Abstract
Aberrant regulation of cellular extrusion can promote invasion and metastasis. Here, we identify molecular requirements for early cellular invasion using a premalignant mouse model of pancreatic cancer with conditional knockout of p120 catenin (Ctnnd1). Mice with biallelic loss of p120 catenin progressively develop high grade PanIN lesions and neoplasia accompanied by prominent acute and chronic inflammatory processes, which is mediated in part through nuclear factor-kB (NF-kB) signaling. Loss of p120 catenin in the context of oncogenic Kras also promotes remarkable apical and basal epithelial cell extrusion. Abundant single epithelial cells exit PanIN epithelium basally, retain epithelial morphology, survive, and display features of malignancy. Similar extrusion defects are observed following p120 catenin knockdown in vitro, and these effects are completely abrogated by activation of S1P/S1pr2 signaling. In the context of oncogenic Kras, p120 catenin loss significantly reduces expression of genes mediating S1P/S1pr2 signaling in vivo and in vitro, and this effect is mediated at least in part through activation of NF-kB. These results provide insight into mechanisms controlling early events in the metastatic process and suggest that p120 catenin and S1P/S1pr2 signaling enhance cancer progression by regulating epithelial cell invasion.
Keywords: pancreatic cancer, epithelial extrusion, p120 catenin, adherens junctions, invasion, pancreatitis
INTRODUCTION
Genetic and epigenetic alterations in genes encoding cell adhesion molecules are a hallmark of many epithelial cancers. For pancreas cancer, homophilic cell adhesion has been categorized as one of twelve core signaling pathways (1). Homophilic cell adhesion in epithelial cells is mediated partly through cadherins and catenins in adherens junctions. Misexpression of the adherens junction protein p120 catenin has been identified in several types of human carcinomas (2). Studies have suggested variable roles for p120 catenin in the pathogenesis of epithelial cancers, to include tumor suppression and metastatic progression (3,4). In human pancreatic cancer, misexpression of p120 catenin in primary tumors is significantly correlated with vascular invasion, metastasis, differentiation, pTNM stage, and poor survival (5–7). Reduction and cytoplasmic relocalization of p120 catenin has also been reported in 100% of solid pseudopapillary tumors of the pancreas (8). A study using a forward genetic screen in mice identified Ctnnd1 as a “candidate cancer gene” in Kras-driven pancreatic neoplasia (7). These data suggest that disruption of CTNND1 in pancreatic tumors has biological relevance to disease, yet, the mechanisms by which p120 catenin contributes to the development and progression of pancreatic cancer are not understood.
Increased occurrence of metastasis with altered p120 catenin expression suggests that p120 catenin may play a role in metastatic progression of pancreatic cancer. A mechanism recently hypothesized to initiate metastasis by mediating invasion is basal epithelial cell extrusion (9,10). Epithelial tissues maintain homeostatic cell numbers by extruding cells through a highly conserved mechanism involving production and secretion of the signaling lipid sphingosine 1-phosphate (S1P). Extracellular S1P binds to S1P receptor 2 (S1pr2) on neighboring cells, which induces contraction of an actomyosin band that extrudes the cell out of the epithelium while preserving barrier function (11–17). Mutations in the tumor suppressor APC and oncogenic Kras have been shown to shift the predominant direction of epithelial cell extrusion from apical to basal, where extruded cells invade the underlying epithelium and survive (18,19). The results presented in this study show that p120 catenin restrains epithelial cell extrusion in the earliest stages of pancreatic neoplastic invasion, via a S1P/S1pr2-dependent mechanism.
MATERIALS AND METHODS
Human pancreatic tissue microarrays
For expression analysis, a labeling score of 0–2 corresponding to absent/low, medium, and high was assigned using immunohistochemistry (IHC), and immunofluorescence (IF) staining or IHC was scored for predominant subcellular localization analysis. p120 catenin expression level and predominant subcellular localization were each scored by 3 independent observers blinded to lesion classification. In this study, predominant is defined as greater than or equal to 60% of the representative staining pattern or expression level.
Mice
Transgenic mouse strains Mist1CreER/+ (CiMist1) (20), Ctnnd1tm1Abre (p120f) (21), and lox-stop-lox; KrasG12D (K) (22) have been previously described. To perform lineage tracing, we used the Rosa26mTmG (G) (23) double fluorescent reporter allele. Transgenic strains Ptf1atm1(cre)Wri (CPtf1a) (24), lox-stop-lox; KrasG12D (K) (22), p53LoxP (P) (25), and R26R-YFP (Y) (26) were used to generate KPCPtf1aY mice, which were sacrificed between 6 and 8 weeks of age and maintained on a mixed genetic background. CiMist1, p120f, K, and G mice were maintained on a C57BL/6J background. To induce Cre recombination, mice were injected with 5mg tamoxifen (Sigma, T5648) subcutaneously once per day for 3 consecutive days. Experimental pancreatitis was elicited as previously described (27). Mice were genotyped by PCR or Transnetyx. All pancreatic pathologies in transgenic mice and humans were classified by a pathologist. For NF-kB inhibition experiments, mice were injected intraperitoneally with 5mg/kg/day SN50 (Santa Cruz, sc-3060). All animal studies were approved by the Animal Care and Use Committees at Johns Hopkins University and University of Texas Health Science Center at Houston.
Histology/immunostaining
Tissues were fixed in 4% paraformaldehyde at 4°C, processed according to standard protocols, and embedded in paraffin. Antigen retrieval was performed using heat-mediated microwave methods and an antigen unmasking solution (Vector Laboratories, H-3300) for all antibodies except rat-anti-CD45, for which Retrievit 6 (BioGenex, BS-1006-00) was used. All sections were blocked in 10% FBS in PBST and primary antibodies were incubated overnight at 4°C. Secondary antibodies, from Jackson Immunoresearch, were used at 1:250 and incubated at room temperature for 2 hours for IF and 30 min for IHC. For IF, slides were stained with IHC-Tek Dapi counterstain solution (IHC World, IW-1404) and mounted in fluorescence mounting medium (Dako, S3023). For IHC, Vectastain Elite ABC kit (Vector Laboratories, PK-6100) and DAB Peroxidase (HRP) Substrate kit (Vector Laboratories, SK-4100) were used. Primary antibodies used in this study are described in Supplemental Table 1.
Premalignant lesion quantification
ADM, PanIN1, PanIN2/3, and fibrostroma were quantified using morphometric analysis on scanned H&E slides in ImageJ. 2 sections per animal sampled at least 400μm apart were analyzed. Quantification of pancreatic area excluded lymph nodes.
CK19 quantification
For quantification of CK19+ basal cell extrusion, CK19+ cells (excluding apically extruded CK19+ cells and normal pancreatic ducts) were counted in 1 scanned section per animal. For quantification of CK19+ apical cell extrusion, CK19+ cells that comprised a luminal pancreatic epithelial structure (lumen sized at least twice the diameter of a cell comprising the epithelial structure and excluding normal ducts) and its associated apically extruded CK19+ cells were counted in 1 scanned section per animal.
Quantification of cerulean-induced pancreatic injury
Pancreatic injury, defined as area containing metaplastic duct lesions and/or associated stroma, was quantified in 1 scanned H&E section per animal using morphometric analysis in ImageJ. Quantification of pancreatic area excluded lymph nodes.
DNA ploidy analysis
DNA ploidy cell cycle analysis of basally extruded single epithelial cells on 4μm thick Feulgen-stained sections was accomplished using OTMIAS Version 2.0 Image Analysis Software by Olive Tree Media, LLC. Serial sections stained by CK19 IHC were used for identification of isolated, basally extruded epithelial cells in Feulgen stained sections. The internal reference control and isolated epithelial cells analyzed were located on the same Feulgen stained section. An aneuploid peak is defined as any distinct peak with a DNA index > 1.25. Abnormal DNA content is defined as any aneuploid peak or any peak ≥5C.
