Abstract
Activating mutations of K-ras are the most common oncogenic alterations found in lung cancer. Unfortunately, attempts to target K-ras mutant lung tumors have thus far failed, clearly indicating the need for new approaches in patients with this molecular profile. We have previously shown NF-κB activation, release of IL-6, and activation of its responsive transcription factor STAT3 in K-ras mutant lung tumors, which was further amplified by the tumor enhancing effect of chronic obstructive pulmonary disease (COPD)-type airway inflammation. These findings suggest an essential role for this inflammatory pathway in K-ras mutant lung tumorigenesis and its enhancement by COPD. Therefore, here we blocked IL-6 using a monoclonal anti-IL-6 antibody in a K-ras mutant mouse model of lung cancer in the absence or presence of COPD-type airway inflammation. IL-6 blockade significantly inhibited lung cancer promotion, tumor cell intrinsic STAT3 activation, tumor cell proliferation, and angiogenesis markers. Moreover, IL-6 inhibition reduced expression of pro-tumor type 2 molecules (Arginase 1, Fizz 1, Mgl, and IDO), number of M2 type macrophages and G-MDSCs, and pro-tumor T-regulatory/T helper 17 cell responses. This was accompanied by increased expression of anti-tumor type 1 molecule (Nos2), and anti-tumor T helper 1/CD8 T cell responses. Our study demonstrates that IL-6 blockade not only has direct intrinsic inhibitory effect on tumor cells, but also re-educates the lung microenvironment toward an anti-tumor phenotype by altering the relative proportion between pro-tumor and anti-tumor immune cells. This information introduces IL-6 as a potential druggable target for prevention and treatment of K-ras mutant lung tumors.
Keywords: Lung Cancer, K-ras, IL-6, STAT3, COPD
Introduction
Lung cancer is the leading cause of cancer mortality worldwide due to its high incidence and low cure rate (1), and cigarette smoke is by far the most common cause of it (2). Activating mutations of K-ras, found in approximately 30% of lung cancer patients, are one of the most prevalent genetic alterations associated with tobacco exposure (1). Unfortunately, pharmacologic attempts to develop targeted therapies to interfere with K-ras activity have shown limited success to date; therefore, alternative strategies are needed to inhibit this oncogenic signaling pathway and bring clinical benefits to lung cancer patients with mutant K-ras. In addition, several studies have found that smokers with chronic obstructive pulmonary disease (COPD) have an increased risk of lung cancer (3 to 10-fold) compared to smokers with comparable cigarette exposure but without COPD (3–5). COPD is a chronic inflammatory disease of the lung which is present in 40–70% of lung cancer patients (6). Importantly, among smokers with COPD, even following withdrawal of cigarette smoke, inflammation persists and lung function continues to deteriorate as does the increased risk of lung cancer (7,8). Furthermore, because of the persistent lung cancer risk among former smokers, and increased diagnosis of early stage lung cancer with the recommended screening method (low dose CT scan) (9), strategies targeting pathways that stop the progression of COPD and early stage lung cancer to advanced lung cancer would also be valuable.
We and other groups have demonstrated that K-ras mutant lung tumors have intrinsic inflammatory characteristics, with activation of the NF-κB pathway, increased levels of the cytokine interleukin 6 (IL-6), which is transcriptionally regulated by NF-κB, and activation of the IL-6-responsive transcription factor STAT3 (10–12). This inflammatory response was further amplified by the tumor enhancing effect of COPD (12,13). These data suggest an essential role for IL-6 in lung cancer pathogenesis. Importantly, they also introduce IL-6 as a potential druggable target for the prevention and treatment of K-ras mutant lung cancer.
In this study, using a K-ras induced lung cancer mouse model, we demonstrate that K-ras activation drives a pro-tumor immune suppressive microenvironment with increased type 2 and reduced type 1 inflammatory signatures via IL-6 signaling, which is amplified in the presence of COPD-type inflammation. Significantly, we show that pharmacologic targeting of IL-6 suppresses K-ras lung tumorigenesis, and re-educates the lung microenvironment toward an anti-tumor immune phenotype.
Methods
Human Studies
STAT3 mRNA expression was determined by array analysis (Illumina v3) of surgically resected lung adenocarcinomas from 150 patients that did not receive neoadjuvant therapy. This cohort was obtained from the Profiling of Resistance patterns and Oncogenic Signaling Pathways in Evaluation of Cancers of the Thorax (PROSPECT) study, developed in 2006 at M.D. Anderson Cancer Center (14). Clinical characteristics of these patients are presented in Table 1. STAT3 mRNA expression from these patients was log2 transformed and median expression was computed. We then dichotomized lung adenocarcinoma patients based on median STAT3 mRNA expression in the manner previously performed (15). Patients with relatively ‘low’ expression displayed lower than the median STAT3 levels whereas patients with relatively ‘high’ expression exhibited greater than the median STAT3 expression levels.
Table 1.
Clinicopathological information of the lung adenocarcinoma PROSPECT set used for STAT3 mRNA analysis
| Covariate | Covariate categories | Frequency (percentage) |
|---|---|---|
| Race | African American | 8 (5.3%) |
| Asian | 6 (4.0%) | |
| Hispanic | 5 (3.3%) | |
| Caucasian | 131 (87.3%) | |
| Tobacco history | No | 19 (12.7%) |
| Yes | 131 (87.3%) | |
| Smoking status | Never | 19 (12.7%) |
| Former | 67 (44.7%) | |
| Current | 64 (42.6%) | |
| Gender | Female | 73 (48.7%) |
| Male | 77 (51.3%) | |
| Stage | I | 90 (60.0%) |
| II | 24 (16.0%) | |
| III | 35 (23.3%) | |
| IV | 1 (0.7%) | |
| KRAS | Wild type | 111 (74.0%) |
| Mutant | 39 (26.0%) | |
| Recurrence | No | 89 (59.3%) |
| Yes | 61 (40.7%) |
Cell Culture and Chemicals
The human NSCLC cell lines NCI-H2030, NCI-H1944, NCI-H647, NCI-838 were kindly provided by Dr. Adi Gazdar (UTSW, Dallas, TX) in 2010, whose lab confirmed their identity by genotype testing. Cell lines were cultured in RPMI supplemented with 10% Fetal Bovine Serum, penicillin/streptomycin/glutamine (Gibco-BRL, USA) at 37°C under 5% CO2. Cisplatin (Teva Pharmaceuticals, Sellersville, PA), and tocilizumab (Roche-Genentech, CA) were obtained from the M.D. Anderson Cancer Center Pharmacy. Siltuximab was kindly provided by Janssen Pharmaceuticals (USA).
MTS Assays
Cytotoxicity to cisplatin, tocilizumab, siltuximab or combinations, were assessed by CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI) according to the manufacturer’s instructions. Absorbance was read at 495nm using Softmax pro (Molecular Devices, CA). Concentrations resulting in cell growth inhibition of 50 (IC50) were calculated for cisplatin. For other treatments, the metabolic state was expressed as a percent of controls (% CT).
