Abstract
The objective of this study was to determine the role of histone deacetylases (HDACs) in regulating HIF-1α protein stability and activity in nucleus pulposus (NP) cells. Treatment of NP cells with pan-HDAC inhibitor TSA resulted in decreased HIF-1α levels under both normoxia and hypoxia in a dose-dependent fashion. TSA-mediated HIF-1α degradation was rescued by concomitant inhibition of not only the 26S proteasome but also PHD2 function. Moreover, TSA treatment of PHD2−/− cells had little effect on HIF-1α levels, supporting the notion that inhibition of PHD2 function by HDACs contributed to HIF-1α stabilization. Surprisingly, class-specific HDAC-inhibitors did not affect HIF-1α protein stability, indicating that multiple HDACs controlled HIF-1α stability by regulating HIF-1α-PHD2 interaction in NP cells. Interestingly, lower-dose TSA that did not affect HIF-1α stability decreased its activity and target gene expression. Likewise, rescue of TSA-mediated HIF-1α protein degradation by blocking proteasomal or PHD activity did not restore HIF-1 activity, suggesting that HDACs independently regulate HIF-1α stability and activity. Noteworthy, selective inhibition of HDAC6 and not of class I and IIa HDACs decreased HIF-1-mediated transcription under hypoxia, to a similar extent as lower-dose TSA, contrasting the reported role of HDAC6 as a transcriptional repressor in other cell types. Moreover, HDAC6 inhibition completely blocked TSA effects on HIF-1 activity. HDAC6 associated with and deacetylated HSP90, an important cofactor for HIF-1 function in NP cells, and HDAC6 inhibition decreased p300 transactivation in NP cells. Taken together, these results suggest that while multiple Class I and Class IIa HDACs control HIF-1 stability, HDAC6, a class IIb HDAC, is a novel mediator of HIF-1 activity in NP cells possibly through promoting action of critical HIF-1 cofactors.
Keywords: Intervertebral disc, nucleus pulposus, HIF-1α, hypoxia, HDAC6, transcription factor
INTRODUCTION
The intervertebral disc, a polyaxial diarthrodial joint, allows for a range of motion of adjacent vertebral bodies (1). The healthy intervertebral disc comprises an outer fibrocartilagenous annulus fibrosus, circumferentially enclosing a gelatinous nucleus pulposus (NP), which is bordered superiorly and inferiorly by cartilaginous endplates. The inner annulus and the notochordally-derived NP are completely avascular and reside within a unique niche (2-4). In this physiologically hypoxic niche, cells are adapted to survive through robust and constitutive expression of the bHLH-PAS family transcription factor HIF-1α (5, 6). Our group has previously shown that conditional deletion of HIF-1α in notochordal NP cells results in complete cell death by birth, due to glycolytic failure, and replacement of NP with a mechanically inferior fibrocartilagenous tissue, indicating the critical importance of HIF-1α in maintaining NP cell survival, phenotype, and function (7, 8).
In most cells in the body, HIF-1α protein stability and activity are independently regulated by non-heme, Fe2+ and oxoglutarate-dependent molecular dioxygenases (9). In the presence of oxygen, hydroxylation of specific proline residues is catalyzed by members of the prolyl-hydroxylase domain (PHD) family. Hydroxylated HIF-1α is recognized and bound by the von Hippel-Lindau (pVHL) tumor suppressor, which acts as an E3 ubiquitin ligase, targeting the protein for proteasomal degradation (10). Additionally, HIF-1α activity is regulated by asparaginyl hydroxylation of the C-terminal transactivation domain (C-TAD), which is catalyzed by factor inhibiting HIF-1 (FIH-1) (11). In NP cells, PHD2, but not PHD3, partially regulates HIF-1α levels and retains enzymatic activity even under hypoxia (12, 13). Additionally, within the NP, FIH-1 plays only a limited role in controlling HIF-1α activity (14). The mechanisms by which HIF-1α is constitutively expressed and PHD2 retains its activity under hypoxia are still unknown.
Coordinated actions of histone deacetylases (HDACs) and histone acetyltransferases (HATs) modulate chromatin structure and its accessibility to transcriptional machinery (15). Because deacetylation of histones results in more compact chromatin, HDACs are characteristically described as transcriptional repressors, while HATs are known to promote transcription. Recent studies, however, have identified numerous non-histone targets of HATs/HDACs, and have reported that the acetylation status of these proteins regulates a wide array of cellular processes (16). For example, several studies using cancer cell lines have described a role of HDACs in regulating stability and activity of HIF-1α (17-21), although the mechanisms of action vary widely based on cell type. The major objective of this study was to determine the role of HDACs in controlling HIF-1α stability and activity in NP cells. We show here for the first time that in NP cells multiple HDACs across different classes promote the stability of HIF-1α through a unique mechanism regulating HIF-1α-PHD2 interaction. In addition, our studies show for the first time that HDAC6, independent of effects on HIF-1α stability, maintains HIF-1 activity through modulation of acetylation status and activity of necessary HIF-1 cofactors HSP90 and p300. Our findings suggest that the cells of the NP are adapted to their hypoxic microenvironmental niche, and that tight regulation of HDAC/HAT balance is critical for maintenance of HIF-1α function and NP cell health.
MATERIALS AND METHODS
Plasmids and Reagents
For transactivation studies of HIF-1α, HIF-2α, and p300, the binary Gal4 reporter plasmids (HIF-1α-N-TAD, aa 530-778; HIF-1α-C-TAD, aa 740-826; HIF-2α-TAD, aa 819-870; p300-N-TAD, aa 1-596; p300-C-TAD, aa 1737-2414) were provided by Dr. Nianli Sang, Drexel University College of Medicine. pFR-Luc (Stratagene) reporter contains the yeast Gal4-binding site upstream of a minimal promoter and the Firefly luciferase gene. Enolase1-WT and Enolase1-HRE-mut promoter were provided by Dr. Gregg Semenza, Johns Hopkins University. HDAC1 expression construct was provided by Dr. Stuart Schreiber, Harvard University (22). HDAC2 and HDAC3 were provided by Dr. Ed Seto, H. Lee Moffitt Cancer Center Research Institute (23, 24). HRE-Luc (#26731) by Navdeep Chandel; HDAC4 (#30485), HDAC6 (#30482) and HDAC6-DC (#30483) by Tso-Pang Yao, and ODD-luciferase-pcDNA3 by William Kaelin (#18956) were obtained from Addgene. pRLTK (Promega) containing the Renilla reniformis luciferase gene was used as an internal transfection control. PHD2f/f;CreER(+) and PHD2+/+;CreER(+) MEFs were a kind gift from Dr. William G. Kaelin of Harvard Medical School (25).
Isolation of NP cells, cell treatments and hypoxic culture
Rat NP cells were isolated and characterized as previously reported (6). Cells were maintained in Dulbecco’s Modification of Eagle’s Medium (DMEM) and 10% FBS supplemented with antibiotics. To investigate the effects of HDAC inhibition, cells were treated with Trichostatin A (TSA; 37.5-500 nM), Tubastatin A (15 μM), MC1568 (20 μM), or pimelic diphenylamide (PD)-106 (10 μM) (Sigma Aldrich) for 4 or 8 hours. To investigate the effects of PHD or proteasomal inhibition, cells were treated with dimethyloxalylglycine (2 mM, Calbiochem) or MG132 (10 μM, Calbiochem) respectively. To investigate the effects of inhibition of protein synthesis, cells were treated with cycloheximide (50 μg/mL, Sigma Aldrich). To investigate effects of HSP90 inhibition on HIF-1α protein levels, cells were treated with 17-(Allylamino)-17-demethoxygeldanamycin (17-AAG; 500 nM, Sigma) for 8 h. Cells were cultured in a Hypoxia Work Station (Invivo2 300, Ruskinn, UK) with a mixture of 1% O2, 5% CO2 and 94% N2. To delete PHD2 through activation of CreER, 4-hydroxytamoxifen (Sigma-Aldrich) was added to the medium at a final concentration of 200 nM for 72 h.