RNA isolation, microarray, and qPCR
Adult pancreatic cells were dissociated as previously described (28). RNA was isolated from sorted GFP+ cells using Arcterus PicoPure RNA isolation kit and gene expression was analyzed using Mouse exon microarray 1.0 ST (Affymetrix). For qPCR experiments, reverse transcription was accomplished using QuantiTect reverse transcription kit (Qiagen, 205311). Complementary DNA was amplified using TaqMan gene expression assays (Life Technologies).
Cell culture/immunostaining
CFPAC-1 and AsPC-1 cells were obtained from and authenticated by ATCC using morphology, karyotyping, and PCR based approaches. Cells were maintained in Dulbecco’s minimum essential medium (DMEM), low glucose, GlutaMAX™ Supplement (Life Technologies, 10567-022) supplemented with 10% FBS (Sigma-Aldrich, F4135-500ML) and 1X penicillin-streptomycin-glutamine (Fisher Scientific, 10378-016) at 5% CO2, 37°C. For culturing CFPAC-1 cells in Matrigel, a single cell suspension was resuspended in 4% Matrigel (BD Biosciences, 356234) at a final concentration of 6 × 104 cells/mL. 300μL cells per well was placed in 24 well glass bottom plates (In Vitro Scientific, P24-1.5H-N) coated with a thin polymerized layer of Matrigel. For immunostaining, cells were fixed in 4% paraformaldehyde at 37°C for 20 minutes, permeabilized for 10 minutes with 0.2% Triton X-100, blocked with 10% FBS in PBST for 30 minutes, and incubated with primary antibodies for 2 hours at RT. Subsequently, cells were incubated with secondary antibodies (Jackson Immunoresearch) at RT for 2 hours. Dapi was used for nuclear staining and cells were mounted in fluorescence mounting medium (Dako, S3023).
Western blotting
Cell extracts were prepared according to standard protocols using cell lysis buffer (Cell Signaling Technology, 9803S) with protease inhibitor cocktail tablets (Roche, 4693159001) and 100 mM PMSF. Membranes were incubated with primary antibodies overnight at 4°C. After RT incubation with the respective HRP-conjugated secondary antibody used at 1:5000 for 1 hour, membranes were developed using either the SuperSignal West Pico Chemiluminescent Substrate (Thermo scientific, 34080) or the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo scientific, 34095). Quantification of Western blot images was performed in ImageJ.
Transfection/drug treatment
CFPAC-1 cells grown in Matrigel formed spheres by Day 2 and were transfected on Day 2 using lipofectamine RNAiMAX (Invitrogen, 56532). The final concentration of siRNA used per well was 5pmol. siRNA against p120 catenin and control siRNA were obtained from Santa Cruz (sc-36139 and sc-37007, respectively). For S1pr2 agonist experiments, cells were treated with 10μM CYM-5520 (Sigma, SML1014-25MG) on Day 4, 48 hours after siRNA transfection. CFPAC-1 spheres were fixed on Day 6, 96 hours after transfection, for analysis. Day 0 is defined as the day of plating the cells in Matrigel. For in vitro SN50 experiments, AsPC-1 cells were transfected using lipofectamine RNAiMAX in a 6 well plate with a final siRNA concentration of 75pmol. 24 hours after siRNA transfection, cells were treated with either dH20 or 18 μM SN50 for 24 hours. 48 hours after siRNA transfection, cells were harvested.
Accession number
The Gene Expression Omnibus accession number for the microarray analysis reported in this paper is GSE68090.
Statistical analysis
Data are presented as mean ± SEM and were analyzed in GraphPad Prism or Microsoft Office Excel. Statistical significance was assumed at a P value of ≤ 0.05. P values were calculated with the unpaired t-test unless indicated otherwise. For interpretation of statistical results from unpaired t-test, * = p value ≤ 0.05, ** = p value ≤ 0.01, *** = p value ≤ 0.001, and **** = p value ≤ 0.0001.
RESULTS
p120 catenin is misexpressed in human premalignant and malignant pancreatic lesions
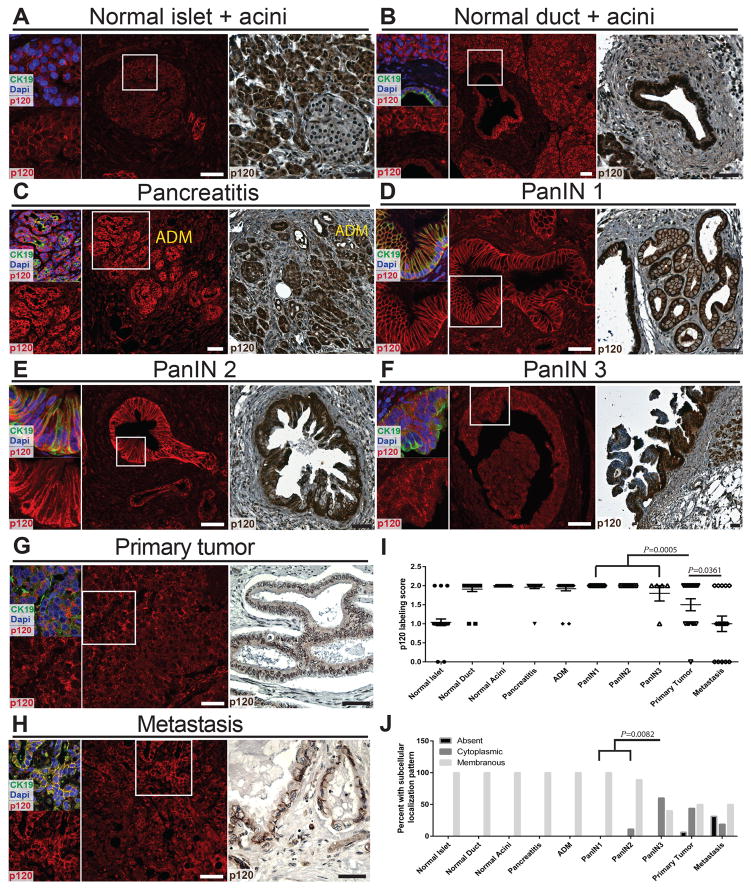
Expression and localization of p120 catenin, a critical cytoskeletal regulator and component of adherens junctions, were rated in human normal pancreas, chronic pancreatitis, acinar to ductal metaplasia (ADM), pancreatic intraepithelial neoplasia (PanIN) 1–3, and primary tumors and metastases. Examples of scores in each category are presented in Figure S1A. p120 catenin is expressed in all normal pancreatic cell types (Figure 1A,B). Localization of p120 catenin was scored as predominantly membranous in 100% of normal pancreatic cell types (n=22–33 patients for each pancreatic cell type) (Figure 1J). Punctate cytoplasmic and nuclear staining were also observed in normal pancreatic cells (Figure 1A,B). p120 catenin expression was rated as high in >91% chronic pancreatitis and ADM (n=25/26 chronic pancreatitis, n=23/25 ADM) (Fig 1C,I). Expression levels of p120 catenin were significantly decreased in primary tumors when compared to PanIN lesions (n=31 PanIN and n=16 primary tumors) (Figure 1D–G,I). Moreover, p120 catenin expression was significantly lower in metastases when compared to primary tumors (n=16 primary tumors and n=16 metastasis) (Figure 1G–I). There was also a significant difference in predominant p120 catenin subcellular localization when comparing PanIN1-2 and PanIN3 (n=26 PanIN1-2 and n=5 PanIN3). Together, these findings demonstrate that altered p120 catenin expression and localization is a distinguishing hallmark of human pancreatic cancer progression.
Figure 1.