Mouse Models
CCSPCre/LSL-K-rasG12D mice (CC-LR) were generated as previously described (12). Briefly, this strain is generated by crossing mice harboring the LSL-K-rasG12D allele with mice containing Cre recombinase inserted into the Club cell secretory protein (CCSP) locus (12). All mice were housed in specific pathogen-free conditions and handled in accordance with the Institutional Animal Care and Use Committee of M.D. Anderson Cancer Center. Mice were monitored daily for evidence of disease or death.
NTHi Lysate Aerosol Exposure
A lysate of Nontypeable Haemophilus influenzae (NTHi) strain 12 was prepared as previously described (12). CC-LR mice were nebulized with the lysate (2.5 mg/ml in phosphate buffered saline, PBS) once a week starting at 6 weeks of age for 8 weeks. The delivery of the lysate to mice was made by aerosolizing a thawed aliquot of NTHi placed in an AeroMist CA-209 nebulizer (CIS-US, Bedford, MA) driven by 10 liter/min of room air supplemented with 5% CO2 for 20 min.
In Vivo IL-6 Blockade
Six week old CC-LR and control mice were injected intraperitoneally (IP) with 20 mg/kg dose of an anti-IL-6 monoclonal (Clone MP5-20F3, R&D, Minneapolis, MN) or IgG1 Isotype control (Clone 43414, R&D, Minneapolis, MN) antibodies twice a week for eight weeks.
Histochemistry
Mice were anesthetized and sacrificed by IP injection of Avertin (Sigma, St. Louis, MO), then the tracheas of euthanized mice were cannulated with PE-50 tubing and sutured into place. The lungs were infused with 10% buffered formalin (Sigma, St. Louis, MO) and then removed and placed in 10% buffered formalin. Tissues then were embedded in paraffin blocks, and sectioned at 5-mm thickness. The sections on glass slides were dried, then were deparaffinized and stained with hematoxylin and eosin (H&E). The H&E-stained slides were examined by a pathologist blinded to genotype and treatment, and the proliferative lesions of the lungs were evaluated in accordance with the recommendations of the Mouse Models of Human Cancer Consortium (16).
Immunostaining
Previously sectioned lung samples on slides were immunohistochemically (IHC) stained and evaluated for expression of P-STAT3 (Tyr705) (1:250; Cell Signaling Technology, MA), Ki-67 (1:200; Abcam, MA), CD31 (1:50, BD Biosciences, CA), VEGF (1:750; Santa Cruz, CA), and MMP-9 (1:500; Santa Cruz, CA). Heat-induced antigen retrieval was performed using 10 mmol/L of citrate buffer (pH 6.0) in a pressure cooker for 20 min. After quenching endogenous peroxidase with 3% hydrogen peroxide, blocking was performed with nonimmune normal serum. Immunoreactivity for immunohistochemistry was detected using biotinylated IgG secondary antibodies specific for each primary antibody followed by incubation with ABC kit (Vector Laboratory, Burlingame, CA), and stained with diaminobenzidine chromogenic substrate. Slides were counter-stained with Harris hematoxylin, followed by dehydration and mounted with cytoseal 60 (ThermoFisher Scientific, Cheshire, UK). Images were obtained by an OLYMPUS BX 60 microscope at 4× or 40× magnification with Image-Pro Plus, version 4.5.1.22. The numbers of labeled positive cells for each marker were quantitated as a fraction of total tumor nuclei per high power field (40×) in 10 fields from three to five mice of each group. Results were expressed as percentage of positive cells ± SE.
Assessment of Lung Tumor Burden and Inflammation
Lung surface tumor numbers were counted, and then bronchoalveolar lavage fluid (BALF) was obtained by sequentially instilling and collecting 2 aliquots of 1 mL PBS through a tracheostomy cannula. The lungs were snap frozen and stored for future RNA and protein analysis. Total leukocyte count was determined using a hemacytometer and cell populations were determined by cytocentrifugation followed by Wright–Giemsa (Sigma, St. Louis, MO) staining (W&G). The remaining BALF was centrifuged at 1,250 × g for 10 min, and supernatants were collected and stored at −80°C for further analysis.
Cytokines/Chemokine Measurement
The levels of IL-6, IL-10, IL-17A, CCL5, CXCL1 and TNFα in the BALF were assessed using the MCYTOMAG-70 K assay (Millipore, St Charles, MO) according to the manufacturer’s instructions. TGF-β was assayed using the TGFB-64 K-01 (Millipore, St Charles, MO) assay. Data were collected using a Luminex 100 (Luminex Corporation, Austin, TX). Standard curves were generated using a 5-parameter logistic curve fitting equation (StarStation V 2.0; Applied Cytometry Systems, SAC, CA). Each sample reading was interpolated from the appropriate standard curve.
Quantitative RT-PCR Analysis
Total RNA was isolated from whole lung according to the TRIzol reagent protocol (Invitrogen, NY) and purified by E.Z.N.A. total RNA kit I (OMEGA, GA). Reverse transcription PCR was performed using the qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD). Quantitative RT-PCR was performed according to a standard protocol using gene-specific primers (Supplementary Table 1). SYBR Green reactions were done using FastMix, Low ROX (Quanta Biosciences, Gaithersburg, MD) and products measured on an ABI Viia 7 PCR system (ABI, Foster City, CA). The expression of individual genes was calculated and normalized with the ΔΔCt method.
Isolation of lung resident mononuclear cells
Lungs were harvested after perfusion with PBS, then were inflated with 150u/mL collagenase IV and 20u/mL DNase1 in PBS for 25 min at 37°C/5%CO2. Single cell suspensions were prepared by mechanical dissociation of lung tissue through a 70-μm nylon mesh. Lung cells were suspended in PBS and layered on lymphocyte separation medium (Lonza, MD). Cells were centrifuged at room temperature for 20 min at 900 × g, and mononuclear cells were harvested from the gradient interphase.
Flow Cytometry and Fluorescence-Activated Cell Sorting (FACS)
Cells were stained with fluorescently labeled antibodies using CD45 (30-F11), CD11b (M1/70), F4/80 (BM8), CD11c (N418), Ly-6C (AL-21), Ly-6G (1A8), and I-A/I-E (M5/114.15.2) markers on ice for 30 minutes (17). Cells were analyzed and gated (Supplementary Figure 1) on an LSRII machine or isolated by FACS (BD Bioscience). Data were further analyzed by FlowJo.
Statistical analysis
Data were analyzed using two-tailed unpaired Student’s t test, and presented as mean ± S.E.M. Graphpad Prism 6 software was used for analysis. Differences were considered significant for P < 0.05. Assessment of disease-free survival was performed using Cox proportional hazard regression analysis and the Kaplan Meier method for estimation of survival probability.
