Abstract
Hedonic substitution, where wheel running reduces voluntary ethanol consumption has been observed in prior studies. Here we replicate and expand on previous work showing that mice decrease voluntary ethanol consumption and preference when given access to a running wheel. While earlier work has been limited mainly to behavioral studies, here we assess the underlying molecular mechanisms that may account for this interaction. From four groups of female C57BL/6J mice (control, access to two-bottle choice ethanol, access to a running wheel, and access to both two-bottle choice ethanol and a running wheel), mRNA-sequencing of the striatum identified differential gene expression. Many genes in ethanol preference quantitative trait loci were differentially expressed due to running. Furthermore, we conducted Weighted Gene Co-expression Network Analysis and identified gene networks corresponding to each effect behavioral group. Candidate genes for mediating the behavioral interaction between ethanol consumption and wheel running include multiple potassium channel genes, Oprm1, Prkcg, Stxbp1, Crhr1, Gabra3, Slc6a13, Stx1b, Pomc, Rassf5, Polr2a, and Camta2. After observing an overlap of many genes and functional groups previously identified in studies of initial sensitivity to ethanol, we hypothesized that wheel running may induce a change in sensitivity, thereby affecting ethanol consumption. A behavioral study examining Loss of Righting Reflex to ethanol following exercise trended toward supporting this hypothesis. These data provide a rich resource for future studies that may better characterize the observed transcriptional changes in gene networks in response to ethanol consumption and wheel running.
Introduction
Although the prevalence of alcohol use disorders remains high, there are relatively few treatment options available (Gunzerath et al., 2011; Rehm et al., 2009; Warren and Hewitt, 2010; WHO | World Health Organization). The concept of hedonic substitution, replacement of one rewarding behavior with another, is a promising area of research. Exercise has been used in the past to help reduce ethanol intake in heavy drinkers (Correia et al., 2005; Murphy et al., 1986; Weinstock, 2010; Werch et al., 2011), and animal studies have shown consistent interaction effects with ethanol consumption (Darlington et al., 2014; Ehringer et al., 2009; Hammer et al., 2010; McMillan, 1978; McMillan et al., 1995; Ozburn et al., 2008; Werme et al., 2002). However, little is known about the neurobiology of this interaction. Our laboratory first attempted to identify transcriptional changes underlying hedonic substitution, and the candidate genes Slc18a2 in the midbrain and Bdnf in the hippocampus were both found to respond differently to ethanol consumption and voluntary wheel running (Darlington et al., 2014; Gallego et al., 2015). The striatum plays an important role in the mesolimbic dopaminergic pathway, processing and integrating input from a number of other brain regions. Therefore, it seems probable that transcriptional events occurring in the striatum may provide further insight into hedonic substitution.
The mesolimbic dopaminergic pathway consists of dopaminergic neurons in the ventral tegmental area (VTA) of the midbrain, which project to the nucleus accumbens (NAc) in the ventral striatum and release dopamine (DA) upon stimulation. Both ingestion of ethanol and voluntary exercise facilitate increased DA levels in the NAc (Di Chiara and Imperato, 1985, 1986; Dishman et al., 2006). However, there is increasing evidence that the whole striatum is involved in the development of addiction (Everitt and Robbins, 2005; Koob and Volkow, 2010). While initial exposure to hedonic stimuli stimulates the shell of the NAc and feeds back to the VTA, interactions between the shell and the core of the NAc induce conditioned reinforcement to the stimuli. Furthermore, animals will respond to direct stimulation of substantia nigra as well as VTA, suggesting an acute nigrostriatal role in hedonia (Prado-Alcalá and Wise, 1984; Wise, 1981, 2009). In heavy drinking humans, there is greater activation in the dorsal striatum than in the ventral striatum when presented with drinking-related cues (Vollstädt-Klein et al., 2010). These studies demonstrate the importance of inclusion of the whole striatum when considering ethanol related changes.
This study was designed to identify candidate genes for hedonic substitution by examining mRNA from striatal tissue using RNA-Sequencing (RNA-seq) to compare transcriptional responses to voluntary ethanol consumption and wheel running. Weighted Gene Co-expression Network Analysis (WGCNA), an agnostic network analysis tool, was used to identify biologically relevant co-expression networks (Langfelder and Horvath, 2008; Zhao et al., 2010). Expression data produced from RNA-Seq and analyzed using WGCNA have been shown to improve network characteristics relative to microarray expression data (Iancu et al., 2012), and both microarrays and RNA-Seq have been used successfully to characterize gene co-expression networks related to ethanol behaviors (Contet, 2012; Darlington et al., 2013; Farris et al., 2014; Iancu et al., 2013; Marballi et al., 2015; McBride et al., 2010, 2013; Mulligan et al., 2011; Vanderlinden et al., 2013, 2015; Zhang et al., 2014). A recent review by Farris et al. (2015) summarizes findings across multiple omics studies. For the current study, we anticipated that we would identify numerous differentially expressed genes, and were interested in overlap with genes located in previously identified quantitative trait loci (QTL) related to ethanol behaviors, including ethanol preference (Belknap and Atkins, 2001; Crabbe et al., 2010; Fehr et al., 2005; Hitzemann et al., 2004; Phillips et al., 1998a), ethanol induced locomotor activation (Palmer et al., 2006; Phillips et al., 1995), loss of righting reflex due to ethanol (Markel et al., 1997), and ethanol acceptance (McClearn et al., 1997).
In addition, we expect to detect novel sets of genes (e.g., pathways) that are co-regulated under these behavioral conditions, and are compared to pathways identified in previous studies of alcohol-related behaviors.
Materials and Methods
Statement on animal care
This study was conducted with approval from the Institutional Animal Care and Use Committee at the University of Colorado, Boulder (Boulder, Colorado) following guidelines established by the Office of Laboratory Animal Welfare. All possible measures were taken to minimize animal discomfort.
Animals for RNA-seq
Adult female C57BL/6J mice (age 65–75 days on day 1 of behavioral testing), bred and housed at the Specific Pathogen Free facility at the Institute for Behavioral Genetics (University of Colorado, Boulder), were used for this study. Mice were group-housed in their home cages on the testing floor for at least 6 days prior to individual housing. On the first day of testing, mice were individually housed in polycarbonate cages with dimensions 30.3cm x 20.6cm x 26cm with cedar wood chips and one bedding square. The room was on a 12-hour light/dark cycle with lights on at 7:00AM. Room temperature and humidity were monitored every day, with temperatures ranging from 23 – 24.5°C and humidity ranging from 20 – 40%. Mice had ad libitum access to both water and standard chow (Harlan Laboratories, Indianapolis, Indiana), and were monitored daily. Body weight and food consumption were measured every four days.
Behavioral paradigm
Mice were tested using a protocol described previously (Darlington et al., 2014), and consistent with methods previously described as producing an hedonic substitution effect (Ehringer et al., 2009). Mice were tested in batches of five animals, staggered every 2 days. Conditions were randomized between staggered groups. Briefly, mice were housed under one of four cage conditions (n=6/condition), including cages with: 1) water only and no wheel (sedentary/water), 2) water and ethanol two-bottle choice and no wheel (sedentary/ethanol, 3) water only with a running wheel (running/water), and 4) water and ethanol two-bottle choice with a running wheel (running/ethanol). Mice housed with running wheels had continuous 24-hour access to the running wheels (diameter 24.2cm, Harvard Apparatus, Holliston, Massachusetts) each day of the 16-day protocol. Running wheel revolutions were measured daily using a magnetic switch (Harvard Apparatus) triggered by a magnet on the wheel. Mice with access to two-bottle choice ethanol and water progressed as follows: water only for days 1–3, water and 3% ethanol (v/v) for days 4–5, water and 7% ethanol for days 6–7, and water and 10% ethanol for days 8–16. To prevent a side preference in the drinking bottles the side of the cage the bottles were on was alternated every two days. Volumes of water and ethanol (if applicable) consumed were measured daily. Bottle leakages were determined using a daily outlier test, with a threshold of 2 standard deviations; however none were observed in this study. One daily wheel revolution count was missing from two mice, due to accidental misalignment of magnetic switch. Those two missing values have been imputed from the average of the two-nearest daily values.
RNA extraction and preparation
Immediately after cervical dislocation, brains were removed and each whole striatum was dissected out and placed in RNALater (Ambion, Foster City, California). Total RNA was extracted and purified using Qiagen RNeasy Midi kits (Qiagen, Valencia, California). Quantity and quality were determined using a NanoDrop™ spectrophotometer (Thermo Fisher Scientific, Wilmington, Delaware) and an Agilent 2100 BioAnalyzer™ (Agilent Technologies, Santa Clara, California). Ratios of absorbance (260nm:280nm) were shown to be excellent (>1.8). RNA Integrity scores were also shown to be excellent (>8.0). For each sample, five μg of total RNA was used, first for ribosomal RNA (rRNA) depletion using Ribo-Zero™ Magnetic kits (Epicentre Biotechnologies, Madison, Wisconsin), then poly-A enrichment using Dynabeads® oligo-dT magnetic beads (Invitrogen, Carlsbad, California), both according to kit specifications.
The preparation of the cDNA libraries was performed using ScriptSeq™ V2 RNA-Seq Library Prep kit (Epicentre Biosystems), which generated strand-specific pair-end libraries for quantitative RNA-Sequencing on Illumina platforms. The protocol followed is described in detail at www.epibio.com. Briefly, 50ng of mRNA was fragmented, and then reverse transcribed to single stranded cDNA. This first strand of cDNA was di-tagged with a 58 nucleotide oligomer, before purification with Agencourt AMPure XP beads (Beckman Coulter, Brea, California). Purified, di-tagged single stranded cDNA was then amplified with 15 cycles by polymerase chain reaction using ScriptSeq™ V2 Index Primers (Epicentre Biosystems), designed to add a 6 nucleotide unique barcode to each cDNA in each sample. After amplification, cDNA was purified using Agencourt AMPure XP beads (Beckman Coulter) and shipped to the University of Colorado, Denver Sequencing Core Facility. Upon arrival, samples were tested for quality and quantity using an Agilent 2100 BioAnalyzer™ (Agilent Technologies) and a Qubit® 2.0 Fluorometer (Invitrogen).