Expression of p120 catenin in human pancreas. A,B) IHC shows high p120 catenin expression in normal acinar and duct cells and medium p120 catenin expression in normal islets. IF labeling showing p120 catenin overlaid with CK19 and Dapi allowed visualization of punctate cytoplasmic and nuclear p120 catenin expression in normal pancreatic cells. C–H) Representative images for pancreatitis, ADM, PanIN1-3, tumor, and metastasis are shown. I,J) Numerical scoring of p120 catenin expression and analysis of predominant p120 catenin subcellular localization are depicted. The P value for (J) was calculated using Fisher’s exact test. Scale bars are 50μm.
Pancreatic loss of p120 catenin in the context of oncogenic Kras results in decreased survival and cachexia
Previously, using CPdx1; p120f/f mice, we reported that p120 catenin is required for proper tubulogenesis and cell-type specification during pancreas development (29). To determine the function of p120 catenin in adult mouse pancreas in the absence of a confounding developmental phenotype, we crossed transgenic mice harboring floxed alleles of p120 catenin (p120f) (21) with Mist1CreER/+ (CiMist1) mice (20). p120f/f, CiMist1; p120wt/wt, CiMist1; p120f/wt, and CiMist1; p120f/f mice displayed normal pancreatic histology 2–4 months post tamoxifen injection (Figure S1B). Similar to what we previously reported for CPdx1; p120f/f mice 10 months of age (29), a subset of CiMist1; p120f/f mice 12 months post tamoxifen injection exhibited pancreatitis and ADM (Figure S1B).
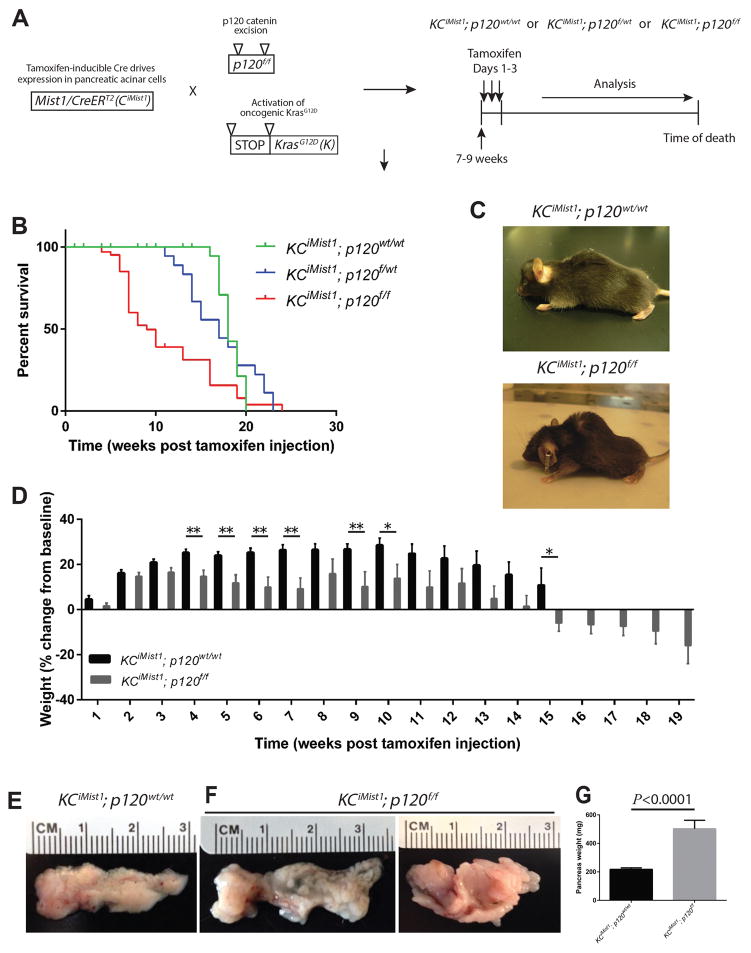
Since we observed mislocalized p120 catenin expression in human PanIN, before the onset of pancreatic cancer, we next sought to determine if p120 catenin plays a functional role in PanIN formation and progression. To this end, we crossed CiMist1; p120f/f mice with lox-stop-lox-KrasG12D (K) mice (22), which resulted in simultaneous ablation of p120 catenin and activation of oncogenic KrasG12D in adult pancreatic acinar cells upon tamoxifen administration (Figure 2A). The KCiMist1 mouse model displays the full spectrum of murine premalignant ADM and PanIN1-3 lesions in a manner that faithfully recapitulates human premalignant pancreatic lesions (20). Survival analysis on cohorts of KCiMist1; p120wt/wt, KCiMist1; p120f/wt, and KCiMist1; p120f/f mice showed significant differences in overall survival (Figure 2B). KCiMist1; p120f/f mice exhibited cachexia, which is also frequently observed in human pancreatic cancer patients (Figure 2C,D). The gross appearance of KCiMist1; p120f/f pancreata was strikingly abnormal and enlarged when compared to KCiMist1; p120wt/wt pancreata (Figure 2E–G).
Figure 2.
Oncogenic Kras and loss of p120 catenin lead to decreased overall survival, cachexia, and increased pancreas size. A) Breeding and tamoxifen administration scheme. B) Kaplan-Meyer survival analysis showed overall median survival was 18 weeks for KCiMist1; p120wt/wt mice (n=9), 17 weeks for KCiMist1; p120f/wt mice (n=18), and 9 weeks for KCiMist1; p120f/f mice (n=37). Log-rank test revealed significantly longer survival of KCiMist1; p120wt/wt (P<0.0001) mice and KCiMist1; p120f/wt (P=0.0035) mice when compared to KCiMist1; p120f/f mice. C) A KCiMist1; p120wt/wt and KCiMist1; p120f/f mouse sacrificed 9 weeks post tamoxifen injection. D) KCiMist1; p120f/f (n=70 total) mice exhibit cachexia when compared to KCiMist1; p120wt/wt (n=62 total) mice. Limited data was available for longer time points for KCiMist1; p120wt/wt mice due to survival. E–G) 1 month post tamoxifen injection, KCiMist1; p120f/f (n=13) pancreata show significantly increased weight when compared to KCiMist1; p120wt/wt (n=13) pancreata.
KCiMist1; p120f/f pancreata show a prominent acute and chronic inflammatory response
As expected, p120 catenin was ubiquitously expressed in KCiMist1; p120wt/wt and KCiMist1; p120f/wt pancreata (Figure S2A,B). Minimal mosaic expression of p120 catenin was observed in KCiMist1; p120f/f pancreatic acini (Figure S2C). KCiMist1; p120f/f pancreata displayed marked acinar cell atrophy, pronounced inflammation, and contained stroma characterized by a unique cellular constitution that differs from the stroma in KCiMist1; p120wt/wt pancreata (Figure S2D–L). KCiMist1; p120f/f pancreata displayed less mucinous lesions than KCiMist1; p120wt/wt and KCiMist1; p120f/wt pancreata, as manifested by Alcian blue staining (Figure S1C). KCiMist1; p120f/f pancreata also showed areas of ductal dilation (Figure S2M–O). Disruption of contiguous basement membrane Laminin expression, a characteristic of human pancreatic cancer (30), was also seen in KCiMist1; p120f/f pancreatic lesions. This was accompanied by cells that escaped intact PanIN epithelium and invaded into the underlying tissue (Figure S2P–R).
