Abstract
Background
The purpose of this study was to evaluate the virulence profiles of Pseudomonas aeruginosa clinical strains recently isolated from patients hospitalized for chronic leg ulcers in the Dermatology Department of Central Military Emergency University Hospital “Carol Davila”, Bucharest, Romania.
Methods
The phenotypic screening evaluated eight soluble virulence factors (haemolysins, lecithinase, lipase, caseinase, gelatinase, amylase, DNase, aesculin hydrolysis), as well as adherence ability (Cravioto adapted method) and invasion capacity on HeLa cells (gentamicin protection assay). Seven virulence genes encoding for protease IV, 3 exoenzymes (exoS, exoT, exoU), two phospholipases plcH- haemolytic phospholipase C and plcN- non-haemolytic phospholipase C) and alginate were investigated by PCR.
Results
The pore forming toxins and enzymes were expressed in variable proportions, the majority of the tested strains producing beta haemolysin (92.3 %), lipase (76.9 %) and lecithinase (61.5 %). The most frequent virulence genes detected in the analyzed strains were the ExoT (100 %) and AlgD (92.3 %) genes, genes codifying for phospholipases (84.6 % each of them) and for protease IV (61.5 %).
Conclusions
This study reveals that correlating virulence profiles and infection clinical outcome is very useful for setting up efficient preventive and therapeutic procedures for hospitalized patients with chronic leg ulcers and positive P. aeruginosa cultures.
Keywords: Pseudomonas aeruginosa, Genetic and phenotypic virulence factors, Chronic leg ulcers
Background
Chronic leg ulcers represent a clinically highly relevant topic, this condition affecting 1–2 % of the general population and being responsible for increased morbidity and impaired quality of life. Pseudomonas aeruginosa, one of the most common bacteria isolated from chronic wounds [1], is an opportunistic pathogen with innate resistance to many antibiotic classes, including antipseudomonal penicillins, carbapenems, aminoglycosides and ciprofloxacin [2, 3]. Treatment options for infections caused by antibiotic resistant P. aeruginosa is limited to carbapenems, but increasing resistance rates mediated in most cases by the production of carbapenemases such as metallo-β-lactamases (MBLs) are reported [4, 5]. In case of P. aeruginosa isolated from chronic wounds there is no consensus on the first-line antibiotic to be used [6].
Besides intrinsic and acquired resistance to many antibiotics, P. aeruginosa expresses many extracellular virulence factors (exotoxins S and T with ADP-ribosylating activity which cause disruption of the host cell cytoskeleton, cytotoxin ExoU with phospholipase activity and cytotoxin Exo Y with an adenylate cyclase activity, phospholipases, protease IV and elastase) and surface associated components, as alginate. All these factors contribute to massive tissue damage, blood infection dissemination, and progression of the disease, and also limit treatment options with currently available antibiotics [7, 8]. A retrospective multicenter evaluation of 970 patients with chronic leg ulcer treated in different dermatologic wound care centers from Germany revealed that P. aeruginosa was found in 31.1 % of the analyzed cases and a positive correlation regarding wound size and duration was observed [9]. Because in many cases chronic leg ulcers are associated with diabetes, and taking into account the high resistance of this species, persistence of P. aeruginosa in these patients was observed. In a longitudinal survey regarding patients with non-healing venous ulcers, P. aeruginosa was isolated ≥ 3 years from the same patients, and a longer wound duration compared to all analyzed patients was revealed [10].
Considering the above mentioned aspects and taking into account the lack of such studies on Romanian strains the purpose of this study was to investigate the phenotypic and genotypic profile of P. aeruginosa strains isolated from Romanian patients with chronic leg ulcers.
Methods
Isolation and identification of bacterial strains
This study was conducted on a total of 12 P. aeruginosa clinical strains isolated during July 2013 - June 2015, from 130 patients hospitalized for chronic leg in the Department of Dermatology of the Central Military Emergency University Hospital “Carol Davila”, Bucharest, Romania. The patient or the legally authorized representative had to be able to read and sign the informed consent. Swab samples were collected for aerobic bacterial culture. The swabs were transported in thioglycolate broth media and then transferred on 5 % sheep blood agar. Cultures were incubated at 37 °C aerobically for 24 h. The identification of microbial isolates was performed using Gram staining, conventional biochemical tests and API BioMerieux systems (20NE).
