Abstract
Patterns of sex‐chromosome differentiation and gonadal development have been shown to vary among populations of Rana temporaria along a latitudinal transect in Sweden. Frogs from the northern‐boreal population of Ammarnäs displayed well‐differentiated X and Y haplotypes, early gonadal differentiation, and a perfect match between phenotypic and genotypic sex. In contrast, no differentiated Y haplotypes could be detected in the southern population of Tvedöra, where juveniles furthermore showed delayed gonadal differentiation. Here, we show that Dmrt1, a gene that plays a key role in sex determination and sexual development across all metazoans, displays significant sex differentiation in Tvedöra, with a Y‐specific haplotype distinct from Ammarnäs. The differential segment is not only much shorter in Tvedöra than in Ammarnäs, it is also less differentiated and associates with both delayed gonadal differentiation and imperfect match between phenotypic and genotypic sex. Whereas Tvedöra juveniles with a local Y haplotype tend to ultimately develop as males, those without it may nevertheless become functional XX males, but with strongly female‐biased progeny. Our findings suggest that the variance in patterns of sex determination documented in common frogs might result from a genetic polymorphism within a small genomic region that contains Dmrt1. They also substantiate the view that recurrent convergences of sex determination toward a limited set of chromosome pairs may result from the co‐option of small genomic regions that harbor key genes from the sex‐determination pathway.
Keywords: Gonadal development, homomorphic sex chromosomes, nonrecombining segment, sex determination, sex races, sex reversal
Introduction
In sharp contrast to the highly differentiated W and Y chromosomes found in most birds and mammals, sex chromosomes are often homomorphic in cold‐blooded vertebrates (Schmid and Steinlein 2001; Devlin and Nagahama 2002; Schmid et al. 2010). Homomorphy may result from occasional XY recombination (Stöck et al. 2011; Guerrero et al. 2012) and/or high rates of sex‐chromosome turnover (Hillis and Green 1990; Schartl 2004; Volff et al. 2007; Evans et al. 2012), two mechanisms possibly stemming from incomplete genetic control over sex determination (Perrin 2009; Grossen et al. 2011). Both XY recombination and sex‐chromosome turnovers have been documented in amphibians (e.g., Stöck et al. 2013; Dufresnes et al. 2015), where approximately 96% of species lack morphologically differentiated sex chromosomes (Schmid et al. 1991; Eggert 2004).
Such is the case of the common frog, Rana temporaria (Fig. 1), a European species widely distributed from Spain to northern Norway. Sex determination in common frogs associates with linkage group 2 (LG2), as initially indicated by sex differences in allele frequencies at a series of microsatellite markers (Matsuba et al. 2008; Alho et al. 2010; Cano et al. 2011). However, genetic differentiation between sex chromosomes was shown to vary among populations along a latitudinal transect across Fennoscandia (Rodrigues et al. 2014). In the northern‐boreal population of Ammarnäs, all males had fixed specific alleles at LG2 markers, forming distinct X and Y haplotypes. In contrast, the same markers failed to identify any sex differentiation in the southern population of Tvedöra: individuals of both sexes harbored the same alleles at similar frequencies, testifying to regular recombination. Intermediate populations displayed a mixed situation: some males had distinct Y haplotypes, while others were genetically indistinguishable from females.
Figure 1.

Mating pair of Rana temporaria in amplexus. Photography credit Andreas Meyer.
Family analyses revealed that the contrast between Ammarnäs and Tvedöra did not stem from differences in sex‐specific patterns of recombination, but in the mechanisms of sex determination (Rodrigues et al. 2015). Juveniles from Ammarnäs families displayed balanced sex ratios already at metamorphosis (a feature characterizing the “differentiated” sex race; Witschi 1929, 1930), and strong associations between phenotypic sex and paternally inherited LG2 haplotypes. In Tvedöra, by contrast, a majority of offspring presented ovaries at metamorphosis (a feature of the “semidifferentiated” sex race); sex ratios were more balanced at the froglet stage, but still variable among families, being male‐biased in some and female‐biased in others. Associations between offspring sex and paternal LG2 haplotype were much weaker than in Ammarnäs, and variable among families, but still highly significant overall, a surprising result given the absence of male‐specific alleles at all LG2 markers investigated. Genotyping of markers from other linkage groups failed to find any sex association outside LG2 in Tvedöra (Rodrigues et al. 2016).
Altogether, these results show that LG2 contributes to sex determination in both populations, but in different ways. In Ammarnäs, alleles at the sex locus associate with early gonadal differentiation (the “differentiated race” syndrome) and strictly genetic sex determination (GSD). Because XY individuals always develop as males (which only recombine in the distal parts of chromosomes; Brelsford et al. 2016a, 2016c), recombination is arrested over most of the sex chromosome, resulting in marked XY differentiation. In Tvedöra, by contrast, alleles at the sex locus associate with delayed gonadal differentiation (the “semidifferentiated race” syndrome) and imperfect match between genetic and phenotypic sex (“leaky GSD”). Occasional events of sex reversal might account for the variance in sex ratios among families (excess of sons in the progeny of XY females, excess of daughters in the progeny of XX males), as well as for the absence of sex‐chromosome differentiation (resulting from XY recombination in XY females – the fountain‐of‐youth model; Perrin 2009; Matsuba et al. 2010).
Importantly (and independent of the underlying mechanisms), the situation in Tvedöra offers a unique opportunity to search for the sex locus. Contrasting with Ammarnäs, where sex chromosomes are differentiated over most of their length, occasional recombination in Tvedöra is expected to regularly restore XY similarity all along the chromosome, except for the immediate neighborhood of the sex‐determining locus. This should greatly facilitate its identification, by narrowing its localization down to a restricted nonrecombining sex‐determining region (SDR) displaying significant XY differentiation.
This study focuses on Dmrt1, an important gene from the sex‐determining cascade mapping to LG2 in R. temporaria (Brelsford et al. 2013). This gene or paralogs participate in sex determination and/or sexual dimorphism throughout the animal kingdom (Beukeboom and Perrin 2014); it plays a central sex‐determining role in birds (Smith et al. 2009), while paralogs take this role in several fish and frogs (Matsuda et al. 2002; Nanda et al. 2002; Yoshimoto et al. 2008). It thus qualifies as a potential candidate sex‐determining gene in our focal species. We identified three polymorphic markers in distinct noncoding parts of Dmrt1 and two more in the genes immediately flanking Dmrt1 in the X. tropicalis genome (namely Kank1 upstream and Dmrt3 downstream) and analyzed them for sex association in adults and families from Ammarnäs and Tvedöra. Our first aim was to test whether these markers showed any sex differentiation in Tvedöra, which would indicate proximity to the sex locus, given the occasional recombination and absence of sex differentiation for all other LG2 markers analyzed so far. In case of a positive result, our second aim was to investigate whether polymorphism at these markers might correlate with the variation in sex‐determination patterns documented among Tvedöra families (Rodrigues et al. 2015), in particular regarding the suggested occurrence of sex‐reversed XX males and XY females.
