Abstract
Apolipoprotein (apo) E4 is the major genetic risk factor for Alzheimer's disease and is associated with poor clinical outcome following traumatic brain injury and other neuropathological disorders. Protein instability and an isoform-specific apoE property called domain interaction are responsible for these neuropathological effects. ApoE4 is the most neurotoxic isoform and can induce neuropathology through various cellular pathways. Neuronal damage or stress induces apoE synthesis as part of the repair response; however, when apoE4 is expressed in neurons, its unique conformation makes it susceptible to proteolysis, resulting in the generation of neurotoxic fragments. These fragments cause pathological mitochondrial dysfunction and cytoskeletal alterations. Here, we review data supporting the hypothesis that apoE4 (> apoE3 > apoE2) has direct neurotoxic effects and highlight studies showing that blocking domain interaction reverses these detrimental effects.
Introduction
Neuronal Injury Induces ApoE Synthesis: A “Hair-Trigger” Mechanism Allows for Rapid Production
Apolipoprotein (apo) E was originally described in the early 1970s as a protein constituent of cholesterol- and triglyceride-rich plasma lipoproteins synthesized by the liver. Its expression is induced by cholesterol-rich diets in a large variety of animals and is enriched in lipoproteins in humans with the genetic disorder type III hyperlipoproteinemia (Mahley, 1988; Mahley and Rall, 2000; Mahley et al., 2009). ApoE circulates in the blood as a protein component of very low density lipoproteins, chylomicron remnants, and a subclass of high density lipoproteins, as well as in the cerebrospinal fluid and central nervous system interstitial fluid on small particles and disks resembling high density lipoproteins. ApoE is responsible for the transport of cholesterol and other lipids, as well as for mediating the clearance of plasma lipoproteins by serving as a critical ligand for lipoprotein uptake by the low density lipoprotein (LDL) receptor and LDL receptor–related protein family members. Furthermore, apoE participates in the redistribution of lipids to cells that require cholesterol and phospholipids for reparative processes throughout the body, including the central nervous system.
Human apoE is a polymorphic protein arising from three alleles at a single gene locus on chromosome 19 (Mahley, 1988; Mahley and Rall, 2000; Mahley et al., 2009). The three major isoforms—apoE2, apoE3, and apoE4—differ from one another by single amino acid interchanges at just two residues; however, these minor changes have profound effects on the structure and function of apoE at both the molecular and cellular levels and, as a consequence, on their association with specific diseases, including Alzheimer's disease (AD).
The pioneering work of Roses and associates during the early 1990s established, through a genetic linkage study, the very strong association between apoE4 and AD (Corder et al., 1993; Saunders et al., 1993; Strittmatter et al., 1993). Expression of the apoE4 allele significantly increases the risk of developing AD during one's lifetime (by 4- to 12-fold compared with apoE3/3 individuals) and decreases the age of onset (by approximately 8 years to 15 years in apoE4 heterozygotes and homozygotes, respectively). It is now established that apoE4 is a major AD gene with semidominant inheritance in apoE4 homozygotes, equivalent to the BRAC1 gene for breast cancer (Genin et al., 2011), making it the strongest genetic risk factor for AD by far (Farrer et al., 1997). Although the data are not as strong as with AD, apoE4 has also been associated with progression or poor clinical outcomes in traumatic brain injury (TBI) (Chamelian et al., 2004; Crawford et al., 2002; Friedman et al., 1999; Gandy and DeKosky, 2012; Mayeux et al., 1995; Nicoll et al., 1996; Teasdale et al., 1997), multiple sclerosis (Chapman et al., 2001; Fazekas et al., 2001), stroke (Alberts et al., 1995; McCarron et al., 1999; Slooter et al., 1997), frontotemporal dementia (Agosta et al., 2009), and Parkinson's disease (Harhangi et al., 2000; Li et al., 2004; Martinez et al., 2005; Parsian et al., 2002). Furthermore, apoE4 is not rare—approximately 25% of all individuals are carriers of this allele—making the potential detrimental effects of apoE4 expression all too common. Indeed, the apoE4 allele is heavily enriched in AD patients, with 65–80% of all AD patients carrying at least one copy (Farrer et al., 1997).
The neuropathological effects of apoE4—the least stable of the three isoforms and the most tightly associated with AD—are myriad and include (for review, see refs. Huang, 2010; Kim et al., 2009; Mahley et al., 2006): 1) impaired neurite outgrowth; 2) cytoskeletal disruption and hyperphosphorylation of tau; 3) mitochondrial dysfunction in neurons, including altered membrane potential, reduced mitochondrial motility, and decreased mitochondrial respiratory enzyme levels and activity; 4) impaired synaptogenesis; 5) increased amyloid β (Aβ) production; 6) increased lysosomal leakage and apoptosis in neurons; 7) brain neuropathology and impaired learning and memory in mice; and 8) altered Aβ peptide clearance and/or deposition.
The premise of this review is that the structural differences among the apoE isoforms determine their roles in the onset and progression of AD and other neurodegenerative diseases, and that modulation of the abnormal structure of apoE4—by converting it to a more apoE3-like (or apoE2-like) structure—will reverse the apoE4-associated detrimental effects in the central nervous system (Mahley and Huang, 2012). First, however, we discuss how apoE may indirectly impact neuropathology in AD through modulation of Aβ metabolism, before moving on to present the apoE hypothesis more fully and the most recent evidence describing the direct effects of apoE (apoE4 > apoE3 > apoE2) in the pathogenesis of neurodegenerative disorders.
ApoE Modulation of the Aβ Pathway
The amyloid hypothesis focuses on the effects of the Aβ peptide and its different assemblies in causing neuropathology, disrupting synaptic connections and forming plaques (Hardy, 2006; Palop et al., 2006; Palop and Mucke, 2010; Selkoe, 2011). Importantly, it is established that there are apoE isoform–specific effects on the Aβ pathway (Huang and Mucke, 2012; Kim et al., 2009; Selkoe, 2011) and that apoE4 expression is associated with a significant increase in amyloid plaques at earlier ages compared with apoE3 or apoE2. Furthermore, apoE4 is known to impair Aβ clearance (Bien-Ly et al., 2011; Castellano et al., 2011; Deane et al., 2008; Kim et al., 2011) and accelerate amyloid synthesis (Ye et al., 2005), as well as amyloid fibril formation and deposition (Bales et al., 1999; Bien-Ly et al., 2011; Sanan et al., 1994; Wisniewski et al., 1995).
ApoE protein levels in the cerebrospinal fluid and brain have been correlated with Aβ levels and related to apoE isoform–specific effects (Beffert et al., 1999). For example, comparison of human apoE3 and apoE4 knock-in mice demonstrated that apoE4 levels were 30–40% lower than apoE3 levels in the cortex, hippocampus, and cerebellum (Ramaswamy et al., 2005). One explanation for this reduction in apoE4 expression was revealed by Zhong et al. (2009), who demonstrated that apoE4 domain interaction activates the endoplasmic reticulum (ER) stress response in astrocytes, which results in the degradation of apoE4. Could this suggest that increasing apoE levels is protective?
Consistent with the postulate that increasing apoE levels could be beneficial, Cramer et al. (2012) demonstrated that induction of mouse apoE expression in an AD mouse model using the RXR agonist bexarotene led to a short-term reduction in soluble Aβ and plaque loads (i.e., within 72 h of initiating treatment). However, after 3 months of oral treatment they observed no change in amyloid burden. While these are potentially important observations, a number of questions remain. First, can one equate the effect of increasing mouse apoE to that of the human apoE isoforms? Mouse apoE is neither structurally nor functionally equivalent to human apoE3 or apoE4 (Zhong and Weisgraber, 2009) and behaves differently from the human isoforms with respect to Aβ clearance (Bien-Ly et al., 2011). Importantly, it was recently reported that genetically increasing either human apoE3 or apoE4 levels increased Aβ accumulation (Bien-Ly et al., 2012; Kim et al., 2011). Second, would it be beneficial to increase apoE4 levels in the brains of patients? As discussed later, numerous studies demonstrate that apoE4 has detrimental effects in the central nervous system (CNS). Finally, bexarotene is known to regulate numerous genes related to lipid metabolism, thus further complicating the interpretation of the data.
Others have emphasized the protective role for apoE3 in the context of amyloid metabolism, postulating that apoE4 lacks the beneficial effects of apoE3. Clearly apoE3 does possess beneficial effects (Kim et al., 2009; Mahley et al., 2006). For example, apoE3 is more effective than apoE4 in mediating Aβ clearance from mouse brains (Kim et al., 2009). It also has been demonstrated that apoE3 suppresses inflammation better than apoE4 (Lynch et al., 2003). In contrast, apoE4 has been shown to stimulate pro-inflammatory cytokines and exacerbate inflammation to a greater extent than apoE3 (Guo et al., 2004). Interestingly, apoE mimetics, which are small peptides corresponding to the apoE receptor–binding region, appear to mimic the anti-inflammatory activity of apoE3 and have been shown to improve cognitive performance and neuronal survival in TBI mouse models (Vitek et al., 2012). However, the mechanism by which these peptides work remains to be defined.
It is now recognized that there are multiple factors acting through various pathways to cause cognitive decline and neurodegeneration in AD (Huang, 2010; Huang and Mucke, 2012; Kim et al., 2009; Mahley et al., 2006). ApoE clearly interacts with the Aβ pathway; however, there is abundant evidence showing that apoE (apoE4 > apoE3 > apoE2) also can directly impact the pathogenesis of AD and other neurodegenerative disorders independent of Aβ. As clinical trials targeting the lowering of amyloid and the Aβ peptide fail to impact AD cognitive decline and neurodegeneration (Selkoe, 2011), alternative mechanisms must be considered, and the critical importance of apoE in pathogenesis further acknowledged.
Direct Effects of ApoE in Causing Neuropathology
Under normal physiological conditions, apoE is synthesized primarily by astrocytes in the brain to support lipid transport and membrane repair processes (Mahley, 1988). In contrast, apoE synthesis in neurons usually occurs in response to neuronal insult or injury. Each of these processes is designed to protect neuronal integrity and promote neuronal repair, respectively, but in fact can be directly injurious when apoE4 is expressed in neurons. Indeed, apoE4 expression is associated with poor clinical outcome or accelerated/more severe progression in numerous neurological disorders. This finding strongly suggests that while apoE participates in general cellular pathway(s) designed to respond adaptively to various environmental, metabolic, or genetic stimuli, its expression can also increase the likelihood of neurodegeneration. Several pathways for this have been suggested.
ApoE is known to exhibit isoform-specific effects on blood–brain barrier (BBB) integrity in mouse models (Bell et al., 2012). In either target replacement mice or glial fibrillary acidic protein promoter transgenic mice, apoE4 expression increases the BBB's susceptibility to injury by activating the proinflammatory cytokine cyclophilin A in pericytes and triggering the NF-κB/matrix metalloproteinase 9 pathway. Interestingly, BBB breakdown is independent of Aβ. The subsequent neuronal damage that occurs appears to result from the leakage of blood-derived proteins—including immunoglobulin G, thrombin, and fibrin—into the brain. The extravasation of blood proteins has been implicated in the activation of numerous neurotoxic pathways. For example, in a mouse model of multiple sclerosis, fibrinogen leakage and fibrin deposition in the brain activates microglia and leads to neuropathology (Adams et al., 2007; Akassoglou et al., 2004; Davalos and Akassoglou, 2012). As will be discussed, BBB leakage could represent an additional insult linked to neuronal injury and the induction of neuronal apoE synthesis and neurotoxicity.
