W1: predefined important confounders (age, sex, obesity, smoking, and history of diabetes); (B) W2: all prespecified confounders; (C) W3: all potential confounders (W2 + the 400 empirical selected variables).
Stabilized weights with a mean of 1 and a small range are desirable for the variance estimation using inverse probability weighting.32 Inverse probability of treatment weights obtained using the most restricted propensity score model were well behaved (Table 5); however, this model was likely to fail to account for many important confounding variables. The moderate propensity score model yielded weights with desired mean of 1 but with much larger maximum value, implying a larger estimated variance for the treatment effect comparing to the variance estimation from the simple propensity model approach. The model adjusting for the full set of covariates generated weights with mean 1.04, again suggesting a near violation of the positivity assumption and possible bias. Moreover, the SD and range of the weights were large, revealing potential poor precision of the effect estimates. In fact, for a fixed outcome model in TMLE, the SE of the estimate increased with increasing number of included covariates.
TABLE 5.
Distributions of the Stabilized Weights from Three Different Propensity Score Models
DISCUSSION
We have demonstrated the implementation and use of TMLE to estimate the average causal effect of statin use on 1-year all-cause mortality using a large administrative dataset, and compared these results to an inverse probability weighted estimator. Our main results showed that, using TMLE, when the full covariate set was included in the outcome model, all estimated ORs were in agreement regardless of the treatment model specification. These estimates are less prone to bias and more precise compared with the inverse probability weighted estimators using the corresponding propensity score models alone, as TMLE offers an additional opportunity for adjustment for confounding via the outcome model, and is also less influenced by practical positivity violations due to extreme propensity scores. This empirically reflects the double robustness property of TMLE, in that when the initial outcome model was consistently estimated, there was little signal in the residual or bias reduction in the updating step via the treatment model (the estimates of the fluctuation parameters were close to 0). However, we were unable to assess the bias of the two methods under a near positivity violation, since the true effect was unknown.
It is important to note that the two methods handle sparse data and near positivity violations differently. Inverse probability weighting methods, which are known to be sensitive to extreme propensity scores and near-positivity violations,34,35 use the estimated propensity score for the treated subjects and 1 minus the estimated propensity score for the untreated subjects. Therefore, only the treated subjects who have estimated propensity scores extremely close to 0 and untreated subjects who receive estimated propensity scores extremely close to 1 will contribute to the extreme weights. On the other hand, in step 3 of the TMLE procedure, updating is done for each subject under treatment and nontreatment. Any propensity score that is extremely close to 1 or 0 inflates the value of the potential clever covariates, which can cause unstable parameter estimates and/or potential bias. However, the use of a non-null outcome model in TMLE is designed to compensate for this instability. Truncation of the propensity score may also prevent poor precision (but at the price of increased bias);31,32 we therefore performed a sensitivity analysis with propensity score truncated at the 1st and 99th percentile for both inverse probability weighting and TMLE. The results are provided in eTable 1 in eAppendix 4 (http://links.lww.com/EDE/B45), showing slightly changed point estimates and improved precision in both inverse probability weighting and TMLE especially in the case of high-dimensional covariates adjustment in the propensity score models.
Our study had some limitations. We excluded censored subjects, assuming noninformative censoring. The estimates would be biased if this assumption was not met. Therefore, we conducted additional sensitivity analysis that incorporated a censoring model. The results were very similar to those from the complete-case analysis, indicating noninformative censoring (results not shown). We therefore only considered the complete-case analysis as our primary analysis. In addition, there was a certain amount of missing values for smoking and obesity in our example (Table 1). We used multiple imputation by chained equations to impute these missing values. In our multiple imputation models, we included all the baseline covariates as well as the exposure and the outcome of interest to make the missing at random assumption more plausible.29 The results might be biased if this assumption was not met. Furthermore, we only included main effects of the covariates in our analysis without considering higher order terms. Residual bias may be still present if the treatment effect is nonlinear. To take advantage of the properties of TMLE, consistent estimation of both the treatment and outcome model are required and thus data-adaptive methods, such as Super Learner,14 are recommended when implementing TMLE. The ability to integrate data adaptive methods while retaining valid inference is a motivation for TMLE. However, we intended to showcase the implementation of TMLE in a relative simple scenario and thus did not consider complex data-adaptive methods.