Results
IL-6/STAT3 pathway as a potential druggable target for K-ras mutant lung tumors
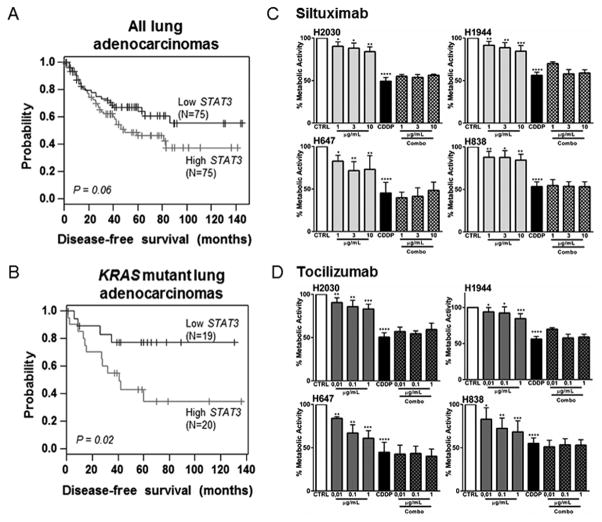
Serum IL-6 level in patients with stage I lung cancer is associated with significantly shorter survival and poorer outcome (18,19). Here we investigated whether STAT3, the principal IL-6-activated signaling molecule, is predictive of clinical outcome in KRAS-mutant human lung adenocarcinomas. Though STAT3 expression was not significantly different between KRAS mutant (mean: 9.137; median: 9.274) and WT (mean: 9.112; median: 9.352) subgroups, we found a trend for association of high STAT3 mRNA expression with poor disease-free survival in all lung adenocarcinomas (hazard ratio = 1.56, P = 0.065) (Fig. 1A). Further analysis demonstrated that in KRAS-mutant lung adenocarcinomas, STAT3 mRNA level was a significant predictor of poor disease-free survival (hazard ratio =3.28, P = 0.02) (Fig. 1B). As STAT3 mRNA amounts can reflect STAT3 activity due to STAT3 autoregulatory function (20), these data insinuate that IL-6/STAT3 pathway activation worsens clinical outcomes of patients with KRAS-mutant lung adenocarcinoma.
Figure 1. IL-6/STAT3 pathway as a potential druggable target for K-ras mutant lung tumors.
(A) All adenocarcinomas (N=152) and (B) KRAS-mutant adenocarcinomas (N=39) from PROSPECT Cohort were stratified based on STAT3 median mRNA expression (high, black; low, grey). Patient subgroups were then analyzed for differences in disease-free survival using the Kaplan-Meier method for estimation of survival probability and the log-rank test. (C–D) The cell lines were incubated with tocilizumab or siltuximab at different concentrations alone or in combination with the respective IC50 of cisplatin (Combo) for 48 hours. Data represent means ± SEM. (n=4 independent experiments) (*p<0.05, **p<0.005, ***p<0.001, ****p<0.0001).
These observations prompted us to block IL-6 using a pharmacologic approach. We first assessed gene expression for IL6, IL6R, and STAT3 in 39 NSCLC cell lines in the Cancer Cell Line Encyclopedia (CCLE) mRNA expression dataset (21). This expression data was plotted using the pheatmap package in R, where dendrograms indicate clustering by standard Ward algorithm (Supplementary Figure. 2A). We then selected (Supplementary Method 1) a representative panel of cells that expressed IL-6 and STAT3 and featured the major histologic subtype and mutational signature found among NSCLC patients (Supplementary Figures 2A, 2B, and 2C). We then tested two FDA approved monoclonal antibodies (Abs) against IL-6 or the IL-6 receptor (IL-6R), siltuximab and tocilizumab, respectively in this representative panel of cell lines. Inhibition of IL-6 or IL-6R resulted in a significant reduction of phosphorylated STAT3, the main IL-6-responsive transcription factor (Supplementary Figure 2D, Supplementary Method 2). This indicates that IL-6 produced by the cancer cell is functionally active and acts through the IL-6R within cancer cells, and these humanized Abs are able to inhibit this process. Surprisingly, IL-6 or IL-6R blockade alone or in combination with cisplatin, while having a mild to moderate effect on tumor cell proliferation, did not show any greater effect than cisplatin alone (Fig .1C, and D). These results suggests a potential role for tumor microenvironment in vulnerability of tumor to respond to targeting inflammatory signals as explored below.
Effects of IL-6 blockade on murine K-ras mutant lung tumors
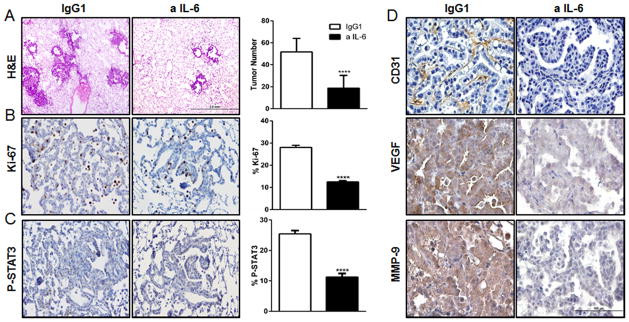
We observed increased levels of IL-6 in a conditional K-ras-induced lung cancer mouse model (CC-LR) compared to controls (LR mice), and during tumor progression (Supplementary Figures 3), which was associated with increased expression of STAT3 mRNA (22), further suggesting an important role for IL6/STAT3 pathway in K-ras mutant lung tumorigenesis. Accordingly, we investigated effects of IL-6 blockade in vivo. Cohorts of 6 week old CC-LR mice were injected IP with a 20 mg/kg dose of monoclonal anti-IL-6 rat IgG1 or IgG1 control antibodies, twice a week, for a period of eight weeks, then their lungs and BALFs were collected at the age of 14 weeks. IL-6 blockade did not cause any significant change in mouse weight compared to control group. As we previously described, main lung lesions in CC-LR mice are epithelial hyperplasia in the early stage, and adenomas in the middle and late stages (12). Lesions that show atypical cytological features and invasive histologic features are identified as adenocarcinomas and are present at late stage. Histopathologic analysis of the lungs from anti-IL-6 treated CC-LR mice revealed fewer and smaller lesions with lower percentage of adenoma/adenocarcinoma lesions (Supplementary Figure 4), and lower number of visible macroscopic lung surface tumors (by 57%) compared to the IgG1 treated control group (Fig. 2A). This was associated with a 46% reduction in tumor cell proliferation, as measured by Ki-67 expression (Fig. 2B). We also observed a significant decrease (by 56%) in STAT3 activation, as judged by tyrosine-phosphorylated STAT3 (pSTAT3) amounts (Fig. 2C). Moreover, targeting IL-6 suppressed angiogenesis in the lungs of CC-LR mice, as measured by decreased expression of the angiogenesis markers CD31, VEGF and MMP-9 (Fig. 2D). These data suggest that IL-6 induces proliferative and angiogenic effects on lung tumors in vivo that might be mediated by STAT3 activation.
Figure 2. Anti-IL-6 treatment reduces lung tumor burden, tumor cell proliferation, tumor angiogenesis, and STAT3 activation.
