Abstract
A family of Pluronic-based, cholesterol end-capped cationic polyrotaxanes (PR+), threaded with 2-hydroxypropyl-β-cyclodextrin (HPCD), was synthesized for efficient and low toxicity plasmid DNA (pDNA) delivery into multiple cell lines. All PR+ formed highly stable, positively charged complexes with pDNA, with diameters of less than 250 nm. The PR+ demonstrated high cellular uptake and pDNA transfection efficiencies that were better than the commercial transfection standards L2K and bPEI, while displaying similar or lower toxicity profiles. We observed that the charge density and threading efficiency of the PR+ had significant impact on the colloidal stability and physical properties of the complexes, which in turn had an effect on their intracellular transfection efficiencies. These complexes showed high transfection efficiencies against a broad range of cell lines, from fibroblast to multiple tumorigenic cell lines. Moreover, confocal analysis experiments further validated the correlation between threading efficiency and intracellular transfection. Taken together, our results suggest that HPCD:Pluronic PR+ can be used as potent vectors for pDNA-based therapeutics.
Keywords: Gene delivery, Polyrotaxane, β-Cyclodextrins, pDNA, Transfection
Graphical abstract
Conceptual diagram showing PR+:pDNA complexation, cellular internalization and endosomal release, resulting in transgene expression.
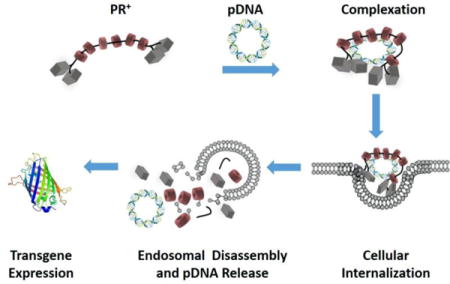
1. Introduction
Cyclodextrins (CD) are a family of cyclic oligosaccharides that are used extensively in pharmaceutical, agrochemical, food, and materials applications1. Their ability to solubilize hydrophobic drugs by encapsulation within their cylindrical cavity, combined with their biocompatibility, makes them widely used as pharmaceutical excipients (e.g., there are more than 45 CD-drug formulations marketed around the world)1. In addition, cationic CD derivatives have been widely used for gene delivery because of their ability to complex nucleic acids as stable nanoparticles, while promoting destabilization of endosomal membranes to enhance their intracellular delivery2. Among the most promising CD-containing materials for non-viral nucleic acid delivery are based on CD polymers3, CD dendrimers4, and CD polyrotaxanes5. For example Davis and co-workers have reported a diverse class of β-CD oligomers coupled via cationic linkers6–10. These agents were successfully used as a condensing agent for siRNA delivery in a clinical trial for treatment of melanoma in humans9. Despite encouraging results, these agents suffered from poor stability in vivo, resulting in premature disassembly of the CD oligomer:siRNA complexes within the glomerular basement membrane (GBM) of the kidney due to the locally high concentrations of heparin sulfate10. Herein, we report the development of polyrotaxanes comprised of 2-hydroxypropyl-β-cyclodextrin (HPCD) and Pluronic block copolymers as potentially stable vehicles for safe and efficient plasmid DNA (pDNA) delivery. Polyrotaxanes (PR) are supramolecular structures derived from the threading of macrocyclic molecules onto a polymer “axle” that is endcapped with bulky groups that prevent dethreading of the macrocyclic “wheels”. The construction of PR from precursors such as HPCDs and Pluronic surfactants that are found in FDA approved pharmaceutical agents makes them highly attractive for biomedical applications5. Consequently, CD-based PR have been used extensively for hydrogel-based drug delivery11, biodegradable drug delivery constructs12, tissue engineering scaffolds13, gene delivery14–24, and as cholesterol mobilizing agents in lysosomal storage disorders25, 26.
Cationic CD PR+s have previously been studied for their DNA complexation and transfection ability in cells, where cationic substituents have been introduced by post-modification reactions after the macrocycle has been threaded onto the polymer backbone14–16. The first generation of these materials employed cationic α-CDs on a central PEG scaffold that was terminated in disulfide bonds, rendering them degradable within the reducing environment of the cell17–20. PR+ have also been synthesized with endcaps that act as targeting ligands to promote uptake via receptor mediated endocytosis21. A recent study described the formation of cationic PR by temperature-activated threading of CDs onto water soluble ionenes and used for pDNA and siRNA delivery22. This technique eliminates the random post-modification step that is often used in attachment of cationic groups to the PR, making them more structurally well-defined and easier to prepare. Our group has previously demonstrated the potential of linear and branched cationic α-CD PR+ as efficient vehicles for siRNA delivery23, 24. We used HPCD instead of β-CD for PR+ with five different PEO-PPO-PEO Pluronics, block copolymers cores due to the lower toxicity, higher solubility, improved renal clearance, and favorable hemolytic profile of HPCD1. The amphiphilic properties of Pluronic were anticipated to be important for these transfection agents, since endcap cleavage and dethreading of the cationic HPCD monomers should release the pDNA cargo and the pluronic core that could promote escape form the endosomal membrane. We chose Pluronics F127, F68, L35, L64 and L81, which differ significantly in their hydrophilic lipophilic balance (HLB), to enable screening for transfection across a wide efficiency range of HLB and PR+ characteristics27–30.
We anticipated that the lateral and rotational mobility of the threaded cationic HPCD on the Pluronic chain would promote enhanced complexation with pDNA to generate potent non-viral vectors at lower N/P ratios than other cyclodextrin polymer constructs. Since a lower transfection complex N/P ratios often translates into improved cell viability, we sought to evaluate these PR+ with respect to their complexation ability, colloidal stability, cell viability, and transfection efficiency in multiple cancer cell lines as well as in fibroblasts and macrophages.
2. Experimental Section
2.1 Materials
PR+ synthesis Materials: The Pluronic triblock copolymers F127 (EO 200, PO 65), F68 (EO 153, PO 29), L35 (EO 22, PO 16), L64 (EO 26, PO 30), and L81 (EO 6, PO 43) were purchased from Sigma-Aldrich and dried by azeotropic distillation from benzene under vacuum before use. 2-Hydroxypropyl-β-cyclodextrin (average degree of hydroxypropyl substitution = 6.8), carbonyldiimidazole (CDI), triethylamine (TEA), tris(2-aminoethyl)amine (TAEA), cholesteryl chloroformate, and N,N-dimethylethylenediamine (DMEDA) were also purchased from Sigma-Aldrich and were used directly. All solvents were reagent grade, purchased from commercial sources, and used without further purification, except for DMF and DCM, which were dried over CaH2, filtered, distilled at reduced pressure and stored under Ar prior to use. Cellulose dialysis membranes were obtained from Spectrum Labs and immersed in deionized water for at least 30 min prior to use. Ultra-pure H2O (resistivity ≈ 18.0 MΩ/cm−1) was generated using a NANOpure Ultrapure H2O system.
