Abstract
Purpose of review
The tight junction (TJ) conductance made of the claudin based paracellular channel is important in the regulation of calcium and magnesium reabsorption in the kidney. This review describes recent findings of the structure, the function, and the physiologic regulation of claudin-14, claudin-16 and claudin-19 channels that through protein interactions confer calcium and magnesium permeability to the TJ.
Recent findings
Mutations in two TJ genes – claudin-16 and claudin-19 cause the inherited renal disorder – familial hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHNC). Recent genome-wide association study (GWAS) has identified claudin-14 as a major risk gene of hypercalciuric nephrolithiasis. Claudin-14 interacts with claudin-16 and inhibits its cation permselectivity. Transgenic overexpression of claudin-14 in the kidney generated the FHHNC-like phenotypes. The crystal structure of claudin-19 has recently been resolved allowing the reconstruction of a claudin assembly model from cis-dimers made of claudin-16 and claudin-19 interaction. MicroRNAs have been identified as novel regulators of the claudin-14 gene. The microRNA-claudin-14 operon is directly regulated by the Ca++ sensing receptor (CaSR) gene in response to hypercalcemia.
Summary
The paracellular pathway in the kidney is particularly important for mineral metabolism. Three claudin proteins – claudin-14, claudin-16 and claudin-19 contribute to the structure and function of this paracellular pathway. Genetic mutations and gene expression changes in these claudins may lead to alteration of the paracellular permeability to calcium and magnesium, ultimately affecting renal mineral metabolism. Claudins can be regulated on several levels including transcription, microRNA repression, trafficking, phosphorylation, and interaction.
Keywords: claudin, tight junction, hypercalciuria, ion channel, microRNA, kidney
Introduction
The tight junction (zonula occludens) is the most apical member of the junctional complex found in vertebrate epithelia responsible for the barrier to movement of ions and molecules between apical and basal compartments, the paracellular pathway [1,2]. The known integral membrane proteins of the tight junction include occludin (a 65 kDa membrane protein bearing four transmembrane domains and two uncharged extracellular loops) [3], the Junctional Adhesion Molecules (JAMs) [4], a four-member group of glycosylated proteins and the claudins. Claudins are tetraspan proteins consisting of a family of at least 22 members [5,6]. They range in molecular mass from 20–28 kD with charged extracellular loops. In renal epithelia, claudins confer ion selectivity to the paracellular pathway resulting in differences in epithelial resistance and paracellular permeabilities [7]. These results led to models of the claudins forming the paracellular channel, a novel class of channels oriented perpendicular to the membrane plane and serving to join two extracellular compartments.
Paracellular reabsorption of Ca++ and Mg++ in the kidney
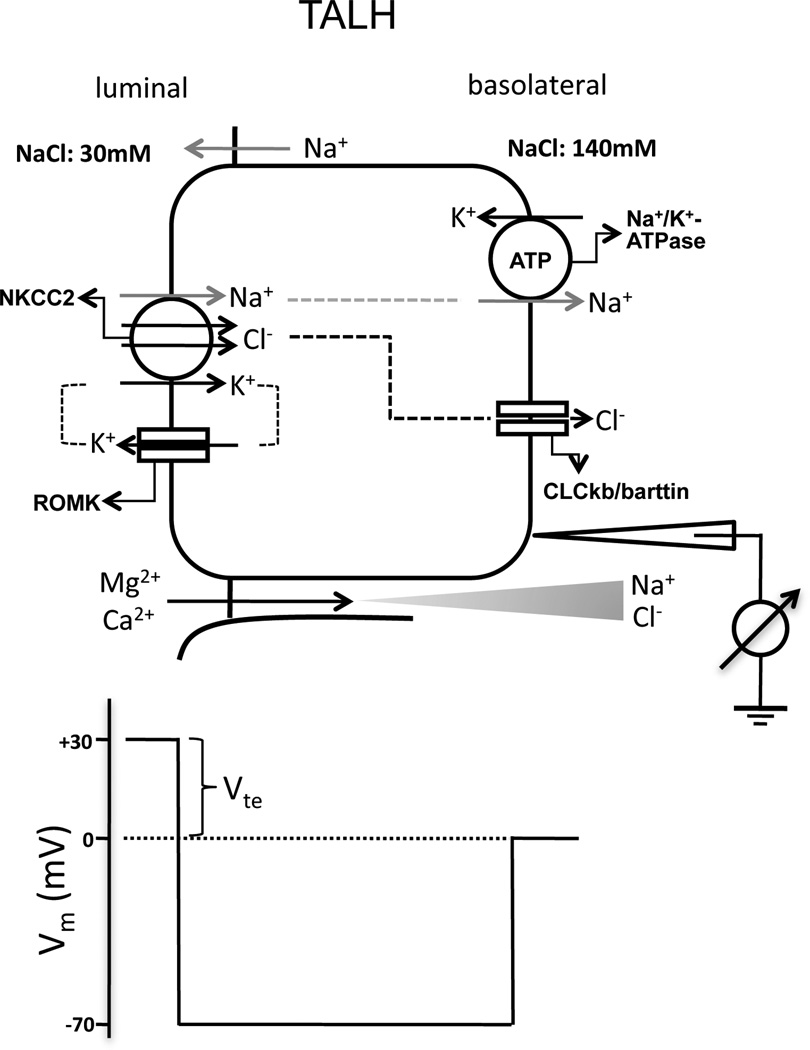
The thick ascending limb of the loop of Henle (TALH) is responsible for reabsorbing 25–40% of filtered Na+ [8], 50–60% of filtered Mg++ [9] and 30–35% of filtered Ca++ [10]. The dissociation of salt and water reabsorption in the TALH results in the dilution of luminal fluid and the generation of an axial corticomedullary osmolality gradient, which then drives water reabsorption in the collecting duct [11]. The dilution of luminal fluid in the TALH also establishes a basolateral-luminal transepithelial salt concentration gradient that results in lumen-positive voltages as the driving force for passive paracellular reabsorption of Ca++ and Mg++ (Figure 1) [12,13].