(A) Histopathologic appearance of lung and lung surface tumor number in CC-LR mice at age of 14 weeks after IgG1 (n=8) or anti-IL-6 treatment (n=12) (4×; Scale bar for, 2mm). (B–C) Representative photomicrographs and quantitative analysis of positive tumor cells for Ki-67, and P-STAT3 in lungs of CC-LR mice at the age of 14 weeks after IgG1 (n=4) or IL-6 (n=5) blockade (20×; Scale bar for, 100μm) (data represent means ± SEM **** p<0,001). (D) Representative photomicrographs of lung tumors stained for CD31, VEGF, and MMP-9 in CC-LR mice at the age of 14 weeks after IgG1 (n=4) or IL-6 (n=5) blockade (40×; Scale bar for, 100μm).
Effect of IL-6 blockade on K-ras mutant lung tumor microenvironment
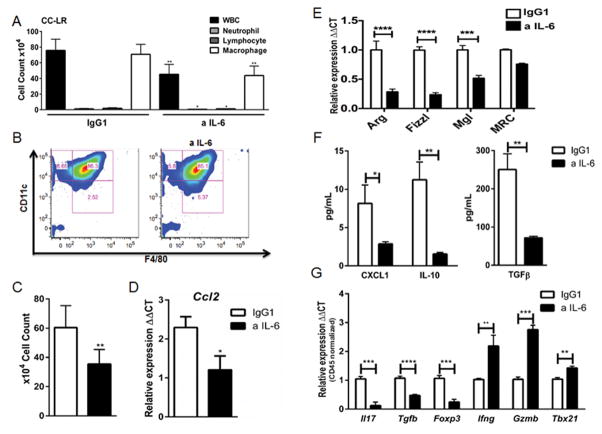
In solid tumors K-ras mutation induces an inflammatory response (10,12), which has been linked to NF-κB activation (11,23). This tumor-promoting inflammation tailors the tumor microenvironment towards a suppressive state which is considered a crucial step for tumor establishment (24), however, IL-6 effects on tumor-infiltrating immune subsets have not been fully characterized. Therefore, we analyzed the BALFs and whole lungs of CC-LR mice with or without IL-6 blockade to determine whether IL-6 affected immune cells within the tumor microenvironment. W&G staining of BALF from CC-LR mice showed that macrophages represent 93% of total WBCs found in lung microenvironment in this model (Fig. 3A). The anti-IL-6 treatment significantly decreased the total WBC number and macrophage population, without major effects on other leukocyte subsets (Fig. 3A). Flow cytometry analysis of BALFs of anti-IL-6 treated CC-LR mice further revealed that there was no change in the percentage of CD45+CD11C+F4/80+ alveolar macrophages compared to IgG1 control group (Fig. 3B). However, we found a 45% decrease in total macrophage number in agreement with our W&G findings (Fig. 3C). We also observed a significant decrease in the expression of the macrophage chemoattractant CCL2 in the lungs of CC-LR mice treated with anti-IL-6 Ab (Fig. 3D), which may explain the reduction in total macrophage number observed.
Figure 3. Anti-IL-6 treatment changes the lung microenvironment in K-ras induced lung tumors.
(A) Total inflammatory cell and lineage-specific leukocyte numbers from BALF of CC-LR mice at the age of 14 weeks after IgG1 (n=5) or IL-6 blockade (n=8). (B) Representative flow cytometry analysis of (live/CD45+) alveolar macrophage population (CD11c+F4/80+) in BALF of CC-LR mice after IgG1 (n=4) or IL-6 blockade (n=6) at the age of 14 weeks. (C) Total (CD11c+F4/80+) cell number in BALF of CC-LR mice after IgG1 (n=4) or IL-6 blockade (n=5) at the age of 14 weeks. (D) Relative expression of Ccl2 mRNA in whole lungs from CC-LR mice after IgG1 (n=4) or IL-6 blockade (n=5) at the age of 14 weeks. (E) Relative expression of type 2 macrophage mRNA signature in BALF cells from CC-LR mice after IgG1 (n=3) or IL-6 blockade (n=3) at the age of 14 weeks. (F) ELISA analysis on BALF of CC-LR mice after IgG1 (n=3) or anti-IL-6 treatment (n=3) at the age of 14 weeks. (G) Relative expression of IL17, Tgfb, Foxp3, Infg, Gzmb and Tbx21 mRNA in whole lungs of CC-LR mice at the age of 14 weeks after IgG1 (n=4) or IL-6 blockade (n=5), normalized by Cd45 expression (data represent means ± SEM *p<0,05, **p<0.005, ****p<0,001).
To study phenotypic changes in macrophage population, we further analyzed the expression of genes related to macrophage polarization in the BALF population from CC-LR mice. We observed a reduction in the expression of macrophage pro-tumor (type 2) genes including Arginase1 (Arg), found in inflammatory zone 1 (FIZZ1), macrophage galactose binding lectin (Mgl) and mannose receptor C, type 1 (Mrc1) with IL-6 blockade (Fig. 3E). We also observed a similar reduction in the expression of Arg1 and FIZZ1 in the whole lung tissue from CC-LR mice with IL-6 blockade (data not shown). We observed an even greater reduction when Arg1 level was normalized to macrophage number and to expression of the hematopoietic-specific marker CD45 (Supplementary Figure 5). Moreover, ELISA analysis of inflammatory mediators in the BALF of CC-LR mice after IL-6 inhibition revealed reduction in CXCL1 expression as well as in the expression of the immunosuppressive cytokines IL-10 and TGFβ (Fig. 3F). Taken together, these findings indicate that IL-6 drives pro-tumor (type 2) macrophage polarization in K-ras mutant lung tumors and suggest that one mechanism by which IL-6 blockade suppresses the tumor progression is by reprograming the lung immune microenvironment.
We have previously demonstrated that Th17 and potentially T regulatory (Treg) cells play a role in K-ras mutant lung tumorigenesis (13). Here, within the lung CD45+ population, we found that IL-6 blockade decreases the expression of interleukin 17 (Il17), transforming growth factor beta (Tgfb), and forkhead box P3 (Foxp3) suggesting a decrease in Th17 and Treg infiltration or differentiation (Fig. 3G). We also detected an increase in interferon gamma (Ifng), granzyme B (Gzmb) and T-bet (Tbx21) expression suggestive of enhanced Th1 differentiation and CD8 T cell cytotoxic activation (Fig. 3G). Together, our data suggest that targeting IL-6 reformats immune subset infiltration and gene expression patterns in the lung microenvironment toward an anti-tumor state.
Pharmacologic targeting of IL-6 as a therapeutic strategy to prevent the tumor promoting effect of COPD on K-ras-induced lung tumorigenesis
We have previously developed a COPD-like mouse model of airway inflammation by repetitive exposure to a lysate of NTHi, a common colonizer of the airways in COPD patients (25). We have shown that this type of airway inflammation has a tumor enhancing effect in CC-LR mice, associated with activation of the NF-κB pathway and increased levels of IL-6 (Supplementary Figure 6) (12,13). COPD patients show high levels of IL-6 in their sputum and there is an inverse correlation between lung function and IL-6 levels in these patients (26,27). IL-6 is also implicated in inflammatory responses in COPD, and its overexpression results in airspace enlargement and inflammation in murine models (28). Therefore, we studied the effect of IL-6 blockade on lung cancer enhancement by COPD-type inflammation.
