Abstract
Engineered variants of rebeccamycin halogenase were used to selectively halogenate a number of biologically active aromatic compounds. Subsequent Pd-catalyzed cross-coupling reactions on the crude extracts of these reactions were used to install aryl, amine, and ether substituents at the halogenation site. This simple, chemoenzymatic method enables non-directed functionalization of C-H bonds on a range of substrates to provide access to derivatives that would be challenging or inefficient to prepare by other means.
Keywords: C-H Functionalization, Halogenase, Cross-Coupling, Chemoenzymatic, Late-Stage Diversification
Graphical Abstract
Late-stage C-H functionalization can be used to optimize molecular function without requiring extensive redesign of synthetic routes used to access initial lead compounds.1 This approach has proven particularly useful for the discovery and optimization of biologically active compounds.2,3 In this context, selective late stage C-H functionalization has been achieved using small molecule catalysts that exploit directing groups4 or subtle differences in the steric,5,6 electronic,7,8 or stereoelectronic9,10 properties of C-H bonds. While separation methods can be used to isolate product isomers resulting from non-selective C-H functionalization reactions,1 selective formation of individual isomers greatly facilitates late stage efforts in which a particular compound is desired.
Enzymes have been catalyzing selective late stage functionalization of C-H bonds on complex natural products for billions of years.11 These catalysts remain underutilized for reactions on non-native substrates, however, due in part to their relatively low activity on such compounds. The same attractive interactions that give rise to highly efficient and selective catalysis can lead to substrate specificity, but directed evolution12 can be used to generate enzymes with expanded substrate scope.13 This is perhaps best illustrated by the extensive use of engineered cytochromes P450 for preparative oxygenation (typically hydroxylation) of a wide range of drugs, natural products, and other compounds.14 The oxygenated products of these reactions often have important biological activity in their own right,15 but the oxygen atom installed can also serve as a handle for subsequent transformations,16 including deoxyfluorination.17 Such chemoenzymatic C-H functionalization methods benefit from the selectivity of enzymatic catalysis and the reaction scope of synthetic chemistry.