NF-kB signaling regulates formation of ADM/PanIN/fibrostroma in KCiMist1; p120f/f mice
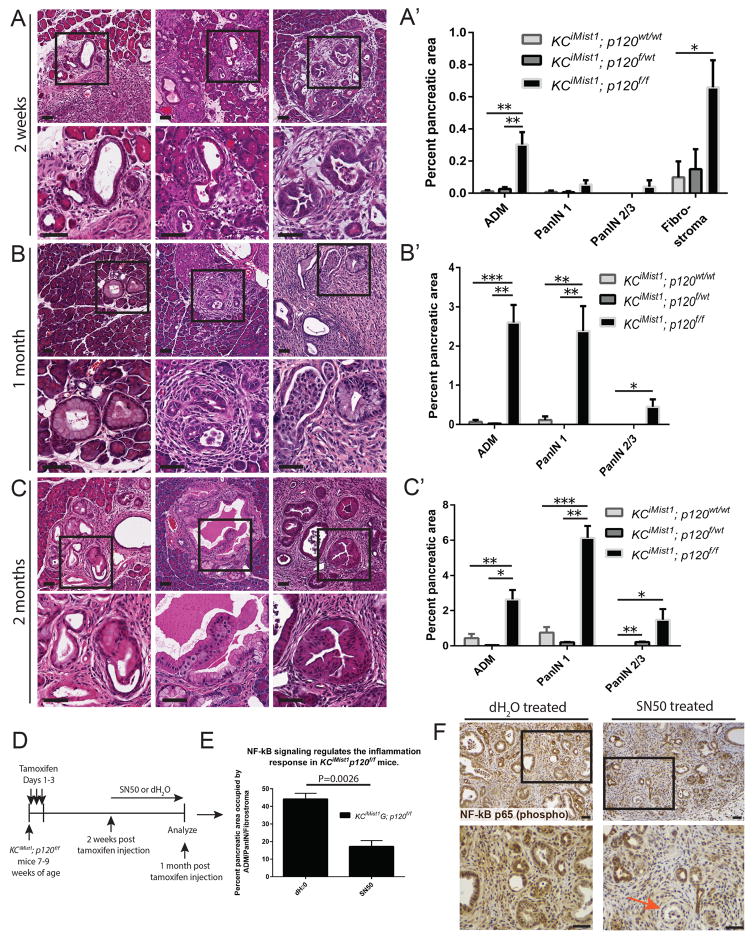
Histologically, significant increases in ADM, low and high grade PanIN formation, and fibrostroma were observed in KCiMist1; p120f/f pancreata when compared to KCiMist1; p120f/wt and KCiMist1; p120wt/wt pancreata beginning 2 weeks post tamoxifen injection (Figures 3A–C′, S1D,D′). Two months post tamoxifen injection, KCiMist1; p120f/f pancreata displayed significantly increased fibrostroma (75.69% ± 9.32% pancreatic area, n=4 mice) when compared to KCiMist1; p120f/wt pancreata (0.55% ± 0.00% pancreatic area, n=2 mice) (P=0.0058) and KCiMist1; p120wt/wt pancreata (1.09% ± 0.54% pancreatic area, n=4 mice) (P=0.0002).
Figure 3.
p120 catenin restrains formation of Kras-induced premalignant pancreatic cancer. A–C′) Representative histology and quantification of ADM, PanIN, and fibrostroma are depicted. N=2–6 for each genotype at each time point. D,E) KCiMist1; p120f/f mice 2 weeks post tamoxifen injection were treated with either SN50 (n=4) or water (n=3) for 2 weeks. F) An orange arrow shows low level nuclear localization of NF-kB p65 (phospho S536) in IHC images of SN50 treated KCiMist1; p120f/f mice. Scale bars are 50μm.
Previously, we and others have demonstrated that p120 catenin loss results in immune cell infiltration and inflammation, which is mediated in part by Nuclear factor-kB (NF-kB) activation (29,31). We next queried whether NF-kB signaling contributed to formation of ADM/PanIN/fibrostroma in KCiMist1; p120f/f mice. To this end, we treated KCiMist1; p120f/f mice with SN50, a potent inhibitor of NF-kB activation, and observed a significant reduction in ADM/PanIN/fibrostroma in SN50 treated KCiMist1; p120f/f mice when compared to controls (Figure 3D–F). These data establish NF-kB signaling as a mechanism by which p120 catenin loss promotes increased ADM, PanIN, and inflammation in KCiMist1; p120f/f mice.
Lineage tracing reveals that pancreatic loss of p120 catenin in cooperation with oncogenic Kras promotes striking cell extrusion
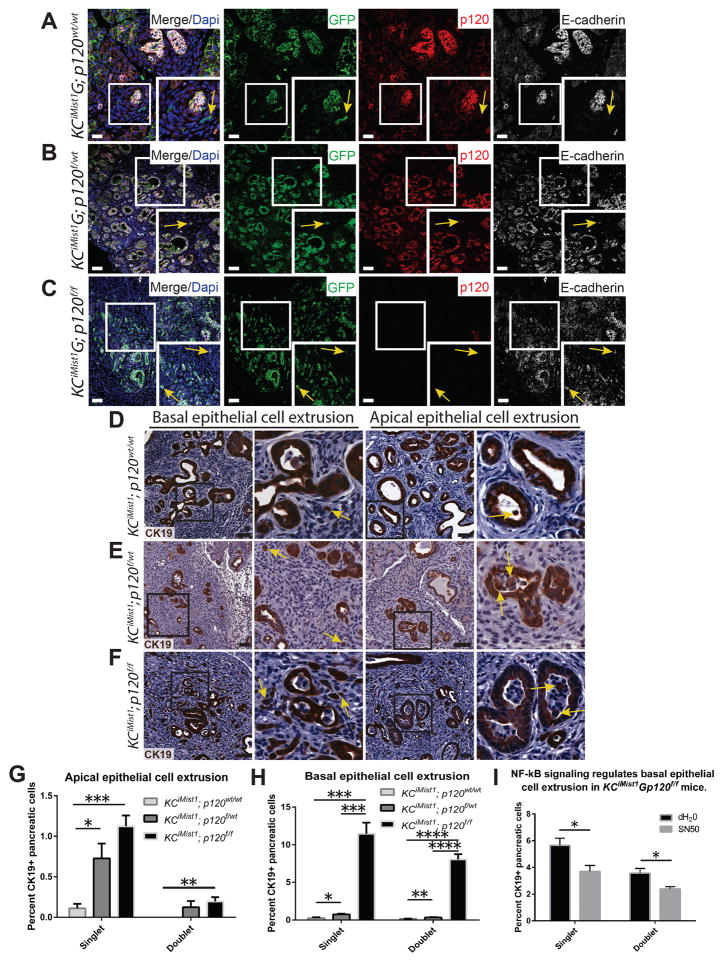
The exit of epithelial cells across the basement membrane of discernable epithelial structures, a process termed delamination, occurs in KPCPdx1Y and KCiMist1Y PanIN mice and has been associated with epithelial-to-mesenchymal transition (EMT) (32). Since loss of p120 catenin in pancreatic cancer cell lines results in increased migration and invasion, also associated with EMT (33), we next sought to determine if p120 catenin regulated delamination in PanIN mice. First, we examined expression of p120 catenin and E-cadherin in delaminated cells in 2 lineage-traced murine models of PanIN: KPCPtf1aY and KCiMist1G (34,35). Delaminated cells in KPCPtf1aY PanIN mice expressed decreased adherens junction proteins p120 catenin and E-cadherin when compared to their surrounding pancreatic epithelia (Figure S3A). KCiMist1G; p120wt/wt PanIN mice showed decreased p120 catenin and E-cadherin expression in non-epithelial delaminated cells, some of which have an elongated fibroblast cell morphology (Figure 4A). These data show that decreased expression of adherens junction proteins p120 catenin and E-cadherin is a manifest feature of delaminated cells in PanIN mice.
Figure 4.