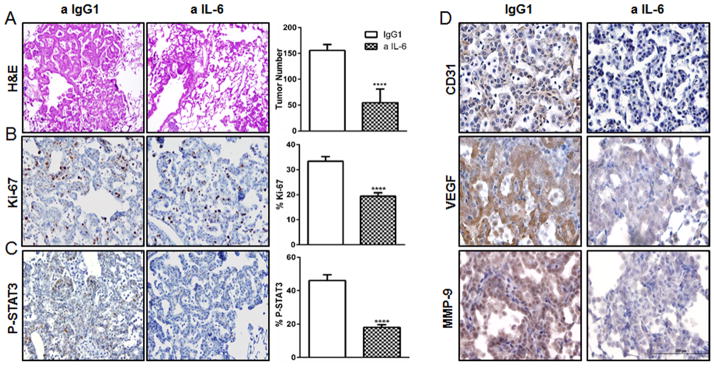
Six weeks old CC-LR mice were treated with anti-IL-6 Ab twice a week while being exposed to aerosolized NTHi lysate once a week, for a period of eight weeks. Histopathologic and macroscopic analysis of the lungs from CC-LR mice after IL-6 blockade and COPD induction revealed fewer lesions (65% reduction) compared to IgG1 treated control group (Fig. 4A). This was associated with a 42% reduction in tumor cell proliferation (Fig. 4B), a 63% reduction in STAT3 phosphorylation (Fig. 4C), and significantly reduced tumor angiogenesis, as judged by angiogenesis marker expression (Fig. 4D). These findings suggest IL-6 blockade directly suppresses enhancement of lung cancer by COPD-type airway inflammation via tumor cell intrinsic effects.
Figure 4. Anti-IL-6 treatment reduces lung tumor cell proliferation, tumor angiogenesis, and STAT3 activation in COPD setting.
(A) Histopathologic appearance of lung and lung surface tumor number in CC-LR mice at the age of 14 weeks after NTHi exposure and IgG1 (n=9) or anti-IL-6 treatment (n=12). (B–C) Representative photomicrographs and quantitative analysis of positive tumor cells for Ki-67, and P-STAT3 in lungs of CC-LR mice at the age of 14 weeks after IgG1 (n=4) or IL-6 (n=5) blockade (data represent means ± SEM **** p<0,001). (D) Representative photomicrographs of lung tumors stained for CD31, VEGF, and MMP-9 in CC-LR mice at the age of 14 weeks after IgG1 (control) (n=4) or IL-6 (n=5) blockade. (40×; Scale bar for, 100μm).
Effect of IL-6 blockade on the COPD-associated tumor microenvironment
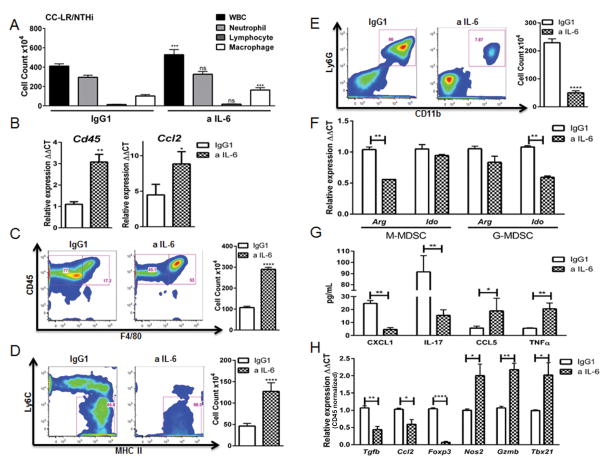
W&G staining of the BALF population from NTHi-exposed CC-LR mice showed that neutrophils comprise 72% of total WBCs in the IgG1 treated control group, similar to BALF of COPD patients (29). Surprisingly, IL-6 blockade induced an increase in total WBCs in BALF, including elevated macrophage numbers without a significant change in total neutrophils (Fig. 5A). To confirm this, we measured expression of Cd45 and Ccl2 in the lungs of CC-LR mice by Q-RT-PCR and we observed a significant increase in the expression of both markers after anti-IL-6 treatment, corroborating our W&G finding (Fig. 5B). In addition, we confirmed the increase in the total macrophage (CD45+F4/80+) population in BALF by flow cytometry (Fig. 5C). Interestingly, the macrophage population showed increased MHCII expression and reduced Ly6C, with a greater proportion of CD45+F4/80+Ly6C−MHCII+ cells (Fig. 5D). Collectively, these results suggest IL-6 blockade induces macrophage infiltration as well as polarization toward an anti-tumor M1 phenotype.
Figure 5. Anti-IL-6 treatment changes the lung microenvironment in K-ras induced lung tumors in COPD setting.
(A) Total inflammatory cell and lineage-specific leukocyte numbers from BALFs of CC-LR mice at the age of 14 weeks after NTHi exposure and IgG1 (n=6) or IL-6 blockade (n=8). (B) Relative expression of Cd45 and Ccl2 mRNA in whole lungs from CC-LR mice after NTHi exposure and IgG1 (n=4) or anti-IL6 treatment (n=5) at the age of 14 weeks. (C) Representative flow cytometry analysis and total number of (live/CD45+) macrophage population (F4/80+) in BALF of CC-LR mice after NTHi exposure and IgG1 (n=3) or IL-6 blockade (n=4) at the age of 14 weeks. (D) Representative flow cytometry data and percentage of (live/CD45+) activated M1 macrophage population (Ly6C−MHCII+) in BALF of CC-LR mice after NTHi exposure and IgG1 (n=3) or IL-6 blockade (n=4) at the age of 14 weeks. (E) Representative flow cytometry data and total number of (live/CD45+) G-MDSC population (Ly6G+CD11b+) in BALF of CC-LR mice after NTHi exposure and IgG1 (n=3) or IL-6 blockade (n=4) at the age of 14 weeks. (F) M-MDSC (CD45+CD11b+Ly6G−) and G-MDSC (CD45+CD11b+Ly6G+) populations in BALF of CC-LR mice after NTHi exposure and IgG1 or IL-6 blockade at the age of 14 weeks were isolated by fluorescence-activated cell sorting, pooled, and relative expression of Arg1 and Ido mRNA were measured and compared in anti-IL-6 (n=3) or IgG1(n=3) treated groups. (G) ELISA analysis of BALFs from CC-LR mice after NTHi exposure and IgG1 (n=3) or IL-6 blockade (n=3) at the age of 14 weeks. (H) Relative expression of Tgfb, Ccl2, Foxp3, Nos2, Gzmb and Tbx21 mRNA in whole lungs of CC-LR mice after NTHi exposure and IgG1 (n=4) or IL-6 blockade (n=5) at the age of 14 weeks normalized by Cd45 expression of anti-IL-6 treated group compared with controls treated with IgG1 (data represent means ± SEM *P < 0.05. **P < 0.05.)