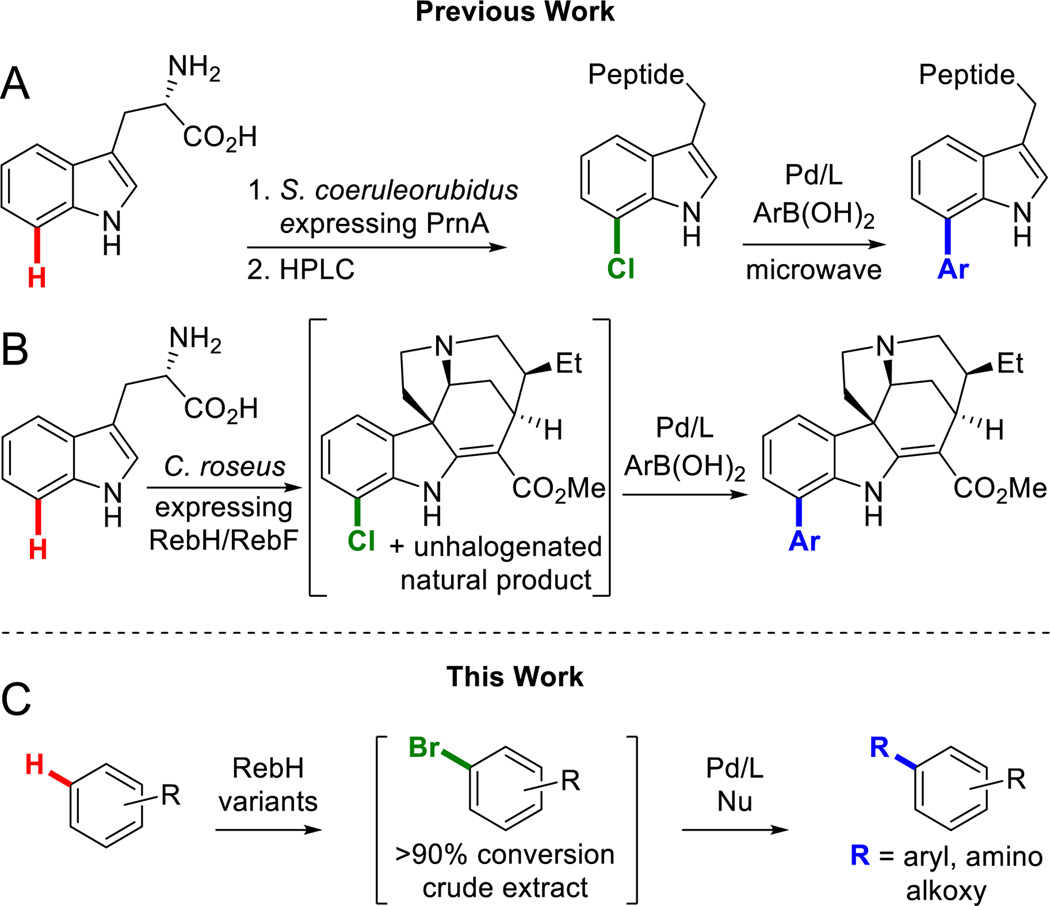
A number of halogenases catalyze selective halogenation of aromatic C-H bonds,18 and this activity has been used in conjunction with Pd-catalysis to enable chemoenzymatic C-H arylation of natural products. For example, Goss demonstrated that PrnA, a FADH2 dependent tryptophan7-halogenase, could be expressed in S. coeruleorubidus to chlorinate tryptophan, which the organism subsequently incorporated into cyclic polypeptides (Scheme 1A).19 These polypeptides were then purified by HPLC and subjected to microwave-mediated Suzuki-Miyaura conditions to furnish arylated polypeptide derivatives. More recently, O’Connor reported that rebeccamycin halogenase (RebH, also a FADH2 dependent tryptophan 7-halogenase) could be expressed in C. roseus to prepare halo-indole alkaloids that could be arylated via Suzuki cross-coupling (Scheme 1B).20 Unlike the P450 examples noted above, however, both of these reports were limited to the native substrate (tryptophan) and selectivity (7-position) of the halogenases used, and compatibility with only a limited range of cross-coupling reactions was demonstrated.
Scheme 1.
Small molecule diversification by enzymatic halogenation/Pd-catalyzed cross-coupling. A/B) Biosynthesis of natural products containing 7-chlorotryptophan followed by Suzuki reaction. C) Halogenation of arenes using engineered halogenases followed by Pd-catalyzed cross-coupling.
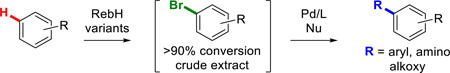
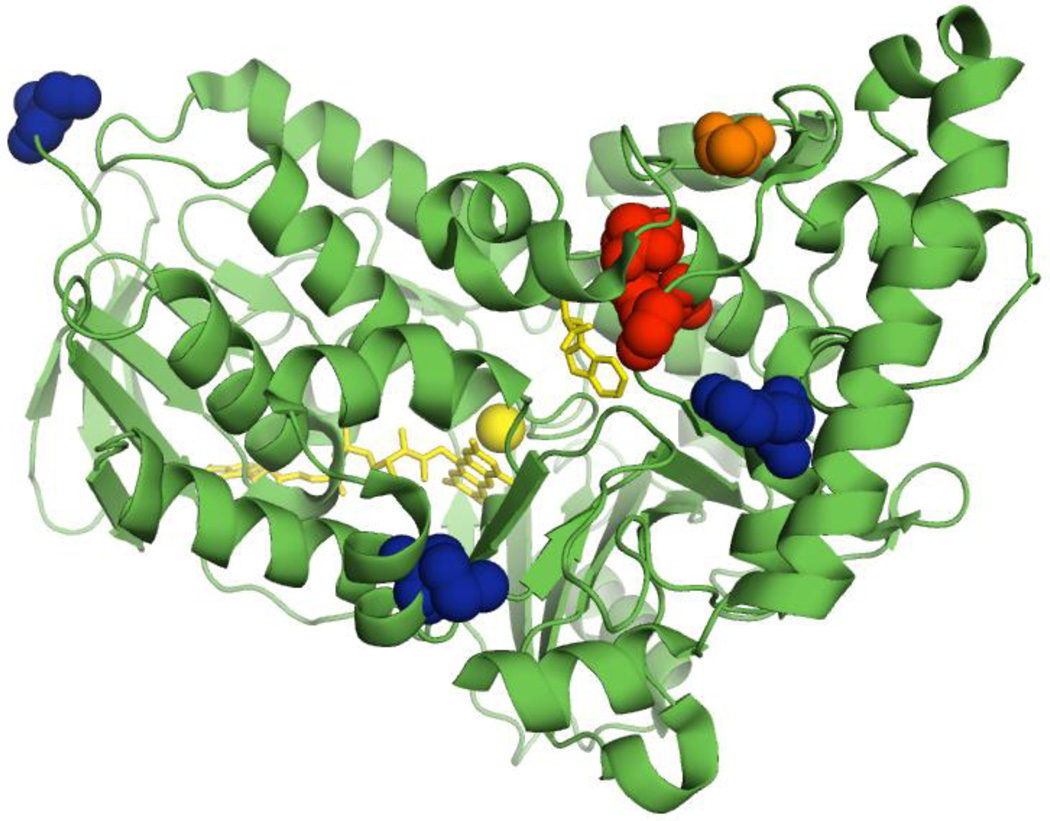
Our group has engineered variants of RebH with improved thermostability,21 expanded substrate scope,22 and altered site-selectivity. These efforts have led to the creation of a panel of halogenases capable of functionalizing arenes beyond the scope of those initially investigated with novel selectivities. Random mutagenesis was used to develop a thermostabilized variant, 1-PVM, which then served as a parent for the evolution of additional variants.21 Through substrate walking, in which substrate scope is expanded by evolving activity on substrates whose structures increasingly depart from the native substrate structure, two variants of particular note were engineered.22 Variant 3-SS was evolved to have higher activity on tryptoline (1) and displayed activity on a range of tryptoline derivatives. Variant 4-V was then evolved from 3-SS to have increased activity on an inhibitor of biofilm formation, deformylflustrabromine, but