Materials and Methods
Field sampling and husbandry
The same samples were used as in Rodrigues et al. (2015). Mating pairs were caught in amplexus during the 2013 breeding season from two Swedish populations: 20 pairs from the northern‐boreal population of Ammarnäs (65°58′12.60″N, 16°12′43.80″E) and 11 pairs from the southern population of Tvedöra (55°42′0.85″N, 13°25′50.91″E). Buccal cells were sampled with sterile cotton swabs before release at the place of capture. Clutches of six pairs from each population (SA1‐SA6 and ST1‐ST6) were reared in outdoor facilities on the campus of the University of Lausanne. Within 1 week of metamorphosis, 40 offspring from each clutch (referred to as “metamorphs”) were anaesthetized and euthanized in 0.2% ethyl3‐aminobenzoate methanesulfonate salt solution (MS222), then dropped in 70% ethanol for preservation at −20°C. The remaining offspring (referred to as “froglets”) were allowed to grow for a few more weeks and similarly euthanized when reaching about 2 cm snout–vent length (Gosner stage 46; Gosner 1960).
Progeny sexing
Metamorphs and froglets were dissected under a binocular microscope in order to determine the phenotypic sex based on gonad morphology. These stages were chosen because “sex races” are defined by their differences in the patterns of gonadal development at metamorphosis (Witschi 1929): contrasting with the “differentiated sex race,” where juveniles present already at metamorphosis testes or ovaries in equal proportions, juveniles from the “semidifferentiated race” mostly present ovaries at this stage (so that discrepancies are expected between genetic and phenotypic sex). Only later in development (at the froglet stage and later) do some of these juveniles replace ovaries by testes (Witschi 1929). Ovaries in common frogs develop from the whole gonadal primordia into a large whitish/yellowish structure with distinct lobes and a characteristic granular aspect conferred by the many oocytes embedded in the cortex (Ogielska and Kotusz 2004). In contrast, testes develop from the anterior part of the gonadal primordia only (the posterior part degenerates) into a small oblong structure, with a smooth cortex covered with melanic spots (Haczkiewicz and Ogielska 2013). As gonads are not always well differentiated externally at metamorphosis, we applied a semiquantitative scale to score individuals along a gradient of apparent maleness. Individuals with distinctive male or female gonads were assigned scores of 1.0 and 0.0, respectively. Individuals identified as “likely” males or females were assigned scores of 0.9 and 0.1, respectively, while others identified as “possibly” males or females were scored as 0.7 and 0.3, respectively. Individuals with undifferentiated gonads were scored as 0.5. Note that only relative score values matter here, because we applied rank statistics (see “Statistical analyses”). All individuals were scored independently by N. Rodrigues and Y. Vuille before genetic analyses (summer 2013), with concordant results (correlation > 0.95).
Marker development
After overnight treatment with 10% proteinase K (Qiagen) at 56°C, DNA was extracted from hindleg tissues (metamorphs and froglets) and buccal swabs (adults) using a Qiagen DNeasy kit and a BioSprint 96 workstation (Qiagen), resulting in a 200 μL Buffer AE (Qiagen) DNA elution.
The cDNA Dmrt1 sequence of Rana chensinensis was downloaded from NCBI gene database. Blasts against the R. temporaria low‐coverage draft genome (Brelsford et al. 2016c) returned five scaffolds as the best hits, each including a full or partial Dmrt1 exon (Appendix S1, Text S1). Exon–intron boundaries were identified by comparing genomic DNA (gDNA) sequences to the cDNA sequences obtained from five froglets (Appendix S1, Text S2). RNA extraction was performed following the standard Trizol protocol. In short, snap frozen froglet samples were individually homogenized in Trizol (Life Technologies), followed by phase separation (using chloroform); after ethanol precipitation of the upper phase, RNA was washed with 70% ethanol twice and collected. cDNA was synthesized using Superscript III Reverse Transcriptase (Life Technologies), after DNAse treatment which removed any gDNA contamination.
Primer pairs (Appendix S2, Table S1) were designed in the intron regions flanking exons (<200 bp each direction); for exons 2 and 5, one flanking region (3′ and 5′, respectively) was missing from the scaffolds, so that the corresponding primers were designed within exons. With these primers, we amplified and sequenced (Microsynth) fragments from 26 individuals (14 from Ammarnäs and 12 from Tvedöra). Ambiguous fragment sequences were cloned before sequencing, using TOPO® TA Cloning® Dual Promoter Kit with One Shot® TOP10 chemically competent E. coli cells, following the protocol provided by the manufacturer. Besides multiple synonymous SNPs within exons, three length‐polymorphic sites were detected in different noncoding regions (Appendix S1, Text S3), corresponding to a microsatellite repeat in the 5′ part of intron 1, an indel in the 3′ part of intron 2, and a single nucleotide repeat (cytosine) in the 3’ UTR region of exon 5 (Fig. 2). Specific fluorescent primers (Appendix S2, Table S2) were designed for all three length‐polymorphic sites.
Figure 2.
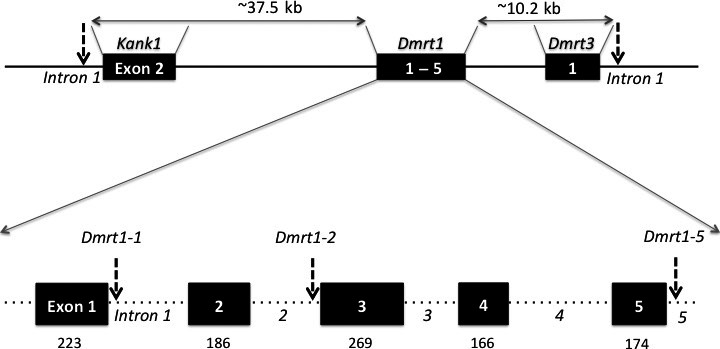
Structure of the genomic region investigated here, with localization of the five length‐polymorphic markers analyzed (arrows). Top: In X. tropicalis, Kank1 is the closest gene upstream of Dmrt1, and Dmrt3 the closest downstream. The distances indicated correspond to X. tropicalis, and might be longer in R. temporaria, because of its larger genome. Bottom: enlargement of Dmrt1; boxes denote the five exons with their respective sizes (in bp) indicated underneath. Dotted lines between exons represent introns of unknown size in R. temporaria.