RNA-Sequencing
Four samples per lane (one per condition, ScriptSeq™ barcodes 4–7) were run on six lanes of an Illumina HiSeq 2000 (Illumina, San Diego, California), and pair-end sequenced to 100 nucleotides. After sequencing, the core facility provided de-barcoded reads. Fastq files were assessed for quality using FastQC (v0.3, Babraham Institute). Using the FASTQ Trimmer (v1.0.0), six nucleotides from the 5′ end of each read were trimmed due to base composition bias at those positions. Trimmed reads were aligned to the mouse reference genome (mm9, Ensembl) using TopHat (v2.0),(Trapnell et al., 2009) allowing for 2 mismatched bases, up to 10 alignments per read, no mismatches in secondary segment alignment, and aligning across known exon junctions. To assemble transcripts and generate read counts per transcript, output from TopHat and the annotated reference genome (mm9, Ensembl) was analyzed using Cufflinks (v2.0.2) to construct the minimum number of transcripts that explain the maximum number of reads (Trapnell et al., 2010). Since the sequenced sample had been rRNA depleted and enriched for poly-A mRNA transcripts, a mask file was used to discriminate against alignments in rRNA, tRNA, and small RNA genes. Read counts per transcript were analyzed with ComBat (v3.18.0) (Johnson et al., 2007) to identify and correct for confounding variables, including cage placement on shelf, RNA extraction date, library preparation date, sequencing date, sequencing lane, and barcode. Corrected read counts were then output to EdgeR (v3.0.8) (Robinson et al., 2010), which was used to test for differential expression. For genes to be included in differential expression testing, there had to be at least one aligned read in each sample for that gene. This minimal threshold is consistent with other studies utilizing RNA-Sequencing/EdgeR (Bottomly et al., 2011; Iancu et al., 2012). EdgeR and ComBat are open-source R packages and provide for the statistical analysis of raw count data. EdgeR is implemented in Bioconductor (Gentleman et al., 2004) and allows for fitting a general linear model, allowing the inclusion of an interaction term, which is important for interpreting this 2x2 experimental design, and relies on the negative binomial distribution to infer differential expression. P-values are corrected using a Benjamini-Hochberg false discovery rate of 5% (FDR<0.05) (Benjamini and Hochberg, 1995).
Weighted Gene Co-expression Network Analysis (WGCNA)
The Weighted Gene Co-expression Network Analysis (WGCNA, v1.25.2) provided an agnostic analysis of patterns of gene expression, regardless of treatment condition (Langfelder and Horvath, 2008; Zhao et al., 2010). Similarly co-expressed genes were clustered into modules, which could then be related to treatment and biological relevance. Raw counts were log2 normalized, then ComBat was used to correct for known batch effects including sequencing lane, barcode, and cage shelf position. Data were then quantile normalized prior to analysis (Supplementary Data). For WGCNA, a signed similarity matrix was generated using Pearson correlation between the expression patterns of each gene across each sample. This was converted to a weighted adjacency matrix by a power function, determined by a scale-free topology model (β=21). The weighted adjacency matrix was converted to topological overlap matrix (TOM), and a measure of dissimilarity was generated by 1 - TOM. Genes were clustered based on hierarchical clustering of TOM-based dissimilarity, with the dynamic tree cutting algorithm cutreeDynamic, and the deepSplit option set to 4. Gene clusters with a minimum of 30 genes were identified using a dynamic tree-cutting algorithm, which identified 88 gene co-expression clusters (modules). Similar gene modules were merged using the mergeCloseModules command, with a dissimilarity threshold of 0.2 (Pearson correlation greater than 0.8). Merging similar modules resulted in 50 remaining modules used in downstream analysis. Notable genes in each module were determined by several categories. First, we ranked each gene by its module membership, calculated by WGCNA. Second, we ranked each gene by individual gene significance for the main effect(s) of the respective module. Third, genes in the top third of each module and previously implicated in ethanol related behavior were also considered notable. Module robustness was tested in three ways. First, average module adjacencies were calculated and compared to the average adjacencies of randomly sampled “modules” of the same size. 100,000 permutations of randomly sampled modules were generated. Modules were considered robust if average module adjacencies were significantly higher than the randomly generated modules. Second, we repeated this permutation test using average topological overlap, similar to Iancu et al. (2012). Third, the intramodular and extramodular connectivity of each module was calculated and scaled according to module size. Modules with higher scaled intramodular connectivity were considered robust.
To identify experimentally relevant co-expression modules, we took the first principle component of the expression data of each module using the moduleEigengenes command from the WGCNA R-package. The resulting module eigengenes are representative of the gene expression levels for each module, as if the module were reduced to a single gene. A two-way analysis of variance of the resulting log2-normalized module eigengene values was used to identify module eigengenes different due to access to a running wheel, access to ethanol, or an interaction effect. Nominal significance was considered when p<0.05, and significant p-values were less than 0.05/50=0.001. Only modules meeting at least nominal significance were further tested for over-representation of functional groups, cell-type enrichment, and ethanol QTL enrichment.
Functional group over-representation
Each set of differentially expressed genes was tested for functional group over-representation using the Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7) (Huang et al., 2009a, 2009b). This tool traverses and incorporates multiple ontologies. We tested for over-representation of functional categories using the DAVID EASE modified Fisher’s exact test and corrected for multiple testing using Benjamini-Hochberg FDR of 0.05.
Using a database of genes over-expressed in cell types—neurons, astrocytes, and oligodendrocytes (Cahoy et al., 2008)—we tested whether these cell-type enriched genes were over-represented in the sets of differentially expressed genes or in each WGCNA module, implementing a one-tailed hypergeometric test in R. Modules were also tested for over-representation of genes found in ethanol-related QTLs, again using a one-tailed hypergeometric test. Modules were only tested if there were at least 2 genes within a category, and significant p-value thresholds were adjusted accordingly (0.05/#tests).
RT-PCR confirmation of gene expression
We selected eleven differentially expressed genes to confirm expression differences in 24 additional C57BL/6J adult female mice (n=6/group) that underwent the same behavioral paradigm as described above. Mice were sacrificed on day 16, and total RNA from the striatum was extracted using EZNA Total RNA Kit II (Omega Bio-tek, Norcross, Georgia). Quality and quantity of RNA were determined by gel electrophoresis and NanoDrop™ spectrophotometer (ThermoFisher Scientific, Waltham, Massachusetts). A260/A280 was determined to be excellent in each case (>1.8). Total mRNA was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, California). Genes selected for quantification were: transthyretin (Ttr), syntaxin 1b (Stx1b), syntaxin 19 (Stx19), opioid receptor mu 1 (Oprm1), potassium inwardly-rectifying channel, subfamily J, member 6 (Kcnj6), guanylate cyclase 1, soluble, alpha 2 (Gucy1a2), Cd24a antigen (Cd24a), coiled-coil domain containing 33 (Ccdc33), coiled-coil domain containing 153 (Ccdc153), B-cell CLL/lymphoma 9-like (Bcl9l), and RIKEN cDNA 1700003M02 (1700003M02Rik). Control genes included Gapdh and the RNA gene Rn18s. Reactions were run as a PCR array, using 384-well plates designed by SABiosciences (Qiagen, Hilden, Germany) and with primers designed by SABiosciences and SYBR Green for detection. Real-time quantitative PCRs were performed using an ABI 7900HT (Applied Biosystems, Foster City, California) running Sequence Detection Systems software (SDS v2.3, Applied Biosystems, Foster City, California). All target genes were normalized using the 2−ΔΔCt method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). Subsequent melting curve analysis of target sequences showed that all primers used in this study generated amplicons that had a single peak, without primer-dimer artifacts.
Loss of righting reflex for differences in acute ethanol sensitivity
Twenty-seven adult female C57BL/6J mice were singly housed in cages as previously described (n=15 with wheel, n=12 no wheel). All mice had access only to water. To determine if 16 days of voluntary wheel running induces differences in acute sensitivity to ethanol’s sedative-hypnotic effects we used the standard Loss of Righting Reflex paradigm (Crabbe et al., 2006). All mice received an intraperitoneal injection of ethanol (4.1 g/kg; vol/vol of a 20% ethanol solution in physiological saline), and were placed on their back in a V-shaped plastic trough when they were unable to right themselves. The length of time until they regained the righting response was monitored. When each mouse was able to right itself three times in 1 minute the mouse was considered recovered. At the recovery time the mice had a 10μL sample of blood drawn from the tail vein for blood ethanol concentration determination. One mouse from the wheel running group did not become sedated after 3min post-injection and was removed from the experiment.
BEC was measured by a modified enzymatic method (Smolen et al., 1986). The 10μL of blood was added to 200μL of ice-cold 0.55M perchloric acid. Samples were neutralized with 200μL 0.6M KOH containing 50mM acetic acid. This solution precipitates the perchlorate anion and buffers the solution to about pH 5. Samples were centrifuged at 1500 rpm for 10 min. The resulting supernatant was used to measure blood ethanol content. A 50μL aliquot of the supernatant solution was transferred into duplicate assay tubes and one blank. The assay solution consisted of 2mM NAD (Roche, Nutley, New Jersey), 8–10 units of yeast alcohol dehydrogenase (ADH, Sigma-Aldrich, St. Louis, Missouri), and 500mM Tris-HCl, pH 8.8, in a total volume of 400μL. Blanks contained no ADH. The reaction mixture was allowed to incubate at 25°C for 30 min prior to measuring the absorbance due to NADH formation at 340nm in a Tecan GENios FL TWT Fluorescence Microplate Reader (Tecan Group Ltd., Morrisville, North Carolina). BEC was calculated from linear regression analysis of a standard curve prepared just prior to BEC analysis.