Evaluation of the soluble enzymatic factors
The virulence phenotypes were assessed by performing enzymatic tests for the expression of some soluble virulence factors. Overnight culture of the strains was evaluated for the following virulence factors expression: haemolysins, other pore forming toxins (lecithinase, lipase), proteases (caseinase, gelatinase), amylase and aesculin hydrolysis. Detection of haemolysin production was performed by spotting the fresh cultures on 5 % sheep blood agar medium and incubation at 37 °C for 24 h. The colorless area around the culture revealed the presence of haemolysis activity. For lipase production the strains were spotted on 1 % Tween 80 agar as a substrate and followed by incubation at 37 °C for 24 h. An opaque (precipitation) zone around the spot was registered as positive reaction; for lecithinase production, the cultures were spotted into 2.5 % yolk agar and incubated at 37 °C for 24 h. A clear zone around the spot indicated the lecithinase production. The protease activity (caseinase and gelatinase) was determined using 15 % soluble casein agar, respectively 3 % gelatin as substrate. The strains were spotted and after incubation at 37 °C for 24 h, a white precipitate surrounding the growth indicated casein proteolysis, and colorless area around culture due to the gelatin hydrolysis, indicated the positive reaction for gelatinase. Amylase was detected using agar with 1 % starch and hydrolysis was revealed after adding Lugol's solution (yellow ring around the culture, while the rest of the plate will be blue). For the aesculin hydrolysis the medium containing Fe 3+ citrate was used and inoculated by spotting, then incubated for 24 h at 37 °C. A black precipitate around culture due to esculetol released under the action of beta-galactosidase was considered positive reaction.
Evaluation of biofilm development on inert substrata was assessed by the microtiter method. Overnight bacterial cultures were grown in 96 multi-well plates containing Tryptic Soy Broth (TSB) for 24 h at 37 °C, then the plate was emptied and washed three times with phosphate buffered saline (PBS). The adherent cells were then fixed with cold methanol, stained with an alkaline 1 % violet crystal solution for 15 min, washed with water and resuspended in a 33 % acetic acid solution. The intensity of the suspension was spectrophotometrically assessed, the amount of adhered biomass being proportional to the absorbance value read at 492 nm.
Evaluation of the bacterial adherence to cellular substrata (HeLa cells)
For the adherence assay, Cravioto’s adapted method was used [11, 12]. Briefly, HeLa cell monolayers were washed with PBS and 1 ml of fresh medium without antibiotics was aseptically added to each well. The suspensions of P. aeruginosa obtained from mid-logarithmic phase cultures grown in nutrient broth were adjusted to 107 colony forming units (CFU)/ml and 1 ml was used for the inoculation of each well. After two hours of incubation at 37 °C the monolayers were washed 3 times with PBS, fixed in ethanol (3 min) and stained with 1:10 v/v Giemsa solution for 20 min. The plates were washed, dried at room temperature overnight, and examined by optic microscopy using wet objective (×1000 magnification), in order to evaluate the adherence indexes and patterns. The adherence indexes were expressed as the ratio between the number of the eukaryotic cells with adhered bacteria and 100 eukaryotic cells counted on the microscopic field. The adherence patterns were defined as: localized adherence (LA) when tight clusters of microorganisms were noticed on the HeLa cell surface, aggregative adherence (AA) when a microbial stacked brick pattern characterized the attachment, diffuse adherence (DA) when the bacteria adhered diffusely, covering the whole surface of the cell [13].