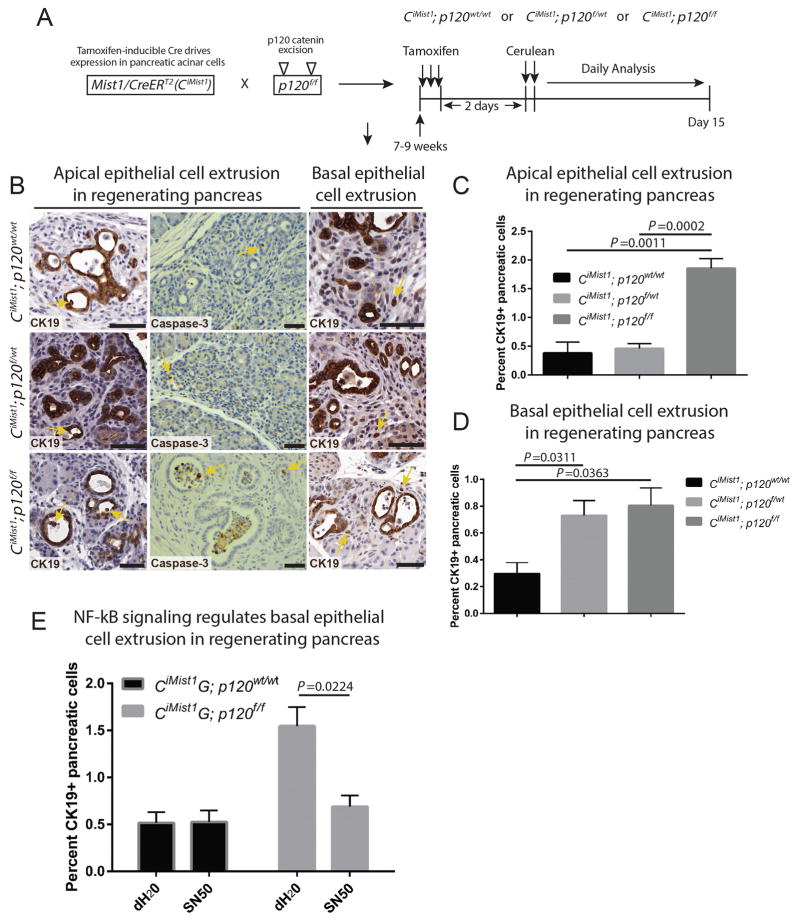
Loss of p120 catenin promotes epithelial cell extrusion in Kras-driven neoplasia. A–C) KCiMist1G; p120wt/wt mice show decreased expression of p120 catenin and E-cadherin in delaminated cells. E-cadherin expression is retained in basally extruded GFP+ cells in KCiMist1G; p120f/wt and KCiMist1G; p120f/f mice. Yellow arrows point to lineage traced delaminated cells. D–H) Yellow arrows show single apically and basally extruded CK19+ cells. Quantification of 7000 CK19+ cells in 4 animals for each genotype at one month post tamoxifen injection revealed significantly increased apically and basally extruded epithelial cells in KCiMist1G; p120f/wt and KCiMist1G; p120f/f mice when compared to KCiMist1G; p120wt/wt mice. I) Mice were treated according to schematic in Figure 3D. N=6124 cells in 3 dH20 treated mice and 6753 cells in 3 SN50 treated mice. Scale bars are 50μm.
To further investigate the role of p120 catenin in delamination in Kras-induced premalignant pancreatic neoplasia, we next examined delaminated cells in lineage-traced KCiMist1G; p120f/wt and KCiMist1G; p120f/f mice. Monoallelic and biallelic loss of p120 catenin resulted in remarkable abundant delamination of GFP+ cells that retained E-cadherin (Figure 4B,C). These data show that p120 catenin is not required for maintenance of E-cadherin localization to cell membranes in the context of oncogenic Kras. Both delamination, also termed basal epithelial cell extrusion (16), as well as apical epithelial cell extrusion were significantly increased in KCiMist1; p120f/wt and KCiMist1; p120f/f pancreata when compared to KCiMist1; p120wt/wt pancreata (Figure 4D–H). Abundant apical and basal epithelial cell extrusion was 100% penetrant in KCiMist1; p120f/wt and KCiMist1; p120f/f pancreata, with the phenotype evident in KCiMist1; p120f/f pancreata by 2 weeks post tamoxifen injection (Figure S3B). Quantification of extruded isolated CK19+ cells revealed 838/7000 in KCiMist1; p120f/f pancreata, 76/7000 in KCiMist1; p120f/wt pancreata, and 19/7000 in KCiMist1; p120wt/wt pancreata (Figure S3C). Lineage tracing in KCiMist1; p120f/f pancreata showed extruded GFP+, CK19+ single cells were negative for Vimentin, a marker of mesenchymal differentiation and EMT (Figure S3D), suggesting that basal epithelial cell extrusion resulting from biallelic p120 catenin loss is not associated with incomplete EMT. Furthermore, treatment of KCiMist1; p120f/f mice with the NF-kB inhibitor SN50 significantly reduced basal epithelial cell extrusion when compared to controls (Figure 4I). These data suggest that NF-kB signaling is a component of the regulatory network that controls basal epithelial cell extrusion in KCiMist1; p120f/f mice.
p120 catenin loss in the context of cerulean-induced pancreatitis promotes epithelial cell extrusion
Biallelic loss of p120 catenin during pancreas development promotes apical epithelial cell extrusion, and luminal cell extrusion has also been reported with p120 catenin loss in developing kidney and MDCK cysts (29,36). Given the abundant apical and basal extrusion observed in KCiMist1; p120f/f pancreata, we next sought to interrogate the extrusion behavior of adult pancreatic cells lacking p120 catenin in an experimental model of acute pancreatitis. Acute pancreatitis was induced in CiMist1; p120wt/wt, CiMist1; p120f/wt, and CiMist1; p120f/f mice (Figure 5A). CiMist1; p120f/f pancreata showed significantly increased susceptibility to injury and inflammation when compared to CiMist1; p120wt/wt pancreata (Figure S4A–J). Since activation of NF-kB signaling augments the severity of pancreatitis (37), we next assessed the role of NF-kB activation in CiMist1; p120f/f and control mice. Inhibition of NF-kB signaling significantly reduced injury in CiMist1; p120f/f pancreata to levels similar to CiMist1; p120wt/wt dH20 treated controls (Figure S4K,L), establishing NF-kB signaling as a direct mechanism by which p120 catenin loss promotes increased susceptibility to injury in CiMist1; p120f/f pancreata.
Figure 5.
p120 catenin regulates apical and basal epithelial cell extrusion in cerulean-induced acute pancreatitis. A) Schematic for tamoxifen injection and induction of experimental acute pancreatitis. B–D) Yellow arrows point to apically and basally extruded cells in CK19 and cleaved Caspase 3 IHC images. A subset of apically extruded cells are positive for cleaved Caspase-3. Quantification of apically extruded CK19+ cells at Days 5 and 7 post cerulean administration revealed a significant increase in CiMist1; p120f/f pancreata (n=8750 cells in 6 pancreata) when compared to CiMist1; p120f/wt pancreata (n=3323 cells in 4 pancreata) and CiMist1; p120wt/wt pancreata (n=3288 cells in 3 pancreata). Quantification of basally extruded CK19+ cells at Days 5 and 7 post cerulean treatment showed a significant increase in CiMist1; p120f/f pancreata (n=10710 cells in 6 pancreata) and CiMist1; p120f/wt pancreata (n=4594 cells in 4 pancreata) when compared to CiMist1; p120wt/wt pancreata (n=6217 cells in 3 pancreata). E) Inhibition of NF-kB signaling significantly reduced basal epithelial cell extrusion in CiMist1; p120f/f pancreata (n=5162 cells in 3 dH20 treated pancreata and 7849 cells in 3 SN50 treated pancreata). For CiMist1; p120wt/wt pancreata, n=2477 cells in 4 dH20 treated pancreata and 1268 cells in 4 SN50 treated pancreata. Scale bars are 50μm.
Quantification of apical and basal epithelial cell extrusion revealed significant increases in CiMist1; p120f/f and CiMist1; p120f/wt pancreata when compared to CiMist1; p120wt/wt pancreata (Figure 5B–D). These data suggest that p120 catenin regulates both apical and basal epithelial cell extrusion in adult mouse pancreas in the context of acute pancreatitis. Furthermore, CiMist1G; p120f/f mice treated with the NF-kB inhibitor SN50 show significantly reduced basal epithelial cell extrusion (Figure 5E), demonstrating that activation of NF-kB regulates extrusion in the setting of p120 catenin loss and experimental pancreatitis.
