Abstract
In previous work, we cloned TaGS5 gene and found the association of TaGS5-A1 alleles with agronomic traits. In this study, the promoter sequence of the TaGS5-A1 gene was isolated from bread wheat. Sequencing results revealed that a G insertion was found in position -1925 bp of the TaGS5-A1 gene (Reference to ATG), which occurred in the Sp1 domain of the promoter sequence. Combined with previous single nucleotide polymorphism (SNP) in the TaGS5-A1 exon sequence, four genotypes were formed at the TaGS5-A1 locus and were designated as TaGS5-A1a-a, TaGS5-A1a-b, TaGS5-A1b-a, and TaGS5-A1b-b, respectively. Analysis of the association of TaGS5-A1 alleles with agronomic traits indicated that cultivars with the TaGS5-A1a-b allele possessed significantly higher thousand-kernel weight (TKW) and lower plant height than cultivars with the TaGS5-A1a-a allele, and cultivars with the TaGS5-A1b-b allele showed higher TKW than cultivars with the TaGS5-A1b-a allele. The differences of these traits between the TaGS5-A1a-a and TaGS5-A1a-b alleles were larger than those of the TaGS5-A1b-a and TaGS5-A1b-b alleles, suggesting that the -1925G insertion plays the more important role in TaGS5-A1a genotypes than in TaGS5-A1b genotypes. qRT-PCR indicated that TaGS5-A1b-b possessed the significantly highest expression level among four TaGS5-A1 haplotypes in mature seeds and further showed a significantly higher expression level than TaGS5-A1b-a at five different developmental stages of the seeds, suggesting that high expression of TaGS5-A1 was positively associated with high TKW in bread wheat. This study could provide a relatively superior genotype in view of TKW in wheat breeding programs and could also provide important information for dissection of the regulatory mechanism of the yield-related traits.
Keywords: bread wheat, TaGS5-A1 gene, yield-related traits, thousand-kernel weight, single nucleotide polymorphism
Introduction
Wheat is one of the most important food crops in the world. However, with the rapid increase in world population, wheat production has been unable to meet the rate of population growth. Therefore, the improvement of wheat yield is crucial for resolution of the food crisis (Bhalla, 2006). Wheat yield is mainly controlled by genetic and environmental factors. To date, numerous yield-related quantitative trail loci (QTLs) have been mapped in bread wheat in the past decades, e.g., QTLs for plant height, spike length, kernel number per spike, kernel weight per spike, kernel length, kernel width, and thousand-kernel weight (TKW; Wang et al., 2009; Zhang et al., 2010; Cui et al., 2014; Liu et al., 2014; Bellucci et al., 2015; Xie et al., 2015).
However, it was very difficult to directly clone yield-related genes in hexaploid wheat so far due to its large genome size (≈ 17.9 Gb; Varshney et al., 2006). Since comparative genomics verified genomic collinearity between wheat and other crops (Gale and Devos, 1998), a few yield-related genes were gradually isolated from bread wheat by in silico cloning, e.g., sucrose synthase 2 orthologous gene (TaSus2; Jiang et al., 2011), putative cytokinin oxidase genes (TaCKX2.1 and TaCKX2.2; Zhang et al., 2011), cell wall invertase gene (TaCwi-A1 and TaCwi-4A; Ma et al., 2010; Jiang et al., 2015), TaGW2 gene (Su et al., 2011; Hong et al., 2014), 1-FEH w3 (fructan exohydrolases) variant, 1-FEH-6B (Van Riet et al., 2008; Zhang et al., 2009, 2015), and TEF-7A (Zheng et al., 2014). Of the yield-related genes, it has been proven that the TaGW2 gene was significantly associated with TKW, heading, and maturity in bread wheat and that it negatively regulated kernel weight by controlling the gene expression level during seed development (Su et al., 2011; Bennarek et al., 2012; Hong et al., 2014; Qin et al., 2014; Simmonds et al., 2014).
To further understand gene function in bread wheat, the polymorphisms of many yield-related genes were examined, and corresponding molecular markers were developed for further utilization of marker-assisted breeding in bread wheat (Chono et al., 2015; Jiang et al., 2015; Zhang et al., 2015). A single nucleotide polymorphism (SNP) of the 1-FEH w3 gene in the -279 bp site of its promoter region was identified to be intimately associated with thousand-kernel weight (TKW) under drought conditions (Zhang et al., 2015). TaGW2-6A, TaGW2-6B, and TaGW2-6D genes were subsequently isolated from bread wheat. At the TaGW2-6A locus, wheat cultivars with Hap-6A-A were significantly associated with wider kernel and higher TKW (Su et al., 2011). At the TaGW2-6B locus, 11 SNPs in the promoter region formed four haplotypes (Hap-6B-1, Hap-6B-2, Hap-6B-3, and Hap-6B-4). Association analysis indicated that the TaGW2-6B has a stronger influence on TKW than TaGW2-6A, and Hap-6B-1 was a relatively favored haplotype in view of grain width and weight (Qin et al., 2014).
More recently, a yield-related gene, TaGS5-A1, was reported to be associated with several agronomic traits including kernel size, TKW, and plant height in two previous studies (Wang et al., 2015; Ma et al., 2016). Two alleles, TaGS5-A1a and TaGS5-A1b, were found in Chinese bread wheat. Furthermore, wheat cultivars with TaGS5-A1b genotypes were associated with lower plant height and higher TKW in Chinese landraces and current cultivars, suggesting that TaGS5-A1b (named as TaGS5-3A-T by Ma et al., 2016) was a relatively preferable genotype and exhibited a larger potential application in wheat high-yield breeding.