Evaluation of the bacterial invasion capacity
To determine the invasion ability the antibiotic protection assay was used [14, 15]. Bacterial suspensions were inoculated in duplicate on HeLa cells grown in 6-well plates and incubated at 37 °C for two hours. The infection assay was performed in the same manner as the adherence assay, but after the incubation period in one of two wells inoculated with each strain 100 μg/ml gentamicin was added in order to kill extracellular bacteria. The plates were further incubated for another hour; after incubation, plates were washed 3 times with PBS and the HeLa cells were permeabilized with 0.1 % Triton X-100 for 10 min, at 37 °C. Serial dilutions of suspended cells harvested were seeded on nutritive agar (three technical replicates for each dilution) in order to establish the invasion indexes, calculated as CFU/ml.
Evaluation of the antibiotic susceptibility
The antibiotic susceptibility testing was performed by Kirby-Bauer standard disk diffusion method (panels of antibiotic disks recommended by CLSI, 2013, 2014, 2015).
PCR assays for virulence genes detection
The genetic support of the virulence factors was investigated by simplex and multiplex PCR, using a reaction mix of 20 or 25 μl (PCR Master Mix 2x, Thermo Scientific) containing 1 μl of bacterial DNA extracted using the alkaline extraction method. In this purpose, 1–5 colonies of bacterial cultures were suspended in 1.5 ml tubes containing 20 μl solution of NaOH (sodium hydroxide) and SDS (sodium dodecyl sulphate) and heated on a thermoblock at 95 °C for 15 min. for the permeabilization of bacterial wall. The following step was the addition of 180 μl of TE buffer (TRIS + EDTA) 1X and centrifugation at 13000 rpm for 3 min. All PCR reactions were performed using the Thermal Cycler machine Corbet. Genomic DNA was used as a template for the PCR screening of 7 virulence genes encoding for protease IV, three exoenzymes – exoS, exoT, exoU, two phospholipases - plcH (haemolytic phospholipase C) and plcN (non-haemolytic phospholipase C) and for alginate. The PCR reactions were initiated with 1 cycle at 95 °C for 2 min, followed by 30 cycles at 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min and a final elongation step at 72 °C for 7 min. The amplification products were visualized by electrophoresis on a 1 % agarose gel, stained with the specific weight marker (100pb, Ladder Bench Top, Promega, USA).
Results and discussion
In the present study, from 130 bacterial strains isolated from chronic leg ulcers of hospitalized patients, only 12 (9.23 %) were P. aeruginosa, accounting for a smaller percentage comparing with the number of cases reported in other studies. For example, in a study of Wolcott et al. (2015) performed on 2,963 patients with chronic wounds, P. aeruginosa represented 25 % from the total number of the isolated strains [16].
The isolated strains expressed between one and six of the seven investigated soluble virulence factors (Table 1). The most frequently expressed soluble virulence factors were caseinase (91.67 %) followed by beta haemolysins and lipase (83.33 %) (Fig. 1). Production of proteases by the majority of the tested strains indicates the ability of these strains to induce tissue lesions and to delay the wound healing. The pore forming enzymes, i.e. beta haemolysins, lecithinase (66.67 %) and lipase play an important role in dissemination of infection and wound extent. Although some studies [17] reported an increased expression of DNase in the strains isolated from wound secretions, in our study only 2 of the 12 strains of P. aeruginosa produced DNase (16.67 %).
Table 1.
Phenotypic and molecular characteristics of Pseudomonas aeruginosastrains
| Strain no. | Strain | DNase | Aesculin hydrolysis | Amylase | Caseinase | Lecithinase | Lipase | Gelatinase | Haemolysin | PLCH | PLCN | ExoU | ExoT | AlgD | ExoS | TCF/TCR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1333(1) | - | + | + | +++ | + | + | + | β | + | + | - | + | + | + | + |
| 2 | 371 | - | + | + | +++ | + | + | + | β | + | + | - | + | + | + | + |
| 3 | c10 | - | - | + | +++ | + | + | + | β | + | + | - | + | + | + | + |
| 4 | 1601(1) | + | - | + | +++ | + | + | + | β | + | + | - | + | + | + | + |
| 5 | 1453(1) | - | - | - | +++ | + | + | + | β | + | + | - | + | + | - | |
| 6 | 2469(1) | - | - | - | +++ | + | + | + | β | - | - | + | + | - | - | - |
| 7 | 5711(1) | + | - | + | +++ | - | + | + | β | + | + | - | + | + | + | + |
| 8 | R2 | - | - | - | +++ | - | + | + | β | + | + | - | + | + | - | + |
| 9 | c03 | - | - | - | +++ | - | + | - | β | + | + | - | + | + | - | - |
| 10 | 2576(1) | - | - | - | +++ | - | + | - | β | + | + | - | + | + | - | + |
| 11 | DU | - | - | - | + | + | - | - | δ | + | + | - | + | + | + | - |
| 12 | DR | - | - | - | - | + | - | - | δ | - | - | - | + | + | - | + |
Fig. 1.
