Abstract
Recently, impressive technical advancements have been made in the isolation and validation of mammary stem cells and cancer stem cells (CSCs), but the signaling pathways that regulate stem cell self-renewal are largely unknown. Further, CSCs are believed to contribute to chemo- and radioresistance. In this study, we used the MMTV-Neu-Tg mouse mammary tumor model to identify potential new strategies for eliminating CSCs. We found that both luminal progenitor and basal stem cells are susceptible to genetic and epigenetic modifications, which facilitate oncogenic transformation and tumorigenic potential. A combination of the DNMT inhibitor 5-azacytidine and the HDAC inhibitor butyrate markedly reduced CSC abundance and increased the overall survival in this mouse model. RNA-seq analysis of CSCs treated with 5-azacytidine plus butyrate provided evidence that inhibition of chromatin modifiers blocks growth-promoting signaling molecules such as RAD51AP1 and SPC25, which play key roles in DNA damage repair and kinetochore assembly. Moreover, RAD51AP1 and SPC25 were significantly overexpressed in human breast tumor tissues and were associated with reduced overall patient survival. In conclusion, our studies suggest that breast CSCs are intrinsically sensitive to genetic and epigenetic modifications and can therefore be significantly affected by epigenetic-based therapies, warranting further investigation of combined DNMT and HDAC inhibition in refractory or drug-resistant breast cancer.
Introduction
Cancer stem cells (CSC), a small subpopulation of cells within tumors, have a characteristic feature of self-renewal, a process that drives tumorigenesis and differentiation contributing to cellular heterogeneity in tumors. CSCs are resistant to chemotherapy and radiation therapy and are considered a major obstacle in cancer treatment (1–3). This results in relapse of breast cancer in about 20–45% of patients within years or decades after treatment. Thus, an effective cancer therapy requires elimination of all tumorigenic cells in the tumor (4). Breast tumors contain a heterogeneous population of cells such as neoplastic epithelial cells, mesenchymal stem cells, infiltrating immune cells, cancer-associated fibroblasts, angiogenic vascular cells, and erythrocytes (5). However, the molecular mechanisms that reprogram normal stem cells into abnormal CSCs are poorly understood.
Stem cells have much longer life span compared to their progeny and therefore, have a greater opportunity to accumulate genetic mutations (6). Hematopoietic stem cells provide the best evidence that normal stem cells could be the target of transforming genetic mutations, which can render them independent of growth signals and undergo uncontrolled proliferation and tumorigenesis. Recent studies have shown that epigenome also plays an important role in cancer initiation and propagation by regulating stem cells (7, 8). For example, ARID1A, a member of SWI/SNF family, is mutated in more than 50% of human cancers; however, this mutation does not directly stimulate tumor formation, rather it determines the epigenetic changes that leads to tumor propagation (9). Thus, the tumorigenic potential of ARID1A resides in its ability to alter the epigenetic profile rather than the DNA sequence. In this context, our recent studies have shown that DNA methyltransferase 1 (DNMT1) plays a critical role in the maintenance of mammary stem/progenitor cells and CSCs (10). Using mammary gland-specific Dnmt1-knockout mice, we have shown that DNMT1 is indispensable for MaSC formation and that Dnmt1 deletion protects mice from mammary tumorigenesis by limiting CSC pool (10). Therefore, targeting the epigenetic modifiers like DNA methylation offers a promising treatment option for human cancers.
Epigenetic modifications represent early events in tumorigenesis (11, 12). Interestingly, unlike genetic mutations, the epigenetic alterations are reversible as proven by the re-expression of tumor suppressor genes by DNMT inhibitors (13). 5-azacytidine (5-AzaC, Vidaza) and 5-aza-2′-deoxycytidine (5-AzaDC, Decitabine) are the most successful epigenetic drugs that are most widely used in clinics (14, 15). However, their use is restricted due to their toxicity and poor stability. Interestingly, combinations of 5-AzaC or 5-AzaDC with histone deacetylation inhibitors (HDACi) have been approved by FDA and European Medicines Agency (EMA) for treatment of hematological malignancies (16). HDACs are upregulated in a wide variety of cancers, and HDACi have long been studied in clinical settings. These inhibitors produce a global effect on the level of acetylation of histone proteins (17). Our recent studies have shown that a combination therapy using 5-AzaC plus butyrate targets CSCs (10). However the impact of this drug combination on CSCs at genome level has not been investigated. In the current study, we provide evidence that a combination of DNMT and HDAC inhibitors not only reduces the tumor mass but also targets CSCs and differentially regulates genes that are involved in tumor growth. Thus, this combination could be considered as an effective therapeutic strategy for breast cancer treatment. Since this combination reduces the pool of drug-resistant CSCs, it can also be used to treat breast cancer patients who have developed resistance to hormone therapies such as trastuzumab.
Materials and Methods
Cell lines
The human breast cancer cell line, CAL51 was obtained from the DSMZ (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures) in October 2011. MCF10A4, an invasive breast cancer cell line, was kindly provided by Dr. Fred Miller at the Barbara Ann Karmanos Cancer Institute, Detroit, MI in April 2009. The mouse mammary tumor cell line 4T1 was obtained from ATCC, Manassas, VA in July 2011. Cell lines from ATCCC and DSMZ have been thoroughly tested and authenticated and uses morphology, karyotyping, and PCR based approaches to confirm the identity of the cell lines. CAL51 cells were grown in DMEM medium supplemented with 20% fetal bovine serum (FBS), glutamine (1%) and penicillin (1%). MCF10A4 cells were grown in DMEM/F12 media supplemented with Donor horse serum (5%), glutamine (1%), penicillin (1%), EGF (20 ng/ml), insulin (10 μg/ml) and hydrocorticosone (500 ng/ml). 4T1 cells were cultured in RPMI medium supplemented with 10% FBS, glutamine (1%) and penicillin (1%). All these cell lines have been routinely tested for mycoplasma contamination using the Universal mycoplasma detection kit obtained from ATCC (Manassas, VA) and the last mycoplasma test was performed in July 2014. Mycoplasma-free cell lines were used in all of our experiments.
Animals
Balb/cAnNCr (Stock #01B05) mice were obtained from the National Cancer Institute. FVB/NJ (Stock #001800), MMTV-Neu-Tg (Stock #005038), athymic Balb/c (Nu/Nu, Stock #007870) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (Stock #00557) mice were obtained from Jackson Laboratories. All these mice were bred and maintained in Augusta University Animal Facility in accordance with the guidelines of the Institutional Animal Care Use Committee.