As we did not aim at characterizing X‐ and Y‐sequences for Kank1 and Dmrt3 (because they do not qualify as candidate sex‐determining genes), we used a simpler procedure to develop length‐polymorphic markers. All scaffolds of the R. temporaria low‐coverage draft genome (Brelsford et al. 2016c) were aligned to the X. tropicalis genome with Blastn. Rana scaffolds mapping to X. tropicalis genes Kank1 and Dmrt3 (Appendix S1, Text S1) were screened for microsatellite markers using the microsatellite identification tool MISA (http://pgrc.ipk-gatersleben.de/misa/), and specific fluorescent primers were designed in the flanking regions of the microsatellite with longest repeat motif for each gene (both are on intron 1, Fig. 2; Appendix S2, Table S2).
Genotyping
All adults and juveniles from Ammarnäs and Tvedöra were then genotyped for these five length‐polymorphic markers. PCRs were performed in a total volume of 10 μL, including 3 μL of extracted DNA, 2.22 μL of Milli‐Q water, 3 μL of Qiagen Multiplex Master Mix, and 0.14–0.3 μL of labeled forward primer and 0.14–0.3 μL of unlabeled reverse primer (in total 1.78 μL of primer mix). PCRs were conducted on Perkin Elmer 2700 machines using the following thermal profile: 15 min of Taq polymerase activation at 95°C, followed by 35 cycles including denaturation at 94°C for 30 sec, annealing at 55°C for 1.5 min and elongation at 72°C for 1 min, ending the PCR with a final elongation of 30 min at 60°C. PCR products for genotyping were run on an automated ABI Prism 3100 sequencer (Applied Biosystems, Foster City, CA), and alleles were scored using GENEMAPPER v. 4.0 (Applied Biosystems).
Statistical analyses
Associations between offspring sex‐phenotype scores and paternally inherited LG2 haplotypes were quantified with Somers’ (1962) Dxy rank correlation (a measure of association between an ordinal variable x and a binary variable y) and tested with nonparametric Wilcoxon–Mann–Whitney (WMW) tests (statistics performed in R, v3.1.1, R Core Team, 2014). Between‐sex F ST values were calculated and tested (10,000 permutations) among adults from Ammarnäs and Tvedöra (FSTAT v2.9.3, updated from Goudet 1995). F ST values for the five markers were compared to those obtained for the 13 LG2 markers genotyped on the same sample by Rodrigues et al. (2015). Family genotypes were also combined with those obtained at these 13 LG2 markers, in order to localize our five markers on the consensus recombination map. Sex‐specific recombination rates were estimated with CRIMAP v2.4 (Green et al. 1990). The twopoint option was used to identify marker pairs with a LOD score exceeding 3.0, the all option to generate loci order, the build option to calculate the distances between loci (centimorgans, cM), and the flip option to test the robustness of loci order. A female consensus recombination map was plotted using MAPCHART v2.2 (Voorrips 2002).
Results
In adults from Ammarnäs, all five markers displayed sex‐diagnostic differences in allele frequencies (Table 1). All 20 males possessed at each locus exactly one copy of a male‐specific allele, not found in any female. As a result, F ST between sexes were high and significant for all five loci (average 0.286, range 0.142–0.514, all P values ~0.0002 after correction for multiple testing; Appendix S2, Table S3). Sibship analyses confirmed that alleles identified as male specific were indeed located on nonrecombining Y haplotypes. The most common haplotype had fixed allele 171 at Kank1, 337 at Dmrt1‐1, 212 at Dmrt1‐2, 296 at Dmrt1‐5, and 291 at Dmrt3. Two other closely related Y haplotypes were found, differing at one or two loci (changes to allele 335 at Dmrt1‐1 and/or 285 at Dmrt3). These analyses also revealed a highly significant association between inheritance of male‐specific Y haplotypes and offspring phenotypic sex, both in metamorphs (n = 240, Somer's Dxy rank correlation = 0.71, P < 2.2 × 10−16, WMW test) and in froglets (n = 31, Dxy = 1.0, P = 4.9 × 10−8, WMW test; Table 2). Correlations were also significant in all families separately (n = 41–49 in each, Dxy varying from 0.60 to 0.95, all P < 10−6), except for family SA6 where gonads were still undifferentiated in metamorphs.
Table 1.
Sex‐specific allele frequencies in Ammarnäs (n = 40) and Tvedöra (n = 22)
| Marker | Allele size | Ammarnäs | Tvedöra | ||
|---|---|---|---|---|---|
| Female | Male | Female | Male | ||
| Kank1 | 165 | 0.00 | 0.00 | 0.23 | 0.14 |
| 168 | 0.00 | 0.00 | 0.00 | 0.05 | |
| 171 | 0.00 | 0.50 | 0.00 | 0.00 | |
| 174 | 1.00 | 0.50 | 0.77 | 0.73 | |
| 178 | 0.00 | 0.00 | 0.00 | 0.09 | |
| Dmrt1‐1 | 291 | 0.73 | 0.43 | 0.09 | 0.14 |
| 292 | 0.28 | 0.08 | 0.64 | 0.41 | |
| 294 | 0.00 | 0.00 | 0.00 | 0.41 | |
| 325 | 0.00 | 0.00 | 0.27 | 0.05 | |
| 335 | 0.00 | 0.05 | 0.00 | 0.00 | |
| 337 | 0.00 | 0.45 | 0.00 | 0.00 | |
| Dmrt1‐2 | 198 | 0.30 | 0.08 | 0.95 | 0.86 |
| 211 | 0.70 | 0.42 | 0.05 | 0.14 | |
| 212 | 0.00 | 0.50 | 0.00 | 0.00 | |
| Dmrt1‐5 | 296 | 0.00 | 0.50 | 0.23 | 0.09 |
| 300 | 0.00 | 0.00 | 0.18 | 0.14 | |
| 301 | 0.08 | 0.00 | 0.55 | 0.64 | |
| 302 | 0.20 | 0.11 | 0.05 | 0.05 | |
| 303 | 0.00 | 0.05 | 0.00 | 0.05 | |
| 304 | 0.73 | 0.34 | 0.00 | 0.00 | |
| 305 | 0.00 | 0.00 | 0.00 | 0.05 | |
| Dmrt3 | 276 | 0.13 | 0.03 | 0.59 | 0.45 |
| 281 | 0.00 | 0.00 | 0.00 | 0.36 | |
| 285 | 0.00 | 0.16 | 0.00 | 0.00 | |
| 287 | 0.05 | 0.00 | 0.00 | 0.00 | |
| 290 | 0.03 | 0.00 | 0.00 | 0.00 | |
| 291 | 0.00 | 0.34 | 0.09 | 0.05 | |
| 293 | 0.00 | 0.03 | 0.00 | 0.00 | |
| 297 | 0.00 | 0.00 | 0.23 | 0.09 | |
| 300 | 0.66 | 0.37 | 0.09 | 0.05 | |
| 303 | 0.00 | 0.03 | 0.00 | 0.00 | |
| 309 | 0.13 | 0.05 | 0.00 | 0.00 | |
Male‐specific alleles are indicated in bold.