Figure 1.
Ion transport mechanism in the thick ascending limb of Henle’s loop (TALH). The membrane voltage (Vm) trace depicts the virtual measurement by an electrode that is pushed from the basolateral side through the cell to the luminal side. In this example, the basolateral membrane voltage is −70mV and the luminal membrane voltage is −100mV, resulting in a transepithelial voltage (Vte) of +30mV with respect to the basolateral side. Vte drives Ca++ and Mg++ transport through the tight junction. NKCC2: Na+/K+/Cl−-cotransporter type 2; ROMK: renal outer medullary K+ channel; CLCkb: Cl− channel type kb.
Human mutations in claudins causing Ca++ and Mg++ imbalance diseases
A monogenic renal disorder, autosomal recessive familial hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHNC; OMIM #248250), is caused by mutations in two claudin genes: claudin-16 [14] and claudin-19 [15]. The claudin-16 and claudin-19 proteins are exclusively localized in the TALH [16]. Hou and colleagues have established that claudin-16 and claudin-19 proteins form a paracellular cation channel through direct protein interactions, which (i) permeates Ca++ and Mg++ [17]; (ii) generates a lumen-positive voltage owing to the transepithelial salt concentration gradient to drive Ca++ and Mg++ reabsorption [18]. A recent genome-wide association study (GWAS) has identified claudin-14 as a major risk gene of hypercalciuric nephrolithiasis [19]. Four common, synonymous variants in the claudin-14 gene locus were significantly associated with kidney stones by genome wide criteria. Two of them are exonic, causing silent mutations in the claudin-14 protein: rs219779[C] (R81R) and rs219780[C] (T229T). Both exonic SNPs show significant association with decreased bone mineral density (BMD). The urinary Ca++ excretion is significantly higher in homozygous carriers of rs219780[C] compared to non-carriers.
The biochemical basis of the paracellular channel made of claudins
The structure and function claudin-16
In low-resistance Madin-Darby canine kidney type II (MDCK-II) cells, Ikari and colleagues have found that transfected claudin-16 increased paracellular Ca++ but not Mg++ flux rates [20]. Using a high-resistance MDCK-C7 cell line, Kausalya and colleagues have revealed a modest effect of claudin-16 increasing paracellular Mg++ permeability [21]. Nevertheless, the effects of claudin-16 on divalent cation permeabilities measured by both studies are so small (<50%) that the theory of claudin-16 forming a divalent channel cannot explain the dramatic phenotype caused by loss of claudin-16 function in FHHNC patients or animal models [14,22,23]. Serendipitously, Hou and colleagues have found that claudin-16 induced a profound increase in paracellular Na+ permeability compared to the modest increase in Mg++ permeability with the low-resistance LLC-PK1 cells [17]. The permeabilities of claudin-16 to other alkali metal cations were also significant and resembled the Eisenman selectivity sequence V-VIII: K+ > Rb+ > Na+ ≈ Li+ [17]. The Eisenman selectivity sequence reflects the charge density within an ion channel pore that will stabilize the permeating ions through electrostatic interaction [24]. Notably, the Eisenman selectivity sequence of claudin-16 is similar to that of claudin-2, a classic cation channel [25]. Mutagenesis studies have identified a locus of negatively charged amino acid residues in the 1st extracellular loop domain of claudin-16 (D104, D105, E119, D126 and E140) that are all required for its cation permeability.