Drug Treatment
For syngeneic tumor transplant studies, 4T1 cells (0.1×106) were injected into the mammary fat pad of Balb/c mice. Twenty-four hour after the tumor cell injection, 5-AzaC (0.5 mg/21 days release) and butyrate (10 mg/21 day release) tablets were implanted using trochar (Innovative Research of America). Similarly, in another group of mice, 24 h after transplantation, salinomycin (5 mg/kg, intraperitoneal injection) treatment was initiated. For in vitro studies, cells were treated with 5-AzaC (1 μg/ml), 5-AzaDC (1 μg/ml), TSA (100 nM), butryate (1 mM), and salinomycin (1 μg/ml).
RNA-seq analysis
RNA samples extracted from mammospheres and tumorospheres were subjected to cDNA library construction (ScriptSeq RNA-Seq Preparation v2 from Epicenter). The quality of the sample was checked using Agilent Bioanalyzer and then subjected to Illumina sequencing - Hiseq 2000, paired end 50 cycles V3. CASAVA1.8.2 was used to generate fastq files. To analyze the genome, we used Top Hat 2.0.1 software and detected the differential expression of transcripts (Cuffdiff/Cufflink 2.2.0). We detected raw mapped reads and normalized reads per kilobase per million mapped reads.
Details for single-cell preparation from mammary gland, generation of mammospheres and tumorospheres, flow cytometry, immunofluorescence staining and microscopic imaging, RNA isolation and real-time PCR, cell cycle, and clonogenic assays are given in Supplementary Materials section.
Statistical analysis
Statistical analysis was done using Student’s t-test with two-tail distribution by Graph Pad Software. Biological function and pathway enrichment analysis were carried out using DAVID (http://david.abcc.ncifcrf.gov/), UCSC Cancer Browser (https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/) and IPA (http://www.ingenuity.com/products/pathways_analysis.html) software. Graph Pad, Sigma Plot and Excel were used to draw figures. Kaplan-Meier analyses were used to determine group differences in tumor-free survival (http://kmplot.com/analysis/). The web interface of GOBO was used (http://co.bmc.lu.se/gobo/) for gene set analysis and co-expressed genes.
Results
Lin−CD49f+CD24+ cells have tumor propagating and metastatic potential
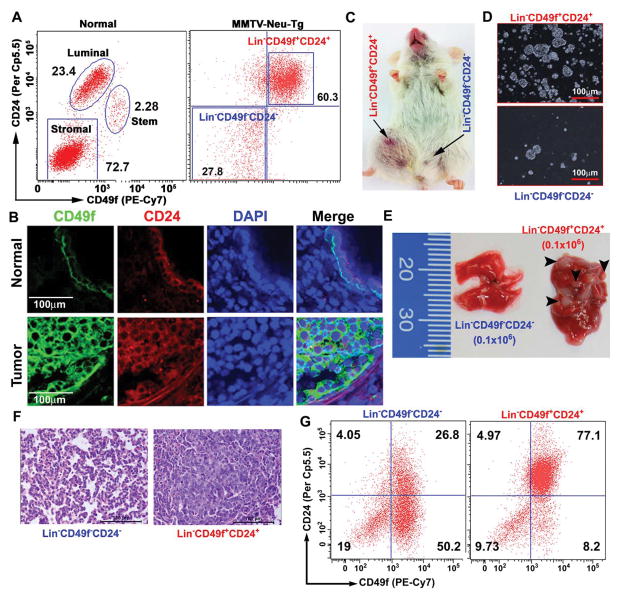
To distinguish normal mammary stem cells (MaSC) and cancer-propagating stem cells (CSC), we isolated mammary stem/progenitor enriched cell populations from normal mammary glands (16-week-old) and CSC cells from tumor tissues of MMTV-Neu-Tg mice (9-month-old) using CD24/CD49f cell-surface markers. We observed three distinct cell populations in normal mammary glands: Lin−CD49fhighCD24+ (basal myoepithelial stem cells), Lin−CD49f+CD24high (luminal progenitor cells), and Lin−CD49f−CD24− (stromal cells). In contrast, there were only two distinct cell populations in tumor tissues: (Lin−CD49f+CD24+ and Lin−CD49f−CD24−) (Fig. 1A). As shown in Fig. 1B, the expression of CD49f and CD24 was dramatically increased in tumor tissues compared to normal mammary glands. Previous studies have shown that only a few cells within the tumor, the CSCs, are tumorigenic and possess the metastatic phenotype (18, 19). Thus, to test the tumor-forming potential of Lin−CD49f+CD24+ and Lin−CD49f−CD24− cells, we injected these cells into mammary fat pad of 12-week-old NOD/SCID mice. As shown in Fig. 1C, Lin−CD49f+CD24+ cells were able to form tumors while the Lin−CD49f−CD24− cells were not. Further, we investigated the self-renewal capacity of Lin−CD49f+CD24+ and Lin−CD49f−CD24− cells, using tumorosphere-forming (primary and secondary) and tumor-forming potentials (serial dilution, 102–105 cells). Lin−CD49f+CD24+ cells were able to form tumorospheres and tumors (as few as 102 cells) but Lin−CD49f−CD24− cells were unable to form either tumorospheres or tumors (Figs. 1D, S1A-C). We also tested the mammosphere-forming potential of the three distinct cell populations that were isolated from the normal mammary glands and found that Lin−CD49fhighCD24+ cells form larger size mammospheres than the Lin−CD49f+CD24low cells, while Lin−CD49f−CD24− cells were unable to form spheres (Fig. S1D).
Figure 1.