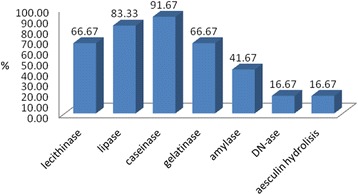
Graphic representation of enzymatic virulence factors expression
Regarding the cell-associated virulence factors, the analyzed strains showed variable rates of adherence to the inert substratum (Fig. 2). This adherence phenotype is correlated with the high capacity of P. aeruginosa strains to develop complex biofilm structures [18], aspect that can explain the persistence of this bacterium in the organism, and also its high antibiotic resistance.
Fig. 2.
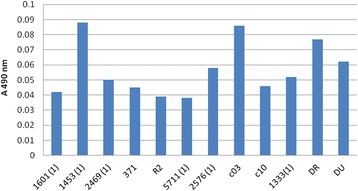
Graphic representation of adherence to inert substratum: all strains exhibited adherence capacity, the highest level being observed in the case of 1453(1), c03 and DR strains. The number of adherent bacteria was determined by measuring the absorbance at 490 nm
P. aeruginosa analyzed strains exhibited different indexes of adherence to the cellular substratum represented by HeLa cells, ranging from 12 to 100 %. All the adherence patterns to HeLa were observed, but mostly the localized pattern was revealed (Fig. 3).
Fig. 3.
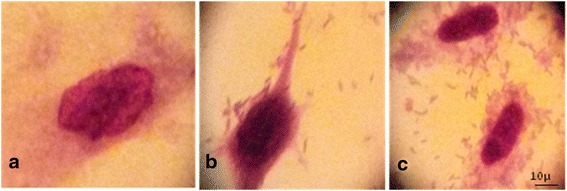
Different adherence patterns of P. aeruginosa analyzed strains: a- control, b- localized pattern, c- aggregative pattern (1000X)
The analyzed strains proved the ability to invade the cellular substratum, which was somehow expected due to the presence of pore forming enzymes that cause pores in the cell membranes, and facilitate internalization of the bacteria in the host cells (Fig. 4, Table 1).
Fig. 4.
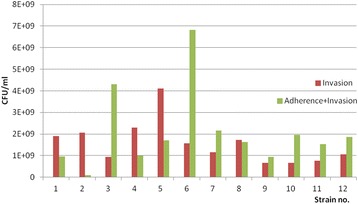
Graphic representation of adherence and invasiveness rates of P. aeruginosa –the number of colony forming units of adherent and invasive bacteria (green) and invasive bacteria (treated with gentamicin) (red) - all the strains adhered to the cellular substrate and also succeeded to invade the cells, some of them showing a high invasive capacity (strains no 1, 2, 4 and 5)
The ability of P. aeruginosa to survive in host cells may explain some aspects of host-pathogen relationship. The ability of bacteria to invade eukaryotic cells could protect them against host defense and antibiotic treatment facilitating the recurrent chronic diseases and long-term colonization process.
P. aeruginosa strains isolated from chronic leg wound were resistant in high proportions to ticarcillin, to third and fourth generation cephalosporins and to ciprofloxacin. Only one strain demonstrated resistance to imipenem and meropenem (Fig. 5). None of the isolated strains demonstrated resistance to colistin. A recent study in Romania [18] identified 4 strains of P. aeruginosa showing comparable resistance rates to antibiotics.