The structure and function of claudin-19
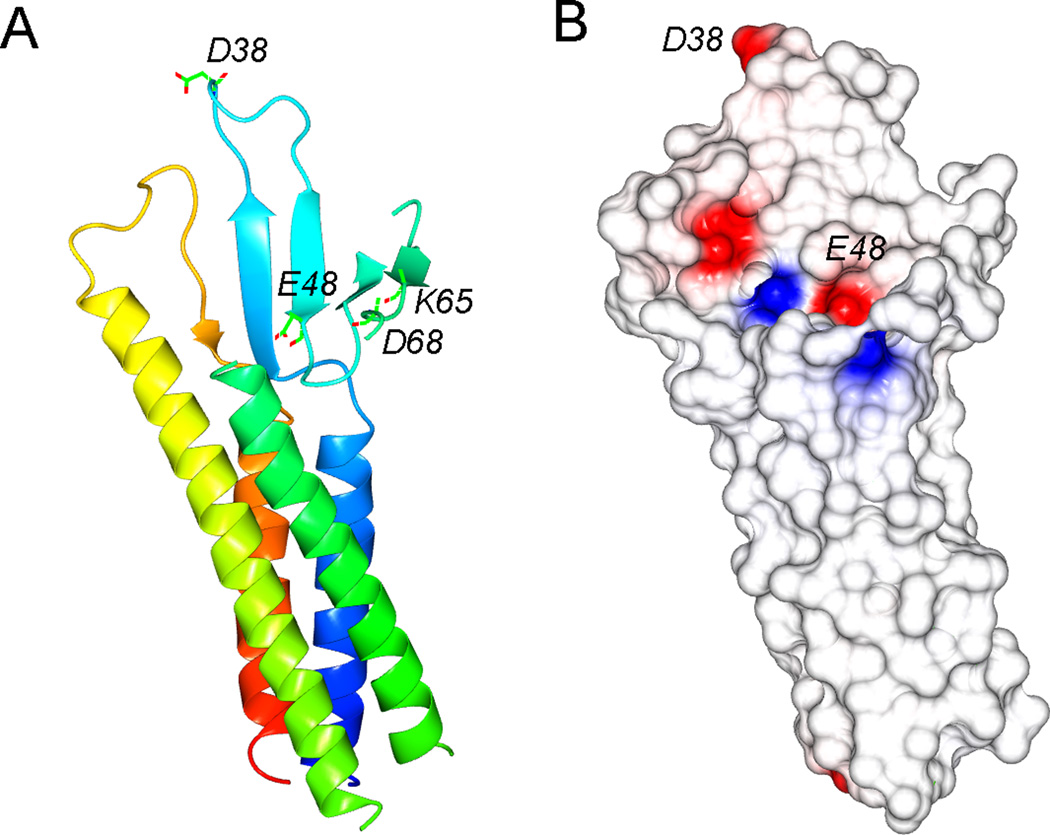
In the cultured LLC-PK1 cells, transfected claudin-19 significantly decreased the paracellular permeability to Cl− without affecting the Na+ or Mg++ permeability [18]. Another study using the MDCK-II cells has found some conflicting results that claudin-19 rather decreased the permeability to Mg++ [26]. Collectively, both studies argue against the hypothetic theory that FHHNC is caused by kidney’s inability to reabsorb Ca++ and Mg++ through a divalent cation channel in the tight junction [14]. Until recently, the 3D crystal structure of claudin-19 has been resolved at 3.7Å resolution [27]. The claudin-19 monomer adopts characteristic β-sheet folds in the extracellular loop domains, which are anchored to four transmembrane α-helical bundles (Figure 2A). A locus of charged amino acids in the 1st extracellular loop domain are purported to interact with the permeating ions by electrostatic field (Figure 2A). The electrostatic potential surface of the 1st extracellular loop domain reveals the location of residues (D38 and E48) potentially important for charge selectivity (Figure 2B).
Figure 2.
Structural insights of claudin-19 protein. (A) Structure of monomeric claudin-19 in ribbon representation. The color changes gradually from the N terminus (blue) to the C terminus (red). The charged residues in the ECL1 are highlighted. (B) Electrostatic potential surface of the extracellular domains viewed in (A), contoured from −2kT/e (red) to +2kT/e (blue). Residues potentially important for charge selectivity are located.
The claudin-16 and claudin-19 interaction
When expressed separately in LLC-PK1 cells, claudin-16 increased paracellular permeability to Na+ while claudin-19 decreased paracellular permeability to Cl− [17,18]. Co-expression of claudin-16 and claudin-19 resulted in more pronounced changes in both Na+ and Cl− permeabilities compared to their each individual levels, therefore suggesting a functional synergy between these two claudins [18]. Using sucrose gradient sedimentation and chemical crosslinking, Gong and colleagues were able to isolate a stable dimer made of claudin-16 and claudin-19 from transfected HEK293 cells and Sf9 cells [28]. The dimerization appeared to be mediated through the cis-association of the transmembrane domain #3 and #4 in both claudin-16 and claudin-19 [28]. When mutations that selectively disrupted the claudin-16 and claudin-19 dimerization were introduced to either claudin, the resultant tight junction dramatically lost its cation permeability, suggesting a mechanism for the role of claudin interaction in the development of FHHNC [28]. Compatible with this hypothesis, in vivo in transgenic animal models, deletion of claudin-16 rendered claudin-19 delocalization from the tight junction and vice versa [16].