Lin−CD49f+CD24+ cells are tumor-propagating cells. A, Representative FACS dot blots showing three distinct cell populations in normal mammary gland: myoepithelial stem cells (Lin−CD49fhighCD24+), luminal progenitor cells (Lin−CD49f+CD24high) and stromal cells (Lin−CD49f−CD24−). However, MMTV-Neu tumor tissues show only two distinct cell populations: tumor-propagating stem cells (Lin−CD49f+CD24+) and non-tumorigenic cells (Lin−CD49f−CD24−) (n=5 mice). B, Representative confocal images (40x) showing increased expression of CD49f and CD24 in tumor tissue compared to the normal mammary tissues (n=3 mice). CD49f (Green) and CD24 (Red) and DAPI (Blue). Scale bars are 100 μm. C, Representative tumor growth with Lin−CD49f+CD24+ cells (102 cells) injected in NOD/SCID mice. No tumor was detected with Lin−CD49f−CD24− cells (105 cells) (n=3 mice). D, Representative images (10x) of tumorospheres generated from Lin−CD49f+CD24+ and Lin−CD49f−CD24− cells. 1,000 cells were used in each group (n=3 mice). E, Representative lung tissue images after tail vein injection of Lin−CD49f+CD24+ and Lin−CD49f−CD24− cells (0.1×106 in each) showing lung metastatic nodules observed in Lin−CD49f+CD24+ cells but not in Lin−CD49f−CD24− cells (n=3 mice in each). F, Representative H&E images (40x) of non-metastatic (Lin−CD49f−CD24−) and lung metastatic tumor (Lin−CD49f+CD24+) tissues (n=3 mice in each). G, Representative FACS dot plots for non-metastatic and metastatic lung tumor tissues showing increased Lin−CD49f+CD24+ cell population (n=3 mice).
To test the metastatic potential of Lin−CD49f+CD24+ cells, we serially diluted these cells and injected into 12-week-old-NOD/SCID mice. As shown in Fig. 1E, the Lin−CD49f+CD24+ cells (0.1×106) were able to form metastatic lesions in the lung. To confirm that the Lin−CD49f+CD24+ cells are indeed the drivers of metastasis to the lung, we prepared single-cell suspensions from the lung tissues and analyzed Lin−CD49f+CD24+ and Lin−CD49f−CD24− cell populations. As shown in Fig. 1G, significantly more Lin−CD49f+CD24+ cells in the lung tissues of mice that were injected with Lin−CD49f+CD24+ cells than in the lung tissues of mice that were injected with Lin−CD49f−CD24− cells. These findings suggest that Lin−CD49f+CD24+ cells could play a critical role in driving tumor metastasis to distant organs. Overall, these results provide evidence that Lin−CD49f+CD24+ cells are tumor-propagating and metastasis-driving CSCs.
Luminal progenitor and basal myoepithelial stem cells equally contribute to mammary tumor development
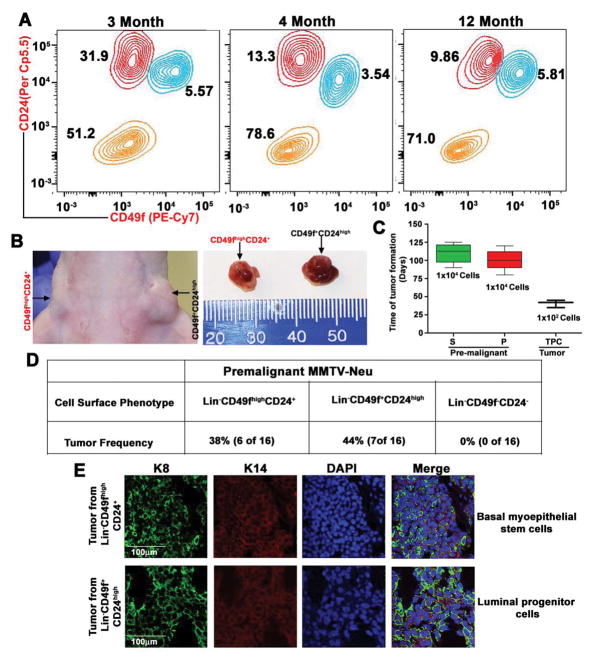
Despite ample evidence showing that cancer arises from populations of self-renewing stem cells, cancer can also arise from progenitor cells that originate from stem cells. For example, a mutation that enhances β-catenin signaling in granulocyte-macrophage progenitor cells causes a blast crisis in patients with chronic myeloid leukemia (20). This suggests that the mutated progenitor cells acquire the ability of self-renewal, a feature thought to be specific to stem cells, and undergo unlimited growth as cancer cells. To test the cell of origin in mammary tumorigenesis, either from basal myoepithelial stem cells or luminal progenitor cells, we used MMTV-Neu-Tg mouse. About 50–60% of MMTV-Neu-Tg mice develop spontaneous mammary tumors after acquiring either additional mutations or epigenetic modifications (21). We analyzed different cell populations in mouse mammary glands of MMTV-Neu-Tg premalignant mice (3 to 12 month-old) with no palpable tumors. Surprisingly, some mice as young as 3-month-old showed an abrupt increase in luminal progenitor population (Lin−CD49f+CD24high) whereas some 12-month-old mice did not show this abnormal phenotype (Fig. 2A), suggesting that these luminal progenitor cells might have undergone genetic/epigenetic modifications.
Figure 2.
Distinct myoepithelial stem cells and luminal progenitor cells represent the cells of origin. A, Representative contour plot of FACS gating shows an abnormal increase in luminal cells and shifting of myoepithelial stem cells toward luminal cells in 3-month-old MMTV-Neu premalignant mice. In contrast, 4- and 12-month-old mice show normal level of stem and luminal cells (n=3 mice in each). B, Tumors in Balb/c mice, driven by basal myoepithelial stem cells and luminal progenitor cells, which were derived from premalignant MMTV-Neu mice (n=3 mice). C, Tumor-forming potential of stem cells and luminal cells from premalignant MMTV-Neu mice, and tumor-propagating cells from MMTV-Neu mouse tumor tissue (n=3 mice). D, Frequency of tumor formation by different cell types, derived from MMTV-Neu premalignant mice. E, Representative confocal images (10x) of keratin 8 (green), keratin14 (red) and DAPI (blue) staining for sections of tumor tissues derived from transformed stem cells and luminal cells (n=3 mice). Scale bars are 100 μm.