Chronic inflammation builds an altered microenvironment with recruitment and accumulation of myeloid derived suppressor cells (MDSCs). These immature cells show nonspecific suppressive capabilities and produce arginase 1 and IDO (indoleamine 2, 3-dioxygenase) that deplete arginine and tryptophan, key amino acids for T cell activation (30,31). In our COPD model, we found that BALFs from CC-LR mice exposed to NTHi have an abundant granulocytic MDSC (G-MDSCs, CD45+CD11b+Ly6G+) population, which was reduced significantly upon anti-IL-6 treatment (Fig. 5E). To study qualitative changes in lung MDSCs, we analyzed gene expression within FACS-purified monocytic (CD45+CD11b+Ly6G−) and granulocytic (CD45+CD11b+Ly6G+) MDSCs from BALF of CC-LR mice after NTHi exposure. We observed decreased expression of Arg1 in the monocytic population and decreased expression of Ido in the granulocytic population upon anti-IL-6 treatment, suggesting skewing of these subsets toward a non-suppressive phenotype (Fig. 5F). Moreover, anti-IL-6 treatment was associated with a decrease in levels of CXCL1 and IL-17 proteins (Fig. 5G), two mediators involved in recruitment and promotion of MDSCs with suppressive activity (32).
We also assayed the T cell differentiation status in the lungs of NTHi-exposed CC-LR mice, and we found that after IL-6 blockade there is an increase in TNFα and CCL5 protein expression (Fig. 5G), a cytokine and a chemokine related to T cell activation and recruitment, respectively (33). We further observed a decrease in Tgfβ and Foxp3 expression within the lung CD45+ population (Fig. 5H), suggestive of reduced Treg differentiation. Moreover, we observed a significant increase in Gzmb and Tbx21 expression (Fig. 5H), signatures of CD8 and Th1 cytotoxic programs, respectively. We also observed a decrease in Ccl2 and increase in Nos2 (Fig. 5H) expression suggestive of M1 macrophage polarization similar to what we observed in non-COPD setting. Overall, these results further suggest IL-6 has a paracrine role in K-ras mutant lung cancer, and its blockade shifts the lung microenvironment towards a non-suppressive anti-tumor (type 1) direction in the presence of COPD as well.
Discussion
Tumor-promoting inflammation is an important cancer hallmark and enabling characteristic (24). It is apparent that the cytokines released during inflammation influence carcinogenesis (34,35). IL-6 is a pleiotropic cytokine that is highly expressed in K-ras mutant lung cancer mouse models (10,12) and human lung cancer, where its expression is associated with lower overall survival (19). Here, we observed that IL-6 induces tumor cell proliferation and orchestrates an immune suppressive lung microenvironment with type 2 macrophage polarization along with MDSCs recruitment putting IL-6 as a potential druggable target for KRAS mutant tumors with a novel paracrine effect. Our data is in agreement with a recently described homeostatic function of IL-6 in limiting inflammation via polarizing monocytes to a suppressive M2 state (36), that could favor tumor immune escape and growth (37).
Strategies to overcome the immunosuppressive state of the tumor microenvironment by re-educating myeloid cells via targeting cytokines have been applied to other solid tumors (38,39) but have not been tested in lung cancer. Targeting of cytokines such as IL-6 is a safe strategy used for treatment of autoimmune diseases (40). We found that anti-IL-6 treatment significantly inhibits K-ras induced lung tumorigenesis in vivo that is correlated with a shift in the lung microenvironment toward a less proliferative, less angiogenic, and less suppressive anti-tumor myeloid phenotype. Tumor inhibition in response to anti-IL-6 Ab treatment was not due to reduced recombination rate as there was no difference in CCSP expression in anti-IL-6 Ab treated groups when compared to non-treated groups (Supplementary Figure 7). Tan et al have also targeted IL-6 using genetic strategy in an Adenovirus Cre dependent K-ras mouse model and found that lack of IL-6 interestingly increases tumor initiation while suppressing tumor growth and increasing survival (41). This was also observed in a model which in IL-6 was targeted conditionally in epithelial cells using the same Adenovirus Cre delivery strategy (42). In our model, IL-6 blockade did not increase tumor initiation. We attribute these differences to the distinct timing and cell-specific effects of mutant K-ras induction and IL-6 blockade, as well as absence or presence of adenoviral infection. Interestingly, using commercially available NSCLC cell lines, while we found a significant STAT3 pathway inhibition by siltuximab (monoclonal Ab against IL-6) or tocilizumab (monoclonal Ab against IL-6 cognate receptor), only a modest effect of these Abs alone or in combination with cisplatin on cell proliferation in vitro was observed. This is similar to what has been recently described with siltuximab alone or in combination with erlotinib in EGFR mutant NSCLC cell lines (43). Thus, our data and the aforementioned study point to the crucial role of myeloid and other cell populations within the tumor microenvironment as being important in the tumor-inhibitory properties of anti-IL-6 Ab.
We have previously shown that COPD-like airway inflammation promotes K-ras induced lung tumorigenesis (12). Here, we further found that this process was associated with robust increase in IL-6 levels, increased STAT3 activation, and an amplified myeloid cell response (M2 type macrophage polarization along with accumulation of neutrophils and G-MDSCs). Interestingly, COPD patients with or without lung cancer show increased MDSC population (44) which is associated with high levels of IL-6 in both group. As expected, treatment with anti-IL-6 Ab significantly reduced the promoting effect of COPD-like airway inflammation on lung tumor cell proliferation and tumor angiogenesis. However, IL-6 blockade surprisingly increased the total lung inflammatory cell number in contrast to what we observed in absence of COPD-like inflammation. This observation was largely due to increased number of lung macrophages with M1 phenotype characterized by high MHC class II expression which could be related to increased expression of CCL2. Interestingly, anti-IL-6 treatment did not increase the absolute number of neutrophils but significantly reduced the number of G-MDSCs thus further pointing to a paracrine effect of IL-6 in modulating tumor microenvironment. This was associated with a decreased level of myeloid cell related chemokine and cytokine, CXCL1 and IL-17, which we have previously shown to have important roles in promotion of K-ras mutant lung cancer by inflammation (45,46).
Phenotypic changes observed in myeloid cells due to anti-IL-6 treatment, accompanied by a significant decrease in expression of metabolic mediators, arginase1 and IDO, two enzymes that deplete L-arginine and tryptophan respectively, which are key amino acids for T cell anabolic functions (47). This was associated with a switch in T cell response from a pro-tumor Treg/Th17 to an anti-tumor Th1/CD8 T cell response. This is in accordance with our previous finding indicating an essential role for Th17 cells in promotion of K-ras induced lung tumors, which was dependent on the presence of MDSCs (13), and reinforces the notion that myeloid cells are key immune cells in K-ras mutant lung tumors.