The claudin-14 and claudin-16 interaction
A recent GWAS study has identified claudin-14 as a major risk gene for hypercalciuric nephrolithiasis [19]. Gong and colleagues have found that claudin-14 protein is predominantly localized in the TALH, thus implicating claudin-14 to be anther candidate with potential roles in paracellular Ca++ and Mg++ transport [29]. In transfected LLC-PK1 cells, claudin-14 reduces the cation permeability of the claudin-16 channel, but not that of the claudin-19 channel, which is further supported by the finding that claudin-14 physically interacts with claudin-16 but not with claudin-19 [29]. Consistent with the hypothesis that claudin-14 acts as a negative regulator of the divalent cation permeation in the TALH, the TALH-specific overexpression of claudin-14 in transgenic animals have generated the FHHNC resembling phenotypes, including hypomagnesemia, hypermagnesiuria and hypercalciuria [30].
The regulatory mechanism of the paracellular channel made of claudins
Regulation of claudin localization in the tight junction
The carboxy-terminal domain of claudin-16 contains a PDZ (postsynaptic density 95/discs large/zonula occludens-1)-binding motif (TRV) that interacts with the PDZ domain of the schaffolding proteins such as ZO-1 [20]. Muller and colleagues identified a mutation in the PDZ-binding motif of claudin-16 (TRV to RRV) from patients with childhood FHHNC, which disrupted the claudin-16 binding to ZO-1 and caused claudin-16 delocalization from the tight junction [31]. Claudin phosphorylation status seemed to affect its interaction with ZO-1 and localization in the tight junction. The Serine 217 in the carboxyl terminus of claudin-16 is phosphorylated by protein kinase A (PKA) and dephosphorylation with PKA inhibitors reduced its interaction with ZO-1 and caused its mislocalization to the lysosome [32]. Extracellular Mg++ levels appear to regulate the tight junction localization of claudin-16 by phosphorylation dependent mechanism. In MDCK cells, low extracellular Mg++ levels reduced the localization of claudin-16 to the tight junction, which can be abolished by the extracellular signal-regulated kinase (ERK) inhibitor – U0126 [33]. With yeast two-hybrid screening approach, Ikari and colleagues have demonstrated direction interaction between claudin-16 and syntaxin-8, an important protein trafficking regulator [34]. Knockdown of syntaxin-8 in renal cells decreased the cell surface localization of claudin-16. On the other hand, the claudin-19 localization to tight junction is determined by its interaction with claudin-16. Deletion of claudin-16 in the mouse kidney caused claudin-19 delocalization from tight junction without affecting its mRNA or protein expression levels [16].
Transcriptional regulation of claudin gene expression
Changes in extracellular Mg++ levels may regulate claudin-16 gene expression as a feedback mechanism to regulate renal Mg++ reabsorption rates. Mice fed with low Mg++ diet had significantly higher expression levels of claudin-16 in the kidney compared to those fed with normal or high Mg++ diet [35]. The 2.5kb proximal promoter of the claudin-16 gene seems to contain sequences that can directly respond to extracellular Mg++ alterations [35]. The promoter activity of claudin-16 can also be regulated by 1,25-(OH)2 vitamin D3. In mice treated with 1,25-(OH)2 vitamin D3, the renal expression levels of claudin-16 were significantly reduced [36]. Luk and colleagues have systematically analyzed the promoter binding sites in the claudin-19 gene and found a strong specific protein 1 (SP1) binding region located from −39 to −108 upstream of the transcription initiation site [37]. SP1 is a zinc finger transcription factor involved in many cellular processes, including chromatin remodeling, DNA damage control and apoptosis. It is not known what kind of extracellular signal regulates SP1 binding to the claudin-19 promoter and its activity. One study has revealed decreased claudin-19 expression levels in human polycystic kidneys [38].