To test whether this abnormal cell population leads to formation of mammary tumor, we injected different cell types (Lin−CD49fhighCD24+, Lin−CD49f+CD24high and Lin−CD49f−CD24−) into mammary fat pad and found that both transformed basal myoepithelial stem cells and luminal progenitor cells were able to form mammary tumors around 120 and 100 days, respectively, whereas the Lin−CD49f−CD24− cells were unable to form mammary tumors (Figs. 2B–D and S2A–B). The transformed luminal progenitor cells exhibit earlier onset of mammary tumorigenesis with accelerated tumor growth than the basal myoepithelial stem cells (Figs. 2B–D and S2B). Interestingly, tumors that developed from MMTV-Neu-Tg basal myoepithelial and luminal progenitor cell types are indistinguishable from the spontaneous tumors that develop in MMTV-Neu-Tg mice, indicating the stem cell-like property of self-renewal (Fig. S2C). To confirm this observation further, we stained the tumors derived from basal myoepithelial stem cells and luminal progenitor cells and found that both tumors express keratin 14, basal epithelial markers, and keratin 8, luminal epithelial markers (Figs. 2E). We confirmed this observation further with additional basal epithelial markers (p63 and Nestin) and luminal epithelial marker (Gata3) (Fig. S2D) (22). These results confirmed that Lin−CD49f+CD24+ cells are tumorigenic and that both transformed basal myoepithelial stem cells and luminal progenitor cells were able to form mammary tumors.
Transformed luminal progenitor and basal myoepithelial stem cells are susceptible to treatment with DNMT and HDAC inhibitors
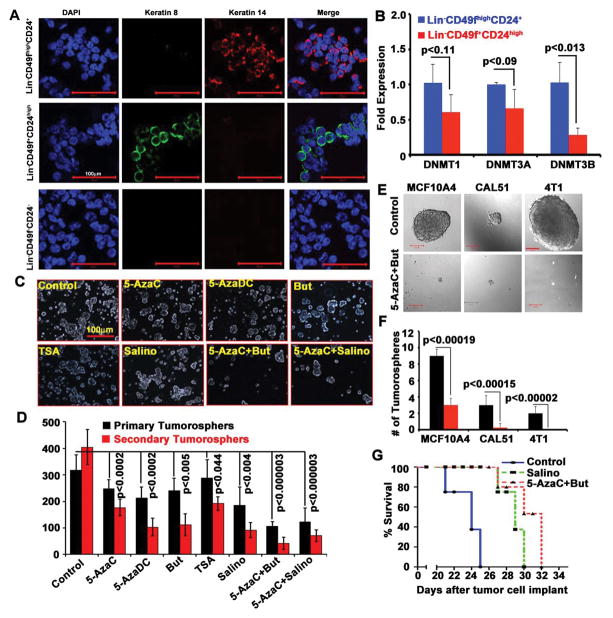
As shown in Fig. 2, both luminal progenitor and basal myoepithelial stem cells contribute to mammary tumor development in MMTV-Neu-Tg mice. However, previous studies have shown that the luminal progenitors are the origin of MMTV-Neu-Tg driven mammary tumor (23) and that basal stem cells are increased during later stages of the mammary tumor development, which contributes to drug resistance and metastasis (24, 25). Further, we have shown that DNMTs, especially DNMT1, play a critical role in regulation of luminal progenitor and basal myoepithelial stem cells and that functional inactivation of DNMT1 significantly reduces both of these populations (10). Therefore, we analyzed the expression of the three major DNMTs (DNMT1, DNMT3A, and DNMT3B) in luminal progenitor and basal stem cells that were isolated from premalignant MMTV-Neu-Tg mice. In order to confirm the purity and free of cross contamination within the specific cell types, we analyzed keratin 14 and keratin 8 in basal, luminal, and stromal cells and found that there was no cross contamination among these cell types (Fig. 3A). Then we analyzed DNMT1, DNMT3A, and DNMT3B in these cell populations and found that both luminal progenitor and basal stem cells express similar levels of DNMT1 and DNMT3A but basal stem cells express significantly more DNMT3B than luminal progenitor cells (Fig. 3B). This suggests that DNMT3B could play a critical role in regulation of basal stem cells. To test whether DNMT inhibitors have any effect in these populations, we generated tumorospheres from the tumor tissues derived from MMTV-Neu-Tg mice and treated with pan-DNMTs inhibitors 5-azacytidine (5-AzaC) and 5-aza-2-deoxycytidine (5-AzaDC). We also used HDACi TSA and butyrate (But), either alone or in combination with DNMT inhibitors. We used Salinomycin (Salino), the known stem cell inhibitor, as a positive control. We found that DNMT and HDACi significantly reduced CSCs (Fig. 3C–D) but the combination of DNMT and HDAC inhibitors (5-AzaC+But) had much more profound effect in reducing CSC populations.
Figure 3.
Treatment with DNMT and HDAC inhibitors restricts CSCs. A, Representative confocal images (25x) of keratin 14 (red), keratin 8 (green) and DAPI (blue) expression in sorted myoepithelial stem cells, luminal progenitor cells, and stromal cells showing free of cross contamination (n=5 mice). B, Real-time PCR showing the relative levels of DNMT1, 3A and DNMT3B expression in myoepithelial stem and luminal progenitor cells. Data represent mean ± SD for 5 mice. C, Representative tumorospheres images (10x) of control and treated (as indicated) for seven days. D, Quantification of primary and secondary tumorospheres derived from control and treated groups. Data represent mean + SD of 3 mice with quadruplicate wells (n=12 samples). E–F, Tumorosphere size and number in MCF10A4, CAL51, and 4T1 cells with or without treatment (5-azaC+But). G, Survival curve for mice: 4T1 cells were injected into mammary fat pad of Balb/c mice and then treated with and without 5-AzaC+But and salinomycin (n=5 mice in each).
We then tested whether the combination of 5-AzaC and butyrate can inhibit CSC signature in human breast cancer cell lines using two metastatic human breast cancer cell lines, MCF10A4 (26) and CAL51 (27), and one mouse metastatic mammary tumor cell line, 4T1 (28). We generated tumorospheres in the presence and absence of 5-AzaC and butyrate. This combination significantly reduced CSC pool (Figs. 3E–F) and colony-formation (Figs. S3A–B). To confirm the efficacy of this combination in vivo, we implanted 4T1 cells into mammary fat pad of Balb/c mice and then treated with 5-AzaC+butyrate as a combination therapy. Salinomycin was used as a positive control. We found that 5-AzaC+butyrate significantly increased the overall survival compared to Salinomycin treatment (Fig. 3G). Overall, these results provide evidence that DNA methylation plays a critical role in the regulation of CSCs in MMTV-Neu-Tg driven mammary tumor and that DNMT and HDAC inhibitors can be used as a combination therapy to reduce CSC pool.