ApoE is also involved in maintaining and regulating synaptic activity and strength in knock-in mice and hippocampal slices. Specifically, apoE4 reduces neuronal cell-surface expression of the apoE receptor 2, as well as the N-methyl-d-aspartate and AMPA receptors, by sequestering them in an intracellular compartment (Chen et al., 2010). It is postulated that the apoE isoform–specific effect on lipoprotein and glutamate receptor trafficking contributes to AD neuropathology by impairing synaptic activity. In addition, other LDL receptor family members have been implicated in AD, owing to their roles in modulating the intracellular trafficking and processing of the amyloid precursor protein (Cam and Bu, 2006).
The effects of astrocyte-derived apoE in the brain are a point of ongoing study, and it remains to be described whether astrocyte-derived apoE impacts neuronal health and pathology differently from the apoE that is synthesized within neurons. The remainder of this review will describe how induction of neuronal apoE (apoE4 > apoE3 > apoE2) in response to injury sets the stage for neuropathology and subsequent neurodegeneration.
The ApoE Hypothesis: Relationship between ApoE Structure, Function, and Expression in Health and Disease
The apoE hypothesis posits that apoE genotype sets the stage for neuropathology in an isoform-dependent manner (apoE4 > apoE3 > apoE2), and “second hits” that directly induce neuronal injury or stress initiate a pathological response to injury when apoE4 is synthesized in neurons (Huang, 2010; Huang and Mucke, 2012; Mahley and Huang, 2012; Mahley et al., 2006). With respect to AD, these second hits could include aging, ischemia, trauma, inflammation, oxidative stress, or toxins like the Aβ peptide and its different assemblies. TBI causes direct damage to neurons, whereas following stroke the second hit may be ischemia. Given the nature of many neurological diseases, where multiple pathologies occur over a protracted period, the possibility for second hits is very high. Other genetic disorders and metabolic disturbances, such as diabetes, can also be injurious factors that contribute to apoE4's neurotoxic effects.
In response to injury, neurons induce the synthesis of apoE, presumably to participate in lipid transport and redistribution for membrane repair and remodeling. However, because of varying degrees of structural instability and tendency to assume domain interaction across the apoE isoforms (which we discuss in greater detail below), apoE can be recognized as structurally abnormal by neurons and undergo proteolytic cleavage (apoE4 > apoE3 > apoE2). The neurotoxic fragments that are generated cause mitochondrial dysfunction and cytoskeletal alterations. In the sections to follow, we describe in more detail the data supporting the apoE hypothesis (Figure 1).
Figure 1. Injurious Agents That Stress or Damage Neurons Induce ApoE Synthesis and Initiate Neuropathology.
ApoE4, because of domain interaction, displays impaired trafficking through the ER and Golgi apparatus compared with apoE3. As a result, it is targeted to a neuron-specific protease that initially cleaves off the C-terminal 27–30 amino acids, generating neurotoxic fragments. These fragments escape the secretory pathway and enter the cytosol, where they cause mitochondrial dysfunction and cytoskeletal alterations, enhance tau phosphorylation, and form NFT-like structures (Mahley et al., 2006). Figure reprinted with permission from Mahley and Huang, Small-molecule structure correctors target abnormal protein structure and function: The structure corrector rescue of apolipoprotein E4–associated neuropathology. J. Med. Chem. 55: 8997–9008, 2012. Copyright 2012 American Chemical Society.
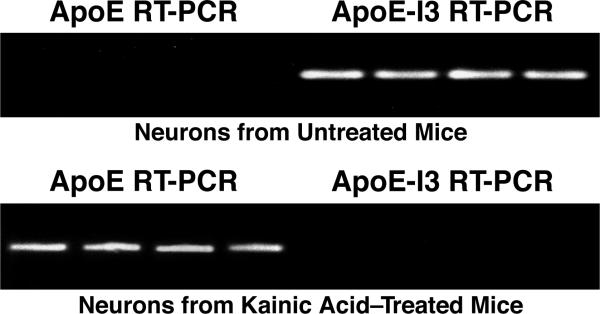
As mentioned previously, in the brain apoE is primarily synthesized by astrocytes under normal physiological conditions (Mahley, 1988) and the neuropathological effects of astrocyte-derived apoE are a point of ongoing study. However, apoE can also be produced by neurons under pathological conditions resulting from neuronal cell injury or stress (Huang, 2010; Huang and Mucke, 2012; Mahley et al., 2006). Xu et al. (2006) established an enhanced green fluorescent protein (EGFP)apoE–reporter mouse model in which EGFP was inserted into one allele of the apoE gene to serve as a reporter of apoE expression. The hippocampus of a normal, uninjured mouse brain showed abundant EGFP expression in astrocytes, but little expression of EGFP in neurons, indicating no or very low neuronal apoE production (Figure 2). However, following kainic acid–induced excitotoxic injury, apoE production was markedly increased in damaged hippocampal neurons, as measured using the EGFP reporter as the readout (Figure 2). In addition, both apoE mRNA and protein were expressed in hippocampal neurons following kainic acid treatment, as shown by in situ hybridization and immunochemistry, respectively (Xu et al., 2006).
Figure 2. EGFP Inserted into One Allele of the ApoE Gene Acts as a Marker for ApoE Expression in Vivo.

Top, schematic of the EGFP-apoE reporter cassette. Bottom, representative images of hippocampal sections taken from EGFPapoE-reporter knockin mice. In the uninjured mouse hippocampus (before kainic acid), astrocytes display abundant apoE synthesis (bottom, left); however, after injury with kainic acid, neurons turn on the synthesis of apoE, as indicated by the colocalization of EGFP (green) and NeuN (red) (bottom, right) (Xu et al., 2006). Modified from Figure 6, Xu, Q., Bernardo, A., Walker, D., Kanegawa, T., Mahley, R.W., and Huang, Y. Profile and regulation of apolipoprotein E (apoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the apoE locus. J. Neurosci. 26: 4985–4994, 2006.
Further studies revealed that the mechanism controlling the synthesis of apoE in neurons is unique and poised for rapid protein production (Xu et al., 2008). These studies found that while little apoE protein is seen in uninjured neurons, the apoE gene is still transcribed; however, in these uninjured neurons intron 3 is typically left intact in the transcribed mRNA sequence, leading to its retention and degradation in the nucleus. However, following kainic acid–induced injury, intron 3 was spliced out, resulting in mature apoE mRNA being transferred out of the nucleus for apoE protein production in neurons. In situ hybridization studies in uninjured mouse brains revealed that hippocampal neurons almost exclusively expressed intron 3–containing apoE mRNA, while hippocampal astrocytes expressed the intron 3–lacking apoE transcript in abundance (Xu et al., 2008). Laser-capture microdissection studies in the hippocampus of uninjured mice also revealed the presence of intron 3–containing apoE mRNA; however, after injury there was a dramatic switch in expression to intron 3–lacking, mature apoE mRNA (Figure 3). This phenomenon is unique to neurons, as apoE intron retention has not been observed in other apoE-synthesizing cell types. In addition, astrocyte-conditioned medium can trigger the synthesis of apoE in neurons, revealing an important “cross-talk” between neurons and glia that is likely to operate during an injury response (Harris et al., 2004b). Thus, neurons possess a unique mechanism whereby they are primed for the rapid production of apoE. The splicing and nuclear export pathways that regulate mRNA and protein production operate ubiquitously in eukaryotic cells and are modulated, in part, through stress; however, these pathways remain to be fully understood (Cullen, 2000; Fox and Lamond, 2010; Galy et al., 2004; Prasanth et al., 2005).
Figure 3. Kainic Acid–Induced Injury Triggers Switch from Intron 3–Containing to Mature ApoE mRNA Synthesis.
EGFPapoE-reporter knockin mice were treated or not with kainic acid, and CA1 hippocampal neurons were isolated by laser-capture microdissection for reverse transcription–polymerase chain reaction (RT-PCR). Neurons from a normal, uninjured mouse demonstrate the presence of apoE mRNA with intron 3 (apoE-I3), whereas after injury there is a dramatic switch to mature apoE mRNA lacking intron 3 (Xu et al., 2008). Modified from Figure 7, Xu, Q., Walker, D., Bernardo, A., Brodbeck, J., Balestra, M.E., and Huang, Y. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J. Neurosci. 28: 1452–1459, 2008.
Why might injured neurons turn on the synthesis of apoE and appear to be primed for apoE secretion? Lipid metabolism is unique in the brain for two reasons. First, apoE is the only apolipoprotein present in the brain that binds to the LDL receptor or members of the LDL receptor family, which are responsible for delivering cholesterol and other complex lipids to central nervous system cells through receptor-mediated endocytosis (Bu, 2009; Herz and Bock, 2002; Mahley and Rall, 2000; Mahley et al., 2009). Second, complex lipids are almost totally synthesized de novo by the various brain cells and very little of these lipids, including cholesterol, are delivered from the periphery by exchange or uptake of plasma lipoproteins across the BBB (Dietschy and Turley, 2001).
The redistribution of complex lipids for membrane repair and other metabolic roles undoubtedly relies on apoE through a process termed secretion-capture (Ji et al., 1994; Mahley and Ji, 1999; Mahley et al., 2009), in which secreted apoE scavenges lipids from the local environment and targets them to cells requiring lipids for normal metabolism or membrane repair. The secretion-capture role for apoE was first demonstrated in peripheral nerve injury and regeneration (Boyles et al., 1989; Ignatius et al., 1987; Mahley, 1988) and later in the CNS following hippocampal injury (Poirier et al., 1991). When the sciatic nerve was injured, macrophages responding to the injury rapidly began secreting very large quantities of apoE (200-fold over the level seen in the uninjured nerve) and “capturing” the lipids in the local environment of the injured nerve. ApoE–lipid complexes were shown to be delivered to the growth cones of the regenerating nerves and to Schwann cells for myelin formation through lipoprotein receptor uptake. The secretion-capture process has been further established in the liver, where apoE captures lipoproteins and targets them for receptor-mediated uptake. In fact, apoE secreted by hepatocytes and macrophages has been shown to bind to cell-surface heparan sulfate proteoglycans where it is available to capture lipids and lipoproteins; the heparan sulfate proteoglycans themselves acting as a receptor or part of a receptor complex (Ji et al., 1994; Mahley and Ji, 1999). Thus, apoE secreted by injured neurons may be serving this critical role in lipid redistribution in the repair process. Alternatively, or in addition, apoE may have a role in cell signaling, as has also been suggested (Hayashi et al., 2007; Herz and Bock, 2002).
Although apoE may play an important role in repairing damaged neuronal membranes, it is also associated with neurodegeneration. This assertion is supported by a vast array of structural, molecular, cellular, and behavioral data showing that the three isoforms of apoE display key variations in their protein structure and stability that, in turn, differentially impact neuropathology.
Structural Differences among the Isoforms Set the Stage for Neuropathology
The single amino acid interchanges that distinguish the apoE isoforms result in differences in protein stability as well as the propensity to display a unique structural property called domain interaction (Dong et al., 1994; Huang, 2010; Mahley et al., 2006; Zhong and Weisgraber, 2009). ApoE2 has a cysteine residue at position 158 whereas apoE3 and apoE4 each have arginine. While this substitution in apoE2 results in defective lipoprotein-receptor binding and the development of the lipid disorder type III hyperlipoproteinemia (Mahley, 1988; Mahley et al., 1999; Mahley and Rall, 2000), it also confers greater protein stability, which likely is associated with its protective effect against Alzheimer's disease (AD).