accepted numerous large indole substrates. The locations of these mutations in the RebH structure are shown in Figure 1. We envisioned that these enzymes, in combination with optimized cross-coupling conditions, could enable late stage chemoenzymatic C-H functionalization of a range of compounds via C-C, C-N, and C-O bond formation without the need for isolation or purification of the halogenated intermediates (Scheme 1C). This approach would mark a significant departure from existing chemoenzymatic halogenation/cross-coupling reports19,20 that are limited to an enzyme’s native substrate and require purification of halogenated intermediates.
Figure 1.
Location of mutations in RebH variants employed in this work. A previously reported crystal structure of wild-type RebH (PDB entry 2OA1)23 with residues that are mutated in variant 1-PVM shown in blue, additional residues mutated in variant 3-SS shown in red, and residue A442 further mutated in variant 4-V shown in orange.22 Bound L-tryptophan, FAD, and chloride are shown in yellow.
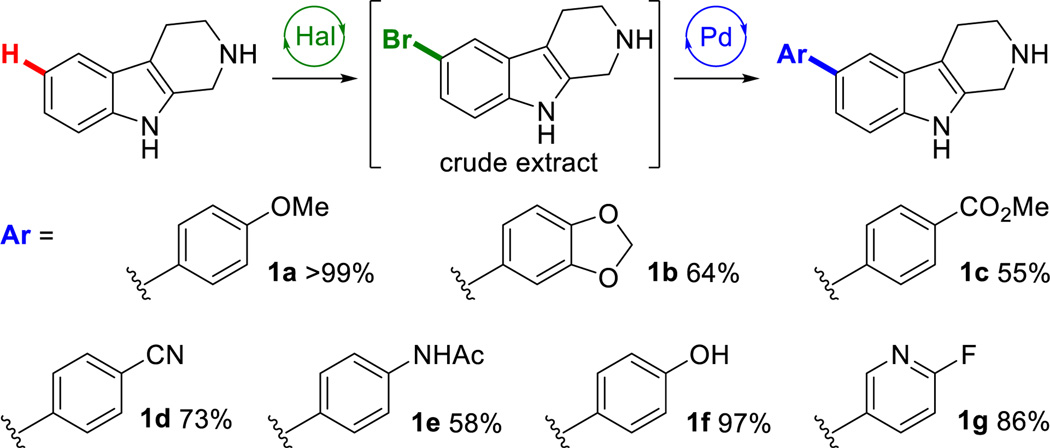
The feasibility of this chemoenzymatic approach was evaluated by submitting the crude organic extracts from analytical scale enzymatic bromination reactions of tryptoline (1) to Suzuki cross-coupling conditions. Tryptoline was chosen as variant 3-SS affords high conversion to 5-halotryptoline at low enzyme loading22 and because of the range of biological activities observed for derivatives of tryptoline.24,25,26 The cross-coupling efficiency of halogenated tryptoline was an early concern due to reported problems with unprotected N-H indoles,27 but a combination of Pd(OAc)2 and water soluble SPhos provided nearly quantitative conversion of crude 6-bromotryptoline to 6-aryltryptoline by LCMS. 10 mg scale bioconversions on 1 were then performed, and the scope of boronic acids for Suzuki reactions performed on the crude bioconversion extract was examined. To our delight, arylated tryptoline derivatives were obtained by reverse phase chromatography in good to high isolated yields (Chart 1). A variety of functional groups including ethers (1a/b), ester (1c), nitrile (1d), amide (1e), hydroxyl (1f), and a substituted pyridine (1g) were tolerated by our protocol, indicating that aryl boronic acids can be coupled to substrates bearing an unprotected indole (N-H) core. Attempts to couple boronic acids bearing strongly-coordinating functional groups (4-pyridyl, 3-aminophenyl) were unsuccessful.