Based on our previous study on TaGS5 genes in bread wheat, we further analyzed the promoter sequence of the TaGS5-A1 gene in different Chinese bread wheat cultivars in this study and discovered a new polymorphism of the TaGS5-A1 gene as well as a relatively superior genotype in view of TKW of bread wheat. This study provides useful information for selection of a relatively preferable genotype in view of TKW in wheat breeding programs as well as provides important information for dissection of the regulatory mechanism of the yield-related traits of bread wheat.
Results
Cloning of the promoter of TaGS5-A1 gene and discovery of an SNP insertion in bread wheat
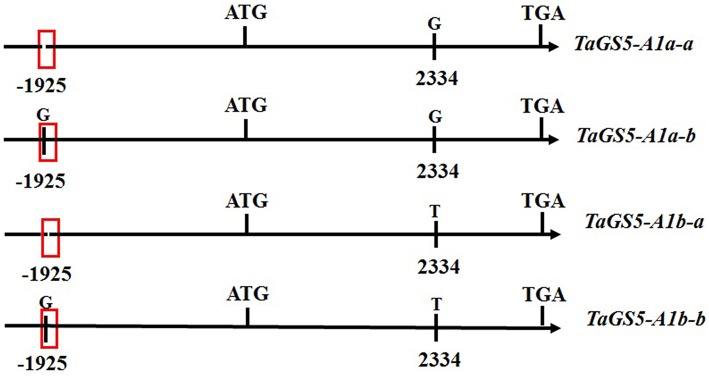
Based on the scaffold 21314 from URGI, a full-length 2287-bp promoter sequence of the TaGS5-A1 gene was successfully cloned from bread wheat. Because TaGS5-A1b was a relatively preferable genotype in previous studies (Wang et al., 2015; Ma et al., 2016), 40 wheat cultivars with the TaGS5-A1b allele, 20 of which relatively large kernel and 20 relatively small kernel, were selected to sequence their promoter sequence of the TaGS5-A1 gene. Finally, a nucleotide G insertion was found at position -1925 bp of the promoter of the TaGS5-A1 gene (Figure S1 and Figure 1) (designated -1925G; BankIt 1916849). Sequencing results confirmed the reliability of this SNP. Furthermore, the prediction of a cis-acting regulatory element of the promoter sequence of TaGS5-A1 gene found that the -1925G insertion was located in the Sp1 domain (light responsive element) using Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Figure 1). Combined with G-T substitution in previous studies (Wang et al., 2015; Ma et al., 2016) and the -1925G insertion, four genotypes were formed and designated as TaGS5-A1a-a (no -1925G insertion and 2334G), TaGS5-A1a-b (-1925G and 2334G), TaGS5-A1b-a (no -1925G insertion and 2334T), and TaGS5-A1b-b (-1925G and 2334T) according to the nomenclature of McIntosh et al. (2007) (Figure 1).
Figure 1.
Four genotypes and predicted Sp1 domain of cis-acting regulatory elements in the promote region of TaGS5-A1 gene.
Association of the -1925G insertion in the promoter region of TaGS5-A1 gene with agronomic traits
Based on PCR amplification with primer set TaGS5−P1 and sequencing results, 27 and 50 out of 77 TaGS5-A1a cultivars belonged to the TaGS5-A1a-a and TaGS5-A1a-b alleles, and 263 and 12 out of 275 cultivars belonged to the TaGS5-A1b-a and TaGS5-A1b-b alleles, respectively. Variance analysis at the TaGS5-A1a locus indicated that the plant height of cultivars with TaGS5-A1a-a alleles was significantly higher than those of cultivars with TaGS5-A1a-b alleles over 3 years. It also showed that panicle length of two genotypes was significantly different in 2014 yet not different in 2013 and 2015 and that internode length below the spike of two genotypes was significantly different in 2013 and 2014 yet not different in 2015 in the Chinese bread wheat cultivars surveyed (Table 1). Moreover, during the 3-year study, wheat cultivars with the TaGS5-A1a-b allele were found to possess significantly higher TKW than those cultivars with the TaGS5-A1a-a allele. Additionally, kernel length and kernel width of two genotypes were significantly different in 2014.
Table 1.
Comparison of agronomic traits of bread cultivars with TaGS5-A1a-a, TaGS5-A1a-b, TaGS5-A1b-a, and TaGS5-A1b-b alleles.