Moreover, we investigated the relative performance of inverse probability weighting and TMLE with a high-dimensional setting in a single-point exposure study on a binary outcome. Our results may not extend to more realistic settings with time-varying treatments and failure time outcomes. We only focused on the high dimensional propensity score algorithm for variable selection, while other alternative data-adaptive procedures are available and could be integrated in TMLE.36–40 In particular, collaborate TMLE was developed to select the best adjustment set in high-dimensional settings and therefore may be more efficient in finite samples.37 In contrast to the univariate approach of high-dimensional propensity score for confounder selection, collaborate TMLE assesses the multivariate effect of covariates on the parameter for the propensity score model. A comparison of these two methods should be considered in future studies.
In summary, we have demonstrated the step-by-step implementation of TMLE and implemented TMLE in a point exposure study, estimating the average effect of post-MI statin use on the 1-year risk of all-cause mortality. The high-dimensional propensity score algorithm can be incorporated naturally in the TMLE process to account for the proxies of unmeasured confounding. Even when using the same modeling approach for the propensity score, TMLE and inverse probability weighting estimators can perform differently due to their different utilization of the propensity score. While TMLE is appealing for its double robustness property, as with many other methods near violations of positivity may be problematic for the estimation process.
ACKNOWLEDGMENTS
The authors thank Dr. Mireille E. Schnitzer for detailed and helpful comments on the manuscript.
Supplementary Material
Footnotes
This study was financially supported by The Canadian Network for Observational Drug Effect Studies (CNODES), a collaborating centre of the Drug Safety and Effectiveness Network (DSEN) that is funded by the Canadian Institutes of Health Research (Grant Number DSE-111845). Dr. Filion holds a New Investigator Award from the Canadian Institutes of Health Research. Dr. Platt is the Albert Boehringer I Chair in Pharmacoepidemiology at McGill University, holds a Chercheur-national (National Scholar) award from the Fonds de Recherche du Québec-Santé (FQR-S) and is a member of the Research Institute of the McGill University Health Centre, who receives core funding from the FQR-S.
Dr. Platt received consulting fees for work unrelated to this project from Pfizer, Amgen, Abbvie, and Novartis. The other authors report no conflicts of interest.
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
REFERENCES
- 1.Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 2.Schneeweiss S, Rassen JA, Glynn RJ, Avorn J, Mogun H, Brookhart MA. High-dimensional propensity score adjustment in studies of treatment effects using health care claims data. Epidemiology. 2009;20:512–522. doi: 10.1097/EDE.0b013e3181a663cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rassen JA, Schneeweiss S. Using high-dimensional propensity scores to automate confounding control in a distributed medical product safety surveillance system. Pharmacoepidemiol Drug Saf. 2012;21(suppl 1):41–49. doi: 10.1002/pds.2328. [DOI] [PubMed] [Google Scholar]
- 4.Rassen JA, Avorn J, Schneeweiss S. Multivariate-adjusted pharmacoepidemiologic analyses of confidential information pooled from multiple health care utilization databases. Pharmacoepidemiol Drug Saf. 2010;19:848–857. doi: 10.1002/pds.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robins JM, Hernán MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology. 2000;11:550–560. doi: 10.1097/00001648-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Resche-Rigon M, Pirracchio R, Robin M, et al. Estimating the treatment effect from non-randomized studies: the example of reduced intensity conditioning allogeneic stem cell transplantation in hematological diseases. BMC Blood Disord. 2012;12:10. doi: 10.1186/1471-2326-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scharfstein DO, Rotnitzky A, Robins JM. Adjusting for nonignorable drop-out using semiparametric nonresponse models. J Am Stat Assoc. 1999;94:1096–1120. [Google Scholar]