2.2. Materials for formulation of complexes and biological assays
AcGFP1 plasmid vector and MTA reagent were purchased from Promega. pDNA was amplified in Escherichia coli and purified according to the supplier’s protocol (Qiagen, Hilden, Germany). The purity and concentration of the purified plasmid DNA was determined by absorption at 260 and 280 nm and by agarose gel electrophoresis. The purified plasmid DNA was resuspended in TE buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA) and kept frozen in aliquots at a concentration of 0.5 mg/mL. Lipofectamine2000 (L2K) transfection reagent, LysoTrackerRed DND-99, SYTOX AADvanced Red Dead Cell Stain, and Wheat Germ Agglutinin-Alexa Fluor680 conjugate were obtained from Life Technologies. Label IT nucleic acid labeling kit (Fluorescein), was obtained from Mirus. Cell culture reagents such as DMEM and RPMI media, FBS, sodium pyruvate, trypsin, and PBS were obtained from Atlanta Biologicals.
2.3. Synthesis of Bis-Cholesterol-End-Capped HPβCD:Pluronic Polyrotaxanes (PR)
The PR were synthesized by modifying a previously reported protocol26. Further details are described in Supplementary Information.
2.4. Synthesis of DMEDA-HPCD:Pluronic Polyrotaxanes (PR+)
The HPCD:Pluronic PR (500 mg) were dissolved in 30 mL dry DMSO and CDI added based on a single equivalent per hydroxyl group in the PR. The reaction mixtures were stirred for 24 h at 20 °C prior to addition of excess DMEDA under Ar, followed by stirring of the reaction mixture for an additional 48 h. The mixture was then dialyzed against DMSO using 6–8 kDa or 12–14 kDa MWCO membranes, depending on the PR molecular weight expected, and then with H2O before lyophilizing to give the DMEDA-HPCD:Pluronic PR+s as white solids. 1H NMR (400 MHz, DMSO-d6): δ = 6.9 ppm (s, H-NCO carbamate), 5.3 ppm (t, 1H, Chl-ethylene H), 5.0 ppm (b, C1-H of CD), 4.5 ppm (b, OH propyl), 3.5–3.8 ppm (m, C3,5,6-H of CD), 3.5 ppm (m, PEG-CH2), 3.1–2.9 ppm (b, DMEDA NH-CH2), 2.7–2.6 ppm (m, 16H, CH2 of TAEA), 2.3–2.2 ppm (b, DMEDA CH2-NMe2), 2.2–2.0 ppm (b, DMEDA N-(CH3)2), 1.0 ppm (d, CH3 of PPG), 0.8–0.6 ppm (m, Chl-CH3).
2.5. Formulation of PR+:pDNA Complexes
The nanoplexes were formulated in TE buffer. The nucleotide consisted of pAcGFP1-1, a plasmid DNA vector (pDNA), coding for green fluorescent protein, (GFP excitation/emission maximum: 475 /505 nm) or FITC-tagged scrambled pDNA (pDNA-FITC). Nanoplexes were formulated by mixing fixed volumes of pDNA or pDNA-FITC (1 μg/mL) and PR+ solution (2 – 10 μg/mL, depending on N/P). The N/P ratios were varied between 5 and 30 to identify the optimum concentration of polymer required for providing both high colloidal stability and good biological performance.
2.6. Particle Size and Zeta (ζ) Potential Measurements
The diameters, size distributions and ζ-potentials of the materials were evaluated by dynamic light scattering (DLS) using a Zetasizer Nano S (Malvern Instruments Ltd.) at 20 °C with a scattering angle of 90º. At least 40 measurements were made and averaged for each sample.
2.7. Atomic Force Microscopy Analysis (AFM)
AFM imaging of the nanoparticles was conducted in tapping mode (MultiMode, Veeco, USA) with a scan rate of 0.5 or 1 Hz using dry samples on mica. Samples were prepared by dropping 2 μL of PR+ and PR+:pDNA solution (2 mg/mL in water) on a mica surface followed by overnight drying at room temperature. The AFM tips (PPP-NCH, Nanoscience Instruments, Inc., USA) had a typical radius of 7 nm or less and a force constant of 46 N/m.
2.8. Gel Retardation Assay
The complexation ability of PR+ with pDNA was determined by 1% agarose (low melting point) gel electrophoresis. The agarose gels were precast in TBE buffer with GelRed dye at 1:10,000 dilution. PR+:pDNA complexes containing 0.2 μg of pDNA at different N/P ratios were loaded onto the gel. A 1:5 dilution of loading dye was added to each well and electrophoresis was carried out at a constant voltage of 55 V for 2 h in TBE buffer. The pDNA bands were then visualized under a UV transilluminator at a wavelength of 365 nm.
2.9. PicoGreen Competitive Binding Assay
Polyplex stability was studied by PicoGreen competition assay. Heparin titration with different polyplexes was performed to study their disassembly in the presence of a negatively charged polymer. Upon heparin challenge, PicoGreen binds to dsDNA that has been released from the pDNA:PR+ complex. Consequently, the increase of fluorescence serves an indication of the level of decomplexation between DNA and polymers. Increasing amounts of heparin were added to the complexes and allowed to incubate for 30 min, followed by addition of the Quant-iT PicoGreen reagent (Invitrogen, Carlsbad, CA) and further incubation for 15 min. The fluorescence was measured in a 96-well plate using a plate reader with an excitation maximum of 480 nm and an emission maximum at 520 nm. The fluorescence intensity was then corrected for background fluorescence. Results from three independent triplicate experiments were analyzed and reported along with the standard error of the measurements.
2.10. Cell Viability Assay
PR+ cytotoxicity relative to bPEI (25 kDa) was evaluated using the MTS assay in NIH 3T3 cells. Cell viabilities were measured as a function of amine content in the PR+. The cells were cultured in complete DMEM medium supplemented with 10% FBS at 37 °C, 5% CO2, and 95% relative humidity. The cells were seeded in 96-well microtiter plates (Nunc, Wiesbaden, Germany) at densities of 10,000 cells/well. After 24 h, the culture media was replaced with serum-free culture media containing increasing concentrations of PR+ and the cells were incubated for 24 h before addition of MTS reagent (15μL) and incubated for 2 h. The absorbance was then measured using a microplate reader (Spectra Plus, TECAN) at a wavelength of 492 nm. The cell viability (%) relative to control cells cultured in media without PR+ was calculated as [A]test/[A]control × 100%, where [A]test is the absorbance of the wells with PR+ and [A]control is the absorbance of the control wells. All cytotoxicity values were measured in triplicate and averaged.