ApoE4, which is strongly linked to AD (Roses, 1996), uniquely possesses an arginine at residue 112, whereas both apoE2 and apoE3 have cysteine at this site. This single amino acid difference in apoE4 is associated with protein instability and domain interaction (Dong et al., 1994; Zhong and Weisgraber, 2009). ApoE3, the most common isoform in humans, has a cysteine residue at position 112 and an arginine at position 158, is more stable than apoE4, and is less likely to display domain interaction than apoE4.
As illustrated in Figure 4, in apoE4 the arginine at residue 112 causes the side chain of arginine-61 to be solvent-exposed, to extend away from the helical bundle in the N-terminal domain, and to interact with glutamic acid–255 in the C-terminal domain through ionic bonding. Although all isoforms may display domain interaction to some extent, the amino acid substitutions of apoE4 encourage domain interaction much more than the apoE3 and apoE2 isoforms (apoE4 > apoE3 > apoE2). Importantly, this apoE4 property has been established biophysically by fluorescence resonance energy transfer (FRET) and electron paramagnetic resonance spectroscopy (Dong et al., 1994; Hatters et al., 2005; Xu et al., 2004; Zhong and Weisgraber, 2009). The distance between arginine-61 and glutamic acid–255 was ~10 Å in apoE4 and greater than 22 Å in apoE3 (Hatters et al., 2005).
Figure 4. Models of the Structures of ApoE3 and ApoE4.

(A) ApoE4 displays a unique property called domain interaction caused by the ionic interaction between arginine-61 in the N-terminal domain with glutamic acid–255 in the C-terminal domain. ApoE3 is significantly less likely to undergo domain interaction than apoE4.
(B) ApoE4 domain interaction can be blocked by a small-molecule apoE4 structure corrector that disrupts the ionic interaction between arginine-61 and glutamic acid–255. This converts apoE4 to an apoE3-like molecule both structurally and functionally (Huang, 2010; Mahley and Rall, 2000; Mahley et al., 2006, 2009). ApoE4SC, apoE4 structure corrector. Figure reprinted with permission from Mahley and Huang, Small-molecule structure correctors target abnormal protein structure and function: The structure corrector rescue of apolipoprotein E4–associated neuropathology. J. Med. Chem. 55: 8997–9008, 2012. Copyright 2012 American Chemical Society.
Recently, the nuclear magnetic resonance structure for full-length apoE3 was determined (Chen et al., 2011b). The structure reveals a unique topology, which is postulated to shield the LDL receptor–binding region by the C-terminal domain and prevent binding to the receptor in the non-lipid form. However, in order to prevent tetramer formation and aggregation, it was necessary to introduce five non-conservative mutations into the C-terminal domain (F257A/W264R/V269A/L279Q/V287E). Unfortunately, these rather severe changes undoubtedly alter the C-terminal domain and likely distort the conformation and intramolecular interactions with this domain. Thus, the nuclear magnetic resonance structure may not represent a physiological and functional structure of apoE.
In all, these data show how relatively minor changes in apoE sequence yield significant differences in apoE's ability to promote neuronal health versus neuronal damage, especially when apoE synthesis is increased after injury. The apoE hypothesis therefore suggests that the isoform-dependent neuronal response to increased apoE expression is a critical step in laying the groundwork for future pathology. In the following sections, we describe the apoE4-specific effects on neuronal function and how they affect neuronal integrity at the cellular level.
Impaired Neuronal Intracellular Trafficking of ApoE (ApoE4 > ApoE3)
Fluorescence recovery after photobleaching experiments revealed impaired intracellular trafficking of apoE4 through the ER and Golgi apparatus in cultured cells (Brodbeck et al., 2011). Neuro-2a cells expressing EGFP-apoE3 or EGFP-apoE4 were photobleached with an argon laser, and the extent and rate of recovery of fluorescence were used as a measure of transit through the secretory pathway. Although the trafficking of apoE4 through the ER and Golgi apparatus was significantly impaired compared with apoE3 (Figure 5A), blocking domain interaction by site-directed mutagenesis (i.e., mutation of arginine-61 to threonine) or by exposure to small-molecule structure correctors restored normal trafficking properties to apoE4 (Figure 5B and 5C) and led to decreased neurotoxic fragment formation. These domain interaction–blocking approaches will be discussed in more detail below. Thus, it is envisioned that 1) the impaired transit of apoE4 occurs because of its abnormal structure, because blocking domain interaction restores the transit, 2) the abnormal structure and trafficking likely target the protein for proteolysis, and 3) small-molecule structure correctors likely target apoE as it is synthesized or soon after entering the ER lumen. Such findings suggest that one way to resolve the negative effects of apoE4 expression is to convert apoE4's structure to be more apoE3-like.
Figure 5. ApoE4 Displays Impaired Trafficking through the Secretory Pathway That Can Be Corrected by Blocking ApoE4 Domain Interaction.
Fluorescence recovery after laser photobleaching experiments were performed on EGFP-apoE3- and EGFP-apoE4-expressing Neuro-2a cells. For each section, the left and center panels show representative recovery curves for the ER and Golgi apparatus, respectively, while the right-hand panel shows quantitative histograms of apoE mobility.
(A) Recovery curves show that apoE4 trafficking is decreased compared with apoE3 in both the ER (left) and Golgi apparatus (center). Right, histogram showing that the percent of the immobile fraction was greater with apoE4 than apoE3.
(B) The impaired trafficking of apoE4 was reversed by site-directed mutagenesis of arginine-61, which blocked domain interaction (apoE4-R61T). This mutation reversed the impairment in apoE4 trafficking and mobility in both the ER and Golgi apparatus.
(C) Treatment of apoE4-expressing cells with a structure corrector (PH-002) restored the trafficking of apoE4 through the secretory pathway. The structure corrector did not affect the trafficking of apoE3 or apoE4-R61T (Brodbeck et al., 2011). Modified from Figures 3, 4, 5 originally published in Brodbeck, J., McGuire, J., Liu, Z., Meyer-Franke, A., Balestra, M.E., Jeong, D.-e., Pleiss, M., McComas, C., Hess, F., Witter, D., Peterson, S., Childers, M., Goulet, M., Liverton, N., Hargreaves, R., Freedman, S., Weisgraber, K., Mahley, R.W., Huang, Y. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J. Biol. Chem. 2011; 286:17217–17226. © the American Society for Biochemistry and Molecular Biology.
ApoE (ApoE4 > ApoE3) Susceptibility to Neuron-Specific Proteolysis and the Generation of Neurotoxic Fragments
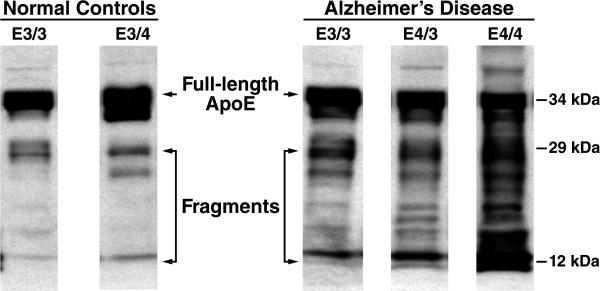
The cellular mechanisms and organelles that promote the clearance of abnormally folded proteins are ubiquitous, and abnormal forms of apoE, especially apoE4, can indeed be targeted for proteolysis. In fact, neurotoxic fragments are generated only by neurons, and not by astrocytes or other apoE-synthesizing cells (Brecht et al., 2004; Harris et al., 2003; Huang et al., 2001). Why, then, are neurons less effective than other cell types at completely degrading and clearing misfolded apoE? It is possible that, because apoE is an avid lipid-binding protein, lipid-based interactions may protect some domains from proteolytic cleavage, thus resulting in the accumulation of a spectrum of neurotoxic fragments. While full-length apoE is 34 kDa, a fragment pattern of bands ranging from 29–30 kDa to 12–14 kDa is consistently seen in extracts from cultured neurons expressing apoE4, apoE4 transgenic mice, and in the brains and cerebrospinal fluid from humans with AD (Brecht et al., 2004; Harris et al., 2003; Huang et al., 2001; Jones et al., 2011). Furthermore, more of these fragments are observed in AD patients expressing the apoE4 allele compared with normal, non-demented apoE4-carrying humans (Figure 6) (Harris et al., 2003; Jones et al., 2011).
Figure 6. ApoE Fragmentation Pattern in Human Temporal Cortex in Non-Demented Controls and in Age-Matched AD Patients.
Compared with full-length apoE (34 kDa), proteolytic cleavage generates an initial fragment with a molecular weight of ~29 kDa. Subsequent to this, fragments of ~12–20 kDa are generated. In AD patients, there is an apoE4 gene–dose effect on apoE fragmentation, whereby apoE4/3 subjects have more fragments than apoE3/3 subjects and the apoE4/4 subjects have the greatest amount of fragments.
Although the unique protease that is responsible for apoE4 fragmentation remains to be identified, it is thought to be a chymotrypsin-like serine protease (Harris et al., 2003). This protease, most likely residing in the ER or Golgi apparatus, generates the unique series of fragments ranging from 29–30 kDa to 12 kDa (Huang, 2010; Huang and Mucke, 2012; Mahley et al., 2006). The 29–30-kDa fragments result from cleavage at methionine-272 and leucine-268, respectively, and subsequent cleavage results in the generation of smaller fragments, primarily in the 12–20 kDa range. All of the neurotoxic fragments lack the C-terminal 27–30 amino acids and contain the lipid-binding region of apoE (residues 240–270) (Brecht et al., 2004; Chang et al., 2005; Harris et al., 2003; Huang et al., 2001; Jones et al., 2011). In addition to containing the lipid-binding region, the neurotoxic fragments also contain the LDL receptor–binding region of apoE (residues 136–150). The secondary cleavage events remove varying lengths of peptide from the N terminus. As mentioned above, these fragments are generated in the ER or Golgi apparatus, and yet many of their effects are seen in the cytosol. The cleavage of the C terminus allows this translocation and several of the subsequent cytosolic effects.
ApoE Fragments Escape the Secretory Pathway and Enter the Cytosol
How do the apoE4 fragments generated by neuron-specific proteolysis leave the ER or Golgi compartments and enter the cytosol? Cleaving off the C-terminal 27–30 amino acids exposes specific regions of apoE that are not accessible in the intact protein. This allows for apoE4 translocation into the cytosol, thereby facilitating mitochondrial localization and causing neurotoxicity (Chang et al., 2005). However, deletion of the lipid-binding region (residues 240–270) in a fragment encompassing residues 1–191 did not inhibit translocation into the cytosol, but this fragment also did not interact with mitochondria or cause neurotoxicity. Finally, removal of the portion of apoE that includes the LDL receptor–binding region (residues 136–150) prevented translocation (Figure 7), as did mutations of critical arginine and lysine residues in this region (Chang et al., 2005).
Figure 7. Regions of ApoE Responsible for Fragment Translocation into the Cytosol, Mitochondrial Targeting, and Neurotoxicity.
