Abstract
Colorectal cancer cells with stem-like properties, referred to as colon cancer initiating cells (CCIC), have high tumorigenic potential. While CCIC can differentiate to promote cellular heterogeneity, it remains unclear whether CCIC within a tumor contain distinct subpopulations. Here we describe the co-existence of fast-cycling and slow-cycling CCIC, which can undergo asymmetric division to generate each other, highlighting CCIC plasticity and interconvertibility. Fast-cycling CCIC express markers such as LGR5 and CD133, relying on MYC for their proliferation, whereas slow-cycling CCIC express markers such as BMI1 and hTERT and are independent of MYC. NOTCH signaling promotes asymmetric cell fate, regulating the balance between these two populations. Overall, our results illuminate the basis for CCIC heterogeneity and plasticity by defining a direct interconversion mechanism between slow-cycling and fast-cycling CCIC.
BACKGROUND
Colorectal tumors, as well as most solid tumors, exhibit considerable intratumoral cellular heterogeneity. Even genetically identical colorectal cancer (CRC) cells from the same tumor demonstrate significant variability with regard to proliferation, invasive potential and chemoresistance (1-3). At least in part, this CRC cellular diversity can be hierarchically organized, with growth driven by a subset of cells with stem-like properties, referred to as colon cancer initiating cells (CCICs) or stem cells (CCSCs) (4-7). Consistent with a role for CCIC in tumor progression, a signature that reflects that of normal colon stem cells is prognostic for higher frequency of CRC relapse (6). These stem cell associated markers expressed in CCICs include CD133, LGR5, BMI1, CD44, and ALDH1 (1, 6, 8, 9). Although LGR5, a co-receptor for the WNT ligand RSPO1, is a marker for fast-cycling intestinal and colon stem cells (10-12), tumor expression of LGR5 is not strongly associated with CRC prognosis (13). However, while associated with a more quiescent stem cell population in the normal intestinal mucosa, BMI expression is also correlated with poor survival and CRC recurrence (14-16), and targeted anti-BMI1 therapy inhibits tumor xenograft growth and self-renewal (1).
CCIC can divide symmetrically to generate two CCIC daughters or asymmetrically to generate a CCIC daughter and a more differentiated daughter cell (8, 17). Disruption of asymmetric division can alter the balance between self-renewal and differentiation in CCIC and consequently, impact tumor growth. Similar observations have been reported in other types of cancer stem cells (18, 19). Importantly, NOTCH signaling, which is well documented to be necessary for both stem cell proliferation as well as lineage allocation in the intestinal mucosa, may be an important determinant that drives asymmetric CCIC daughter fate (17). In mouse models of CRC, Notch signaling is elevated in tumorigenesis (20). Furthermore, suppression of NOTCH signaling induces differentiation of adenoma cells into goblet cells, and targeted deletion of the Notch ligand JAG-1 decreases intestinal tumor volumes in APCMin/+ mice (21, 22). Also consistent with the pro-tumorigenic potential of NOTCH signaling is the high expression of the downstream effectors of NOTCH in human adenomas and early stage tumors compared to late stage adenocarcinomas (20, 23). Furthermore, NOTCH also promotes CRC chemoresistance (24) and metastasis (25).
Here, we demonstrate co-existence of fast- and slow-cycling CCIC populations in the same tumors with fast-cycling cells expressing LGR5, CD133, and CD44, and slow-cycling CCICs expressing BMI1, hTERT, and HOPX. The two populations can directly interconvert via asymmetric division, which simultaneously generates a fast-cycling daughter cell and a slow-cycling daughter cell. Fast-cycling CCICs depend on MYC for proliferation, but slow-cycling CCICs are less dependent on MYC. NOTCH signaling promotes such asymmetric cell fate and regulates the balance between the two CCIC populations. Maintaining both fast- and slow-cycling stem cells may provide a growth and survival strategy for neoplastic tissue.
METHODS
Antibodies
Frozen human normal colonic and CRC tissues were stained with anti-α-TUBULIN (ab6160), anti-BMI1 (ab14389), anti-LGR5 (ab75732), anti-Ki67 (ab15580), anti-NOTCH1 (ab44986) antibodies purchased from Abcam, anti-NUMB (2756) purchased from Cell Signaling, anti-MYC (sc-40) anti-PARD3A (sc-79577) purchased from SCBT. Antibody concentrations and standard immunofluorescence procedures (IF) are described in Supplemental Methods.
Microscopy
Frozen sections of normal human colonic tissue or tissue from various stages of colon cancer (Normal colon: n = 20, CRC: n = 20 (n = 5 per CRC stage)) embedded in O.C.T were stained for Hematoxylin and Eosin (H&E) and IF. The fraction of dividing BMI1+/LGR5+/α-TUBULIN+ asymmetric pairs was quantified in 500 α-TUBULIN+ dividing pairs per specimen. Images were acquired on a Zeiss LSM 510 confocal microscope using an Apo 63× 1.40 oil objective and analyzed with ZEN confocal software.
CCIC Isolation and Culture
CCIC lines (CCIC-1, CCIC-2) were derived from patients (ages 51 or 57 years) with early stage (Stage I-II), well-differentiated CRC resections lacking p53 and KRAS mutations and cultured as described in 2013 (17) under protocols approved by Weill Cornell Medical College. CRC tumors were washed in PBS, minced into 1-2 mm fragments, incubated in collagenase at 37°C for 45 minutes, and strained through a 40μm filter to isolate single cells for CCIC culture. CCIC lines have been tested in sphere propagation and serial dilution assays and were last authenticated in late 2015. Exome sequencing data was collected for these lines and analyzed in 2015 (26). CCICs were cultured in ultra low-attachment flasks in serum free DMEM:F12 medium (Invitrogen) supplemented with minimal nonessential amino acids (Thermo Fisher), sodium pyruvate (Thermo Fisher), N2 supplement (Invitrogen), B27 supplement (Invitrogen), 4 μg/ml heparin (Sigma), antibiotic-antimycotic (Invitrogen), 20 ng/ml basic fibroblast growth factor (R&D Systems) and 40 ng/ml epidermal growth factor (R&D Systems) at 37°C and 5% CO2. In order to passage CCICs, cell suspension was gently centrifuged, suspended in 1X Trypsin-EDTA (Invitrogen) for 5 minutes at 37°C, and mechanically dissociated into single cells before final resuspension in culture medium.
CCIC Studies
Single cells from CCIC-1 and CCIC-2 lines were plated in collagen-coated chamber slides in serum free culture medium for pair-cell assays (17). Live cell imaging was performed using a Nikon BioStation CT at a 40X objective for 48 hours to track single cell divisions. Slides were incubated at 37°C and 5% CO2 for 24-48 hours followed by IF. For pair-cell assays, CCIC were treated with one of the following conditions: DMSO or DAPT (Sigma) in culture medium. Lentiviral constructs containing NUMB-shRNA (Sigma), NUMB-SYM (gift from Zhong labatory), JAG-1 shRNA (Sigma), NICD-OE (Cell Biolab), or TCF-EGFP reporter (Addgene) were used to infect CCIC to assay asymmetric cell pairs. CCICs were also transduced with lentivirus containing a BMI promoter-mCherry construct (GeneCopoeia) with a puromycin selection marker in order to isolate BMI1+ and LGR5+ cells using FACS. The procedure for lentiviral transduction is described in Supplemental Methods. BrdU incorporation was performed as previously described (17) where single CCICs were plated and grown for 16 hours. Following this division, cells were treated for 3 hours with 10mM BrdU (Sigma), fixed in cold 70% ethanol, followed by incubation for 1 hour in 2M HCl. Cells were washed in PBS, incubated for with 100mM Na2B4O7 for two minutes and stained using standard IF protocol. Primers and antibodies for RT-PCR and Western blotting are described in Supplemental Methods.