Results
Behavior
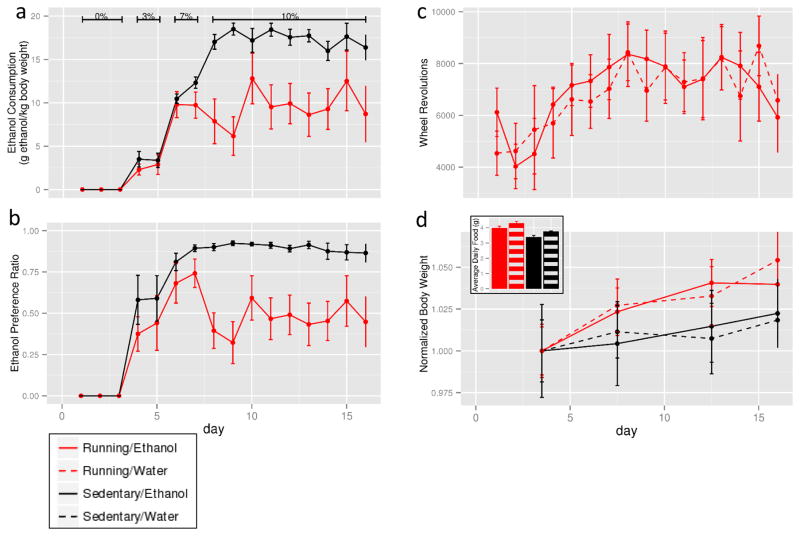
Over the course of the 16-day protocol, mice with access to a running wheel consumed less ethanol than sedentary mice. Exercising mice consumed 8.47±0.8 g/kg/day compared to 14.3±0.4 g/kg/day (F12,120=10.2, p<0.001, repeated measures ANOVA, Figure 1a), and also showed less preference for ethanol than sedentary mice, with preference ratios of 0.49±0.05 and 0.84±0.02, respectively (F12,120=27.9, p<0.001, repeated measures ANOVA, Figure 1b). There was no effect of access to ethanol on daily running wheel revolutions (Figure 1c). Mouse body weights increased, with no effects of access to ethanol or wheel running (19.5 ±0.2 baseline, 20.2 ±0.2 on day 16, F3,60=9.4, p<0.001, repeated measures ANOVA, Figure 1d). There was a main effect of both access to a running wheel and access to ethanol on food consumption as measured by two-way ANOVA (Figure 1d, inset). Running mice consumed slightly more food than sedentary mice. Running mice consumed 4.13±0.1 g/day compared to 3.57±0.08 g/day for sedentary mice (F1,20=27, p<0.001). Mice with access to ethanol consumed slightly less food (3.68±0.1 g/day) than water only mice (4.01±0.1 g/day, F1,20=9.7, p<0.01).
Figure 1.
Behavioral outcomes of sixteen days of voluntary ethanol consumption and voluntary wheel running. Mice with access to running wheel are represented in red, while mice with access to ethanol are denoted by solid lines/bars. Running mice consumed less ethanol compared to sedentary mice when normalized to body weight (A) and when computed as a preference ratio (B). There was no difference in daily running wheel revolutions between mice consuming ethanol and mice only consuming water (C). Body weights for all mice increased over the course of the experiment (D). Mice consuming ethanol consumed less food than water-only mice, and running mice consumed more food than sedentary mice (inset).
Differential expression analysis
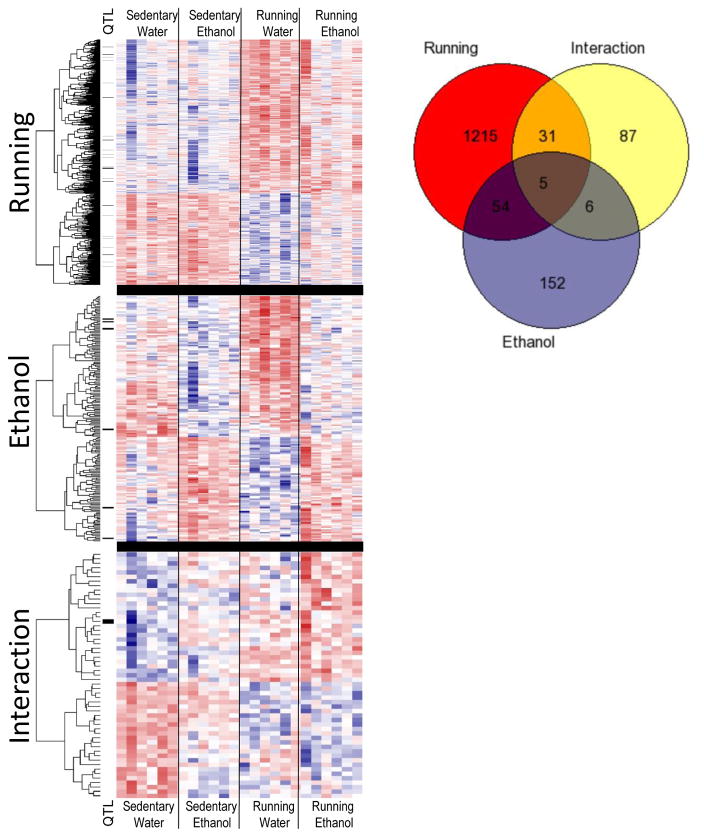
We quantitatively sequenced mRNA from 24 striatum samples from 4 groups of mice on the Illumina HiSeq 2000 platform. We generated paired-end reads, 100 nucleotides long, and after trimming 6 bases from the 5′ end, aligned them to the mouse reference genome, masked to preferentially align to protein coding genes. Alignment statistics from the sequencing and alignment are shown in Supplementary Data. 14,919 genes were expressed across the samples, meeting the minimum threshold and tested for differential expression. Using the general linear model in EdgeR with a False Discovery Rate (FDR<0.05), there were 1,305 differentially expressed genes (DEGs) due to the access to running wheel. 217 genes were differentially expressed due to ethanol consumption, and 129 genes showed an interaction effect (Figure 2). Fifty-nine genes were differentially expressed due to both main effects, ethanol consumption and wheel running. Details about all differentially expressed genes are summarized in Supplementary Data. Notable genes previously implicated in ethanol related behaviors include protein kinase c gamma (Prkcg), opioid receptor mu 1 (Oprm1), pro-opiomelanocortin (Pomc), GABA A receptor alpha 3 (Gabra3), GABA transporter (Slc6a13), eighteen potassium channel genes (Kcna2, Kcna3, Kcnb1, Kcnc1, Kcnc3, Kcnd1, Kcnd3, Kcng3, Kcng4, Kcnh4, Kcnh5, Kcnh7, Kcnj6, Kcnj10, Kcnj16, Kcnk1, Kcnq2, Kcnq3), Ras association domain family member 5 (Rassf5), syntaxin 1b (Stx1b), corticotropin releasing hormone receptor 1 (Crhr1), protein tyrosine phosphatase, receptor type N (Ptprn), and multiple genes located in previously identified ethanol related QTL regions. There were also many highly significant differentially expressed genes that have not been previously implicated in ethanol behaviors.
Figure 2.
Heatplots and dendrograms generated from log2 count per million expression of differentially expressed genes (DEGs) across all four groups of mice (n=6/group). The three heatplots show genes differentially expressed due to a main effect of wheel running, ethanol consumption and the interaction of wheel running and ethanol consumption. Red signifies higher expression, blue denotes lower expression values. Genes residing within ethanol-related quantitative trait loci are shown with a black bar next to heatplot. The Venn diagram displays the overlap between genes in each group.
We used DAVID to test for over-representation of functional groups. The set of ethanol responsive differentially expressed genes was enriched for genes involved in apoptosis (4 genes, p=0.01), and response to calcium signaling (5 genes, p=3.9x10-4). Genes differentially expressed due to wheel running were enriched for numerous functional groups, including potassium signaling, synaptic transmission, among others. Genes differentially expressed due to the interaction of ethanol and wheel running were enriched for functional groups involved in drug metabolism (3 genes, p=0.005) and neural cell adhesion molecule signaling (4 genes, p=0.003). The complete list of over-represented functional groups is shown in Supplemental materials.
We tested for over-representation of genes highly expressed in either neurons, astrocytes, or oligodendrocytes, based on a study of genes over-expressed at least 1.5-fold in these cell types (Cahoy et al., 2008) compared to at least one of the other cell-types. The set of genes differentially expressed due to running were enriched for each cell-type (neurons, 213 genes, p=3.8x10-15; astrocytes, 206 genes, p=1.6x10-4; oligodendrocytes, 175 genes, p=3.3x10-4). We further tested for enrichment of genes contained in ethanol related QTLs. No QTLs were enriched in the sets of interaction or ethanol consumption DEGs. The set of wheel running DEGs were enriched for genes in the alcohol preference 1 QTL (Ap1q) on chromosome 1 (16 genes, p=0.029), and trended towards enrichment of genes in the loss of righting reflex due to ethanol 4 QTL (Lore4) on chromosome 11 (6 genes, p=0.058).