Fig. 5.
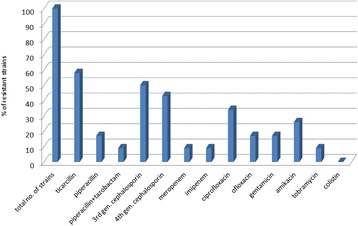
Antibiotic resistance profiles (%) among isolated Pseudomonas strains
On the other hand, international data demonstrate variable resistance: one study revealed strains isolated from chronic wounds resistant to piperacillin/tazobactam and aztreonam [18], another study found P. aeruginosa strains 100 % susceptible only to imipenem and meropenem, followed by ceftazidime (86 %) and amikacin, aztreonam, cefepime, ciprofloxacin, and piperacillin tazobactam (57 % each) [19]; opposite, another study found only 11.5 % resistance to imipenem (out of a collection of 512 P. aeruginosa strains), 66 % of the strains resistant to streptomycin, and 25.2 % and 28.5 % respectively were resistant to aminoglycoside antibiotics ciprofloxacin and gentamicin, respectively, polymyxin B having the greatest antibacterial activity (4.9 % resistant strains to this antibiotic) [20].
The molecular analysis through PCR arrays showed that all the analyzed strains revealed the ExoT gene, 92.3 % of the isolates expressed AlgD gene, 84.6 % of the strains revealed plcH and plcH genes, 61.5 % of P. aeruginosa expressed the gene codifying for protease IV (TCF/TCR) and only 1 strain expressed the ExoU gene (Fig. 6).
Fig. 6.
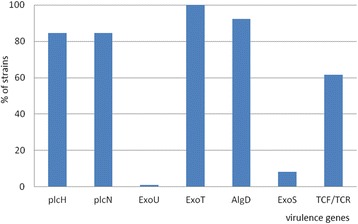
Percentage distribution of virulence genes in analyzed strains
Multiple bacterial virulence factors impact the pathogenesis of P. aeruginosa infections. The combination of virulence factors expressed by each P. aeruginosa strain tends to determine the outcome of an infectious process. In the hospitals units, it is often difficult to distinguish between colonization and infection, and no diagnostic tool is available to assess the virulence potential of a given isolate [21, 22]. Among P. aeruginosa isolates three soluble proteins are responsible for invasion: a rhamnolipid and two phospholipases C (haemolytic phospholipase C (plcH), and non-haemolytic phospholipase C (plcN)) [23]. These two phospholipases could work synergistically, plcH would promote degradation of the erythrocyte membrane (phospholipids components of the outer leaflet: phosphatidylcholine and sphingomyelin), exposing the inner leaflet. Plc-N could then hydrolyze phosphatidylserine present in the inner leaflet. Our results of genotyping analysis showed that 84.6 % of the P. aeruginosa isolates possess phospholipases genes (Figs. 6 and 7) compared with other studies from our country in which only the plcH gene was revealed in the strains isolated from blood cultures and wound secretions [17]. The phospholipase gene expression is sustained at phenotypic level, the same strains being positive for lipase production.
Fig. 7.
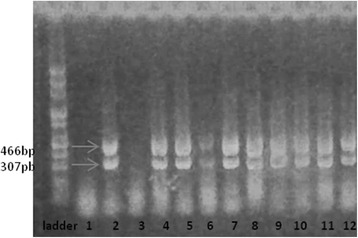
Electrophoresis gel for PlcH (466pb) and PlcN (307pb) genes: all the tested strains except no. 2469 and DR revealed the two phospholipases. Lines: PCR Marker (Promega) - 100pb, 1 - 2469, 2- DU, 3- DR, 4- 571(1), 5- 1453, 6- C03, 7- 1333, 8- 2576, 9- 371, 10-C10, 11- 601, 12- R2
Our PCR results concerning the presence of algD gene showed that 92.3 % of the isolates express this gene (Fig. 8). The algD gene encodes GDP-mannose dehydrogenase, which acts as a rate-limiting enzyme in mucoid strains by catalyzing the conversion of GDP-mannose to GDP-mannuronic acid, thereby promoting the cell to alginate production [24], which demonstrates the involvement of these strains in infections with biofilm formation [23]. And indeed, the high level of AlgD gene expression correlates with phenotypic results, adherence to inert substratum being also observed among tested strains (Table 1).