Xenograft Assays
1 × 106 unsorted, LGR5+ and BMI1+ CCICs transduced with MYC-shRNA or scrambled shRNA were injected s.c into 8-week-old NOD/SCID mice (n = 5 mice/condition) in a 200ul volume containing 1:1 culture medium: Matrigel mixture. Tumors were propagated for 4 weeks. Additionally, 1 × 106 unsorted CCICs also were also injected s.c into NOD/SCID mice (n = 6) to form tumors after 4 weeks; tumors were directly injected over 3 days with DMSO or DAPT delivered at a dose of 200 mg/kg every 12 hours (27). Tumor dimensions were measured using a caliper. Tumors were minced into 1-2 mm fragments and incubated in Accumax/EDTA solution (Millipore) for 30 minutes at room temperature while shaking to obtain a single cell suspension for FACS. FlowJo software was used to gate populations according to 7-AAD (Molecular Probes) viability, and forward and side scattering. Cutoff thresholds were provided by unstained cells as a negative control.
Statistical Analysis
The data displayed are represented as mean ± SD. Statistical comparisons between two groups was made using Student t-test or one-way ANOVA for multiple groups. P < 0.05 was used to establish statistical significance.
RESULTS
Asymmetric BMI1+/LGR5+ cell pairs in CRC
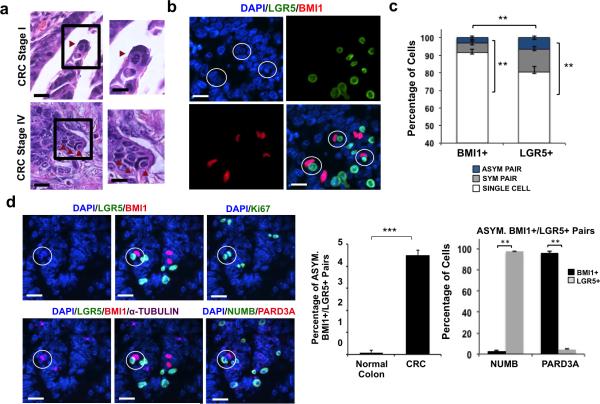
Histopathological review of stage I through IV primary CRC tumors consistently identified a distinctive paired cell morphology: a cell with a long, slender nucleus situated adjacent to a cell with a smaller, rounded nucleus (Fig. 1a). This paired cell morphology was probed by immunofluorescence (IF) in patient normal colon and CRC specimens using antibodies against human LGR5 and BMI1 (see Methods) (28-30). This revealed co-localization of a larger BMI1+ cell with a slender nucleus adjacent to a smaller, rounder LGR5+ cell. Multiple such BMI1+/LGR5+ pairs could be detected in the same magnification field of CRC tumors (Fig. 1b). Whether the paired BMI1/LGR5+ cells in these structures were generated by cell division was examined using α-TUBULIN staining for mitotic spindle labeling and Ki67 staining as a proliferation marker. Analysis of primary CRC tissue showed that approximately 3.5% of BMI1+ cells or 5.4% of LGR5+ cells were associated as asymmetrically dividing BMI1/LGR5 pairs (Fig. 1c; p = 0.01; Student t-test). In multiple BMI1+/LGR5+ pairs in primary CRC tissue, α-TUBULIN staining is consistent with the configuration of microtubules in telophase, the final mitotic phase, in which the midbody is at the division plane during cytokinesis and asters are at the poles. Ki67 expression was also detected, confirming that these pairs arose from cell division. Quantitative analysis revealed that the frequency of dividing BMI1+/LGR5+ cell pairs increased in neoplastic tissue compared to normal colon tissue from the same patient (Fig. 1d; p = 0.001; Student t-test). Additionally, we examined CRC tissue for classical markers of asymmetric division, including cell fate determinant NUMB and polarity protein PARD3A. Consistently, NUMB localized to the LGR5+ cell and PARD3A localized to the BMI1+ daughter in dividing BMI1/LGR5 pairs (Fig. 1d; p = 0.01; Student t-test). These data suggest that BMI1+/LGR5+ pairs could result from asymmetric cell division and are associated with CRC.
Figure 1. Primary CRC tumors contain BMI1+/LGR5+ cell pairs.
(a) H&E images of human Colorectal Cancer (CRC) Stage I (top row) and Stage IV (bottom row) at low (left) and high (right) magnifications. Scale bar: 50 μm (left), 25 μm (right). (b) Representative co-IF from CRC stage IV showing BMI1+(red)/LGR5 (green) asymmetric CCIC pairs. DAPI (blue) labels nuclei; scale bar: 25μm. (c) Percent of BMI1+ or LGR5+ cells associated as single cells, symmetrically dividing (α-TUBULIN+) pairs, or BMI1+/LGR5+ asymmetrically dividing cell pairs in CRC tumor tissue based on co-IF. Data represents mean ± SD of n = 20 tumors from 500 cells/specimen (**, p = 0.01; Student t-test). (d) Left: Representative co-IF of CRC specimen showing LGR5 (green), BMI1 (red), and α-TUBULIN (purple) expression. Also shown is NUMB (green) and PARD3A (red) expression. Ki67 staining (green); DAPI (blue); scale bar: 25 μm. Middle: Frequency of dividing (α-TUBULIN+) BMI1+/LGR5+ asymmetric cell pairs in normal colon vs. CRC tumor tissue based on co-IF. Data represents mean ± SD of n = 20 normal or CRC specimens from 500 TUBULIN+ pairs/specimen(***, p = 0.001; Student t-test). Right: Percent of BMI1+ or LGR5+ cells expressing NUMB or PARD3A in asymmetric BMI1+/LGR5+ pairs. The data represents the mean ± SD from n = 20 tumors (**, p = 0.01; Student t-test).
Asymmetric BMI1+/LGR5+ CCIC daughter pairs
To understand the lineage relationship between BMI1+ and LGR5+ cells in asymmetric pairs, we derived and characterized CCIC lines (CCIC-1 and CCIC-2) from two CRC patients as previously described (4, 26) and performed a pair-cell assay (17, 31). Single CCICs were monitored through approximately 36 hours followed by IF of cell pairs (Supplementary Fig. 1A) for markers, including BMI1, LGR5, NOTCH1 and others. Co-IF for LGR5 and BMI1 confirmed that CCICs could divide into asymmetric BMI1+/LGR5+ daughter cell pairs; additionally, a notable difference was observed in the longest nuclear diameters of the BMI1+ CCIC and its LGR5+ counterpart (Supplementary Fig. 1B). The presence of these BMI1+/LGR5+ asymmetric cell pairs in CCIC is consistent with what we observed in primary CRCs. In approximately 5% of newly divided pairs, asymmetric division was identified producing daughter cell pairs that exhibited the specific cellular and nuclear morphology of a crescent-shaped CCIC enveloping another CCIC (Supplementary Fig. 1C). Using co-IF for PARD3A (a polarity cell fate marker for asymmetric division) and LGR5 expression, we confirmed a similar 4.5% frequency of asymmetric cell division in CCICs (Supplementary Fig. 1D). In contrast, HCT116 cells, a CRC line that does not have CCIC, showed no evidence of asymmetric division as determined by specific nuclear morphology and PARD3A/LGR5 expression (Supplementary Fig. 1D).