Real Time RT-PCR Analysis
Total RNA was extracted from NP cells using RNAeasy mini columns (Qiagen). Before elution from the column, RNA was treated with RNase-free DNase I (Qiagen). Purified, DNA-free RNA was converted to cDNA using EcoDry™ Premix (Clontech). Template cDNA and gene-specific primers were added to the SYBR Green master mixture (Applied Biosystems) and mRNA expression was quantified using the Step One Plus Real-time PCR System (Applied Biosystems). HPRT was used to normalize gene expression. Melting curves were analyzed to verify the specificity of the RT-PCR and the absence of primer dimer formation. Each sample was analyzed in duplicate and included a template- free control. All primers used were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Protein extraction, Immunoprecipitation, and Western Blotting
Cells were placed on ice immediately following treatment and washed with ice-cold PBS. Wash buffer and lysis buffer contained 1x protease inhibitor cocktail (Thermo Scientific), NaF (4 mM), Na3VO4 (20 mM), NaCl (150 mM), β-glycerophosphate (50 mM), and DTT (0.2 mM). Nuclear and cytosolic proteins were prepared using the CellLytic NuCLEAR extraction kit (Sigma Aldrich). Immunoprecipitation was performed using Protein A/G Plus Agarose beads (Pierce) following manufacturer’s protocol using anti-HIF-1α (Abcam), anti-HDAC6 (Cell Signaling), and anti-acetyl-lysine (Cell Signaling). Total cell proteins were resolved on 8-10% SDS-polyacrylamide gels and transferred to PVDF membranes (Fisher Scientific). Membranes were blocked with 5% nonfat dry milk in TBST (50 mM Tris pH 7.6, 150 mM NaCl, 0.1% Tween 20) and incubated overnight at 4°C in 5% nonfat dry milk in TBST with the anti-HIF-1α (1:500, R&D Systems; 1:1000, Abcam); anti-HIF-2α (1:200, R&D Systems); anti-HDAC6 (1:1000), anti-HSP90 (1:1000), anti-acetylated-α-tubulin (1:1000) all from Cell Signaling; anti-β-tubulin (1:5000, Developmental Studies Hybridoma Bank); anti-α-tubulin (1:2000, Abcam); or anti-Lamin A/C (1:1000, Cell Signaling). Immunolabeling was detected using ECL reagent (LAS4000, GE Life Sciences). Densitometric analysis was performed using ImageQuant TL (GE Life Sciences).
Transfections and dual luciferase assay
Cells were transferred to 48-well plates at a density of 2 × 104 cells/well one day before transfection. Lipofectamine 2000 (Invitrogen) was used as a transfection reagent. For each transfection, plasmids were premixed with the transfection reagent. For measuring the effect of hypoxia or HDAC inhibitors on HRE reporter and HIF-TAD activity, the cells in some wells were treated with HDAC inhibitor or moved to the hypoxia workstation 24 h after transfection. Cells were harvested 48-72 h after transfection. Dual-Luciferase Reporter Assay System (Promega) was used for sequential measurements of Firefly and Renilla luciferase activities. Quantification of luciferase activities and calculation of relative ratios were carried out using a TD-20/20 luminometer (Turner Biosystems).
Statistical analysis
All measurements were performed at least in biological triplicate. Data are presented as mean ± S.E.M. Differences between groups were analyzed by Student’s t-test or one-way ANOVA with post-hoc Tukey’s HSD test for multiple comparisons where appropriate; *p< 0.05.
RESULTS
Pan-HDAC inhibition in NP cells decreases HIF-1α protein stability
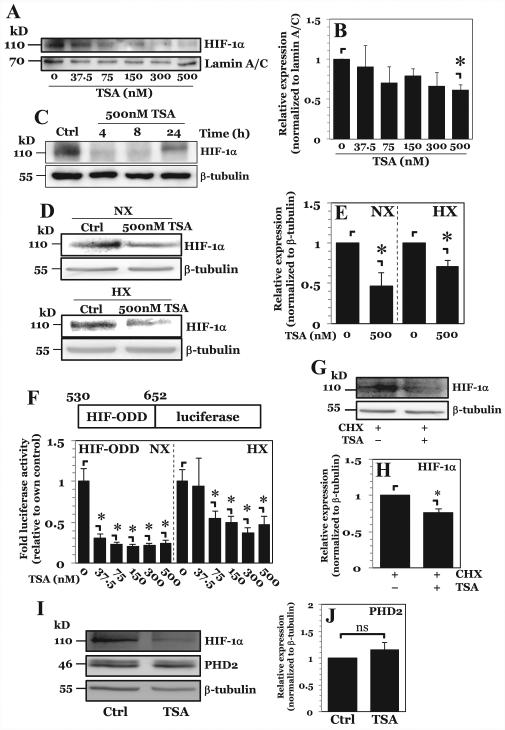
To investigate the role of HDACs in control of HIF-1α, rat NP cells were treated with pan-HDAC inhibitor TSA. TSA treatment under hypoxia results in a dose-dependent decrease in nuclear levels of HIF-1α, with significant decline seen only at the highest dose tested (500 nM) (Fig. 1A, B). Additionally, HIF-1α levels remain suppressed for at least 8 h following TSA treatment (Fig. 1C) and this effect is seen under both normoxia and hypoxia to a similar extent (Fig. 1D, E). No decrease in cell viability was seen at any of the doses or time points used (data not shown). To determine whether the decrease of HIF-1α is due to increased protein turnover, cells were transfected with a HIF-1α-ODD luciferase fusion construct, consisting of HIF-1α amino acids 530-652 fused in-frame to Firefly luciferase. This construct has previously been shown both in vitro and in vivo to be sensitive to PHD/VHL-mediated degradation, with amount/activity of luciferase being directly proportional to endogenous HIF-1α stability and turnover, not HIF-1α protein activity (26). Similar to full-length protein, HIF-1α-ODD is destabilized after TSA treatment under both normoxia and hypoxia, suggesting that HIF-1α-ODD is necessary and sufficient for destabilization of HIF-1α protein following pan HDAC inhibition (Fig. 1F). Interestingly, unlike full-length native HIF-1α protein, HIF-1α-ODD levels decline even when lower doses of TSA are used. This result suggests that the synthetic ODD domain is more sensitive to TSA-mediated degradation than full-length HIF-1α. Although these results suggest an ODD-dependent mechanism of increased HIF-1α turnover, it was important to ascertain whether there was decreased translation of HIF-1α after TSA treatment. We treated NP cells with cycloheximide to inhibit protein synthesis, and then treated cells with 500 nM TSA. TSA treatment resulted in decreased HIF-1α protein, even in the presence of cycloheximide (Fig. 1G, H). This finding suggests that pan-HDAC inhibition promotes degradation of HIF-1α in NP in an ODD-dependent fashion with minor, or no effect on its synthesis.
Figure 1. Pan-HDAC inhibition in NP cells decreases HIF-1α protein stability in a dose dependent fashion.