Treatment with DNMT and HDAC inhibitors differentially regulates genes that are involved in basal stem cell driven breast cancer
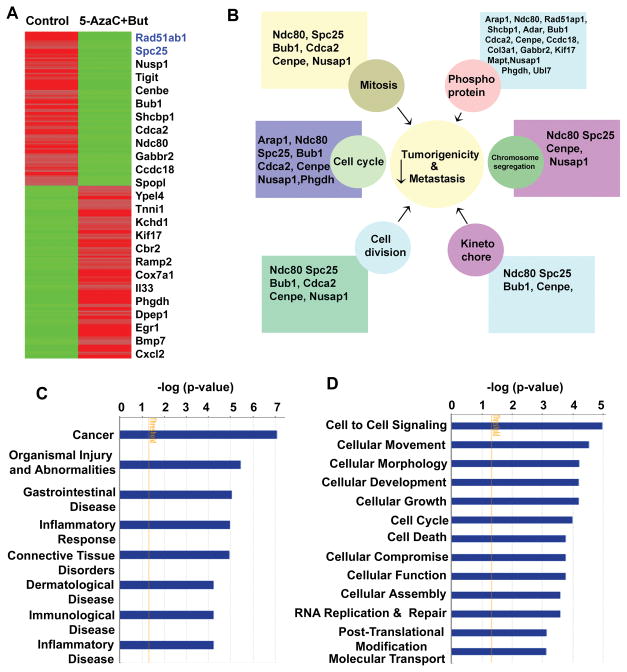
To test the efficacy of 5-AzaC+butyrate on CSCs, we isolated Lin−CD49f+CD24+ cells from MMTV-Neu-Tg mouse and injected into NOD/SCID mice (Fig. S4). Single cell suspension prepared from the secondary tumors, which were derived from these mice, were then used to generate 3D cultures. These 3D cultures were treated with 5-AzaC (1 μg/ml) plus butyrate (1 mM) for 72 h. This provided a unique opportunity to study the therapeutic efficacy of 5-AzaC and butyrate on self-renewing CSCs (Fig. S4). We isolated RNA and performed RNA-seq analysis. We found that 5-AzaC+butyrate combination differentially regulated genes that are involved in cell cycle, cell division, kinetochore formation, chromosome segregation and mitosis (Figs. 4A and B). Using the Ingenuity System Database (IPA) software, we analyzed signaling pathways using the differential gene expression that were altered significantly at or above p<0.05 level between control and treated groups. This analysis identified cancer and organismal injury and abnormalities as the top two disease and disorders pathways that were altered significantly (Fig. 4C). Since IPA showed a close association of genes that are affected in cancer, we performed in-depth molecular and functional analyses that are related to cancer. Interestingly, the top molecular functions altered were cell-to-cell signaling, cellular movement and cell morphology, cell cycle regulation, programmed cell death, DNA replication, post-translational modification, and molecular transport (Fig. 4D).
Figure 4.
Treatment with DNMT and HDAC inhibitors differentially regulates genes that are involved various signaling pathways. A, Heat map, generated from the RNA-seq analysis, showing differential gene expression between control and treated groups (n=3 mice). B, Network map created by IPA software using differentially regulated genes and their function. C, IPA analysis showing the top disease and disorders based genes that are differentially expressed between control and treated groups. D, Molecular function of genes altered by combination of 5-azaC+But.
We then investigated gene expression signature in different breast cancer subtypes using UCSC cancer genome browser to create heat map for genes that were differentially expressed between untreated and treated (5-AzaC+butyrate) groups. Interestingly, genes that were downregulated by 5-AzaC+butyrate were significantly increased in human breast cancer samples, whereas genes that were induced by 5-AzaC+butyrate were decreased in different breast cancer samples (Fig. S5). Further, we found that expression of RAD51AP1, NUSAP1 and SPC25 was very high in basal breast cancer subtype compared to other subtypes. This suggests that these genes could play a critical role in basal stem cells-driven breast cancer. To validate these observations, we selected two genes (RAD51AP1 and SPC25) and analyzed the expression of these genes in treated and untreated groups. Combination of 5-AzaC+butyrate inhibited RAD51AP1 gene expression (Fig. 4A and Table S1). Previous studies have shown that RAD51AP1 is associated with DNA double strand break repair and homologous recombination (29, 30). Many cancer drugs produce DNA lesions at replication fork; cancer cells might use RAD51, RAD51AP1 and other homologous recombination proteins to repair double strand breaks and maintain the cellular integrity. It has been shown that cell’s ability to repair double strand breaks significantly affects the outcome of cancer treatment whereas cells deficient in homologous recombination and DNA damage repair mechanisms are hypersensitive to drug-induced cell death, resulting in a better chemotherapeutic response and outcome (31).
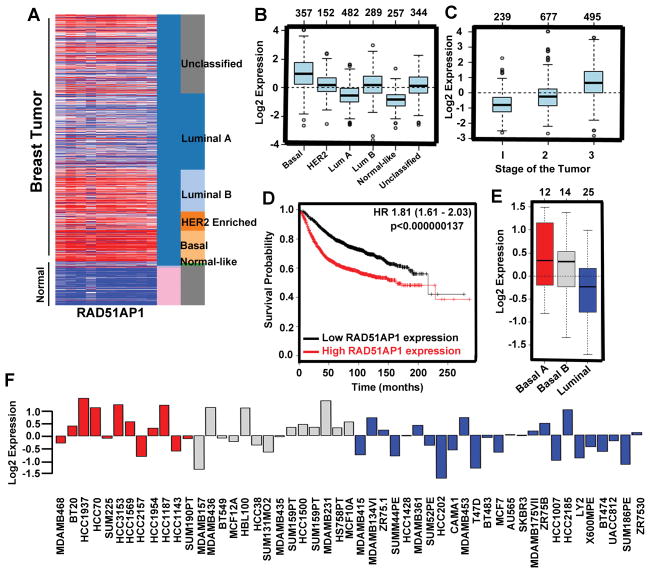
RAD51AP1 is highly expressed in basal breast cancer
Using the UCSC cancer genome browser and GOBO gene enrichment application, we investigated RAD51AP1 expression in normal and various breast cancer subtypes. We found that RAD51AP1 expression was significantly higher in breast cancer, especially in the basal subtype, than in normal mammary tissue (Fig. 5A). Further detailed analysis of GOBO gene enrichment analysis revealed that RAD51AP1 gene expression is higher in triple negative basal cell types and in HER-2 positive breast cancer in comparison to hormone-responsive luminal subtypes (Figs. 5B and S6A). Next, we investigated RAD51AP1 gene expression in human tumor subtypes and PAM50 tumor subtypes and found that basal cells express higher levels of this gene (Fig. 5B and S6B). ER-negative human breast tumor subtype expresses high levels of RAD51AP1 (Fig. S6C). GSA tumor analysis of RAD51AP1 gene indicates high expression of this gene in grade III histological tumor types (Fig. 5C). To understand the prognostic value of RAD51AP1 in overall survival, we investigated the Kaplan-Meier plotter integrative bioinformatics interface analysis and found a significant correlation between high RAD51AP1 expression and poor disease-free survival (Fig. 5D). To analyze the expression status of RAD51AP1 in human breast cancer cell lines, we investigated RAD51AP1 mRNA expression levels across different breast cancer cell lines using the GOBO gene enrichment cell line application. We found that triple-negative breast cancer (TNBC) cell lines expressed higher levels of this gene than other cell lines (Fig. 5F). Overall, these findings suggest that the combination of 5-AzaC+butyrate targets RAD51AP1, which in turn affects basal stem cell formation and reduces tumor growth.