2.11. Cellular Uptake Studies
NIH 3T3 cells were used to study the uptake of the complexes by plating 100,000 cells per well in 24-well plates and incubating for 24 h before the experiment. Complexes of pDNA-FITC:PR+ at different N/P ratios were incubated with cells for 4 h at 37 °C in 10% serum-supplemented media. After 4 h, the spent media was removed and the cells were washed with PBS and trypsinized. These cells were then collected and analyzed by flow cytometry using the FL1 channel.
2.12. In Vitro Transfection/Cell Viability Experiments
NIH 3T3, HeLa, MCF7, and MB49 cells were cultured in complete DMEM medium, while H441 and 3D431 cells were cultured in RPMI medium supplemented with 10% FBS. All cells were incubated at 37 °C, 5% CO2, and 95% relative humidity, respectively, at a cell density of 100,000 cells/well in 24-well plates. After 24 h, the culture media was replaced with serum-supplemented media containing 1μg pDNA complexes with PR+ at N/P ratios of 10, 20 and 30. The cells were incubated with the complexes for 4 h, after which the spent media was aspirated and fresh serum-supplemented media was added. After an additional 32 h incubation, the media was aspirated and the cells were washed with PBS, trypsinized and added to FACS analysis tubes. SYTOX AADvanced dead cell stain (1 μL) was added to each sample and incubated on ice for 15 min before analysis by FACS. FL1 channel was used for the GFP fluorescence and FL4 for the SYTOX AADvanced dead stain (red) fluorescence. %GFP mean fluorescence intensity was calculated relative to L2K, which was considered as 100%.
2.13. Intracellular Trafficking of PR+:pDNA Complexes
Confocal microscopy was used to study the intracellular trafficking of PR+:pDNA complexes. The plasma membrane was stained using AlexaFluor 680 WGA and the acidic compartments in the cytoplasm were stained using LysoTracker Red DND. NIH 3T3 cells were cultured in complete DMEM medium supplemented with 10% FBS at 37 °C, 5% CO2, and 95% relative humidity at a cell density of 30,000 cells/well in 4 chambered cover glass slides. After 24 h, the culture media was replaced with DMEM media containing the PR+:pDNA complexes labeled with 0.5 μg FITC-pDNA at an N/P ratio of 20. The cells were incubated with the complexes for 2, 4, and 9 h, after which they were stained with Alexa 689 WGA (1 μL) for 5 min, washed with PBS (3×), and then counterstained with Lysotracker DND 99 (2 μL) for 10 min. The cells were then washed with PBS three times before observation using a 60X silicone oil objective using a Nikon A1R MP Multiphoton microscope.
3. Results and Discussion
3.1. Synthesis of Bis-Cholesterol-endcapped DMEDA-HPCD: Pluronic PR+
A family of HPCD:Pluronic PR+ was synthesized by modifying a previously reported procedure using heterogeneous conditions26. 1H NMR spectroscopy was used to quantify the number of cyclodextrins threaded onto the Pluronic axle by comparing the integral intensities of the HPCD C1-H (5.05 ppm) and PPG CH3 (1.0 ppm) signals. The coverage ratio was calculated based on the assumption that one CD molecule was capable of including two PPG units within its cavity. The proton peak at ~1.0 ppm is assigned to the PPG methyl groups on the copolymer, whereas the proton signals in the 3 – 3.5 ppm region are attributed to the methylene units (CH2) of the PEG and C3-H, C5-H and C6-H of the HPCD protons. The broad signal displayed in the 4.5 – 5.0 ppm region is assigned to the HPCD C1-H proton as well as the OH-8 proton of the HPCD modification. The broadening of peaks in this region confirms that the Pluronic copolymers were successfully threaded with HPCD units (Figures S1–S5). Post-modification of these HPCD:Pluronic PR was achieved via CDI activated coupling with DMEDA in DMSO, giving the final cationic PR+ with random carbamate-linked DMEDA modifications. The reaction mixtures were then exhaustively dialyzed against DMSO and H2O to remove excess DMEDA and other low molecular weight impurities. Finally, the dialyzed products were lyophilized and characterized by 1H NMR to calculate the average DMEDA density. The characterization was carried out by comparing the integral intensities of the DMEDA N-(CH3)2 peak at 2.2–2.0 ppm with the PPG CH3 (1.0 ppm) signals (Table 1, Figure 1, S6–S10). Even though the threading and post modification conditions were similar, each PR+ exhibited variable HPCD threading efficiency, most likely due to the lengths of PEO and PPO blocks of the Pluronic.
Table 1.
Composition of PR+: The threading efficiency was calculated based on a presumed 1:2 HPCD:PO unit ratio. # CD refers to the number of HPCD molecules threaded onto the Pluronic core and DMEDA represents umber of DMEDA attached to each PR+ as determined by 1H NMR integration. * Molecular weight estimated from proton NMR integration.
| PR+ | # CD | DMEDA | DMEDA/CD | HLBa | Threading Efficiencyb | Mw of PR+ (kDa)c |
|---|---|---|---|---|---|---|
| F127 | 15 | 63 | 4.2 | 22 | 46% | 42.9 |
| L81 | 11 | 95 | 8.6 | 2 | 52% | 32.4 |
| L64 | 8 | 71 | 8.8 | 15 | 53% | 23.9 |
| F68 | 3 | 10 | 3.3 | 29 | 22% | 19.5 |
| L35 | 3 | 20 | 6.6 | 19 | 37% | 11.3 |
HLB (hydrophilic-lipophilic balance) of the copolymers were determined by the manufacturer.
The threading efficiency was calculated based on a presumed 1:2 HPCD:PO unit ratio. # CD refers to the number of HPCD molecules threaded onto the Pluronic core and DMEDA represents umber of DMEDA attached to each PR+ as determined by 1H NMR integration.
Molecular weight estimated from proton NMR integration.
Figure 1.