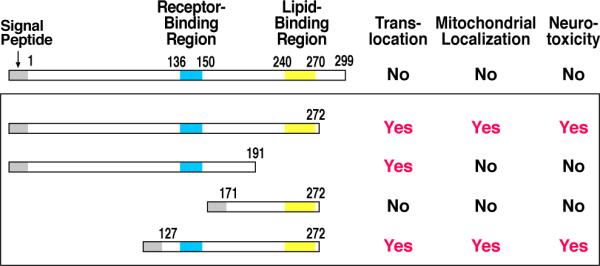
Fragments containing the receptor-binding region (residues 136–150) and the lipid-binding region (residues 240–270) represent the minimal structure required for translocation, mitochondrial localization, and neurotoxicity (Mahley et al., 2006). Modified from Figure 4, Mahley, R.W., Weisgraber, K.H., and Huang, Y. Apolipoprotein E4: A causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc. Natl. Acad. Sci. USA 103: 5644–5651, 2006. © by the National Academy of Sciences.
These studies show that a minimal structure supporting translocation, mitochondrial localization, and neurotoxicity requires the presence of both the receptor- and lipid-binding regions of apoE (Chang et al., 2005). The charged arginine and lysine residues in the 136–150 region are critical for translocation, a region that is similar to the protein-translocation domains of other proteins, including viral proteins. The hydrophobicity of the lipid-binding region (residues 240–270) is certainly involved in mitochondrial interaction and subsequent neurotoxicity, because mutation of critical conserved residues in this region, or deletion of this region altogether, blocked mitochondrial localization. Importantly, these truncation variants generated in the laboratory are likely counterparts to the spectrum of toxic fragments observed in the brain (Figure 6) and cerebrospinal fluid of human AD patients, making the results highly relevant to our understanding of human AD pathology.
ApoE4-Associated Neuronal Mitochondrial Dysfunction
Mitochondrial dysfunction is a hallmark of several neurodegenerative diseases, including AD (Atamna and Frey II, 2007; Parihar and Brewer, 2007). Functional brain imaging with fluorodeoxy-glucose (18F), a radiotracer used to study glucose utilization as a surrogate measure of mitochondrial activity, revealed that the cerebral metabolic rate was reduced in both AD patients and in cognitively normal apoE4 carriers in their 20s and 30s, several decades before the typical onset of dementia (Reiman, 2007; Reiman et al., 2004). Gene-expression profiles in postmortem human brain tissue showed a significant association between apoE genotype and the expression levels of multiple mitochondrial respiratory enzymes (Conejero-Goldberg et al., 2011), specifically, a reduction in the expression of electron transport–chain genes in apoE4 carriers (Liang et al., 2008). These results suggest that in humans, apoE genotype can influence brain metabolism and possibly mitochondrial function, and that alterations in these processes may be linked to the onset and/or progression of AD.
We have shown that primary neurons from apoE4 transgenic mice had reduced levels of numerous mitochondrial respiratory enzymes compared with apoE3-expressing cells (Chen et al., 2011a). However, there were no differences in the mitochondrial complexes in astrocytes expressing apoE4 driven by the glial fibrillary acidic protein promoter, revealing that apoE4's effects are neuron-specific. In addition, apoE4 expression in Neuro-2a cells led to reduced levels of mitochondrial complexes I, IV, and V, as well as a reduction in functional respiratory capacity (Chen et al., 2011a). The addition of an uncoupling agent led to a 98% increase in the maximal oxygen-consumption rate in apoE3-expressing cells compared with only a 50% increase in the apoE4-expressing cells (Chen et al., 2011a).
In cells transfected with the apoE4 variant lacking the C-terminal 27 amino acids, the fragment was found to be expressed in a granular distribution that localized preferentially to mitochondria (Chang et al., 2005); this led to mitochondrial dysfunction, as demonstrated by loss of membrane integrity and electropotential. Again, the active fragments that altered mitochondrial membrane electropotential contained the receptor- and lipid-binding regions. Nakamura et al. (2009) demonstrated that apoE4 lacking the C-terminal 27 amino acids bound directly to several components of the electron transport–chain enzymes and reduced respiratory activity. Likewise, apoE4, and especially the apoE4 fragments, have been shown to impair mitochondrial dynamics and synaptogenesis (Brodbeck et al., 2011; Chen et al., 2012). For example, mitochondrial motility was reduced by 35% and 57% by apoE4 and apoE4 fragments lacking the C-terminal 27 amino acids, respectively, in transfected PC12 cells. Furthermore, dendritic spine densities were reduced in apoE4-expressing primary neurons by 26% and in apoE4 fragment–expressing neurons by 46%, compared with apoE3 (Brodbeck et al., 2008). These abnormalities, which could be related to mitochondrial dysfunction, were domain-interaction dependent, as these effects could be reversed by either blocking domain interaction by site-directed mutagenesis (apoE4-R61T) or by treatment with small-molecule structure correctors.
How and where the apoE fragments interact with mitochondria remains to be determined. As discussed, the mitochondrial binding of apoE fragments requires the absence of the C terminus and the presence of the lipid-binding region. The unique lipid composition of mitochondria could allow the fragments to interact directly with the mitochondrial surface through a hydrophobic lipid interaction. On the other hand, there are numerous mitochondrial outer membrane proteins, the function of which could be altered by an interaction with apoE fragments. For example, apoE fragment interaction with the voltage-dependent anion channel (also known as mitochondrial porin), which controls the entry and exit of mitochondrial metabolites, could disrupt multiple functions ascribed to this channel (Shoshan-Barmatz et al., 2010).
A link between apoE and a specific mitochondrial protein has been suggested. In AD patients, Roses (2010) demonstrated an age-of-onset-associated polymorphism in the translocase of the outer mitochondrial–membrane (TOMM40) gene, which is in the region of the apoE locus and is in strong linkage disequilibrium. Variable-length poly-T polymorphisms appear to alter the age-of-onset of AD. For example, apoE3, in the context of the longer TOMM40 poly-T repeats, is associated with an earlier age-of-onset than apoE3 individuals with shorter repeats. Such polymorphisms could modulate the apoE isoform–specific effects on AD. TOMM40 is a part of the mitochondrial machinery that controls protein translocation into the mitochondria (Kutik et al., 2007; Pfanner and Wiedemann, 2002; Rapaport, 2005). Specific pre- or internal sequences within a protein, or interactions with transfer chaperones—such as HSP90- and HSP70-class chaperones—participate in the recognition and translocation of proteins into the mitochondria. It has been postulated that apoE–TOMM40 protein interactions may alter mitochondrial function, possibly causing cytochrome c release and apoptosis (Roses, 2010). This observation has been confirmed in some studies but not in others (Cruchaga et al., 2011; Maruszak et al., 2012). In fact, a recent large study of more than 11,000 AD patients and 10,000 cognitively normal controls from 15 genome-wide association studies demonstrated that the apoE alleles (ε2, ε3, and ε4) accounted for essentially all the risk and age-of-onset of AD (Jun et al., 2012). The inherited susceptibility was not associated with neighboring genes, including TOMM40 and apoC1. These data suggest that the genetic role of TOMM40 should be reassessed; thus, the mechanism whereby apoE alters mitochondrial function remains to be determined.
ApoE-Associated Alterations in the Neuronal Cytoskeleton
The neuronal cytoskeleton is composed of microtubules, neurofilaments, and microfilaments. Microtubules, polymeric structures composed of α- and β-tubulin, are critical for neurite extension and organelle trafficking, including the distribution of mitochondria to the sites of newly forming synapses. They are associated with a heterogeneous set of microtubule-associated proteins, including tau, that modulate their structure and function. Until recently, the major function of tau was thought to be its ability to stabilize microtubules (Lee et al., 2001; Morris et al., 2011; Sydow et al., 2011). Now, it appears that its role may be more dynamic, possibly participating in intracellular signal transduction, among other roles. Importantly, tau is highly susceptible to hyperphosphorylation and the formation of intracellular NFTs, both of which are hallmark neuropathologies that critically promote the damage observed in AD and TBI (McKee et al., 2009; Morris et al., 2011).
ApoE4 Alters Neuronal Microtubules and Impairs Neurite Outgrowth
ApoE4 has been shown to directly alter microtubule structure and to stimulate tau hyperphosphorylation and NFT formation (Huang, 2010; Huang and Mucke, 2012). Exposure of neuronal cultures to exogenous apoE4-lipid complexes caused significant cytoskeletal disruption and impaired neurite outgrowth compared with apoE3-containing complexes (Bellosta et al., 1995; Nathan et al., 1994; Nathan et al., 1995). ApoE4-treated Neuro-2a cells also had fewer intracellular microtubules, as identified by immunocytochemical localization of β-tubulin and by electron microscopy (Nathan et al., 1995), and these effects were associated with impaired neurite outgrowth in apoE4-treated cells. These results were replicated in apoE3- or apoE4-transfected Neuro-2a cells expressing nanogram quantities of apoE (Bellosta et al., 1995).
ApoE4 Stimulates Tau Phosphorylation and NFT-like Formation
Increased tau phosphorylation has been observed in transgenic mice expressing apoE4 in neurons, but not in those expressing apoE4 in astrocytes, suggesting a cellular source–dependent effect of apoE4 on tau phosphorylation (Harris et al., 2003; Tesseur et al., 2000), occurring in parallel with the generation of apoE fragments in neurons (Andrews-Zwilling et al., 2010; Harris et al., 2003). There is evidence that apoE4 stimulates tau phosphorylation by activating the extracellular signal–regulated protein kinase pathway in the hippocampus (Harris et al., 2004a), although other signaling pathways may also be involved. Intraneuronal phospho-tau inclusions are prominent in the hippocampus and form NFT-like structures composed of apoE4, phosphotau, and neurofilaments; by electron microscopy, these inclusions are visualized as tightly packed, straight filaments that closely associate with mitochondria (Harris et al., 2003). In addition to such insights into the cell biological impacts of apoE fragments, transgenic animal experiments are uncovering how such neurotoxicity at the cellular level corresponds to neuronal function and behavior.
Neurotoxic Effects of ApoE Fragments
The expression of truncated apoE4 in transgenic mice provides insights into how and where the fragments cause neurotoxicity. Transgenic mice expressing a variant of apoE4 that lacks the C-terminal 27 amino acids [apoE4(1–272)], driven by the Thy1.2 promoter, had significant hippocampal neurodegeneration and neuronal loss (Harris et al., 2003). However, not every fragment of apoE is toxic, because expression of apoE4(1–240)—the form lacking the lipid-binding region—was not found to trigger hippocampal neurodegeneration. The loss of hippocampal neurons in the apoE4(1–272) mice was found to correlate with impaired learning and memory behaviors as early as 6 months of age, as assessed by the Morris water maze test (Andrews-Zwilling et al., 2010; Harris et al., 2003). Likewise, when full-length apoE4 was specifically expressed in neurons (NSE-apoE4 transgenic mice), these mice displayed early-onset impaired learning and memory (Buttini et al., 1999; Raber et al., 1998; Raber et al., 2000) that correlated with multiple neuropathological effects, including loss of synaptic connections and neurodegeneration (Buttini et al., 1999). These effects are likely mediated through apoE4 fragment generation (Brecht et al., 2004). In addition to neurodegenerative changes in the brain, the apoE4(1–272) mice display other pathologies, including neurofibrillary tangle (NFT)–structures in the hippocampus rich in hyperphosphorylated tau, which were elevated by about sixfold over levels seen in nontransgenic mice (Andrews-Zwilling et al., 2010; Harris et al., 2003).