| Year | Trait/genotype | TaGS5-A1a-a | TaGS5-A1a-b | TaGS5-A1b-a | TaGS5-A1b-b |
|---|---|---|---|---|---|
| Sample no. | 27 | 50 | 263 | 12 | |
| Plant height (cm) | 89.9 ± 4.76a | 75.8 ± 2.01b | 71.5 ± 1.02c | 73.4 ± 3.08bc | |
| Panicle length (cm) | 10.8 ± 0.42a | 10.0 ± 0.21ab | 10.3 ± 0.09ab | 9.8 ± 0.45b | |
| Internode length below spike (cm) | 30.2 ± 1.34a | 25.3 ± 0.76b | 25.0 ± 0.33b | 24.9 ± 1.69b | |
| Spikelet number per spike | 19.2 ± 0.39b | 18.7 ± 0.27b | 19.3 ± 0.14a | 19.6 ± 0.63a | |
| 2013 | Kernel number per spike | 45.4 ± 3.44a | 43.7 ± 1.68ab | 42.8 ± 0.62b | 44.6 ± 1.95ab |
| Kernel length (mm) | 6.80 ± 0.13a | 6.97 ± 0.06a | 6.85 ± 0.03a | 6.88 ± 0.10a | |
| Kernel width (mm) | 3.20 ± 0.05a | 3.24 ± 0.03a | 3.29 ± 0.01a | 3.33 ± 0.04a | |
| Kernel length/kernel width ratio | 2.13 ± 0.04a | 2.16 ± 0.03a | 2.0 ± 0.019a | 2.08 ± 0.04a | |
| Thousand-kernel weight(g) | 39.5 ± 1.67c | 42.1 ± 1.01b | 43.0 ± 0.43b | 46.7 ± 1.54a | |
| Sample no. | 27 | 50 | 263 | 12 | |
| Plant height (cm) | 99.8 ± 5.27a | 78.5 ± 2.48b | 77.6 ± 1.03b | 80.1 ± 5.65b | |
| Panicle length (cm) | 11.6 ± 0.58a | 9.4 ± 0.24b | 9.1 ± 0.10b | 9.4 ± 0.47b | |
| Internode length below spike (cm) | 35.0 ± 1.67a | 29.2 ± 1.00b | 29.8 ± 0.95b | 29.0 ± 2.44b | |
| Spikelet number per spike | 19.14 ± 0.36a | 19.1 ± 0.26a | 19.9 ± 0.11a | 19.8 ± 0.40a | |
| 2014 | Kernel number per spike | 51.0 ± 1.76bc | 47.8 ± 0.26c | 52.94 ± 0.68ab | 56.7 ± 2.31a |
| Kernel length (mm) | 6.83 ± 0.12b | 7.03 ± 0.07a | 6.93 ± 0.03ab | 7.05 ± 0.12a | |
| Kernel width (mm) | 3.32 ± 0.07b | 3.55 ± 0.04a | 3.73 ± 0.13a | 3.65 ± 0.10a | |
| Kernel length/kernel width ratio | 2.07 ± 0.05a | 2.01 ± 0.03ab | 1.92 ± 0.01b | 1.94 ± 0.10b | |
| Thousand-kernel weight(g) | 45.6 ± 1.87c | 51.4 ± 1.01ab | 50.7 ± 0.37b | 52.3 ± 1.64a | |
| Sample no. | 27 | 50 | 263 | 12 | |
| Plant height (cm) | 108.6 ± 4.35a | 85.1 ± 1.95c | 92.0 ± 1.95b | 95.1 ± 4.39b | |
| Panicle length (cm) | 11.5 ± 0.39a | 10.7 ± 0.25b | 10.4 ± 0.10b | 10.5 ± 0.33b | |
| Internode length below spike (cm) | 35.0 ± 1.24a | 31.3 ± 1.78ab | 30.3 ± 0.38b | 30.5 ± 1.78b | |
| Spikelet number per spike | 19.5 ± 0.51a | 19.0 ± 0.33a | 19.3 ± 0.12a | 18.8 ± 0.51a | |
| 2015 | Kernel number per spike | 47.3 ± 2.13a | 47.8 ± 1.41a | 48.1 ± 1.41a | 48.2 ± 2.38a |
| Kernel length (mm) | 6.79 ± 0.10a | 7.03 ± 0.07a | 6.82 ± 0.03a | 6.88 ± 0.10a | |
| Kernel width (mm) | 3.32 ± 0.04a | 3.41 ± 0.04a | 3.42 ± 0.01a | 3.33 ± 0.07a | |
| Kernel length/kernel width ratio | 2.05 ± 0.03a | 2.07 ± 0.03a | 2.00 ± 0.01a | 2.08 ± 0.05a | |
| Thousand-kernel weight(g) | 37.1 ± 1.51c | 43.4 ± 1.22b | 42.34 ± 0.46b | 46.71 ± 3.04a |
Letters after numbers showed significant (P < 0.05) differences.
Variation analysis at the TaGS5-A1b locus showed that plant height, panicle length, internode length below spike, spikelet number per spike, kernel number per spike, kernel length and kernel width, KL/KW ratio did not show any significant difference between cultivars with TaGS5-A1b-a and TaGS5-A1b-b alleles. However, cultivars with TaGS5-A1b-b possessed a significantly higher TKW than cultivars with TaGS5-A1b-a for this study of over 3 years of Chinese wheat cultivars. Furthermore, cultivars with TaGS5-A1b-b possessed the significantly highest TKW among cultivars with four TaGS5-A1 alleles.
Therefore, the -1925G insertion contributed to a higher TKW at both TaGS5-A1a and TaGS5-A1b loci in the Chinese bread wheat cultivars surveyed. Further analysis indicated that the difference of the agronomic traits between cultivars with and without the -1925G insertion sharply reduced at the TaGS5-A1b locus when compared with the TaGS5-A1a locus. This finding suggests that the -1925G insertion played the more important role in modulating the TKW of cultivars with TaGS5-A1a than in cultivars with TaGS5-A1b. It could be that the TaGS5-A1 gene has played the more important role in cultivars with the TaGS5-A1b allele than in cultivars with the TaGS5-A1a allele according to previous studies (Wang et al., 2015; Ma et al., 2016). Looking at the combined results of previous studies by Ma et al. (2016), and Wang et al. (2015), TaGS5-A1b-b was a relatively preferable genotype in view of TKW in bread wheat and could be considered for improvement of TKW in the Chinese wheat breeding program (Table 1). However, TKW showed significantly negative correlation with kernel number per spike (Table 2), suggesting that TaGS5-A1b-b genotype might be negatively associated with kernel number per spike in the Chinese bread wheat cultivars surveyed.
