Abstract
DNA-based machines that walk by converting chemical energy into controlled motion could be of use in applications such as next generation sensors, drug delivery platforms, and biological computing. Despite their exquisite programmability, DNA-based walkers are, however, challenging to work with due to their low fidelity and slow rates (~1 nm/min). Here, we report DNA-based machines that roll rather than walk, and consequently have a maximum speed and processivity that is three-orders of magnitude greater than conventional DNA motors. The motors are made from DNA-coated spherical particles that hybridise to a surface modified with complementary RNA; motion is achieved through the addition of RNase H, which selectively hydrolyses hybridised RNA. Spherical motors move in a self-avoiding manner, whereas anisotropic particles, such as dimerised particles or rod-shaped particles travel linearly without a track or external force. Finally, we demonstrate detection of single nucleotide polymorphism by measuring particle displacement using a smartphone camera.
Inspired by biological motors, a number of synthetic motors have been developed as model systems employing a wide range of chemical reactions.1-6 DNA-based machines that walk along a track have shown the greatest promise in recapitulating the properties of biological motor proteins.1, 7-14 However, the maximum distance traveled by the most processive DNA-based motors is 1 μm.8, 9, 12 The velocity of these walkers is also limited to rates of ~1 nm/min due to a fundamental trade-off between motor endurance and speed. The problem is that DNA-walkers tend to spontaneously dissociate off their tracks, and while increasing motor multivalency improves track affinity and motor endurance (the mean distance travelled), this leads to a significant decrease in motor velocity. For example, increasing the multivalency of a DNAzyme-based motor from two anchors to six led to complete abrogation of motor transport.15
Yet, in order for DNA based machines to transition beyond an elegant model system for motor proteins and truly achieve their potential, both motor efficiency and velocity need to be enhanced by orders of magnitude. In addition, the sophisticated instrumentation currently required to observe DNA walkers needs to be greatly simplified to render DNA machines as useful. To overcome these challenges, we designed a highly multivalent motor that moves through a cog-and-wheel mechanism, allowing μm or nm sized particles to roll rather than walk. Rolling represents a fundamentally distinct type of movement that overcomes the documented tradeoffs inherent to multivalent molecular motors.15, 16 In principle, this is because rolling reduces the DNA motor footprint, reducing unproductive sampling interactions, and thus increasing velocity.
Design and synthesis of spherical RNase H powered motors
The motor consists of a DNA-coated spherical particle (5 μm or 0.5 μm diameter particles) that hybridises to a surface modified with complementary RNA. The particle moves upon addition of RNase H, which selectively hydrolyses hybridised RNA but not single stranded RNA (Figure 1). Since the driving force for movement is derived from the free energy of binding new single stranded RNA that biases Brownian motion away from consumed substrate (Figure 1a), this type of motion is often described as a “burnt-bridge Brownian ratchet”. Note that molecular walkers also employ a burnt bridge mechanism, where oligonucleotide hybridisation is followed by DNAzyme/endonuclease hydrolysis of the fuel strand. The main difference between the molecular walkers and our system is the massive multivalency of the DNA coated particles – molecular walkers typically employ 2-6 anchor points while our particle-based motor employs thousands of anchoring strands. This equates to 100-1000 fold greater contact area or 10-100 fold greater contact diameter with the surface when compared to molecular spiders. Hence, the micron sized length scale of the particle significantly increases the number of contacts with the surface, which should lead to collective and emergent properties not expected for DNA-based walkers. Importantly, we expected that our particles would overcome the diminished velocities due to high multivalency, as is predicted by theory, because the RNase H hydrolysis rate is approximately two orders of magnitude greater than that of DNAzymes and endonucleases. 14, 17, 18 Moreover, highly multivalent motors are expected to display greater processivity, thus potentially addressing a major limitation of DNA walkers. Finally, the spherical particle template allows for the potential to roll, which is a fundamentally different mode for translocation of DNA based machines.
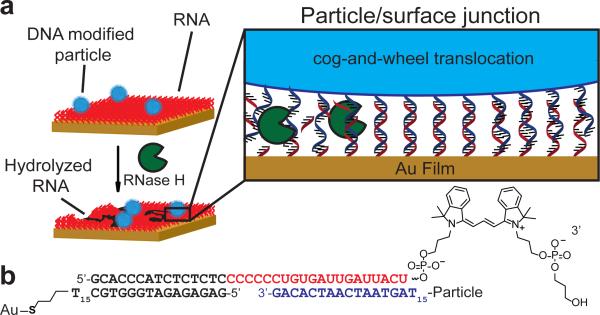
Figure 1. Approach for generating RNA-fueled, enzyme catalysed autonomous DNA motors.
a, DNA-modified particles were hybridised to an RNA monolayer presenting a complementary strand. Particles were immobile until RNase H was added, which selectively hydrolyses RNA duplexed to DNA. b, Schematic representation showing the hybridised oligonucleotide sequences at the particle-substrate junction. Note that there are hundreds of duplexes within this junction. RNA bases are shown in red, while the particle DNA bases are shown in blue.
We first generated an RNA-monolayer on a substrate by immobilising a DNA anchor strand to a thin gold film and then hybridising a fluorescently labeled RNA-DNA chimera strand to the surface. A Cy3 fluorophore at the 3’ RNA terminus was used to optimise RNA density and to detect RNA hydrolysis using fluorescence microscopy (Fig. 1b, Fig. 2a, b, Supplementary Fig. 1). Using the optimised conditions, we achieved a maximum RNA density of 50,000 molecules/μm2, equivalent to an average molecular footprint of 20 ± 6 nm2 per RNA strand (Supplementary Fig. 2). This RNA density was maintained for at least 4 hrs in 1×PBS and 10 μM DTT, a thiol reducing agent necessary for maintaining RNase H activity (Supplementary Fig. 3). In the absence of DTT, surfaces were stable for weeks in 1×PBS.