The proliferative, survival, and angiogenic effects of IL-6 on epithelial (tumor) cells are mediated by the STAT3 pathway activation (48–50). In a previously published study by Qu et al, upregulation of STAT3 and its target genes were observed in lung tissues from both smokers and non-smokers with COPD (51). We have also found that relatively increased STAT3 expression is significantly associated with worse disease-free survival in patients with K-ras mutant lung tumors. This suggests that aberrant STAT3 expression impacts clinical outcome among early-stage lung adenocarcinoma patients with KRAS mutations, which can be attributed to cross talk between KRAS and STAT3 signaling. We further found that IL-6 blockade inhibits epithelial STAT3 activation and subsequent tumor cell proliferation in our K-ras mutant mouse which is in agreement with recently published data (42,52). Chronic STAT3 activation could also inhibit the expression of mediators necessary for immune activation against tumor cells (53), and contributes to the accumulation of MDSCs with immunosuppressive properties (54,55), which we observed in our study as well. Thus, our results suggest that chronic IL-6/STAT3 signals may contribute to both observed cell intrinsic and immunosuppressive mechanisms of tumor promotion which were all significantly reduced in our model after IL-6 blockade.
Collectively, our data show that K-ras mutation besides its cell intrinsic proliferative and angiogenic effects, drives an immunosuppressive pro-tumor microenvironment with a M2 TAM polarization, recruitment of MDSCs, and increases Treg/Th17 response, which is probably orchestrated by IL-6 in both autocrine and paracrine fashion. To our knowledge, these are novel functions for IL-6 and novel findings in pathogenesis of K-ras-induced lung tumorigenesis. Importantly, we further showed that IL-6 blockade not only has direct inhibitory effect on tumor (epithelial) cells but also could skew pro-tumor immunosuppressive environment toward an anti-tumor phenotype. Therefore, we propose using anti-IL-6 treatment in combination with targeting downstream effectors of K-ras (e.g. MEK) or immune checkpoint molecules (e.g. PD-1, PDL-1) as an alternative strategy to subvert undruggable K-ras mutant lung cancer.
Supplementary Material
Acknowledgments
Financial support: This work was supported by grant [LCD-114696-N] from the American Lung Association/LUNGevity Foundation, and [RSG-11-115-01-CNE] from American Cancer Society awarded to Seyed Javad Moghaddam, a pilot grant award from the Center for Inflammation and Cancer, UT M. D. Anderson Cancer Center to Stephanie S. Watowich, UT Lung Specialized Programs of Research Excellence grant [P50CA70907] to Ignacio I. Wistuba, and M. D. Anderson Institutional Tissue Bank award [2P30CA016672] from the NIH National Cancer Institute.
We thank Dr. Burton F. Dickey, Dr. Michael J. Tuvim, and Ms. Muge Celiktas for their scientific advices and technical assistance in the completion of this project.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest.
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. The New England journal of medicine. 2008;359(13):1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toll BA, Rojewski AM, Duncan LR, Latimer-Cheung AE, Fucito LM, Boyer JL, et al. “Quitting smoking will benefit your health”: the evolution of clinician messaging to encourage tobacco cessation. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(2):301–9. doi: 10.1158/1078-0432.CCR-13-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mannino DM, Aguayo SM, Petty TL, Redd SC. Low lung function and incident lung cancer in the United States: data From the First National Health and Nutrition Examination Survey follow-up. Archives of internal medicine. 2003;163(12):1475–80. doi: 10.1001/archinte.163.12.1475. [DOI] [PubMed] [Google Scholar]
- 4.de-Torres JP, Wilson DO, Sanchez-Salcedo P, Weissfeld JL, Berto J, Campo A, et al. Lung cancer in patients with chronic obstructive pulmonary disease. Development and validation of the COPD Lung Cancer Screening Score. American journal of respiratory and critical care medicine. 2015;191(3):285–91. doi: 10.1164/rccm.201407-1210OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takiguchi Y, Sekine I, Iwasawa S, Kurimoto R, Tatsumi K. Chronic obstructive pulmonary disease as a risk factor for lung cancer. World journal of clinical oncology. 2014;5(4):660–6. doi: 10.5306/wjco.v5.i4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young RP, Hopkins RJ, Christmas T, Black PN, Metcalf P, Gamble GD. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. The European respiratory journal. 2009;34(2):380–6. doi: 10.1183/09031936.00144208. [DOI] [PubMed] [Google Scholar]
- 7.Rutgers SR, Postma DS, ten Hacken NH, Kauffman HF, van Der Mark TW, Koeter GH, et al. Ongoing airway inflammation in patients with COPD who do not currently smoke. Thorax. 2000;55(1):12–8. doi: 10.1136/thorax.55.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapperre TS, Postma DS, Gosman MM, Snoeck-Stroband JB, ten Hacken NH, Hiemstra PS, et al. Relation between duration of smoking cessation and bronchial inflammation in COPD. Thorax. 2006;61(2):115–21. doi: 10.1136/thx.2005.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England journal of medicine. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji H, Houghton AM, Mariani TJ, Perera S, Kim CB, Padera R, et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene. 2006;25(14):2105–12. doi: 10.1038/sj.onc.1209237. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer discovery. 2014;4(4):452–65. doi: 10.1158/2159-8290.CD-13-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moghaddam SJ, Li H, Cho SN, Dishop MK, Wistuba II, Ji L, et al. Promotion of lung carcinogenesis by chronic obstructive pulmonary disease-like airway inflammation in a K-ras-induced mouse model. American journal of respiratory cell and molecular biology. 2009;40(4):443–53. doi: 10.1165/rcmb.2008-0198OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang SH, Mirabolfathinejad SG, Katta H, Cumpian AM, Gong L, Caetano MS, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(15):5664–9. doi: 10.1073/pnas.1319051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardnell RJ, Behrens C, Diao L, Fan Y, Tang X, Tong P, et al. An Integrated Molecular Analysis of Lung Adenocarcinomas Identifies Potential Therapeutic Targets among TTF1-Negative Tumors, Including DNA Repair Proteins and Nrf2. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(15):3480–91. doi: 10.1158/1078-0432.CCR-14-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(23):6556–65. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer research. 2004;64(7):2307–16. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 17.Zaynagetdinov R, Sherrill TP, Kendall PL, Segal BH, Weller KP, Tighe RM, et al. Identification of myeloid cell subsets in murine lungs using flow cytometry. American journal of respiratory cell and molecular biology. 2013;49(2):180–9. doi: 10.1165/rcmb.2012-0366MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan BM, Pine SR, Chaturvedi AK, Caporaso N, Harris CC. A combined prognostic serum interleukin-8 and interleukin-6 classifier for stage 1 lung cancer in the prostate, lung, colorectal, and ovarian cancer screening trial. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2014;9(10):1494–503. doi: 10.1097/JTO.0000000000000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrera L, Montes-Servin E, Barrera A, Ramirez-Tirado LA, Salinas-Parra F, Banales-Mendez JL, et al. Cytokine profile determined by data-mining analysis set into clusters of non-small-cell lung cancer patients according to prognosis. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2015;26(2):428–35. doi: 10.1093/annonc/mdu549. [DOI] [PubMed] [Google Scholar]