Next, asymmetric CCIC daughter cell pairs were characterized based on expression of stem cell markers and signaling pathway components (Supplementary Figs. 1E-1G). These studies revealed that the BMI1+ CCIC also expressed high levels of NOTCH1 receptor (NOTCH1), HES5 (a downstream marker of active NOTCH signaling), hTERT, and the TGF-β pathway markers nuclear phospho-SMAD1/5 (32). In asymmetric cell pairs, the partnered LGR5+ daughter also expressed the WNT marker nuclear β-CATENIN, NOTCH ligand DLL4, and the stem cell markers CD44, ALDH1, and CD133 (5, 32-34). Quantitative analysis showed that ALDH1 and CD133 labeled LGR5+ daughters in over 95% of BMI1/LGR5 asymmetric pairs (Supplementary Fig. 1F; p = 0.01; Student t-test). Several intestinal stem cell markers (10) were also informative in asymmetric cell pairs: BMI1+ daughters were HOPX+ and hTERT+ while LGR5+ daughters were OLFM4+ and ASCL2+ (Supplementary Fig. 1G; p = 0.01; Student t-test).
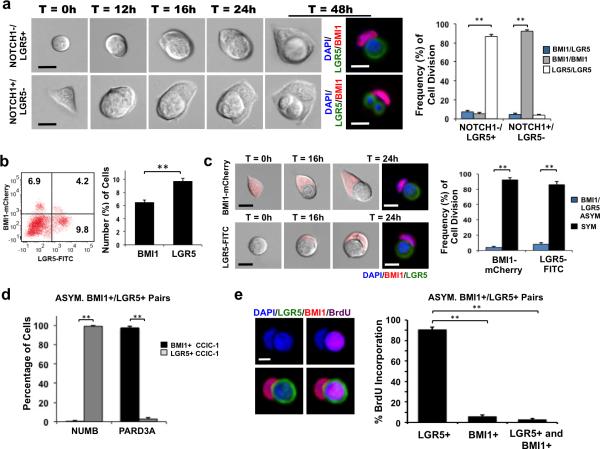
We then investigated which type of CCIC (BMI1+ or LGR5+) gives rise to the BMI1+/LGR5+ pair. Because BMI1 is an intracellular protein, the cell surface NOTCH1 receptor was used as a surrogate membrane marker based on the observation above that BMI1+ CCIC is NOTCH1+ and LGR5+ CCIC is NOTCH1−. Time-lapse imaging of NOTCH1/LGR5 double sorted cells was recorded over 48 hours followed by fixation and IF (Fig. 2a). A round, NOTCH1-/LGR5+ CCIC produced an elongated, BMI1+ daughter cell in ~7.2% of cell divisions (p = 0.01; Student t-test). Reciprocally, a crescent-shaped, NOTCH1+/LGR5- CCIC generated a rounder daughter cell, which further divided into two smaller, rounder LGR5+ daughter cells in 4.1% of cell divisions (Fig. 2a; p = 0.01; Student t-test). Next, CCIC were lentivirally transduced with a BMI1-mCherry promoter reporter construct and sorted to directly isolate BMI1+ (~6.2% of CCIC) and LGR5+ (~9.5% of CCIC) populations (Fig. 2b; p = 0.01; Student t-test). RT-PCR analysis of BMI1+ CCIC showed enrichment of HOPX, NOTCH1, and hTERT gene expression while LGR5+ CCIC had elevated ALDH1, ASCL2, CD44, CD133, and OLFM4 gene expression, consistent with our earlier findings (Supplementary Fig. 1H; p = 0.01; Student t-test). Similarly, time-lapse imaging of single BMI1+ or LGR5+ CCIC followed by IF demonstrated both were capable of asymmetric division to produce BMI1+/LGR5+ pairs with a frequency of 4.2% or 7.5%, respectively (Fig. 2c; p = 0.01; Student t-test). Therefore, both LGR5+ and BMI1+ CCICs generated asymmetric BMI1+/LGR5+ pairs through division.
Figure 2. Asymmetric BMI1+/LGR5+ CCIC-1 daughter pairs.
(a) Left: Time-lapse series of single NOTCH1−/LGR5+ CCIC (top panel) and NOTCH1+/LGR5− CCIC (bottom panel) undergoing asymmetric division over 48 hours. Co-IF after fixation at 48 hours: LGR5 (green), BMI1 (red), DAPI (blue); scale bar: 10μm. Right: Frequency of asymmetric BMI1+/LGR5+ (blue), symmetric BMI1+/BMI1+ (grey), or symmetric LGR5+/LGR5+ (white) cell pairs in sorted NOTCH1/LGR5 CCIC. Data represents the mean ± SD from 3 independent experiments with n = 100 LGR5+ cells/replicate or n = 1000 NOTCH1+(BMI1+) cells/replicate (**, p = 0.01; Student t-test). (b) CCICs were lentivirally transduced with a BMI1-mCherry promoter reporter construct. Left: FACS plot (left) and the percent of BMI1+ and LGR5+ CCIC determined by FACS (right). Data represents mean ± SD from 3 independent experiments. (**, p =0.01; Student t-test). (c) Left: Time-lapse series of single BMI1+ CCIC (top panel) and LGR5+ CCIC (bottom panel) undergoing asymmetric division over 24 hours. BF: BMI1-mCherry (red). Co-IF antibody staining at 24 hours: BMI1 (red); LGR5 (green); DAPI (blue); scale bar: 10μm. Right: Frequency of asymmetric BMI1+/LGR5+ (blue) or symmetric (black) cell pairs in sorted BMI1+ or LGR5+ CCIC. The data represents the mean ± SD from 3 independent experiments with n = 100 LGR5+ cells/replicate or n = 1000 BMI1+ cells/replicate (**, p = 0.01; Student t-test). (d) Quantitative analysis of the percent of BMI1+ or LGR5+ daughters expressing NUMB or PARD3A in asymmetric BMI1+/LGR5+ CCIC pairs. The data represents mean ± SD from 3 independent experiments with n = 500 pairs/replicate (**, p = 0.01; Student t-test). (e) BrdU incorporation following division of CCIC. Left: LGR5 (green), BMI1 (red), and BrdU (purple) staining. DAPI (blue); scale bar: 10μm. Right: Quantitative analysis in asymmetric BMI1+/LGR5+ pairs with percentage of BrdU incorporation in LGR5+ CCIC, BMI1+ CCIC or both LGR5+ and BMI1+ CCIC daughters indicated (**, p = 0.004, one-way ANOVA). The data represents mean ± SD of 3 independent experiments with n = 500 cell pairs/replicate.
BMI1 and LGR5 CCICs were capable of both symmetric and asymmetric divisions (Supplementary Fig. 2A). For asymmetric division, PARD3A polarizes to the side of the BMI1 daughter compartment during cytokinesis, as confirmed by α-TUBULIN staining of midbody at the division plane, supporting intrinsic asymmetric division of CCICs to generate BMI1+ and LGR5+ daughters (Supplementary Fig. 2B). Further quantification in BMI1+/LGR5+ CCIC pairs showed that PARD3A labeled BMI1+ daughters in ~97% of pairs, while NUMB localized to the LGR5+ daughter in over 98% of pairs (Fig. 2d, Supplementary Fig. 2C; p=0.01; Student t-test). The three-dimensional morphology of cell fate asymmetry was analyzed using confocal imaging and z-stack acquisition, revealing that BMI1+(PARD3A+) CCIC physically surrounds the smaller LGR5+(NUMB+) CCIC daughter cell in 3 dimensions (Supplementary Movie 1).