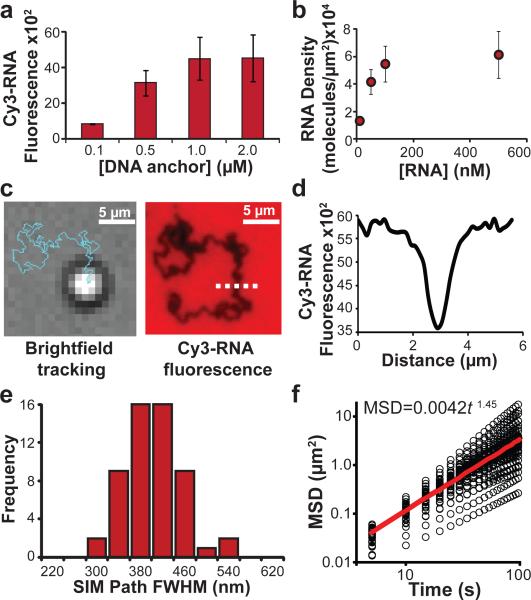
Figure 2. Characterising RNase H-driven particle motion.
a, Bar graph showing how the DNA anchor strand incubation concentration alters the Cy3-RNA fluorescence intensity which is directly proportional to the RNA surface density. The maximum RNA surface density was achieved when the DNA anchor strand incubation concentration was equal to or greater than 1 μM. Error bars represent the standard deviation in the average fluorescence intensity from at least 5 regions across each channel. b, Plot showing the RNA surface density as a function of RNA concentration during Cy3-RNA hybridisation with surface immobilised DNA anchor strand. The RNA density was maximised when RNA was hybridised at a concentration of at least 100 nM. Error bars represent the standard deviation in the average fluorescence intensity from at least 5 regions across each channel. c, Representative BF image and trajectory (blue line) from a time-lapse video tracking a single microparticle 30 min following RNase H addition. The same region was then imaged in the Cy3 fluorescence channel, revealing the location of depleted Cy3 signal. d, Line scan plot of dashed white line from c showing the depletion track from the widefield fluorescence image. e, Histogram analysis of FWHM of the depletion path width acquired using structured illumination microscopy. f, MSD versus log (time) analysis from individual particle trajectories (n = 43), which is shown with black circles. The red line indicates the average slope derived from all the individual particle trajectories.
Given that particle motion is intimately connected with RNase H efficiency and enzyme rates vary when substrates are immobilised, we initially measured hydrolysis kinetics for a DNA-RNA duplex monolayer. We found that PEG passivation was critical, since RNase H was inhibited when the surface was incompletely passivated (Supplementary Fig. 4). 19, 20 For optimally passivated surfaces, we measured a kcat = 25 min−1 for RNase H catalysed RNA hydrolysis. This rate is 6,000 fold greater than the turn-over number for nanoparticle-immobilised DNAzymes, thus supporting our expectation that RNase H powered rolling motors may achieve rapid velocities (Supplementary Fig. 5).17, 18
DNA-functionalised particles with a density of ~91,000 molecules/μm2 (footprint of 11 ± 3 nm2 per molecule) (Supplementary Fig. 6) were then synthesised and hybridised to a substrate presenting the complementary RNA strand. Importantly, the DNA density matched that of the RNA density on the planar substrate to ensure a high degree of polyvalency (~104 contacts/μm2), therefore minimising motor detachment from the substrate and maximising run processivity. Particles remained immobile until RNase H was added, which led to rapid translocation of particles across the substrate. This was quantitatively tracked by finding the centroid of the particles in time-lapse brightfield (BF) microscopy at 5 sec intervals (Fig. 2c and Supplementary Fig. 7).21 Importantly, the BF-generated tracks matched the widefield fluorescence depletion tracks (FWHM of 720 ± 110 nm), confirming that the particle motion was associated with continuous RNA hydrolysis (Fig. 2c, d, Supplementary Fig. 8). The line scan analysis of the fluorescence depletion indicates ~50% of the RNA underneath the particle is hydrolysed (Fig. 2d). Structured illumination microscopy (SIM), a super resolution microscopy technique with ~110 nm resolution revealed a more accurate footprint of the particle substrate junction corresponding to an average track width of 380 ± 50 nm (n = 55 tracks). This footprint indicates a maximum of ~5,500 DNA-RNA surface contacts at the motor-substrate junction (Fig. 2e, Supplementary Fig. 9, Supplementary Fig. 10). Importantly, substrates comprised of DNA did not lead to any translocation upon addition of RNase H (Supplementary Fig. 7), confirming that particle motion is specific to RNA hydrolysis at the particle-substrate junction.
Motors travel in a self-avoiding fashion
In principle, the particle motion should be biased away from consumed RNA substrate and toward new regions that had not been previously sampled. To investigate this model, particle motion was analysed by plotting the mean squared displacement (MSD) against time t, since MSD is known to be proportional to tα.22 However, we found that the MSD versus time did not have a linear dependence, as is the case for random diffusion (α= 1), but rather a power dependence where α= 1.45 ± 0.20, indicating that particle motion is not well described by random diffusion (Fig. 2f). A histogram summarising the distribution of diffusional exponents is shown in Supplementary Fig. 11. Moreover, random diffusion is known to have displacement distributions in the form of P(x) ~ exp(−A|x|ν) where ν= 2, yet we measured ν= 4.3 ± 1 (Supplementary Fig. 12). Again, further supporting that motion is biased away from hydrolysed substrate.
We then asked whether particle motion can be described by a self-avoiding random walk, where a particle cannot cross its own path but otherwise moves randomly. These types of self-avoiding random walks have only been theorised, and never experimentally realised. Because self-avoiding diffusion is typically studied using polymer chains,23 we conducted a multivalent stochastic simulation of hundreds of particles to predict the values for α and v associated with self-avoiding random diffusion (Supplementary Discussion 1). This modeling revealed that over short distances (short timescales) particles exhibit tethered diffusion (MSD ~ tα, with α= 1), and over long distances particles exhibit the scaling MSD ~ t log(t) which is theoretically expected for self-avoiding behaviour (Supplementary Discussion 1).24, 25 The rate of data acquisition falls in the transition region between these two time scales, where simulations predict a power-law MSD scaling with α= 1.49 ± 0.05 and a displacement distribution with v= 3.8 ± 0.1, both in agreement with the measured values (Supplementary Discussion 1). These findings demonstrate that DNA-powered particle motors execute motion that is statistically consistent with self-avoiding behaviour. To the best of our knowledge, rolling DNA-based motors provide the first experimental realisation of the “true” self-avoiding walker (TSAW) statistical process.24, 26
Mechanism of motor translocation
Particle motion could occur through three plausible mechanisms: a) walking/sliding, b) hopping, and c) rolling (Supplementary Fig. 13). The hopping mechanism was immediately ruled out upon examination of the continuous fluorescence depletion tracks (Fig. 2c). To differentiate between the two remaining mechanisms, particles were hybridised to an RNA substrate, and the unbound DNA on the particle was blocked by hybridisation with a complementary DNA strand (Fig. 3a). If motion primarily occurs through a walking/sliding mechanism, we expected that particles would move in a processive fashion leaving behind an RNA depletion track. However, upon RNase H addition, particles diffused randomly, producing an α= 0.99 ± 0.22 and a v= 1.8 ± 0.8, and no corresponding RNA depletion tracks were observed (Fig. 3b, c, Supplementary Discussion 1, Supplementary Fig. 11). By ruling out the hopping and walking/sliding mechanisms of motion, we conclude that particles primarily translocate by rolling, in a monowheel or cog-and-wheel like fashion. The rolling mechanism of motion was further confirmed through two other sets of experiments (vide infra). This is the first example of a DNA-based autonomous rolling motor, and we will subsequently refer to these machines as monowheels.