WGCNA
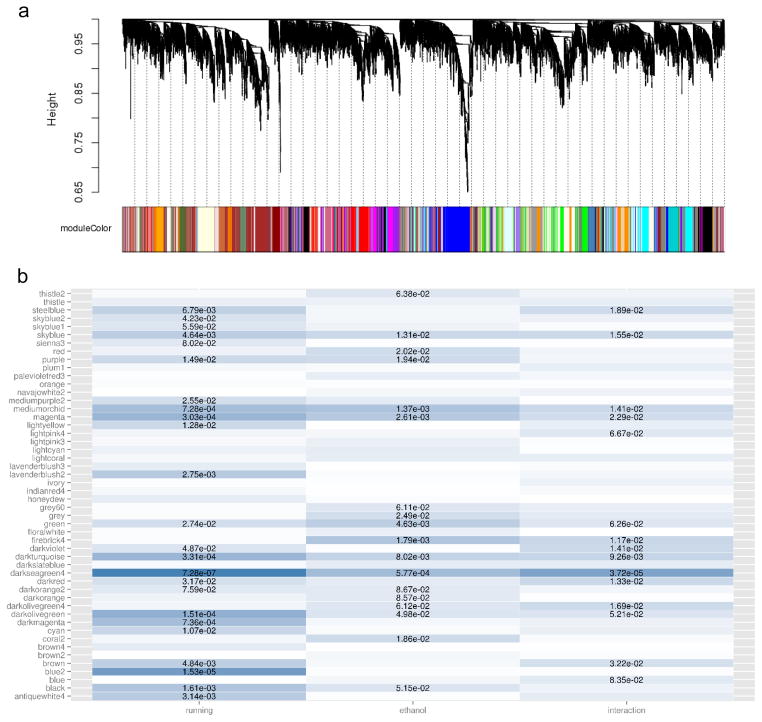
A single WGCNA for all 24 samples produced 50 distinct clusters (modules) of co-expressed genes (Figure 3, Table 1). Each module was arbitrarily named after a color, and 27 genes were assigned to the ‘grey’ module, indicating failure to fit a co-expression pattern consistent with the other 50 modules. The number of genes per module other than grey ranged from 42 to 1191. Robustness testing confirmed that each module has: greater average adjacency than random, greater average topological overlap than random, and greater scaled intramodular connectivity than scaled extramodular connectivity. The grey module failed each test of robustness (Supplemental Data).
Figure 3.
Results of the WGCNA. Hierarchical clustering and dynamic tree cutting are shown in (A). The y-axis represents a dissimilarity measurement based on topological overlap, with the distance between the branches signifying dissimilarity. Each branch of the dendrogram represents one gene. Branches of the dendrogram are dynamically “pruned” into modules, corresponding to each color in the bottom row. In (B), the log2 normalized moduleEigengenes from each module, y-axis, were tested by 2-way ANOVA for main effects of running, ethanol, or the interaction, x-axis. Darker colors represent lower p-values, and p-values less than 0.1 are presented in text.
Table 1.
| Main Effect (p-value)a | Module | genes | Cell Type Enrichment (p-value)b | QTL Enrichment (p-value)c | Functional Enrichmentd | Notable Genese |
|---|---|---|---|---|---|---|
| Ethanol (0.019) | coral2 | 77 | Neuron (0.046) | Lore5 (0.022) Aaq1 (0.0081) |
ribosome, acetylation | Pmm1, Rangap1 |
| Ethanol (0.02) | red | 629 | acetylation, phosphoprotein, nucleotide binding | Kirrel | ||
| Interaction (0.000037)* | darkseagreen4 | 498 | Lore4 (0.045) Lore1 (0.0083)* |
alternative splicing, metal ion binding | Jph4, Ptprn, Acr | |
| Interaction (0.012) | firebrick4 | 61 | Neuron (6.6x10-12)* | Lore5 (0.012) Aaq1 (0.0042)* |
ion channel activity | Baiap3, Wdr6, Gdap1l1, Adcy8, Oxtr, Crh, Stx12, Chgb, Trhr, Oprm1 |
| Interaction (0.013) | darkred | 181 | Astrocyte (0.0055) Oligodendrocyte (0.035) |
Ap1q (0.0033)* | extracellular matrix, secreted protein | Slc2a12, Gpx8, Stk38l |
| Interaction (0.014) | darkviolet | 56 | apoptosis | Snx5, Nkap, Negr1 | ||
| Interaction (0.016) | skyblue | 160 | Ap1q (0.039) | nucleus, RNA processing | Plxnb2, Luzp1 | |
| Interaction (0.017) | darkolivegreen4 | 59 | Neuron (0.00065)* Astrocyte (0.0076) |
ribonucleotide binding | Sag, Acvr2a, Stx7 | |
| Interaction (0.019) | steelblue | 154 | Oligodendrocyte (p<1x10-13)* Astrocyte (0.017) |
Etact1 (0.016) | amino acid transport/metabolism, myelin sheath | Mdk, Nkain1, Ppap2c, Rassf2, Hapln2, Trf, Carhsp1 |
| Interaction (0.032) | brown | 1191 | Ap3q (0.023) Etact2 (0.015) |
transcriptional regulation, signaling (neurotrophin, Wnt, MAPK, ErbB, Notch), ion channels | Nedd4l, Polr2a | |
| Running (0.00002)* | blue2 | 57 | Oligodendrocyte (0.000036)* | Ap3q (0.0035)* | ion transport, metabolic processes | Mal |
| Running (0.00015)* Ethanol (0.05) |
darkolivegreen | 149 | metal ion binding, alternative splicing | Chrna7, Cacna1e, Siae | ||
| Running (0.0003)* Interaction (0.023) |
magenta | 450 | ribosome, translation, neurodegenerative diseases | Hyal2, Aurkaip1, Ncaph2, Ssb | ||
| Running (0.00033)* Interaction (0.0093) |
darkturquoise | 408 | Etact2 (0.035) | transcriptional regulation | Nln, Ramp3, Fcgrt | |
| Running (0.00073)* Interaction (0.014) |
mediumorchid | 75 | acetylation, nuclear lumen | Uchl1, Myl12a, Lamb2 | ||
| Running (0.00074)* | darkmagenta | 149 | Astrocyte (0.0059) | Ap3q (0.046) | phosphoprotein, acetylation | Aldh3b1, Syt1 |
| Running (0.0016) | black | 803 | Astrocyte (0.0024) | Etact1 (0.019) Lore1 (0.03) |
nucleus, ribonucleotide binding | Ogfr, Ramp1 |
| Running (0.0028) | lavenderblush2 | 261 | Neuron (0.00055)* | Lore4 (0.0053)* | phosphoprotein, plexin, synaptosome | Nav1, Syn1, Camta2, Bsn, Vamp2, Kcnj10, Rab6b, Cabin1, St8sia1, Prrg3, Crhr1, Prkcg |
| Running (0.0031) | antiquewhite4 | 359 | protein transport, mitochondria | Eif3e, Cdkn2c | ||
| Running (0.011) | cyan | 605 | mitochondrial processes, neurodegenerative disease | Fbln2, Tmem159 | ||
| Running (0.013) | lightyellow | 555 | metal ion binding, alternative splicing, cell junction | Lonrf2, Nyap2, Thrb, Mdm4, Kcnq3, Myo9a | ||
| Running (0.015) Ethanol (0.019) |
purple | 316 | RNA processing, transcriptional regulation | Crabp1. Ssrp1, S1pr5, Dnm2, Ntng1, Hdac8 | ||
| Running (0.026) | mediumpurple2 | 117 | Neuron (0.0015)* | Ap1q (0.014) Aaq1 (0.025) |
membrane, vesicle, synapse | Itga4, Dcx, Lrfn5, Rasgrp1, Dzank1, Cadm2, Sorbs2, Slc19a2 |
| Running (0.027) Ethanol (0.0046) |
green | 539 | Lore4 (0.00053)* | membrane, transport, transmembrane region | Stx16, Kcnh4, Nrros, Nrsn2, Stx3, Slc25a11, Ntrk3, Slc6a8 | |
| Running (0.04) | sienna3 | 246 | Neuron (8.4x10-13)* | Ap5q (0.0068) | synapse, cell junction, axon guidance, potassium ion transport, ion channel activity | Etl4, Sncb, Stxbp1, Rassf5, Slc4a8, Mast1, Kcnc1 |
| Running (0.04) | skyblue2 | 74 | Oligodendrocyte (0.038) | ubiquitination | Pla2g4a, Drd1, Hes5 |
Module eigengene significance for main effects and/or interaction, determined by ANOVA of log2 normalize module eigengene.
Over-represented cell-types, using data from Cahoy et al (2008). P-values bases on one tail hypergeometric test.
Over-represented ethanol related quantitative trait loci. P-values bases on one tail hypergeometric test.
Over-represented functional groups. Assessed using DAVID.
Notable genes were determined by high module membership, gene-trait significance, brain expression, or ethanol QTL.
p-values significant when corrected for number of tests.