The WNT activity status of BMI1+(NOTCH1+)/LGR5+ cell pairs was evaluated by infecting CCICs with a lentiviral TCF-GFP reporter construct and treatment with WNT3A ligand. This revealed that the LGR5+ CCIC daughter displayed active WNT signaling (Supplementary Fig. 2D). Furthermore, based on FACS analysis, TCF-GFP reporter and CD133 expression overlapped primarily with the LGR5+/BMI1− CCIC population (Supplementary Fig. 2E).
LGR5+ CCIC daughter is faster-cycling than BMI1+ daughter
The proliferative capacity of the LGR5+ and BMI1+ daughter cells in asymmetric pairs was analyzed by BrdU incorporation in the pair-cell assay. Single cells were plated and allowed to divide once in proliferative medium for 24 hours (1st division) before treatment of BrdU for 3 hours to label cells entering the 2nd division (18). Co-IF showed that the smaller, rounder LGR5+ daughter was positive for BrdU (Fig. 2e). Quantitative analysis in BMI1+/LGR5+ asymmetric pairs showed that BrdU incorporation occurred most frequently in LGR5+ daughters (p = 0.004, one-way ANOVA), consistent with faster-cycling LGR5+ CCIC vs. slower-cycling BMI1+ CCIC.
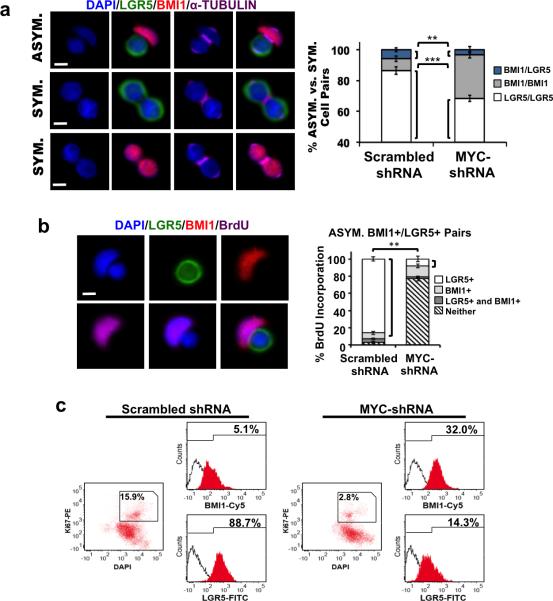
Because LGR5+ daughters have higher WNT signaling levels than BMI1+ daughters (Supplementary Figs. 1E, 2D, 2E), and WNT upregulates MYC, a transcription factor that promotes proliferation and growth (35), we examined the potential role of MYC in promoting differential proliferative capacity by establishing a knockdown model using MYC-shRNA lentiviral transduction. Efficient MYC knockdown was confirmed by Western blot analysis (Supplementary Fig. 3A), but did not alter Notch signaling. NICD expression (Supplementary Fig. 3A) and RT-PCR for NOTCH effectors Hes1 and Hes5 (Supplementary Fig. 3B) showed that CCIC with MYC knockdown maintained similar levels of NOTCH signaling activity compared to control (scrambled shRNA-transduced CCICs). Furthermore, decreased expression of the MYC target genes Cyclin D2 and Ornithine Decarboxylase (ODC), and increased expression of p21 in MYC-shRNA treated CCICs was observed, consistent with the anti-proliferative effect of MYC knockdown (Supplementary Fig. 3B). While overall CCIC proliferation was suppressed by MYC knockdown, co-IF for BMI1, LGR5 and α-TUBULIN expression showed that division of LGR5+ cells was disproportionally affected (Fig. 3a; **, p =0.01; ***, p=0.001; Student t-test). MYC shRNA knockdown also switched BrdU incorporation from the LGR5+ daughter cell to the BMI1+ daughter in asymmetric CCIC pairs (Fig. 3b). Furthermore, quantitative analysis of asymmetric BMI1+/LGR5+ pairs showed that, while overall BrdU incorporation was significantly reduced by MYC knockdown, BrdU incorporation then occurred most frequently in BMI1+ daughters (p = 0.005, one-way ANOVA) (Fig. 3b). This reversal of relative proliferation between BMI1+ and LGR5+ daughter CCICs suggests that LGR5+ CCIC proliferation shows a greater dependence on MYC. We then analyzed how MYC knockdown affects the timing of division in BMI1+ vs. LGR5+ CCIC populations following BrdU incorporation. This showed that the absolute number of proliferating LGR5+ CCIC decreased substantially (p = .002, Student t-test) while the number of dividing BMI1+ CCICs was similar to the control (Supplementary Fig. 3C). FACS analysis confirmed that MYC-shRNA resulted in a smaller population of Ki67+ dividing CCICs compared to the control, and this gated population showed a greater percentage of BMI1+ vs. LGR5+ CCICs (Fig. 3c). LGR5-FITC+ CCIC and CCIC expressing a BMI1-mCherry promoter reporter were infected with scrambled (Sc.) shRNA or MYC-shRNA. Consistent with our earlier observations, asymmetric proliferation and the number of LGR5+ daughters produced was significantly reduced in MYC-shRNA CCIC compared to the control (Supplementary Fig. 3D; p=0.01; Student t-test). Overall, these experiments were consistent with MYC as a determining factor in the differential asymmetric proliferation potential between LGR5+ and BMI1+ CCICs.
Figure 3. MYC knockdown in BMI1+/LGR5+ CCIC-1 daughter pairs.
(a) Single CCIC were lentivirally transduced with scrambled shRNA (Sc. shRNA) or MYC-shRNA. Left: CCIC daughter pairs undergoing BMI1+/LGR5+ asymmetric, LGR5+ symmetric, or BMI1+ symmetric division. LGR5 (green), BMI1 (red), α-TUBULIN (purple), DAPI (blue), scale bar: 10μm. Right: Percentage of BMI1+/LGR5+ asymmetric (blue), BMI1+/BMI1+ symmetric (grey), or LGR5+/LGR5+ symmetric (white) CCIC pairs determined by co-IF for LGR5, BMI1, and α-TUBULIN expression. The data represents mean ± SD from three independent experiments with n = 500 TUBULIN+ pairs/replicate (**, p = 0.0033, one-way ANOVA). (b) BrdU incorporation following division of a single CCIC transduced with MYC-shRNA. Left: co-IF: LGR5 (green), BMI1 (red), BrdU (purple), DAPI (blue); scale bar: 10μm. Right: Quantitative analysis in asymmetric BMI1+/LGR5+ pairs with percentage of BrdU incorporation in LGR5+ CCIC, BMI1+ CCIC, both LGR5+ and BMI1+ CCICs, or neither indicated (**, p = 0.005, one-way ANOVA). The data represents mean ± SD of 3 independent experiments with n = 500 cells/replicate. (c) FACS analysis of CCIC transduced with scrambled shRNA (left) or MYC-shRNA (right) showing expression of LGR5 and BMI1 within the gated Ki67+ population.
