Abstract
Non-small cell lung cancer (NSCLC) is the major cause of cancer death worldwide. Increasing evidences show that long non coding RNAs (lncRNAs) are widely involved in the development and progression of NSCLC. The lncRNA HOTTIP has been identified as an oncogene in several human cancers, but its role in NSCLC remains unknown. The present study was to determine the expression and function of HOTTIP in NSCLC. Quantitative real-time PCR was used to detect the expressions of HOTTIP in 53 paired NSCLC tissues and cell lines. Furthermore, RNA interference (RNAi) and over-expression approaches were used to investigate the biological function of HOTTIP in lung cancer cell line. The impacts of HOTTIP on cell migration, proliferation and apoptosis were analyzed using wound scratch assay, MTT and flow cytometry, respectively. The results revealed that the HOTTIP expression was significantly up-regulated in NSCLC tissues and cells when compared with corresponding adjacent normal tissues and normal bronchial epithelial cells (p<0.05). Furthermore, knock-down of HOTTIP significantly inhibited cell proliferation, migration and induced cell apoptosis in vitro, while over-expression of HOTTIP led to the opposite effects. In addition, we identified HOTTIP as a transcriptional regulator of HOXA13 in lung cancer cell. Ectopic expression of HOTTIP suppressed the endogenous level of HOXA13, while knock-down of HOTTIP increased HOXA13 expression. Knock-down of HOXA13 by RNA interference (siHOXA13) revealed that HOTTIP promoted lung cell proliferation, migration, and inhibited apoptosis, at least partly by regulating HOXA13. The present study is the first to identify that HOTTIP functions as an oncogene by regulating HOXA13 in NSCLC, which may represent a new biomarker and a potential therapeutic target for NSCLC intervention.
Keywords: NSCLC, HOTTIP, oncogene, HOXA13
Introduction
Lung cancer is the most common cause of cancer death, with more than 226,000 new cases in the United States in 2012. The majority of lung cancer cases are non-small cell lung cancer (NSCLC), which accounts for approximately 80% of lung cancer [1,2]. The majority of NSCLC patients are diagnosed at advanced stages as they are usually asymptomatic at early stages [3]. Despite the enormous improvements made in chemotherapy and radiotherapy over the past few decades, the outlook for patients with NSCLC was dismal, with only slightly more than 15% of patients alive 5 years after diagnosis [4]. Therefore, it is vital to reveal the molecular mechanisms of the progression of NSCLC for the development of effective therapies.
Long non-coding RNAs (lncRNAs) are evolutionarily conserved non coding RNAs that are longer than 200 nucleotides in length with no protein-coding capacity [5]. These protein-non-coding sequences account for the majority of human genome while protein-coding gene only about 2% [6]. Recent studies demonstrated that lncRNAs are differentially expressed in normal cells and tumor cells, and since lncRNAs are a significant regulatory factor of gene expression, their aberrant expression will inevitably lead to abnormalities in gene expression and tumorigenesis. LncRNA disorders are also a feature of several types of cancer and promote the development, invasion and metastasis of tumors by a variety of mechanisms [7-9].
Previous studies have demonstrated that lncRNAs are involved in the development and progression of NSCLC. However, research into lncRNAs in NSCLC is in its infancy and only a small number of NSCLC-associated lncRNAs have been identified, including lncRNA HOTAIR [10], lncRNA H19 [11], lncRNA ANRIL [12], lncRNA MALAT1 [13], lncRNA SCAL1 [14] and lncRNA AK126698 [15].
A recently identified long non-coding RNA, HOTTIP, was located in the 7p15.2 gene desert region [16]. HOTTIP was identified as the most significantly up-regulated lncRNA in human HCCs, which negatively regulated by miR-125b [17]. However, the role of HOTTIP in NSCLC has not been researched. Previous sequencing for lncRNAs expression showed HOTTIP was up-regulated in NSCLC [18,19]. Thus, there is a need to explore the expression and function of HOTTIP in NSCLC. In the present study, we explored HOTTIP expression pattern in NSCLC tissues cell lines. Meanwhile, the oncogenic role and molecular mechanism of HOTTIP was investigated in NSCLC cell line.
Materials and methods
Sample collection
The study was undertaken with the understanding and written consent of each patient. All patients recruited to this study did not receive any pre-operative treatments. This study was approved by the Human Ethics Committee of The First Affiliated Hospital at Harbin Medical University (Harbin, China). 53 paired NSCLC and adjacent non-tumor lung tissues (≥3 cm away from tumor) were obtained from patients who underwent surgery at The First Affiliated Hospital between 2010 and 2013 and were diagnosed with NSCLC based on histopathological evaluation. The clinic-pathological information of the patients is shown in Table 1. Each sample was snap-frozen in liquid nitrogen and stored at -80°C prior to RNA isolation.