- 8.Van der Laan MJ. Targeted maximum likelihood based causal inference: part I. Int J Biostat. 2010;6 doi: 10.2202/1557-4679.1211. Article 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenblum M, van der Laan MJ. Targeted maximum likelihood estimation of the parameter of a marginal structural model. Int J Biostat. 2010;6 doi: 10.2202/1557-4679.1238. Article 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore KL, van der Laan MJ. Covariate adjustment in randomized trials with binary outcomes: targeted maximum likelihood estimation. Stat Med. 2009;28:39–64. doi: 10.1002/sim.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stitelman OM, De Gruttola V, van der Laan MJ. A general implementation of TMLE for longitudinal data applied to causal inference in survival analysis. Int J Biostat. 2012;8 doi: 10.1515/1557-4679.1334. [DOI] [PubMed] [Google Scholar]
- 12.Rose S, van der Laan M. A double robust approach to causal effects in case-control studies. Am J Epidemiol. 2014;179:663–669. doi: 10.1093/aje/kwt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruber S, van der Laan MJ. An application of targeted maximum likelihood estimation to the meta-analysis of safety data. Biometrics. 2013;69:254–262. doi: 10.1111/j.1541-0420.2012.01829.x. [DOI] [PubMed] [Google Scholar]
- 14.van der Laan MJ, Rose S. Targeted Learning: Causal Inference for Observational and Experimental Data. Berlin, Germany: Springer Science & Business Media; 2011. [Google Scholar]
- 15.Gruber S, van der Laan MJ. An application of collaborative targeted maximum likelihood estimation in causal inference and genomics. Int J Biostat. 2010;6 doi: 10.2202/1557-4679.1182. Article 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter KE, Gruber S, van der Laan MJ, Sekhon JS. The relative performance of targeted maximum likelihood estimators. Int J Biostat. 2011;7 doi: 10.2202/1557-4679.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porter KE. The relative performance of targeted maximum likelihood estimators under violations of the positivity assumption. 2011. http://escholarship.org/uc/item/3hp4r33n.pdf. Accessed 27 April 2016. [DOI] [PMC free article] [PubMed]
- 18.Schnitzer ME, Moodie EE, Platt RW. Targeted maximum likelihood estimation for marginal time-dependent treatment effects under density misspecification. Biostatistics. 2013;14:1–14. doi: 10.1093/biostatistics/kxs024. [DOI] [PubMed] [Google Scholar]
- 19.Neugebauer R, Schmittdiel JA, Zhu Z, Rassen JA, Seeger JD, Schneeweiss S. High-dimensional propensity score algorithm in comparative effectiveness research with time-varying interventions. Stat Med. 2015;34:753–781. doi: 10.1002/sim.6377. [DOI] [PubMed] [Google Scholar]
- 20.Patorno E, Glynn RJ, Hernández-Díaz S, Liu J, Schneeweiss S. Studies with many covariates and few outcomes: selecting covariates and implementing propensity-score-based confounding adjustments. Epidemiology. 2014;25:268–278. doi: 10.1097/EDE.0000000000000069. [DOI] [PubMed] [Google Scholar]
- 21.Toh S, García Rodríguez LA, Hernán MA. Confounding adjustment via a semi-automated high-dimensional propensity score algorithm: an application to electronic medical records. Pharmacoepidemiol Drug Saf. 2011;20:849–857. doi: 10.1002/pds.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garbe E, Kloss S, Suling M, Pigeot I, Schneeweiss S. High-dimensional versus conventional propensity scores in a comparative effectiveness study of coxibs and reduced upper gastrointestinal complications. Eur J Clin Pharmacol. 2013;69:549–557. doi: 10.1007/s00228-012-1334-2. [DOI] [PubMed] [Google Scholar]
- 23.Gruber S, van der Laan MJ. tmle: An R Package for Targeted Maximum Likelihood Estimation. 2011. Available at http://biostats.bepress.com/ucbbiostat/paper275/. Accessed 22 May 2014.