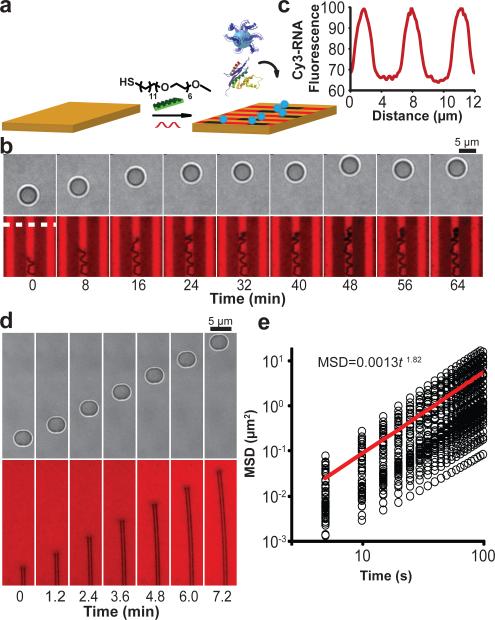
Figure 3. Elucidating the mechanism of particle motion and determining factors influencing particle velocity.
a, Schematic showing the strategy used to test whether particles roll during translocation by blocking the free DNA of the particle by hybridising with a blocking DNA strand (green). b, Representative BF image and trajectory (blue) taken from a time-lapse video tracking a single particle that had been blocked with DNA and treated with RNase H. The same region was imaged using the Cy3 fluorescence channel, indicating the lack of a RNA hydrolysis track. Note that a small transient spot with lower fluorescence intensity (see centre of fluorescence image) is typically observed under particles and is not due to RNA hydrolysis. c, MSD versus log(time) plot of particle diffusion for the blocked particles. The black circles represent individual data points from n = 32 particles, while the red line indicates the average of these plots. d, Histogram analysis of particle velocity for each 5 s interval as a function of [KCl]; 38 mM (red, n = 43 particles (15,480 occurrences)) and 150 mM (blue, n = 52 particles (18,720 occurrences)). e, Histogram analysis of particle velocity for each 5 s interval as a function of pH; 8.0 (blue, n = 43 particles (15,480 occurrences)), 7.5 (red, n = 50 particles (18,000 occurrences)), and 7.0 (green, n = 26 particles (9,360 occurrences)). Inset compares RNase H kcat (red) and average particle velocity (blue) as a function of pH. f, Representative BF image and trajectory (blue and red) taken from a time-lapse video tracking a single particle for 30 min where the red section indicates when the particle becomes entrapped. g, Representative velocity histogram of an individual particle when the particle is entrapped (red) or not entrapped (blue). Entrapment leads to significant decrease in particle velocity. h, Plot showing the particle velocity dependence on RNase H concentration. Note that in the absence of RNase H, the particles do not move. Error bars represent the standard deviation from n = 3 experiments. Most experiments were performed with [RNase H] = 144 nM.
For particles to move in a cog-and-wheel mechanism, the following sequence of reactions must occur: 1) RNA hydrolysis, 2) DNA unbinding (koff) from the hydrolysed substrate, 3) tethered particle diffusion (rolling), and 4) DNA hybridisation to new RNA substrate (kon) (Supplementary Fig. 14). To help determine what parameters affect particle velocity, we measured particle translocation as a function of monovalent and divalent salt concentration, pH, and RNase H concentration. Under high salt concentration we expected that the rate of hybridisation (kon) would increase, while the rate of dehybridisation (koff) would decrease and under decreasing pH, kcat would decrease. We found that particle velocity is highly dependent on salt concentration and decreases under higher ionic strengths. In contrast, particle speed is only sensitive to kcat when kcat is reduced below 0.4 per min (Fig. 3d, eSupplementary Fig. 15). Taken together, these results suggest that koff acts as the kinetic bottleneck for particle motion at pH 7.5 or greater (Supplementary Fig. 14).
Interestingly, the particle speed histogram contains two populations (Fig. 3d). Upon further analysis of individual particle velocities and accounting for stage drift, the two populations correspond to two states for each particle, a slow/stalled state and a fast state, as opposed to two types of particles or contributions owing to stage drift (Supplementary Fig. 16). The slower state is due to transient stalling of the particle, which may be attributed to factors that include surface defects leading to non-specific particle binding, inactive enzyme bound to the particle-substrate junction, and particle self-entrapment. Upon detailed analysis of individual particle trajectories, we found that stalling mostly correlated to particle entrapment (Fig. 3f, g). To determine whether the enzyme concentration used, [RNase H] = 140 nM, saturated the available substrate binding sites, monowheel velocity was monitored as a function of enzyme concentration (Fig. 3h and Supplementary Fig. 17). At 10 fold greater enzyme concentration, only a slight increase in average velocity was observed, whereas 10 fold and 100 fold dilution of RNase H led to near abolition of monowheel motion. The particle speed histograms for decreasing RNase H concentration (Supplementary Fig. 17) show a gradual decrease in velocity as opposed to a shift to the low velocity population, thus confirming that multiple RNase H enzymes are operating in parallel.