Fig. 8.
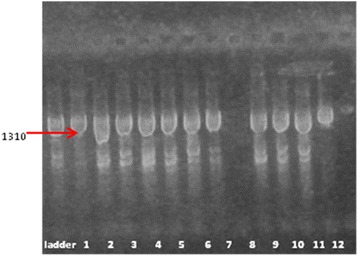
Electrophoresis gel of AlgD gene. The figure shows that all the analyzed strains except one (the strain DR) are positive for the respective gene. Lines: PCR Marker (Promega) - 100pb, 1-R2, 2-1333(1); 3-5711, 4-1453, 5-2576, 6-C10, 7-2469, 8-DR, 9-1601, 10-371, 11-C03, 12-DU
It is known that the exotoxins ExoS, ExoY, and ExoT inhibit invasion, while ExoU confers cytotoxicity [25–27]. ExoT and ExoS act on a number of host small G proteins, thus altering the cytoskeleton and signaling pathways [28]. The results of Shaver and Hauser indicate that ExoU has the greatest effect on virulence of the type III secreted proteins [29]. The concomitant presence of ExoU and ExoS was also revealed by one isolate of P. aeruginosa (Figs. 9 and 10).
Fig. 9.
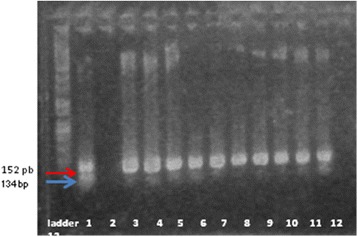
Electrophoresis gel of ExoU and ExoT genes: the figure shows that all the isolates revealed the ExoT. 2469 expressed the ExoU gene. Lines: PCR Marker (Promega) - 100pb, 1-2469, 2-DR, 3-5711, 4-1453, 5-C03, 6-1333(1), 7-2576, 8-371, 9-C10, 10-R2, 11-DU, 12-1601
Fig. 10.
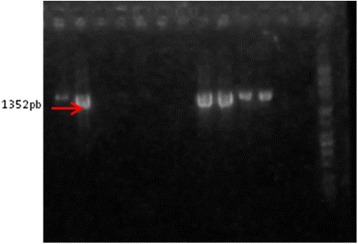
Electrophoresis gel of ExoS gene: the positive strains are 1333, 5711, 1601, 371, c10, DU. PCR Marker (Promega) - 100pb, 1-R2, 2-1333(1); 3-5711, 4-1453, 5-2576, 6-C10, 7-2469, 8-DR, 9-1601, 10-371, 11-C03, 12-DU
The isolated strains of P. aeruginosa exhibited a high frequency of protease IV (TCF/TCR), which plays a role in tissue injuries (Fig. 11). Holban A.M. et al. [17] reported the highest frequency of plcH and protease IV genes in P. aeruginosa strains analyzed in their study.
Fig. 11.
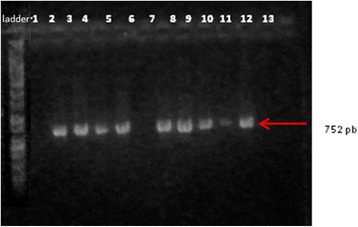
Electrophoresis gel of protease IV (TCF/TCR) the positive strains are no. 1333, 371, C10, 2576, 5711, R2, 1601, DR. Lines: PCR Marker (Promega) - 100pb, 1-1453, 2-DU, 3-2576, 4-5711, 5-R2, 6-1601, 7-2469, 8-DR, 9-1333(1), 10-371, 11-C03, 12-C10
Conclusions
Our phenotypic and molecular results demonstrate that the analyzed P. aeruginosa strains express virulence features which explain the survival and implication of these strains in the poor prognosis of chronic leg wounds. The phenotypic virulence markers were correlated with some specific virulence genes profiles, revealing that the isolates could adapt easily to the microenvironment encountered within the host by modulating the expression of these genes. Furthermore, the diversity of virulence factors expression explains the large panel of clinical manifestations in Pseudomonas aeruginosa infections. All the data resulting from this study, the first of its kind in our country, could help clinicians to achieve correlations between clinical manifestations and the virulence of the involved strain and to adjust the therapeutic approach, consequently.