Table 2.
Numbers of metamorphs and froglets per maleness score (from 0 to 1) as a function of presence (+) or absence (−) of a Y‐specific Dmrt1‐1 allele (294 in Tvedöra, 335 or 337 in Ammarnäs) in six families from Ammarnäs (SA1 to SA6) and Tvedöra (ST1 to ST6). Also provided are Somers rank correlation values (Dxy), sample sizes (n), and P values from Wilcoxon–Mann–Whitney tests (P)
| Family | Metamorphs | Froglets | Total | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.1 | 0.3 | 0.5 | 0.7 | 0.9 | 1 | Dxy | n | P | 0 | 0.1 | 0.3 | 0.5 | 0.7 | 0.9 | 1 | Dxy | n | P | Dxy | n | P | ||
| SA1 | − | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0.75 | 40 | 1.2e‐08*** | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 9 | 0.009* | 0.8 | 49 | 1.2e‐10*** |
| + | 1 | 0 | 0 | 6 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | ||||||||||
| SA2 | − | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0.58 | 40 | 1.9e‐09*** | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0.48 | 0.6 | 43 | 4.7e‐10*** |
| + | 0 | 0 | 0 | 13 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||||||||||
| SA3 | − | 16 | 0 | 1 | 9 | 0 | 0 | 0 | 0.93 | 40 | 2.1e‐07*** | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 7 | 0.03* | 0.93 | 47 | 3.3e‐09*** |
| + | 0 | 0 | 0 | 2 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | ||||||||||
| SA4 | − | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0.92 | 40 | 9.4e‐10*** | 1 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 1 | NA | 0.92 | 41 | 5.5e‐10*** |
| + | 0 | 0 | 0 | 1 | 1 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||||
| SA5 | − | 17 | 0 | 0 | 1 | 0 | 0 | 0 | 0.95 | 40 | 1.4e‐09*** | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 7 | 0.046* | 0.95 | 47 | 6.4e‐11*** |
| + | 0 | 1 | 0 | 1 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | ||||||||||
| SA6 | − | 0 | 0 | 0 | 18 | 0 | 0 | 0 | NA | 40 | NA | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0.19 | NA | 44 | 0.04* |
| + | 0 | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||||
| Ammarnäs | − | 98 | 0 | 1 | 28 | 0 | 0 | 0 | 0.71 | 240 | <2.2e‐16*** | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 31 | 4.9e‐08*** | 0.74 | 271 | <2.2e‐16*** |
| + | 1 | 1 | 0 | 45 | 4 | 0 | 62 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | ||||||||||
| ST1 | − | 40 | 0 | 0 | 0 | 0 | 0 | 0 | NA | 40 | NA | 10 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 11 | NA | 0 | 51 | NA |
| + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||||
| ST2 | − | 4 | 0 | 0 | 20 | 0 | 0 | 0 | 0.62 | 40 | 0.046* | 0 | 0 | 0 | 0 | 0 | 0 | 4 | NA | 7 | NA | 0.12 | 47 | 0.18 |
| + | 0 | 0 | 0 | 15 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | ||||||||||
| ST3 | − | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0.58 | 40 | 3.0e‐05*** | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0.58 | 15 | 0.04* | 0.51 | 55 | 1.2e‐06*** |
| + | 7 | 2 | 5 | 0 | 1 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 11 | ||||||||||
| ST4 | − | 23 | 1 | 0 | 0 | 0 | 0 | 1 | 0.42 | 40 | 0.04* | 4 | 0 | 0 | 0 | 0 | 0 | 2 | 0.13 | 22 | 0.22 | 0.37 | 62 | 0.0007** |
| + | 10 | 0 | 1 | 1 | 0 | 0 | 3 | 4 | 0 | 0 | 4 | 0 | 0 | 8 | ||||||||||
| ST5 | − | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0.52 | 40 | 2.8e‐05*** | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0.89 | 20 | 3.0e‐05*** | 0.59 | 60 | 2.7e‐08*** |
| + | 7 | 3 | 3 | 2 | 0 | 0 | 9 | 0 | 0 | 0 | 1 | 0 | 0 | 11 | ||||||||||
| ST6 | − | 13 | 0 | 1 | 0 | 0 | 0 | 0 | 0.39 | 40 | 0.0006** | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 8 | 0.016* | 0.44 | 48 | 0.0001*** |
| + | 9 | 3 | 1 | 7 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | ||||||||||
| Tvedöra | − | 117 | 1 | 1 | 20 | 0 | 0 | 1 | 0.59 | 240 | 3.8e‐15*** | 26 | 0 | 0 | 1 | 0 | 0 | 8 | 0.56 | 83 | 2.2e‐08*** | 0.51 | 323 | 2.2e‐16*** |
| + | 34 | 8 | 10 | 25 | 1 | 3 | 19 | 5 | 0 | 0 | 5 | 0 | 0 | 38 | ||||||||||
NA, not applicable. *, P < 0.05; **, P < 0.001; ***, P < 0.0001.
In Tvedöra, male‐specific alleles were found at Dmrt1‐1 and Dmrt3 (alleles 294 and 281, respectively, Table 1), both of which were however missing in two males of 11. F ST values for these markers reached 0.167 and 0.084, respectively (with P values < 0.004 and 0.087 after correction for multiple testing, Appendix S2, Table S3). Although the F ST value associated with Dmrt3 is only close to significance after correction, the exact probability for the observed distribution of the male‐specific allele can be computed from combinatorial statistics as the ratio of 28 × 11!/(8! × 3!) = 42,240 (number of combinations of eight copies of allele 281 among 11 males, one copy each) over 44!/(8! × 36!) = 177,232,627 (number of combinations of these eight copies among 44 copies of Dmrt3), which amounts to P ~ 2.4 × 10−4. If we furthermore account for the fact that these copies only occurred in males that otherwise possess allele 294 at Dmrt1‐1, the probability becomes P ~ 1.3 × 10−5. The three other loci did not show significant sex differences in allele frequencies. Between‐sex F ST values averaged 0.042 over the five markers (as compared to −0.0005 over all other LG2 markers; Rodrigues et al. 2015). Locus‐specific F ST values are plotted along the consensus female recombination map in Figure 3, showing the contrasted patterns of sex differentiation between populations, and localizing the small differential segment in Tvedöra, identified through Dmrt1‐1 and Dmrt3. From this recombination map, Dmrt1 clearly has much tighter linkage with Dmrt3 than with Kank1 (~1 cM vs. 25 cM), suggesting that Kank1 and Dmrt1 lie much further apart on the physical map than expected (e.g., as a result of an inversion), or are separated by a strong recombination hotspot.