Epithelial cells that extrude basally in KCiMist1; p120wt/wt, KCiMist1; p120f/wt, and KCiMist1; p120f/f pancreata survive
We next sought to determine the fate of both apically and basally extruded epithelial cells in KCiMist1; p120wt/wt, KCiMist1; p120f/wt, and KCiMist1; p120f/f pancreata. Since epithelial cells can extrude apically by a mechanism involving S1P/S1pr2 signaling and activation of cleaved Caspase-3 (11,14), we next examined cleaved Caspase-3 expression in extruded CK19+ cells. Consistent with this previously reported mechanism of apical cell extrusion, we observed cleaved Caspase-3 expression in apically extruded CK19+ cells, but not in basally extruded CK19+ cells in KCiMist1; p120wt/wt, KCiMist1; p120f/wt, and KCiMist1; p120f/f pancreata (Figure S5A–C), suggesting that basally extruded CK19+ cells exit intact pancreatic epithelium and remain viable.
Basally extruded isolated epithelial cells in KCiMist1; p120f/f pancreata display aneuploidy and nuclear enlargement
Because we observed prominent nucleoli and nuclear enlargement of basally extruded epithelial cells in KCiMist1; p120f/f pancreata, we next analyzed the DNA content of isolated CK19+ cells in KCiMist1; p120f/f pancreata using OTMIAS Image Analysis Software on Feulgen stained slides (Figure S6A–C). A population of pancreatic cells was observed with a DNA index of 1.5, which was indicative of aneuploidy (Figure S6C). In addition, 8.3% (21/253) pancreatic cells analyzed showed abnormal DNA content with a DNA index of ≥ 2.5 (Figure S6C). The histology of KCiMist1; p120f/f pancreata thus comprises a very unique phenotype with overall benign neoplasia and reactive stroma containing isolated epithelial cells that display features of malignancy including enlarged, hyperchromatic and pleomorphic nuclei, prominent nucleoli, aneuploidy and occasional binuclear cells. These findings suggest that p120 catenin loss in the context of oncogenic Kras promotes formation of invasive pancreatic neoplasia (Figure S6D).
Isolated epithelial cells in human PDA misexpress p120 catenin
As we identified that loss of p120 catenin in cooperation with oncogenic Kras promotes invasion of epithelial cells displaying characteristics of malignancy in pancreatic neoplasia, we next examined expression of p120 catenin in isolated epithelial cells in human pancreatic ductal adenocarcinoma (PDA). Single malignant epithelial cells in human PDA are depicted in Figure S7A,B. Quantification of p120 catenin subcellular localization in 253 isolated epithelial cells from 17 patients with PDA showed a sparse 4.74% of cells with normal membrane labeling and 95.26% of cells with predominant cytoplasmic or absent p120 catenin localization (Figure S7C,D). These data show that altered p120 catenin subcellular localization is a distinctive feature of isolated malignant epithelial cells in human PDA.
KCiMist1; p120f/f pancreata display a unique transcriptome signature and a downregulated S1P biosynthetic pathway
As a means to identify a molecular basis by which p120 catenin ablation in cooperation with oncogenic Kras promotes epithelial cell extrusion, we performed whole transcriptome analysis on fluorescence activated cell sorted (FACS) GFP+ pancreatic cells in KCiMist1G; p120wt/wt and KCiMist1G; p120f/f mice 2 weeks post tamoxifen injection (Figure S8A). IPA analysis showed 56 statistically significant differentially expressed pathways, several of which are related to actin cytoskeleton signaling, the inflammatory response, and cell adhesion and migration (Figure S8B). We next sought to validate these results by examining expression of select genes in each of these categories using IHCIHC. Previously, we showed that actin cytoskeleton organization was disrupted in CPdx1; p120f/f pancreata, and that this observation was associated with increased cytoplasmic PKCζ, a known modulator of actin cytoskeleton dynamics (29). Here, we similarly find that KCiMist1; p120f/f pancreata show increased cytoplasmic PKCζ when compared to KCiMist1; p120f/wt and KCiMist1; p120wt/wt pancreata (Figure S8C–E). Expression of adherens junction components E-cadherin and β-catenin was also reduced in KCiMist1; p120f/f pancreata (Figure S8F–K). In addition, IHC confirmed intrinsic activation of NF-kB in KCiMist1; p120f/f pancreata (Figure S8L–N).
Since defective S1P/S1pr2 mediated cell extrusion has been shown to shift the predominant direction of cell extrusion from apical to basal (19), we next queried whether p120 catenin regulated S1P/S1pr2 signaling. We performed qPCR on FACS-sorted GFP+ pancreatic cells at 1 month post tamoxifen injection, which showed an overall decrease in expression of genes mediating S1P/S1pr2 signaling in KCiMist1G; p120f/f mice when compared to KCiMist1G; p120wt/wt mice (Figure 6A). The expression of genes involved in the biosynthetic pathway of S1P, Sphk1 and Sphk2, was significantly decreased −5.83 fold and −3.96 fold, respectively, suggesting that p120 catenin regulates biosynthesis of S1P in a mutant Kras-dependent context.
Figure 6.
Loss of p120 catenin induces significant transcriptional changes in genes affecting S1P/S1pr2 signaling. A) qPCR showed significant downregulation of genes involved in the biosynthesis of S1P in KCiMist1; p120f/f pancreata (n=3 pancreata) when compared to KCiMist1; p120wt/wt pancreata (n=4 pancreata). B,C) Western blot images and quantification shown are representative of 3 independent experiments. D) IF images show effective p120 catenin knockdown in siRNA treated CFPAC-1 spheres. E) Western blot depicts expression of p120 catenin and S1pr2 in CFPAC-1 cells. F) Quantification shown represents n=3 experiments for each condition. Scale bars are 50μm.
We next sought to understand how p120 catenin regulated expression of S1P/S1pr2 pathway members. Since we observed activation of NF-kB in KCiMist1; p120f/f mice, we hypothesized that this regulation may occur though NF-kB signaling. Similar to our observations in KCiMist1; p120f/f mice, AsPC-1 cells treated with p120 catenin siRNA showed significantly reduced expression of S1pr2 (Figure 6B,C). Inhibition of NF-kB activation with SN50 completely restored expression of S1pr2 in p120 catenin siRNA treated AsPC-1 cells, suggesting that p120 catenin regulates expression of S1pr2 through activation of NF-kB (Figure 6B,C).
We next investigated the relationship between p120 catenin loss and epithelial cell extrusion mediated by S1P/S1pr2 signaling using an epithelial, Kras mutant, human pancreatic cancer cell line. CFPAC-1 cells express both p120 catenin and S1pr2 and form spheres (CFPAC-1 spheres) that can extrude cells basally when grown in Matrigel (Figure 6D,E). The percentage of CFPAC-1 spheres extruding epithelial cells basally increased significantly following p120 catenin knockdown (Figure 6F). Furthermore, the specific S1pr2 agonist CYM-5520 (38) significantly decreased the frequency of basal extrusion in p120 catenin-deficient spheres to levels similar to those observed in p120 catenin-expressing spheres (Figure 6D). The ability of restored S1P/S1pr2 signaling to rescue the p120 catenin loss of function phenotype indicates a direct mechanism by which p120 catenin loss promotes increased basal epithelial cell extrusion.
DISCUSSION
The critical role of the cell-cell adhesion apparatus during tumorigenesis of epithelial cancers is firmly established (1,39). The association of p120 catenin with E-cadherin at epithelial cell membranes is crucial for formation and maintenance of adherens junctions (40). p120 catenin loss or mislocalization can destabilize E-cadherin and affect the adhesive repertoire of the cell and its signal transduction status. In KCiMist1; p120f/f mice, simultaneous p120 catenin and E-cadherin loss likely destabilizes cell adhesion and promotes migration and invasion.