Table 1.
Clinical and pathologic characteristics of all analyzed samples
| Factors | Number |
|---|---|
| Age (years) | |
| ≤65 | 42 |
| >65 | 11 |
| Gender | |
| Male | 35 |
| Female | 18 |
| Pathology | |
| Squamous cell carcinoma | 32 |
| Adenocarcinoma | 21 |
| TNM Stage | |
| Ia+Ib | 34 |
| IIa+IIb | 12 |
| IIIa | 7 |
| Tumor size | |
| ≤5 cm | 37 |
| >5 cm | 16 |
| Lymph node metastasis | |
| Negative | 28 |
| Positive | 25 |
Cell culture and transfection
Three NSCLC adenocarcinoma cell lines (A549, SPC-A1, NCI-H1975), a NSCLC squamous carcinomas cell line (SK-MES-1), and a normal human bronchial epithelial cell line (16HBE) were purchased from the American Type Culture Collection (ATCC, USA). All cell lines were routinely maintained in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin sodium, and 100 mg/ml streptomycin sulfate at 37°C in a humidified air atmosphere containing 5% CO2. Cells were used when they were in the logarithmic growth phase.
The HOTTIP and HOXA13 specific siRNA oligonucleotides were purchased from Qiagen (Hilden, Germany). The sequences were as follows: HOTTIP siRNA: 5’-UGGGAACCCGCUAUUUCACUCUAUU-3’, negative control: 5’-UGACAACUCUUAGGGACCUA-3’; HOXA13 siRNA: 5’-CAGAACUCGUUGCUUUGCCCA-3’, negative control: 5’-UAAACUCCUAACCGCAGUGC-3’. The HOTTIP and HOXA13 over-expression plasmids, TRIM52-AS1-pcDNA3.1+ and HOXA13-pcDNA3.1+, were synthesized in Life Technology (Invitrogen, Carlsbad, CA, USA). Cells were grown on six-well plates to 70% confluence and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
RNA isolation and qRT-PCR assay
Total RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed in a final volume of 20 µl using random primers under standard conditions for the PrimeScript RT reagent Kit (TaKaRa, Dalian, China). We used the SYBR Premix Ex Taq (TaKaRa, Dalian, China) to detect the expression of HOTTIP and HOXA13 levels, following the manufacturer’s instructions. Results were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and calculated according to the Applied Biosystems Comparative Ct Method [20]. The specific primers used as follows: HOTTIP forward primer: 5’-CACACTCACATTCGCACACT-3’, reverse primer: 5’-TCCAGAACTAAGCCAGCCATA-3’; HOXA13 forward primer: 5’-CGCTTCAGAACTCGTTGCTTTGC-3’, reverse primer: 5’-CGGAAGAACTGGCAGTCTTTACCT-3’. GAPDH (internal control) forward primer: 5’-AGTGGCAAAGTGGAGATT-3’, reverse primer: 5’-GTGGAGTCATACTGGAACA-3’.
Cell migration assay
Wound scratch assay was used to assess the migratory ability of A549 cell in vitro. Approximately 2×106 cells were seed per 6-well dish and transfected with HOTTIP siRNA and HOTTIP-pcDNA3.1+ vector or HOXA13 siRNA and HOXA13-pcDNA3.1+ vector 24 h later after transfected Lipofectamine 2000. After 5 h of transfection, a vertical horizontal wound was made with a sterile 10 μl pipette tip and markers were made to allow observation of cells at the same point. The cell was then rinsed with PBS and cultured in the incubator at 37°C. Images of the wounds were acquired with a digital camera system 0 and 12 h after the wounds were made at the same points. Wound widths (μm) were measured using a standard caliper and the experiments were performed in triplicate, repeated at least three times.
Cell proliferation assay
To determine cell growth, 2×103 A549 cell were seeded in 96-well plate and transfected with HOTTIP siRNA and HOTTIP-pcDNA3.1+ vector or HOXA13 siRNA and HOXA13-pcDNA3.1+ vector. The cell proliferation ability was determined using a MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) according to the manufacturer’s protocol. The fluorescence intensity was measured using a fluorescence microplate reader [Model 680 (BIO-RAD)] and absorbance was measured at 570 nm. Three independent experiments (3 replicates in each) were performed.