Table 2.
Correlation analysis of agronomic traits in the Chinese bread wheat cultivars.
| Traits | Plant height (cm) | Panicle length (cm) | Internode length below spike (cm) | Spikelet number per spike | Kernel number per spike | Kernel length (mm) | Kernel width (mm) | Kernel length/kernel width ratio |
|---|---|---|---|---|---|---|---|---|
| Panicle length (cm) | 0.25** | |||||||
| Internode length below spike | 0.65** | 0.21** | ||||||
| Spikelet number per spike | −0.13** | 0.27** | −0.15** | |||||
| Kernel number per spike | −0.13** | 0.22** | −0.13** | 0.59** | ||||
| Kernel length (mm) | −0.08 | 0.23** | 0.01 | −0.05 | −0.21* | |||
| Kernel width (mm) | −0.12* | −0.05 | −0.09 | 0.02 | 0.02 | 0.04 | ||
| KL/KW ratio | 0.16** | 0.26** | 0.18** | −0.11* | −0.18** | 0.66** | −0.36** | |
| Thousand-kernel weight(g) | −0.24** | −0.03 | −0.13** | −0.08 | −0.28** | 0.43** | 0.18** | −0.12* |
after numbers showed extreme (P < 0.01) and significant (P < 0.05) differences, respectively.
Expression analysis of TaGS5-A1b-a and TaGS5-A1b-b genotypes
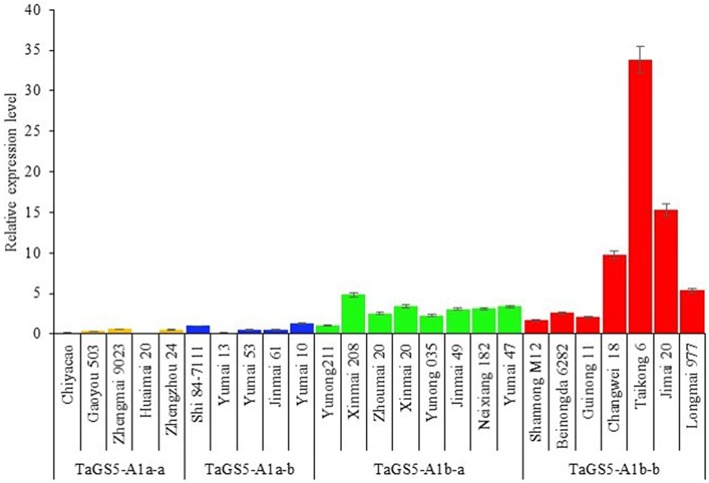
The mature seeds of 25 Chinese cultivars (5 cultivars with TaGS5-A1a-a, 5 cultivars with TaGS5-A1a-b, 8 cultivars with TaGS5-A1b-a and 7 cultivars with TaGSS5-A1b-b) were used to analyze relative expression levels of TaGS5-A1 gene. qRT-PCR results showed that relative expression levels of eight cultivars with TaGS5-A1b-a and seven cultivars with TaGS5-A1b-b were significantly higher than those of cultivars with TaGS5-A1a-a and TaGS5-A1a-b (Figure 2). Furthermore, 4 out of 7 cultivars with TaGS5-A1b-b showed significantly higher expression levels than those of cultivars with TaGS5-A1b-a. Furthermore, two cultivars Yunong 211 with TaGS5-A1b-a and Jimai 20 were selected to identify expression levels at five different developmental stages of the seeds (Figure 3). qRT-PCR results indicated that the relative expression level of TaGS5-A1b-b was significantly higher than that of TaGS5-A1b-a at all five of the stages (Figure 3), indicating that the -1925G insertion most likely improved the expression level of the TaGS5-A1 gene.
Figure 2.
Relative expression levels of 25 Chinese cultivars with different TaGS5-A1 alleles in mature seeds.
Figure 3.
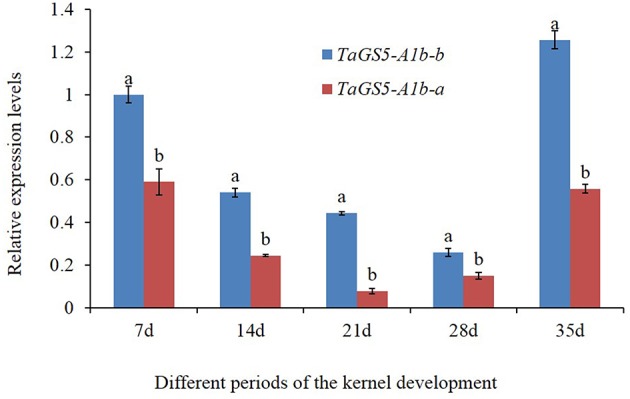
Relative expression levels of TaGS5-A1b-a and TaGS5-A1b-b alleles in the seeds of different developmental stages. Different letters on the top of the bars indicated the significant difference at 5% probability level.