A, B) Western blot analysis of HIF-1α (A) and corresponding densitometric quantification of at least 3 independent experiments (B) following exposure of NP cells to increasing doses of pan-HDAC inhibitor Trichostatin A (TSA) under hypoxia. Level of nuclear HIF-1α protein decreases only at highest dose of 500 nM of TSA. C) Western blot analysis of HIF-1α after higher-dose TSA treatment for 4-24 hours. D, E) Western blot analysis (D) and corresponding densitometric quantification of at least 3 independent sets (E) of HIF-1α after 8hr higher-dose TSA treatment under both normoxia and hypoxia. TSA treatment decreases HIF-1α expression under both conditions. F) Stability of HIF-ODD-luciferase fusion construct after increasing doses of TSA under both normoxia and hypoxia. G, H) Western blot (G) and corresponding densitometry (H) of HIF-1α in NP cells after 30 minute pretreatment with cycloheximide (CHX, 50 μg/mL) followed by treatment with 500 nM TSA. TSA treatment still results in increased HIF-1α degradation in the presence of CHX. I, J) Western blot (I) and corresponding densitometry (J) of PHD2 in NP cells after TSA treatment showing no change in PHD2 expression upon pan-HDAC inhibition. Quantitative data is represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3); *, p < 0.05.
PHD2 is the major isoform controlling HIF-1α in NP as well as other cell types (12, 13). The results using cycloheximide therefore also have another important implication—it suggests that TSA-mediated increase in HIF-1α turnover is independent of changes in PHD2 protein synthesis. To confirm this, we analyzed PHD2 expression by Western blot after TSA treatment. No appreciable changes in PHD2 protein expression were seen after TSA treatment (Fig. 1I, J), suggesting that increased HIF-1a turnover is unlikely due to increased PHD2 expression.
HDACs stabilize HIF-1α through controlling HIF-PHD2 interaction independently of regulating HIF-1α activity
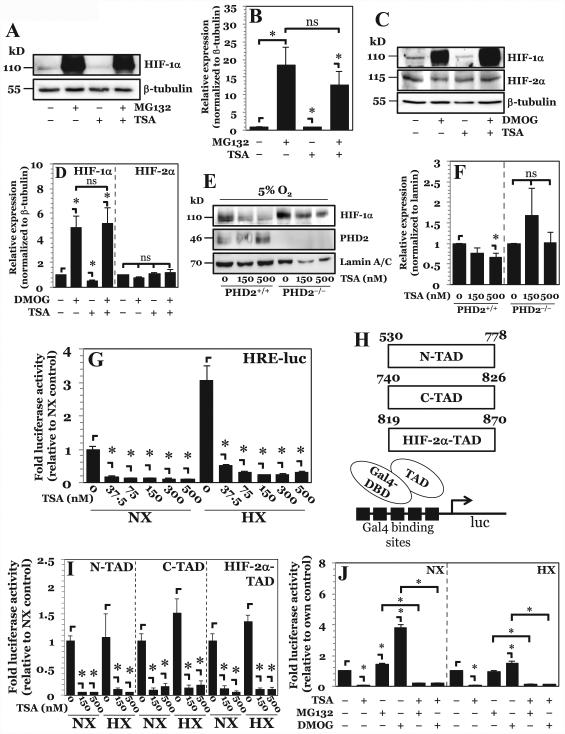
In NP cells PHD2 maintains its enzymatic activity even under hypoxic conditions and controls HIF-1α levels (12, 13). Since TSA was able to decrease HIF-1α protein levels independent of oxygen tension (Fig. 1D, E), and because the HIF-1α-ODD was involved in TSA-mediated destabilization of HIF-1α (Fig. 1F), we investigated whether the TSA-mediated decrease in HIF-1α was dependent on the canonical PHD2 and 26S proteasome pathway. Inhibition of the 26S proteasome using MG132 rescues the decrease in HIF-1α protein caused by pan-HDAC inhibition (Fig. 2A, B). Importantly, no difference in HIF-1α protein levels is seen between the MG132-alone and MG132 with TSA groups suggesting a major contribution of the 26S proteasome to this process. We then investigated the effects of PHD inhibition on TSA-mediated HIF-1α destabilization by using dimethyloxalylglycine (DMOG). DMOG treatment, similar to MG132, was able to abrogate the decrease in HIF-1α protein levels incurred by TSA treatment (Fig. 2C, D). As expected, treatment with DMOG alone caused accumulation of HIF-1α protein. Notably, no significant differences are seen in HIF-1α protein levels between the DMOG-alone and DMOG with TSA groups. Since our previous studies have shown that HIF-2α in NP cells is resistant to PHD-mediated degradation (12), we examined the effect of TSA treatment on this isoform. In line with our earlier reports, we found that DMOG treatment had no effect on HIF-2α protein expression; likewise, TSA treatment showed no effect on HIF-2α protein levels (Fig. 2C, D). Taken together, these results support the role of PHDs in TSA-mediated degradation of HIF-1α in NP cells.
Figure 2. Higher-dose TSA treatment results in increased HIF-1α turnover through PHD-26S proteasome pathway while lower-dose TSA treatment independently decreases HIF-1α activity.
A, B) Western blot analysis of HIF-1α (A) and corresponding densitometric quantification of at least 3 independent experiments (B) after treatment of NP cells with either proteasomal inhibitor MG132 (10 μM) or TSA (500 nM), or both. Decrease in HIF-1α protein seen with higher-dose TSA treatment is rescued by proteasomal inhibition. C, D) Western blot analysis (C) and corresponding densitometry (D) of HIF-1α in NP cells after exposure to PHD inhibitor dimethyloxalylglycine (DMOG, 2 mM) TSA (500 nM), or both. Decrease in HIF-1α protein levels seen with higher-dose TSA treatment is rescued by PHD inhibition. E, F) Western blot (E) and corresponding densitometry of at least 3 independent blots (F) of HIF-1α in PHD2+/+ and PHD2−/− MEFs after exposure to TSA. HIF-1α shows no degradation following higher-dose TSA treatment in PHD2−/− cells. G) HRE-luciferase reporter activity under both normoxia and hypoxia following treatment with increasing doses of TSA. HRE activity decreases with all doses of TSA irrespective of oxygen tension. H) Schematic of Gal-4-TAD constructs and the assay system used for studies described in (I). I) HIF-1α-N-TAD, -C-TAD, and HIF-2α-TAD activity under normoxia and hypoxia after TSA treatment. Both lower dose and higher dose TSA decrease TAD activity under both normoxia and hypoxia. J) HRE-luc activity in NP cells under NX or HX after treatment with MG132, DMOG, and TSA. Data is represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3); *, p < 0.05.
To ascertain the specific contribution of PHD2 to TSA-mediated HIF-1α destabilization, we utilized PHD2−/− MEFs, a cell type that was shown to mirror some aspects of PHD2 biology in NP cells (27). MEFs were cultured under 5% O2 to promote limited HIF-1α accumulation while still preserving partial PHD2 function. Pan-HDAC inhibition in PHD2+/+ cells results in destabilization of HIF-1α protein. As the same effect is not seen with PHD2−/− cells, it supported the hypothesis that HDACs stabilize HIF-1α by controlling HIF-1α-PHD2 interaction (Fig. 2E, F).
We then investigated the effect of TSA treatment on HIF-α activity using an HRE-luciferase reporter construct. Surprisingly, in absence of appreciable change in HIF-1α protein levels and independent of oxygen tension, HIF activity is strongly decreased with lower doses of TSA (Fig. 2G). To further investigate the mechanism of decreased HIF activity, we used constructs consisting of Gal4-DBD fused to different transactivation (TAD) domains of HIF-1α or HIF-2α (Fig. 2H). Lower-dose and higher-dose TSA treatment results in decreased transactivation potential of both the N-terminal TAD (N-TAD) and C-terminal TAD (C-TAD) of HIF-1α, as well as the full length TAD of HIF-2α (Fig. 2I). This result suggests that HDACs may independently control HIF-1α expression and activity in NP cells. To confirm this, we again utilized MG132 and DMOG inhibitors. Surprisingly, while both MG132 and DMOG are able to rescue HIF-1α degradation by TSA (Fig. 2A-D), HIF-1α activity remained significantly lower (Fig. 2J). Thus, accumulated HIF-1α protein destined for proteasomal degradation following TSA treatment was largely inactive, indicating that HDACs have independent roles in NP in regulating both HIF-1α stability and activity.