Flow cytometric analysis
For apoptosis assay, A549 cell was seeded at a density of 1×106 cells/well and cultured in 6-well plates at 37°C and transfected with HOTTIP siRNA and HOTTIP-pcDNA3.1+ vector or HOXA13 siRNA and HOXA13-pcDNA3.1+ vector at a confluence of approximately 70%. After 48 h of transfection, cells including floating cells were harvested and washed twice with cold PBS and re-suspended in 1×binding buffer (Invitrogen, Carlsbad, CA, USA). Subsequently, 10 μl of Annexin V-FITC (Invitrogen, USA) and 5 μl of propidium iodide (PI) were added to each cell suspension. The fluorescence of stained cells was then analyzed by flow cytometry (Beckman, Irvine, CA, USA) using 488 nm excitation within 30 min of staining, according to the manufacturer’s protocol.
Statistical analysis
All statistical analyses were performed using SPSS 17.0 (SPSS, Chicago, USA). The gene expression level of HOTTIP in tumors was compared with adjacent normal tissues utilizing the paired-sample t test. The expression differences between cell lines, cell migration, proliferation and apoptosis assays were analyzed using independent-samples t test. All data were presented as mean ± standard error from three independent experiments with each measured in triplicate. A two-sided p value of less than 0.05 was considered to be statistically significant.
Results
Expression profile of HOTTIP in NSCLC tissues and cell lines
Previous sequencing for lncRNAs expression showed HOTTIP was up-regulated in NSCLC [18,19]. To confirm the result of sequencing, qRT-PCR was used to quantify the HOTTIP expression in 53 matched NSCLC tissues and adjacent normal tissues. Relative expression of HOTTIP [log2 (T/N)] was showed in Figure 1A. The results demonstrated HOTTIP expression in RCC tissues was significantly higher compared with adjacent normal tissues (p<0.01). Furthermore, we determined its expression level in lung cancer cell lines including adenocarcinoma and squamous carcinoma subtypes utilizing qRT-PCR. When normalized to normal bronchial epithelial cell line (16HBE), the expression level of HOTTIP was also up-regulated in SPC-A1, A549, NCI-H1975 and SK-MES-1 cell lines (Figure 1B).
Figure 1.
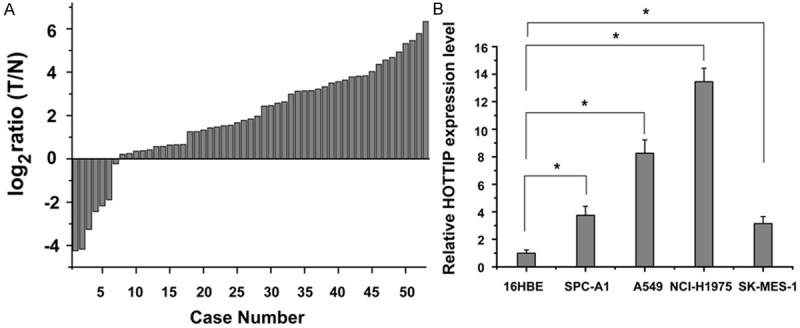
HOTTIP was up-regulated in NSCLC tissues and cell lines. A. Quantitative RT-PCR of HOTTIP expression relative to GAPDH expression in NSCLC tumor samples (T) compared to adjacent normal lung tissue (N) of 53 renal cancer patients. 2-ΔΔCT method was used to analyze the data, and the data are shown in log2 ratio (T/N). B. qRT-PCR analysis of HOTTIP expression levels in NSCLC cell lines (SK-MES-1, SPC-A1, NCI-H1975 and A549) compared with the normal bronchial epithelial cell line (16HBE). Data were expressed as the mean ± SD (n=3), and bars marked with asterisks showed significant differences (p<0.05).
The effects of HOTTIP on the migration of NSCLC cell
To investigate the effect of HOTTIP on NSCLC cells migration, wound scratch assay was performed. The results showed that knock-down the expression of HOTTIP levels significantly impeded the migration of A549 cell compared with negative control (Figure 2A), while over-expression of HOTTIP increased the migration level in A549 cell (Figure 2B).
Figure 2.