- 20.Narimatsu M, Maeda H, Itoh S, Atsumi T, Ohtani T, Nishida K, et al. Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells. Molecular and cellular biology. 2001;21(19):6615–25. doi: 10.1128/MCB.21.19.6615-6625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochoa CE, Mirabolfathinejad SG, Ruiz VA, Evans SE, Gagea M, Evans CM, et al. Interleukin 6, but not T helper 2 cytokines, promotes lung carcinogenesis. Cancer prevention research (Philadelphia, Pa) 2011;4(1):51–64. doi: 10.1158/1940-6207.CAPR-10-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang DJ, Ratnam NM, Byrd JC, Guttridge DC. NF-kappaB functions in tumor initiation by suppressing the surveillance of both innate and adaptive immune cells. Cell reports. 2014;9(1):90–103. doi: 10.1016/j.celrep.2014.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Moghaddam SJ, Clement CG, De la Garza MM, Zou X, Travis EL, Young HW, et al. Haemophilus influenzae lysate induces aspects of the chronic obstructive pulmonary disease phenotype. American journal of respiratory cell and molecular biology. 2008;38(6):629–38. doi: 10.1165/rcmb.2007-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donaldson GC, Seemungal TA, Patel IS, Bhowmik A, Wilkinson TM, Hurst JR, et al. Airway and systemic inflammation and decline in lung function in patients with COPD. Chest. 2005;128(4):1995–2004. doi: 10.1378/chest.128.4.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celli BR, Locantore N, Yates J, Tal-Singer R, Miller BE, Bakke P, et al. Inflammatory biomarkers improve clinical prediction of mortality in chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine. 2012;185(10):1065–72. doi: 10.1164/rccm.201110-1792OC. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn C, 3rd, Homer RJ, Zhu Z, Ward N, Flavell RA, Geba GP, et al. Airway hyperresponsiveness and airway obstruction in transgenic mice. Morphologic correlates in mice overexpressing interleukin (IL)-11 and IL-6 in the lung. American journal of respiratory cell and molecular biology. 2000;22(3):289–95. doi: 10.1165/ajrcmb.22.3.3690. [DOI] [PubMed] [Google Scholar]
- 29.Barnes PJ. Cellular and molecular mechanisms of chronic obstructive pulmonary disease. Clinics in chest medicine. 2014;35(1):71–86. doi: 10.1016/j.ccm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Meyer C, Sevko A, Ramacher M, Bazhin AV, Falk CS, Osen W, et al. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(41):17111–6. doi: 10.1073/pnas.1108121108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nature reviews Immunology. 2012;12(4):253–68. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma S, Cheng Q, Cai Y, Gong H, Wu Y, Yu X, et al. IL-17A produced by gammadelta T cells promotes tumor growth in hepatocellular carcinoma. Cancer research. 2014;74(7):1969–82. doi: 10.1158/0008-5472.CAN-13-2534. [DOI] [PubMed] [Google Scholar]
- 33.Thakur A, Schalk D, Tomaszewski E, Kondadasula SV, Yano H, Sarkar FH, et al. Microenvironment generated during EGFR targeted killing of pancreatic tumor cells by ATC inhibits myeloid-derived suppressor cells through COX2 and PGE2 dependent pathway. Journal of translational medicine. 2013;11:35. doi: 10.1186/1479-5876-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin EY, Pollard JW. Role of infiltrated leucocytes in tumour growth and spread. British journal of cancer. 2004;90(11):2053–8. doi: 10.1038/sj.bjc.6601705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauer J, Chaurasia B, Goldau J, Vogt MC, Ruud J, Nguyen KD, et al. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nature immunology. 2014;15(5):423–30. doi: 10.1038/ni.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerkar SP, Restifo NP. Cellular constituents of immune escape within the tumor microenvironment. Cancer research. 2012;72(13):3125–30. doi: 10.1158/0008-5472.CAN-11-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. The Journal of clinical investigation. 2006;116(8):2132–41. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer research. 2014;74(18):5057–69. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheinecker C, Smolen J, Yasothan U, Stoll J, Kirkpatrick P. Tocilizumab. Nature reviews Drug discovery. 2009;8(4):273–4. doi: 10.1038/nrd2863. [DOI] [PubMed] [Google Scholar]
- 41.Tan X, Carretero J, Chen Z, Zhang J, Wang Y, Chen J, et al. Loss of p53 attenuates the contribution of IL-6 deletion on suppressed tumor progression and extended survival in Kras-driven murine lung cancer. PloS one. 2013;8(11):e80885. doi: 10.1371/journal.pone.0080885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu Z, Sun F, Zhou J, Li L, Shapiro SD, Xiao G. Interleukin-6 Prevents the Initiation but Enhances the Progression of Lung Cancer. Cancer research. 2015;75(16):3209–15. doi: 10.1158/0008-5472.CAN-14-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song L, Smith MA, Doshi P, Sasser K, Fulp W, Altiok S, et al. Antitumor efficacy of the anti-interleukin-6 (IL-6) antibody siltuximab in mouse xenograft models of lung cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2014;9(7):974–82. doi: 10.1097/JTO.0000000000000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baraldo S, Pinton L, Ballarin A, Mandruzzato S, Bazzan E, Falisi E, et al. Myeloid derived suppressor cells in the crosstalk between COPD and lung cancer. European Respiratory Journal. 2011;38(Suppl 55) [Google Scholar]
- 45.Moghaddam SJ, Barta P, Mirabolfathinejad SG, Ammar-Aouchiche Z, Garza NT, Vo TT, et al. Curcumin inhibits COPD-like airway inflammation and lung cancer progression in mice. Carcinogenesis. 2009;30(11):1949–56. doi: 10.1093/carcin/bgp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong L, Cumpian AM, Caetano MS, Ochoa CE, De la Garza MM, Lapid DJ, et al. Promoting effect of neutrophils on lung tumorigenesis is mediated by CXCR2 and neutrophil elastase. Molecular cancer. 2013;12(1):154. doi: 10.1186/1476-4598-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews Cancer. 2012;12(4):252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer cell. 2009;15(2):103–13. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. European journal of cancer (Oxford, England : 1990) 2005;41(16):2502–12. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 51.Qu P, Roberts J, Li Y, Albrecht M, Cummings OW, Eble JN, et al. Stat3 downstream genes serve as biomarkers in human lung carcinomas and chronic obstructive pulmonary disease. Lung cancer (Amsterdam, Netherlands) 2009;63(3):341–7. doi: 10.1016/j.lungcan.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Qu Z, Yan S, Sun F, Whitsett JA, Shapiro SD, et al. Differential roles of STAT3 in the initiation and growth of lung cancer. Oncogene. 2015;34(29):3804–14. doi: 10.1038/onc.2014.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nature reviews Immunology. 2007;7(1):41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 54.Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. The Journal of experimental medicine. 2008;205(10):2235–49. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. Journal of immunology (Baltimore, Md : 1950) 2009;182(9):5693–701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.