HDAC6 controls HIF-1 transcriptional activity in NP cells independent of HIF-1 protein stability
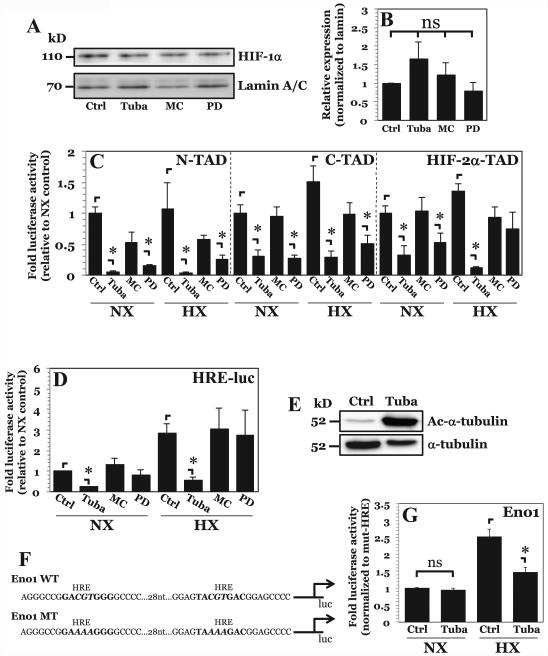
To further investigate the mechanism by which HDAC inhibition results in decreased HIF-1α levels and activity, we used class specific HDAC inhibitors, MC1568 (class IIa HDACs), PD-106 (class I HDACs) and Tubastatin A (specific for HDAC6). Interestingly, use of the specific inhibitors had no effect on levels of nuclear HIF-1α, and could not recapitulate the decrease in HIF-1α levels seen with TSA-mediated pan-HDAC inhibition in NP cells (Fig. 3A, B). This result suggests that there is likely redundancy among multiple classes of HDACs in maintaining HIF-1α stabilization. Considering the unequivocal importance of HIF-1α in NP cell survival (7), the HDAC redundancy was not surprising.
Figure 3. HDAC6 inhibition results in decreased HIF-1α activity independent of effects on protein stability.
A, B) Western blot of HIF-1α (A) and corresponding densitometric analysis of multiple independent experiments (B) after NP cell treatment with inhibitors Tubastatin A (15 μM), MC1568 (20 μM), or PD-106 (10 μM). Selective inhibition of HDAC6, class IIa HDACs, or class I HDACs had no effect on HIF-1α protein stability. C, D) HIF-1α-N-TAD, HIF-1α-C-TAD, and HIF-2α-TAD and D) HRE-luciferase reporter activity activity under normoxia and hypoxia after Tubastatin A, MC1568, or PD-106 treatment. A decrease in HIF-1α-N-TAD and HIF-1α-C-TAD activity was seen with inhibition of HDAC6 and Class I HDACs. Endogenous HIF-1α activity, however, was decreased only by HDAC6 inhibition under both normoxia and hypoxia. E) Western blot analysis of α-tubulin, a known target of HDAC6, shows significantly enhanced acetylation (Ac-α-tubulin) after treatment with Tubastatin A. F) Schematic of enolase-1 promoters used in studies. G) Enolase promoter activity, normalized to activity of promoter with mutated HRE sites, after treatment with Tubastatin A under normoxia and hypoxia. Effects of HDAC6 inhibition on promoter activity are only seen under hypoxia. Data is represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3); *, p < 0.05.
Since multiple classes of HDACs control HIF-1α stability, it was next important to determine the mechanism by which HDACs independently maintain HIF-1α activity in NP cells, and ascertain which specific HDAC was required. Using Gal4-TAD reporters, as described previously (14, 28, 29), we measured the effect of different HDAC inhibitors on HIF-1 activity. Inhibition of HDAC6 as well as class I HDACs resulted in a decrease in N-TAD and C-TAD activity under both normoxia and hypoxia (Fig. 3C). Likewise, HDAC6 inhibition decreased HIF-2α-TAD activity under normoxia and hypoxia. Class I HDAC inhibition by PD-106 decreased HIF-2α-TAD activity only under normoxia. Interestingly, inhibition of class IIa HDACs using MC1568 had no effect on HIF-TAD activity. We then measured changes in endogenous HIF activity using the highly sensitive HRE-luciferase reporter. Selective inhibition of HDAC6 resulted in decreased HIF activity under both normoxia and hypoxia (Fig. 3D). Inhibition of class I HDACs had no effect on reporter activity, in spite of suppressive effect on HIF-1-TAD activities. This result suggests that although class I HDAC inhibition is able to repress synthetic TAD construct activity, it does not have the same effect on full-length endogenous HIF-1α in NP cells. This is likely due to a conformational effect of the full-length HIF-1α protein. Noteworthy, unlike reports using other cell types (17, 30, 31), inhibition of class IIa HDACs had no effect on HIF-1 activity in NP cells. Since HDAC6 is known to deacetylate α-tubulin, the level of acetylated α-tubulin was used to validate the potency of Tubastatin A as an inhibitor of HDAC6 activity in NP cells. As expected, a robust increase in levels of acetylated α-tubulin is seen after Tubastatin A treatment (Fig. 3E). We were of course cognizant that Tubastatin A exerts an inhibitory effect on HDAC8 and that HDAC8, a class I HDAC, is inhibited also by PD-106. Given the observation that PD-106 had little effect on endogenous HIF activity in NP cells (Fig. 3D) it is concluded that the effects of Tubastatin A on HIF-1α activity is wholly or at least mostly due to specific inhibition of HDAC6.
In addition to using a prototypic HRE reporter, we measured the activity of the Enolase1 (Eno1) promoter containing two well-characterized HRE sites as an indicator of HIF-1α activity (Fig. 3F) (32). Interestingly, HDAC6 inhibition had little effect on Eno1 promoter under normoxia, but hypoxic induction of the promoter was partially abrogated by Tubastatin A (Fig. 3G). This result suggests that HDAC6 promotes hypoxic induction of HIF-1 target genes and that Eno1 promoter, unlike the highly sensitive HRE reporter, is less responsive to changes in HIF-1 activity in normoxia.