- 24.Robins J. A new approach to causal inference in mortality studies with a sustained exposure period—application to control of the healthy worker survivor effect. Math Model. 1986;7:1393–1512. [Google Scholar]
- 25.Schwab J, Lendle S, Petersen M, van der Laan M, Gruber S. Ltmle: Longitudinal Targeted Maximum Likelihood Estimation. 2014. Available at http://cran.r-project.org/web/packages/ltmle/index.html. Accessed 7 December 2014.
- 26.Ray WA. Evaluating medication effects outside of clinical trials: new-user designs. Am J Epidemiol. 2003;158:915–920. doi: 10.1093/aje/kwg231. [DOI] [PubMed] [Google Scholar]
- 27.Rassen JA, Doherty M, Huang W, Schneeweiss S. Pharmacoepidemiology Toolbox. Available at http://www.drugepi.org/dope-downloads/. Accessed 27 April 2016. [Google Scholar]
- 28.van Buuren S, Groothuis-Oudshoorn K. MICE: multivariate imputation by chained equations in R. J Stat Softw. 2011;45:1–67. [Google Scholar]
- 29.White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med. 2011;30:377–399. doi: 10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- 30.Pang M, Kaufman JS, Platt RW. Studying noncollapsibility of the odds ratio with marginal structural and logistic regression models. Stat Meth Med Res. doi: 10.1177/0962280213505804. forthcoming. [DOI] [PubMed] [Google Scholar]
- 31.Petersen ML, Porter KE, Gruber S, Wang Y, van der Laan MJ. Diagnosing and responding to violations in the positivity assumption. Stat Meth Med Res. 2012;21:31–54. doi: 10.1177/0962280210386207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole SR, Hernán MA. Constructing inverse probability weights for marginal structural models. Am J Epidemiol. 2008;168:656–664. doi: 10.1093/aje/kwn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruber S, van der Laan MJ. A targeted maximum likelihood estimator of a causal effect on a bounded continuous outcome. Int J Biostat. 2010;6 doi: 10.2202/1557-4679.1260. Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neugebauer R, van der Laan M. Why prefer double robust estimators in causal inference? J Stat Plan Inference. 2005;129:405–426. [Google Scholar]
- 35.Ertefaie A, Stephens DA. Comparing approaches to causal inference for longitudinal data: inverse probability weighting versus propensity scores. Int J Biostat. 2010;6 doi: 10.2202/1557-4679.1198. Article 14. [DOI] [PubMed] [Google Scholar]
- 36.van der Laan MJ, Polley EC, Hubbard AE. Super learner. Stat Appl Genet Mol Biol. 2007;6:Article25. doi: 10.2202/1544-6115.1309. [DOI] [PubMed] [Google Scholar]
- 37.van der Laan MJ, Gruber S. Collaborative double robust targeted maximum likelihood estimation. Int J Biostat. 2010;6 doi: 10.2202/1557-4679.1181. Article 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westreich D, Lessler J, Funk MJ. Propensity score estimation: neural networks, support vector machines, decision trees (CART), and meta-classifiers as alternatives to logistic regression. J Clin Epidemiol. 2010;63:826–833. doi: 10.1016/j.jclinepi.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCaffrey DF, Ridgeway G, Morral AR. Propensity score estimation with boosted regression for evaluating causal effects in observational studies. Psychol Methods. 2004;9:403–425. doi: 10.1037/1082-989X.9.4.403. [DOI] [PubMed] [Google Scholar]
- 40.Sinisi SE, Van Der Laan MJ. Loss-based cross-validated deletion/substitution/addition algorithms in estimation. Berkeley Division of Biostatistics Working Paper Series. 2004 [Google Scholar]