ApoE Fragments Induce Neuropathology in Specific Neuronal Subtypes
Andrews-Zwilling et al. (2010) showed that apoE4 knock-in mice display an age-dependent decrease in GABAergic interneurons selectively in the hilus of the hippocampus, and that this decrease was associated with impaired learning and memory behaviors in these mice. Transgenic expression of an apoE4 truncation mutant lacking the C-terminal 27 amino acids led to an even more marked decrease in GABAergic interneuron levels, along with pronounced hyperphosphorylation of tau in the hippocampus (Andrews-Zwilling et al., 2010). Studies by Li et al. (2009 have also revealed a link between apoE4 expression and the impaired generation of new neurons, demonstrating that apoE4 knock-in mice have reduced hilar GABAergic interneurons, leading to impaired hippocampal neurogenesis. Based on these data, it has been postulated that the parallel decreases in interneuron levels and hippocampal neurogenesis are responsible for the behavioral impairments seen in apoE4 transgenic mice (Andrews-Zwilling et al., 2010; Huang and Mucke, 2012; Li et al., 2009). In support of this hypothesis, optogenetic manipulations of hilar GABAergic interneurons confirmed that functional inhibition of this specific neuronal population results in spatial learning and memory deficits, as seen in apoE4 knock-in mice (Andrews-Zwilling et al., 2012).
These effects on hippocampal GABAergic neurons and memory behaviors are consistent across studies, but nonetheless reveal interesting differences depending on whether apoE4 is expressed selectively in neurons versus ubiquitously in the brain. For example, apoE4 knock-in mice showed impaired learning and memory behaviors, but at a later time point (16 months of age) compared with neuron-specific (i.e., Thy1.2- or NSE-promoter-driven) apoE4 mice (Andrews-Zwilling et al., 2010). Although not directly proven, it is reasonable to speculate that the delayed impairment in the apoE4 knock-in mice reflects the age-related and comparatively delayed accumulation of neurotoxic fragments in neurons, while neuron-specific apoE-expressing mice accumulate these fragments much earlier.
Having uncovered many of the isoform-specific detrimental effects of apoE, new findings examine whether such effects can be reversed by correcting apoE4's abnormal protein structure, an approach that can potentially open new avenues for therapeutics.
Domain Interaction In Vitro and In Vivo
Interestingly, all animal species except humans have threonine at the site equivalent to residue 61 (Mahley and Rall, 2000; Weisgraber, 1994; Zhong and Weisgraber, 2009). Lower species do not display isoforms, and almost all—including mice and rats—have an arginine at the site equivalent to residue 158, making it human apoE4–like in sequence. However, because these apoE orthologs expressed in lower species lack an arginine-61 equivalent, they also lack domain interaction. Thus, they are not equivalent to either apoE3 or apoE4 structurally or functionally (Mahley and Rall, 2000; Weisgraber, 1994; Zhong and Weisgraber, 2009).
Raffaï et al. (2001) created a “humanized” apoE mouse line, in which the threonine located at the residue equivalent to 61 in human apoE was replaced by gene targeting with an arginine, thus allowing mouse apoE to have domain interaction. Importantly, these mice developed some characteristics resembling human apoE4 knock-in mice, including loss of synaptic marker immunolabeling in the hippocampus and a mild memory deficit in the Morris water maze test (Zhong et al., 2008).
We identified several small-molecule structure correctors that block apoE4 domain interaction using cellular FRET assays (Chen et al., 2012; Mahley and Huang, 2012). Additional assays were also used to establish the downstream functional effects of blocking apoE4 domain interaction with structure correctors (Brodbeck et al., 2011; Chen et al., 2011a; Chen et al., 2012).
The FRET assay to measure domain interaction relied on the creation of human apoE variants tagged at both the N and C termini with fluorescent constructs. When we tagged either apoE3 or apoE4 with yellow fluorescent protein (YFP) at the N terminus and cyan fluorescent protein (CFP) at the C terminus and transfected these constructs into Neuro-2a cells, we observed significantly enhanced FRET signal in the apoE4-expressing cells compared with their apoE3-expressing counterparts (Figure 8) (Xu et al., 2004), confirming that this FRET approach can detect apoE4 domain interaction, and that domain interaction is a property of apoE4 that occurs intracellularly.
Figure 8. ApoE4-Transfected Cells Demonstrate Intracellular FRET Signal.
ApoE constructs were generated to express YFP and CFP at the N and C termini, respectively, to act as reporter FRET fluorophores. When expressed in Neuro-2a cells, YFP-apoE4-CFP displays significantly more FRET signal compared to YFP-apoE3-CFP-expressing cells. This reflects the closer interaction between the N- and C-terminal domains in apoE4 and demonstrates that the biophysical property of domain interaction occurs in cells (Xu et al., 2004). Modified from Figure 1, Xu, Q., Brecht, W.J., Weisgraber, K.H., Mahley, R.W., and Huang, Y. Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J. Biol. Chem. 279: 25511–25516, 2004. © the American Society for Biochemistry and Molecular Biology.
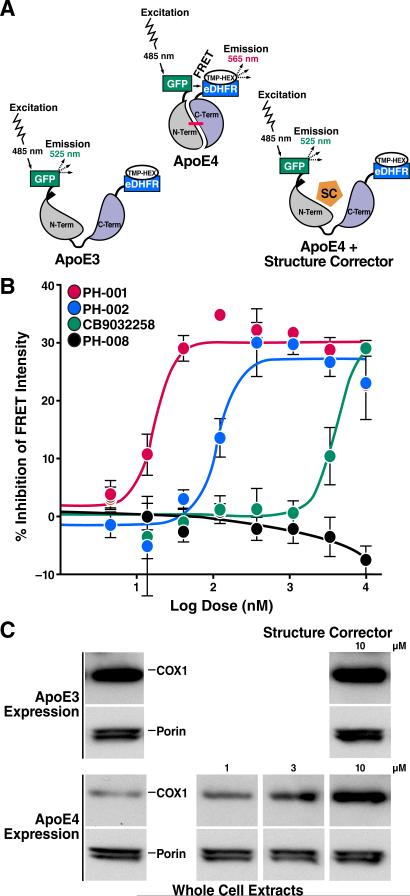
In a complementary set of assays (Chen et al., 2012), apoE4 was instead tagged with GFP at the N terminus and E. coli dihydrofolate reductase (eDHFR) at the C terminus. The construct was expressed in Neuro-2a cells, and an eDHFR high-affinity ligand (trimethoprim) conjugated with hexachlorofluorescein served as the acceptor fluorophore. The GFP-apoE4-eDHFR gave a significantly higher FRET signal than the apoE3 construct, reflecting a closer proximity of the N- and C-terminal domains in apoE4 (Chen et al., 2012). Furthermore, a screen for small molecules capable of blocking domain interaction identified several that dose-dependently decreased the FRET signal (Figures 9A and 9B) (Chen et al., 2012), serving as proof-of-concept that apoE4 is a promising target for the development of small molecule–based therapeutics.
Figure 9. FRET Assay Identifies ApoE4 Structure Correctors.
(A) The FRET signal is generated between the donor and acceptor fluorophores conjugated to the N and C termini of apoE. ApoE4 gives a higher FRET signal because of the closeness of the N- and C-terminal domains. The apoE4 signal is reduced by the small-molecule apoE structure corrector (SC) by disrupting domain interaction (Chen et al., 2012).
(B) Dose-response analysis reveals the relative potencies of three active and one inactive structure corrector (Chen et al., 2012).
(C) Mitochondrial cytochrome c oxidase (COX1) is reduced in Neuro-2a cells expressing apoE4 compared with apoE3. A small-molecule structure corrector (CB9032258) corrects the COX1 deficiency in the apoE4-expressing cells, but does not significantly affect apoE3-expressing cells.
(A) and (B). Modified from Figure 2, Chen, H.-K., Liu, Z., Meyer-Franke, A., Brodbeck, J., Miranda, R.D., McGuire, J.G., Pleiss, M.A., Ji, Z.-S., Balestra, M.E., Walker, D.W., Xu, Q., Jeong, D.-e., Budamagunta, M.S., Voss, J.C., Freedman, S.B., Weisgraber, K.H., Huang, Y., Mahley, R.W. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J. Biol. Chem. 2012; 287:5253–5266. © the American Society for Biochemistry and Molecular Biology.
Detrimental Effects of ApoE4 Can Be Reversed by Blocking Domain Interaction
Blocking domain interaction in apoE4 reverses many of its detrimental effects, both in vitro and in vivo (Mahley and Huang, 2012). This can be accomplished by site-directed mutagenesis in which arginine-61 is exchanged for threonine, thereby preventing the ionic interaction, or by small-molecule structure correctors that interact with apoE in the vicinity of arginine-61 to prevent or retard domain interaction.
Importantly, blocking domain interaction by site-directed mutagenesis or small-molecule structure correctors markedly reduced proteolysis and fragment formation. Mitochondrial dysfunction was no longer observed in cells expressing an apoE4 variant that lacked the ability to undergo domain interaction (apoE4-R61T). Furthermore, a small-molecule structure corrector restored the level of complex IV mitochondrial cytochrome c oxidase in apoE4-expressing cells to levels seen in apoE3-expressing cells (Figure 9C) (Chen et al., 2012). These studies were expanded to identify potent apoE4 structure correctors that could restore the level of mitochondrial cytochrome c oxidase with the potential to be used in vivo. A class of such small-molecule compounds that displays a significant structure-activity relationship has been identified (Chen et al., 2012).
As described, blocking apoE4 domain interaction restores neurite outgrowth, mitochondrial motility, and synaptic density (Brodbeck et al., 2011; Chen et al., 2011a). Thus, apoE4 domain interaction is a critical structural element that modulates both the physiological and pathophysiological functions of apoE4 (Mahley and Huang, 2012).
Hypothesis: ApoE Sets the Stage and Response to Neuronal Injury Triggers Neuropathology
The studies reviewed here, which comprise only a subset of the work done on apoE4 in the central nervous system, overwhelmingly point to a critical direct role for apoE4 in AD-mediated neurodegeneration. Based upon these studies, we propose the following model (Figure 10) to illustrate this hypothesis.
Figure 10. ApoE Sets the Stage and Response to Neuronal Injury Triggers Neuropathology.
(1) Injury to neurons induces the synthesis of apoE. ApoE (apoE4 > apoE3) is susceptible to proteolytic cleavage in neurons, and the neurotoxic fragments that are generated escape the secretory pathway and cause mitochondrial dysfunction and cytoskeletal alterations. This is most likely to occur when apoE4 is expressed (apoE4 > apoE3) due to its abnormal protein conformation (instability and domain interaction). (2) Exogenous apoE, primarily from astrocytes, could cause neuronal injury and could generate neurotoxic fragments by being shunted to the ER/Golgi apparatus, where proteolysis could occur. In addition, exogenous apoE does impact Aβ clearance/deposition. (3) Aβ expression can be induced by injured/stressed neurons, and together with other injurious agents could perpetuate the toxic cycle of injury in neurons. This would include apoE synthesis followed by proteolytic cleavage, toxic fragment formation, and neuropathology.