A general design rule for creating processive DNA machines necessitates that the rate of cleaving contacts and leg disassociation from the track is slower than the rate of forming new track contacts. The stochastic nature of association and disassociation rates implies that molecular DNA walkers will occasionally disassociate from the track. This is reflected in the relatively low fidelity of molecular walkers.1, 7-11 Higher multivalency buffers against spontaneous disassociation, thus increasing run processivity.15, 16, 27 Our analysis suggests that the monowheels are highly multivalent with at least ~1000 DNA-RNA duplex contacts present at any given time. This estimate is based on the RNA depletion track analysis, which indicates that 50% of RNA is hydrolysed within the ~400 nm wide track. Given the RNase H turnover rate of 25 min−1 and the RNA surface density (1×104 RNA/μm2), the monowheels must have at least ~1000 RNA-DNA duplexes that are occupied by RNase H to maintain a velocity of 2 μm/min. Fewer contacts would result in lower particle speeds. This is a lower bound estimate of the particle multivalency because this calculation assumes every duplex is bound by active enzyme, which is unlikely. Therefore, monowheels display 2-3 orders of magnitude greater multivalency than molecular walkers, which affords significant improvement in motor processivity.
Another important difference between molecular walkers and our monowheels is the effect of gravity on μm-sized particles. Albeit small, the force of gravity (~0.6 pN for a 5 μm silica particle in water) may bias the observed processivity by limiting particle-surface dissociation. To determine the role of gravity on particle motion, we inverted the sample geometry such that monowheels were upside down, operating against the force gravity (Supplementary Fig. 18). Under identical conditions, 80% of the inverted monowheels displayed similar speeds and processivity (distance travelled before stalling) as that of the upright samples. Note that at [KCl] = 38 mM, 20% of particles detached upon flipping the sample, whereas at [KCl] = 75 mM, none of the observed particles detached from the surface and monowheels displayed slightly enhanced velocities to that observed at [KCl] = 38 mM (either when the sample was upright or flipped). Therefore, the primary contributor to monowheel processivity is the high multivalency of the system rather than the force of gravity.
To simplify analysis and quantification, we used 5 μm diameter particles for the majority of experiments. However, note that the rolling mechanism of translocation can be recapitulated with 0.5 μm diameter particles (Supplementary Fig. 19). In principle, these smaller particles reduce the maximum number of DNA-RNA contacts by one of order of magnitude to the range of several hundred. Similar maximum velocities up to 5 μm/min and average velocities of 1.8 ± 0.4 and 1.9 ± 0.5 μm/min were observed for both 0.5 and 5 μm particles, respectively, showing that the fundamental cog-and-wheel mechanism of rolling is independent of cargo size within the range tested (Supplementary Fig. 19). This was initially surprising since the smaller particles have fewer contacts with the surface and reduced drag, resulting in a faster koff, thus smaller particles were expected to translocate more quickly. Upon further analysis, we noted that smaller particles are less multivalent, and therefore fewer RNA-DNA duplexes are available for hydrolysis at any given time. In other words, there is reduced hydrolysis of RNA in parallel when using smaller particles, which is expected to slow down the absolute RNA hydrolysis rate and thus reduce particle velocity. These opposing effects create a complex relationship between motor speed and particle size, and when comparing the 5 μm beads with the 500 nm particles the effect of particle size effectively cancels out, leading to similar observed velocities. Although as expected, the less multivalent 0.5 μm particles roll for shorter average run lengths compared to 5 μm diameter particles (~3 μm versus ~200 μm), which continue processively moving throughout the 30 min video and even continue moving for over 5 hrs (Supplementary Fig. 20). The decrease in run length for the 0.5 μm particles is due to the lower multivalency (fewer contacts) with the surface, thus increasing the probability of detachment. Nonetheless, increasing the KCl and Mg concentrations to 75 mM and 3 mM, respectively, enhances 0.5 μm particle endurance such that the majority of particles display processive motion for the entire 30 min video. This provides the 0.5 μm particles with an average run length of greater than 25 μm. Despite much work in the area of DNA-based machines, this class of multivalent rolling motor exceeds the velocity and processivity of previous DNA-motors by three-orders of magnitude, and approaches the efficiency of biological enzymes, with velocities on the order of tens of μm's per second and run lengths of hundreds of μm's.
Track-guided unidirectional motion
To achieve unidirectional transport resembling motor protein motion along a filament, RNA was spatially micro-patterned into 3 μm wide tracks (Fig. 4a). Particles were then hybridised to the patterned RNA, and RNase H was added to initiate motion (Fig. 4b). Using BF time-lapse tracking and RNA fluorescence depletion, we observed that a subset of particles moved along the 3 μm substrate corral unidirectionaly deflecting away from the PEG-printed regions. Note that many particles became entrapped, partially because of RNA cross contamination into the PEG-passivated regions (Fig. 4c) and self-entrapment in consumed substrate corrals. It is likely that generating well-passivated nanoscale RNA tracks commensurate in size to the particle-substrate junction width (~400 nm) would lead to an increase yield of linear trajectories.8 Nonetheless, this provides a proof-of-concept demonstration of autonomous translocation using self-assembled components without an external field, akin to the motion of motor proteins.
Figure 4. Directional motor translocation from self-avoiding to ballistic.
a, Schematic showing the strategy used to generate RNA micro-tracks by using microcontact printing. SH-PEG barriers were directly printed onto the gold film, which was backfilled with RNA. b, BF and fluorescence time-lapse images of a particle moving along a 3 μm wide track following the addition of RNase H. c, Line scan analysis of the region highlighted in the fluorescence channel with a dotted white line showing the track dimensions and the effectiveness of the PEG barriers. d, Representative BF and fluorescence images taken from a time-lapse movie that tracked a dimerised particle following RNase H addition. The BF-analysed trajectory as well as the two parallel fluorescence depletion tracks showed near linear particle motion. e, MSD versus log(time) analysis of the dimerised particle motion. Slope of this plot shows an average power log dependence of 1.82± 0.13, confirming that the particle dimer traveled in a ballistic, linear fashion.