Studies have shown that p120 catenin can function as a bona fide tumor suppressor in murine oral cavity, esophagus, and forestomach (3). In contrast, targeted knockout of p120 catenin in murine salivary gland (21), intestine (31,41), brain (42), mammary gland (43), kidney (36), eye (44), epidermis (31,45), and lung (46) is not sufficient to promote formation of cancer. It is therefore likely that the requirement for p120 catenin in development and progression of cancer is tissue-specific. For pancreas, ablation of p120 catenin in pancreatic progenitor cells in CPdx1; p120f/f mice (29) and somatic knockout of p120 catenin in adult pancreatic acinar cells in CiMist1; p120f/f mice does not result in development of pancreatic cancer, so evidence for a bona fide tumor suppressor role for p120 catenin in pancreas is lacking.
The inflammation suppression function of p120 catenin is well documented (47). Loss of p120 catenin generates a microenvironment disposed to chronic inflammation in normal, injured, and neoplastic pancreas. We show that formation of ADM/PanIN/fibrostroma in KCiMist1; p120f/f mice and injury in CiMist1G; p120f/f mice is mediated in part through activation of NF-kB. Dexamethasone, which has anti-inflammatory and immunosuppressant effects (including inhibition of NF-kB activation), has been shown to reduce ADM and PanIN formation in mice (32), suggesting a potential link between inflammation and premalignant lesion formation.
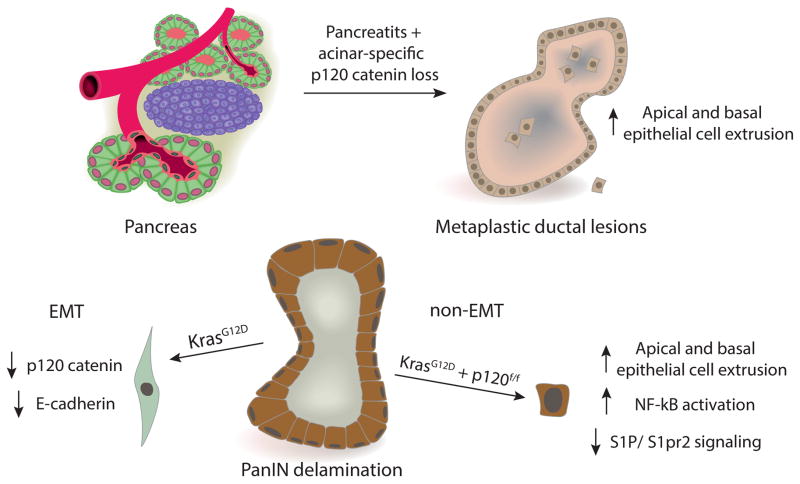
We show loss of p120 catenin in delaminated cells associated with EMT and non-EMT in PanIN mice. While loss of p120 catenin precedes non-EMT associated delamination in KCiMist1G; p120f/f pancreata, it is unclear if loss of p120 catenin precedes or occurs after delamination associated with EMT in KPCPtf1aY and KCiMist1G; p120wt/wt pancreata. Actin cytoskeleton remodeling events are necessary for EMT, and loss of cytoplasmic p120 catenin before occurrence of delamination may prevent cytoskeletal restructuring required for delamination associated with EMT. Biallelic loss of p120 catenin results in significant downregulation of genes mediating S1P/S1pr2 signaling in PanIN mice and AsPC-1 cells, an observation which we demonstrate in vitro is mediated partly through activation of NF-kB. Activation of S1pr2 in vitro completely rescues increased basal epithelial cell extrusion seen with p120 catenin loss. These data suggest that p120 catenin loss in the context of oncogenic Kras may promote neoplastic epithelial cell invasion in part by altering S1P/S1pr2 signaling through activation of NF-kB (Figure 7).
Figure 7.
Illustration depicting p120 catenin regulation of epithelial cell extrusion. a) p120 catenin loss in adult pancreatic acinar cells in the context of experimental pancreatitis promotes apical and basal epithelial cell extrusion. b) In the context of oncogenic Kras, PanIN epithelial cells extruding basally through delamination associated with EMT express less adherens junction proteins p120 catenin and E-cadherin. Mutant Kras PanIN epithelial cells deficient for p120 catenin retain epithelial morphology after extruding basally and display features of malignancy.
Few mechanisms for non-EMT associated delamination are described, yet the evidence that EMT is not absolutely required for invasion, dissemination, and metastasis is emerging (48–50). In summary, we have created a model in which cooperating genetic insults have unraveled a new mechanism for neoplastic epithelial cell invasion in premalignant pancreatic cancer. The evidence that p120 catenin regulates epithelial cell extrusion through activation of NF-kB is compelling, as p120 catenin loss affects this biologic process in injured and neoplastic pancreata. Further studies are needed to clarify the metastatic potential associated with monoallelic and biallelic p120 catenin loss in different pancreatic cell types and how these characteristics determine the evolution of PDA.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Melissa Pruski, Kanchan Singh, Neal C. Jones, Ka Liu, Anzer Habibulla, Mara Swaim, Jeffrey Roeser, and Qingfeng Zhu for their indispensable help in maintaining mouse colonies, genotyping, and technical assistance. The authors thank Florencia McAllister for helpful intellectual discussions. We would also like to thank Conover Talbot Jr. for crucial aid with analysis of microarray gene expression. This work was supported by NIH P01 CA134292, RO1 DK097087, and R21 CA158898 (to S.D.L.). Jennifer M. Bailey was supported by an AACR/Pancreatic Cancer Action Network Pathway to Leadership Award and by Texas Medical Center Digestive Diseases Center grant (DK56338).
Footnotes
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
A.M.H. carried out experiments, drafted the manuscript, and was involved together with J.M.B. and S.D.L. in design and analysis of experiments. A.M.H., Y.J.W., and J.M.B. evaluated p120 catenin expression in human tissues. J.A. and K.J.L. contributed intellectually to the study. H.Z. assisted in the design and analysis of FACS experiments. I.A. aided with cachexia quantification and survival analysis. S.G.S. and M.A.P. assisted with analysis of regeneration data. Y.C., K.P. and K.S. helped with Western blot experiments. M.G. provided human PanIN and pancreatic cancer TMAs. N.R. and M.H. provided tissue from KPCPtf1aY mice. A.B.R. provided Ctnnd1tm1Abre (p120f) mice. M.Y. performed DNA ploidy analysis on Feulgan stained slides. C. ID. provided matched human pancreatic cancer and metastasis TMA. C. I-D., A.M., and M.Y. carried out pathologic interpretation of histology. S.D.L. and J.M.B. supervised all aspects of the work.
The authors declare no competing financial interests.