Discussion
The TaGS5 gene has previously been cloned and was further identified to have at least two alleles, designated as TaGS5-A1a and TaGS5-A1b, at the TaGS5-A1 locus, and the TaGS5-A1b genotype has been considered to be associated with a relatively lower plant height, wider kernel width, and higher KTW (Wang et al., 2015; Ma et al., 2016). In this study, a new polymorphism was discovered at the TaGS5-A1 locus which was also intimately associated with kernel size and other agronomic traits. Furthermore, TaGS5-A1a-b and TaGS5-A1b-b genotypes showed significantly higher TKW at TaGS5-A1a and TaGS5-A1b loci, respectively, and TaGS5-A1b-b genotype showed the highest TKW among four TaGS5-A1 genotypes. Based on the distribution of four TaGS5-A1 genotypes in this study, the TaGS5-A1b-b genotype with the highest TKW showed the lowest percentage in the Chinese bread wheat cultivars. Therefore, TaGS5-A1b-b could be considered as a relatively preferred genotype for potential utilization in view of TKW in the current Chinese wheat breeding program. However, the increased TKW probably would cause the reduction of kernel number per spike due to their negative correlation in this study.
A previous study indicated that the TaGS5-A1b (TaGS5-3A-T) genotype has a higher enzymatic activity than that of the TaGS5-A1a (TaGS5-3A-G) genotype (Ma et al., 2016). TaGS5-A1b also exhibited a higher expression level than TaGS5-A1a at different stages of seed development (Wang et al., 2015). In this study, TaGS5-A1a-b genotype possessed significantly higher TKW and showed higher expression levels at TaGS5-A1a locus, and TaGS5-A1b-b genotype exhibited the highest TKW and the highest expression level in mature seeds among four haplotypes. Overexpression of the TaGS5-A1b genotype in transgenic rice showed larger kernel size and higher TKW than that of TaGS5-A1a (TaGS5-3A-G; Ma et al., 2016). Combining the results all studies thus far on TaGS5-A1, the expression level of TaGS5-A1 has been positively associated with higher TKW in both wheat and rice. Therefore, overexpression or improving the enzymatic activity of the TaGS5-A1 gene could possibly result in higher TKW and could be potentially used in a wheat breeding program.
At the TaGS5-A1a locus, TaGS5-A1a-b was a relatively preferred genotype when compared to TaGS5-A1a-a and possessed higher TKW and lower plant height, panicle length, and internode length below spike. Therefore, higher TKW and larger kernel size of cultivars with the TaGS5-A1a-b allele possibly resulted from its relatively lower plant height, shorter panicle length, and internode length below spike. However, further analysis indicated that the difference of the agronomic traits between cultivars with and without -1925G insertion sharply reduced at the TaGS5-A1b locus when compared with the TaGS5-A1a locus, and plant height, panicle length, and internode length below spike did not show a significant difference between cultivars with TaGS5-A1b-a and TaGS5-A1b-b alleles even though the TaGS5-A1b-b genotype still possessed a significantly higher TKW than the TaGS5-A1b-a genotype. The results suggested that the -1925G insertion in the TaGS5-A1 gene played a more important role in modulating the TKW of cultivars with TaGS5-A1a than in cultivars with TaGS5-A1b. The reason was potentially because the TKW of bread wheat can also be modulated by other loci and cultivars with TaGS5-A1b had possessed a relatively higher expression at the TaGS5-A1 locus and had showed a higher TKW. Therefore, it is possibly more effective for cultivars with TaGS5-A1a to select the -1925G insertion in the TaGS5-A1 gene in view of TKW in a high-yield wheat breeding program. For cultivars with TaGS5-A1b, consideration of other yield-related loci was possibly more effective in improving TKW than the -1925G insertion in the TaGS5-A1 gene. However, more work need to further understand the influence of the -1925G insertion on expression of the TaGS5-1 gene as well as agronomic traits.
The polymorphism commonly occurred in the promoter region of yield-related genes in crops and thus resulted in diverse agronomic traits. For example, the GS5 gene was firstly cloned in rice, and the polymorphism of the promoter region of the OsGS5 gene caused variation of TKW by regulating kernel width and filling (Li et al., 2011). The TaGW2 gene was cloned earlier in bread wheat by in silico cloning, and the polymorphism of its promoter region was identified to be intimately associated with TKW (Su et al., 2011; Qin et al., 2014; Simmonds et al., 2014). A yield-related gene, TaCWI, was identified as having four SNPs and two InDels in its promoter region and thus formed two haplotypes (Hap-5D-G and Hap-5D-C). Hap-5D-C genotypes showed lower plant height, earlier heading and maturity date, and higher TKW in Chinese modern wheat cultivars (Jiang et al., 2015). In this study, the -1925G insertion was discovered in the promoter region of the TaGS5 gene and was intimately associated with high expression of the TaGS5 gene as well as increased TKW, and it changed other agronomic traits. It was possible that the -1925G insertion might be able to activate expression of the TaGS5-A1 gene and resulted in higher expression of the TaGS5-A1 gene. More recently, two SNPs (GS5-1 and GS5-2) in light-responsive elements of the promoter of GS5 gene of rice were identified to be responsible for alteration of the response to light induction, leading to higher expression of GS5-2 than GS5-1 in leaves (Xu et al., 2015). Because the -1925G insertion was in the Sp1 domain (light responsive element) of the TaGS5-A1 gene, therefore, this study will also be useful for further analyzing the role of the Sp1 domain in gene function as a cis element.