Dimer and rod particles travel ballistically
An alternate strategy to achieve linear motion would be to limit lateral particle motion by incorporating multiple monowheels on the body of a single chassis. This has been investigated previously by Diehl et al., but given the unique rolling mechanism for our system, we speculated that multiple motors could only result in ballistic or linear motion.27 By happenstance, we noticed that a 1-10% subset of our particles were fused forming dimers, a common byproduct in silica particle synthesis. We found that these particles travelled linearly for distances that spanned hundreds of μm's at a velocity of ~0.6 ±0.5 μm/min, n = 68 dimer particles (Fig. 4d). Not surprisingly, a plot of MSD versus t for particle dimers showed a power-law scaling of α= 1.82 ±0.13, confirming that particle motion was nearly linear (α= 2). In addition, we also discovered that 50% of single spherical particles displayed a transient component of their trajectory that is linear and associated with wider tracks; linear motion was correlated with wider ~1.0 ±0.1 μm tracks or multiple contact points (Supplementary Fig. 8 and 9). The ballistic (linear) motion observed for what appears to be wider tracks may be due to particles possessing multiple contact points that cannot be resolved or due to particles rolling along imperfections along the surface. Following these observations, we synthesised DNA-coated microrods and anticipated similar behaviour. Microrods showed nearly linear motion, which demonstrates that the rolling mechanism is not limited to spherical particles (Supplementary Fig. 21). These are the first examples of directional motion without the need of a patterned track or external electromagnetic field, which is only afforded due to the unique cog-and-wheel translocation mechanism.
SNP detection by measuring particle displacement
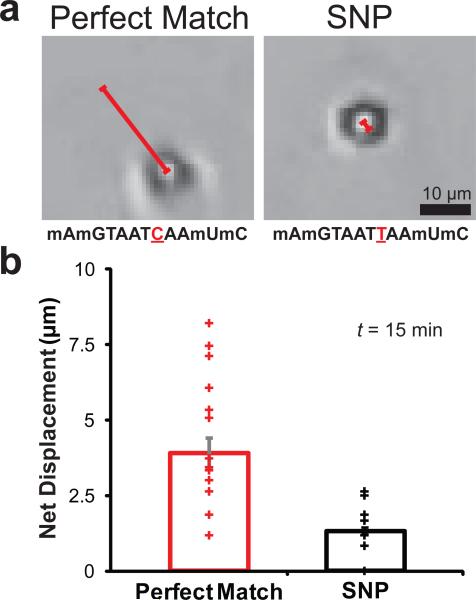
Since monowheel motion is exquisitely sensitive to kon, koff and kcat, we anticipated that particle motion could provide a readout of molecular recognition. As a proof-of-concept, we aimed to detect a single nucleotide polymorphism (SNP) mutation, which is a biomedically relevant and a challenging mutation to identify. Particles displaying the SNP (5'mAmGTAATTAAmUmC3’) traveled ~60% slower (0.3 μm/min) than identical particles with a perfect match (5'mAmGTAATCAAmUmC3’) (Supplementary Fig. 22). This difference in velocity can be attributed to a slower rate of hydrolysis for RNase H to hydrolyse duplexes possessing a single base mismatch.28 Due to the μm-sized cargo and large distances travelled, even a smartphone camera equipped with an inexpensive plastic lens could detect motion associated with this SNP by recording particle displacement within a short time interval (t = 15 min) (Fig. 5a and b). SNP detection could also be achieved using unmodified DNA (Supplementary Fig. 23); although maximum discrimination required shortening the RNase H recognition sequence. Most importantly, the size and efficiency of the motors allows for rapid and simple SNP sensing and represents a new application for DNA-based motors.
Figure 5. Single nucleotide polymorphism (SNP) detection.
a, Representative bright field images collected by using a smartphone microscope. The images show particles and their net displacement for perfect match and SNP sequences after RNase H addition (t = 15 min). The sequences are illustrated below each image in the 5’ to 3’ orientation where the single base difference between sequences is underlined and indicated in red and the non-natural 2’ methyl ether RNA modification is indicated by the letter m before the base (scale bar = 5 μm). Note that due to the shorter recognition sequence, higher salt concentrations were used in the reaction buffer (75 mM Tris (pH = 8), 110 mM KCl and 4.5 mM MgCl2). b, A scatter plot displaying particle net displacements 15 minutes following RNase H addition. The mean particle displacement is shown as the height of the bar. Error bars represent standard deviation from n = 42 particles total (p < 0.0001, as determined by a t-Test with unequal variances).
In conclusion, monowheels take advantage of multivalent substrate interactions to produce emergent collective properties that are difficult to predict a priori from the properties of the individual biomolecules.29-31 Monowheels also achieve velocities up to ~5 μm/min, a speed which is ~5,000 fold greater than the fastest known synthetic DNA walker and approaching the velocities of biological motor proteins.8 Monowheels use the same chemistry as molecular walkers - specific DNA-RNA hybridisation followed by RNA hydrolysis. Moreover, the leg to leg spacing (DNA-DNA spacing) for the walkers is similar to the spacing between DNA strands for monowheels (~5 nm). However, the monowheels differ from the DNA walkers because of their massive multivalency, which leads to collective behaviours such as track free linear motion and rolling. Although highly multivalent motors typically display diminished speeds, monowheels break this trend due to a reduction in unproductive DNA-surface contacts enabled by rolling and the parallel activity of RNase H in a highly dense DNA-RNA contact (as suggested by modelling). Further experiments with anisotropic particles and different enzyme-substrate pairs will elucidate the role of these contributions. At best, the efficiency of converting chemical energy into mechanical work for monowheels is approximately 0.001%, which is on par with chemically powered synthetic motors32 and may be further optimised in the future (Supplementary Discussion 2). Due to the multi-μm sized chassis, and rapid translocation rates, real-time tracking of movement can be achieved with conventional optical microscopy and even a smartphone-based microscope, thus circumventing the need for AFM and electron microscopy-based analysis. Because of the highly sensitive cog-and-wheel rolling mechanism of motion, monowheels represent an important step in bringing together the field of DNA-based sensing with the emerging area of DNA machines and motors. For example, motor velocity can likely be modulated by aptamer-analyte interactions, suggesting that monowheels will become a new and powerful tool in label-free sensing assays.