MicroRNA based post-transcriptional regulation of claudin
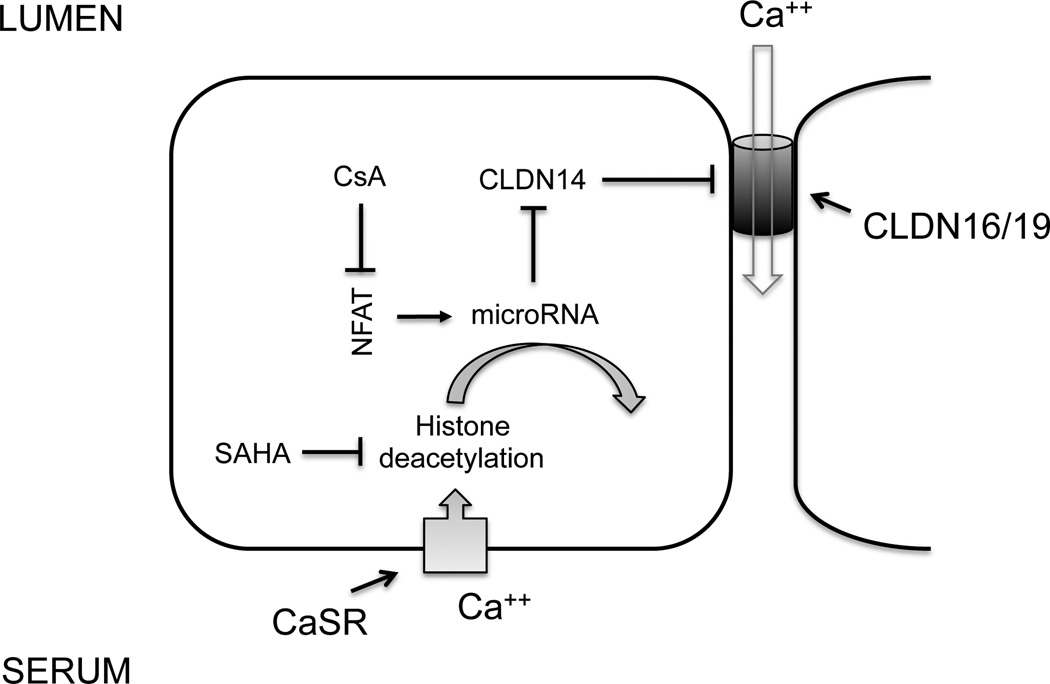
MicroRNAs are key regulatory molecules that regulate gene expression on the post-transcriptional level by inducing target mRNA decay or translational repression [39]. Gong and colleagues have identified two microRNA molecules from the kidney – miR-9 and miR-374, both of which recognize complimentary binding sites located in the 3’-UTR of the claudin-14 mRNA [29]. Experiments using antisense oligonucleotide against miR-9 or miR-374 revealed that both microRNAs suppressed claudin-14 translation and induced its mRNA decay in a synergistic manner [29]. In vivo in the mouse kidney, anti-miR-374 oligonucleotide injection caused pronounced renal wasting of Ca++ and Mg++ due to unopposed expression of claudin-14 [40]. High Ca++ intake increases while low Ca++ intake decreases the expression levels of miR-9 and miR-374 in the kidney, which in turn causes a reciprocal increase in the claudin-14 expression level [29]. Toka and colleagues found that kidney specific deletion of the Ca++ sensing receptor (CaSR) caused pronounced reduction in the claudin-14 gene expression levels but the authors did not detect any change in the kidney microRNA level [41]. Using CaSR specific agonist (calcimimetic) or antagonist (calcilytic), Gong and colleagues further demonstrated that CaSR activation caused decreases in miR-9 and miR-374 expression levels but increases in claudin-14 expression levels, while CaSR inhibition exerted the opposite effects on microRNA and claudin-14 via histone deacetylation dependent epigenetic mechanisms [30,40]. These results suggest that the microRNA-claudin-14 molecular pathway is under direct regulation of CaSR and forms an essential part of the CaSR signaling cascade in the kidney (Figure 3).
Figure 3.
CaSR signaling cascade in the kidney. A feedback control loop of CaSR regulation in the TALH includes the following key components: microRNAs (miR-9 and miR-374) and claudin-14. Claudin-14 negatively regulates claudin-16 and -19 permeability via direct interaction. MicroRNAs regulate claudin-14 mRNA stability and translational efficacy. The gene transcription of microRNAs themselves is regulated by the transcriptional factor – NFAT and via deacetylation of nearby histone molecules. NFAT: nuclear factor of activated T cells; CsA: cyclosporine A; SAHA: suberanilohydroxamic acid.
Open questions
The previous macroscopic recordings of claudin-14, -16 and -19 conductance reflect the aggregates of thousands, or even millions of channel conductive events, with current density reaching µA/cm2. Clearly, these measurements lack the resolution on the molecular level. Chen and colleagues developed a novel tool based on the Scanning Ion Conductance Microscopy (SICM) [42] to record the claudin-2 channel in MDCK cells with the conductance resolution reaching pA and the spatial resolution reaching nanometer [43]. Gong and colleagues applied this tool to resolve the TJ conductance difference made of the cis- and trans-interactions between claudin-16 and -19 [28]. However, single-channel conductance for claudin based paracellular channel has yet been captured using this approach. The major difficulty is how to separate the TJ conductance from nearby membrane conductance. Zhou and colleagues have proposed a novel route to independent measurements of TJ conductance by simultaneously neutralizing the apical membrane currents [44]. PTH and CaSR are two primary signaling mechanisms for extracellular Ca++ homeostasis. Both signals have been shown to directly regulate the renal reabsorption of Ca++ through the tight junction [45,46]. While claudin-14 has been found as a downstream effector of CaSR, it remains unknown how PTH regulates the tight junction permeability to Ca++ and what the underlying molecules are. In addition to microRNA-based mechanism, phosphorylation, palmitoylation, and trafficking, may also exist for claudin-14, -16, and -19 that may explain the changes in paracellular Ca++ transport induced by PTH [45,46]. Finally, how claudins assemble into functional TJ architecture remains to be a major mystery. Based upon the spatial arrangement of claudin-15 within its crystal lattice, Suzuki and colleagues proposed a cis-dimeric assembly model through interactions of the β4 domain of the extracellular loop 1 (ECL1) of claudin-15 [47]. Using a systematic mutagenesis approach, Gong and colleagues suggested a similar cis-dimeric assembly model for claudin-16 and -19 that featured anti-parallel association between the 3rd and 4th transmembrane domains of the two claudins [28]. To resolve the real TJ architecture from its native lipid environment, nevertheless, may need methodological breakthroughs in protein structural biology.