Figure 10 (1): What is well established is that neuronal injury or stress, caused by a variety of injurious agents, induces the synthesis of apoE by neurons. The structural properties of each apoE isoform dictate its propensity to undergo domain interaction (apoE4 > apoE3 > apoE2), which leads to apoE isoform–dependent proteolysis and the generation of neurotoxic fragments. In turn, these fragments cause mitochondrial dysfunction and cytoskeletal alterations, leading to neurodegeneration (Huang, 2010; Huang and Mucke, 2012; Mahley et al., 2006).
Figure 10 (2): What is clearly more speculative is whether there is a direct effect of exogenous apoE4, i.e., astrocyte-derived apoE, on neurons and neuropathology. In some studies, exogenous apoE4 can have direct detrimental effects on neurons, e.g., by inhibiting neurite outgrowth and mitochondrial motility (Chen et al., 2012; Nathan et al., 1994). Hartman et al. (2001) demonstrated that apoE4, when produced by astrocytes, impaired working memory but did not induce AD-like neuropathology in mice. Whether internalized apoE that is sequestered in the endosomes escapes lysosomal degradation, is transferred to the ER/Golgi apparatus compartment, and undergoes proteolysis and toxic fragment formation—or whether exogenous apoE4 acts through another mechanism independent of intracellular apoE fragment generation—is unknown. For example, in hepatocytes and macrophages, it has been established that apoE can escape lysosomal degradation and re-enter the secretory pathway (Farkas et al., 2003; Fazio et al., 2000; Zhu et al., 2005). It is possible that the detrimental effects of apoE4 observed by Chen et al. (2010) in neurons reflect the sequestration of apoE4 in endosomes and recycling to the ER/Golgi apparatus, in addition to its effects on N-methyl-d-aspartate receptors. Alternatively, astrocyte-derived apoE4 might directly damage the integrity of the BBB, allowing blood-derived proteins to enter the brain and injure neurons, and setting up the detrimental response to injury (Akassoglou et al., 2004; Bell et al., 2012). What is not speculative, however, is that astrocytic apoE4 can negatively impact Aβ clearance and/or deposition (Bien-Ly et al., 2011; Castellano et al., 2011; Kim et al., 2011) and thus induce the apoE neurotoxic pathway.
Figure 10 (3): Although much is yet to be learned about the regulation of Aβ and its potential roles in normal neuronal biology, it appears that Aβ production can be stimulated by injury to neurons through oxidative stress (Misonou et al., 2000; Paola et al., 2000; Tamagno et al., 2002) and energy deprivation (O'Connor et al., 2008; Velliquette et al., 2005). For example, exposure to low concentrations of 4-hydroxy-2,3-nonenal increased intracellular Aβ production by 2–6-fold in NT2-differentiated cells (Paola et al., 2000). Furthermore, glucose deprivation in cultured cells and in vivo in transgenic mice induces the phosphorylation of eIF2α (a translation initiation factor), which increases BACE1 activity (the rate-limiting enzyme involved in Aβ production) and elevates Aβ levels (O'Connor et al., 2008). In addition, it is well established that Aβ injures neurons (Huang and Mucke, 2012; Palop and Mucke, 2010; Selkoe, 2011), which, based on our hypothesis, would stimulate apoE production, induce apoE neurotoxic fragment formation, and further perpetuate the toxic cycle.
Conclusion
Although much remains to be understood about how apoE function affects both health and disease states, it is clear that apoE plays a critical role in the pathogenesis of many different neurodegenerative diseases. This assertion is no surprise, given apoE's fundamental role in cellular lipid transport and metabolism. However, its widespread expression also highlights how apoE is exceptionally well poised to induce or accelerate neuronal damage in apoE4-carrying individuals. The significant risk posed by apoE4 expression, combined with its widespread presence in the population and the ever-increasing average lifespan in which apoE4 carriers may suffer from its detrimental effects—in AD, TBI, and possibly other neuropathological disorders—underscore the enormous value that can come from developing therapies to counter its neurotoxic effects.
Acknowledgments
We thank the authors's laboratory members for many stimulating discussions on the topics covered in this review. We also thank Sylvia Richmond for manuscript preparation, Anna Lisa Lucido and Gary Howard for editorial assistance, and John C. W. Carroll for graphics. This work was supported in part by National Institutes of Health grants P01 AG022074 and R01 AG028793 and a gift from the Stephen D. Bechtel, Jr. Foundation.
Abbreviations used
- AD
Alzheimer's disease
- Aβ
amyloid β
- apo
apolipoprotein
- BBB
blood–brain barrier
- eDHFR
E. coli dihydrofolate reductase
- CFP
cyan fluorescent protein
- CNS
central nervous system
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- FRET
fluorescence resonance energy transfer
- LDL
low density lipoproteins
- NFT
neurofibrillary tangle
- TBI
traumatic brain injury
- YFP
yellow fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams RA, Bauer J, Flick MJ, Sikorski SL, Nuriel T, Lassmann H, Degen JL, Akassoglou K. The fibrin-derived γ377-395 peptide inhibits microglia activation and suppresses relapsing paralysis in central nervous system autoimmune disease. J. Exp. Med. 2007;204:571–582. doi: 10.1084/jem.20061931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F, Vossel KA, Miller BL, Migliaccio R, Bonasera SJ, Filippi M, Boxer AL, Karydas A, Possin KL, Gorno-Tempini ML. Apolipoprotein E ε4 is associated with disease-specific effects on brain atrophy in Alzheimer's disease and frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 2009;106:2018–2022. doi: 10.1073/pnas.0812697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akassoglou K, Adams RA, Bauer J, Mercado P, Tseveleki V, Lassmann H, Probert L, Strickland S. Fibrin depletion decreases inflammation and delays the onset of demyelination in a tumor necrosis factor transgenic mouse model for multiple sclerosis. Proc. Natl. Acad. Sci. USA. 2004;101:6698–6703. doi: 10.1073/pnas.0303859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts MJ, Graffagnino C, McClenny C, DeLong D, Strittmatter W, Saunders AM, Roses AD. ApoE genotype and survival from intracerebral haemorrhage. Lancet. 1995;346:575. doi: 10.1016/s0140-6736(95)91411-0. [DOI] [PubMed] [Google Scholar]
- Andrews-Zwilling Y, Bien-Ly N, Xu Q, Li G, Bernardo A, Yoon SY, Zwilling D, Yan TX, Chen L, Huang Y. Apolipoprotein E4 causes age- and tau-dependent impairment of GABAergic interneurons, leading to learning and memory deficits in mice. J. Neurosci. 2010;30:13707–13717. doi: 10.1523/JNEUROSCI.4040-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Zwilling Y, Gillespie AK, Kravitz AV, Nelson AB, Devidze N, Lo I, Yoon SY, Bien-Ly N, Ring K, Zwilling D, et al. Hilar GABAergic interneuron activity controls spatial learning and memory retrieval. PLoS One. 2012;7:e40555. doi: 10.1371/journal.pone.0040555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna H, Frey WH., II Mechanisms of mitochondrial dysfunction and energy deficiency in Alzheimer's disease. Mitochondrion. 2007;7:297–310. doi: 10.1016/j.mito.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Bales KR, Verina T, Cummins DJ, Du Y, Dodel RC, Saura J, Fishman CE, DeLong CA, Piccardo P, Petegnief V, et al. Apolipoprotein E is essential for amyloid deposition in the APPV717F transgenic mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 1999;96:15233–15238. doi: 10.1073/pnas.96.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Cohn JS, Petit-Turcotte C, Tremblay M, Aumont N, Ramassamy C, Davignon J, Poirier J. Apolipoprotein E and β-amyloid levels in the hippocampus and frontal cortex of Alzheimer's disease subjects are disease-related and apolipoprotein E genotype dependent. Brain Res. 1999;843:87–94. doi: 10.1016/s0006-8993(99)01894-6. [DOI] [PubMed] [Google Scholar]
- Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z, Holtzman DM, Betsholtz C, Armulik A, Sallstrom J, et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485:512–516. doi: 10.1038/nature11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosta S, Nathan BP, Orth M, Dong L-M, Mahley RW, Pitas RE. Stable expression and secretion of apolipoproteins E3 and E4 in mouse neuroblastoma cells produces differential effects on neurite outgrowth. J. Biol. Chem. 1995;270:27063–27071. doi: 10.1074/jbc.270.45.27063. [DOI] [PubMed] [Google Scholar]
- Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Wang C, Huang Y. C-terminal-truncated apolipoprotein (apo) E4 inefficiently clears amyloid-β (Aβ) and acts in concert with Aβ to elicit neuronal and behavioral deficits in mice. Proc. Natl. Acad. Sci. USA. 2011;108:4236–4241. doi: 10.1073/pnas.1018381108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Ly N, Gillespie AK, Walker D, Yoon SY, Huang Y. Reducing human apolipoprotein E levels attenuates age-dependent Aβ accumulation in mutant human amyloid precursor protein transgenic mice. J. Neurosci. 2012;32:4803–4811. doi: 10.1523/JNEUROSCI.0033-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, Hui DY, Mahley RW, Gebicke-Haerter PJ, Ignatius MJ, Shooter EM. A role for apolipoprotein E, apolipoprotein A-I, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J. Clin. Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht WJ, Harris FM, Chang S, Tesseur I, Yu G-Q, Xu Q, Fish JD, Wyss-Coray T, Buttini M, Mucke L, et al. Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 2004;24:2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck J, Balestra ME, Saunders AM, Roses AD, Mahley RW, Huang Y. Rosiglitazone increases dendritic spine density and rescues spine loss caused by apolipoprotein E4 in primary cortical neurons. Proc. Natl. Acad. Sci. USA. 2008;105:1343–1346. doi: 10.1073/pnas.0709906104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck J, McGuire J, Liu Z, Meyer-Franke A, Balestra ME, Jeong D.-e., Pleiss M, McComas C, Hess F, Witter D, et al. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J. Biol. Chem. 2011;286:17217–17226. doi: 10.1074/jbc.M110.217380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu G. Apolipoprotein E and its receptors in Alzheimer's disease: Pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttini M, Orth M, Bellosta S, Akeefe H, Pitas RE, Wyss-Coray T, Mucke L, Mahley RW. Expression of human apolipoprotein E3 or E4 in the brains of Apoe–/– mice: Isoform-specific effects on neurodegeneration. J. Neurosci. 1999;19:4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam JA, Bu G. Modulation of β-amyloid precursor protein trafficking and processing by the low density lipoprotein receptor family. Mol. Neurodegener. 2006;1:8. doi: 10.1186/1750-1326-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci. Transl. Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamelian L, Reis M, Feinstein A. Six-month recovery from mild to moderate traumatic brain injury: The role of APOE-ε4 allele. Brain. 2004;127:2621–2628. doi: 10.1093/brain/awh296. [DOI] [PubMed] [Google Scholar]