Materials
All chemicals were purchased from Sigma-Aldrich unless otherwise noted. Stock solutions were made using Nanopure water (Barnstead Nanopure system, resistivity = 18.2 MΩ), herein referred to as DI water. 5-μm aminated silica beads were purchased from Bangs Laboratory (Product #: SA06N). The DNA fluorescence assay kit (Quant-iT™ OliGreen® ssDNA kit) was acquired from Life Technologies (formerly Invitrogen, Carlsbad, CA) and was used to quantify DNA density on the particle surface. All oligonucleotides were custom synthesised by Integrated DNA Technologies (IDT) and are summarised in SI Table 1. RNase H was obtained from Takara Clontech (Product #: 2150A). Thin Au films were generated by using a home-built thermal evaporator system. All Au surface functionalisation was carried out using an IBIDI sticky-Slide VI0.4 17 × 3.8 × 0.4 mm channels.
Methods
Optical microscopy
Brightfield and fluorescence images were acquired on a fully automated Nikon Eclipse Ti TIRF microscope controlled using the Elements software package (Nikon). Images were collected using the Evolve electron multiplying charge-coupled device (EMCCD; Photometrics) through a CFI Apo 100× 1.49 NA objective or a Plan Fluor 20× 0.50 NA objective. Fluorescence images of Cy3 were collected using an Intensilight Hg lamp epifluorescence source (Nikon) and a TRITC filter cube set supplied by Chroma. An interferometric lock-in focus system (Perfect focus system (PFS)) was used during acquisition of time lapse movies to minimise focus drift.
Super resolution imaging of the fluorescence depletion tracks
Structured illumination microscopy (SIM) images were acquired on a Nikon N-SIM system, which is equipped with a CFI Apo 100× 1.49 NA objective and Andor iXon EMCCD (60nm/pixel). For each N-SIM image, 9 images of 3'Cy3-RNA sample were acquired in different phases using a 561 nm laser as an excitation source and was reconstructed using Nikon Element software package.
Thermal evaporation of gold films
A number 1.5 glass slide (25 mm × 75 mm) was cleaned by sonication in DI water for 5 min. The sample was then subjected to a second sonication in fresh DI water for 5 min. Finally, the slide was sonicated in propanol for 5 min, which was subsequently dried under a stream of N2. The cleaned slide was then introduced into a home built thermal evaporator chamber and the pressure was reduced to 50 × 10−3 Torr. The chamber was purged with N2 three times and the pressure was reduced to 1-2 × 10−7 Torr by using a turbo pump and cooling the chamber using a liquid N2 trap. Once the desired pressure was achieved, a 1.5 nm film of Cr was deposited onto the slide at a rate of 0.2 Å/sec, which was determined by a quartz crystal microbalance. After the Cr adhesive layer was deposited, 4 nm of Au was deposited at a rate of 0.4 Å/sec. The Au-coated samples were used within one week of evaporation.
Fabrication of RNA monolayers
An IBIDI sticky-Slide VI0.4 flow chamber was adhered to the Au-coated slide to produce six channels (17 × 3.8 × 0.4 mm dimensions). Prior to surface functionalisation, each channel was washed with ~5 ml of DI water. Next, a densely packed DNA monolayer was assembled onto the Au surface by using published protocols where 40 μL of a 1 μM 3’ disulfide modified DNA strand (anchoring strand) was incubated with the surface for 12 hrs under high ionic strength buffer (1 M KHPO4).33 The chamber was then sealed by Parafilm to prevent evaporation. After incubation, excess DNA was removed from the channel using a ~5 ml DI water rinse. To block any bare gold sites and to maximise hybridisation of RNA to the DNA anchoring strand, the surface was backfilled with 100 μL of a 100 μM SH(CH2)11(OCH2CH2)6OCH3 (SH-PEG) solution in ethanol for 6 hrs. Excess SH-PEG was removed by ~5 ml rinses with ethanol and water each. Lastly, the RNA substrate was immobilised to the surface through hybridisation of 100 μL of a complementary RNA/DNA chimera (100 nM) in 1x PBS (phosphate buffered saline) for 12 hrs. The wells were sealed with Parafilm for each step to prevent evaporation and the resulting RNA monolayer remained stable for ~1-2 weeks, as determined based on fluorescence imaging.
Determining RNA surface density
RNA surface density was determined by releasing the Cy3-tagged RNA from the surface by adding 100 μL of RNase A (100 μg/ml in 1 xPBS) and then quantifying the Cy3 fluorescence intensity in solution using a calibration curve obtained using the fluorescence microscope.
μ-contact printing of RNA tracks
To generate RNA tracks, SH-PEG barriers were initially patterned onto the thin gold film using established protocols.34 Briefly, 30 μL of a 1 mM SH-PEG solution in ethanol was applied to a PDMS stamp with 3 μm wide parallel lines spaced at a 3 μm pitch. Next, the SH-PEG solution was dried under a stream of N2 for 1 min and the remaining solution was wicked away. The coated stamp was then brought into contact with the gold film surface for 30 sec. After contact, the stamp was peeled away from the substrate and used to print repeatedly without re-applying additional SH-PEG ink. After patterning the SH-PEG barriers onto the surface, the IBIDI sticky-Slide VI0.4 flow chamber was adhered to the gold slide, and the substrate was prepared as described above in the “Fabrication of RNA monolayers” section. Note that the RNA track preparation protocol used a 6 hr incubation with the DNA anchor solution rather than 12 hrs.
Synthesis of azide functionalised particles
First, azide-functionalised particles were synthesised by mixing 1 mg of 5-μm aminated silica beads (Bangs Laboratory) with 1 mg of N-hydroxysuccinimidyl azide heterobifunctional linker. This mixture was subsequently diluted in 100 μL of DMSO and 1 μL of a 10x diluted TEA (triethylamine) stock in DMSO. The reaction proceeded overnight for 24 hrs and the azide-modified particles were purified by adding 1 mL of DI water and centrifuging down the particles at 15,000 rpm for 5 min. The supernatant was discarded and the resulting particles were resuspended in 1 mL of DI water. This process was repeated 7 times, and during the last centrifugation step, the particles were resuspended in 100 μL of DI water, yielding an azide-modified particle stock. The azide-modified particles were stored at 4 °C in the dark and were used within one month of preparation.