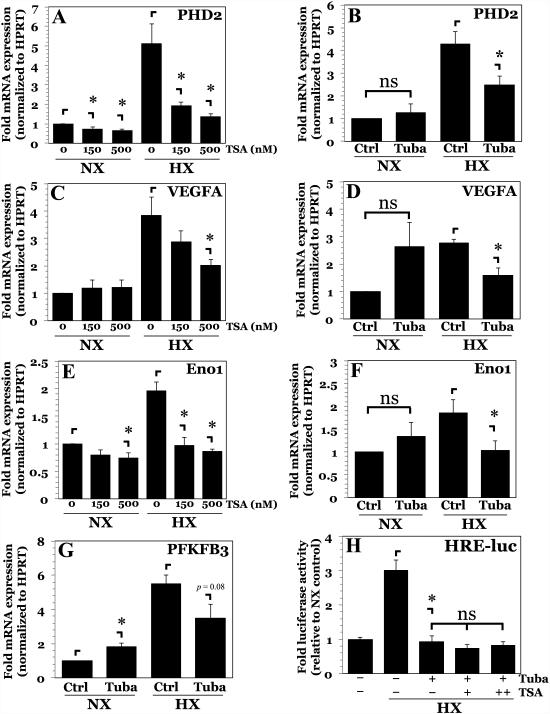
We then investigated the effect of HDAC inhibition on mRNA expression of classical HIF-1 target genes in NP cells. TSA treatment at both 150 nM and 500 nM had a small effect on PHD2, VEGF-A, and Eno1 mRNA expression under normoxia (Fig. 4A, C, E). This was an interesting finding, considering that 500 nM TSA treatment in normoxia decreased HIF-1α protein by ~50% (Fig. 1D, E). As such, these results were consistent with what is known of Eno1 promoter activity (Fig. 3F), suggesting that either normoxic expression of these transcripts in NP cells is not entirely controlled by HIF-1, or alternatively, compensatory mechanisms under normoxia exist to maintain gene expression. All three genes, PHD2, VEGFA, and Eno1, showed increase in transcript levels under hypoxia (Fig. 4A, C, E). This hypoxic induction was abrogated by pan-HDAC inhibition. Inhibition of HDAC6 with Tubastatin A evoked a similar change in mRNA expression (Fig. 4B, D, F). We also examined mRNA expression of PFKFB3, another known HIF-1 target gene. Similar to the other targets, inhibition of HDAC6 under hypoxia showed a trend towards decreased PFKFB3 expression (Fig. 4G).
Figure 4. Inhibition of HDAC6 in NP cells abrogates hypoxic induction of HIF-1 target genes.
A, B) mRNA expression of PHD2 measured by qPCR after TSA (A) or Tubastatin A (B) treatment. C, D) mRNA expression of VEGFA measured by qPCR after TSA (C) or Tubastatin A (D) treatment. E, F) mRNA expression of Eno1 measured by qPCR after TSA (E) or Tubastatin A (F) treatment. G) mRNA expression of PFKFB3 measured by qPCR after Tubastatin A treatment. H) HRE-reporter activity in NP cells pretreated with Tubastatin A following addition of increasing doses of TSA (37.5-150 nM) under hypoxia. Data is represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3); *, p < 0.05.
Our results suggested that HDAC6 was largely responsible for promoting HIF-1 activity in hypoxia. To determine the exclusivity of HDAC6 in controlling HIF-1 activity under hypoxia, NP cells pretreated with Tubastatin A were then treated with TSA, and HIF activity determined. No further decrease in HIF-1 activity was evident when compared to Tubastatin A alone (Fig. 4H). This observation suggests that the effect of lower-dose TSA on HIF-1 activity in NP cells is due to inhibition of HDAC6.
HDAC6 promotes HIF-1α activity in NP cells by modulating HIF-1 cofactors
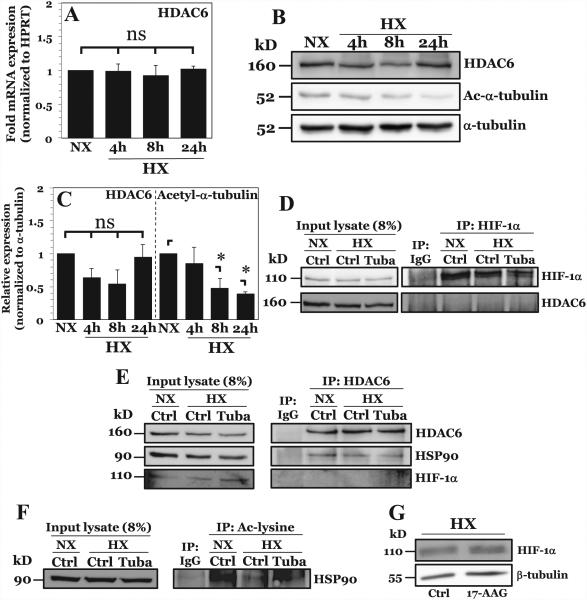
We investigated the effect of hypoxia on HDAC6 expression and activity in NP cells. Both HDAC6 HDAC6 mRNA expression (Fig. 5A) and protein (Fig. 5B, C) were unaffected by hypoxia. The finding that HDAC6 activity, as measured by deacetylation of α-tubulin, increased under hypoxia in a time-dependent fashion provides further support for the notion that HDAC6 regulates hypoxic induction of HIF-1 target gene expression (Fig. 5B, C).
Figure 5. HDAC6 promotes HIF-1 activity in NP cells under hypoxia through positive effects on HIF-1 transcriptional cofactor HSP90.
A) mRNA expression of HDAC6 measured by qPCR after culture in HX for 4-24 hours. B, C) Western blot analysis of HDAC6 and acetylated α-tubulin (Ac-α-tubulin) (B) and corresponding densitometric quantification of experiments (C) following NP cell culture in hypoxia (HX) for 4-24 h. HDAC6 activity, but not expression, increases under hypoxia. D) Immunoprecipitation of HIF-1α in NP cells under normoxia and hypoxia. HDAC6 was unable to be co-precipitated with HIF-1α. E) Immunoprecipitation of HDAC6 in NP cells with or without Tubastatin A treatment. HSP90, but not HIF-1α, was immunoprecipitated with HDAC6 under both normoxia and hypoxia. F) Immunoprecipitation of acetyl-lysine residues in NP cells under normoxia and hypoxia with or without Tubastatin A. Acetylation of HSP90 decreases under hypoxia and increases with HDAC6 inhibition. G) Western blot analysis of HIF-1α after HSP90 inhibition by 8 hr treatment with 17-AAG (500 nM). HSP90 inhibition results in no appreciable change in HIF-1α protein levels. Quantitative data is represented as mean ± S.E. of three independent experiments performed in triplicate (n = 3); *, p < 0.05.
To investigate the mechanism by which HDAC6 regulates HIF-1 target gene expression under hypoxia, we performed co-immunoprecipitation assays. Surprisingly, we were unable to pull down HDAC6 with HIF-1α in NP cells (Fig. 5D), suggesting that HDAC6 may indirectly affect HIF-1α transcriptional activity. In NP cells as well as in other cell types, HIF-1 activity is maintained through recruitment of transcriptional cofactors. Co-immunoprecipitation experiments indicated that, in common with other cell types, HSP90 associated with HDAC6 under both normoxia and hypoxia in NP cells (Fig. 5E). We were again unable to pull down HIF-1α with HDAC6, further supporting an indirect mechanism through effects on HSP90, possibly by regulation of its acetylation status. To confirm whether HSP90 is a direct target of HDAC6 in NP, we performed co-immunoprecipitation studies using an antibody targeting acteyl-lysine residues (Fig. 5F). Our results show that overall levels of acetylated HSP90 decreases in hypoxia in NP cells similar to acetylated α-tubulin, supporting the notion that HDAC6 enzymatic activity increases under hypoxia. Noteworthy, acetylated HSP90 increased upon treatment with Tubastatin A, confirming that deacetylation of HSP90 under hypoxia in NP is regulated by HDAC6. It is worthwhile noting that in other cell types HSP90 promotes HIF-1α protein stability through competitive binding with HSP70, preventing VHL-independent degradation pathways (19, 33). Treatment of NP cells with 17-AAG, a specific HSP90 inhibitor, had no effect on HIF-1α protein stability (Fig. 5G). Our group has previously reported that HSP90 inhibition decreases HIF-1α activity in NP cells (34). Taken together, these results, in stark contrast to previous reports in other cell types (19), further support the role of HDAC6/HSP90 as positive regulators of HIF-1α activity in NP cells independent of effects on protein stability.