- Chang S, Ma TR, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipidand receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc. Natl. Acad. Sci. USA. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J, Vinokurov S, Achiron A, Karussis DM, Mitosek-Szewczyk K, Birnbaum M, Michaelson DM, Korczyn AD. APOE genotype is a major predictor of long-term progression of disability in MS. Neurology. 2001;56:312–316. doi: 10.1212/wnl.56.3.312. [DOI] [PubMed] [Google Scholar]
- Chen H-K, Ji Z-S, Dodson SE, Miranda RD, Rosenblum CI, Reynolds IJ, Freedman SB, Weisgraber KH, Huang Y, Mahley RW. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J. Biol. Chem. 2011a;286:5215–5221. doi: 10.1074/jbc.M110.151084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-K, Liu Z, Meyer-Franke A, Brodbeck J, Miranda RD, McGuire JG, Pleiss MA, Ji Z-S, Balestra ME, Walker DW, et al. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J. Biol. Chem. 2012;287:5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA. 2011b;108:14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Durakoglugil MS, Xian X, Herz J. ApoE4 reduces glutamate receptor function and synaptic plasticity by selectively impairing ApoE receptor recycling. Proc. Natl. Acad. Sci. USA. 2010;107:12011–12016. doi: 10.1073/pnas.0914984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conejero-Goldberg C, Hyde TM, Chen S, Dreses-Werringloer U, Herman MM, Kleinman JE, Davies P, Goldberg TE. Molecular signatures in postmortem brain tissue of younger individuals at high risk for Alzheimer's disease as based on APOE genotype. Mol. Psychiatry. 2011;16:836–847. doi: 10.1038/mp.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cramer PE, Cirrito JR, Wesson DW, Lee CYD, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, et al. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford FC, Vanderploeg RD, Freeman MJ, Singh S, Waisman M, Michaels L, Abdullah L, Warden D, Lipsky R, Salazar A, Mullan MJ. APOE genotype influences acquisition and recall following traumatic brain injury. Neurology. 2002;58:1115–1118. doi: 10.1212/wnl.58.7.1115. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Nowotny P, Kauwe JSK, Ridge PG, Mayo K, Bertelsen S, Hinrichs A, Fagan AM, Holtzman DM, Morris JC, Goate AM. Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch. Neurol. 2011;68:1013–1019. doi: 10.1001/archneurol.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Connections between the processing and nuclear export of mRNA: Evidence for an export license? Proc. Natl. Acad. Sci. USA. 2000;97:4–6. doi: 10.1073/pnas.97.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin. Immunopathol. 2012;34:43–62. doi: 10.1007/s00281-011-0290-8. [DOI] [PubMed] [Google Scholar]
- Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, Holtzman DM, Zlokovic BV. ApoE isoform–specific disruption of amyloid β peptide clearance from mouse brain. J. Clin. Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Curr. Opin. Lipidol. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- Dong L-M, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- Farkas MH, Swift LL, Hasty AH, Linton MF, Fazio S. The recycling of apolipoprotein E in primary cultures of mouse hepatocytes: Evidence for a physiologic connection to high density lipoprotein metabolism. J. Biol. Chem. 2003;278:9412–9417. doi: 10.1074/jbc.M208026200. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, Van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. J. Am. Med. Assoc. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Fazekas F, Strasser-Fuchs S, Kollegger H, Berger T, Kristoferitsch W, Schmidt H, Enzinger C, Schiefermeier M, Schwarz C, Kornek B, et al. Apolipoprotein E ε4 is associated with rapid progression of multiple sclerosis. Neurology. 2001;57:853–857. doi: 10.1212/wnl.57.5.853. [DOI] [PubMed] [Google Scholar]
- Fazio S, Linton MF, Swift LL. The cell biology and physiologic relevance of apoE recycling. Trends Cardiovasc. Med. 2000;10:23–30. doi: 10.1016/s1050-1738(00)00033-5. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman G, Froom P, Sazbon L, Grinblatt I, Shochina M, Tsenter J, Babaey S, Yehuda AB, Groswasser Z. Apolipoprotein E-ε4 genotype predicts a poor outcome in survivors of traumatic brain injury. Neurology. 1999;52:244–248. doi: 10.1212/wnl.52.2.244. [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Gandy S, DeKosky ST. APOE ε4 status and traumatic brain injury on the gridiron or the battlefield. Sci. Transl. Med. 2012;4:134ed134. doi: 10.1126/scitranslmed.3004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, et al. APOE and Alzheimer disease: A major gene with semi-dominant inheritance. Mol. Psychiatry. 2011;16:903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, LaDu MJ, Van Eldik LJ. A dual role for apolipoprotein E in neuroinflammation. Anti- and pro-inflammatory activity. J. Mol. Neurosci. 2004;23:205–212. doi: 10.1385/JMN:23:3:205. [DOI] [PubMed] [Google Scholar]
- Hardy J. Alzheimer's disease: The amyloid cascade hypothesis: An update and reappraisal. J. Alzheimers Dis. 2006;9(Suppl. 3):151–153. doi: 10.3233/jad-2006-9s317. [DOI] [PubMed] [Google Scholar]
- Harhangi BS, de Rijk MC, van Duijn CM, Van Broeckhoven C, Hofman A, Breteler MMB. APOE and the risk of PD with or without dementia in a population-based study. Neurology. 2000;54:1272–1276. doi: 10.1212/wnl.54.6.1272. [DOI] [PubMed] [Google Scholar]
- Harris FM, Brecht WJ, Xu Q, Mahley RW, Huang Y. Increased tau phosphorylation in apolipoprotein E4 transgenic mice is associated with activation of extracellular signal-regulated kinase: Modulation by zinc. J. Biol. Chem. 2004a;279:44795–44801. doi: 10.1074/jbc.M408127200. [DOI] [PubMed] [Google Scholar]
- Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, Fish JD, Masliah E, Hopkins PC, Scearce-Levie K, et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. USA. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris FM, Tesseur I, Brecht WJ, Xu Q, Mullendorff K, Chang S, Wyss-Coray T, Mahley RW, Huang Y. Astroglial regulation of apolipoprotein E expression in neuronal cells. Implications for Alzheimer's disease. J. Biol. Chem. 2004b;279:3862–3868. doi: 10.1074/jbc.M309475200. [DOI] [PubMed] [Google Scholar]
- Hartman RE, Wozniak DF, Nardi A, Olney JW, Sartorius L, Holtzman DM. Behavioral phenotyping of GFAP-apoE3 and -apoE4 transgenic mice: ApoE4 mice show profound working memory impairments in the absence of Alzheimer's-like neuropathology. Exp. Neurol. 2001;170:326–344. doi: 10.1006/exnr.2001.7715. [DOI] [PubMed] [Google Scholar]
- Hatters DM, Budamagunta MS, Voss JC, Weisgraber KH. Modulation of apolipoprotein E structure by domain interaction. Differences in lipid-bound and lipid-free forms. J. Biol. Chem. 2005;280:34288–34295. doi: 10.1074/jbc.M506044200. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Campenot RB, Vance DE, Vance JE. Apolipoprotein E-containing lipoproteins protect neurons from apoptosis via a signaling pathway involving low-density lipoprotein receptor-related protein-1. J. Neurosci. 2007;27:1933–1941. doi: 10.1523/JNEUROSCI.5471-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annu. Rev. Biochem. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- Huang Y. Aβ-independent roles of apolipoprotein E4 in the pathogenesis of Alzheimer's disease. Trends Mol. Med. 2010;16:287–294. doi: 10.1016/j.molmed.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, Mahley RW. Apolipoprotein E fragments present in Alzheimer's disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc. Natl. Acad. Sci. USA. 2001;98:8838–8843. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MJ, Shooter EM, Pitas RE, Mahley RW. Lipoprotein uptake by neuronal growth cones in vitro. Science. 1987;236:959–962. doi: 10.1126/science.3576212. [DOI] [PubMed] [Google Scholar]
- Ji Z-S, Fazio S, Lee Y-L, Mahley RW. Secretion-capture role for apolipoprotein E in remnant lipoprotein metabolism involving cell surface heparan sulfate proteoglycans. J. Biol. Chem. 1994;269:2764–2772. [PubMed] [Google Scholar]
- Jones PB, Adams KW, Rozkalne A, Spires-Jones TL, Hshieh TT, Hashimoto T, von Armin CAF, Mielke M, Bacskai BJ, Hyman BT. Apolipoprotein E: Isoform specific differences in tertiary structure and interaction with amyloid-β in human Alzheimer brain. PLoS One. 2011;6:e14586. doi: 10.1371/journal.pone.0014586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Vardarajan BN, Buros J, Yu C-E, Hawk MV, Dombroski BA, Crane PK, Larson EB, Alzheimer's Disease Genetics Consortium. Mayeux R, et al. Comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch. Neurol. 2012;69:1270–1279. doi: 10.1001/archneurol.2012.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jiang H, Park S, Eltorai AE, Stewart FR, Yoon H, Basak JM, Finn MB, Holtzman DM. Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-β amyloidosis. J. Neurosci. 2011;31:18007–18012. doi: 10.1523/JNEUROSCI.3773-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Guiard B, Meyer HE, Wiedemann N, Pfanner N. Cooperation of translocase complexes in mitochondrial protein import. J. Cell Biol. 2007;179:585–591. doi: 10.1083/jcb.200708199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VM-Y, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Li G, Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Ring K, Halabisky B, Deng C, Mahley RW, Huang Y. GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell. 2009;5:634–645. doi: 10.1016/j.stem.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Hauser MA, Scott WK, Martin ER, Booze MW, Qin XJ, Walter JW, Nance MA, Hubble JP, Koller WC, et al. Apolipoprotein E controls the risk and age at onset of Parkinson disease. Neurology. 2004;62:2005–2009. doi: 10.1212/01.wnl.0000128089.53030.ac. [DOI] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl. Acad. Sci. USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JR, Tang W, Wang H, Vitek MP, Bennett ER, Sullivan PM, Warner DS, Laskowitz DT. APOE genotype and an apoE-mimetic peptide modify the systemic and central nervous system inflammatory response. J. Biol. Chem. 2003;278:48529–48533. doi: 10.1074/jbc.M306923200. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Huang Y. Small-molecule structure correctors target abnormal protein structure and function: The structure corrector rescue of apolipoprotein E–associated neuropathology. J. Med. Chem. 2012;55:8997–9008. doi: 10.1021/jm3008618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Huang Y, Rall SC., Jr. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia): Questions, quandaries, and paradoxes. J. Lipid Res. 1999;40:1933–1949. [PubMed] [Google Scholar]
- Mahley RW, Ji Z-S. Remnant lipoprotein metabolism: Key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J. Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- Mahley RW, Rall SC., Jr. Apolipoprotein E: Far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: A causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: Structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J. Lipid Res. 2009;50:S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Brice A, Vaughan JR, Zimprich A, Breteler MMB, Meco G, Filla A, Farrer MJ, Bétard C, Singleton A, et al. Apolipoprotein E4 is probably responsible for the chromosome 19 linkage peak for Parkinson's disease. Am. J. Med. Genet. 2005;136B:72–74. doi: 10.1002/ajmg.b.30196. [DOI] [PubMed] [Google Scholar]