In this work, the promoter region of the TaGS5-A1 gene was successfully cloned from the bread wheat, and a -1925G insertion was identified in this region. Further analysis indicated that the -1925G insertion was intimately associated with high TKW as well as plant height, spike length, and internode length below spike in Chinese bread wheat. In addition, the -1925G insertion played a greater role in the TaGS5-A1a genotype than in the TaGS5-A1b genotype. Furthermore, cultivars with the TaGS5-A1b-b allele, showing the highest expression at the TaGS5-A1 locus in the mature seeds, exhibited relatively higher TKW. Therefore, this study provides a relatively preferable genotype at the TaGS5-A1 locus in view of TKW for a wheat breeding program and could also provide useful information for understanding the molecular and genetics basis of yield-related traits in bread wheat.
Materials and methods
Plant materials and investigation of agronomic traits
In this study, a total of 352 wheat cultivars were used to analyze the effect of the -1925G insertion in the promoter region of TaGS5-A1 gene on agronomic traits. These cultivars were mainly from the Yellow and Huai wheat regions of China, and were planted at the experimental field of Zhengzhou Scientific Research and Education Center of Henan Agricultural University (N34.9°, E113.6°) in 2012–2013, 2013–2014, and 2014–2015 cropping seasons. A randomized complete block design was adopted, and two replicates were performed. A specific experimental planting method was performed according to the method of Wang et al. (2015). A number of agronomic traits were investigated in the fields before harvesting, i.e., plant height, spike length, internode length below spike (ILBS), and spikelet number of ten spikes of each cultivar. After harvesting, kernel number of ten spikes, kernel length (KL), kernel width (KW), KL/KW ratio and thousand-kernel weight (TKW) of each cultivar surveyed were investigated using naturally dried seeds.
DNA extraction, PCR amplification parameters, and sequencing
Genomic DNA was extracted from all wheat cultivars surveyed according to the method of Chen et al. (2010). DNA concentration and quality were detected using a Thermo Scientific NanoDrop 2000. PCR reaction system containing 25 μL. It mainly consisted of 0.5 μL primer sets (10 pmol/μl), 100 ng genomic DNA, 2.5 μL 10 × Taq buffer (Mg2+ plus), 0.5 μL dNTP (2.5 mM), and 1.25 U Taq DNA polymerases (Tiangen Biotech, Beijing Co. LTD.). PCR amplifications were performed using BioRad-S1000 or ABI 9700 thermal cyclers. The PCR reaction included three stages: 94°C for 5 min, 35 cycles (94°C for 30 s, 55–59°C annealing for 30 s and 72°C for 1–2 min), and 72°C for 7 min. PCR products were analyzed and separated on 1.0–1.5% (w/v) agarose gels, stained with ethidium bromide, and visualized with UV light.
The expected PCR products were purified using the SanPrep Column DNA Gel Extraction Kit (Shanghai Biological Technology Co., Ltd., Shanghai, China). The purified PCR products were ligated into pMD18-T vector (TaKaRa Biotechnology Co., Ltd., Dalian, China) and were transformed into cells of the Escherichia coli DH-5α strain. Plasmids containing targeted fragments were extracted by the Plasmid Rapid Isolation Kit (Shanghai Biological Technology Co., Ltd.) and 12 sub-clones for each sample were sequenced from both directions by Shanghai Sangon Biotech Co., Ltd. The sequenced results were analyzed using software DNAMAN Version 6.0 (http://www.lynnon.com/). The reliability of the sequencing results was checked by examining the sequence chromatograms using Chromas Version 1.4.5 and FinchTV 1.5.0 (http://www.geospiza.com/Products/finchtv.shtml).
Primer design and identification of the SNP
Based on the URGI database (Unité de Recherche Génomique Info: https://urgi.versailles.inra.fr/) and the TaGS5-A1 gene we cloned previously (Wang et al., 2015), a scaffold 21314 containing the TaGS5-A1 gene was obtained from the URGI database. Based on the sequence of this scaffold, four pairs of primer sets (TaGS5−P1, TaGS5−P2, TaGS5−P3, TaGS5−P4 in Table 3) were designed to amplify the promoter sequence of the TaGS5-A1 gene in Chinese Spring. Finally, a 2287-bp promoter sequence of the TaGS5-A1 gene was successfully amplified in Chinese Spring. All primers (Table 3) in this study were designed by Premier Primer 3.0 software (http://primer3.ut.ee/) and Premier Primer 5.0 software, and were synthesized by Shanghai Biological Technology Co., Ltd. (http://www.sangon.com/).
Table 3.