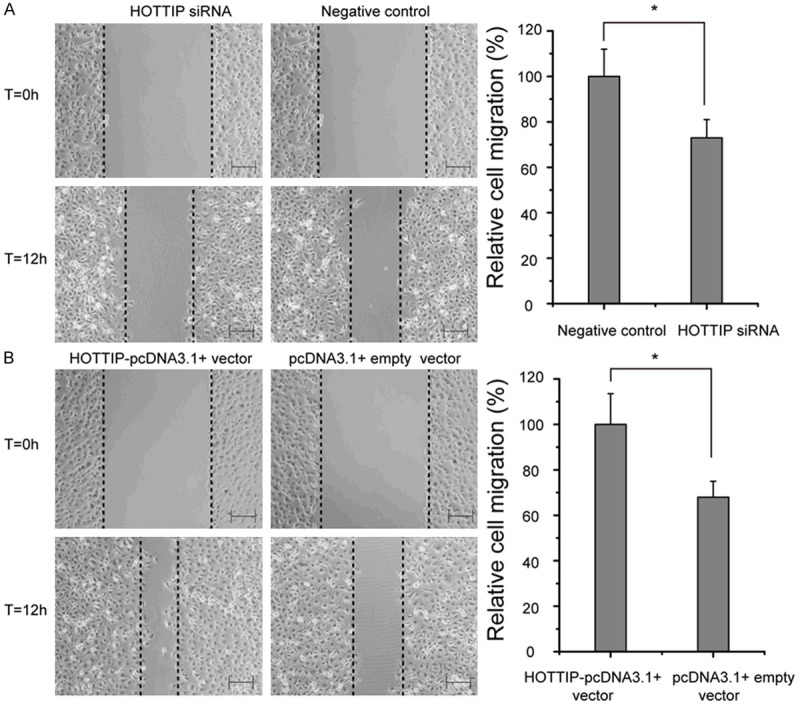
Effects of the HOTTIP on cell migration in A549 cell line. A. A549 cell transfected with HOTTIP siRNA and negative control. B. A549 cell transfected with HOTTIP-pcDNA3.1+ vector and pcDNA3.1+ empty vector. All experiments were performed three times, and a representative picture is shown. *p<0.05. Bar=200 μm.
The effects of HOTTIP on the proliferation of NSCLC cell
As shown in Figure 3, MTT assays revealed that A549 cell growth was inhibited after transient transfected with HOTTIP siRNA compared with negative control. Meanwhile, over-expression of HOTTIP could increase the ability of cell growth.
Figure 3.
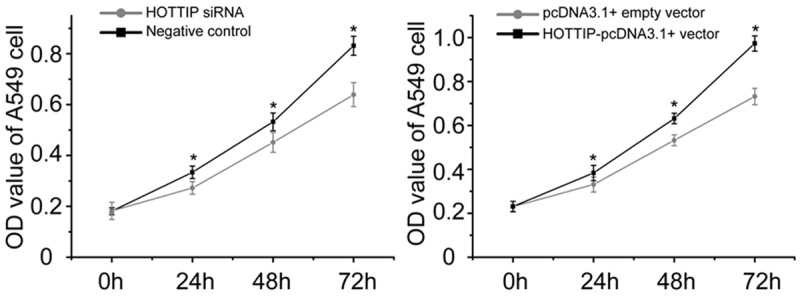
MTT assay for the proliferation of A549 cell transfected with HOTTIP siRNA and negative control or HOTTIP-pcDNA3.1+ vector and pcDNA3.1+ empty vector. The experiments were performed in duplicate and repeated three times. *p<0.05.
The effects of HOTTIP on the apoptosis of NSCLC cell
Some lncRNAs have been reported to play an important role in tumor cell apoptosis, especially in the apoptosis escape of cancer cells [12,21,22]. To determine the effect of HOTTIP on lung cancer cell apoptosis, flow cytometry assay was performed to detect the apoptosis rate of A549 cell after 48 h of transfection. As shown in Figure 4, the apoptosis rate of A549 transfected with HOTTIP siRNA and negative control were 12.44% verse 6.58% (p<0.05), while the apoptosis rates of A549 transfected with HOTTIP-pcDNA3.1+ vector and pcDNA3.1+ empty vector were 3.29% verse 6.23% (p<0.05). These prompted that the up-regulation of HOTTIP inhibited NSCLC cell apoptosis.
Figure 4.