Acknowledgements
The results presented in this study were supported by the Ideas 1716/2011 grant (contract no. 154/05.10.2011) and Human Resources (PN-II-RU-TE- 2014-4-2037).
Declaration
Publication of this supplement was funded by the Prof. Dr. Matei Balș Foundation, Bucharest, Romania.
Footnotes
Competing interests
No conflict of interests to declare.
Authors’ contributions
MG, IG, CB and CI performed the experiments and drafted the article. MC designed the experiment and VL read and corrected the manuscript. All authors approved the final version of the manuscript.
Contributor Information
Mihaela Georgescu, Email: georgescucmihaela@gmail.com.
Irina Gheorghe, Email: IRYNA_84@yahoo.com.
Carmen Curutiu, Email: carmeniordache78@yahoo.com.
Veronica Lazar, Email: veronica.lazar2009@gmail.com.
Coralia Bleotu, Email: cbleotu@yahoo.com.
Mariana-Carmen Chifiriuc, Email: carmen_balotescu@yahoo.com.
References
- 1.Serra R, Grande R, Butrico L, Rossi A, Settimio UF, Caroleo B, et al. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther. 2015;13:605–13. doi: 10.1586/14787210.2015.1023291. [DOI] [PubMed] [Google Scholar]
- 2.Dundar D, Otkun M. In-vitro efficacy of synergistic antibiotic combinations in multidrug resistant Pseudomonas aeruginosa strains. Yonsei Med J. 2010;51:111–6. doi: 10.3349/ymj.2010.51.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahid M, Malik A, Sheeba Multidrug resistant Pseudomonas aeruginosa strains harbouring R-plasmids and Amp C β-lactamases isolated from hospitalized burn patients in tertiary care hospital of North India. FEMS Lett. 2003;228:181–6. doi: 10.1016/S0378-1097(03)00756-0. [DOI] [PubMed] [Google Scholar]
- 4.Gupta V. Metallo beta lactamases in Pseudomonas aeruginosa and Acinetobacter baumannii species. Expert Opin Investig Drugs. 2008;17:131–43. doi: 10.1517/13543784.17.2.131. [DOI] [PubMed] [Google Scholar]
- 5.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–25. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzika E, Ferrara D, Boehncke WH, Trellu LT, Barouti N. Chronic leg ulcers and Pseudomonas aeruginosa infection. Rev Med Suisse. 2015;11:768–72. [PubMed] [Google Scholar]
- 7.Breidenstein EB, Fuente-Nunez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19:419–26. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73:338–44. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Jockenhöfer F, Gollnick H, Herberger K, Isbary G, Renner R, Stücker M, et al. Bacteriological pathogen spectrum of chronic leg ulcers: Results of a multicenter trial in dermatologic wound care centers differentiated by regions. J Dtsch Dermatol Ges. 2013;11:1057–63. doi: 10.1111/ddg.12170. [DOI] [PubMed] [Google Scholar]
- 10.Renner R, Sticherling M, Rüger R, Simon J. Persistence of bacteria like Pseudomonas aeruginosa in non-healing venous ulcers. Eur J Dermatol. 2012;22:751–7. doi: 10.1684/ejd.2012.1865. [DOI] [PubMed] [Google Scholar]
- 11.Saviuc C, Grumezescu AM, Holban A, Chifiriuc C, Mihaescu D, Lazar V. Hybrid nanostructurated material for biomedical applications. Biointerface Res App Chem. 2011;1:64–71. [Google Scholar]
- 12.Saviuc C, Grumezescu AM, Oprea E, Radulescu V, Dascalu L, Chifiriuc MC, et al. Antifungal activity of some vegetal extracts on Candida biofilms developed on inert substratum. Biointerface Res App Chem. 2011;1:15–23. [Google Scholar]