Figure 5.
RAD51AP1 is upregulated in basal breast cancer. A, Heat map generated from the TCGA database showing relative expression of RAD51AP1 gene in normal and different breast cancer subtypes. B, Representative box plot of RAD51AP1 gene expression in different breast cancer subtypes. C, Box plot represents RAD51AP1 gene expression for tumor samples stratified according to histological grade. D, Kaplan-Meier plots represent overall survival of breast cancer patients in whole data sets for breast cancer patients categorized according to RAD51AP1 gene expression. The P-value was calculated using a log rank test. E, GSA tumor Box plots showing RAD51AP1 gene expression in human breast cancer cell lines categorized according to the basal A (red), basal B (grey) and luminal (blue) subgroups. F, Gene expression analysis of RAD51AP1 in 51 breast cancer cell lines.
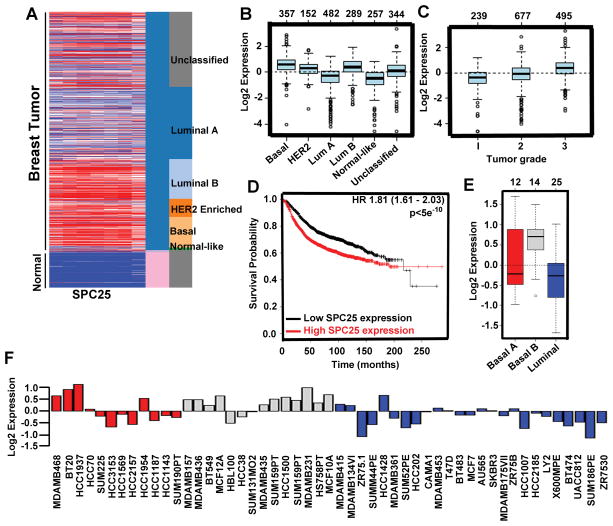
SPC25 expression is high in basal breast cancer
To establish the rationale for targeting genes that are involved in basal stem cell driven breast cancer, we also investigated the functional significance of SPC25 in human breast cancer and how the expression of this gene responds to 5-AzaC+butyrate treatment (Table S1 and S2). SPC25 is an essential component of kinetochore-associated NDC80 complex (32). It has been well established that kinetochore function is important for chromosome segregation during mitosis and for maintenance of chromosome stability (33). Further, chromosomal instability is one of the major causes of heterogeneity observed in various human cancers (34). We first tested the expression profile of SPC25 between normal and various breast cancer subtypes using UCSC cancer genome browser and GOBO gene enrichment application. We found that SPC25 expression was significantly increased in HER2-positive and basal tumors compared to normal counterparts (Fig. 6A). SPC25 expression was particularly high in basal tumor types, which represents more stem-like cells (Fig. 6B). Similar to RAD51AP1 gene, SPC25 gene is also highly expressed in basal, ER-negative tumors (Fig. 6B and S7A). GSA tumor analysis shows higher histological grade and poor prognosis with higher expression of SPC25 (Fig. 6C). To understand the prognostic value of SPC25 in overall survival, we used the Kaplan-Meier plotter analysis and found a significant positive correlation between high SPC25 expression and poor disease-free survival (Fig. 6D). Finally, we checked the expression of SPC25 across the human breast cancer cell lines. We found that TNBC cell lines had high level of SPC25 expression (Fig.6E and F). Though RAD51AP1 and SPC25 expressions were high in human breast tumor tissues and cell lines and their expression downregulated by 5-AzaC and butyrate, we do not have data to show that RAD51AP1 and SPC25 expressions are directly regulated by DNA methylation or histone acetylation. Further studies are needed to address this issue.
Figure 6.
SPC25 is upregulated in breast cancer. A, Heat map generated from the TCGA database showing relative expression of SPC25 gene in normal and different breast cancer subtypes. B, Box plot represents SPC25 gene expression in different breast cancer subtypes. C, Representative box plot of SPC25 gene expression according to histological grade. D, Kaplan-Meier plots represent overall survival of breast cancer patients in whole data sets for breast cancer patients stratified according to SPC25 gene expression. The P-value was calculated using log rank test. E, Box plots represents SPC25 gene expression in human breast cancer cell lines classified according to the basal A (red), basal B (grey) and luminal (blue) subgroups. F, SPC25 gene expression in 51 breast cancer cell lines.
Discussion
Breast CSCs have been successfully isolated from human breast tumor tissues and from tumors from various spontaneous mouse mammary tumor models. In our studies, we used CD24 and CD49f cell-surface markers mainly because these two markers are commonly expressed in both human and murine cancers and also in various spontaneous mouse mammary tumor models. Though these two markers do not yield pure luminal progenitor and basal stem cell populations, our studies provide evidence that CD49fhighCD24+ cells exclusively express K14 (a marker for basal myoepithelial stem cells) and that CD49f+CD24high cells express K8 (a marker for luminal progenitor cells) without any cross contamination (Fig. 2E). In our recently published studies, we also used CD61 marker to differentiate mature myoepithelial, mature luminal, and luminal progenitor cells (10). However, all these three cell surface markers (CD49f, CD24 and CD61) are unable to distinguish the normal MaSCs and CSC. Additional studies are warranted to find new cell-surface markers that are capable of distinguishing between MaSCs and CSCs.