Chart 1.
Boronic acid scope of chemoenzymatic arylation of tryptoline.a
aHal: 0.01 equiv. 3-SS, 0.0005 equiv. MBP-RebF, 9 U/mL GDH, 35 U/mL catalase, 20 equiv. NaBr, 40 equiv. glucose, 0.2 equiv. NAD & FAD, 3.5% v/v i-PrOH/phosphate buffer (25 mM, pH 8.0), 22 °C, 16h (See Figure S1). Pd: 0.05 equiv. Pd(OAc)2 & SPhos-SO3Na, 1.5 equiv. ArB(OH)2, 50% v/v i-PrOH/phosphate buffer (170 mM, pH 8.5), 90 °C, 1h.
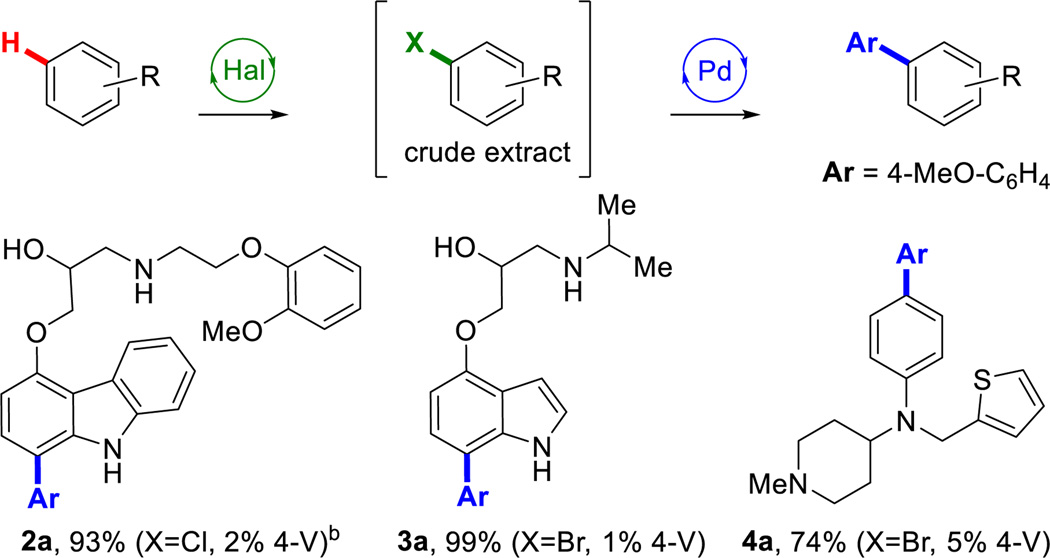
Arene scope was then evaluated by functionalizing different biologically active arenes (Chart 2). Previous work in our lab focused on engineering RebH to halogenate carbazole- and indole-containing small molecules, including carvedilol (2a), and pindolol (3a),22 two nonselective beta-blockers widely used for the treatment of hypertension.28,29 In addition, RebH variant 4-V also accepted the antihistamine thenalidine (4a), a substrate not previously demonstrated to undergo halogenation, which does not bear an indole moiety but instead is functionalized at the para position of its aniline core. The crude extracts from 10 mg scale halogenation reactions of each of the aforementioned arenes were submitted to Pd-catalyzed Suzuki cross-coupling conditions to furnish the corresponding arylated analogues. The site selectivities of the halogenation reactions were identical to those previously reported by our group,22 and the subsequent Suzuki coupling did not alter this selectivity. Notably, the cross-coupling step tolerated the installation of either bromine or chlorine, both of which can be incorporated by RebH in the halogenation step without a significant change in the yield,30 further highlighting the flexibility of this methodology.