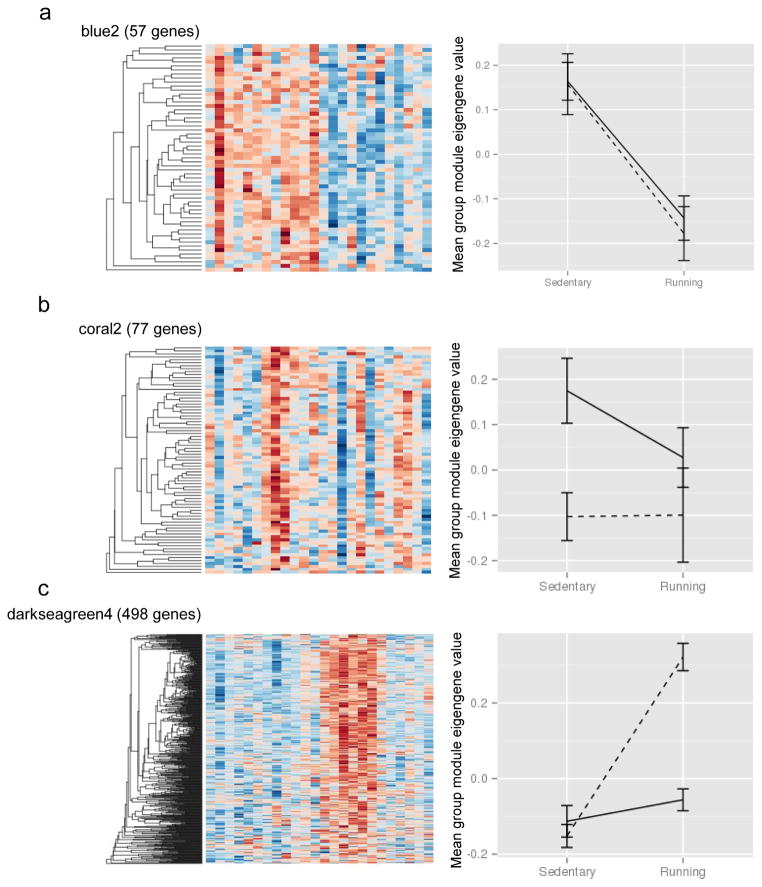
To identify potentially biologically relevant modules, the first principle component of the expression data for each module was calculated using the moduleEigengenes command in R. The calculated module eigengene was representative of the expression pattern of each gene in the module across all samples. A two-way ANOVA (main effects access to ethanol and access to wheel running) suggested several modules corresponded with access to wheel running (Figure 3). Six modules were significant when accounting for multiple testing (darkolivegreen, p=0.00015; blue2, p=0.00002; magenta, p=0.0003; darkturquoise, p=0.00033; mediumorchid, p=0.00073; and darkmagenta, p=0.00074). Ten other modules were nominally significant. No modules were significant for ethanol consumption when adjusted for multiple testing, but 5 modules were nominally significant. The darkseagreen4 module reached significance for an interaction effect (p=0.000037), and 10 other modules were nominally significant. Representative modules for each main effect are shown in Figure 4.
Figure 4.
Representative modules corresponding to each effect. Heat maps depict log2 normalized expression levels for each gene (rows) in the module, blue colors represent lower expression, red colors represent higher expression. Each column represents one mouse sample. Line graphs are plots of corresponding module eigenGene, grouped by treatment. Dashed lines are water-only mice, solid lines are ethanol-consuming mice. The blue2 module is representative of a main effect of running (A). The coral2 module shows a main effect of ethanol consumption (B). The darkseagreen4 module displays an interaction effect (C).
Functional over-representation analysis using DAVID showed significant functional group enrichment for many of the modules. Several modules did not have any functional groups meet significance after correcting the p-value according to a Benjamini-Hochberg false discovery rate of 0.05, although this seems likely to be a function of module size, as they are all among the smaller modules. While too numerous to name, the most common significantly enriched functional groups were related to transcriptional regulation and RNA processing. Modules were also enriched for signaling pathways (brown), ion channels (brown, sienna3, firebrick4), synapse (mediumpurple2, lavenderblush2, sienna3), and neurodegenerative disease (cyan, magenta).
Several modules were enriched for genes shown to be overexpressed in neurons (firebrick4, p=6.6x10-12; darkolivegreen4, p=6.5x10-4; mediumpurple2, p=1.5x10-3; lavenderblush2, p=5.5x10-4; and sienna3, p=8.4x10-13) and oligodendrocytes (steelblue, p<1.0x10-13; and blue2, p=3.6x10-5). Modules were also enriched for genes contained within ethanol related QTLs. Firebrick4 was enriched for genes in alcohol acquisition 1 QTL (Aaq1) on chromosome 15 (p=0.0042); darkred was enriched for genes contained in alcohol preference 1 QTL (Ap1q) on chromosome 1 (p=0.0033); darkseagreen4 was enriched for genes in loss of righting reflex due to ethanol 1 (Lore1) on chromosome 1 (p=0.0083); blue2 was enriched for genes in alcohol preferences 3 QTL (Ap3q) on chromosome 4; and both green (p=0.00053) and lavenderblush2 (p=0.0053) were enriched for genes in loss of righting reflex due to ethanol 4 (Lore4) on chromosome 11.
RT-PCR confirmation of gene expression
Using an additional 24 mice undergoing the same behavioral paradigm, we attempted to confirm gene expression changes in the striatum using quantitative real-time polymerase chain reaction. In all cases, the direction of change in gene expression was consistent with observations from RNA-Seq, however not every difference reached statistical significance (Supplementary Data).
Loss of righting reflex for differences in acute ethanol sensitivity
We used the loss-of-righting-reflex due to ethanol paradigm to test whether wheel running would lead to increased acute sensitivity to ethanol. There were no group differences in duration of LORR, although exercising mice tended to have longer durations (113.9±10 minutes compared to 99.5±9.5 minutes in the sedentary group, one-sided t23.9=1.04, p=0.15, Cohen’s d=0.41, Figure 5). There was no difference between blood ethanol concentrations, measured at the time mice regained righting reflex, between groups. Exercising mice consumed more water and more food over the course of the 16 day experiment.
Figure 5.
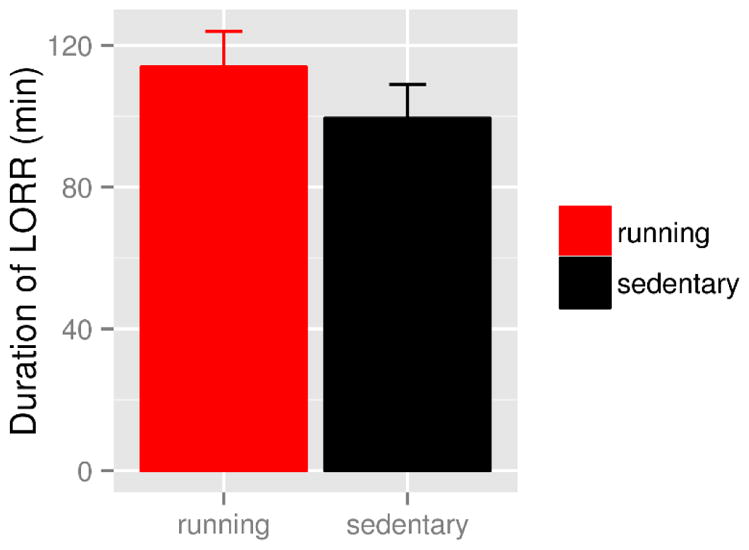
Duration of loss of righting reflex (LORR) due to intraperitoneal injection of 4.0 g/kg ethanol. There was a non-significant trend that mice with 16 days of access to a running wheel (red bar) had a longer duration of LORR (113.9±10 minutes) than mice without access to running wheel (black bar, 99.5±9.5 minutes, one-sided t23.9=1.04, p=0.15, Cohen’s d=0.41).
Discussion
Over the course of 16 days, voluntary wheel running reduced the amount of ethanol consumed by mice, supporting the hypothesis that under certain conditions, hedonic stimulus from one behavior can substitute for stimulus from another behavior. Transcriptional changes in the striatum, a major component of the mesolimbic dopaminergic reward pathway, were examined in order to provide insight into the mechanisms underlying reduced ethanol preference. Wheel running in rodents has been shown to induce a myriad of behavioral responses related to tests of stress, anxiety, and depression (Adlard and Cotman, 2004; Brené et al., 2007; Clark et al., 2015; Dishman et al., 2006; Greenwood et al., 2011, 2013; Hare et al., 2014; Lapmanee et al., 2013; Loughridge et al., 2013; Mika et al., 2015; Sciolino et al., 2015; Sierakowiak et al., 2014). Furthermore, previous studies have demonstrated wheel running was sufficient to reduce voluntary intake of amphetamine (Kanarek et al., 1995), cocaine (Cosgrove et al., 2002; Smith et al., 2011), as well as nicotine-seeking during extinction (Sanchez et al., 2013; Smith and Lynch, 2011). Taken together with the effects of wheel running on ethanol behaviors, observed across multiple species (Darlington et al., 2014; Ehringer et al., 2009; Gallego et al., 2015; Hammer et al., 2010; McMillan et al., 1995; Weinstock, 2010; Werme et al., 2002), these data show clear evidence of hedonic substitution. Understanding the neurobiological components of hedonic substitution will provide for further comprehension of the addiction process, and additional strategies for combating AUDs.
Only female mice were used due to the increased hedonic substitution compared to male mice (Ehringer et al., 2009). Unfortunately, attempts to control for estrous cycle, using daily vaginal swabs (Goldman et al., 2007), abolished any effect of running on ethanol consumption (not shown), and therefore was not included in the study. However, with a cycle length of 3–4 days, any effect of hormonal fluctuation on ethanol consumption should be minimal.
This study validates and extends the findings from our laboratory (Darlington et al., 2014; Ehringer et al., 2009; Gallego et al., 2015) both behaviorally and at the level of gene expression. Previously, striatal Drd1a showed reduced expression due to access to a running wheel (Darlington et al., 2014). In the currently study using RNA-Seq we observed reduced expression due to wheel running, although not quite significant when corrected for multiple testing (p=0.06). We further identified additional differentially expressed genes (DEGs) including 1305 wheel running-responsive DEGs, 217 ethanol-responsive DEGs, and 129 DEGs that are significant for the interaction between wheel running and ethanol consumption. In addition to differential expression testing, we utilized WGCNA to identify 50 gene co-expression modules, of which several exhibited expression patterns consistent with behavioral condition. Many of these modules were enriched for genes known to be highly expressed in either neurons or oligodendrocytes, as well as functional group enrichment.