Conclusion
The concept of paracellular ionic pathway has been known for decades. Its importance in renal mineral metabolism has received growing attention because of the recent genetic, biochemical and biophysical analyses of the claudins that caused mineral imbalance diseases. Newly discovered regulatory mechanisms acting upon the claudins may reveal novel therapeutic directions to correct these mineral imbalance diseases.
Bullet points.
Paracellular reabsorption of Ca++ and Mg++ is driven by a lumen positive potential through a cation selective paracellular pathway.
Three claudin proteins – claudin-14, claudin-16 and claudin-19 make the paracellular pathway for Ca++ and Mg++ permeation.
Genetic mutations or gene expression changes in all three claudins cause mineral imbalance diseases.
The paracellular pathway made of claudins can be regulated by extracellular Ca++ levels via the Ca++ sensing receptor (CaSR).
MicroRNAs represent a novel regulatory mechanism for claudins.
Acknowledgments
None
Financial support and sponsorship
This work was supported by the National Institutes of Health Grants RO1DK084059 and P30 DK079333, and American Heart Association Grant 0930050N.
Footnotes
Conflict of interest
The author declares no conflict of interest.
Reference
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukita S, Furuse M. Pores in the wall: claudins constitute tight junction strands containing aqueous pores. J Cell Biol. 2000;149:13–16. doi: 10.1083/jcb.149.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebnet K, Suzuki A, Ohno S, Vestweber D. Junctional adhesion molecules (JAMs): more molecules with dual functions? J Cell Sci. 2004;117:19–29. doi: 10.1242/jcs.00930. [DOI] [PubMed] [Google Scholar]
- 5.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and-2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou J, Rajagopal M, Yu AS. Claudins and the kidney. Annu Rev Physiol. 2013;75:479–501. doi: 10.1146/annurev-physiol-030212-183705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giebisch G, Klose RM, Windhager EE. MICROPUNCTURE STUDY OF HYPERTONIC SODIUM CHLORIDE LOADING IN THE RAT. Am J Physiol. 1964;206:687–693. doi: 10.1152/ajplegacy.1964.206.4.687. [DOI] [PubMed] [Google Scholar]
- 9.Cole DE, Quamme GA. Inherited disorders of renal magnesium handling. J Am Soc Nephrol. 2000;11:1937–1947. doi: 10.1681/ASN.V11101937. [DOI] [PubMed] [Google Scholar]
- 10.Hebert SC. Calcium and salinity sensing by the thick ascending limb: a journey from mammals to fish and back again. Kidney Int Suppl. 2004:S28–S33. doi: 10.1111/j.1523-1755.2004.09105.x. [DOI] [PubMed] [Google Scholar]
- 11.Han JS, Thompson KA, Chou CL, Knepper MA. Experimental tests of three-dimensional model of urinary concentrating mechanism. J Am Soc Nephrol. 1992;2:1677–1688. doi: 10.1681/ASN.V2121677. [DOI] [PubMed] [Google Scholar]
- 12.Hebert SC, Culpepper RM, Andreoli TE. NaCl transport in mouse medullary thick ascending limbs. I. Functional nephron heterogeneity and ADH-stimulated NaCl cotransport. Am J Physiol. 1981;241:F412–F431. doi: 10.1152/ajprenal.1981.241.4.F412. [DOI] [PubMed] [Google Scholar]
- 13.Hebert SC, Culpepper RM, Andreoli TE. NaCl transport in mouse medullary thick ascending limbs. II. ADH enhancement of transcellular NaCl cotransport; origin of transepithelial voltage. Am J Physiol. 1981;241:F432–F442. doi: 10.1152/ajprenal.1981.241.4.F432. [DOI] [PubMed] [Google Scholar]