Chart 2.
Arene scope of chemoenzymatic arylationa
aHal: as in Chart 1 with the halide source and RebH variant/loading indicated. Pd: as in Chart 1 using 1.5 equiv. 4-MeO-C6H4-B(OH)2. b0.05 equiv. Pd(OAc)2 & SPhos, 1.5 equiv. 4-MeO-C6H4-B(OH)2, 2 equiv. K3PO4, 20% v/v H2O/dioxane, 100 °C, 12h.
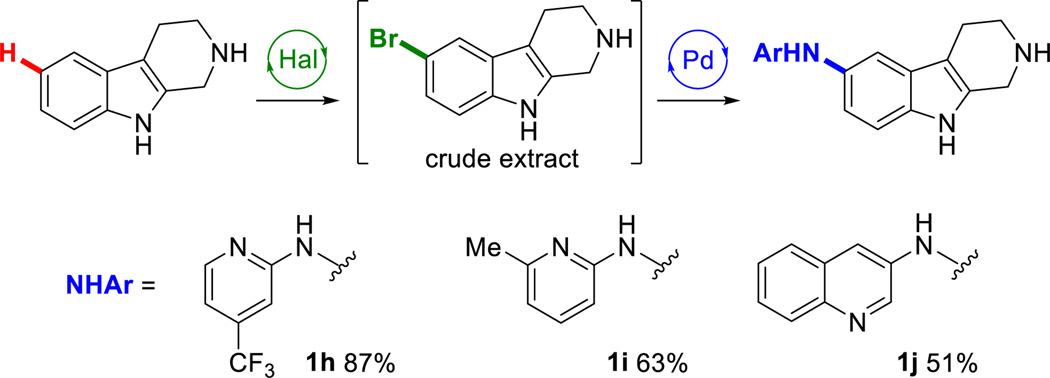
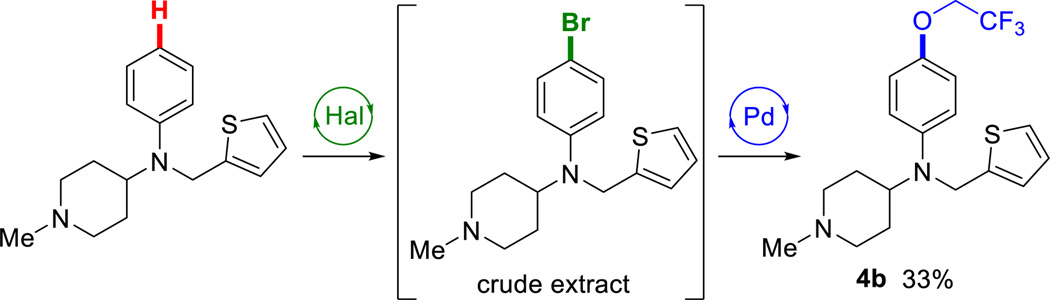
To expand the reaction scope beyond C-C bond formation, crude extracts from enzymatic halogenation reactions were also subjected to Buchwald-Hartwig amination31 and alkoxylation32 conditions. Tryptoline underwent smooth two-step C-H amination with aminopyridine derivatives to afford secondary amines (Chart 3, 1h–j). Attempts to perform a C-H alkoxylation on tryptoline, however, were unsuccessful in our hands. To our knowledge, there are no reports of Pd-catalyzed alkoxylation reactions on unprotected indoles, and our results corroborate its difficulty. Fortunately, thenalidine underwent two-step C-H alkoxylation providing trifluoroethoxythenalidine (4b) in useful, albeit low, yield (Scheme 2), illustrating that substrates compatible with alkoxylation can be used in our chemoenzymatic procedure.
Chart 3.
Chemoenzymatic amination of tryptolinea
aHal: as in Chart 1. Pd: 0.03 equiv. Pd(OAc)2, 0.03 equiv. BrettPhos, 3 equiv. ArNH2, 6 equiv. NaOt-Bu, dioxane, 100 °C, 14h.
Scheme 2.
Chemoenzymatic alkoxylation of thenalidine.a
aHal: as in Chart 1 using 0.05 equiv. 4-V. Pd: 0.005 equiv. [(allyl)PdCl]2, 0.015 equiv. RockPhos, 2 equiv. CF3CH2OH & Cs2CO3, PhMe, 90 °C, 14h.