NOTCH regulates asymmetric BMI1+/LGR5+ CCIC fate in vitro
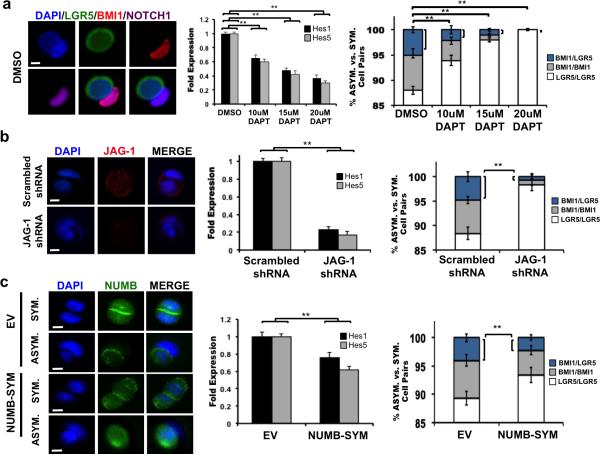
The role of NOTCH in modulating asymmetric CCIC daughter fate in vitro (Fig. 4a) was studied using the pair-cell assay in which CCICs were treated with DMSO or increasing concentrations of DAPT. Co-IF of DMSO-treated CCICs showed the presence of asymmetric BMI1+(NOTCH1+)/LGR5+ cell pairs. Next, RT-PCR for Hes1 and Hes5 confirmed DAPT suppression of NOTCH activity. Increasing concentrations of DAPT reduced CCIC BMI1+/LGR5+ cell pairs compared to the control (p = 0.01; Student t-test). Additionally, asymmetric pairs as determined by nuclear morphology and BMI1/LGR5 expression was not detected in HCT116 or RKO cells under any condition (Supplementary Fig. 4A). In order to confirm that NOTCH affects cell fate determination, we performed NOTCH1, LGR5, and α-TUBULIN co-staining of CCIC pairs just before completion of cell division in DMSO- and DAPT-treated cells (Supplementary Fig. 4B). Asymmetric distribution of NOTCH1 and LGR5 was observed in the control group compared to symmetric expression of both markers with DAPT treatment, suggesting that higher NOTCH signaling promotes asymmetric BMI1/LGR5 daughter cell fates. FACS analysis following a short, 24-hour DAPT treatment also showed that NOTCH suppression decreased the BMI1+/LGR5+ double positive population containing the asymmetric pairs and reduced the ratio of BMI1+ vs. LGR5+ CCIC populations (Supplementary Fig. 4C). Finally, LGR5-FITC+ CCIC and CCIC expressing a BMI1-mCherry promoter reporter were treated with DMSO or DAPT. Consistent with our earlier observations, asymmetric proliferation and the number of BMI1+ daughters produced was significantly reduced in DAPT-treated CCIC compared to the control (Supplementary Fig. 4D; p=0.01; Student t-test).
Figure 4. NOTCH suppression decreases BMI1+/LGR5+ CCIC-1 daughter pairs.
(a) Pair-cell assay of CCIC treated with DMSO vs. DAPT. Left: NOTCH1 (purple) expression in BMI1+(red)/LGR5+(green) pair. DAPI labels nuclei; scale bar: 10μm. Middle: RT-qPCR measurements of Hes1 and Hes5 expression. Right: Fraction of BMI1+/LGR5+ asymmetric (blue), BMI1+/BMI1+ symmetric (grey) and LGR5+/LGR5+ symmetric (white) cell pairs for each treatment condition (**, p = 0.01; one-way ANOVA). (b) Left: JAG-1 (red) staining of CCIC daughter pairs infected with scrambled shRNA (control) or JAG-1 shRNA. Scale bar: 10μm. Middle: JAG-1 knockdown decreased Hes1 and Hes5. Right: JAG-1 shRNA reduced the frequency of asymmetric BMI1+/LGR5+ pairs (blue) in CCICs (**, p = 0.0028; Student t-test). (c) Left: NUMB (green) staining in empty vector (EV) or NUMB-SYM-transduced CCIC. Scale bar: 10μm. Middle: Hes1 and Hes5 expression measured by RT-qPCR. Right: Fraction of asymmetric BMI1+/LGR5+ cell pairs (blue) decreased in NUMB-SYM expressing CCICs (**, p = 0.0022; Student t-test). In all panels, RT-PCR was performed in triplicate and is presented as mean ± SD; data from cell pair assays represents mean ± SD from three independent experiments with n = 500 TUBULIN+ pairs/replicate.
Next, the NOTCH ligand JAG-1 was targeted for knockdown. CCICs were infected with JAG-1 shRNA or with scrambled shRNA (control) lentiviral particles prior to performing the pair cell assay (Fig. 4b). IF, Western blot (Supplementary Fig. 4E), and gene expression analysis for NOTCH effectors Hes1 and Hes5 confirmed the knockdown efficiency for JAG-1. JAG-1 shRNA significantly decreased asymmetric BMI1+/LGR5+ daughter cell pairs to less than 1% (p = 0.0028; Student t-test) compared to the control group (~5%). Finally, LGR5-FITC+ CCIC and CCIC expressing a BMI1-mCherry promoter reporter were also infected with scrambled shRNA or JAG-1 shRNA. We observed asymmetric proliferation and the number of BMI1+ daughters produced decreased in JAG-1 shRNA CCIC compared to the control (Supplementary Fig. 4E; p=0.01; Student t-test).
In other cases of NOTCH mediated asymmetric cell fate determination, NUMB, an inhibitor of NOTCH, localizes to one side of the dividing cell and causes degradation of membrane-bound NOTCH receptors and NICD (36), which creates differential NOTCH signaling levels in daughter cells (37). We tested a mutant NUMB (NUMB-SYM), which suppresses NOTCH signaling but does not partition differentially among daughters (38). Lentiviral expression of NUMB-SYM and subsequent pair cell assay analysis using IF for NUMB localization confirmed overexpression of NUMB segregation in both symmetric and asymmetric NUMB-SYM expressing CCIC daughter pairs compared to the control group (Fig. 4c). Western blotting confirmed an increase of NUMB protein (Supplementary Fig. 4F) and RTPCR analysis of Hes1 and Hes5 validated decreased expression from the NUMB-SYM expressing CCICs vs. empty vector-transduced (control) CCICs. Similar to NOTCH inhibition, NUMB-SYM led to a decrease in the number of BMI1+/LGR5+ cell pairs from approximately 4.5% (control) to 2% (p = 0.0022; Student t-test). LGR5-FITC+ CCIC and CCIC expressing a BMI1-mCherry promoter reporter were also transduced with an empty vector control or NUMB-SYM. We observed asymmetric proliferation and the number of BMI1+ daughters produced decreased in NUMB-SYM CCIC compared to the control (Supplementary Fig. 4F; p=0.01; Student t-test). The suppression of NOTCH signaling by NUMB-SYM is consistent with higher NOTCH activity promoting asymmetric CCIC daughter fates.
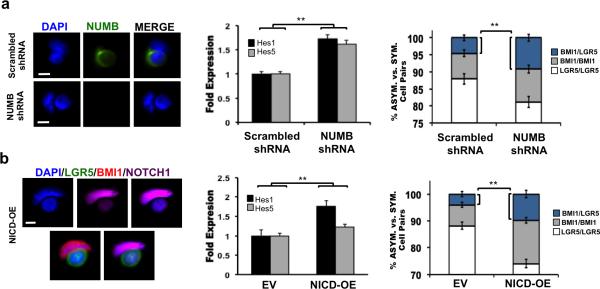
The effect of NUMB knockdown on asymmetric BMI1+/LGR5+ CCIC pairs was also examined (Fig. 5a). CCICs were infected with lentivirus to stably express NUMB shRNA or scrambled shRNAs (control). Consistent with NUMB knockdown, IF microscopy showed diminished NUMB expression, western blotting confirmed reduced NUMB protein levels (Supplementary Fig. 5A), and qPCR analysis of NOTCH signaling targets showed increased Hes1 and Hes5 expression. Targeted knockdown of NUMB increased the number of asymmetric BMI1+/LGR5+ cell pairs from 5% (control) to 9.5% (p = 0.008; Student t-test). LGR5-FITC+ CCIC and CCIC expressing a BMI1-mCherry promoter reporter were also infected with scrambled shRNA or NUMB shRNA. We observed asymmetric proliferation and the number of BMI1+ daughters produced increased in NUMB shRNA CCIC compared to the control (Supplementary Fig. 5A; p=0.01; Student t-test). Thus, NUMB is not required for CCIC asymmetric cell fate determination to generate BMI1+ and LGR5+ daughter pairs.