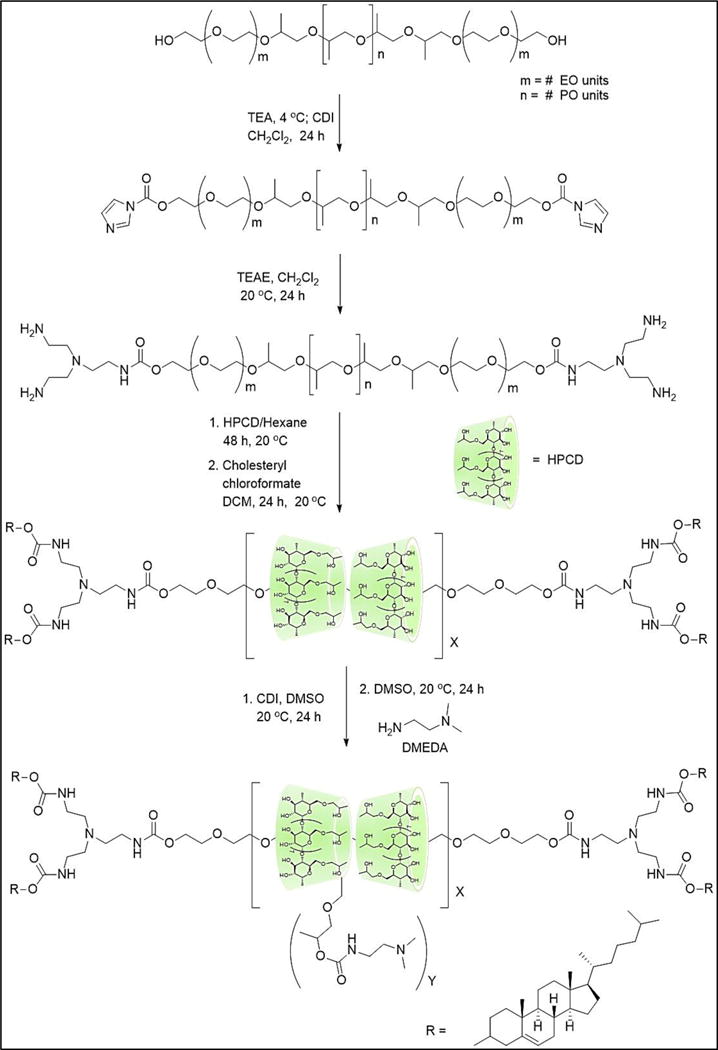
Synthesis scheme of DMEDA-HPCD:Pluronic Polyrotaxanes (PR+)
3.2. Physical Characterization of PR+:pDNA Complexes
The abilities of the PR+ materials to form pDNA complexes was initially evaluated by analyzing their particle diameters and surface charges at different N/P ratios. Dynamic light scattering (DLS) was used to determine the number-averaged diameters produced using the different PR+:pDNA complexes. Our data shows that the PR+ formed complexes with diameters between 150 – 250 nm. At low N/P ratios (e.g., 5), complexes in the 300 – 400 nm size regime were produced; however, the observed diameters dropped below 250 nm when N/P ≥ 10, showing no appreciable change in diameter as the N/P ratio increased further (Figure 2A). This is in agreement with similar observations of bPEI:pDNA complexes wherein the particle diameters become smaller when N/P ≥ 5. The PDIs for all the complexes were in the range of of 0.2–0.35.
Figure 2.
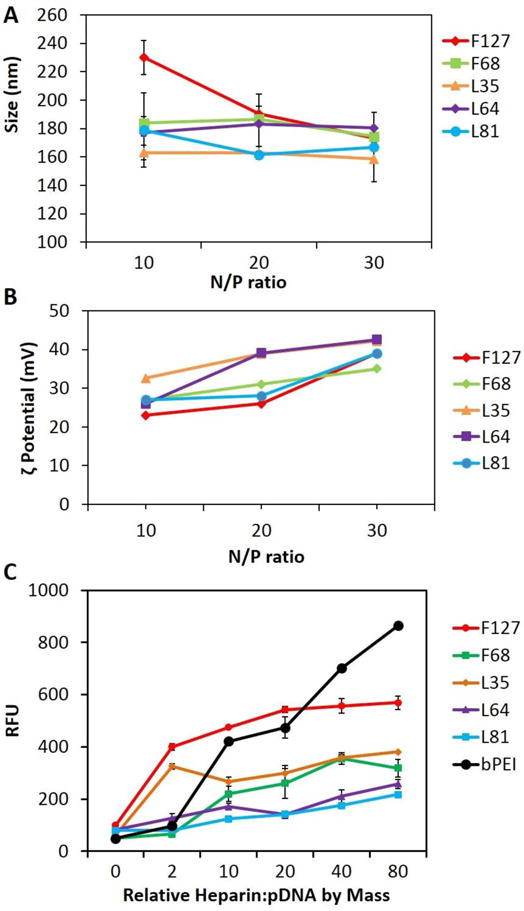
Physical Characterization of PR+:pDNA Complexes: Size, ζ-Potential, and Colloidal Stability as a Function of PR+ Type. (A) Particle diameter, determined by dynamic light scattering (DLS) analysis, as a function of N/P; (B) ζ-potentials of PR+:pDNA complexes formulated at various N/P ratios; and (C) PicoGreen competitive binding assay in the presence of added heparin showing colloidal stability of PR+:pDNA complexes and bPEI:pDNA complexes at N/P = 10. For DLS and ζ-potential analysis, PR+:pDNA particles were formulated and measured using Zetasizer Nano-S at 20 °C with a scattering angle of 90º. At least 40 measurements were made and averaged for each sample for ζ-potential as well as size determination. For the PicoGreen binding assay, 100 μL of PR+:pDNA complexes were incubated with increasing amounts of heparin for 30 min in 96 well plates at room temperature, followed by addition of the Quant-iT PicoGreen reagent and further incubation for 15 min. The fluorescence was measured using a Spectra Plus microplate reader with an excitation maximum of 480 nm and an emission maximum at 520 nm.
The ζ-potentials of the complexes were measured to determine the extent of charge development on the PR+:pDNA transfection complexes (Figure 2B). We found that all the complexes produced an increase in the observed ζ-potential as the N/P ratio increased, with observed ζ-potentials in the range of 20–40 mV. It was also observed that increasing N/P ratio or changing the PR+ type did not affect the ζ-potential significantly, such that the PR+ complexes all displayed displayed ζ-potentials between 35 – 40 mV at N/P = 30. In general, positive ζ-potential promote efficient in vitro cellular uptake; however, it adversely affects the in vivo performance of the complexes due to non-specific uptake, macrophage clearance, and mononuclear phagocytic system (MPS) opsonization. Consequently, the formation of transfection complexes with modest postive ζ-potentials may contribute to their improved in vivo performance.
The size and shape of the PR+:pDNA complexes were further validated using atomic force microscopy (AFM). Nanoparticles were observed with diameters of the range of 150–250 nm, consistent with our DLS observations (Figure S11).