Synthesis of high DNA density silica particles
High density DNA functionalised particles were synthesised by adding 5 nmoles (5 μl) of alkyne modified DNA stock solution to 5 μL of azide functionalised particles. The particles and DNA were diluted with 25 μL of DMSO and 5 μL of a 2 M triethyl ammonium acetate buffer (pH 7.0). Next, 4 μL from a 5 mM stock solution of ascorbic acid was added to the reaction as a reducing agent. Cycloaddition between the alkyne modified DNA and azide functionalised particles was initiated by adding 2 μL from a 10 mM Cu-TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine) stock solution in 55 vol % DMSO.35 The reaction was incubated for 24 hrs, where the resulting DNA functionalised particles were purified by centrifugation. Specifically, the particles were centrifuged at 15,000 rpm for five min; after which, the supernatant was discarded and the particles were resuspended in 1 mL of a 1x PBS and 10% Triton-X solution. This process was repeated 7 times, where the particles were resuspended in 1 mL 1x PBS only for the 4th-6th centrifugations. During the final centrifugation/wash, the particles were resuspended in 50 μL of 1x PBS. The high density DNA functionalised particles were stored at 4 °C and protected from light.
Determining DNA particle surface density
DNA surface density on the silica particles was determined by releasing the DNA using HF etching followed by quantifying the DNA concentration with a fluorescence assay. Initially, 50 μL of DNA functionalised particles were diluted with 45 μL of PBS and 5 μL of concentrated (50% by vol) HF. After 30 min of etching, DNA was assumed to be fully released from the surface based on brightfield characterisation of particle whittling (Supplementary Fig. 6). Subsequently, the DNA concentration was quantified using a commercial fluorescence assay for detecting single stranded DNA (Quant-iT OliGreen ssDNA kit, Invitrogen) following the published product protocol. The final DNA density was calculated by determining the concentration of particle stock using a hemocytometer.
Determination of RNase H surface kinetics
A monolayer of RNA/DNA duplexes was constructed by hybridising a soluble complementary DNA strand to the RNA monolayer. Briefly, 100 μL of a DNA stock solution (100 μM) in 1x PBS was added to the ssRNA surface. After 1 hr of hybridisation, the excess DNA was washed away by rinsing the channel with 5 mL 1xPBS, which was exchanged for 100 μL of RNase H reaction buffer (25 mM Tris pH 8.0, 8 mM NaCl, 37.5 mM KCl, 1.5 mM MgCl2, 10 vol % formamide, and 0.75% (g/mL) Triton X). The RNA hydrolysis reaction was initiated by the addition of 2 μL (5 units) of RNase H from an enzyme stock of 7.2 μM (2.5 units/μL) enzyme, 500 μM DTT, and 1x PBS. Reaction progress was monitored by measuring the loss in surface RNA fluorescence over time. Since kinetic conditions satisfy Michaelis-Menten requirements with substrate RNA concentration at a ~60 fold excess over enzyme (1.2 nM), we determined kcat by dividing the initial rate of reaction (Vmax) by enzyme concentration (Vmax = kcat × [enzyme]).36
RNase H powered particle translocation
Initially, RNA substrate surfaces were washed with 5 mL of PBS to wash away excess unbound RNA. Next, DNA functionalised particles were hybridised to the RNA substrate. Briefly, 5 μL of DNA functionalised particles were diluted with 45 μL of PBS, which was added to the RNA substrate. Hybridisation between the particles and the complementary RNA monolayer occurred over a thirty minute incubation period. After hybridisation, particle translocation was initiated by buffer exchange with 100 μL of RNase H reaction buffer (25 mM Tris pH 8.0, 8 mM NaCl, 37.5 mM KCl, 1.5 mM MgCl2, 10% formamide, and 0.75% (g/mL) Triton X) and 2 μL (5 units) of RNase H from an enzyme stock of 7.2 μM enzyme, 500 μM DTT, and 1x PBS. The microchannel was sealed with Parafilm to prevent evaporation. Particle tracking was achieved through brightfield imaging acquired at 5 sec intervals using MOSAIC ImageJ plugin.21 High-resolution epifluorecence images (100x) of fluorescence depletion tracks were acquired to verify particle motion was due to processive RNA hydrolysis. For high-resolution movies (100x), both brightfield and epifluorescence images were recorded at 5 sec intervals. For control (blocked) particles, after DNA particle hybridisation with the surface, the remaining ssDNA on the particle was hybridised with a blocking strand (100 μM) in 1x PBS for 30 minutes. Prior to buffer exchange, the particles were carefully washed with 2 mL PBS to remove excess DNA blocking strand. For particle motion along RNA patterned tracks, a higher salt concentration of 150 mM KCl was used.
Smartphone based SNP detection
Two focusing lenses (obtained from a toy laser pointer, $2) were taped together and adhered to the cell phone camera via double sided tape. The smartphone was placed onto a lab jack, which was used to focus onto the sample. The sample was placed onto an optically transparent plastic sheet, where a battery powered LED was positioned underneath for illumination. Tape was used to immobilise both the smartphone to the jack and the sample to the plastic sheet in order to minimise drift. Using the smartphone application Lapse It Pro, images were acquired at the desired time intervals, where the displacements were subsequently measured using ImageJ image processing software. The reaction buffer required higher salt concentrations (110 mM KCl and 4.5 mM MgCl2) due to the lower stability of the shorter sequences used in the SNP assay.
Supplementary Material
Acknowledgements
K.S. is grateful for support from the NIH through R01-GM097399, the Alfred P. Sloan Research Fellowship, the Camille-Dreyfus Teacher-Scholar Award, and the NSF CAREER Award (1350829). K.Y. thanks the ARCS foundation for their support and Victor Pui-Yan Ma for generating Figure 1. We also thank Sergei Urazhdin for access to the thermal evaporator and Martha Grover and Daniel Stabley for helpful discussions. E.R.W. was funded by NSF (CMMI-1250235) and Skanda was funded by Emory. This research project was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core.
Footnotes
Author contribution section: K.Y. conducted all experiments and analysis, A.M. performed simulations and theoretical validation, S.V. helped in data analysis and validation of theoretical model, Y.Z. helped with particle functionalisation, Y.L. collected SIM data, K.S. and K.Y. wrote the manuscript with input from A.M. and E.R.W. K.S. oversaw all aspects of the work, E.R.W. supervised and discussed experiments with S.V.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at www.nature.com/reprints.