Previous studies have shown that a large majority of mammary tumors, including the mammary tumors that develop in MMTV-Neu-Tg mouse (19), are derived from the luminal progenitor cell type (18, 21) rather than from the basal cell type. However, a recent study has shown that mammary tumors that develop in MMTV-Neu-Tg mouse can originate from both luminal and basal cell types (35). Though our present studies reinforce the previous observations, our findings provide further evidence that tumors originating from the basal cell types are relatively slow growing and are responsible for metastatic progression. Further, our studies provide evidence that both luminal and basal cells require additional mutations to activate the unactivated Neu-Tg to become the tumorigenic phenotype (Figs. 2A–C and S2A–B). This suggests that a small portion of basal stem cell population may acquire mutations or epigenetic modifications and serve as the cancer-initiating stem cells (CISCs) in Neu-Tg-induced mammary tumor and that such cells are capable of dedifferentiating into luminal cells. This is supported by the observation that tumors derived from the basal cell type express both K8 and K14, suggesting a transition of basal stem cells into early luminal progenitors (Fig. 2E).
One of the most interesting and unexpected findings of our study is that basal stem and luminal progenitor cells isolated from the premalignant mammary gland of younger mice (3-month-old) were able to propagate mammary tumor in NOD/SCID mice while same cell types isolated from older mice (12-month-old) were unable to form tumors. This suggests that, irrespective of the age, genetic/epigenetic events lead to activation of unactivated Neu-Tg into a transformed mammary tumor forming phenotype along with reprogramming of the stem/progenitor cells into tumor propagating cancer stem cells. This is surprising because aging is the single biggest risk factor for tumor development; however, our studies provide evidence that genetic/epigenetic modifications reprogram the basal myoepithelial stem cells and luminal progenitor cells into tumor propagating cancer stem cells and play a critical role in tumor development. Further, tumors derived from both luminal and basal cell types of pre-malignant MMTV-Neu-Tg mice showed similar self-renewing tumor-propagating CSC phenotype (Fig. 2B–D and S2C). In addition, tumors developed from these two cell types were indistinguishable from the spontaneous tumors that developed in MMTV-Neu-Tg mice, suggesting that basal cell type has similar tumor propagating capability as luminal cell type and that this tumor propagating potential of basal cell type could have a great impact in drug resistance in HER2-positive breast cancer patients treated with trastuzumab. Trastuzumab targets only HER2-positive luminal progenitor cells but not HER2-negative basal myoepithelial stem cells, which are multipotent and quiescent in tumor. This raises the possibility that during chemotherapy, especially in trastuzumab therapy, the tumorigenic basal myoepithelial stem cells escape or becomes unresponsive to the therapy and then enter into circulation and ultimately lead to tumor metastasis. This is further supported by the previous findings that demonstrated the increased basal cell populations in HER2-resistant tumor after chemotherapy (23, 36). Therefore, it is obligatory to target both transformed luminal progenitor cells and basal myoepithelial stem cells when selecting chemotherapy for breast cancer treatment.
Our findings also provide evidence that Lin−CD49f+CD24+ cells are metastatic. We used 4T1 cell line, a metastatic cancer cell line that mimics grade IV human breast cancer (36), to test the metastatic potential of Lin−CD49f+CD24+ cells. 4T1 cell line was established from the myoepithelial cells of a spontaneous mutation-driven mouse mammary tumor (37). Previous studies showed that basal myoepithelial cells are leader cells and that the basal myoepithelial enrichment program helps in the process of metastasis (38, 39). These findings suggest that a few basal myoepithelial stem cells dedifferentiate into tumor-propagating luminal cell type. Based on these observations, we speculate that, in addition to epithelial-to-mesenchymal transition (EMT), the basal tumorigenic cell types play a key role in drug resistance, tumor recurrence, and metastasis. Further, our findings provide strong evidence that the basal myoepithelial stem cells play a critical role in metastatic spread, either by themselves or by dedifferentiating into tumor-propagating luminal cell type. Thus, 5-AzaC+butyrate combination could be a novel therapeutic strategy to target basal cell gene signature.
The enhanced expression of RAD51AP1 in a majority of breast cancers compared to normal counterparts suggests the oncogenic role of this gene in human breast cancer. Previous studies have shown that RAD51AP1 expression is upregulated in hepatocellular carcinomas (40), acute myeloid leukemia with complex karyotypic abnormalities (41), and aggressive mantle cell lymphoma (42). Furthermore, downregulation of RAD51AP1 by gene-specific siRNA resulted in growth suppression of cholangiocarcinoma cells. These results indicate that RAD51AP1 expression is essential for the growth of tumor cells and that its inhibition may be a novel therapeutic option for the treatment of breast cancer. On the other hand, CpG methylation of its promoter region leads to the reduced expression of RAD51AP1 in prostate cancer cells (43). Therefore, the tumorigenic potential of RAD51AP1 may be cell type-dependent. Similarly, SPC25 is highly expressed in human breast tumor tissues and in breast cancer cell lines when compared to normal controls. Though the role of SPC25 in tumorigenesis is unknown, studies have shown that it forms a complex with NDC80, NUF2, and SPC24, localized at the kinetochore outer plate after the G2 phase (44–47), and plays a critical role in microtubule-kinetochore attachment and spindle assembly checkpoint in mitosis (48). Further, genetic stability depends primarily on accurate chromosome segregation during the cell cycle and kinetochores are essential for this process. Thus, SPC25 function is required for tumor cells to sustain the genomic stability by maintaining mitotic spindle assembly for their eternal growth. Overall, our studies demonstrate that tumor cell of origin and tumor-propagating cells are regulated by DNA methylation and that inhibition of DNMTs reverses the abnormal self-renewal properties of tumor-propagating cells. Further, our studies unequivocally show that the combination of 5-AzaC and butyrate efficiently blocks mammary tumorigenesis and reduces tumorosphere-forming potential of tumor-propagating cells by reactivating the tumor suppressor genes. Therefore, 5-AzaC/butyrate combination could be an effective adjuvant therapy for treatment of breast cancer. This combination would not only provide an effective strategy to prevent tumor growth but also prevent relapse of the breast cancer.