All primers used for cloning the promoter sequence of TaGS5 gene and qRT-PCR.
| Primer | Primer sequence(5′-3′) | Annealing Temp (°C) | Size of PCR fragment (bp) |
|---|---|---|---|
| TaGS5−P1 | Forward:TCATACACACATAATCCAGTCGA Reverse:GATCGTGGGTGTTGCATCTAT |
55 | 800 |
| TaGS5−P2 | Forward:GACTTAGAACCACGACAGCC Reverse:CGTAGCATCCATCGGCATG |
57 | 1086 |
| TaGS5−P3 | Forward:GAGCACAAGAGTGAAGCGAGATGG Reverse:CGTTGTTGGCGTATGCGTCTGA |
59 | 1400 |
| TaGS5−P4 | Forward:AAGGTCGGGCAAAGTCTATG Reverse:CGAGGAGAAAGAGAGCAAGGA |
56 | 1000 |
| 18s | Forward:CCTGCGGCTTAATTGACTC Reverse: GTTAGCAGGCTGAGGTCTCG |
56 | 150 |
| TaGS5−P5 | Forward:TAGAGCCTCAAACTGGACCG Reverse: AGATGCTGATGATGTTTGTCCA |
56 | 127 |
Quantitative RT-PCR of TaGS5-A1b-a and TaGS5-A1b-b alleles
Total RNA was extracted by a Trizol reagent and was reverse-transcripted to cDNA by a PrimeScript RT reagent kit with gDNA eraser (TaKaRa Biotechnology Co. Ltd, Dalian, China) per the kit instructions. Gene-specific primer set TaGS5−P5 were designed to examine the expression levels of TaGS5-A1b-a and TaGS5-A1b-b alleles. An 18s gene was selected as the internal control with the primer 18s (Table 3). The mature seeds of 25 cultivars with different TaGS5-A1 alleles (5 for TaGS5-A1a-a, 5 for TaGS5-A1a-b, 8 for TaGS5-A1b-a, and 7 for TaGS5-A1b-b) were used to analyze the relative expression levels of four TaGS5-A1 alleles. Two wheat cultivars, Yunong 211 (193 days to flowering) with TaGS5-A1b-a allele and Jimai 20 (190 days to flowering) with TaGS5-A1b-b allele, were selected to compare expression levels of TaGS5-A1b-a and TaGS5-A1b-b alleles at the different stages of seed development. The immature seeds from Yunong 211 and Jimai 20 at the five stages (7, 14, 21, 28, and 35 d) after anthesis were sampled in the experimental field conducted at Zhengzhou Scientific Research and Education Center of Henan Agricultural University during the 2014-2015 cropping season for qRT-PCR. Quantitative RT-PCR was performed using the Bio-Rad iQ5 Sequence Detection System with the SYBR Premix Ex TaqII [TaKaRa Biotechnology Co., Ltd., Dalian, China]. The PCR conditions consisted of an initial denaturation step for 2 min at 94°C followed by 40 cycles of 10 s at 95°C, 10 s at 56°C, 20 s at 72°C, and a final extension of 10 min at 72°C. The 18s gene was selected as the internal control. The 2−ΔΔCT method was used to normalize and calibrate transcript values relative to the endogenous 18s control. Eight independent samples with triplicate repeats were analyzed.
Statistical analysis
Coefficient of correlation among agronomic traits were performed by Excel 2013. Analysis of variance was conducted by PROC MIXED in the Statistical Analysis System (SAS Institute Inc., 2000) with genotype classes as categorical variables to derive the means of agronomic traits for each class and test the significant level for the four classes. The differences in agronomic traits among cultivars with different genotypes were tested by Tukey Honestly Significant Difference (HSD) using SPSS 19.0 for multiple comparisons.
Author contributions
FC designed the project. SW and XY performed experiment. YW, HL, and DC investigated agronomic traits. FC and SW wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by the 973 projects (2014CB160303 and 2014CB138105), Program for New Century Excellent Talents in University (NCET-13-0776) of China and Program for Innovative Research Team (in Science and Technology) in University of Henan Province (14IRTSTHN010).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00783
References
- Bellucci A., Torp A. M., Bruun S., Magid J., Andersen S. B., Rasmussen S. K. (2015). Association mapping in scandinavian winter wheat for yield, plant height, and traits important for second-generation bioethanol production. Front. Plant Sci. 6:1046. 10.3389/fpls.2015.01046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennarek J., Boulaflous A., Girousse C., Ravel C., Tassy C., Barret P., et al. (2012). Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 16, 5945–5955. 10.1093/jxb/ers249 [DOI] [PubMed] [Google Scholar]
- Bhalla P. L. (2006). Genetic engineering of wheat – current challenges and opportunities. Trends Biotechnol. 24, 305–311. 10.1016/j.tibtech.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Chen F., Xu H. X., Zhang F. Y., Xia X. C., He Z. H., Wang D. W., et al. (2010). Physical mapping of puroindoline b-2 genes and molecular characterization of a novel variant in durum wheat (Triticum turgidum L.). Mol. Breed. 28, 153–161. 10.1007/s11032-010-9469-2 [DOI] [Google Scholar]
- Chono M., Matsunaka H., Seki M., Fujita M., Kiribuchi-Otobe C., Oda S., et al. (2015). Molecular and genealogical analysis of grain dormancy in Japanese wheat varieties, with specific focus on MOTHER OF FT AND TFL1 on chromosome 3A. Breed. Sci. 65, 103–109. 10.1270/jsbbs.65.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Zhao C., Ding A., Li J., Wang L., Li X., et al. (2014). Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor. Appl. Genet. 127, 659–675. 10.1007/s00122-013-2249-8 [DOI] [PubMed] [Google Scholar]