Figure 3.
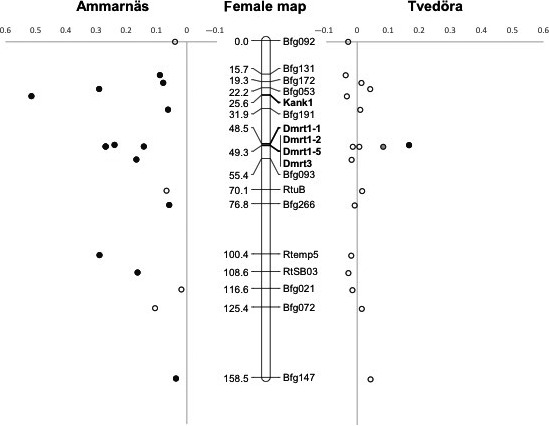
Consensus female recombination map based on all 12 families from Ammarnäs and Tvedöra. Between‐sex F ST values are indicated for each marker, either left (Ammarnäs) or right (Tvedöra). Indicated in bold are the five markers developed here. Loci with significant F ST values are indicated by black symbols, and Dmrt3 in Tvedöra (with a distribution of the male‐specific allele that departs significantly from random) by a gray symbol.
Sibship analyses confirmed that the Dmrt1‐1 and Dmrt3 alleles identified as male specific in Tvedöra were indeed located on nonrecombining Y haplotypes. The most common Y haplotype had fixed allele 174 at Kank1, 294 at Dmrt1‐1, 198 at Dmrt1‐2, 301 at Dmrt1‐5, and 281 at Dmrt3. Three other closely related Y haplotypes differed at one or two loci (changes to allele 165 or 178 at Kank1, 302 at Dmrt1‐5, and/or 276 at Dmrt3). These analyses also revealed a highly significant association between inheritance of a male‐specific Y haplotype and offspring phenotypic sex (Table 2), both in metamorphs (n = 240, Dxy = 0.59, P = 3.8 × 10−15) and in froglets (n = 83, Dxy = 0.56; P = 2.2 × 10−8). Among the six families analyzed, five turned out to possess a Y haplotype, which correlated significantly with offspring maleness score, although with some variation among families (n = 47–60 each, Dxy ranging 0.12–0.59). The only family lacking a Y haplotype (ST1) displayed an extremely female‐biased sex ratio (50 daughters vs. one son).
In both populations, the male specificity of local Y haplotypes, as measured by Dxy, increased from the juvenile to the adult stages: In Ammarnäs, sex association was imperfect among metamorphs (Dxy = 0.71; Fig. 4A), mostly due to some offspring with undifferentiated gonads and two XY females, but perfect in both froglets and adults (Dxy = 1.0). In Tvedöra, Dxy was below 0.60 in juveniles (Fig. 4B), mostly due to frequent XY individuals with ovaries, but reached 0.82 in adults, where no female had a Y haplotype, while two males lacked it.
Figure 4.
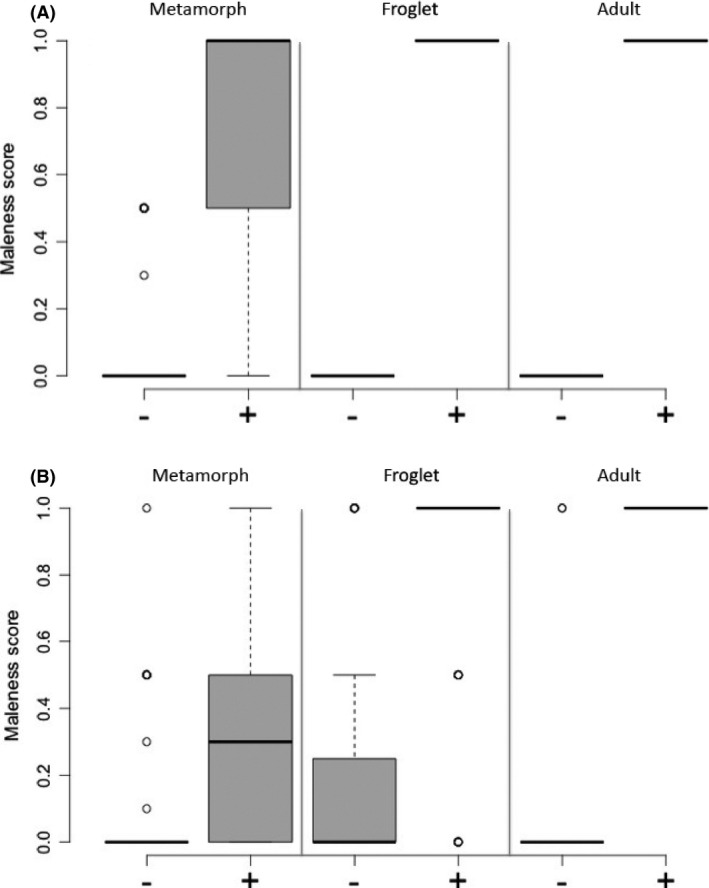
Boxplots of maleness scores for individuals with (+) or without (−) the local Y‐specific Dmrt1‐1 alleles in metamorphs, froglets, and adults from Ammarnäs (A) and Tvedöra (B).
Discussion
The first and main aim of this study was to identify a small sex‐linked region on LG2 in a population from the “semidifferentiated race,” in which previous studies had failed to find any XY differentiation despite strong evidence for a role of this linkage group in sex determination. This aim was entirely fulfilled: our genotyping of adult males and females from Tvedöra uncovered a small nonrecombining segment on LG2 that displays significant XY differentiation (Fig. 3). Male‐specific alleles were identified at Dmrt1‐1 and Dmrt3 but not at Dmrt1‐2 and Dmrt1‐5, which lie in‐between (and thus necessarily also belong to the nonrecombining segment) but had fixed alleles on the Y haplotype that also segregate on the X chromosomes. Sex association was further confirmed by sibship analyses, which showed a strong association between offspring phenotypic sex and inheritance of the local Y haplotype (Fig. 4). This result constitutes an important step toward the identification of the sex locus, given that all other LG2 markers investigated so far showed no differentiation.