3.3. Complexation Properties and Colloidal Stability of PR+:pDNA Complexes
The complexation properties and colloidal stabilities of the PR+:pDNA complexes were determined by gel retardation and PicoGreen competitive binding assays, respectively. Gel retardation assay results showed that the complexation ability of PR+ with pDNA increases with increasing N/P ratio for all PR+ species. Although the polyrotaxanes displayed modest condensation ability at N/P = 5, it improved significantly at N/P ratios of 10, 20 and 30, where pDNA complexation ability was comparable to the commercial transfection standard bPEI. Interestingly, no significant differences were observed in the complexation ability of the different polymers, with all essentially being very similar with regard to their gel shift behavior (Figure S12). Moreover, to compare the complexation ability of the present material with that of previously reported cationic-CD based compounds, we chose the Admantane-PVA-PEG pendant polymer system (Ad-PP, which showed the best complexation ability among pendant polymer family member) and F68-PR+ (which showed intermediate complexation ability among the PR+ family members) for gel shift analysis (Figure S13)31. We found that F68+showed much better complexation ability than Ad-PP at all N/P ratios, which could be due to increased mobility of the threaded cationic HPCD via rotation and translation along the Pluronic chain compared to fixed facial of the pendant polymer system, resulting in enhanced complexation ability with pDNA.
The PicoGreen competitive binding assay was performed to study the colloidal stability of complexes in the presence of the negatively-charged polymer, heparin, as an example of a biogenic polyanion that can promote decomplexation in vivo. Since PicoGreen displays exceptionally high fluorescence enhancement when it selectively binds to free dsDNA, we used this property to screen for complex disassembly in the presence of increasing amounts of heparin at weight ratios of 0, 2, 10, 20, 40, and 80 relative to the pDNA content in the complexes. Under the experimental conditions employed, the fluorescence increased with added heparin due to release of free dsDNA from the PR+:pDNA complexes (Figure 2C, S013).
We found that, among all the PR+ evaluated, L81- and L64-based PR+:pDNA complexes had the highest colloidal stability across N/P ratios of 10, 20 and 30, with negligible disassembly observed even at the very high heparin:DNA ratio of 80:1 (Figure 2C and S13). We attribute this observation to the higher DMEDA/CD ratios of the L81 and L64 PR+ relative to the other PR+ family members. We infer from these findings that increased charge density and smaller PEO block lengths on the PR leads to stronger pDNA complexation. It was also observed that complexes formed by all the PR+, except F127, had greater colloidal stability than complexes formed by the commercial transfection standard bPEI. The low colloidal stability of F127 PR+ compared to others PR+ family members can be attributed to its low DMEDA/CD ratio and the fact that it possesses the larger PEO block length (Table 1). The superior colloidal stability of the PR+:pDNA in the presence of a negatively charge polymer such as heparin suggests that these PR+:pDNA polyplexes may be stable in serum and more stable toward glomerular basement membrane disassembly in the kidney, where the high concentration of heparin sulfate is known to disassemble lower MW cationic cyclodextrin oligomer:siRNA complexes.28
3.4. Biological Performance of PR+:pDNA Complexes
The in vitro cell viability of PR+:pDNA complexes were evaluated in NIH 3T3 cells as a function of N/P ratio (Figure 3A) to provide a direct comparison of the cytotoxicity arising from the different transfection agents as a function of their molecular weights and charge densities. Commercial transfection standards bPEI and L2K were used as the benchmarks for these experiments.
Figure 3.
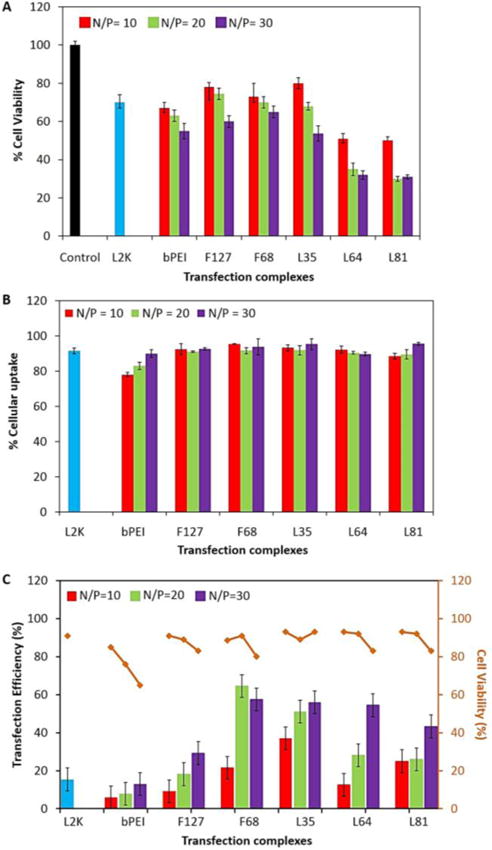
Biological Performance of PR+:pDNA Complexes in Mouse Embryonic Fibroblasts (NIH 3T3). (A) MTS cell viability assay as a function of N/P ratio after 24 h exposure to the complexes; (B) Cell associated fluorescence of PR+:pDNA, L2K:pDNA, and bPEI:pDNA complexes after incubation for 4 h, as a percentage of all cells exposed to transfection complexes, at N/P ratios of 10, 20 and 30; and (C) Two-color flow-cytometric analysis of transfection efficiency and SYTOX AADvanced dead cell stain (red) cytotoxicity after exposure to PR+:pDNA complexes at N/P ratios of 10, 20 and 30, with bPEI and L2K as positive controls. For MTS assay, 75000 cells were plated in 96 well plates and incubated with PR+:pDNA complexes for 24 h at 37 ºC. After the incubation period, 15 μL of MTS reagent was added to each sample (triplicates) and further incubated for 2 h. Following the incubation period, the absorbance was measured using a Spectra Plus microplate reader at a wavelength of 492 nm. Cellular uptake was performed using PR+:pDNA-FITC (1μg/mL) complexes that were incubated with cells in 24 well plate for 4 h. Following the incubation time, cells were washed with PBS, trypsinized, and analyzed using flow-cytometry. The percentage of the parent cell population (10,000 cells) displaying fluorescein fluorescence was analyzed using the FL1 channel and compared with controls. A similar protocol was performed for the transfection experiments using PR+:pDNA, where after removal of particles, the cells were further grown for 36 h, trypsinized, and 5 μL of SYTOX AAdvanced RED was added to each sample followed by incubation for 15 min on ice. After incubation, the samples were analyzed in tandem; the percentage of cells expressing GFP was analyzed using the FL1 channel and the fluorescence due to intercalation of SYTOX AADvanced RED with nucleic acid (in membrane compromised cells) was analyzed using the FL4 channel.