As noted previously, we observed no difference in running behavior due to ethanol consumption, but did observe a replicable significant difference in ethanol consumption due to running. These findings suggest that at these levels of exposure to wheel running and ethanol, the latter exerts the dominant effect. Therefore, it seems reasonable to suggest that wheel running affects transcription of genes involved in regulating ethanol behaviors. This is supported by the subset of DEGs, as well as genes in behaviorally associated co-expression modules, located in previously identified ethanol behavior QTLs on chromosomes 1, 2, 4, 9, 11 and 15, (Belknap and Atkins, 2001; Crabbe et al., 2010; Fehr et al., 2005; Hitzemann et al., 2004; Markel et al., 1997; McClearn et al., 1997; Palmer et al., 2006; Phillips et al., 1998a, 1998b). In particular, there were 16 DEGs in an ethanol preference QTL on chromosome 1 (Ap1q1), including Rassf5, one of the top genes in the sienna3 co-expression module. Multiple wheel running DEGs were found in loss-of-righting-reflex due to ethanol QTL regions (6 in Lore4 and 9 in Lore5). In Lore4, DNA-directed RNA polymerase II polypeptide A (Polr2a) is a brain expressed gene enriched in oligodendrocytes. Running led to increased expression of Polr2a while ethanol consumption decreased expression. Although there was no significant interaction, when tested as an individual gene, Polr2a was a top gene in the large brown module, nominally significant for an interaction effect. While Polr2a has not been previously associated with ethanol behaviors, the opposing effects of wheel running and ethanol consumption suggest that it may play a role in mediating this behavioral interaction.
Previously identified as a candidate gene for ethanol preference, the expression of opioid receptor mu 1 (Oprm1) was increased due to an interaction effect of ethanol consumption and wheel running, consistent with its inclusion in the firebrick4 module. Running alone increased expression of Oprm1, but this effect was eliminated in the presence of ethanol. Naltrexone, an approved pharmacological treatment for AUDs (Johnson, 2010), is an antagonist of the μ-opioid receptor. To speculate, an increase in expression of Oprm1 under running conditions suggests possible compensation for a reduction in receptor sensitivity, similar to the effect of antagonizing the receptor.
In mapping an ethanol preference QTL on mouse chromosome 2, syntaxin binding protein 1 (Stxbp1) was identified as a candidate gene for ethanol preference (Fehr et al., 2005). Recently, it was identified as a hub gene in a co-expression module corresponding to lifetime ethanol consumption in humans (Farris et al., 2014). Although Stxbp1 was not differentially expressed after correction for multiple testing, it was identified as a top gene in the sienna3 module. Furthermore, syntaxin 1b (Stx1b) was significantly differentially expressed, with higher expression due to wheel running. These two proteins have been shown to interact, and facilitate neurotransmitter release (Mishima et al., 2014). Stx1b was shown to be more highly expressed in the prefrontal cortex of ethanol preferring P rats compared to non-preferring NP rats (Worst et al., 2005), suggesting regional differences in expression patterns in response to wheel running. Also in sienna3, Ras association domain family member 5 (Rassf5) is located in an alcohol preference QTL region on chromosome 1 (Ap1q1). This neuronally expressed gene has been shown to interact with alcohol dehydrogenase 6 (ADH6) (Wang et al., 2011) as well as Ras association domain family member 2 (RASSF2) (Ortiz-Vega et al., 2002), previously implicated in differences in the loss of righting reflex due to ethanol (Darlington et al., 2013; MacLaren et al., 2006). Rassf5 was also shown to be upregulated in the amygdala and nucleus accumbens in several rat lines bred for high consumption of ethanol (McBride et al., 2013).
A previous study of gamma-protein kinase c (Prkcg) showed the differences in ethanol consumption between null mutant and wildtype mice could be correlated with differences in the expression of transthyretin (Ttr) (Smith et al., 2006). While we previously showed a similar difference in Ttr expression between strains of mice selected for ethanol sensitivity, the current study does not support this. In fact, we see a significant increase in the expression of Prkcg in running mice, with no changes in Ttr expression. As a neuronally expressed gene, Prkcg seems a more likely candidate for a role in ethanol behaviors, especially given evidence of decreased expression in rats bred for high ethanol consumption (McBride et al., 2013). Contained within the brown module, Prkcg was co-expressed in the lavenderblush2 module with several other neuronally expressed genes, including synapsin 1 (Syn1), cadherin, EGF LAG seven-pass G-type receptor 2 (Celsr2), and calmodulin binding transcription activator 2 (Camta2, also in Lore4). Also in this module, corticotropin releasing hormone receptor 1 (Crhr1), significantly up-regulated due to wheel running, has been shown to mediate GABAergic signaling in response to ethanol consumption (Chen et al., 2010; Funk et al., 2007), was up-regulated in rats bred for high ethanol consumption (McBride et al., 2013), and down-regulated after ethanol consumption in mice (Marballi et al., 2015). The corticotrophin signaling pathway plays an important role in the addiction process (Koob, 2008).
Multiple potassium channel subunits have been shown to be direct targets of ethanol, especially the G-protein gated inward rectifying receptors (Aryal et al., 2009; Federici et al., 2009). Kcnq2 and Kcnq3 have been shown to mediate the release of dopamine after exposure to ethanol (McGuier et al., 2015). Furthermore, recent studies have shown differential regulation in ethanol behaviors of Kcnb1 (Farris et al., 2014; Marballi et al., 2015), Kcnj6 and Kcnj16 (McBride et al., 2013), and Kcnk1 (Marballi et al., 2015). The brown (8 potassium channel subunit genes including Kcnq2) and sienna3 (6 potassium channel subunit genes) co-expression modules were enriched for potassium signaling genes. Likewise, the gamma aminobutyric acid (GABA) receptor A3 (Gabra3), a ligand gated ion channel, has been shown to mediate the response to ethanol (Blednov et al., 2013; Farris et al., 2014). The GABA neurotransmitter system plays an important role in the physiological response to ethanol (Davies, 2003; Lobo and Harris, 2008), and another GABA gene, the GABA transporter (Slc6a13) was also differentially expressed. Both Gabra3 and Slc6a13 were co-expressed in the floralwhite module, which as a module was not significant for any behavioral association.
At first, when compared to the effects of 16 days of wheel running, it was surprising how few genes were differentially regulated from 16 days of ethanol consumption. The important point may not be the small number of genes affected by ethanol, but the myriad of transcriptional changes due to wheel running, and the suggestion that running can influence other behaviors at the molecular level. Therefore, it is reasonable that a number of DEGs and top module genes have no obvious connection to ethanol behaviors, both functionally and based on tissue expression. This could be due to the limited data available on these genes, although it is also likely that these genes respond to wheel-running with or without corresponding changes to other behaviors besides ethanol consumption. As additional evidence accumulates through future transcriptome-wide studies, the functional connection between these genes and ethanol behaviors may become apparent. It is beyond the scope of this paper to speculate on the role of each identified gene. This study can serve as a replication of previous findings, as well as adding evidence of new genes involved in the mediation of ethanol consumption and wheel running behaviors.
Furthermore, the similarities of differentially expressed genes (Prkcg, opioid signaling genes, Rassf5, potassium channel subunits, Lore QTL genes) and gene networks (MAPK signaling pathway, oligodendrocyte enrichment, Lore1 and Lore4 QTL enrichment) between this study and prior studies using other models of alcohol behaviors were noteworthy (Darlington et al., 2013; Smith et al., 2006). This led us to propose that wheel running induced an increase in sensitivity to ethanol, thereby decreasing ethanol consumption. This would be consistent with previous findings that wheel running does not change ethanol metabolic rates (Ehringer et al., 2009), thereby suggesting a metabolic mechanism rather than hedonic compensation. We tested this hypothesis using a model of ethanol sensitivity, the loss of righting reflex due to ethanol. We found that 16 days of wheel running was insufficient to alter the sensitivity to acute ethanol using the LORR protocol. However, the effect of this trend was in the predicted direction. It remains possible that other tests of ethanol sensitivity may identify wheel running-induced differences, but most other tests of sensitivity like rotarod or balance beam, involve motor coordination, that could be confounded by the presence of exercise in one group.
These data represent the highest resolution of striatal transcriptome responses to ethanol consumption and wheel running. We identified many wheel running-responsive genes that have been previously implicated in ethanol behaviors (Oprm1, Prkcg, potassium channels, Stxbp1, Crhr1, Gabra3, Slc6a13), are associated with previously implicated genes (Stx1b, Pomc, Rassf5), or reside in ethanol behavior QTLs (Polr2a, Camta2).
Supplementary Material
Acknowledgments
Support for this research came from internal University of Colorado funds and T32 DA017637.