This differential segment is much shorter in Tvedöra than in Ammarnäs, with an estimated length on the female recombination map ranging between 0.8 cM (distance between Dmrt1‐1 and Dmrt3) and 23 cM (distance between Bfg191 and Bfg093), as compared to a minimal length of 143 cM in Ammarnäs (distance between Bfg131 and Bfg147). It is also less differentiated, with an F ST of 0.061 as compared to 0.230 in Ammarnäs for this specific region (averages over the Dmrt markers). The Tvedöra and Ammarnäs Y haplotypes differ in fact markedly, bearing distinct alleles at each of the four Dmrt markers (as opposed to the X‐linked alleles that are largely shared).
This smaller and less differentiated SDR associates with weaker masculinizing effects. The five Tvedöra families with a Y haplotype displayed lower Dxy values than Ammarnäs families, mostly due to a high number of XY individuals presenting ovaries at the metamorph and froglet stages. Interestingly, these discrepancies between phenotypic and genotypic sex decreased between the juvenile and adult stages, suggesting that sex differentiation can be delayed beyond the froglet stage in the “semidifferentiated race.” Occasional XY females that reach reproductive age might actually account for the overall absence of XY differentiation in Tvedöra, as recombination patterns in frogs seem to depend on phenotypic rather than genotypic sex (the fountain‐of‐youth hypothesis; Perrin 2009; Matsuba et al. 2010). Reciprocally, X‐specific haplotypes in Tvedöra seemingly have weaker feminizing effects, as shown by the occurrence of XX males. The progeny of one of the two males (of 11) that lacked a Y haplotype could be analyzed and revealed an extreme female bias (50 daughters for one son), further supporting an XX paternal genotype. This result confirms that sex reversals account for some of the variance in sex ratios among families and provides further support for a sex‐determining role of the Y haplotypes identified here.
It is obviously of interest that the small nonrecombining segment in Tvedöra encompasses Dmrt1, a gene from the sex‐determining cascade that plays a key role in sex determination and sexual dimorphism throughout all metazoans. Whether this gene is directly involved in the patterns documented here (i.e., is the sex locus), or only turned out by chance to be trapped in the nonrecombining segment, is an open question. The classical paradigm of sex‐chromosome evolution predicts absence of Y polymorphism in the SDR (as a result of complete arrest of XY recombination and ensuing strong genetic drift and Hill‐Robertson interferences), which does not fit with the Dmrt1 polymorphism documented here. However, this classical paradigm was specifically developed to account for the highly differentiated sex chromosomes documented in lineages with purely GSD such as mammals, birds, and Drosophila; it has little relevance for systems with homomorphic sex chromosomes such as found in many fish, amphibians, and nonavian reptiles, where nongenetic effects may also contribute to sex determination. Sex reversals and occasional XY recombination are expected to refuel the genetic variance at the SDR. In the specific case of R. temporaria, furthermore, the patterns of sex determination and gonadal differentiation are known to be polymorphic both within and among populations (Witschi 1929, 1930; Rodrigues et al. 2013, 2014, 2015); sex determination varies from entirely genetic in some families to entirely nongenetic in others (e.g., Brelsford et al. 2016c). Hence, some polymorphism is indeed expected at the SDR.
This issue will clearly not be settled with data in hand, but our results do suggest further investigations that might help to clarify this point. Extension of analyses in Tvedöra to genomic regions between Dmrt1 and Kank1 (which does not seem to belong to the SDR), and downstream of Dmrt3 (which is apparently involved), might help evaluate more precisely the extent of the SDR and possibly identify alternative candidate genes. Similar analyses in Ammarnäs would not be informative, given that most of the sex chromosome belongs to the nonrecombining SDR. Although the strongly masculinizing/feminizing effects of sex‐specific haplotypes in Ammarnäs might possibly stem from the distinct Dmrt1 alleles segregating in this population, linkage with other genes from the sex‐determining pathway located on the same chromosome (such as Amh) is expected to contribute as well.
Investigations of polymorphisms in this genomic region should also be extended to a broader geographic scale. The “differentiated sex race” occurs in both alpine and boreal climates (Witschi 1930). It would be worth checking whether the same Dmrt1 Y haplotypes as in Ammarnäs are found in Alpine populations, or whether different Y haplotypes independently evolved in these distinct geographic areas. Similarly, populations from the “undifferentiated sex race,” spread in milder climates (from southern England, Netherlands, and central Germany down to the Jura mountains; Witschi 1930) should be investigated for the same markers. If sex determination in the undifferentiated sex race is purely nongenetic, as hypothesized by Rodrigues et al. (2015), then we predict a complete absence of sex differentiation in the genomic region surrounding Dmrt1. On a broader scale, the question arises whether the “sex races” described in other species of Ranidae (e.g., Pflüger 1881; Swingle 1926; Hsü and Liang 1970; Gramapurohit et al. 2000) also differ in the size and differentiation of nonrecombining segments on their sex chromosomes.
It is worth noting that the chromosome pair under focus, corresponding to X. tropicalis scaffold 1, has been independently co‐opted for sex determination in different lineages of amphibians, including species of Bufonidae, Hylidae and Ranidae (e.g., Sumida and Nishioka 2000; Miura 2007; Brelsford et al. 2013; Dufresnes et al. 2015). Recent investigations on four European species of tree frogs from the Hyla arborea group have furthermore shown these species to share a small SDR that also contains Dmrt1 (Brelsford et al. 2016b). Hence, our results substantiate the view that such recurrent convergences of sex determination toward a limited set of chromosome pairs might result from the co‐option of small genomic regions that harbor key genes from the sex‐determination pathway (Graves and Peichel 2010; O'Meally et al. 2012; Brelsford et al. 2013).
Conflict of Interest
None declared.
Data Accessibility
Genotyping data of all Dmrt1 markers are deposited in Dryad database, doi:10.5061/dryad.kp296.
Supporting information
Appendix S1.
Text S1. Seven scaffolds from the draft genome of Rana temporaria, containing, respectively, the five Dmrt1 exons, Kank1 intron 1, and Dmrt3 intron 1.
Text S2. Transcript sequences of R. temporaria Dmrt1 in five froglets.