- Maruszak A, Pepłońska B, Safranow K, Chodakowska-Żebrowska M, Barcikowska M, Żekanowski C. TOMM40 rs10524523 polymorphism's role in late-onset Alzheimer's disease and in longevity. J. Alzheimers Dis. 2012;28:309–322. doi: 10.3233/JAD-2011-110743. [DOI] [PubMed] [Google Scholar]
- Mayeux R, Ottman R, Maestre G, Ngai C, Tang M-X, Ginsberg H, Chun M, Tycko B, Shelanski M. Synergistic effects of traumatic head injury and apolipoprotein-ε4 in patients with Alzheimer's disease. Neurology. 1995;45:555–557. doi: 10.1212/wnl.45.3.555. [DOI] [PubMed] [Google Scholar]
- McCarron MO, Delong D, Alberts MJ. APOE genotype as a risk factor for ischemic cerebrovascular disease: A meta-analysis. Neurology. 1999;53:1308–1311. doi: 10.1212/wnl.53.6.1308. [DOI] [PubMed] [Google Scholar]
- McKee AC, Cantu RC, Nowinski CJ, Hedley-Whyte ET, Gavett BE, Budson AE, Santini VE, Lee H-S, Kubilus CA, Stern RA. Chronic traumatic encephalopathy in athletes: Progressive tauopathy after repetitive head injury. J. Neuropathol. Exp. Neurol. 2009;68:709–735. doi: 10.1097/NEN.0b013e3181a9d503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Morishima-Kawashima M, Ihara Y. Oxidative stress induces intracellular accumulation of amyloid β-protein (Aβ) in human neuroblastoma cells. Biochemistry. 2000;39:6951–6959. doi: 10.1021/bi000169p. [DOI] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Watanabe A, Fujino T, Hosono T, Michikawa M. Apolipoprotein E4 (1–272) fragment is associated with mitochondrial proteins and affects mitochondrial function in neuronal cells. Mol. Neurodegener. 2009;4:35. doi: 10.1186/1750-1326-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan BP, Bellosta S, Sanan DA, Weisgraber KH, Mahley RW, Pitas RE. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- Nathan BP, Chang K-C, Bellosta S, Brisch E, Ge N, Mahley RW, Pitas RE. The inhibitory effect of apolipoprotein E4 on neurite outgrowth is associated with microtubule depolymerization. J. Biol. Chem. 1995;270:19791–19799. doi: 10.1074/jbc.270.34.19791. [DOI] [PubMed] [Google Scholar]
- Nicoll JAR, Roberts GW, Graham DI. Amyloid β-protein, APOE genotype and head injury. Ann. N.Y. Acad. Sci. 1996;777:271–275. doi: 10.1111/j.1749-6632.1996.tb34431.x. [DOI] [PubMed] [Google Scholar]
- O'Connor T, Sadleir KR, Maus E, Velliquette RA, Zhao J, Cole SL, Eimer WA, Hitt B, Bembinster LA, Lammich S, et al. Phosphorylation of the translation initiation factor eIF2α increases BACE1 levels and promotes amyloidogenesis. Neuron. 2008;60:988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Mucke L. A network dysfunction perspective on neurodegenerative diseases. Nature. 2006;443:768–773. doi: 10.1038/nature05289. [DOI] [PubMed] [Google Scholar]
- Palop JJ, Mucke L. Amyloid-β–induced neuronal dysfunction in Alzheimer's disease: From synapses toward neural networks. Nat. Neurosci. 2010;13:812–818. doi: 10.1038/nn.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paola D, Domenicotti C, Nitti M, Vitali A, Borghi R, Cottalasso D, Zaccheo D, Odetti P, Strocchi P, Marinari UM, et al. Oxidative stress induces increase in intracellular amyloid β-protein production and selective activation of βI and βII PKCs in NT2 Cells. Biochem. Biophys. Res. Commun. 2000;268:642–646. doi: 10.1006/bbrc.2000.2164. [DOI] [PubMed] [Google Scholar]
- Parihar MS, Brewer GJ. Mitoenergetic failure in Alzheimer disease. Am. J. Physiol. Cell Physiol. 2007;292:C8–C23. doi: 10.1152/ajpcell.00232.2006. [DOI] [PubMed] [Google Scholar]
- Parsian A, Racette B, Goldsmith LJ, Perlmutter JS. Parkinson's disease and apolipoprotein E: Possible association with dementia but not age at onset. Genomics. 2002;79:458–461. doi: 10.1006/geno.2002.6707. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N. Mitochondrial protein import: Two membranes, three translocases. Curr. Opin. Cell Biol. 2002;14:400–411. doi: 10.1016/s0955-0674(02)00355-1. [DOI] [PubMed] [Google Scholar]
- Poirier J, Hess M, May PC, Finch CE. Astrocytic apolipoprotein E mRNA and GFAP mRNA in hippocampus after entorhinal cortex lesioning. Mol. Brain Res. 1991;11:97–106. doi: 10.1016/0169-328x(91)90111-a. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Raber J, Wong D, Buttini M, Orth M, Bellosta S, Pitas RE, Mahley RW, Mucke L. Isoform-specific effects of human apolipoprotein E on brain function revealed in ApoE knockout mice: Increased susceptibility of females. Proc. Natl. Acad. Sci. USA. 1998;95:10914–10919. doi: 10.1073/pnas.95.18.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raber J, Wong D, Yu G-Q, Buttini M, Mahley RW, Pitas RE, Mucke L. Apolipoprotein E and cognitive performance. Nature. 2000;404:352–354. doi: 10.1038/35006165. [DOI] [PubMed] [Google Scholar]
- Raffaï RL, Dong L-M, Farese RV, Jr., Weisgraber KH. Introduction of human apolipoprotein E4 “domain interaction” into mouse apolipoprotein E. Proc. Natl. Acad. Sci. USA. 2001;98:11587–11591. doi: 10.1073/pnas.201279298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy G, Xu Q, Huang Y, Weisgraber KH. Effect of domain interaction on apolipoprotein E levels in mouse brain. J. Neurosci. 2005;25:10658–10663. doi: 10.1523/JNEUROSCI.1922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D. How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane? J. Cell Biol. 2005;171:419–423. doi: 10.1083/jcb.200507147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM. Linking brain imaging and genomics in the study of Alzheimer's disease and aging. Ann. N.Y. Acad. Sci. 2007;1097:94–113. doi: 10.1196/annals.1379.011. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc. Natl. Acad. Sci. USA. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roses AD. Apolipoprotein E alleles as risk factors in Alzheimer's disease. Annu. Rev. Med. 1996;47:387–400. doi: 10.1146/annurev.med.47.1.387. [DOI] [PubMed] [Google Scholar]
- Roses AD. An inherited variable poly-T repeat genotype in TOMM40 in Alzheimer disease. Arch. Neurol. 2010;67:536–541. doi: 10.1001/archneurol.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanan DA, Weisgraber KH, Russell SJ, Mahley RW, Huang D, Saunders A, Schmechel D, Wisniewski T, Frangione B, Roses AD, Strittmatter WJ. Apolipoprotein E associates with β amyloid peptide of Alzheimer's disease to form novel monofibrils. Isoform apoE4 associates more efficiently than apoE3. J. Clin. Invest. 1994;94:860–869. doi: 10.1172/JCI117407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, St George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Resolving controversies on the path to Alzheimer's therapeutics. Nat. Med. 2011;17:1060–1065. doi: 10.1038/nm.2460. [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol. Aspects Med. 2010;31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Slooter AJC, Tang M-X, van Duijn CM, Stern Y, Ott A, Bell K, Breteler MMB, Van Broeckhoven C, Tatemichi TK, Tycko B, et al. Apolipoprotein E ε4 and the risk of dementia with stroke. A population-based investigation. J. Am. Med. Assoc. 1997;277:818–821. doi: 10.1001/jama.277.10.818. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: High-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydow A, Van der Jeugd A, Zheng F, Ahmed T, Balschun D, Petrova O, Drexler D, Zhou L, Rune G, Mandelkow E, et al. Tau-induced defects in synaptic plasticity, learning, and memory are reversible in transgenic mice after switching off the toxic tau mutant. J. Neurosci. 2011;31:2511–2525. doi: 10.1523/JNEUROSCI.5245-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagno E, Bardini P, Obbili A, Vitali A, Borghi R, Zaccheo D, Pronzato MA, Danni O, Smith MA, Perry G, Tabaton M. Oxidative stress increases expression and activity of BACE in NT2 neurons. Neurobiol. Dis. 2002;10:279–288. doi: 10.1006/nbdi.2002.0515. [DOI] [PubMed] [Google Scholar]
- Teasdale GM, Nicoll JAR, Murray G, Fiddes M. Association of apolipoprotein E polymorphism with outcome after head injury. Lancet. 1997;350:1069–1071. doi: 10.1016/S0140-6736(97)04318-3. [DOI] [PubMed] [Google Scholar]
- Tesseur I, Van Dorpe J, Spittaels K, Van den Haute C, Moechars D, Van Leuven F. Expression of human apolipoprotein E4 in neurons causes hyperphosphorylation of protein tau in the brains of transgenic mice. Am. J. Pathol. 2000;156:951–964. doi: 10.1016/S0002-9440(10)64963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velliquette RA, O'Connor T, Vassar R. Energy inhibition elevates β-secretase levels and activity and is potentially amyloidogenic in APP transgenic mice: Possible early events in Alzheimer's disease pathogenesis. J. Neurosci. 2005;25:10874–10883. doi: 10.1523/JNEUROSCI.2350-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek MP, Christensen DJ, Wilcock D, Davis J, Van Nostrand WE, Li FQ, Colton CA. APOE-mimetic peptides reduce behavioral deficits, plaques and tangles in Alzheimer's disease transgenics. Neurodegener. Dis. 2012;10:122–126. doi: 10.1159/000334914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisgraber KH. Apolipoprotein E: Structure–function relationships. Adv. Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- Wisniewski T, Lalowski M, Golabek A, Vogel T, Frangione B. Is Alzheimer's disease an apolipoprotein E amyloidosis? Lancet. 1995;345:956–958. doi: 10.1016/s0140-6736(95)90701-7. [DOI] [PubMed] [Google Scholar]
- Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, Huang Y. Profile and regulation of apolipoprotein E (apoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the apoE locus. J. Neurosci. 2006;26:4985–4994. doi: 10.1523/JNEUROSCI.5476-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Brecht WJ, Weisgraber KH, Mahley RW, Huang Y. Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J. Biol. Chem. 2004;279:25511–25516. doi: 10.1074/jbc.M311256200. [DOI] [PubMed] [Google Scholar]
- Xu Q, Walker D, Bernardo A, Brodbeck J, Balestra ME, Huang Y. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J. Neurosci. 2008;28:1452–1459. doi: 10.1523/JNEUROSCI.3253-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Huang Y, Müllendorff K, Dong L, Giedt G, Meng EC, Cohen FE, Kuntz ID, Weisgraber KH, Mahley RW. Apolipoprotein (apo) E4 enhances amyloid β peptide production in cultured neuronal cells: ApoE structure as a potential therapeutic target. Proc. Natl. Acad. Sci. USA. 2005;102:18700–18705. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N, Ramaswamy G, Weisgraber KH. Apolipoprotein E4 domain interaction induces endoplasmic reticulum stress and impairs astrocyte function. J. Biol. Chem. 2009;284:27273–27280. doi: 10.1074/jbc.M109.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N, Scearce-Levie K, Ramaswamy G, Weisgraber KH. Apolipoprotein E4 domain interaction: Synaptic and cognitive deficits in mice. Alzheimers Dement. 2008;4:179–192. doi: 10.1016/j.jalz.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N, Weisgraber KH. Understanding the association of apolipoprotein E4 with Alzheimer disease: Clues from its structure. J. Biol. Chem. 2009;284:6027–6031. doi: 10.1074/jbc.R800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M-Y, Hasty AH, Harris C, Linton MF, Fazio S, Swift LL. Physiological relevance of apolipoprotein E recycling: Studies in primary mouse hepatocytes. Metabolism. 2005;54:1309–1315. doi: 10.1016/j.metabol.2005.04.019. [DOI] [PubMed] [Google Scholar]