The relative cell viability profiles of the PR+:pDNA, bPEI:pDNA, and L2K:pDNA complexes were evaluated by MTS assay in NIH 3T3 cells (Figure 3A). F127-, F68-, and L35- based PR+ had cell viability profiles that were comparable to both L2K and bPEI, whereas L64- and L81-based PR+ complexes showed higher toxicity, with only 30% – 50% viable cells at higher N/P ratios. The cell viability profiles of the PR+ are correlated with their charge densities. Since L64- and L81-based PR+ complexes have higher charge densities per molecule than those found in F127-, F68-, and L35-based PR+, we attribute the higher observed cytotoxicity to the increased charge densities on the L64- and L81-based PR+ and their respective dethreaded DMEDA-HPCD monomer fragments. These results are in agreement with other cationic transfection reagents that have been used for pDNA delivery, wherein higher charge densities are responsible for higher cytotoxicities32, 33.
The in vitro cellular uptake profile of PR+:pDNA-FITC complexes were assessed in NIH 3T3 cells at different N/P ratios in 10% serum-supplemented media (Figure 3B, S14 &15) under conditions that were optimized for L2K complexes. It was observed that L2K complexes had an uptake efficiency of almost 90%, which is in good agreement with literature reports.27 All the PR+ complexes had comparable or better cellular uptake efficiency (≥ 90%) relative to L2K and bPEI at all N/P ratios studied. We attribute these observations to the enhanced stability of PR+:pDNA complexes in the presence of biogenic polyanions such that improved stability under cell culture conditions leads to higher cellular association and uptake.
The transfection efficiency of the complexes was assessed in NIH 3T3 cells in the presence of 10% serum-supplemented medium by a two-color flow cytometry assay. The transfection experiment was run in tandem with a cell viability assay to enable the measurement of both these parameters in the same experiment. The experiment was performed with L2K and bPEI as positive controls (Figure 3C, S16–19).
The experiments in NIH 3T3 cells reveal that the transfection efficiency was dependent on the N/P ratio as well as the member of the PR+ family used. However, regardless of the PR+ employ, our findings reveal that PR+:pDNA complexes had transfection efficiencies that were superior to both bPEI and L2K. F68-, L35-, and L64-based PR+ had the highest transfection efficiencies, displaying between 35% – 50% greater transgene expression than either L2K or bPEI under conditions that were optimized for these standard reagents. The F127-based PR+ was comparable to L2K and bPEI, showing 25% transfection at an N/P ratio of 30, making it the least efficient member of the PR+ family. F68-based PR+ complexes had the highest transfection efficiencies, with ~70% transfection at N/P = 20, whereas L35 based PR+ complexes produced similar transfection efficiencies of ~60% for N/P = 30. The transfection efficiencies of each member of the PR+ family improved with increasing N/P ratio. Cell viabilities, measured in tandem with transfection efficiencies using SYTOX AADvanced to label the viable cells, showed that cells treated with the PR+ complexes had viabilities between 80% – 90%, regardless of the PR+ used, although the cell viability dropped from 90% to 80% when the N/P ratio was increased from 10 to 30. These cell viability profiles were similar to the commercial reagent L2K, used as a standard in these experiments. bPEI displayed much higher toxicity than the PR+ complexes, with only 50% viable cells at N/P = 30.
Interestingly, the PR+ family members displaying the highest transfection efficiencies (i.e., F68-, L35-, and L64- based PR+) also possessed the best colloidal stability as measured by the PicoGreen competitive binding assay. The F127- based PR+ complexes demonstrated the lowest transfection efficiency, likely due to its poor colloidal stability as demonstrated by the PicoGreen assay. The poor colloidal stability of F127 PR+:pDNA complexes may result in their premature disassembly within serum supplemented media, thereby resulting in lower transfection efficiency. bPEI also demonstrated poor colloidal stability and demonstrated comparable transfection efficiency to the F127-based PR+:pDNA complexes. These findings suggests that the colloidal stability of the PR+:pDNA complexes is a key physical property to evaluate in order to correlate relative efficiencies of cellular transfection reagents.
3.5. Intracellular Trafficking of PR+:pDNA Complexes
Cellular uptake and intracellular trafficking of pDNA complexes formed with F68- and F127- based PR+ was studied as a function of time by confocal microscopy (Figure 4). These two PR+:pDNA formulations were selected since they displayed the highest and the lowest transfection efficiencies in NIH 3T3 cells, respectively (Figure 4C). For both F68- and F127- based PR+:pDNA complexes, it was observed that very few transfection complexes co-localized with the plasma membrane at 2 h. After 4 h, however, it was observed that a much larger population of complexes localized with the plasma membrane, including a small number of complexes that were internalized and colocalized with the lysosomes in F68-based PR+ complexes (Figure 4, top), while negligible internalization was observed for F127 (Figure 4, bottom).
Figure 4.
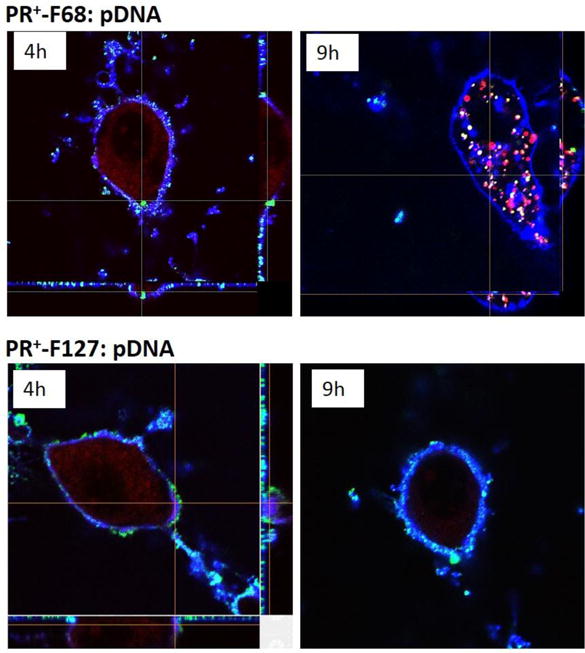
Confocal Microscopy z-stack Images of PR+:pDNA-FITC Complexes @ N/P = 20 Incubated at 4 h and 9 h. Blue – Alexa 689 WGA labeled plasma membrane; Red – Lysotracker DND 99 labeled lysosomes, Green – transfection complexes. NIH 3T3 cells plated in 4 well confocal chambers and were grown for 24 h at 37 ºC before the experiment. PR+:pDNA-FITC complexes were incubated with the cells for 4 h, washed, and observed under the confocal microscope immediately or after a total incubation time of 9 h. Cells were also labeled with 1 μL of Alexa 689 WGA (blue) for 5 min, as a plasma membrane label washed with PBS (3x) and followed by incubation with 2 μL of Lysotracker DND 99 (red) for 10 min as a late endosome/lysosome marker and before washing and observation using a 60X silicone oil objective.