Text S3. Concatenated sequences of three Dmrt1 polymorphic sites for 26 individuals from Ammarnäs and Tvedöra.
Appendix S2
Table S1. Primer pairs and PCR conditions for amplifying Dmrt1 transcript and individual exons.
Table S2. Primers pairs and PCR conditions for genotyping.
Table S3. Between‐sex F ST values in Ammarnäs and Tvedöra.
Acknowledgments
J. Loman and Y. Vuille contributed to the field sampling, Y. Vuille to the rearing and sexing of froglets, and J. Leuenberger to the marker development. We benefited from useful comments by Paris Veltsos on a previous version of the manuscript. The computations were performed at the Vital‐IT (http://www.vital-it.ch) Center for high‐performance computing of the SIB Swiss Institute of Bioinformatics. Capture permits were delivered by the prefectures of Skåne and Västerbotten counties for Tvedöra (522‐363‐2013) and Ammarnäs (522‐3396‐2013). An additional permit was delivered for Ammarnäs as part of the nature reserve of Vindelfjällen (521‐3407‐2013). Ethical permits were delivered by the Swedish Board of Agriculture for Tvedöra (M 19‐13) and Ammarnäs (A 10‐13) and by the Veterinary Office of the Canton Vaud, Switzerland (authorization 2287). This work was supported by the Swiss National Science Foundation (grants 31003A_147091 to NP; and Sinergia grant CRSII3_147625 to NP, John Pannell and Mark Kirkpatrick).
References
- Alho, J. S. , Matsuba C., and Merilä J.. 2010. Sex reversal and primary sex ratios in the common frog (Rana temporaria). Mol. Ecol. 19:1763–1773. [DOI] [PubMed] [Google Scholar]
- Beukeboom, L. W. , and Perrin N.. 2014. The evolution of sex determination. Oxford Univ. Press, Oxford, UK. [Google Scholar]
- Brelsford, A. , Stöck M., Betto‐Colliard C., Dubey S., Dufresnes C., Jourdan‐Pineau H., et al. 2013. Homologous sex chromosomes in three deeply divergent anuran species. Evolution 67:2434–2440. [DOI] [PubMed] [Google Scholar]
- Brelsford, A. , Dufresnes C., and Perrin N.. 2016a. High‐density sex‐specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity 116:177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelsford, A. , Dufresnes C., and Perrin N.. 2016b. Trans‐species variation in Dmrt1 is associated with sex determination in four European tree‐frog species. Evolution 70:840–847. [DOI] [PubMed] [Google Scholar]
- Brelsford, A. , Rodrigues N., and Perrin N.. 2016c. High‐density linkage maps fail to detect any genetic component to sex determination in a Rana temporaria family. J. Evol. Biol. 29:220–225. [DOI] [PubMed] [Google Scholar]
- Cano, J. M. , Li M. H., Laurila A., Vilkki J., and Merilä J.. 2011. First‐generation linkage map for the common frog Rana temporaria reveals a sex linkage group. Heredity 107:530–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, R. H. , and Nagahama Y.. 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208:191–363. [Google Scholar]
- Dufresnes, C. , Borzée A., Horn A., Stöck M., Ostini M., Sermier R., et al. 2015. Sex‐chromosome homomorphy in Palearctic tree frogs results from both turnovers and X‐Y recombination. Mol. Biol. Evol. 32:2328–2337. [DOI] [PubMed] [Google Scholar]
- Eggert, C. 2004. Sex determination: the amphibian models. Reprod. Nutr. Dev. 44:539–549. [DOI] [PubMed] [Google Scholar]
- Evans, B. J. , Pyron R. A., and Wiens J. J.. 2012. Polyploidization and sex chromosome evolution in amphibians Pp. 385–410 in Soltis P. S. and Soltis D. E., eds. Polyploidy and genome evolution. Springer, Heidelberg. [Google Scholar]
- Gosner, K. L. 1960. A simplified table for staging anuran embryos and larvae with notes on identification. Herpetologica 16:183–190. [Google Scholar]
- Goudet, J. 1995. FSTAT (version 1.2): a computer program to calculate F‐statistics. J. Hered. 86:485–486. [Google Scholar]
- Gramapurohit, N. P. , Shanbhag B. A., and Saidapur S. K.. 2000. Pattern of gonadal sex differentiation, development, and onset of steroidogenesis in the frog, Rana curtipes . Gen. Comp. Endocrinol. 119:256–264. [DOI] [PubMed] [Google Scholar]
- Graves, J. A. M. , and Peichel C. L.. 2010. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 11:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P. , Falls K., and Crook S.. 1990. Documentation for CRIMAP, version 2.4. St. Louis, MO: Washington Univ. School of Medicine. [Google Scholar]
- Grossen, C. , Neuenschwander S., and Perrin N.. 2011. Temperature‐dependent turnovers in sex‐determination mechanisms: a quantitative model. Evolution 65:64–78. [DOI] [PubMed] [Google Scholar]
- Guerrero, R. , Kirkpatrick M., and Perrin N.. 2012. Cryptic recombination in the ever‐young sex chromosomes of Hylid frogs. J. Evol. Biol. 25:1947–1954. [DOI] [PubMed] [Google Scholar]
- Haczkiewicz, K. , and Ogielska M.. 2013. Gonadal differentiation in frogs: how testes become shorter than ovaries. Zoolog. Sci. 30:125–134. [DOI] [PubMed] [Google Scholar]
- Hillis, D. M. , and Green D. M.. 1990. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J. Evol. Biol. 3:49–64. [Google Scholar]
- Hsü, C. Y. , and Liang H. M.. 1970. Sex races of Rana catesbeiana in Taiwan. Herpetologica 26:214–221. [Google Scholar]
- Matsuba, C. , Miura I., and Merilä J.. 2008. Disentangling genetic vs. environmental causes of sex determination in the common frog, Rana temporaria . BMC Genet. 9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba, C. , Alho J. S., and Merilä J.. 2010. Recombination rate between sex chromosomes depends on phenotypic sex in the common frog. Evolution 64:3634–3637. [DOI] [PubMed] [Google Scholar]
- Matsuda, M. , Nagahama Y., Shinomiya A., Sato T., Matsuda C., Kobayashi T., et al. 2002. DMY is a Y‐specific DM‐domain gene required for male development in the medaka fish. Nature 417:559–563. [DOI] [PubMed] [Google Scholar]
- Miura, I. 2007. An evolutionary witness: the frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex. Dev. 1:323–331. [DOI] [PubMed] [Google Scholar]