Figure 5. NOTCH signaling promotes BMI1+/LGR5+ CCIC-1 daughter pairs.
(a) Left: NUMB (green) staining of CCIC daughter pairs infected with scrambled shRNA (control) or NUMB-shRNA. Scale bar: 10μm. Middle: NUMB knockdown increased Hes1 and Hes5 expression. Right: Frequency of asymmetric BMI1+/LGR5+ pairs (blue) increased upon NUMB knockdown (**, p = 0.008; Student t-test). (b) CCICs were infected with an ectopic NICD expression (NICD-OE) construct. Left: Co-IF of NICD-OE BMI1+(red)/ LGR5+(green) CCIC pair, indicating differential asymmetric NOTCH1 (purple) expression. Middle: Hes1 and Hes5 levels measured by RT-qPCR. Right: NICD-OE increased the frequency of asymmetric BMI1+/LGR5+ pairs (blue) (**, p = 0.0019; Student t-test). In all panels, RT-PCR was performed in triplicate and is presented as mean ± SD; data from cell pair assays represents mean ± SD from three independent experiments with n = 500 TUBULIN+ pairs/replicate.
Finally, the NOTCH intracellular domain (NICD), which can translocate to the nucleus to activate expression of NOTCH effector genes in CCICs, was overexpressed (Fig. 5b). Following NICD-OE, co-IF of BMI1/LGR5+ CCIC pairs showed differential NOTCH1 expression levels in which the BMI1+ daughter was NOTCH1-high compared to the LGR5+ (NOTCH1-low) daughter. CCICs that incorporated the NICD-OE construct showed correspondingly increased NICD as well as HES1 and HES5 through Western blotting (Supplementary Fig. 5B) and RTPCR analyses. NICD overexpression increased asymmetric BMI1+/LGR5+ cell pairs to approximately 10% compared to 4.5% in the empty vector-transduced (control) group (p = 0.0029; Student t-test). Next, LGR5-FITC+ CCIC and CCIC expressing a BMI1-mCherry promoter reporter were also transduced with an empty vector control or NICD-OE. We observed asymmetric proliferation and the number of BMI1+ daughters produced increased in NICD-OE CCIC compared to the control (Supplementary Fig. 5B; p=0.01; Student t-test). BMI1 expression was elevated in NICD-CCICs (Supplementary Fig. 5C), consistent with previous reports showing BMI1 as a downstream target of NOTCH in T cells (39) and murine intestinal cells (40). NOTCH1, LGR5, and α-TUBULIN co-staining of NICD-OE CCIC pairs showed that differential asymmetric distribution of NOTCH1 preceded the end of cell division (Supplementary Fig. 5D). These data suggest a mechanistic role for NOTCH signaling in driving generation of BMI1+/LGR5+ CCIC asymmetric daughter cell pairs.
BMI1+ and LGR5+ CCICs interconvert in xenograft tumors
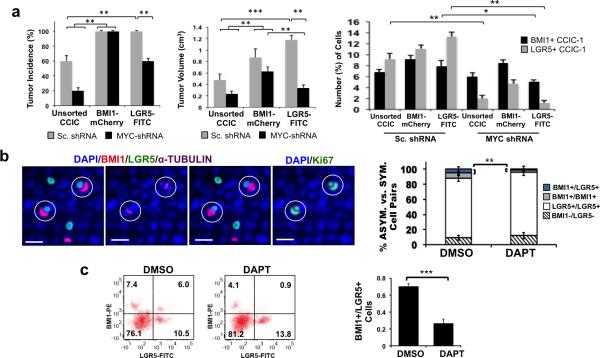
We then examined the functional role of BMI1+ and LGR5+ CCICs in xenograft tumors. Using the model we established earlier, LGR5-FITC+ CCIC and CCIC expressing a BMI1-mCherry promoter reporter were infected with scrambled (Sc.) shRNA or MYC-shRNA. Subsequently, 1×106 unsorted, BMI1+, or LGR5+ single CCICs expressing Sc. shRNA or MYC-shRNA were injected subcutaneously (s.c.) into NOD/SCID mice to form tumors, which were evaluated after 4 weeks (Fig. 6a; Supplementary Fig. 6A). FACS analysis revealed that tumors derived from BMI1+ or LGR5+ CCICs infected with the Sc. control contained both BMI1+ and LGR5+ subfractions with similar ratios. Conversely, MYC knockdown disproportionately decreased tumor incidence and tumor size derived from LGR5+ CCIC compared to the Sc. shRNA control (Fig. 6a; ***, p =0.0004; **, p=0.01; one-way ANOVA). FACS analysis of dissociated tumors similarly showed that the percentage of LGR5+ cells (~13.1%) in LGR5+ CCIC-Sc. shRNA tumors was significantly reduced to ~1.5% in MYC-shRNA tumors. Likewise, asymmetric proliferation potential to produce BMI1+ cells from LGR5+ CCIC tumors decreased with MYC knockdown compared to the control (**, p=0.01; *, p=0.02; one-way ANOVA). Testing another primary CCIC line (CCIC-2) by the same FACS and xenograft assays (Supplementary Fig. 6B) was consistent with these data, suggesting that BMI1+ CCICs and LGR5+ CCICs co-exist, are approximately balanced within a tumor, and can generate each other potentially through asymmetric division. Overall, these findings show MYC-dependent LGR5+ proliferation in CCIC.
Figure 6. Xenograft tumors derived from CCIC-1 line.
(a) CCIC-1 cells expressing a BMI1-mcherry promoter reporter were lentivirally transduced with scrambled shRNA (Sc. shRNA) or MYC-shRNA. 1×106 unsorted, BMI1+, or LGR5+ sorted CCICs were injected s.c. into NOD/SCID mice (n = 5) to develop tumors for 4 weeks. Tumor incidence (left), tumor volume (middle), and the percentage of BMI1+ or LGR5+ CCIC from tumors (right) are indicated. The data represents mean ± SD. (***, p =0.0004; **, p=0.01; *, p=0.02; one-way ANOVA). (b) 1×106 unsorted CCICs were injected s.c. into NOD/SCID mice (n = 6) to develop tumors for 4 weeks; tumors were then injected with DMSO or DAPT over 72 hours. Left: Tumor co-IF: BMI1+(red), LGR5+(green), α-TUBULIN (purple), Ki67 (green), DAPI (blue); scale bar: 25 μm. Right: Frequency of BMI1+/LGR5+ asymmetric (blue), BMI1+/BMI1+ symmetric (grey), or LGR5+/LGR5+ symmetric (white) cell pairs in xenograft tumors determined by co-IF for LGR5, BMI1, and α-TUBULIN. The data represent mean ± SD of n = 500 TUBULIN+ dividing pairs/mouse (**, p = 0.003, Student t-test). (c) RepresentativeFACS plots (left) and the ratio of BMI1+/LGR5+ cells determined by FACS (right) from tumors described in (f). Data represents mean ± SD of n = 3 mice/condition. (***, p =0.0002; Student t-test).