- 14.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 15.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk JM, Becker C, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79:949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou J, Renigunta A, Gomes AS, Hou M, Paul DL, Waldegger S, Goodenough DA. Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci U S A. 2009;106:15350–15355. doi: 10.1073/pnas.0907724106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou J, Paul DL, Goodenough DA. Paracellin-1 and the modulation of ion selectivity of tight junctions. J Cell Sci. 2005;118:5109–5118. doi: 10.1242/jcs.02631. [DOI] [PubMed] [Google Scholar]
- 18.Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, Waldegger S, Goodenough DA. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest. 2008;118:619–628. doi: 10.1172/JCI33970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorleifsson G, Holm H, Edvardsson V, Walters GB, Styrkarsdottir U, Gudbjartsson DF, Sulem P, Halldorsson BV, de Vegt F, d'Ancona FC, et al. Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet. 2009;41:926–930. doi: 10.1038/ng.404. [DOI] [PubMed] [Google Scholar]
- 20.Ikari A, Hirai N, Shiroma M, Harada H, Sakai H, Hayashi H, Suzuki Y, Degawa M, Takagi K. Association of paracellin-1 with ZO-1 augments the reabsorption of divalent cations in renal epithelial cells. J Biol Chem. 2004;279:54826–54832. doi: 10.1074/jbc.M406331200. [DOI] [PubMed] [Google Scholar]
- 21.Kausalya PJ, Amasheh S, Gunzel D, Wurps H, Muller D, Fromm M, Hunziker W. Disease-associated mutations affect intracellular traffic and paracellular Mg2+ transport function of Claudin-16. J Clin Invest. 2006;116:878–891. doi: 10.1172/JCI26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou J, Shan Q, Wang T, Gomes AS, Yan Q, Paul DL, Bleich M, Goodenough DA. Transgenic RNAi depletion of claudin-16 and the renal handling of magnesium. J Biol Chem. 2007;282:17114–17122. doi: 10.1074/jbc.M700632200. [DOI] [PubMed] [Google Scholar]
- 23.Will C, Breiderhoff T, Thumfart J, Stuiver M, Kopplin K, Sommer K, Gunzel D, Querfeld U, Meij IC, Shan Q, et al. Targeted deletion of murine Cldn16 identifies extra- and intrarenal compensatory mechanisms of Ca2+ and Mg2+ wasting. Am J Physiol Renal Physiol. 2010;298:F1152–F1161. doi: 10.1152/ajprenal.00499.2009. [DOI] [PubMed] [Google Scholar]
- 24.Eisenman G, Horn R. Ionic selectivity revisited: the role of kinetic and equilibrium processes in ion permeation through channels. J Membr Biol. 1983;76:197–225. doi: 10.1007/BF01870364. [DOI] [PubMed] [Google Scholar]
- 25.Yu AS, Cheng MH, Angelow S, Gunzel D, Kanzawa SA, Schneeberger EE, Fromm M, Coalson RD. Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J Gen Physiol. 2009;133:111–127. doi: 10.1085/jgp.200810154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelow S, El-Husseini R, Kanzawa SA, Yu AS. Renal localization and function of the tight junction protein, claudin-19. Am J Physiol Renal Physiol. 2007;293:F166–F177. doi: 10.1152/ajprenal.00087.2007. [DOI] [PubMed] [Google Scholar]
- 27. Saitoh Y, Suzuki H, Tani K, Nishikawa K, Irie K, Ogura Y, Tamura A, Tsukita S, Fujiyoshi Y. Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science. 2015;347:775–778. doi: 10.1126/science.1261833. ••This report showed the hight resolution crystal structure of claudin-19 in complex with the Clostridium perfringens enterotoxin and demonstrated how Clostridium perfringens enterotoxin may break the tight junction architecture.
- 28. Gong Y, Renigunta V, Zhou Y, Sunq A, Wang J, Yang J, Renigunta A, Baker LA, Hou J. Biochemical and biophysical analyses of tight junction permeability made of claudin-16 and claudin-19 dimerization. Mol Biol Cell. 2015 doi: 10.1091/mbc.E15-06-0422. ••This study revealed the fundamental dimeric structure made of claudin-16 and claudin-19 and described a novel recording approach for resolving claudin-16 and claudin-19 interaction based upon the scanning ion conductance microscopy.