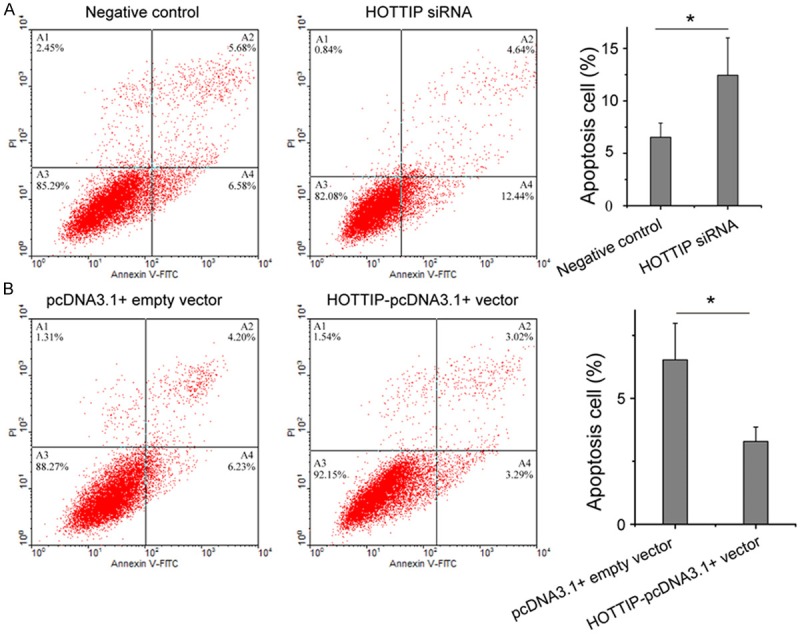
Flow cytometry for the apoptosis of A549 cell transfected with HOTTIP siRNA and negative control (A) or HOTTIP-pcDNA3.1+ vector and pcDNA3.1+ empty vector (B). We repeated each experiment three times with independent individuals. *p<0.05.
HOXA13 is regulated by HOTTIP in NSCLC cell
Because many emerging evidences suggested that HOXA13 was involved in cancer progression and development [23,24], we hypothesized that HOTTIP might regulate the biological behavior of NSCLC cell via regulation of the HOXA13 expression. To confirm this speculation, we first evaluated the effect of HOTTIP knockdown on the expression of HOXA13 by qRT-PCR and Western blot. As shown in Figure 5, depletion of HOTTIP increased the expression of HOXA13 in A549 cell. Inhibition of HOXA13 levels was further confirmed after ectopic expression of HOTTIP (Figure 5).
Figure 5.
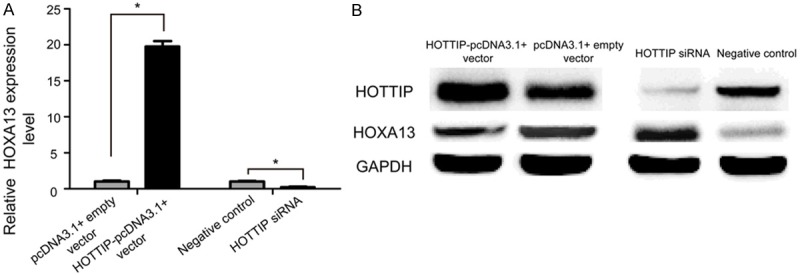
HOXA13 is regulated by HOTTIP in NSCLC cell. The expression levels of HOXA13 was evaluated by qRT-PCR (A) and Western blot (B) 48 h after transfection in A549 cell.
HOXA13 partly mediates the effect of HOTTIP on NSCLC cell biology
In the above section, we identified that the strongest regulatory effect of HOTTIP on HOXA13 expression. To determine whether HOTTIP promotes NSCLC progression by regulating HOXA13, we next employed HOXA13 siRNA and HOTTIP-pcDNA3.1+ vector to specifically silence and prompt the expression of HOXA13 in A549 cell. Interesting, knock-down of HOXA13 increased A549 cell growth (Figure 6), migration (Figure 7A), and arrested apoptosis (Figure 8A), while restoration of HOXA13 inhibited A549 cell growth (Figure 6), migration (Figure 7B), and induced apoptosis (Figure 8B). Taken together, these results suggested that the carcinogenesis of HOTTIP in NSCLC cell biology act at least in part, by regulating HOXA13.
Figure 6.
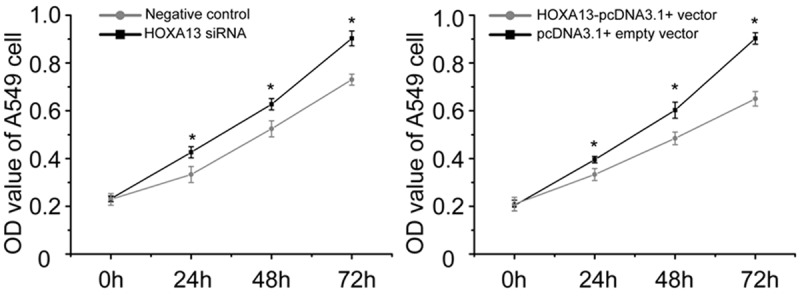
MTT assay for the proliferation of A549 cell transfected with HOXA13 siRNA and negative control or HOXA13-pcDNA3.1+ vector and pcDNA3.1+ empty vector. *p<0.05.