- Gale M. D., Devos K. M. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. U.S.A. 95, 1971–1974. 10.1073/pnas.95.5.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Chen L., Du L. P., Su Z., Wang J., Ye X., et al. (2014). Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct. Integr. Genomics 14, 341–349. 10.1007/s10142-014-0380-5 [DOI] [PubMed] [Google Scholar]
- Jiang Q., Hou J., Hao C., Wang L., Ge H., Dong Y., et al. (2011). The wheat (T. aestivum) sucrose synthase 2 gene (TaSus2) active in endosperm development is associated with yield traits. Funct. Integr. Genomics 11, 49–61. 10.1007/s10142-010-0188-x [DOI] [PubMed] [Google Scholar]
- Jiang Y., Jiang Q., Hao C., Hou J., Wang L., Zhang H., et al. (2015). A yield-associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis. Theor. Appl. Genet. 128, 131–143. 10.1007/s00122-014-2417-5 [DOI] [PubMed] [Google Scholar]
- Liu G., Jia L., Lu L., Qin D., Zhang J., Guan P., et al. (2014). Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor. Appl. Genet. 127, 2415–2432. 10.1007/s00122-014-2387-7 [DOI] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Ma D., Yan J., He Z., Wu L., Xia X. (2010). Characterization of a cell wall invertase gene TaCwi-A1 on common wheat chromosome 2A and development of functional markers. Mol. Breed. 29, 43–52. 10.1007/s11032-010-9524-z [DOI] [Google Scholar]
- Ma L., Li T., Hao C., Wang Y., Chen X., Zhang X. (2016). TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 14, 1269–1280. 10.1111/pbi.12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh R. A., Devos K. M., Dubcovsky J., Rogers W. J., Morris C. F., Appels R., et al. (2007). Catalogue of Gene Symbols for Wheat: 2007 Supplement. Available online at: http://wheat.pw.usda.gov/ggpages/wgc/2007upd.html
- Qin L., Hao C., Hou J., Wang Y., Li T., Wang L., et al. (2014). Homologous haplotypes, expression, genetic effects and geographic distribution of the wheat yield gene TaGW2. BMC Plant Biol. 14:107. 10.1186/1471-2229-14-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds J., Scott P., Leverington-Waite M., Turner A. S., Brinton J., Korzun V., et al. (2014). Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat(Triticum aestivum L.). BMC Plant Biol. 14:191. 10.1186/s12870-014-0191-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Hao C., Wang L., Dong Y., Zhang X. (2011). Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 122, 211–223. 10.1007/s00122-010-1437-z [DOI] [PubMed] [Google Scholar]
- Van Riet L., Altenbach D., Vergauwen R., Clerens S., Kawakami A., Yoshida M., et al. (2008). Purification, cloning and functional differences of a third fructan 1-exohydrolase (1-FEHw3) from wheat (Triticum aestivum). Physiol. Plant. 133, 242–253. 10.1111/j.1399-3054.2008.01070.x [DOI] [PubMed] [Google Scholar]
- Varshney R. K., Hoisington D. A., Tyagi A. K. (2006). Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 24, 490–499. 10.1016/j.tibtech.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Wang R. X, Hai L., Zhang X. Y, You G. X., Yan C. S., Xiao S. H. (2009). QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai x Yu8679. Theor. Appl. Genet. 118, 313–325. 10.1007/s00122-008-0901-5 [DOI] [PubMed] [Google Scholar]
- Wang S., Zhang X. F., Chen F., Cui D. Q. (2015). A single-nucleotide polymorphism of TaGS5 gene revealed its association with kernel weight in Chinese bread wheat. Front. Plant Sci. 6:1166. 10.3389/fpls.2015.01166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Mayes S., Sparkes D. L. (2015). Carpel size, grain filling, and morphology determine individual grain weight in wheat. J. Exp. Bot. 66, 6715–6730. 10.1093/jxb/erv378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Liu Y., Li Y., Xu X., Li X., Xiao J., et al. (2015). Differential expression of GS5 regulates grain size in rice. J. Exp. Bot. 66, 2611–2623. 10.1093/jxb/erv058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Dell B., Conocono E., Waters I., Setter T., Appels R. (2009). Water deficits in wheat: fructan exohydrolase (1-FEH) mRNA expression and relationship to soluble carbohydrate concentrations in two varieties. New Phytol. 181, 843–850. 10.1111/j.1469-8137.2008.02713.x [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu W., Yang X., Gao A., Li X., Wu X., et al. (2011). Isolation and characterization of two putative cytokinin oxidase genes related to grain number per spike phenotype in wheat. Mol. Biol. Rep. 38, 2337–2347. 10.1007/s11033-010-0367-9 [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu Y., Chen W., Dell B., Vergauwen R., Biddulph B., et al. (2015). A wheat 1-FEH w3 variant underlies enzyme activity for stem WSC remobilization to grain under drought. New Phytol. 205, 293–305. 10.1111/nph.13030 [DOI] [PubMed] [Google Scholar]
- Zhang L. Y., Liu D. C., Guo X. L., Yang W. L., Sun J. Z., Wang D. W., et al. (2010). Genomic distribution of quantitative trait loci for yield and yield-related Traits in Common. J. Integr. Plant Biol. 52, 996–1007. 10.1111/j.1744-7909.2010.00967.x [DOI] [PubMed] [Google Scholar]
- Zheng J., Liu H., Wang Y., Wang L., Chang X, Jing, R., et al. (2014). TEF-7A, a transcript elongation factor gene, influences yield-related traits in bread wheat (Triticum aestivum L.). J. Exp. Bot. 65, 5351–5365. 10.1093/jxb/eru306 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.