NOTCH signaling balances BMI1+ and LGR5+ CCICs in vivo
NOTCH levels are higher in the slow-cycling (BMI1+) daughter than in the fast-cycling (LGR5+) daughter, raising the possibility that NOTCH signaling may play a role in regulating asymmetric BMI1+/LGR5+ cell fate. We first studied the effect of NOTCH suppression on generating BMI1+/LGR5+ asymmetric daughters in xenograft tumors. 1 × 106 unsorted CCICs were injected s.c. into NOD/SCID mice to form tumors; after 4 weeks, tumors were directly injected with DMSO or NOTCH inhibitor DAPT over 72 hours before being harvested and assayed. The short treatment duration was chosen to evaluate effects on cell fate determination while avoiding potential longer term effects on senescence or apoptosis. Western blot analysis confirmed NICD suppression and RT-PCR data showed decreased expression of NOTCH downstream targets Hes1 and Hes5 upon DAPT treatment (Supplementary Fig. 6C). Co-IF for BMI1, LGR5, and α-TUBULIN as well as Ki67 staining confirmed the existence of asymmetric BMI1+/LGR5+ dividing pairs in tumor sections. DAPT treatment reduced the frequency of BMI1+/LGR5+ pairs to 0.6% compared to 5.2% in DMSO-treated tumors (p = 0.003; Student t-test) (Fig. 6b). FACS analysis of dissociated xenograft tumors showed that DAPT treatment also reduced the ratio of BMI1+ vs. LGR5+ CCIC populations (p =0.0002; Student t-test) within 72 hours (Fig. 6c). These results indicate NOTCH signaling promotes asymmetric CCIC daughter fate and balances BMI1+ and LGR5+ populations.
DISCUSSION
CCICs from the same tumor are often treated as a homogenous population. Here we demonstrate that fast- and slow-cycling subfractions can co-exist and CCICs can asymmetrically divide to generate both types in a tumor, expanding our understanding of CCICs. Specifically, we show that NOTCH and MYC are important mechanisms in regulating the balance between the two cell populations, although other signaling pathways may additionally impact slow- and fast-cycling stem cell pools (41, 42). NOTCH-dependent asymmetric cell fate determination may help to establish CCIC diversity by maintaining both slow-cycling, MYC-independent BMI1+ CCICs and fast-cycling, MYC-dependent LGR5+ CCICs.
Despite their relatively low frequency, slow-cycling cells may still serve an important role as reserve stem cells. In the context of CRC, CCICs may leverage this mechanism to generate a slow-cycling, long-lasting, more chemo-resistant reserve population, establishing an important link to tumor growth and repopulation. Promotion of slow-cycling BMI1+ CCICs by NOTCH signaling may support chemoresistance (43), although their exact role in CRC remains unknown. The ability of both BMI1+ and LGR5+ CCICs to undergo asymmetric division in order to generate each other highlights plasticity among CCICs, suggesting also that these lineages may be somewhat unique in cancer. This direct interconversion process may provide CRCs with a growth and survival strategy to promote self-renewal, tumorigenicity, and chemoresistance. Chemotherapies that focus on targeting fast-proliferating cells and shrinking tumor volume may inadvertently enrich the slow-cycling CCIC population, giving rise to recurrence and chemoresistance.
Supplementary Material
Footnotes
The authors declare no conflicts of interest.
SUPPLEMENTAL INFORMATION
Supplemental information contains additional methods, six figures and one movie.
REFERENCES
- 1.Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nature medicine. 2014;20(1):29–36. doi: 10.1038/nm.3418. doi: 10.1038/nm.3418. PubMed PMID: 24292392. [DOI] [PubMed] [Google Scholar]
- 2.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–807. doi: 10.1182/blood-2008-08-077941. doi: 10.1182/blood-2008-08-077941. PubMed PMID: 19064739. [DOI] [PubMed] [Google Scholar]
- 3.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–37. doi: 10.1038/nature12624. doi: 10.1038/nature12624. PubMed PMID: 24048065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer research. 2010;70(4):1469–78. doi: 10.1158/0008-5472.CAN-09-2557. doi: 10.1158/0008-5472.CAN-09-2557. PubMed PMID: 20145124; PubMed Central PMCID: PMC4010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(24):10158–63. doi: 10.1073/pnas.0703478104. doi: 10.1073/pnas.0703478104. PubMed PMID: 17548814; PubMed Central PMCID: PMC1891215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell stem cell. 2011;8(5):511–24. doi: 10.1016/j.stem.2011.02.020. doi: 10.1016/j.stem.2011.02.020. PubMed PMID: 21419747. [DOI] [PubMed] [Google Scholar]
- 7.Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nature cell biology. 2014;16(3):268–80. doi: 10.1038/ncb2910. doi: 10.1038/ncb2910. PubMed PMID: 24561623. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y, et al. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer cell. 2012;21(6):777–92. doi: 10.1016/j.ccr.2012.04.036. doi: 10.1016/j.ccr.2012.04.036. PubMed PMID: 22698403. [DOI] [PubMed] [Google Scholar]
- 9.Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal Cancer Stem Cells: From the Crypt to the Clinic. Cell stem cell. 2014;15(6):692–705. doi: 10.1016/j.stem.2014.11.012. doi: 10.1016/j.stem.2014.11.012. PubMed PMID: 25479747. [DOI] [PubMed] [Google Scholar]
- 10.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nature reviews Molecular cell biology. 2014;15(1):19–33. doi: 10.1038/nrm3721. doi: 10.1038/nrm3721. PubMed PMID: 24326621. [DOI] [PubMed] [Google Scholar]
- 11.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11452–7. doi: 10.1073/pnas.1106083108. doi: 10.1073/pnas.1106083108. PubMed PMID: 21693646; PubMed Central PMCID: PMC3136304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO reports. 2011;12(10):1055–61. doi: 10.1038/embor.2011.175. doi: 10.1038/embor.2011.175. PubMed PMID: 21909076; PubMed Central PMCID: PMC3185347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziskin JL, Dunlap D, Yaylaoglu M, Fodor IK, Forrest WF, Patel R, et al. In situ validation of an intestinal stem cell signature in colorectal cancer. Gut. 2013;62(7):1012–23. doi: 10.1136/gutjnl-2011-301195. doi: 10.1136/gutjnl-2011-301195. PubMed PMID: 22637696. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Yoon SY, Kim CN, Joo JH, Moon SK, Choe IS, et al. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer letters. 2004;203(2):217–24. doi: 10.1016/j.canlet.2003.07.009. PubMed PMID: 14732230. [DOI] [PubMed] [Google Scholar]
- 15.Li DW, Tang HM, Fan JW, Yan DW, Zhou CZ, Li SX, et al. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. Journal of cancer research and clinical oncology. 2010;136(7):997–1006. doi: 10.1007/s00432-009-0745-7. doi: 10.1007/s00432-009-0745-7. PubMed PMID: 20024662. [DOI] [PubMed] [Google Scholar]
- 16.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem cells. 2012;30(3):372–8. doi: 10.1002/stem.1035. doi: 10.1002/stem.1035. PubMed PMID: 22252887. [DOI] [PubMed] [Google Scholar]