Initial attempts to conduct both the halogenation and cross-coupling steps contempoaneously were unsuccessful in our hands.33 Inhibition of the halogenase by boronic acids, inhibition of the Pd catalyst by the halogenase (but not all enzymes), and formation of hydrogen peroxide over the course of the halogenation reaction all led to difficulties in this regard. Efforts are underway in our lab to address these issues and to expand the scope of arenes halogenated by RebH to enable functionalization of an even more diverse range substrates.
Chemoenzymatic halogenation/cross-coupling complements conventional methods for C-H functionalization methods in terms of selectivity and reaction scope. Setting the selectivity of the C-H functionalization process by enzymatic halogenation offers a rare example of remote C-H bond cleavage without the need for a directing group.11 This is especially useful for making substituted tryptolines that typically are prepared de-novo via Fischer indole synthesis. In addition to novel selectivity, enzymatic halogenation installs a useful synthetic handle that allows for subsequent formation of a wide range of different bond types (C-C, C-N, C-O) using robust Pd-catalyzed protocols. This expands beyond conventional metal-catalyzed C-H functionalization in which a catalyst system is typically limited to one class of substrates and one type of new bond formed.
In summary, an operationally simple, two-step C-H halogenation/Pd-catalyzed cross-coupling was developed and used to diversify biologically active small molecules. Engineered variants of RebH enabled site-selective halogenation of structurally diverse arenes, and subsequent Pd-catalyzed cross-coupling with boronic acids, amines, and trifluoroethanol furnished compounds with new C-C, C-N, and C-O bonds. These results illustrate how chemoenzymatic processes can be used to exploit the selectivity of enzymatic catalysis and the substrate and reaction scope of small-molecule catalysis. While the reactions described in this work were performed using engineered variants of RebH, this method should be applicable to the derivatization of the diverse products of other halogenases, such as PrnA,34 Rdc2,35 and wild-type RebH.30,36 Continued efforts in our21,22 and other20,37,38 laboratories to engineer halogenase variants with expanded scope, altered selectivity, and improved activity will further increase the utility of this approach.
Supplementary Material
Acknowledgments
This work was supported by The University of Chicago Louis Block Fund for Basic Research and Advanced Study and by a NIH Chemistry and Biology Interface training grant to J.T.P. (T32 GM008720). We also wish to thank The Novartis Institutes for Biomedical Research for the donation of compounds to the CCHF and for partial financial support of this work.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental procedures and compound characterization are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Krska SW, Vachal P, Welch CJ, Smith GF. WO 2014052174 A1. Compound Diversification Using Late Stage Functionalization. 2014
- 2.Cernak T, Dykstra KD, Tyagarajan S, Vachal P, Krska SW. Chem. Soc. Rev. 2016 doi: 10.1039/c5cs00628g. Advance Article. [DOI] [PubMed] [Google Scholar]
- 3.Giuliano MW, Miller SJ. Top. Curr. Chem. 2015:1–45. doi: 10.1007/128_2015_653. [DOI] [PubMed] [Google Scholar]
- 4.Engle KM, Mei T, Wasa M, Yu J. Acc. Chem. Res. 2012;45:788–802. doi: 10.1021/ar200185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishiyama T, Miyaura N. Pure Appl. Chem. 2006;78:1369–1375. [Google Scholar]
- 6.Gormisky PE, White MC. J. Am. Chem. Soc. 2013;135:14052–14055. doi: 10.1021/ja407388y. [DOI] [PubMed] [Google Scholar]
- 7.O’Hara F, Blackmond DG, Baran PSJ. J. Am. Chem. Soc. 2013;135:12122–12134. doi: 10.1021/ja406223k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia C, Kitamura T, Fujiwara Y. Acc. Chem. Res. 2001;34:633–639. doi: 10.1021/ar000209h. [DOI] [PubMed] [Google Scholar]