Footnotes
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
References
- Adlard Pa, Cotman CW. Voluntary exercise protects against stress-induced decreases in brain-derived neurotrophic factor protein expression. Neuroscience. 2004;124:985–92. doi: 10.1016/j.neuroscience.2003.12.039. [DOI] [PubMed] [Google Scholar]
- Aryal P, Dvir H, Choe S, Slesinger PA. A discrete alcohol pocket involved in GIRK channel activation. Nat Neurosci. 2009;12:988–995. doi: 10.1038/nn.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- Blednov YA, Benavidez JM, Black M, Chandra D, Homanics GE, Rudolph U, et al. Linking GABA(A) receptor subunits to alcohol-induced conditioned taste aversion and recovery from acute alcohol intoxication. Neuropharmacology. 2013;67:46–56. doi: 10.1016/j.neuropharm.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly D, Walter NaR, Hunter JE, Darakjian P, Kawane S, Buck KJ, et al. Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PloS One. 2011;6:e17820. doi: 10.1371/journal.pone.0017820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brené S, Bjørnebekk A, Aberg E, Mathé Aa, Olson L, Werme M. Running is rewarding and antidepressive. Physiol Behav. 2007;92:136–40. doi: 10.1016/j.physbeh.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci Off J Soc Neurosci. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ACH, Manz N, Tang Y, Rangaswamy M, Almasy La, Kuperman S, et al. Single-nucleotide polymorphisms in corticotropin releasing hormone receptor 1 gene (CRHR1) are associated with quantitative trait of event-related potential and alcohol dependence. Alcohol Clin Exp Res. 2010;34:988–96. doi: 10.1111/j.1530-0277.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Ethanol preferentially stimulates dopamine release in the nucleus accumbens of freely moving rats. Eur J Pharmacol. 1985;115:131–2. doi: 10.1016/0014-2999(85)90598-9. [DOI] [PubMed] [Google Scholar]
- Di Chiara G, Imperato a. Preferential stimulation of dopamine release in the nucleus accumbens by opiates, alcohol, and barbiturates: studies with transcerebral dialysis in freely moving rats. Ann N Y Acad Sci. 1986;473:367–81. doi: 10.1111/j.1749-6632.1986.tb23629.x. [DOI] [PubMed] [Google Scholar]
- Clark PJ, Amat J, McConnell SO, Ghasem PR, Greenwood BN, Maier SF, et al. Running Reduces Uncontrollable Stress-Evoked Serotonin and Potentiates Stress-Evoked Dopamine Concentrations in the Rat Dorsal Striatum. PloS One. 2015;10:e0141898. doi: 10.1371/journal.pone.0141898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contet C. Gene expression under the influence: transcriptional profiling of ethanol in the brain. Curr Psychopharmacol. 2012;1:301. doi: 10.2174/2211556011201040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia CJ, Benson Ta, Carey KB. Decreased substance use following increases in alternative behaviors: a preliminary investigation. Addict Behav. 2005;30:19–27. doi: 10.1016/j.addbeh.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Cosgrove KP, Hunter RG, Carroll ME. Wheel-running attenuates intravenous cocaine self-administration in rats: sex differences. Pharmacol Biochem Behav. 2002;73:663–71. doi: 10.1016/s0091-3057(02)00853-5. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Ponomarev I, Prescott Ca, Wahlsten D. Effects of genetic and procedural variation on measurement of alcohol sensitivity in mouse inbred strains. Behav Genet. 2006;36:536–52. doi: 10.1007/s10519-006-9067-6. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Belknap JK. The complexity of alcohol drinking: studies in rodent genetic models. Behav Genet. 2010;40:737–50. doi: 10.1007/s10519-010-9371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TM, Ehringer MA, Larson C, Phang TL, Radcliffe RA. Transcriptome analysis of Inbred Long Sleep and Inbred Short Sleep mice. Genes Brain Behav. 2013;12:263–74. doi: 10.1111/gbb.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TM, McCarthy RD, Cox RJ, Ehringer MA. Mesolimbic transcriptional response to hedonic substitution of voluntary exercise and voluntary ethanol consumption. Behav Brain Res. 2014;259:313–20. doi: 10.1016/j.bbr.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. The role of GABAA receptors in mediating the effects of alcohol in the central nervous system. J Psychiatry Neurosci JPN. 2003;28:263–274. [PMC free article] [PubMed] [Google Scholar]
- Dishman RK, Berthoud HR, Booth FW, Cotman CW, Edgerton VR, Fleshner MR, et al. Neurobiology of exercise. Obes Silver Spring Md. 2006;14:345–56. doi: 10.1038/oby.2006.46. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, Hoft NR, Zunhammer M. Reduced alcohol consumption in mice with access to a running wheel. Alcohol. 2009;43:443–52. doi: 10.1016/j.alcohol.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat Neurosci. 2005;8:1481–9. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. [Accessed November 24, 2015];Mol Psychiatry. 2014 doi: 10.1038/mp.2014.159. Available at: http://www.nature.com/mp/journal/vaop/ncurrent/full/mp2014159a.html. [DOI] [PMC free article] [PubMed]
- Farris SP, Pietrzykowski AZ, Miles MF, O’Brien MA, Sanna PP, Zakhari S, et al. Applying the new genomics to alcohol dependence. Alcohol. 2015 doi: 10.1016/j.alcohol.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici M, Nisticò R, Giustizieri M, Bernardi G, Mercuri NB. Ethanol enhances GABAB-mediated inhibitory postsynaptic transmission on rat midbrain dopaminergic neurons by facilitating GIRK currents. Eur J Neurosci. 2009;29:1369–1377. doi: 10.1111/j.1460-9568.2009.06700.x. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Crabbe JC, Belknap JK, Buck KJ, Phillips TJ. The Syntaxin Binding Protein 1 Gene (Stxbp1) Is a Candidate for an Ethanol Preference Drinking Locus on Mouse Chromosome 2. Alcohol Clin Exp Res. 2005;29:708–720. doi: 10.1097/01.ALC.0000164366.18376.EF. [DOI] [PubMed] [Google Scholar]
- Funk CK, Zorrilla EP, Lee MJ, Rice KC, Koob GF. Corticotropin-releasing factor 1 antagonists selectively reduce ethanol self-administration in ethanol-dependent rats. Biol Psychiatry. 2007;61:78–86. doi: 10.1016/j.biopsych.2006.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego X, Cox RJ, Funk E, Foster RA, Ehringer MA. Voluntary exercise decreases ethanol preference and consumption in C57BL/6 adolescent mice: Sex differences and hippocampal BDNF expression. Physiol Behav. 2015;138:28–36. doi: 10.1016/j.physbeh.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JM, Murr AS, Cooper RL. The rodent estrous cycle: characterization of vaginal cytology and its utility in toxicological studies. Birth Defects Res B Dev Reprod Toxicol. 2007;80:84–97. doi: 10.1002/bdrb.20106. [DOI] [PubMed] [Google Scholar]
- Greenwood BN, Foley TE, Le TV, Strong PV, Loughridge AB, Day HEW, et al. Long-term voluntary wheel running is rewarding and produces plasticity in the mesolimbic reward pathway. Behav Brain Res. 2011;217:354–62. doi: 10.1016/j.bbr.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood BN, Spence KG, Crevling DM, Clark PJ, Craig WC, Fleshner M. Exercise-induced stress resistance is independent of exercise controllability and the medial prefrontal cortex. Eur J Neurosci. 2013;37:469–78. doi: 10.1111/ejn.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzerath L, Hewitt BG, Li TK, Warren KR. Alcohol research: past, present, and future. Ann N Y Acad Sci. 2011;1216:1–23. doi: 10.1111/j.1749-6632.2010.05832.x. [DOI] [PubMed] [Google Scholar]
- Hammer SB, Ruby CL, Brager AJ, Prosser RA, Glass JD. Environmental modulation of alcohol intake in hamsters: effects of wheel running and constant light exposure. Alcohol Clin Exp Res. 2010;34:1651–8. doi: 10.1111/j.1530-0277.2010.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare BD, Beierle JA, Toufexis DJ, Hammack SE, Falls WA. Exercise-associated changes in the corticosterone response to acute restraint stress: evidence for increased adrenal sensitivity and reduced corticosterone response duration. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2014;39:1262–1269. doi: 10.1038/npp.2013.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B, et al. On the Integration of Alcohol-Related Quantitative Trait Loci and Gene Expression Analyses. Alcohol Clin Exp Res. 2004;28:1437–1448. doi: 10.1097/01.ALC.0000139827.86749.DA. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki Ra. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iancu OD, Kawane S, Bottomly D, Searles R, Hitzemann R, McWeeney S. Utilizing RNA-Seq data for de novo coexpression network inference. Bioinforma Oxf Engl. 2012;28:1592–7. doi: 10.1093/bioinformatics/bts245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Oberbeck D, Darakjian P, Metten P, McWeeney S, Crabbe JC, et al. Selection for drinking in the dark alters brain gene coexpression networks. Alcohol Clin Exp Res. 2013;37:1295–303. doi: 10.1111/acer.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Ba. Medication treatment of different types of alcoholism. Am J Psychiatry. 2010;167:630–9. doi: 10.1176/appi.ajp.2010.08101500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kanarek RB, Marks-Kaufman R, D’Anci KE, Przypek J. Exercise attenuates oral intake of amphetamine in rats. Pharmacol Biochem Behav. 1995;51:725–9. doi: 10.1016/0091-3057(95)00022-o. [DOI] [PubMed] [Google Scholar]