- Nanda, I. , Kondo M., Hornung U., Asakawa S., Winkler C., Shimizu A., et al. 2002. A duplicated copy of DMRT1 in the sex‐determining region of the Y chromosome of the medaka, Oryzias latipes . Proc. Natl Acad. Sci. USA 99:11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogielska, M. , and Kotusz A.. 2004. Pattern and rate of ovary differentiation with reference to somatic development in anuran amphibians. J. Morphol. 259:41–54. [DOI] [PubMed] [Google Scholar]
- O'Meally, D. , Ezaz T., Georges A., Sarre S. D., and Graves J. A. M.. 2012. Are some chromosomes particularly good at sex? Insights from amniotes Chromosome Res. 20:7–19. [DOI] [PubMed] [Google Scholar]
- Perrin, N. 2009. Sex reversal: a fountain of youth for sex chromosomes? Evolution 63:3043–3049. [DOI] [PubMed] [Google Scholar]
- Pflüger, E. 1881. Über das geschlechtsbestimmenden Ursachen und die Geschlechtsverhältnisse der Frösche. Arch. F. Gesamte Phys. 29:13–40. [Google Scholar]
- R Core Team . 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Rodrigues, N. , Betto‐Colliard C., Jourdan‐Pineau H., and Perrin N.. 2013. Within‐population polymorphism of sex‐determination systems in the common frog (Rana temporaria). J. Evol. Biol. 26:1569–1577. [DOI] [PubMed] [Google Scholar]
- Rodrigues, N. , Merilä J., Patrelle C., and Perrin N.. 2014. Geographic variation in sex‐chromosome differentiation in the common frog (Rana temporaria). Mol. Ecol. 23:3409–3418. [DOI] [PubMed] [Google Scholar]
- Rodrigues, N. , Vuille Y., Loman J., and Perrin N.. 2015. Sex‐chromosome differentiation and ‘sex races’ in the common frog (Rana temporaria). Proc. R. Soc. B. 282:20142726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, N. , Yuille Y., Bresford A., and Perrin N.. 2016. The genetic contribution to sex determination and number of sex chromosomes vary among populations of common frogs (Rana temporaria). Heredity. doi:10.1038/hdy.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl, M. 2004. Sex chromosome evolution in non‐ mammalian vertebrates. Curr. Opin. Genet. Dev. 14:634–641. [DOI] [PubMed] [Google Scholar]
- Schmid, M. , and Steinlein C.. 2001. Sex chromosomes, sex‐linked genes, and sex determination in the vertebrate class Amphibia Pp. 143–176 in Scherer G., Schmid M., eds. Genes and mechanisms in vertebrate sex determination. Birkhäuser Verlag, Basel. [DOI] [PubMed] [Google Scholar]
- Schmid, M. , Nanda I., Steinlein C., Kausch K., and Haaf T.. 1991. Sex‐determining mechanisms and sex chromosomes in amphibian Pp. 393–430 in Green D. M. and Sessions S. K., eds. Amphibian cytogenetics and evolution. Academic Press, San Diego. [Google Scholar]
- Schmid, M. , Steinlein C., Bogart J. P., Feichtinger W., León P., La Marca E., et al. 2010. The chromosomes of terraranan frogs. Insights into vertebrate cytogenetics. Cytogenet. Genome Res. 129:1–4. [DOI] [PubMed] [Google Scholar]
- Smith, C. A. , Roeskler K. N., Ohnesorg T., Cummins D. M., Farlie P. G., Doran T. J., et al. 2009. The avian Z‐linked gene DMRT1 is required for male sex determination in the chicken. Nature 461:267–271. [DOI] [PubMed] [Google Scholar]
- Somers, R. H. 1962. A new asymmetric measure of association for ordinal variables. Am. Sociol. Rev. 27:799–811. [Google Scholar]
- Stöck, M. , Horn A., Grossen C., Lindtke D., Sermier R., Betto‐Colliard C., et al. 2011. Ever‐young sex chromosomes in European tree frogs. PLoS Biol. 9:e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöck, M. , Savary R., Betto‐Colliard C., Biollay S., Jourdan‐Pineau H., and Perrin N.. 2013. Low rates of X‐Y recombination, not turnovers, account for homomorphic sex chromosomes in several diploid species of Palearctic green toads (Bufo viridis subgroup). J. Evol. Biol. 26:674–682. [DOI] [PubMed] [Google Scholar]
- Sumida, M. , and Nishioka M.. 2000. Sex‐linked genes and linkage maps in amphibians. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126:257–270. [DOI] [PubMed] [Google Scholar]
- Swingle, W. W. 1926. The germ cells of anurans. II. An embryological study of sex differentiation in Rana catesbeiana . J. Morphol. 41:441–546. [Google Scholar]
- Volff, J. N. , Nanda I., Schmid B., and Schartl M.. 2007. Governing sex determination in fish: regulatory putches and ephemeral dictators. Sex. Dev. 1:85–99. [DOI] [PubMed] [Google Scholar]
- Voorrips, R. E. 2002. MapChart: software for graphical presentation of linkage maps and QTLs. J. Hered. 93:77–78. [DOI] [PubMed] [Google Scholar]
- Witschi, E. 1929. Studies on sex differentiation and sex determination in amphibians. III. Rudimentary hermaphroditism and Y chromosome in Rana temporaria . J. Exp. Zool. 54:157–223. [Google Scholar]
- Witschi, E. 1930. Studies on sex differentiation and sex determination in amphibians. IV. The geographical distribution of the sex races of the European grass frog (Rana temporaria, L.). J. Exp. Zool. 56:149–165. [Google Scholar]
- Yoshimoto, S. , Okada E., Umemoto H., Tamura K., Uno Y., Nishida‐Umehara C., et al. 2008. A W‐linked DM‐domain gene, DM‐W, participates in primary ovary development in Xenopus laevis . Proc. Natl Acad. Sci. USA 105:2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1.
Text S1. Seven scaffolds from the draft genome of Rana temporaria, containing, respectively, the five Dmrt1 exons, Kank1 intron 1, and Dmrt3 intron 1.
Text S2. Transcript sequences of R. temporaria Dmrt1 in five froglets.
Text S3. Concatenated sequences of three Dmrt1 polymorphic sites for 26 individuals from Ammarnäs and Tvedöra.
Appendix S2
Table S1. Primer pairs and PCR conditions for amplifying Dmrt1 transcript and individual exons.
Table S2. Primers pairs and PCR conditions for genotyping.
Table S3. Between‐sex F ST values in Ammarnäs and Tvedöra.
Data Availability Statement
Genotyping data of all Dmrt1 markers are deposited in Dryad database, doi:10.5061/dryad.kp296.