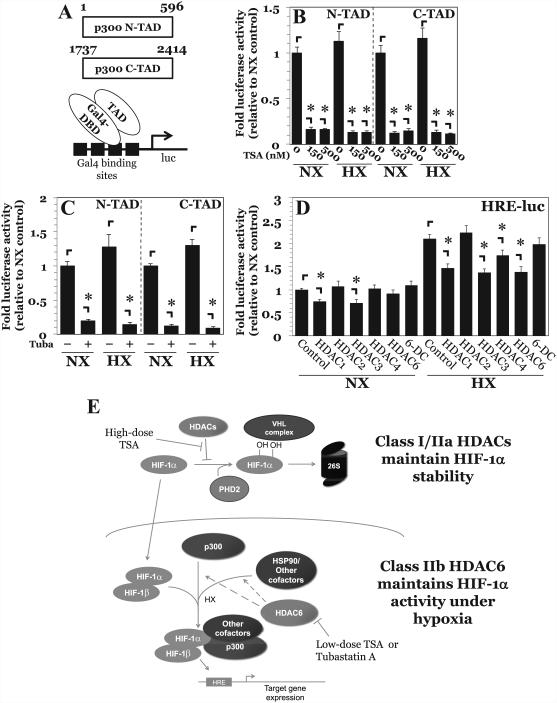
We also examined the effect of HDAC6 inhibition on the activity of the necessary cofactor p300, which contains both an N-TAD and a C-TAD that cooperate in HIF-1-mediated transcription (Fig. 6A) (35, 36). Surprisingly, both p300 N-TAD and C-TAD activity were decreased upon treatment with lower- and higher-dose TSA (Fig. 6B), as well as with Tubastatin A (Fig. 6C), under both normoxia and hypoxia. These studies clearly suggest that in NP cells, HDAC6 may promote hypoxic induction of HIF-1 activity by enhancing the function and transactivation of important cofactors.
Figure 6. HDAC6 inhibition decreases p300 activity in NP cells.
A) Schematic of p300-TAD constructs and the assay system. B, C) p300-N-TAD and p300-C-TAD activity under normoxia and hypoxia after treatment with TSA (150-500 nM) (B) and Tubastatin A (15 μM) (C). Treatment with either TSA or Tubastatin decreased p300 transactivation potential under both normoxia and hypoxia. D) HRE-luciferase activity after transient overexpression of various HDAC isoforms. Overexpression of HDAC1 and HDAC3 irrespective of oxygen tension, and of HDAC4 and HDAC6 under hypoxia decreased HIF-1 activity. E) Proposed model of regulation of HIF-1α stability and activity by HDACs in NP cells. Multiple Class I and Class IIa HDACs promote HIF-1α stability in NP cells by regulating PHD2-HIF-1 interaction. In contrast, HDAC6, independently of regulating HIF-1α stability, promotes hypoxic induction of HIF-1 activity through positive effects on multiple cofactors including p300 and chaperone HSP90.
Because HDAC6 inhibition resulted in decreased HIF-1 activity and multiple HDACs control HIF-1α stability, we investigated the effect of overexpression of HDAC6 as well as other HDAC isoforms on HIF-1 function in NP cells. Irrespective of oxygen tension, overexpression of class I HDACs HDAC1 and HDAC3 resulted in decreased HIF-1 activity (Fig. 6D). This result was not surprising, since HDACs canonically serve as transcriptional repressors. Interestingly, while overexpression of HDAC6 under normoxia had no effect, overexpression under hypoxia resulted in decreased HIF-1 activity. In contrast, overexpression of a catalytically inactive HDAC6 mutant had no effect on HIF-1 activity under either condition. The selective effect seen under hypoxia is consistent with the earlier observation that HDAC6 in NP is more active under hypoxia (Fig. 5B, C, F). Importantly, this suggests that overexpression of HDAC6 causes it to act as a traditional transcriptional repressor like other HDACs, and that HDAC6 levels in NP cells are tightly regulated in order to maintain maximal HIF-1 activity and promote proper NP cell function.
DISCUSSION
The NP represents the largest hypoxic tissue in the body, and hence cell survival within this niche is critically dependent on the expression and activities of HIF-1α (3, 7, 8). We have recently shown that this transcription factor is uniquely regulated, such that it shows stabilized expression irrespective of oxygen tension (6). Related to this observation, PHD2 in NP cells is able to partially regulate HIF-1α levels and maintains enzymatic activity even under hypoxic conditions (12, 13). These observations suggest that PHD2-mediated degradation of HIF-1α is tightly controlled in NP cells and may involve other accessory proteins. Our studies show that multiple Class I and Class IIa HDACs are concerned with the control of HIF-1 stability. Most importantly, we report that Class IIb HDAC6 serves as a novel mediator of HIF-1 activity in NP cells by promoting the recruitment of critical cofactors that include HSP90 as well as p300. These observations provide a specific mechanism by which NP cells can maintain adequate levels of HIF-1α and thereby enhance cell survival in a hypoxic tissue niche.
While other studies have drawn attention to the effect of HDACs on the expression and stabilization of HIF-1α (17-20, 37, 38), our results clearly show that in NP cells pan-HDAC inhibition by TSA at 500 nM destabilizes HIF-1α dependent on the HIF-ODD, and the decrease in HIF-1α can be rescued by blocking PHD or 26S proteasome function. Our results DMOG and PHD2−/− cells suggest that HDAC modulation of HIF-1α-PHD2 interaction is at least partially responsible for the unique and characteristic type of HIF-1α stabilization seen in NP cells. More specifically, this observation helps explain in part why HIF-1α protein levels in NP cells are stably maintained in both normoxia and hypoxia, despite the presence of functional PHD2. This PHD2-dependent mechanism is novel and unique to cells of the NP, and is substantially different from results in cancer cell lines that implicate a PHD-VHL-independent process for HIF-1α destabilization incurred by HDAC inhibition (18, 19), or repression of HIF-1α translation caused by HDAC inhibition (20).
Use of class-specific inhibitors revealed that there is redundancy among multiple HDACs in promoting HIF-1α stability in NP cells. Previous reports in cancer and other cell types have implicated that specific inhibition of Class I HDAC1 and HDAC3 (36), or HDAC2 (37), or Class II HDAC4 (17, 18, 29) or HDAC6 (18, 19) decreases HIF-1α stability. We present here a novel characteristic of NP cells in which multiple HDACs across different classes collectively provide a mechanism to maintain requisite levels of HIF-1α for survival (7), even when there is a change in the activity of specific HDAC classes or isoforms.