In the case of F68-based PR+ complexes, most of the particles were colocalized with the endosome/lysosome compartment after 9 h, with very few complexes were still associated with the plasma membrane. This was in distinct contrast with the findings for F127-based PR+ complexes wherein most of the particles were still associated with the plasma membrane and no detectable complexes were found in the endosome/lysosomes compartments. Interestingly, the extensive cell associated fluorescence observed by flow cytometry at 4 h are in agreement with the confocal images obtained at the same time point; however, the lack of F127-based PR+ complex internalization as observed at longer circulation periods (9 h) via confocal microcopy is consistent with the observed poor transfection efficiency of these complexes. Taken together, we infer from these findings that the lower colloidal stability and slower internalization kinetics of F127-based PR+ complexes accounts for their poorer transfection performance. Furthermore, we further conclude that the similar ζ-potentials observed for all PR+:pDNA based complexes results in high cellular association, although the colloidal stability of the complexes appears to be the primary determinant for eventual cellular internalization and transgene expression.
3.6. Effect of Cell Line on PR+ Transfection Performance
Cellular transfection and cell viability assays were performed using the entire PR+ family in HeLa, MCF7, MB49, H441, and 3D4-31 cell lines to evaluate their relative performance with respect to transgene expression. The transfection levels and cell viability profiles of the PR+ materials in HeLa cells (Figure 5A) were similar to those observed in NIH 3T3 (Figure 3C). In the case of MB49, a murine bladder cancer cell line (Figure 5B), the transfection efficiencies were similar to those observed in NIH 3T3 (Figure 3C) and HeLa (Figure 5A) cells; however, the cell viabilities were as low as 60–80% for F68- and L64- based PR+ complexes. Cell viabilities were higher (~90%) in 3D4-31 macrophages relative to MB49 cells for all members of the PR+ family (Figure 5 C); however, the transfection levels were significantly lower than those observed in NIH 3T3, HeLa, and MB49. Since macrophages are known to be difficult to transfect, it is interesting to note that all the PR+, with the exception of F127-based PR+, showed transfection efficiencies that were slightly lower than optimized L2K formulations in this cell line. H441 cells, a human lung cancer cell line, showed improved performance relative to L2K for all PR+ except the F127-and F68- based materials (Figure 5D), although the higher transfection efficiencies of L35-, L64- and L81-based PR+ complexes were also accompanied by higher cytotoxicities of these complexes at N/P ratios >10. L35-based complexes appeared to provide the best compromise in H441 cells, with a transfection efficiency of ~50% and minimal cytotoxicity (~10 %). Finally, none of the PR+ reagents or the bPEI and L2K standards were effective towards transfection of MCF-7 cells (Figure 5E).
Figure 5.
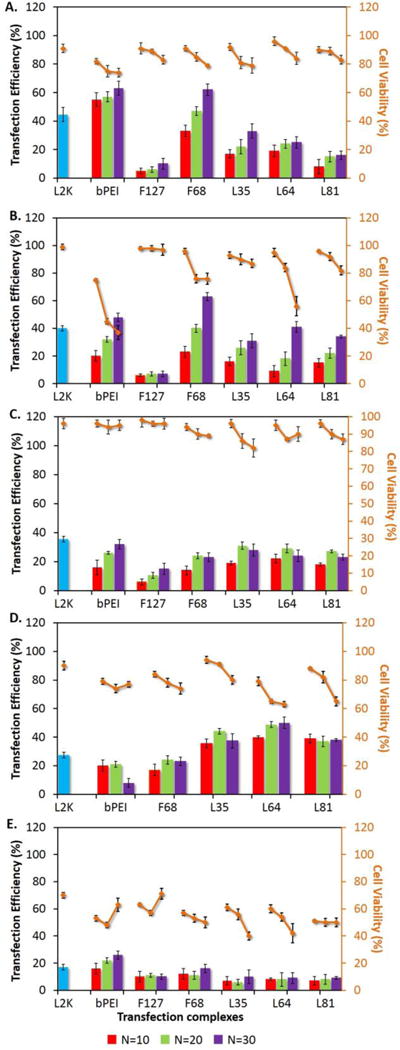
Biological Performance of PR+:pDNA Complexes in (A) HeLa, (B) MB49, (C) 3D4-31 (D) H441, and (E) MCF7 cells. Two-color flow-cytometry analysis of intracellular transfection efficiency and SYTOX AADvanced dead stain (red) cytotoxicity was employed after exposure of the cells to PR+:pDNA complexes formulated with N/P ratios of 10, 20 and 30, with bPEI and L2K as controls. PR+:pDNA complexes were incubated with cells in 24 well plates for 4 h and following the incubation time, complexes were aspirated and the cells were washed with PBS and were further grown for an additional 32 h. After this incubation period, cells were trypsinized and 5 μL of SYTOX AADvanced RED was added to each sample, followed by incubation for 15 min on ice. After incubation, samples were analyzed in tandem; the percentage of cells expressing GFP was analyzed in FL1 channel and the fluorescence due to intercalation of SYTOX AADvanced RED with nucleic acid (in membrane compromised cells) was analyzed in FL4 channel.
4. CONCLUSIONS
In summary, a family of five HPCD:PluronicPR+ reagents were synthesized and evaluated for their ability to form stable and efficient pDNA transfection complexes. It was observed that all PR+ formed pDNA nanocomplexes less than 250 nm in size at N/P ratios above 5, with colloidal stability properties that were superior to bPEI. In general, the PR+:pDNA complexes demonstrated transfection efficiencies and cytotoxicity profiles that were similar, or superior in many cases, to the commercial transfection standards, L2K and bPEI. It was also observed that the charge density and threading efficiency of the polyrotaxanes had a significant impact on the colloidal stability and physical properties of the transfection complexes, which in turn influenced their transfection efficiencies. F68-, L35-, and L64-based PR+:pDNA complexes displayed the highest colloidal stabilities and greatest transfection efficiencies. The superior performance of these agents was observed in several cell lines (e.g., HeLa, MB49, 3D4-31, and H441), showing transfection efficiencies that were comparable or better than L2K. These results suggest that DMEDA-HPCD:Pluronic-based PR+ may be effective vectors for pDNA-based therapeutics. Since the PR+:pDNA complexes are stable towards heparin exposure, they may potentially be stable towards disassembly in the glomerular basement membrane of the kidney during blood circulation, where the high concentration of heparin sulfate has degraded other cationic cyclodextrin oligomeric complexes with siRNA [10]. Experiments in progress are designed to evaluate this opportunity.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the support of NIH RO1 GM087016 and R21 CA15196. The NMR, MS and flow cytometry data were acquired using facilities that are supported by a grant to Purdue University of Center for Cancer Research (P30 CA023168). The assistance of Jeffrey Woodliff with the flow cytometry experiments and Aaron Taylor with the confocal microscopy experiments is also gratefully acknowledged.