Figure 7.
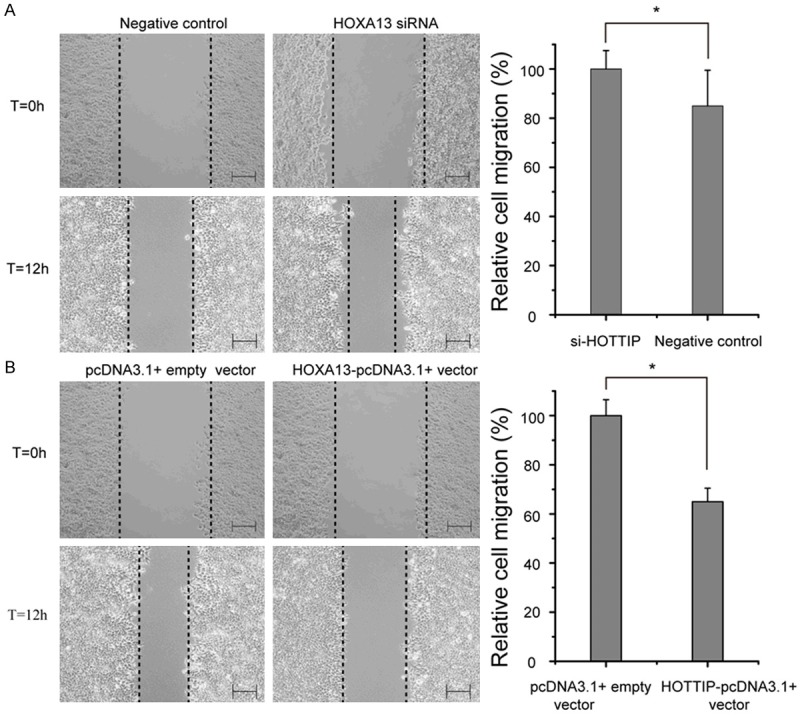
Effects of the HOXA13 on cell migration in A549 cell line. A. A549 cell transfected with HOXA13 siRNA and negative control. B. A549 cell transfected with HOXA13-pcDNA3.1+ vector and pcDNA3.1+ empty vector. *p<0.05. Bar=200 μm.
Figure 8.
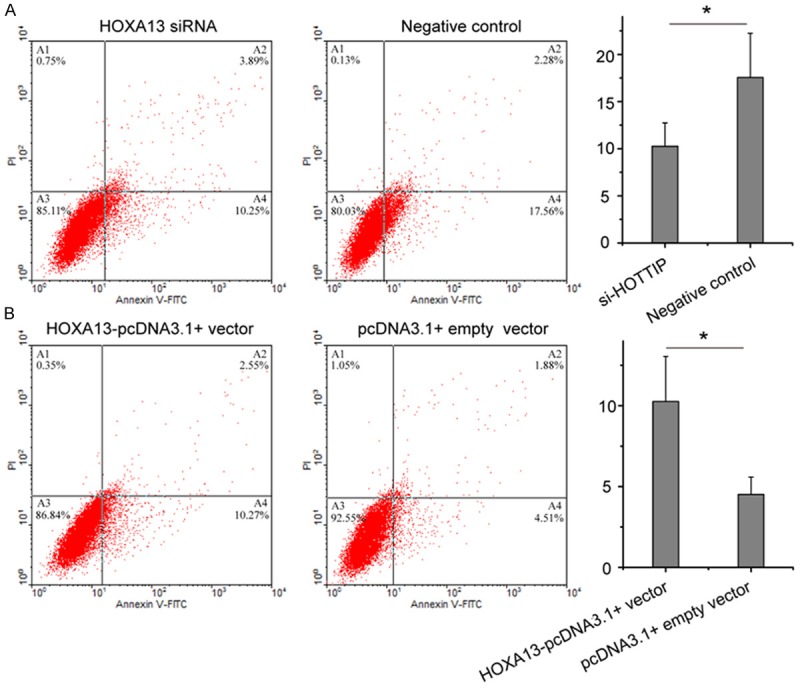
Flow cytometry for the apoptosis of A549 cell transfected with HOXA13 siRNA and negative control (A) or HOXA13-pcDNA3.1+ vector and pcDNA3.1+ empty vector (B). *p<0.05.