- 29.Gong Y, Renigunta V, Himmerkus N, Zhang J, Renigunta A, Bleich M, Hou J. Claudin-14 regulates renal Ca(+)(+) transport in response to CaSR signalling via a novel microRNA pathway. Embo j. 2012;31:1999–2012. doi: 10.1038/emboj.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Y, Hou J. Claudin-14 Underlies Ca++-Sensing Receptor-Mediated Ca++ Metabolism via NFAT-microRNA-Based Mechanisms. J Am Soc Nephrol. 2014;25:745–760. doi: 10.1681/ASN.2013050553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller D, Kausalya PJ, Claverie-Martin F, Meij IC, Eggert P, Garcia-Nieto V, Hunziker W. A novel claudin 16 mutation associated with childhood hypercalciuria abolishes binding to ZO-1 and results in lysosomal mistargeting. Am J Hum Genet. 2003;73:1293–1301. doi: 10.1086/380418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikari A, Matsumoto S, Harada H, Takagi K, Hayashi H, Suzuki Y, Degawa M, Miwa M. Phosphorylation of paracellin-1 at Ser217 by protein kinase A is essential for localization in tight junctions. J Cell Sci. 2006;119:1781–1789. doi: 10.1242/jcs.02901. [DOI] [PubMed] [Google Scholar]
- 33.Ikari A, Kinjo K, Atomi K, Sasaki Y, Yamazaki Y, Sugatani J. Extracellular Mg(2+) regulates the tight junctional localization of claudin-16 mediated by ERK-dependent phosphorylation. Biochim Biophys Acta. 2010;1798:415–421. doi: 10.1016/j.bbamem.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Ikari A, Tonegawa C, Sanada A, Kimura T, Sakai H, Hayashi H, Hasegawa H, Yamaguchi M, Yamazaki Y, Endo S, et al. Tight junctional localization of claudin-16 is regulated by syntaxin 8 in renal tubular epithelial cells. J Biol Chem. 2014;289:13112–13123. doi: 10.1074/jbc.M113.541193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efrati E, Hirsch A, Kladnitsky O, Rozenfeld J, Kaplan M, Zinder O, Zelikovic I. Transcriptional regulation of the claudin-16 gene by Mg2+ availability. Cell Physiol Biochem. 2010;25:705–714. doi: 10.1159/000315090. [DOI] [PubMed] [Google Scholar]
- 36.Kladnitsky O, Rozenfeld J, Azulay-Debby H, Efrati E, Zelikovic I. The claudin-16 channel gene is transcriptionally inhibited by 1,25-dihydroxyvitamin D. Exp Physiol. 2015;100:79–94. doi: 10.1113/expphysiol.2014.083394. [DOI] [PubMed] [Google Scholar]
- 37.Luk JM, Tong MK, Mok BW, Tam PC, Yeung WS, Lee KF. Sp1 site is crucial for the mouse claudin-19 gene expression in the kidney cells. FEBS Lett. 2004;578:251–256. doi: 10.1016/j.febslet.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Lee NP, Tong MK, Leung PP, Chan VW, Leung S, Tam PC, Chan KW, Lee KF, Yeung WS, Luk JM. Kidney claudin-19: localization in distal tubules and collecting ducts and dysregulation in polycystic renal disease. FEBS Lett. 2006;580:923–931. doi: 10.1016/j.febslet.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 40. Gong Y, Himmerkus N, Plain A, Bleich M, Hou J. Epigenetic regulation of microRNAs controlling CLDN14 expression as a mechanism for renal calcium handling. J Am Soc Nephrol. 2015;26:663–676. doi: 10.1681/ASN.2014020129. ••This study revealed an epigenetic program in the kidney that regulated the microRNA and the claudin-14 gene expression levels in order to cause calciuretic effects.
- 41.Toka HR, Al-Romaih K, Koshy JM, DiBartolo S, 3rd, Kos CH, Quinn SJ, Curhan GC, Mount DB, Brown EM, Pollak MR. Deficiency of the calcium-sensing receptor in the kidney causes parathyroid hormone-independent hypocalciuria. J Am Soc Nephrol. 2012;23:1879–1890. doi: 10.1681/ASN.2012030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansma PK, Drake B, Marti O, Gould SA, Prater CB. The scanning ion-conductance microscope. Science. 1989;243:641–643. doi: 10.1126/science.2464851. [DOI] [PubMed] [Google Scholar]
- 43.Chen CC, Zhou Y, Morris CA, Hou J, Baker LA. Scanning ion conductance microscopy measurement of paracellular channel conductance in tight junctions. Anal Chem. 2013;85:3621–3628. doi: 10.1021/ac303441n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou L, Zeng Y, Baker LA, Hou J. A proposed route to independent measurements of tight junction conductance at discrete cell junctions. Tissue Barriers. 2015;3:e1105907. doi: 10.1080/21688370.2015.1105907. •This method article described a novel approach to independently record the tight junction conductance by neutralizing the apical membrane conductance.
- 45.Motoyama HI, Friedman PA. Calcium-sensing receptor regulation of PTH-dependent calcium absorption by mouse cortical ascending limbs. Am J Physiol Renal Physiol. 2002;283:F399–F406. doi: 10.1152/ajprenal.00346.2001. [DOI] [PubMed] [Google Scholar]
- 46.Loupy A, Ramakrishnan SK, Wootla B, Chambrey R, de la Faille R, Bourgeois S, Bruneval P, Mandet C, Christensen EI, Faure H, et al. PTH-independent regulation of blood calcium concentration by the calcium-sensing receptor. J Clin Invest. 2012;122:3355–3367. doi: 10.1172/JCI57407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki H, Tani K, Tamura A, Tsukita S, Fujiyoshi Y. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J Mol Biol. 2015;427:291–297. doi: 10.1016/j.jmb.2014.10.020. [DOI] [PubMed] [Google Scholar]