Footnotes
Supporting Information is available online from the Wiley Online Library or from the author.
References
- 1.Davis ME, Brewster ME. Nat Rev Drug Discov. 2004;3(12):1023–1035. doi: 10.1038/nrd1576. [DOI] [PubMed] [Google Scholar]
- 2.Huang HL, Tang GP, Wang QQ, Li D, Shen FP, Zhou J, Yu H. Chem Commun. 2006;(22):2382–2384. doi: 10.1039/b601130f. [DOI] [PubMed] [Google Scholar]
- 3.Lee CC, MacKay JA, Frechet JMJ, Szoka FC. Nat Biotechnol. 2005;23(12):1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 4.Helms B, Meijer EW. Science. 2006;313(5789):929–930. doi: 10.1126/science.1130639. [DOI] [PubMed] [Google Scholar]
- 5.Li JJ, Zhao F, Li J. Appl Microbiol Biot. 2011;90(2):427–443. doi: 10.1007/s00253-010-3037-x. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez H, Hwang SJ, Davis ME. Bioconjugate Chem. 1999;10(6):1068–1074. doi: 10.1021/bc990072j. [DOI] [PubMed] [Google Scholar]
- 7.Reineke TM, Davis ME. Bioconjugate Chem. 2003;14(1):247–254. doi: 10.1021/bc025592k. [DOI] [PubMed] [Google Scholar]
- 8.Reineke TM, Davis ME. Bioconjugate Chem. 2003;14(1):255–261. doi: 10.1021/bc025593c. [DOI] [PubMed] [Google Scholar]
- 9.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464(7291):1067–U140. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuckerman JE, Choi CHJ, Han H, Davis ME. P Natl Acad Sci USA. 2012;109(8):3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Li J. J Biomed Mater Res A. 2008;86A(4):1055–1061. doi: 10.1002/jbm.a.31710. [DOI] [PubMed] [Google Scholar]
- 12.Moon C, Kwon YM, Lee WK, Park YJ, Yang VC. J Control Release. 2007;124(1–2):43–50. doi: 10.1016/j.jconrel.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichi T, Watanabe J, Ooya T, Yui N. Biomacromolecules. 2001;2(1):204–210. doi: 10.1021/bm005617n. [DOI] [PubMed] [Google Scholar]
- 14.Yang C, Wang X, Li HZ, Goh SH, Li J. Biomacromolecules. 2007;8(11):3365–3374. doi: 10.1021/bm700472t. [DOI] [PubMed] [Google Scholar]
- 15.Yang CA, Li HZ, Wang X, Li J. J Biomed Mater Res A. 2009;89A(1):13–23. doi: 10.1002/jbm.a.31976. [DOI] [PubMed] [Google Scholar]
- 16.Yang C, Wang X, Li HZ, Tan E, Lim CT, Li J. J Phys Chem B. 2009;113(22):7903–7911. doi: 10.1021/jp901302f. [DOI] [PubMed] [Google Scholar]
- 17.Ooya T, Choi HS, Yamashita A, Yui N, Sugaya Y, Kano A, Maruyama A, Akita H, Ito R, Kogure K, Harashima H. J Am Chem Soc. 2006;128(12):3852–3853. doi: 10.1021/ja055868+. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita A, Kanda D, Katoono R, Yui N, Ooya T, Maruyama A, Akita H, Kogure K, Harashima H. J Control Release. 2008;131(2):137–144. doi: 10.1016/j.jconrel.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Yamada Y, Nomura T, Harashima H, Yamashita A, Yui N. Biomaterials. 2012;33(15):3952–3958. doi: 10.1016/j.biomaterials.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 20.Yamada Y, Nomura T, Harashima H, Yamashita A, Katoono R, Yui N. Biol Pharm Bull. 2010;33(7):1218–1222. doi: 10.1248/bpb.33.1218. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Wang H, Wang CX, Li YS, Lu WF, Chen SF, Luo JD, Jiang YN, Chen JH. Mol Pharmaceut. 2012;9(5):1067–1076. doi: 10.1021/mp200315c. [DOI] [PubMed] [Google Scholar]
- 22.Dandekar P, Jain R, Keil M, Loretz B, Muijs L, Schneider M, Auerbach D, Jung G, Lehr CM, Wenz G. J Control Release. 2012;164(3):387–393. doi: 10.1016/j.jconrel.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni A, DeFrees K, Schuldt RA, Vlahu A, VerHeul R, Hyun SH, Deng W, Thompson DH. Integr Biol-Uk. 2013;5(1):115–121. doi: 10.1039/c2ib20107k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulkarni A, DeFrees K, Schuldt RA, Hyun SH, Wright KJ, Yerneni CK, VerHeul R, Thompson DH. Mol Pharmaceut. 2013;10(4):1299–1305. doi: 10.1021/mp300449t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins CJ, McCauliff LA, Hyun SH, Zhang ZR, Paul LN, Kulkarni A, Zick K, Wirth M, Storch J, Thompson DH. Biochemistry-Us. 2013;52(19):3242–3253. doi: 10.1021/bi3010889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondjinou YA, McCauliff LA, Kulkarni A, Paul L, Hyun SH, Zhang Z, Wu Z, Wirth M, Storch J, Thompson DH. Biomacromolecules. 2013;14(12):4189–97. doi: 10.1021/bm400922a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batrakova EV, Kabanov AV. J Control Release. 2008;130(2):98–106. doi: 10.1016/j.jconrel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang CX, Zhang JL, Li W, Feng XY, Hou MQ, Han BX. J Colloid Interf Sci. 2008;327(1):157–161. doi: 10.1016/j.jcis.2008.07.046. [DOI] [PubMed] [Google Scholar]
- 29.Kabanov A, Zhu JA, Alakhov V. Adv Genet. 2005;53:231–261. [PubMed] [Google Scholar]
- 30.Huang SJ, Hsu ZR, Wang LF. Rsc Adv. 2014;4(60):31552–31563. [Google Scholar]
- 31.Kulkarni A, Badwaik V, DeFrees K, Schuldt RA, Gunasekera DS, Powers C, Vlahu A, VerHeul R, Thompson DH. Biomacromolecules. 2014;15(1):12–9. doi: 10.1021/bm401096v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv HT, Zhang SB, Wang B, Cui SH, Yan J. J Control Release. 2006;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Hunter AC. Adv Drug Deliver Rev. 2006;58(14):1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