- 13.Fleiszik SMJ, Zaidi TS, Preston MJ, Grout M, Evans DJ, Pier GB. The relationship between cytotoxicity and epithelial cell invasion by corneal isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64:2288–94. doi: 10.1128/iai.64.6.2288-2294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lannote P, Watt S, Mereghetti L, Dartigueslongue N, Rastegar-Lari A, Goudeau A, et al. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol. 2004;53:73–81. doi: 10.1099/jmm.0.05324-0. [DOI] [PubMed] [Google Scholar]
- 15.Zaas DW, Duncan MJ, Li G, Wright JR, Abraham SN. Pseudomonas invasion of type I pneumocytes is dependent on the expression and phosphorylation of caveolin-2. J Biological Chem. 2005;280:4864–72. doi: 10.1074/jbc.M411702200. [DOI] [PubMed] [Google Scholar]
- 16.Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2015 doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
- 17.Holban AM, Chifiriuc MC, Cotar AI, Bleotu C, Grumezescu AM, Banu O, et al. Virulence markers in Pseudomonas aeruginosa isolates from hospital acquired infections occurred in patients with underlying cardiovascular disease. Rom Biotechnol Letters. 2013;18:8843–54. [Google Scholar]
- 18.Mihai MM, Holban AM, Giurcăneanu C, Popa LG, Buzea M, Filipov M, et al. Identification and phenotypic characterization of the most frequent bacterial etiologies in chronic skin ulcers. Rom J Morphol Embryol. 2014;55:1401–8. [PubMed] [Google Scholar]
- 19.Wong SY, Manikam R, Muniandy S. Prevalence and antibiotic susceptibility of bacteria from acute and chronic wounds in Malaysian subjects. J Infect Dev Ctries. 2015;9:936–44. doi: 10.3855/jidc.5882. [DOI] [PubMed] [Google Scholar]
- 20.Gelatti LC, Bonamigo RR, Becker AP, Eidt LM, Ganassini L, d’Azevedo PA. Phenotypic, molecular and antimicrobial susceptibility assessment in isolates from chronic ulcers of cured leprosy patients: a case study in Southern Brazil. An Bras Dermatol. 2014;89:404–8. doi: 10.1590/abd1806-4841.20142688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balarjishvili NS, Kvachadze LI, Kutateladze MI, Meskhi TS, Pataridze TK, Berishvili TA, et al. New virulent bacteriophages active against multiresistant Pseudomonas aeruginosa strains. App Biochem Microbiol. 2015;51:674–82. doi: 10.1134/S0003683815060034. [DOI] [PubMed] [Google Scholar]
- 22.Sadikot RT, Blackwell TS, Christman JW, Prince AS. Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171:1209–23. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotar AI, Chifiriuc MC, Banu O, Lazar V. Molecular characterization of virulence patterns in Pseudomonas aeruginosa strains isolated from respiratory and wound samples. Biointerface Res Appl Chem. 2013;3:551–8. [Google Scholar]
- 24.Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–22. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 25.Fink-Barbancon V, Goranson J, Zhu LJ, Sawa T, Wiener-Kronish JP, Fleiszik SMJ, et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–57. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 26.Cowell BA, Chen DY, Frank DW, Vallisa AJ, Fleiszik SM. ExoT of cytotoxic Pseudomonas aeruginosa prevents uptake by corneal epithelial cells. Infect Immun. 2000;68:403–6. doi: 10.1128/IAI.68.1.403-406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiol. 2001;147:2659–69. doi: 10.1099/00221287-147-10-2659. [DOI] [PubMed] [Google Scholar]
- 28.Krall RG, Schmidt KA, Barbierij T. Pseudomonas aeruginosa ExoT is a Rho GTPase activating protein. Infect Immun. 2000;68:6066–8. doi: 10.1128/IAI.68.10.6066-6068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72:6969–77. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