Using HRE reporter and Gal4-TAD constructs we were able to show that TSA at concentrations lower than 500 nM reduced HIF-1 activity in NP cells with minimal effect on HIF-1α protein levels. Noteworthy, we observed that although PHD2 inhibition rescued TSA-mediated HIF-1α degradation, it did not restore HIF-1 activity. These experiments suggested that HDACs regulate HIF-1 activity through an additional mechanism independent of modulating HIF-1α-PHD2 interaction. Using Tubastatin A, a specific inhibitor of HDAC6 (IC50 ≈ 15 nM), it was evident that HDAC6 promotes HIF-1 activity in NP cells. Noteworthy, Tubastatin A also inhibits HDAC8, a class I HDAC, but to a much lesser extent (IC50 ≈ 900 nM), Related to this observation, as the class I specific inhibitor PD-106 had no effect on HIF activity measured by HRE reporter, it is likely that the effects of Tubastatin A on HIF activity are through HDAC6. Interestingly, while HDAC6 inhibition decreased both HRE and TAD reporter activities under conditions of both normoxia and hypoxia, expression of mRNA transcripts of HIF-1 target genes were only hypoxia-sensitive. Similar to Tubastatin A, TSA at all doses exerted an inhibitory effect on HIF reporter activities. Remarkably, in normoxia although higher-dose TSA caused a ~50% reduction in HIF-1α protein levels, a comparable effect on mRNA of HIF-1 targets PHD2, VEGFA, and Eno1 was not seen. On the other hand, expression of HIF-1 target genes under hypoxia was affected by TSA regardless of dose, and to a similar degree as Tubastatin A treatment Slightly larger effects seen with high-dose TSA for some HIF-1 target genes can be attributed to the combination of decreased activity as well as decreased level of HIF-1α protein. Importantly, the inability of TSA to further significantly reduce HIF-1 activity in cells pretreated with Tubastatin A clearly suggested that HDAC6 is the major isoform that promotes HIF-1 activity under hypoxia, although minor contributions from other isoforms cannot be ruled out. These results suggest that in NP cells, when HDAC6 activity is inhibited, either a compensatory mechanism controls normoxic expression of HIF-1 target transcripts, or their basal levels in normoxia are somewhat insensitive to changes in HIF-1 activity. Nonetheless, hypoxic induction of these targets is primarily driven by the increase in HIF-1 activity (28). Thus, although HDAC6 in NP cells controls HIF-1 activity irrespective of oxygen tension, its effects are only realized under hypoxia, where HIF-1 exclusively drives transcription of target genes.
Relevant to this discussion is the observation that HDAC6 catalytic activity increases under hypoxia in NP cells. This finding is in line with a similar observation in fibrosarcoma cells (39), and suggests that HDAC6 activity correlates with HIF-1 signaling and likely drives the hypoxic increase in HIF-1 activity. We observed that HDAC6 inhibition in NP cells did not affect nuclear localization and stability of HIF-1α under hypoxia. Additionally, co-IP studies in NP cells were unable to reveal an interaction between HDAC6 and HIF-1α, unlike in other cell types (19). It has been previously reported in other cell types that pan-HDAC inhibition decreases HIF-1α C-TAD activity independently of acetylation of HIF-1α itself (21). Furthermore, in our study we observed a decrease in both HIF-1α-N-TAD and C-TAD activity after HDAC6 inhibition, suggesting that the effects are indirect. One clue to a possible mechanism involved the activity of cofactors that are required for enhanced HIF-1 signaling under hypoxia (40, 41). Since HSP90 is a known HIF-1 cofactor, a substrate of HDAC6, and seeing as the acetylation status of HSP90 governs its function (42), we asked the question does HDAC6 promote HIF-1 signaling in NP cells through HSP90. Our studies clearly demonstrate an interaction between HDAC6 and HSP90 in NP cells, and that hypoxic deacetylation of HSP90 by HDAC6 is abrogated by Tubastatin A. Moreover, we have previously shown that inhibition of HSP90 using a specific inhibitor 17-AAG results in decreased HIF-1 activation (34), and we show here that 17-AAG does not affect HIF-1α protein levels. While a previous report implicates the HSP90-HSP70 axis in stability of HIF-1α through an HDAC6-dependent mechanism (19), our results suggest that rather in NP cells, HDAC6 and HSP90 contribute to maintenance of HIF-1α transcriptional activity independent of effects on protein stability.
p300 is another cofactor required for HIF-1-mediated transcription (40) and activates both the N-TAD (35) and C-TAD (36) of HIF-1α. In other cell types, HDAC4 and HDAC5, both class IIa HDACs, have been reported to promote HIF-1 activity through differential recruitment of p300 and FIH-1 (31). NP cells clearly do not use this specific mechanism, as inhibition of class IIa HDACs had no effect on HIF-1 activity, and as HIF-1α is refractory to regulation by endogenous FIH-1 (14). Nonetheless, p300 was an attractive target due to the effects of HDAC6 inhibition on activity of both HIF-1α C-TAD and N-TAD. Surprisingly, treatment with TSA as well as Tubastatin A resulted in a decrease in p300-TAD activity. This novel observation in NP cells directly contrasts with previous studies showing that HDAC inhibition promotes p300 hyper-acetylation and increases its transactivation (43). This implies that in NP cells HDAC6 promotes HIF-1α activity through positive effects on p300 in addition to HSP90 (Fig. 6E).
NP cells are finely attuned to maintain HIF-1 activity in their physiologically hypoxic niche. Our group has recently shown that HIF-1α in NP cells is refractory to regulation by FIH-1 that modulates recruitment of p300 to HIF-1α-C-TAD, despite FIH-1 being functional under hypoxia similar to PHD2, likely by preferential binding of FIH-1 by Notch-ICDs under hypoxia (14). Taken together, our results suggest that in the hypoxic intervertebral disc, competitive inhibition of FIH-1 by Notch and increased HDAC6 activity serve together to promote p300-HIF-1 interaction, maintaining the function of NP cells. Interestingly, two recent reports demonstrate that acetylation of HDAC6 by p300 decreases both its nuclear import and its deacetylase activity (44, 45). Taken in conjunction with our results, this finding provides an attractive plausible feedback loop in NP cells, through which HDAC6 activity promotes p300-HIF-1 transactivation potential, and p300 acetylase activity, in turn, inhibits HDAC6 through a feedback mechanism. Our study also indicates that, due to the critical importance of HIF-1α in NP cells, there must be a tight regulation of HDAC/HAT activity in order to maintain proper cell function and promote disc health. This conclusion is supported by the observation that overexpression of several different HDAC isoforms, including HDAC6, can negatively impact HIF-1 activity. Because of the critical importance of HIF-1 in maintaining NP health, future studies are aimed at investigating the potential role of HDAC/HAT imbalance in the pathogenesis of disc degeneration in vivo. However, it is important to recognize that NP cells in rodents are notochordal in nature and may not completely capture the phenotypic and functional diversity of cells in human NP. Taken together, due to the requirement of proper maintenance of acetylation status for NP cell function, it is not unreasonable to speculate that drugs restoring HDAC/HAT activity can be used as a potential therapeutic strategy for combating disc aging and degeneration.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grants R01AR055655, R01AR064733, and R01AR050087. ZRS is supported by T32AR052273 and F30AR066506.
Footnotes
DISCLOSURES
Authors have no disclosures to declare.
Author contributions: Study design: IMS and MVR. Data collection: ZRS. Data analysis: ZRS. Data interpretation: ZRS, MVR, and IMS. Drafting manuscript: ZRS, MVR, and IMS. ZRS takes responsibility for the integrity of the data analysis.