Discussion
NSCLC is among the leading causes of cancer-related death. Our understanding of NSCLC pathogenesis has improved through the identification of activating mutations in and amplifications of oncogenes, including KRAS [25], EGFR [26], CRABP1 [27], and inactivating mutations in tumor-suppressive genes p53 [28]. However, the mechanism of NSCLC progression, including the role of cell proliferation, migration, metastasis, and apoptosis resistance, has not been clarified. Accumulating evidences have highlighted the importance of lncRNAs in cancer [7,29-32].
LncRNA dysregulation contributes to a range of biological functions and provides a cellular growth advantage, resulting in progressive and uncontrolled tumor growth [7,31,33,34]. Effective control of both cell proliferation and migration is critical to the prevention of oncogenesis and successful cancer therapy. Therefore, identification of NSCLC-associated lncRNAs and investigation of their clinical significance and functions may provide a missing piece of the well-known oncogenic and tumor suppressor network puzzle. Although literature about lncRNA is increasing, only a small proportion of lncRNAs have been characterized in NSCLC. Our study provided evidence that HOTTIP, a new lncRNA, was clinically and functionally relevant to the progression of NSCLC.
LncRNA HOTTIP (ID: 100316868) is a new lncRNA, which located in the 7p15.2 gene desert region. Previous sequencing for lncRNAs expression showed HOTTIP was up-regulated in NSCLC [18,19]. It remains unclear whether HOTTIP plays an oncogenic role in NSCLC. In this study, we found that the expression of HOTTIP was up-regulated in NSCLC tissues compared with normal tissues. Interesting, its expression was also up-regulated in lung cancer cell lines.
To further investigate the biological role of HOTTIP in NSCLC, we explored the effects of loss or gain of function of HOTTIP on various aspects of NSCLC cell biology. The NSCLC cell line (A549) that was commonly chosen to investigate the effect of lncRNAs on lung cancer. RNAi-mediated suppression of HOTTIP in A549 cell led to a significant inhibition of cell proliferation, migration and induced cell apoptosis. Conversely, introducing HOTTIP into A549 cell induced malignant tumor cell behaviors. Thus indicated that HOTTIP act as oncogene and might represent a promising target for NSCLC treatment.
HOXA13 is a marker of gut primordial posteriorization during progression, which has been shown to play critical role in tumorigenesis of the bladder, liver and esophageal carcinoma [35-37]. Here, we found that siRNA-mediated HOXA13-knockdown induced the proliferation, migration and inhibited apoptosis of NSCLC cell, which was consistent with the functional changes that occurred after restoration the expression of HOTTIP in NSCLC cell. In addition to the HOTTIP-targeted regulation of HOXA13 expression, we also observed reduced HOXA13 levels upon siRNA-mediated knockdown of HOTTIP in A549 cell line. Our data was also consistent with a pioneering research demonstrating that knock-down of HOTTIP expression led to a reduction of HOXA13 expression in hepatocellular carcinoma cell lines [38]. Thus, our work demonstrated that the regulatory passage between HOTTIP and its target, HOXA13, was also preserved during NSCLC tumorigenesis.
Taken together, our present study demonstrated that the up-regulation of HOTTIP characterized as an oncogene in NSCLC progression. Our results also showed that HOTTIP exerted its function in NSCLC at least partly by regulating HOXA13. These findings furthered the understanding of NSCLC pathogenesis and progression, and facilitated the development of lncRNA-directed diagnostics and therapeutics against NSCLC.