- 9.Chen K, Eschenmoser A, Baran PS. Angew. Chem. Int. Ed. 2009;48:9705–9708. doi: 10.1002/anie.200904474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen MS, White MC. Science. 2010;327:566–571. doi: 10.1126/science.1183602. [DOI] [PubMed] [Google Scholar]
- 11.Lewis JC, Coelho PS, Arnold FH. Chem. Soc. Rev. 2011;40:2003–2021. doi: 10.1039/c0cs00067a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero PA, Arnold FH. Nat. Rev. Mol. Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 14.Fasan R. ACS Catal. 2012;2:647–666. [Google Scholar]
- 15.Sawayama AM, Chen MMY, Kulanthaivel P, Kuo M-S, Hemmerle H, Arnold FH. Chemistry. 2009;15:11723–11729. doi: 10.1002/chem.200900643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis JC, Bastian S, Bennett CS, Fu Y, Mitsuda Y, Chen MM, Greenberg WA, Wong C-H, Arnold FH. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16550–16555. doi: 10.1073/pnas.0908954106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentmeister A, Arnold FH, Fasan R. Nat. Chem. Biol. 2009;5:26–28. doi: 10.1038/nchembio.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem. Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 19.Roy AD, Grüschow S, Cairns N, Goss RJM. J. Am. Chem. Soc. 2010;132:12243–12245. doi: 10.1021/ja1060406. [DOI] [PubMed] [Google Scholar]
- 20.Runguphan W, O’Connor SE. Org. Lett. 2013;15:2850–2853. doi: 10.1021/ol401179k. [DOI] [PubMed] [Google Scholar]
- 21.Poor CB, Andorfer MC, Lewis JC. Chem Bio Chem. 2014;15:1286–1289. doi: 10.1002/cbic.201300780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payne JT, Poor CB, Lewis JC. Angew. Chem. Int. Ed. 2015;54:4226–4230. doi: 10.1002/anie.201411901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bitto E, Huang Y, Bingman CA, Singh S, Thorson JS, Phillips GN. Proteins. 2008;70:289–293. doi: 10.1002/prot.21627. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka M, Li X, Hikawa H, Suzuki T, Tsutsumi K, Sato M, Takikawa O, Suzuki H, Yokoyama Y. Bioorg. Med. Chem. 2013;21:1159–1165. doi: 10.1016/j.bmc.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 25.Pähkla R, Harro J, Rägo L. Pharmacol. Res. 1996;34:73–78. doi: 10.1006/phrs.1996.0066. [DOI] [PubMed] [Google Scholar]
- 26.Song H, Liu Y, Liu Y, Wang L, Wang Q. J. Agric. Food Chem. 2014;62:1010–1018. doi: 10.1021/jf404840x. [DOI] [PubMed] [Google Scholar]
- 27.Duefert a, Billingsley KL, Buchwald SL. J. Am. Chem. Soc. 2013;135:12877–12885. doi: 10.1021/ja4064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aellig W. Br. J. Clin. Pharmacol. 1982;13:187S–192S. doi: 10.1111/j.1365-2125.1982.tb01909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stafylas PC, Sarafidis PA. Vasc. Health Risk Manag. 2008;4:23–30. doi: 10.2147/vhrm.2008.04.01.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payne JT, Andorfer MC, Lewis JC. Angew. Chem. Int. Ed. 2013;52:5271–5274. doi: 10.1002/anie.201300762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maiti D, Fors BP, Henderson JL, Nakamura Y, Buchwald SL. Chem. Sci. 2011;2:57. doi: 10.1039/C0SC00330A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Fors BP, Buchwald SL. Angew. Chem. Int. Ed. 2011;50:9943–9947. doi: 10.1002/anie.201104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gröger H, Hummel W. Curr. Opin. Chem. Biol. 2014;19:171–179. doi: 10.1016/j.cbpa.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Hölzer M, Burd W, Reißig H-U, Pée K-H van. Adv. Synth. Catal. 2001;343:591–595. [Google Scholar]
- 35.Zeng J, Lytle AK, Gage D, Johnson SJ, Zhan J. Bioorg. Med. Chem. Lett. 2013;23:1001–1003. doi: 10.1016/j.bmcl.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frese M, Guzowska PH, Voß H, Sewald N. Chem Cat Chem. 2014;6:1270–1276. [Google Scholar]
- 37.Frese M, Sewald N. Angew. Chem. Int. Ed. 2015;54:298–301. doi: 10.1002/anie.201408561. [DOI] [PubMed] [Google Scholar]
- 38.Shepherd SA, Karthikeyan C, Latham J, Struck A-W, Thompson ML, Menon BRK, Styles MQ, Levy C, Leys D, Micklefield J. Chem. Sci. 2015;6:3454–3460. doi: 10.1039/c5sc00913h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.