Supplementary Material
Acknowledgments
We thank Dr. Chang and Dr. Kitamura, Integrated Genomic core facility, Augusta University Cancer Center, for their help in RNA-Seq analysis. This work was supported by grants from the National Institutes of Health 5R01CA131402 (M. T) and 5R01CA15677-07 (B. L. L), Department of Defense (BC074289) and Augusta University Intramural Pilot Study grant, Start-up and Bridge funds. G. M. is a fellow of Department of Biotechnology (DBT), India sponsored overseas fellowship program.
Footnotes
Author contribution: R. P. and M. T. designed experiments and analyzed data; R. P. performed most of the experiments; H. S., R. K. and J-H. C. contributed in RNA-seq analysis; S. R., P. T. and P. D. P. contributed in mouse xenograft studies; S. M., G. M. and P. T. contributed in MaSC and CSC isolation and generation of mammospheres and tumorospheres; R. K. and S. S. contributed in tumor morphometric analysis; R. P., B. L. L., V. G. and M. T. wrote the manuscript.
All authors declare no conflict of interest.
References
- 1.Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 2.Park CY, Tseng D, Weissman IL. Cancer stem cell-directed therapies: recent data from the laboratory and clinic. Mol Ther. 2009;17:219–230. doi: 10.1038/mt.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis SJ, Sinkevicius KW, Li D, et al. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome--components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 8.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brower V. Epigenetics: Unravelling the cancer code. Nature. 2011;471:S12–13. doi: 10.1038/471S12a. [DOI] [PubMed] [Google Scholar]
- 10.Pathania R, Ramachandran S, Elangovan S, et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun. 2015;6:6910. doi: 10.1038/ncomms7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 13.Ramchandani S, Bhattacharya SK, Cervoni N, et al. DNA methylation is a reversible biological signal. Proc Natl Acad Sci U S A. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flotho C, Claus R, Batz C, et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- 15.Daskalakis M, Nguyen TT, Nguyen C, et al. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. Blood. 2002;100:2957–2964. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 16.Gros C, Fahy J, Halby L, et al. DNA methylation inhibitors in cancer: recent and future approaches. Biochimie. 2012;94:2280–2296. doi: 10.1016/j.biochi.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 17.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shackleton M, Quintana E, Fearon ER, et al. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson CH, Ailles LE, Dylla SJ, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. New Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 21.Guy CT, Webster MA, Schaller M, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 23.Lo PK, Kanojia D, Liu X, et al. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene. 2012;31:2614–2626. doi: 10.1038/onc.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveras-Ferraros C, Vazquez-Martin A, Martin-Castillo B, et al. Pathway-focused proteomic signatures in HER2-overexpressing breast cancer with a basal-like phenotype: new insights into de novo resistance to trastuzumab (Herceptin) Int J Oncol. 2010;37:669–678. doi: 10.3892/ijo_00000716. [DOI] [PubMed] [Google Scholar]
- 25.Martin-Castillo B, Oliveras-Ferraros C, Vazquez-Martin A, et al. Basal/HER2 breast carcinomas: integrating molecular taxonomy with cancer stem cell dynamics to predict primary resistance to trastuzumab (Herceptin) Cell cycle. 2013;12:225–245. doi: 10.4161/cc.23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santner SJ, Dawson PJ, Tait L, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 27.Turner N, Lambros MB, Horlings HM, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller FR, Miller BE, Heppner GH. Characterization of metastatic heterogeneity among subpopulations of a single mouse mammary tumor: heterogeneity in phenotypic stability. Invasion & Metastasis. 1983;3:22–31. [PubMed] [Google Scholar]
- 29.Dunlop MH, Dray E, Zhao W, et al. Mechanistic insights into RAD51-associated protein 1 (RAD51AP1) action in homologous DNA repair. J Biol Chem. 2012;287:12343–12347. doi: 10.1074/jbc.C112.352161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiese C, Dray E, Groesser T, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31:955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- 32.Janke C, Ortiz J, Lechner J, et al. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuen KW, Montpetit B, Hieter P. The kinetochore and cancer: what’s the connection? Curr Opin Cell Biol. 2005;17:576–582. doi: 10.1016/j.ceb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Tan W, Wu X, et al. A NIK-IKKalpha module expands ErbB2-induced tumor-initiating cells by stimulating nuclear export of p27/Kip1. Cancer cell. 2013;23:647–659. doi: 10.1016/j.ccr.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Rijn M, Perou CM, Tibshirani R, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–1996. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 38.Cheung KJ, Gabrielson E, Werb Z, et al. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–1651. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheridan C, Kishimoto H, Fuchs RK, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song LJ, Zhang WJ, Chang ZW, et al. PU. 1 Is Identified as a Novel Metastasis Suppressor in Hepatocellular Carcinoma Regulating the miR-615-5p/IGF2 Axis Asian Pacific. J Cancer Prevent. 2015;16:3667–3671. doi: 10.7314/apjcp.2015.16.9.3667. [DOI] [PubMed] [Google Scholar]
- 41.Schoch C, Kern W, Kohlmann A, et al. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chrom Cancer. 2005;43:227–238. doi: 10.1002/gcc.20193. [DOI] [PubMed] [Google Scholar]
- 42.Henson SE, Tsai SC, Malone CS, et al. Pir51, a Rad51-interacting protein with high expression in aggressive lymphoma, controls mitomycin C sensitivity and prevents chromosomal breaks. Mut Res. 2006;601:113–124. doi: 10.1016/j.mrfmmm.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Yu Q, Cho AH, et al. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia. 2005;7:748–760. doi: 10.1593/neo.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hori T, Haraguchi T, Hiraoka Y, et al. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- 45.MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–11337. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- 46.McCleland ML, Kallio MJ, Barrett-Wilt GA, et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol. 2004;14:131–137. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- 47.DeLuca JG, Dong Y, Hergert P, et al. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Bio Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun SC, Lee SE, Xu YN, et al. Perturbation of Spc25 expression affects meiotic spindle organization, chromosome alignment and spindle assembly checkpoint in mouse oocytes. Cell cycle. 2010;9:4552–4559. doi: 10.4161/cc.9.22.13815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.