References
- 1.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Hengel J, van Roy F. Diverse functions of p120ctn in tumors. Biochimica et biophysica acta. 2007;1773(1):78–88. doi: 10.1016/j.bbamcr.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 3.Stairs DB, Bayne LJ, Rhoades B, Vega ME, Waldron TJ, Kalabis J, et al. Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell. 2011;19(4):470–83. doi: 10.1016/j.ccr.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schackmann RC, Klarenbeek S, Vlug EJ, Stelloo S, van Amersfoort M, Tenhagen M, et al. Loss of p120-catenin induces metastatic progression of breast cancer by inducing anoikis resistance and augmenting growth factor receptor signaling. Cancer research. 2013;73(15):4937–49. doi: 10.1158/0008-5472.CAN-13-0180. [DOI] [PubMed] [Google Scholar]
- 5.Mayerle J, Friess H, Buchler MW, Schnekenburger J, Weiss FU, Zimmer KP, et al. Up-regulation, nuclear import, and tumor growth stimulation of the adhesion protein p120 in pancreatic cancer. Gastroenterology. 2003;124(4):949–60. doi: 10.1053/gast.2003.50142. [DOI] [PubMed] [Google Scholar]
- 6.Fei Y, Cheng Z, Liu S, Liu X, Ge Z, Wang F, et al. Expression and clinical significance of p120 catenin mRNA and protein in pancreatic carcinoma. Bosnian journal of basic medical sciences/Udruzenje basicnih mediciniskih znanosti = Association of Basic Medical Sciences. 2009;9(3):191–7. doi: 10.17305/bjbms.2009.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(16):5934–41. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chetty R, Jain D, Serra S. p120 catenin reduction and cytoplasmic relocalization leads to dysregulation of E-cadherin in solid pseudopapillary tumors of the pancreas. American journal of clinical pathology. 2008;130(1):71–6. doi: 10.1309/FEYD99TXC4LMYVA5. [DOI] [PubMed] [Google Scholar]
- 9.Slattum GM, Rosenblatt J. Tumour cell invasion: an emerging role for basal epithelial cell extrusion. Nature reviews Cancer. 2014;14(7):495–501. doi: 10.1038/nrc3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu Y, Shea J, Slattum G, Firpo MA, Alexander M, Mulvihill SJ, et al. Defective apical extrusion signaling contributes to aggressive tumor hallmarks. eLife. 2015;4:e04069. doi: 10.7554/eLife.04069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenblatt J, Raff MC, Cramer LP. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Current biology: CB. 2001;11(23):1847–57. doi: 10.1016/s0960-9822(01)00587-5. [DOI] [PubMed] [Google Scholar]
- 12.Andrade D, Rosenblatt J. Apoptotic regulation of epithelial cellular extrusion. Apoptosis: an international journal on programmed cell death. 2011;16(5):491–501. doi: 10.1007/s10495-011-0587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slattum G, McGee KM, Rosenblatt J. P115 RhoGEF and microtubules decide the direction apoptotic cells extrude from an epithelium. The Journal of cell biology. 2009;186(5):693–702. doi: 10.1083/jcb.200903079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Y, Forostyan T, Sabbadini R, Rosenblatt J. Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. The Journal of cell biology. 2011;193(4):667–76. doi: 10.1083/jcb.201010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien CB, Morcos PA, et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484(7395):546–9. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Y, Rosenblatt J. New emerging roles for epithelial cell extrusion. Current opinion in cell biology. 2012;24(6):865–70. doi: 10.1016/j.ceb.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenhoffer GT, Rosenblatt J. Bringing balance by force: live cell extrusion controls epithelial cell numbers. Trends in cell biology. 2013;23(4):185–92. doi: 10.1016/j.tcb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall TW, Lloyd IE, Delalande JM, Nathke I, Rosenblatt J. The tumor suppressor adenomatous polyposis coli controls the direction in which a cell extrudes from an epithelium. Mol Biol Cell. 2011;22(21):3962–70. doi: 10.1091/mbc.E11-05-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slattum G, Gu Y, Sabbadini R, Rosenblatt J. Autophagy in oncogenic K-Ras promotes basal extrusion of epithelial cells by degrading S1P. Current biology: CB. 2014;24(1):19–28. doi: 10.1016/j.cub.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(48):18913–8. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis MA, Reynolds AB. Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev Cell. 2006;10(1):21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 23.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nature genetics. 2002;32(1):128–34. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 25.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 26.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siveke JT, Lubeseder-Martellato C, Lee M, Mazur PK, Nakhai H, Radtke F, et al. Notch signaling is required for exocrine regeneration after acute pancreatitis. Gastroenterology. 2008;134(2):544–55. doi: 10.1053/j.gastro.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Alsina J, Leach S, Bailey J. Incorporation of polaxamers during tissue dissociation allows isolation of high quality RNA from FACS-sorted pancreatic cells. Pancreapedia: The Exocrine Pancreas Knowledge Base. 2013 [Google Scholar]
- 29.Hendley AM, Provost E, Bailey JM, Wang YJ, Cleveland MH, Blake D, et al. p120 Catenin is required for normal tubulogenesis but not epithelial integrity in developing mouse pancreas. Developmental biology. 2015;399(1):41–53. doi: 10.1016/j.ydbio.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan S, Chen R, Reimel BA, Crispin DA, Mirzaei H, Cooke K, et al. Quantitative proteomics investigation of pancreatic intraepithelial neoplasia. Electrophoresis. 2009;30(7):1132–44. doi: 10.1002/elps.200800752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. p120-catenin mediates inflammatory responses in the skin. Cell. 2006;124(3):631–44. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamada S, Satoh K, Miura S, Hirota M, Kanno A, Masamune A, et al. miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. Journal of cellular physiology. 2013;228(6):1255–63. doi: 10.1002/jcp.24280. [DOI] [PubMed] [Google Scholar]
- 34.McAllister F, Bailey JM, Alsina J, Nirschl CJ, Sharma R, Fan H, et al. Oncogenic Kras Activates a Hematopoietic-to-Epithelial IL-17 Signaling Axis in Preinvasive Pancreatic Neoplasia. Cancer Cell. 2014;25(5):621–37. doi: 10.1016/j.ccr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, et al. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. 2014;146(1):245–56. doi: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marciano DK, Brakeman PR, Lee CZ, Spivak N, Eastburn DJ, Bryant DM, et al. p120 catenin is required for normal renal tubulogenesis and glomerulogenesis. Development. 2011;138(10):2099–109. doi: 10.1242/dev.056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Liu Y, Daniluk J, Gaiser S, Chu J, Wang H, et al. Activation of nuclear factor-kappaB in acinar cells increases the severity of pancreatitis in mice. Gastroenterology. 2013;144(1):202–10. doi: 10.1053/j.gastro.2012.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satsu H, Schaeffer MT, Guerrero M, Saldana A, Eberhart C, Hodder P, et al. A sphingosine 1-phosphate receptor 2 selective allosteric agonist. Bioorg Med Chem. 2013;21(17):5373–82. doi: 10.1016/j.bmc.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nature reviews Cancer. 2004;4(2):118–32. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 40.Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, et al. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell. 2010;141(1):117–28. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 41.Smalley-Freed WG, Efimov A, Short SP, Jia P, Zhao Z, Washington MK, et al. Adenoma formation following limited ablation of p120-catenin in the mouse intestine. PLoS One. 2011;6(5):e19880. doi: 10.1371/journal.pone.0019880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elia LP, Yamamoto M, Zang K, Reichardt LF. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006;51(1):43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurley SJ, Bierie B, Carnahan RH, Lobdell NA, Davis MA, Hofmann I, et al. p120-catenin is essential for terminal end bud function and mammary morphogenesis. Development. 2012;139(10):1754–64. doi: 10.1242/dev.072769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian H, Sanders E, Reynolds A, van Roy F, van Hengel J. Ocular anterior segment dysgenesis upon ablation of p120 catenin in neural crest cells. Investigative ophthalmology & visual science. 2012;53(9):5139–53. doi: 10.1167/iovs.12-9472. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Moreno M, Song W, Pasolli HA, Williams SE, Fuchs E. Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15399–404. doi: 10.1073/pnas.0807301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chignalia AZ, Vogel SM, Reynolds AB, Mehta D, Dull RO, Minshall RD, et al. p120-Catenin Expressed in Alveolar Type II Cells Is Essential for the Regulation of Lung Innate Immune Response. The American journal of pathology. 2015 doi: 10.1016/j.ajpath.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu G. p120-Catenin: a novel regulator of innate immunity and inflammation. Critical reviews in immunology. 2012;32(2):127–38. doi: 10.1615/critrevimmunol.v32.i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Huang H, Remmers N, Hollingsworth MA. Loss of E-cadherin and epithelial to mesenchymal transition is not required for cell motility in tissues or for metastasis. Tissue barriers. 2014;2(4):e969112. doi: 10.4161/21688362.2014.969112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuji T, Ibaragi S, Hu GF. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer research. 2009;69(18):7135–9. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shamir ER, Pappalardo E, Jorgens DM, Coutinho K, Tsai WT, Aziz K, et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. The Journal of cell biology. 2014;204(5):839–56. doi: 10.1083/jcb.201306088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.