References
- 1.He Y, Liu DR. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nat Nanotechnol. 2010;5:778–782. doi: 10.1038/nnano.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewandowski B, et al. Sequence-specific peptide synthesis by an artificial small-molecule machine. Science. 2013;339:189–193. doi: 10.1126/science.1229753. [DOI] [PubMed] [Google Scholar]
- 3.von Delius M, Geertsema EM, Leigh DA. A synthetic small molecule that can walk down a track. Nature chemistry. 2010;2:96–101. doi: 10.1038/nchem.481. [DOI] [PubMed] [Google Scholar]
- 4.Paxton WF, Sundararajan S, Mallouk TE, Sen A. Chemical locomotion. Angewandte Chemie. 2006;45:5420–5429. doi: 10.1002/anie.200600060. [DOI] [PubMed] [Google Scholar]
- 5.Pavlick RA, Sengupta S, McFadden T, Zhang H, Sen A. A polymerization-powered motor. Angewandte Chemie. 2011;50:9374–9377. doi: 10.1002/anie.201103565. [DOI] [PubMed] [Google Scholar]
- 6.Orozco J, et al. Artificial enzyme-powered microfish for water-quality testing. ACS nano. 2013;7:818–824. doi: 10.1021/nn305372n. [DOI] [PubMed] [Google Scholar]
- 7.Gu H, Chao J, Xiao SJ, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lund K, et al. Molecular robots guided by prescriptive landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wickham SF, et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nat Nanotechnol. 2011;6:166–169. doi: 10.1038/nnano.2010.284. [DOI] [PubMed] [Google Scholar]
- 10.Omabegho T, Sha R, Seeman NC. A bipedal DNA Brownian motor with coordinated legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickham SF, et al. A DNA-based molecular motor that can navigate a network of tracks. Nat Nanotechnol. 2012;7:169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- 12.Cha TG, et al. A synthetic DNA motor that transports nanoparticles along carbon nanotubes. Nat Nanotechnol. 2014;9:39–43. doi: 10.1038/nnano.2013.257. [DOI] [PubMed] [Google Scholar]
- 13.Yin P, Choi HMT, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–U314. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 14.Bath J, Green SJ, Turberfield AJ. A free-running DNA motor powered by a nicking enzyme. Angew Chem Int Edit. 2005;44:4358–4361. doi: 10.1002/anie.200501262. [DOI] [PubMed] [Google Scholar]
- 15.Pei R, et al. Behavior of polycatalytic assemblies in a substrate-displaying matrix. Journal of the American Chemical Society. 2006;128:12693–12699. doi: 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]
- 16.Perl A, et al. Gradient-driven motion of multivalent ligand molecules along a surface functionalized with multiple receptors. Nature chemistry. 2011;3:317–322. doi: 10.1038/nchem.1005. [DOI] [PubMed] [Google Scholar]
- 17.Fang S, Lee HJ, Wark AW, Kim HM, Corn RM. Determination of ribonuclease H surface enzyme kinetics by surface plasmon resonance imaging and surface plasmon fluorescence spectroscopy. Analytical chemistry. 2005;77:6528–6534. doi: 10.1021/ac051283m. [DOI] [PubMed] [Google Scholar]
- 18.Yehl K, et al. Catalytic deoxyribozyme-modified nanoparticles for RNAi-independent gene regulation. ACS nano. 2012;6:9150–9157. doi: 10.1021/nn3034265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Yehl K, Narui Y, Salaita K. Tension Sensing Nanoparticles for Mechano-Imaging at the Living/Nonliving Interface. Journal of the American Chemical Society. 2013;135:5320–5323. doi: 10.1021/ja401494e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Nanoparticle Tension Probes Patterned at the Nanoscale: Impact of Integrin Clustering on Force Transmission. Nano Lett. 2014;14:5539–5546. doi: 10.1021/nl501912g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Gal N, Lechtman-Goldstein D, Weihs D. Particle tracking in living cells: a review of the mean square displacement method and beyond. Rheol Acta. 2013;52:425–443. [Google Scholar]
- 23.Domb C, Gillis J, Wilmers G. On Shape and Configuration of Polymer Molecules. P Phys Soc Lond. 1965;85:625. [Google Scholar]
- 24.Amit DJ, Parisi G, Peliti L. Asymptotic-Behavior of the True Self-Avoiding Walk. Phys Rev B. 1983;27:1635–1645. [Google Scholar]
- 25.Obukhov SP, Peliti L. Renormalization of the True Self-Avoiding Walk. J Phys a-Math Gen. 1983;16:L147–L151. [Google Scholar]
- 26.Family F, Daoud M. Experimental Realization of True Self-Avoiding Walks. Phys Rev B. 1984;29:1506–1507. [Google Scholar]
- 27.Diehl MR, Zhang K, Lee HJ, Tirrell DA. Engineering cooperativity in biomotor-protein assemblies. Science. 2006;311:1468–1471. doi: 10.1126/science.1122125. [DOI] [PubMed] [Google Scholar]
- 28.Ostergaard ME, et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berna J, et al. Macroscopic transport by synthetic molecular machines. Nat Mater. 2005;4:704–710. doi: 10.1038/nmat1455. [DOI] [PubMed] [Google Scholar]
- 30.Eelkema R, et al. Nanomotor rotates microscale objects. Nature. 2006;440:163–163. doi: 10.1038/440163a. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, et al. Linear artificial molecular muscles. Journal of the American Chemical Society. 2005;127:9745–9759. doi: 10.1021/ja051088p. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Chiang TY, Velegol D, Mallouk TE. Understanding the Efficiency of Autonomous Nano- and Microscale Motors. Journal of the American Chemical Society. 2013;135:10557–10565. doi: 10.1021/ja405135f. [DOI] [PubMed] [Google Scholar]
- 33.Peterson AW, Heaton RJ, Georgiadis RM. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan L, Zhao XM, Whitesides GM. Patterning a preformed, reactive SAM using microcontact printing. J Am Chem Soc. 1998;120:6179–6180. [Google Scholar]
- 35.Zhang Y, Ge CH, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat Commun. 2014;5 doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson KA, Goody RS. The Original Michaelis Constant: Translation of the 1913 Michaelis-Menten Paper. Biochemistry-Us. 2011;50:8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.