REFERENCES
- [1].Shapiro IM, Vresilovic EJ, Risbud MV. Is the spinal motion segment a diarthrodial polyaxial joint: What a nice nucleus like you doing in a joint like this? Bone. 2012;50:771–6. doi: 10.1016/j.bone.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gruber HE, Ashraf N, Kilburn J, et al. Vertebral endplate architecture and vascularization: application of micro-computerized tomography, a vascular tracer, and immunocytochemistry in analyses of disc degeneration in the aging sand rat. Spine (Phila Pa 1976) 2005;30:2593–600. doi: 10.1097/01.brs.0000187877.30149.83. [DOI] [PubMed] [Google Scholar]
- [3].Risbud MV, Schipani E, Shapiro IM. Hypoxic regulation of nucleus pulposus cell survival: from niche to notch. Am J Pathol. 2010;176:1577–83. doi: 10.2353/ajpath.2010.090734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Johnson ZI, Shapiro IM, Risbud MV. Extracellular osmolarity regulates matrix homeostasis in the intervertebral disc and articular cartilage: Evolving role of TonEBP. Matrix Biol. 2014;40:10–6. doi: 10.1016/j.matbio.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rajpurohit R, Risbud MV, Ducheyne P, Vresilovic EJ, Shapiro IM. Phenotypic characteristics of the nucleus pulposus: Expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 2002;308:401–7. doi: 10.1007/s00441-002-0563-6. [DOI] [PubMed] [Google Scholar]
- [6].Risbud MV, Guttapalli A, Stokes DG, et al. Nucleus pulposus cells express HIF-1alpha under normoxic culture conditions: A metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem. 2006;98:152–9. doi: 10.1002/jcb.20765. [DOI] [PubMed] [Google Scholar]
- [7].Merceron C, Mangiavini L, Robling A, et al. Loss of HIF-1α in the Notochord Results in Cell Death and Complete Disappearance of the Nucleus Pulposus. PLoS One. 2014;9:e110768. doi: 10.1371/journal.pone.0110768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Risbud MV, Schoepflin ZR, Mwale F, et al. Defining the phenotype of young healthy nucleus pulposus cells: Recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J Orthop Res. 2015;33:283–93. doi: 10.1002/jor.22789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–60. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- [11].Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–71. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fujita N, Chiba K, Shapiro IM, Risbud MV. HIF-1α and HIF-2α degradation is differentially regulated in nucleus pulposus cells of the intervertebral disc. J Bone Miner Res. 2012;27:401–12. doi: 10.1002/jbmr.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fujita N, Markova D, Anderson DG, et al. Expression of Prolyl Hydroxylases (PHDs) is selectively controlled by HIF-1 and HIF-2 proteins in nucleus pulposus cells of the intervertebral disc: Distinct roles of PHD2 and PHD3 proteins in controlling HIF-1α activity in hypoxia. J Biol Chem. 2012;287:16975–86. doi: 10.1074/jbc.M111.334466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hirose Y, Johnson ZI, Schoepflin ZR, et al. FIH-1-Mint3 axis does not control HIF-1α transcriptional activity in nucleus pulposus cells. J Biol Chem. 2014;289:20594–605. doi: 10.1074/jbc.M114.565101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- [17].Geng H, Harvey CT, Pittsenbarger J, et al. HDAC4 protein regulates HIF1α protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286:38095–102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Qian DZ, Kachhap SK, Collis SJ, et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1α. Cancer Res. 2006;66:8814–21. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- [19].Kong X, Lin Z, Liang D, Fath D. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1α. Mol Cell Biol. 2006;26:2019–28. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hutt DM, Roth DM, Vignaud H, Cullin C, Bouchecareilh M. The Histone Deacetylase Inhibitor, Vorinostat, Represses Hypoxia Inducible Factor 1 Alpha Expression through Translational Inhibition. PLoS One. 2014;9:e106224. doi: 10.1371/journal.pone.0106224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fath DM, Kong X, Liang D, et al. Histone deacetylase inhibitors repress the transactivation potential of hypoxia-inducible factors independently of direct acetylation of HIF-α. J Biol Chem. 2006;281:13612–9. doi: 10.1074/jbc.M600456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hassig CA, Tong JK, Fleischer TC, et al. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci U S A. 1998;95:3519–24. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–7. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- [24].Wen YD, Perissi V, Staszewski LM, et al. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci U S A. 2000;97:7202–7. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Minamishima YA, Moslehi J, Padera RF, Bronson RT, Liao R, Kaelin WG. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol. 2009;29:5729–41. doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Safran M, Kim WY, O'Connell F, Flippin L, Günzler V, Horner JW, Depinho RA, Kaelin WG. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci U S A. 2006;103(1):105–10. doi: 10.1073/pnas.0509459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li J, Yuan W, Jiang S, et al. Prolyl-4-hydroxylase Domain Protein 2 Controls NF-κB/p65 Transactivation and Enhances the Catabolic Effects of Inflammatory Cytokines on cells of the Nucleus Pulposus. J Biol Chem. 2015;290:7195–207. doi: 10.1074/jbc.M114.611483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Agrawal A, Guttapalli A, Narayan S, Albert TJ, Shapiro IM, Risbud MV. Normoxic stabilization of HIF-1alpha drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk. Am J Physiol Cell Physiol. 2007;293:C621–31. doi: 10.1152/ajpcell.00538.2006. [DOI] [PubMed] [Google Scholar]
- [29].Agrawal A, Gajghate S, Smith H, et al. Cited2 modulates hypoxia-inducible factor-dependent expression of vascular endothelial growth factor in nucleus pulposus cells of the rat intervertebral disc. Arthritis Rheum. 2008;58:3798–808. doi: 10.1002/art.24073. [DOI] [PubMed] [Google Scholar]
- [30].Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–9. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Seo HW, Kim EJ, Na H, Lee MO. Transcriptional activation of hypoxia-inducible factor-1α by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett. 2009;583:55–60. doi: 10.1016/j.febslet.2008.11.044. [DOI] [PubMed] [Google Scholar]
- [32].Semenza GL, Jiang BH, Leung SW, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase a gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–37. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- [33].Liu YV, Baek JH, Zhang H, Diez R, Cole RN, Semenza GL. RACK1 Competes with HSP90 for Binding to HIF-1α and Is Required for O2-Independent and HSP90 Inhibitor-Induced Degradation of HIF-1α. Mol Cell. 2007;25:207–17. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gogate SS, Fujita N, Skubutyte R, Shapiro IM, Risbud MV. Tonicity enhancer binding protein (TonEBP) and hypoxia-inducible factor (HIF) coordinate heat shock protein 70 (Hsp70) expression in hypoxic nucleus pulposus cells: Role of Hsp70 in HIF-1a degradation. J Bone Miner Res. 2012;27:1106–17. doi: 10.1002/jbmr.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ruas JL, Berchner-Pfannschmidt U, Malik S, et al. Complex regulation of the transactivation function of hypoxia-inducible factor-1α by direct interaction with two distinct domains of the creb-binding protein/p300. J Biol Chem. 2010;285:2601–9. doi: 10.1074/jbc.M109.021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Freedman SJ, Sun Z-YJ, Poy F, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002;99:5367–72. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim S-H, Jeong J-W, Park JA, et al. Regulation of the HIF-1alpha stability by histone deacetylases. Oncol Rep. 2007;17:647–51. [PubMed] [Google Scholar]
- [38].Yang F, Liu M, Yang F, et al. VPA inhibits renal cancer cell migration by targeting HDAC2 and down-regulating HIF-1α. Mol Biol Rep. 2014;41:1511–8. doi: 10.1007/s11033-013-2996-2. [DOI] [PubMed] [Google Scholar]
- [39].Arsenault D, Brochu-Gaudreau K, Charbonneau M, Dubois CM. HDAC6 Deacetylase Activity Is Required for Hypoxia-Induced Invadopodia Formation and Cell Invasion. PLoS One. 2013;8:e55529. doi: 10.1371/journal.pone.0055529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Arany Z, Huang LE, Eckner R, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93:12969–73. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–61. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- [42].Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–34. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- [43].Stiehl DP, Fath DM, Liang D, Jiang Y, Sang N. Histone deacetylase inhibitors synergize p300 autoacetylation that regulates its transactivation activity and complex formation. Cancer Res. 2007;67:2256–64. doi: 10.1158/0008-5472.CAN-06-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu Y, Peng L, Seto E, Huang S, Qiu Y. Modulation of histone deacetylase 6 (HDAC6) nuclear import and tubulin deacetylase activity through acetylation. J Biol Chem. 2012;287:29168–74. doi: 10.1074/jbc.M112.371120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Han Y, Jeong HM, Jin YH, et al. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2009;383:88–92. doi: 10.1016/j.bbrc.2009.03.147. [DOI] [PubMed] [Google Scholar]