- 17.Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell stem cell. 2013;12(5):602–15. doi: 10.1016/j.stem.2013.03.002. doi: 10.1016/j.stem.2013.03.002. PubMed PMID: 23642368; PubMed Central PMCID: PMC3646336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer cell. 2011;20(3):328–40. doi: 10.1016/j.ccr.2011.08.011. doi: 10.1016/j.ccr.2011.08.011. PubMed PMID: 21907924; PubMed Central PMCID: PMC3297490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji A, et al. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. Epub 2011/09/02. doi: cddis201180 [pii] 10.1038/cddis.2011.80. PubMed PMID: 21881602; PubMed Central PMCID: PMC3186899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(15):6309–14. doi: 10.1073/pnas.0900427106. Epub 2009/03/03. doi: 0900427106 [pii] 10.1073/pnas.0900427106. PubMed PMID: 19251639; PubMed Central PMCID: PMC2649205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–63. doi: 10.1038/nature03659. doi: 10.1038/nature03659. PubMed PMID: 15959515. [DOI] [PubMed] [Google Scholar]
- 22.Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(15):6315–20. doi: 10.1073/pnas.0813221106. Epub 2009/03/28. doi: 0813221106 [pii] 10.1073/pnas.0813221106. PubMed PMID: 19325125; PubMed Central PMCID: PMC2669348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reedijk M, Odorcic S, Zhang H, Chetty R, Tennert C, Dickson BC, et al. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncol. 2008;33(6):1223–9. doi: 10.3892/ijo_00000112. PubMed PMID: 19020755; PubMed Central PMCID: PMC2739737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng RD, Shelton CC, Li YM, Qin LX, Notterman D, Paty PB, et al. gamma-Secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer research. 2009;69(2):573–82. doi: 10.1158/0008-5472.CAN-08-2088. doi: 10.1158/0008-5472.CAN-08-2088. PubMed PMID: 19147571; PubMed Central PMCID: PMC3242515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonoshita M, Aoki M, Fuwa H, Aoki K, Hosogi H, Sakai Y, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer cell. 2011;19(1):125–37. doi: 10.1016/j.ccr.2010.11.008. doi: 10.1016/j.ccr.2010.11.008. PubMed PMID: 21251616. [DOI] [PubMed] [Google Scholar]
- 26.Chen HJ, Sun J, Huang Z, Hou H, Jr., Arcilla M, Rakhilin N, et al. Comprehensive models of human primary and metastatic colorectal tumors in immunodeficient and immunocompetent mice by chemokine targeting. Nat Biotechnol. 2015;33(6):656–60. doi: 10.1038/nbt.3239. doi: 10.1038/nbt.3239. PubMed PMID: 26006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(51):20558–63. doi: 10.1073/pnas.0710156104. doi: 10.1073/pnas.0710156104. PubMed PMID: 18077357; PubMed Central PMCID: PMC2154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan XS, Wu HY, Yu HP, Zhou Q, Zhang YF, Huang Q. Expression of Lgr5 in human colorectal carcinogenesis and its potential correlation with beta-catenin. International journal of colorectal disease. 2010;25(5):583–90. doi: 10.1007/s00384-010-0903-z. doi: 10.1007/s00384-010-0903-z. PubMed PMID: 20195621. [DOI] [PubMed] [Google Scholar]
- 29.Simon E, Petke D, Boger C, Behrens HM, Warneke V, Ebert M, et al. The spatial distribution of LGR5+ cells correlates with gastric cancer progression. PloS one. 2012;7(4):e35486. doi: 10.1371/journal.pone.0035486. doi: 10.1371/journal.pone.0035486. PubMed PMID: 22530031; PubMed Central PMCID: PMC3329462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganguli-Indra G, Wasylyk C, Liang X, Millon R, Leid M, Wasylyk B, et al. CTIP2 expression in human head and neck squamous cell carcinoma is linked to poorly differentiated tumor status. PloS one. 2009;4(4):e5367. doi: 10.1371/journal.pone.0005367. doi: 10.1371/journal.pone.0005367. PubMed PMID: 19399189; PubMed Central PMCID: PMC2671404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129(20):4843–53. doi: 10.1242/dev.129.20.4843. PubMed PMID: 12361975. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds A, Wharton N, Parris A, Mitchell E, Sobolewski A, Kam C, et al. Canonical Wnt signals combined with suppressed TGFbeta/BMP pathways promote renewal of the native human colonic epithelium. Gut. 2014;63(4):610–21. doi: 10.1136/gutjnl-2012-304067. doi: 10.1136/gutjnl-2012-304067. PubMed PMID: 23831735; PubMed Central PMCID: PMC3963552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer research. 2009;69(8):3382–9. doi: 10.1158/0008-5472.CAN-08-4418. doi: 10.1158/0008-5472.CAN-08-4418. PubMed PMID: 19336570; PubMed Central PMCID: PMC2789401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gracz AD, Fuller MK, Wang F, Li L, Stelzner M, Dunn JC, et al. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem cells. 2013;31(9):2024–30. doi: 10.1002/stem.1391. doi: 10.1002/stem.1391. PubMed PMID: 23553902; PubMed Central PMCID: PMC3783577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espada J, Calvo MB, Diaz-Prado S, Medina V. Wnt signalling and cancer stem cells. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2009;11(7):411–27. doi: 10.1007/s12094-009-0380-4. PubMed PMID: 19574199. [DOI] [PubMed] [Google Scholar]
- 36.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. The Journal of biological chemistry. 2003;278(25):23196–203. doi: 10.1074/jbc.M302827200. doi: 10.1074/jbc.M302827200. PubMed PMID: 12682059. [DOI] [PubMed] [Google Scholar]
- 37.Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes & development. 2009;23(23):2675–99. doi: 10.1101/gad.1850809. doi: 10.1101/gad.1850809. PubMed PMID: 19952104; PubMed Central PMCID: PMC2788323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong W, Jiang MM, Weinmaster G, Jan LY, Jan YN. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 1997;124(10):1887–97. doi: 10.1242/dev.124.10.1887. PubMed PMID: 9169836. [DOI] [PubMed] [Google Scholar]
- 39.Schaller MA, Logue H, Mukherjee S, Lindell DM, Coelho AL, Lincoln P, et al. Delta-like 4 differentially regulates murine CD4 T cell expansion via BMI1. PloS one. 2010;5(8):e12172. doi: 10.1371/journal.pone.0012172. doi: 10.1371/journal.pone.0012172. PubMed PMID: 20808960; PubMed Central PMCID: PMC2923143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Arribillaga E, Rodilla V, Pellegrinet L, Guiu J, Iglesias M, Roman AC, et al. Bmi1 regulates murine intestinal stem cell proliferation and self-renewal downstream of Notch. Development. 2015;142(1):41–50. doi: 10.1242/dev.107714. doi: 10.1242/dev.107714. PubMed PMID: 25480918. [DOI] [PubMed] [Google Scholar]
- 41.Dey-Guha I, Alves CP, Yeh AC, Salony, Sole X, Darp R, et al. A mechanism for asymmetric cell division resulting in proliferative asynchronicity. Molecular cancer research : MCR. 2015;13(2):223–30. doi: 10.1158/1541-7786.MCR-14-0474. doi: 10.1158/1541-7786.MCR-14-0474. PubMed PMID: 25582703; PubMed Central PMCID: PMC4336804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dey-Guha I, Wolfer A, Yeh AC, J GA, Darp R, Leon E, et al. Asymmetric cancer cell division regulated by AKT. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(31):12845–50. doi: 10.1073/pnas.1109632108. doi: 10.1073/pnas.1109632108. PubMed PMID: 21757645; PubMed Central PMCID: PMC3150943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 2013;34(7):1420–30. doi: 10.1093/carcin/bgt127. doi: 10.1093/carcin/bgt127. PubMed PMID: 23585460; PubMed Central PMCID: PMC3697894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.