- Koob GF. Alcoholism, corticotropin-releasing factor, and molecular genetic allostasis. Biol Psychiatry. 2008;63:137–8. doi: 10.1016/j.biopsych.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2010;35:217–38. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapmanee S, Charoenphandhu J, Charoenphandhu N. Beneficial effects of fluoxetine, reboxetine, venlafaxine, and voluntary running exercise in stressed male rats with anxiety- and depression-like behaviors. Behav Brain Res. 2013;250:316–325. doi: 10.1016/j.bbr.2013.05.018. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lobo Ia, Harris RA. GABA(A) receptors and alcohol. Pharmacol Biochem Behav. 2008;90:90–4. doi: 10.1016/j.pbb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughridge AB, Greenwood BN, Day HEW, McQueen MB, Fleshner M. Microarray analyses reveal novel targets of exercise-induced stress resistance in the dorsal raphe nucleus. Front Behav Neurosci. 2013;7:37. doi: 10.3389/fnbeh.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren EJ, Bennett B, Johnson TE, Sikela JM. Expression profiling identifies novel candidate genes for ethanol sensitivity QTLs. Mamm Genome Off J Int Mamm Genome Soc. 2006;17:147–156. doi: 10.1007/s00335-005-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marballi K, Genabai NK, Blednov YA, Harris RA, Ponomarev I. Alcohol consumption induces global gene expression changes in VTA dopaminergic neurons. Genes Brain Behav. 2015 doi: 10.1111/gbb.12266. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel PD, Bennett B, Beeson M, Gordon L, Johnson TE. Confirmation of quantitative trait loci for ethanol sensitivity in long-sleep and short-sleep mice. Genome Res. 1997;7:92–99. doi: 10.1101/gr.7.2.92. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, et al. Gene expression within the extended amygdala of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Alcohol. 2013;47:517–529. doi: 10.1016/j.alcohol.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, Schultz Ja, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol Fayettev N. 2010;44:171–83. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClearn GE, Tarantino LM, Rodriguez LA, Jones BC, Blizard DA, Plomin R. Genotypic selection provides experimental confirmation for an alcohol consumption quantitative trait locus in mouse. Mol Psychiatry. 1997;2:486–489. doi: 10.1038/sj.mp.4000320. [DOI] [PubMed] [Google Scholar]
- McGuier NS, Griffin WC, Gass JT, Padula AE, Chesler EJ, Mulholland PJ. Kv7 channels in the nucleus accumbens are altered by chronic drinking and are targets for reducing alcohol consumption: Kv7 channels and alcohol. Addict Biol. 2015 doi: 10.1111/adb.12279. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DE. Effects of access to a running wheel on ethanol intake in rats under schedule-induced polydipsia. Curr Alcohol. 1978;3:221–235. [Google Scholar]
- McMillan DE, McClure GY, Hardwick WC. Effects of access to a running wheel on food, water and ethanol intake in rats bred to accept ethanol. Drug Alcohol Depend. 1995;40:1–7. doi: 10.1016/0376-8716(95)01162-5. [DOI] [PubMed] [Google Scholar]
- Mika A, Bouchet CA, Bunker P, Hellwinkel JE, Spence KG, Day HEW, et al. Voluntary exercise during extinction of auditory fear conditioning reduces the relapse of fear associated with potentiated activity of striatal direct pathway neurons. Neurobiol Learn Mem. 2015;125:224–235. doi: 10.1016/j.nlm.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T, Fujiwara T, Sanada M, Kofuji T, Kanai-Azuma M, Akagawa K. Syntaxin 1B, but not syntaxin 1A, is necessary for the regulation of synaptic vesicle exocytosis and of the readily releasable pool at central synapses. PloS One. 2014;9:e90004. doi: 10.1371/journal.pone.0090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Harris RA, Ponomarev I. Molecular Profiles of Drinking Alcohol to Intoxication in C57BL/6J Mice. Alcohol Clin Exp Res. 2011;35:659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TJ, Pagano RR, Marlatt Ga. Lifestyle modification with heavy alcohol drinkers: effects of aerobic exercise and meditation. Addict Behav. 1986;11:175–86. doi: 10.1016/0306-4603(86)90043-2. [DOI] [PubMed] [Google Scholar]
- Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang X, Dammann R, Pfeifer GP, et al. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21:1381–1390. doi: 10.1038/sj.onc.1205192. [DOI] [PubMed] [Google Scholar]
- Ozburn AR, Harris RA, Blednov Ya. Wheel running, voluntary ethanol consumption, and hedonic substitution. Alcohol Fayettev N. 2008;42:417–24. doi: 10.1016/j.alcohol.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AA, Lessov-Schlaggar CN, Ponder CA, McKinnon CS, Phillips TJ. Sensitivity to the locomotor-stimulant effects of ethanol and allopregnanolone: a quantitative trait locus study of common genetic influence. Genes Brain Behav. 2006;5:506–517. doi: 10.1111/j.1601-183X.2005.00198.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK, Buck KJ, Cunningham CL. Genes on mouse chromosomes 2 and 9 determine variation in ethanol consumption. Mamm Genome Off J Int Mamm Genome Soc. 1998a;9:936–41. doi: 10.1007/s003359900903. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Huson MG, McKinnon CS. Localization of genes mediating acute and sensitized locomotor responses to cocaine in BXD/Ty recombinant inbred mice. J Neurosci Off J Soc Neurosci. 1998b;18:3023–34. doi: 10.1523/JNEUROSCI.18-08-03023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Huson M, Gwiazdon C, Burkhart-Kasch S, Shen EH. Effects of Acute and Repeated Ethanol Exposures on the Locomotor Activity of BXD Recombinant Inbred Mice. Alcohol Clin Exp Res. 1995;19:269–278. doi: 10.1111/j.1530-0277.1995.tb01502.x. [DOI] [PubMed] [Google Scholar]
- Prado-Alcalá R, Wise Ra. Brain stimulation reward and dopamine terminal fields. I Caudate-putamen, nucleus accumbens and amygdala. Brain Res. 1984;297:265–73. doi: 10.1016/0006-8993(84)90567-5. [DOI] [PubMed] [Google Scholar]
- Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373:2223–33. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Moore CF, Brunzell DH, Lynch WJ. Effect of wheel-running during abstinence on subsequent nicotine-seeking in rats. Psychopharmacology (Berl) 2013;227:403–11. doi: 10.1007/s00213-012-2964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sciolino NR, Smith JM, Stranahan AM, Freeman KG, Edwards GL, Weinshenker D, et al. Galanin mediates features of neural and behavioral stress resilience afforded by exercise. Neuropharmacology. 2015;89:255–264. doi: 10.1016/j.neuropharm.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierakowiak A, Mattsson A, Gómez-Galán M, Feminía T, Graae L, Aski SN, et al. Hippocampal morphology in a rat model of depression: the effects of physical activity. Open Neuroimaging J. 2014;9:1–6. doi: 10.2174/1874440001509010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Bowers BJ, Radcliffe Ra, Wehner JM. Microarray analysis of the effects of a gamma-protein kinase C null mutation on gene expression in striatum: a role for transthyretin in mutant phenotypes. Behav Genet. 2006;36:869–81. doi: 10.1007/s10519-006-9083-6. [DOI] [PubMed] [Google Scholar]
- Smith Ma, Lynch WJ. Exercise as a potential treatment for drug abuse: evidence from preclinical studies. Front Psychiatry. 2011;2:82. doi: 10.3389/fpsyt.2011.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Ma, Walker KL, Cole KT, Lang KC. The effects of aerobic exercise on cocaine self-administration in male and female rats. Psychopharmacology (Berl) 2011;218:357–69. doi: 10.1007/s00213-011-2321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen A, Marks MJ, Smolen TN, Collins AC. Dose and route of administration alter the relative elimination of ethanol by long-sleep and short-sleep mice. Alcohol Clin Exp Res. 1986;10:198–204. doi: 10.1111/j.1530-0277.1986.tb05071.x. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma Oxf Engl. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams Ba, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden LA, Saba LM, Bennett B, Hoffman PL, Tabakoff B. Influence of sex on genetic regulation of “drinking in the dark” alcohol consumption. Mamm Genome. 2015;26:43–56. doi: 10.1007/s00335-014-9553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden LA, Saba LM, Kechris K, Miles MF, Hoffman PL, Tabakoff B. Whole Brain and Brain Regional Coexpression Network Interactions Associated with Predisposition to Alcohol Consumption. PLoS ONE. 2013;8:e68878. doi: 10.1371/journal.pone.0068878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollstädt-Klein S, Wichert S, Rabinstein J, Bühler M, Klein O, Ende G, et al. Initial, habitual and compulsive alcohol use is characterized by a shift of cue processing from ventral to dorsal striatum. Addict Abingdon Engl. 2010;105:1741–9. doi: 10.1111/j.1360-0443.2010.03022.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Huo K, Ma L, Tang L, Li D, Huang X, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2011;7:536. doi: 10.1038/msb.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren KR, Hewitt BG. NIAAA: Advancing Alcohol Research for 40 Years. Alcohol Res Health J Natl Inst Alcohol Abuse Alcohol. 2010;33:5–17. [PMC free article] [PubMed] [Google Scholar]
- Weinstock J. A review of exercise as intervention for sedentary hazardous drinking college students: rationale and issues. J Am Coll Health J ACH. 2010;58:539–44. doi: 10.1080/07448481003686034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werch CE, Bian H, Carlson JM, Moore MJ, Diclemente CC, Huang IC, et al. Brief integrative multiple behavior intervention effects and mediators for adolescents. J Behav Med. 2011;34:3–12. doi: 10.1007/s10865-010-9281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werme M, Lindholm S, Thorén P, Franck J, Brené S. Running increases ethanol preference. Behav Brain Res. 2002;133:301–8. doi: 10.1016/s0166-4328(02)00027-x. [DOI] [PubMed] [Google Scholar]
- WHO | World Health Organization. Available at: www.who.int.
- Wise Ra. Intracranial self-stimulation: mapping against the lateral boundaries of the dopaminergic cells of the substantia nigra. Brain Res. 1981;213:190–4. doi: 10.1016/0006-8993(81)91260-9. [DOI] [PubMed] [Google Scholar]
- Wise RA. Roles for nigrostriatal--not just mesocorticolimbic--dopamine in reward and addiction. Trends Neurosci. 2009;32:517–24. doi: 10.1016/j.tins.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, et al. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res. 2005;80:529–38. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang F, Xu H, Liu Y, Liu J, Zhao H, et al. Differentially co-expressed genes in postmortem prefrontal cortex of individuals with alcohol use disorders: influence on alcohol metabolism-related pathways. Hum Genet. 2014;133:1383–1394. doi: 10.1007/s00439-014-1473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat. 2010;20:281–300. doi: 10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.