Acknowledgements
This work was supported by the Heilongjiang Province Scientific Research Foundation for Returners (LC2013C40).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 3.Smith RA, Manassaram-Baptiste D, Brooks D, Cokkinides V, Doroshenk M, Saslow D, Wender RC, Brawley OW. Cancer screening in the United States, 2014: a review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2014;64:30–51. doi: 10.3322/caac.21212. [DOI] [PubMed] [Google Scholar]
- 4.Pastorino U. Lung cancer screening. Br J Cancer. 2010;102:1681–1686. doi: 10.1038/sj.bjc.6605660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–151. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 10.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12:607–616. [PubMed] [Google Scholar]
- 12.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long Noncoding RNA ANRIL Promotes Non-Small Cell Lung Cancer Cell Proliferation and Inhibits Apoptosis by Silencing KLF2 and P21 Expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 13.Weber DG, Johnen G, Casjens S, Bryk O, Pesch B, Jockel KH, Kollmeier J, Bruning T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49:204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess DJ. Non-coding RNA: HOTTIP goes the distance. Nat Rev Genet. 2011;12:300. doi: 10.1038/nrg2992. [DOI] [PubMed] [Google Scholar]
- 17.Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, Wong CM. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35:1597–606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 18.Yu H, Xu Q, Liu F, Ye X, Wang J, Meng X. Identification and Validation of Long non-coding RNA Biomarkers in Human Non-Small Cell Lung Carcinomas. J Thorac Oncol. 2015;10:645–54. doi: 10.1097/JTO.0000000000000470. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Xu G, Chen W, Pan Q, Huang K, Pan J, Zhang W, Chen J. Detection of long-chain non-encoding RNA differential expression in non-small cell lung cancer by microarray analysis and preliminary verification. Mol Med Rep. 2015;11:1925–1932. doi: 10.3892/mmr.2014.2944. [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Feng G, Wang Y, Yue Y, Zhao W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: implications for TAA pathogenesis. Int J Clin Exp Pathol. 2014;7:7643–7652. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Zou Y, Wang W, Zuo Q, Jiang Z, Sun M, De W, Sun L. Down-Regulated Long Non-Coding RNA MEG3 and its Effect on Promoting Apoptosis and Suppressing Migration of Trophoblast Cells. J Cell Biochem. 2015;116:542–50. doi: 10.1002/jcb.25004. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang Y, Zeng J, Song Y, Gao W, Zheng S, Zhuang B, Chen H, Li W, Li H, Fu Z, Chen R. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan TT, Jia WD, Yao QY, Sun QK, Ren WH, Huang M, Ma J, Li JS, Ma JL, Yu JH, Ge YS, Liu WB, Zhang CH, Xu GL. Overexpression of HOXA13 as a potential marker for diagnosis and poor prognosis of hepatocellular carcinoma. Tohoku J Exp Med. 2014;234:209–219. doi: 10.1620/tjem.234.209. [DOI] [PubMed] [Google Scholar]
- 25.Kim M, Chen X, Chin LJ, Paranjape T, Speed WC, Kidd KK, Zhao H, Weidhaas JB, Slack FJ. Extensive sequence variation in the 3’ untranslated region of the KRAS gene in lung and ovarian cancer cases. Cell Cycle. 2014;13:1030–1040. doi: 10.4161/cc.27941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonicelli A, Cafarotti S, Indini A, Galli A, Russo A, Cesario A, Lococo FM, Russo P, Mainini AF, Bonifati LG, Nosotti M, Santambrogio L, Margaritora S, Granone PM, Dutly AE. EGFR-targeted therapy for non-small cell lung cancer: focus on EGFR oncogenic mutation. Int J Med Sci. 2013;10:320–330. doi: 10.7150/ijms.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Favorskaya I, Kainov Y, Chemeris G, Komelkov A, Zborovskaya I, Tchevkina E. Expression and clinical significance of CRABP1 and CRABP2 in non-small cell lung cancer. Tumour Biol. 2014;35:10295–10300. doi: 10.1007/s13277-014-2348-4. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi F, Kugawa S, Tateno H, Kokubu F, Fukuchi K. Analysis of EGFR, KRAS and P53 mutations in lung cancer using cells in the curette lavage fluid obtained by bronchoscopy. Lung Cancer. 2012;78:201–206. doi: 10.1016/j.lungcan.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 31.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, Logothetis CJ, Araujo JC, Pisters LL, Tewari AK, Canman CE, Knudsen KE, Kitabayashi N, Rubin MA, Demichelis F, Lawrence TS, Chinnaiyan AM, Feng FY. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 33.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 (INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 35.Gu ZD, Shen LY, Wang H, Chen XM, Li Y, Ning T, Chen KN. HOXA13 promotes cancer cell growth and predicts poor survival of patients with esophageal squamous cell carcinoma. Cancer Res. 2009;69:4969–4973. doi: 10.1158/0008-5472.CAN-08-4546. [DOI] [PubMed] [Google Scholar]
- 36.Mohamadkhani A. Long Noncoding RNAs in Interaction With RNA Binding Proteins in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18794. doi: 10.5812/hepatmon.18794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo B, Che T, Shi B, Guo L, Yin Y, Li L, Wang J, Yan D, Chen Y. Screening and identification of specific markers for bladder transitional cell carcinoma from urine urothelial cells with suppressive subtractive hybridization and cDNA microarray. Can Urol Assoc J. 2011;5:E129–137. doi: 10.5489/cuaj.09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, Andersen JB, Hammerle M, Tornillo L, Heim MH, Diederichs S, Cillo C, Terracciano LM. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]


